Abstract
A collection of 12 monoclonal antibodies (MAbs) raised against porcine reproductive and respiratory syndrome (PRRS) virus was used to study the antigenic structure of the virus nucleocapsid protein (N). The full-length N gene, encoded by open reading frame 7, was cloned from the Canadian PRRS virus, PA-8. Deletions were introduced into the N gene to produce a series of nine overlapping protein fragments ranging in length from 25 to 112 amino acids. The individual truncated genes were cloned as glutathione S-transferase fusions into a eukaryotic expression vector downstream of the T7 RNA polymerase promoter. HeLa cells infected with recombinant vaccinia virus expressing T7 RNA polymerase were transfected with plasmid DNA encoding the N protein fragments, and the antigenicity of the synthesized proteins was analyzed by immunoprecipitation. Based on the immunoreactivities of the N protein deletion mutants with the panel of N-specific MAbs, five domains of antigenic importance were identified. MAbs SDOW17, SR30, and 5H2.3B12.1C9 each identified independent domains defined by amino acids 30 to 52, 69 to 123, and 37 to 52, respectively. Seven of the MAbs tested specifically recognized the local protein conformation formed in part by the amino acid residues 52 to 69. Furthermore, deletion of 11 amino acids from the carboxy terminus of the nucleocapsid protein disrupted the epitope configuration recognized by all of the conformation-dependent MAbs, suggesting that the carboxy-terminal region plays an important role in maintaining local protein conformation.
Porcine reproductive and respiratory syndrome (PRRS) is presently one of the most important infectious diseases of swine. The disease, which first appeared in North America in 1987, is characterized by severe reproductive failure and respiratory distress (1). The specific viral etiology of PRRS was originally established in The Netherlands (26) and was given the name Lelystad virus. In 1992, the first North American isolate responsible for PRRS was identified (3) and designated VR-2332. Although North American and European isolates of PRRS virus have morphological and structural similarities, they display significant molecular and antigenic variation, which suggests that they represent two distinct serotypes (17, 25).
PRRS virus consists of a plus-sense polyadenylated RNA genome (15 kb) surrounded by a cubical nucleocapsid core and a lipoprotein envelope. Nucleotide sequence analysis of the entire Leylstad virus genome (14) and the bulk of the VR-2332 genome (4) identified eight overlapping open reading frames (ORFs). ORFs 1a and 1b, which are expressed from genomic RNA, occupy more than two-thirds of the genome and encode the viral RNA polymerase. ORFs 2 to 7, located downstream of ORF 1b, encode the structural proteins. ORFs 2 through 5 code for membrane glycoproteins, ORF 6 encodes a nonglycosylated membrane protein, and ORF 7 encodes a highly basic nucleocapsid (N) protein. These genes are expressed from a 3′-coterminal nested set of functionally monocistronic subgenomic mRNAs (19). Based on genome organization, replication strategy, and propensity for infecting macrophages, PRRS virus has been classified in the genus Arterivirus in the newly proposed family Arteriviridae (2). Other members of this family include lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus (SHFV).
The products of ORFs 5, 6, and 7 constitute the major structural viral proteins (13). These proteins elicit discrete populations of functional antibodies (Abs) at various times during infection (15), with the N protein evoking the greatest immune response (20). Comparative analysis of the predicted N protein amino acid sequence indicates that this protein is well conserved among PRRS virus isolates within a given genotype, often displaying between 96 and 100% amino acid identity (11, 12). Furthermore, the antigenic character of the N protein appears to be largely conserved. This was illustrated by the results of a recent study in which 89% of 300 North American PRRS virus isolates tested reacted positively with a panel of 11 well-characterized N-specific monoclonal Abs (MAbs) (18). By virtue of its antigenic properties and sequence homology, the N protein has been used for detection of PRRS virus-specific antibodies in swine sera as well as for production of diagnostic reagents (24). Moreover, N-specific MAbs have been utilized to differentiate North American and European isolates of PRRS virus (17). Therefore, information regarding the antigenic properties of the N protein will facilitate classification of diagnostic MAbs according to their epitope specificities.
In the present study, a series of MAbs raised against several PRRS virus isolates was used to study the antigenic structure of the nucleocapsid protein. DNA fragments constituting discrete segments of the N protein were derived from the N coding region (ORF 7) of the PA-8 PRRS virus. The immunoreactivities of the N-specific MAbs with each of the fusion protein fragments produced in mammalian cells was analyzed by radioimmunoprecipitation (RIP) and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The amino acid sequences responsible for generating epitopes recognized by the MAbs in question were mapped to five independent regions of the N protein.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
HeLa cells were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 10% heat-inactivated fetal bovine serum (CanSera; Rexdale, Ontario, Canada). MARC-145 cells were similarly maintained in Dulbecco’s modified Eagle’s medium supplemented with 4% heat-inactivated fetal bovine serum (10). Recombinant vaccinia virus, vTF7-3, encoding T7 RNA polymerase was previously constructed by Fuerst et al. (7). To prepare vTF7-3 virus stock, HeLa cells were infected at a multiplicity of infection (MOI) of 0.1 PFU/cell. When cytopathic effects were evident, approximately 48 h postinfection, cells and media were harvested and centrifuged at 500 × g for 10 min. The cell pellet was subjected to one cycle of freeze-thaw to release virus particles. Cellular debris was removed by centrifugation at 500 × g for 10 min, and the clarified supernatant was used as crude virus stock. To prepare the PRRS virus strain PA-8 (kindly provided by J. Cho, Animal Disease Research Institute, Lethbridge, Alberta, Canada), MARC-145 cells were infected at an MOI of 5 PFU/cell. At 2 days postinfection, cells and supernatant were harvested and collected by centrifugation for 10 min at 500 × g. The clarified supernatant was used as crude virus stock. Rabbit anti–glutathione S-transferase (GST) polyclonal Ab (immunoglobulin G [IgG] fraction) (Sigma, St. Louis, Mo.) was used in immunoprecipitation experiments to identify expression of GST fusion proteins. A porcine hyperimmune serum raised against PRRS virus was used as a positive control to examine the antigenicity of N protein deletion mutants. A collection of 12 N-specific MAbs was used for immunoprecipitation of both the full-length and truncated N protein fragments. The strains of PRRS virus utilized in the production of the N-specific MAbs, as well as relevant references, are listed in Table 1.
TABLE 1.
Properties of MAbs specific for PRRS virus N protein
| MAb | PRRSV isolate used for MAb production | Ab isotype | RIP off:
|
Reference or source | |||
|---|---|---|---|---|---|---|---|
| Western blotf | 15 kDaa | PA-8 15 kDab | GST-N 41 kDac | ||||
| SDOW17 | VR-2332 | IgG | − | + | + | + | 16 |
| VO17 | 92-1509 | IgG1 | − | + | + | + | 16 |
| EP147 | 92-1509 | IgG1 | − | + | + | + | 16 |
| SR30 | 92-23983 | IgG1 | − | + | + | + | 17 |
| MR40 | 92-23983 | IgG1 | − | + | + | + | 17 |
| JP25 | 92-23983 | IgG1 | − | + | + | + | 17 |
| CF163 | 92-23983 | IgM | − | + | − | − | 17 |
| NS99 | Lelystad | IgG1 | + | + | − | − | 16 |
| 1D2 | MN-1b | IgG2b | − | + | + | + | D. Deregtd |
| 2G7 | MN-1b | IgG2b | − | + | + | + | D. Deregt |
| 7C10 | MN-1b | IgG2b | − | + | + | + | D. Deregt |
| 5H2 | LVHA933 | IgG | + | + | + | + | R. Magare |
Cell lysates infected with the autologous PRRS virus used in MAb production.
Cell lysates infected with PA-8 isolate.
Cell lysates infected with recombinant GST-N fusion protein.
Animal Disease Research Institute, Lethbridge, Alberta, Canada.
Laboratoire d’Hygiène Vétérinaire et Alimentaire, St. Hyacinth, Québec, Canada.
+, positive result; −, negative result.
RNA purification and cDNA cloning of ORF 7.
Culture supernatant was harvested from virus-infected cells 2 days postinfection, and cell debris was clarified. Virus was pelleted through a 30% sucrose cushion by ultracentrifugation using an SW28 rotor at 25,000 rpm for 2 h. The virus pellet was resuspended in Tris-EDTA (TE) buffer, and viral genomic RNA was prepared by phenol extraction. The extracted RNA was used for cloning of the N gene. A pair of forward (5′-CGGATC CCCTTGTCAAATATGCCAA-3′) and reverse (5′-AGAATGCCAGCCCATCA-3′) primers was fashioned for N gene amplification using Vent DNA polymerase (New England Biolabs, Mississauga, Ontario, Canada). The PCR product was cloned directly into the SmaI site of pGEM3zf(+) (Promega, Madison, Wis.) in the opposite orientation to the lacZ gene, allowing the convenient retrieval of the N gene through BamHI digestion. The resulting plasmid, named pGEM3zf-ORF7, represents the parental plasmid from which the N gene derivatives were constructed. The gene was sequenced in both directions by chain termination methods using an ABI automated sequencer.
Construction of recombinant plasmid expression vectors.
Fragments of the N gene were cloned in frame, downstream of the T7 promoter, into the mammalian expression vector pCITE-2a (Novagen, Madison, Wis.). Recombinant plasmids were constructed and used to transform the Escherichia coli strain JM105 following the methods of Sambrook et al. (23). Strategies for the construction of the N protein deletion mutants are illustrated in Fig. 1. The carboxy-terminal deletion mutants C-11, C-50, C-86, and C-98 were generated by digestion of pGEM3zf-ORF7 with restriction enzymes HgaI, AvaII, MfeI, and PflMI, respectively. For C-11, C-50, and C-86, the 5′ overhang was filled in with the Klenow fragment of DNA polymerase I, and for blunting of the 3′ overhang present in C-98, T4 DNA polymerase was used. Gel-purified fragments were digested with BamHI and subcloned into pGEX-3X (Pharmacia, Uppsala, Sweden) at the BamHI-SmaI site. To construct the C-66 deletion mutant, the full-length N gene was first cloned into the BamHI site of pGEX-3X. This construct was subsequently digested with BfaI, blunted with the Klenow fragment of DNA polymerase I, and redigested with BamHI. The BamHI-BfaI fragment was gel purified and subcloned back into the original pGEX-3X plasmid. The carboxy-terminal deletion mutants (C-11, C-50, C-66, C-86, and C-98) were amplified from pGEX-3X as GST fusion proteins by using the forward primer 5′-ATTTCCATGGTCATGTCCCCTATACTAGGTT-3′ and the reverse primer 5′-ATTACCATGGAACGCGCGAGGCAG-3′. PCR amplifications were carried out with 20 ng of template DNA, 40 pmol of each primer, and 2 U of Vent DNA polymerase. The samples were subjected to 30 cycles of amplification under the following conditions: denaturation at 94°C for 30 s, primer annealing at 55°C for 1 min, primer extension at 72°C for 2 min, and final extension at 72°C for 5 min. The PCR products were digested with NcoI and subcloned into pCITE-2a. For the amino-terminal deletion mutants, truncated gene fragments were cloned directly into pCITE2a-GST. To construct the plasmid pCITE2a-GST, the GST coding sequence, obtained by PCR amplification using the forward primer 5′-ATTTCCATGGTCATGTCCCCTATACTAGGTT-3′ and the reverse primer 5′-ATTACCATGAATTCCCGGGGA-3′, was digested with NcoI and subcloned into pCITE-2a. Plasmid pGEM3zf-ORF7 was used as template DNA for the PCR amplification of the N-18, N-30, N-52, and N-69 gene fragments. Individual fragments were amplified by using the following forward primers: 5′-CGTAGATCTGTCAATCAGCTGTGCCAGATG-3′ for N-18, 5′-GGCAGATCTATCGTTCAGCAAAACCAGTCC-3′ for N-30, 5′-CCGGCAGATCTCCCCATTTTCTTCTAGCGACT-3′ for N-52, and 5′-CGGAGATCTAGTGAGCGGCAATTGTGTCTG-3′ for N-69 (in combination with the reverse primer 5′-GAATACTCAAGCTTGCATGCCTG-3′). The PCR products were digested with restriction enzymes BglII and PstI and subcloned into pCITE2a-GST.
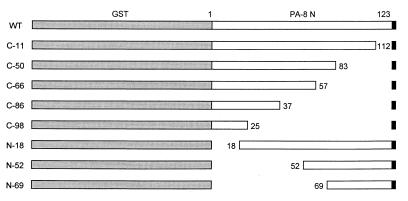
FIG. 1.
Schematic presentation of the N protein deletion mutants. The deletions and truncations of the N protein were constructed as described in Materials and Methods. Locations of the mutant proteins relative to the N protein are indicated by positions of amino acids. Shaded areas represent the GST coding sequence, and the black areas represent the translation termination.
Protein expression and radiolabelling.
Plasmid DNA was prepared using plasmid purification columns (Qiagen Inc., Santa Clarita, Calif.) according to the manufacturer’s recommended procedures. HeLa cells grown to 90% confluence in 100-mm-diameter dishes were infected at an MOI of 5 to 10 PFU/cell with vaccinia virus vTF7-3 and allowed to adsorb for 1 h at 37°C with occasional rocking. Five milliliters of fresh medium containing 10% fetal calf serum was added, and incubation continued for 1 h at 37°C. The plasmid DNA-LipofectACE (Gibco BRL, Burlington, Ontario, Canada) solutions were incubated in OPTI-MEM serum-reduced medium (Gibco BRL) at room temperature for 45 min prior to transfection. The inoculum was removed, and the cells were transfected for approximately 8 h with 10 μg of plasmid DNA and 40 μl of LipofectACE in 6.5 ml of OPTI-MEM. The supernatant was removed at 10 h postinfection, and the cells were labelled for 16 h with Easy Tag EXPRESS protein labelling mix (50 μCi/ml) consisting of [35S]methionine and [35S]cysteine (specific activity, 407 MBq/ml; New England Nuclear, Boston, Mass.) in methionine-free Eagle’s minimal essential media (Sigma) supplemented with 2% fetal calf serum. The cells were harvested, rinsed with cold phosphate-buffered saline, and resuspended in 600 μl of lysis buffer (0.1% Triton X-100 and 10 mM Tris-HCl, pH 7.4). After incubation for 10 min on ice, cell lysates were centrifuged at 14,000 rpm in an Eppendorf microcentrifuge for 10 min. The supernatant containing the cytoplasmic fraction was collected for immunoprecipitation experiments.
Immunoprecipitation and SDS-PAGE analysis.
Aliquots (20 μl) of labelled cell lysates were adjusted with RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.1% SDS) to a final volume of 100 μl and incubated for 2 h at room temperature with 1 μl of MAb or polyclonal Ab. The immune complexes were adsorbed to 10 mg of protein A-Sepharose CL-4B beads (Pharmacia) for 16 h at 4°C in 800 μl of RIPA buffer containing 0.3% SDS. The precipitates collected by centrifugation at 6,000 rpm for 2 min were washed two times with RIPA buffer and once with wash buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). Pellets were resuspended in 20 μl of SDS sample buffer (10 mM Tris-HCl [pH 6.8], 25% glycerol, 10% SDS, 10% β-mercaptoethanol, and 0.12% [wt/vol] bromophenol blue) and heated for 5 min at 95°C. After centrifugation at 10,000 rpm for 5 min, the clarified samples were analyzed by SDS–12% PAGE and visualized by autoradiography.
Nucleotide sequence accession number.
The sequence reported in this work has been deposited to the GenBank database under accession no. AF066068.
RESULTS
MAb reactivities with N protein of PA-8.
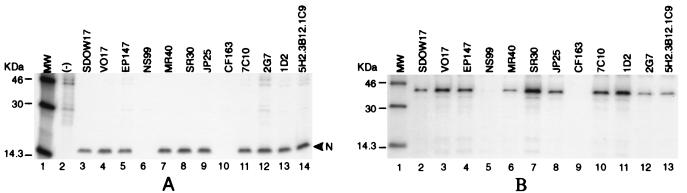
Panels of MAbs specific for the N protein have been developed by using various PRRS isolates (5a, 13). With the exception of MAb 5H2.3B12.1C9, which was derived from immunization with formalin-fixed PRRS virus, these MAbs were all produced by immunizing with viable virus. Characteristics of those MAbs, including virus isolates used in immunization, immunoglobulin isotype, Western blot, and RIP reactivities, are summarized in Table 1. RIP was used to determine the specificity of all the MAbs with the N protein, whereas 5H2.3B12.1C9 was tested by Western blotting. The former MAbs reacted negatively on Western blots, suggesting that they were specific for discontinuous conformational epitopes on the N protein. To examine the reactivity of those MAbs to the N protein of the PA-8 isolate, immunoprecipitations were performed with lysates from PA-8 virus-infected cells with each of the 12 MAbs on the panel (Fig. 2A). Each of the other MAbs specifically precipitated a protein of 15 kDa, while MAbs NS99 (lane 6) and CF163 (lane 10) did not recognize the PA-8 N protein by immunoprecipitation. No specific proteins were detected in the uninfected cells with a mixture of all 12 MAbs (lane 2).
FIG. 2.
(A) Specificity of the MAbs for the N protein of PA-8 virus. MARC-145 cells were infected with the PA-8 strain of PRRS virus at an MOI of 5 PFU/cell. Virus-infected cells were labelled with [35S]methionine at 50 μCi/ml, and cell extracts were prepared 48 h postinfection. Immunoprecipitation was performed with each of the 12 N-specific MAbs. Immune complexes were collected with protein A-Sepharose and resolved by SDS–12% PAGE as described in Materials and Methods. Uninfected cell lysate precipitated with a mixture of all 12 MAbs on the panel was used as a negative control (lane 2). The band migrating at 15 kDa representing the PA-8 N protein is marked with an arrowhead. (B) Specificity of the MAbs toward the recombinant GST-N fusion protein. HeLa cells infected with vaccinia virus expressing T7 RNA polymerase were transfected with plasmid DNA encoding the GST-N fusion protein under the control of the T7 promoter. Radiolabelled cell lysates were collected and immunoprecipitated with individual MAbs. The protein band (lanes 2 to 13) migrating at a molecular mass of 41 kDa represents the GST-N fusion protein. MW, molecular mass markers.
In order to study the antigenic structure required for those MAbs, the complete coding sequence for the N protein of PA-8 virus was cloned and sequenced (GenBank accession no. AF066068). The cloned N gene was able to encode a polypeptide of 123 amino acids. The predicted PA-8 protein sequence was compared to those of six isolates of PRRS virus from which the N-specific MAbs used in our mapping studies were derived (Table 1). Sequence homology ranged from 94.3 to 98.4% with respect to the North American strains and only 55% for the European strain. The VR-2332 N protein showed the highest degree of homology to that of PA-8, with 98.4% sequence identity and substitutions at positions 31 (Val to Ala) and 56 (Leu to Pro). These substitutions were found among all of the isolates compared. Isolate MN-1b showed the greatest variability, with additional substitutions at positions 9 (Thr to Gln), 10 and 11 (Glu to Arg), and 89 and 91 (Thr to Ile), resulting in an overall identity of 94.3% (data not shown).
We attempted to express the N gene in mammalian cells by the vaccinia T7 expression system. The N coding sequence was first cloned into the pGEX-3X expression vector in frame with the GST coding sequence. The coding sequence for GST-N fusion protein was subsequently inserted into the mammalian expression vector pCITE-2a downstream of the T7 RNA polymerase promoter. A stretch of spacer sequence of 30 amino acids was inserted to separate the N protein sequence from the GST sequence to reduce potential deleterious interactions which might occur during the formation of the epitopes. Expression of the N protein was confirmed by immunoprecipitation using anti-GST antibody (data not shown). Subsequently, immunoreactivities of the recombinant N protein with individual MAbs were examined. Similar to the results obtained from immunoprecipitation studies with PA-8-infected cell lysates, the 41-kDa GST-N fusion protein was recognized by all the MAbs examined except NS99 and CF163 (Fig. 2B, lanes 5 and 9, respectively). Collectively, our data show that 10 of the 12 MAbs specifically recognize the authentic N protein of the PA-8 isolate of PRRS virus in addition to the recombinant GST-N protein expressed in cells. Due to lack of reactivity with the PA-8 N protein, MAbs NS99 and CF163 were eliminated from subsequent experiments.
Expression of GST-coupled N protein deletions in HeLa cells.
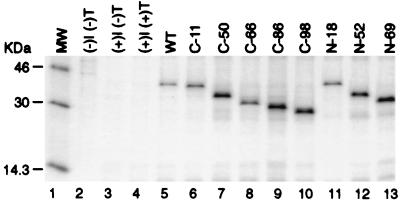
For our mapping studies, eight deletions and truncations were constructed to represent overlapping fragments of the N protein ranging from 25 to 112 amino acids (Fig. 1). These mutants were created by deleting sequences progressively from either the 5′ or 3′ terminus of the N coding sequence. Since our mutant constructs represent small fragments of the N protein, it was necessary to fuse the constructs with GST protein to facilitate visualization of the precipitated proteins. HeLa cells, infected with recombinant vaccinia virus vTF7-3, were transfected with plasmids encoding the deletion mutants. The transfected cells were radiolabelled and subjected to immunoprecipitation with anti-GST antibody. The resulting proteins expressed in HeLa cells are shown in Fig. 3. A band migrating in agreement with the expected molecular weight for each deletion mutant was identified (lanes 5 to 13). Expression of the deletion mutants synthesized as GST fusion proteins was also confirmed by reactivity with hyperimmune antisera from swine immunized with PRRS virus (data not shown). Results from these experiments indicated that the N deletion proteins were synthesized as fusion proteins at sufficient levels with which to examine the N-specific MAb reactivities.
FIG. 3.
Expression of the GST-N fusion constructs immunoprecipitated with anti-GST antibody. HeLa cells infected with vaccinia virus expressing T7 RNA polymerase (vTF7-3) were transfected with plasmid DNA. Cells labelled with [35S]methionine were collected 26 h postinfection, immunoprecipitated with anti-GST antibody, and resolved by SDS–12% PAGE. Lane 1, molecular mass markers (MW); lane 2, uninfected and untransfected cells; lane 3, vTF7-3-infected, untransfected cells; lane 4, vTF7-3-infected, control plasmid DNA (pCITE2a)-transfected cells; lanes 5 to 13, lysates from cells expressing each of the N fusion proteins.
Immunoreactivity of the N-specific MAbs with N protein fragments.
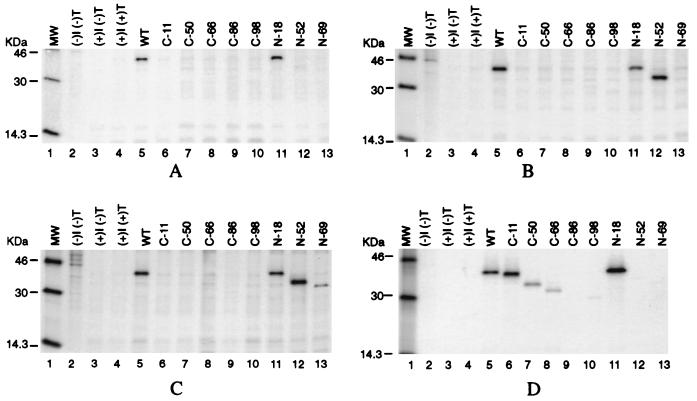
To localize the antigenically important domains present on the N protein of PRRS virus, the reactivities of the deletion mutants were examined with a collection of 10 N-specific MAbs. MAb SDOW17 reacted with the deletion mutant N-18, which migrated at the molecular mass of 40 kDa (Fig. 4A, lane 11), but failed to recognize either N-52 and N-69 (lanes 12 and 13), or any of the carboxy-terminal deletion mutants (lanes 6 to 10). The MAb EP147 reacted with both N-18 and N-52 (Fig. 4B, lanes 11 and 12), but failed to precipitate the N-69 deletion mutant (lane 13) or any of the carboxy-terminal truncation mutants (lanes 6 to 10). MAbs VO17, MR40, JP25, 1D2, 2G7, and 7C10 reacted similarly to EP147 in our immunoprecipitation assays (data not shown). The deletion mutant N-69 is composed of amino acids 69 to 123; therefore, it is conceivable that the region pertaining to amino acids 52 to 69 constitutes an antigenically important domain for this group of MAbs. The MAb SR30 reacted with all three amino-terminal deletion mutants—N-18, N-52, and N-69 (Fig. 4C, lanes 11 to 13)—but failed to recognize any of the carboxy-terminal deletion mutants (lanes 6 to 10). The binding pattern observed for MAb SR30 suggests that the epitope it recognizes lies within the carboxy-terminal region of the N protein, more specifically between amino acids 69 and 123. In contrast to other MAbs, 5H2.3B12.1C9 recognized the carboxy-terminal deletion mutants C-11, C-50, and C-66 (Fig. 4D, lanes 6 to 8) in addition to the N-18 deletion mutant (Fig. 4D, lane 11).
FIG. 4.
Immunoprecipitation of the GST-N protein deletion mutants with individual MAbs. HeLa cells were infected with vTF7-3 and transfected with plasmids encoding the N protein deletion mutants. Cells were radiolabelled with [35S]methionine (50 μCi/ml) for 16 h. Cell lysates containing the cytoplasmic fraction were immunoprecipitated with each of the N-specific MAbs, and the immune complexes were resolved by SDS–12% PAGE followed by autoradiography. (A) Immunoprecipitation with MAb SDOW17; (B) immunoprecipitation with MAb EP147 (representative also of the results observed for MAbs VO17, MR40, JP25, 1D2, 2G7, and 7C10); (C) immunoprecipitation with MAb SR30; (D) immunoprecipitation with MAb 5H2.3B12.1C9. Lane 1, molecular mass markers (MW); lane 2, uninfected, untransfected HeLa cells; lane 3, vTF7-3-infected, untransfected cells; lane 4, vTF7-3-infected, control plasmid DNA (pCITE2a)-transfected cells; lanes 5 to 13, lysates of cells expressing each of the N protein mutants.
DISCUSSION
Diagnosis of PRRS virus infection on the basis of clinical signs is generally considered unreliable because symptoms often vary among herds. Clinical diagnosis is further complicated in instances when secondary infections produce PRRS-like symptoms. Consequently, serological techniques including enzyme-linked immunosorbent assay (ELISA), indirect immunofluorescence assay, and immunoperoxidase monolayer assay are routinely used in the diagnosis of PRRS virus infection. Many of these procedures rely on the highly immunogenic nature of the PRRS virus N protein, both for antibody production and antibody detection. The following examples illustrate this point. A highly specific indirect ELISA utilizing baculovirus-expressed N protein as the antigen was recently developed for detecting antibodies against PRRS virus in swine sera (5). This technique proved to be considerably more specific than conventional ELISAs using whole-virus antigen and less expensive and time-consuming by virtue of its use of a single recombinant protein rather than infectious virus. MAbs derived from PRRS virus, which are most often specific for the 15-kDa N protein, can be conjugated to fluorescein isothiocyanate to allow detection of PRRS virus in swine tissue (24). Furthermore, N-specific MAbs can be used for differential diagnosis of PRRS virus infection (9). MAbs capable of distinguishing between field and vaccine strains of PRRS virus are of particular importance due to the fact that an attenuated live PRRS virus vaccine is now widely administered. Moreover, multiple viral strains exist within the swine population, and these do not frequently cross-react serologically. Thus, knowledge about the location of epitopes on the N protein as well as serological classification of N-specific MAbs will serve to enhance the specificity of many serology-based diagnostic tests.
The expression of subgenomic fragments in eukaryotic cells is frequently used to localize antigenic domains on proteins (21, 27). This approach to epitope mapping offers an advantage over other methods involving oligopeptides and/or E. coli-produced protein fragments, because viral proteins expressed in eukaryotic cells will undergo the necessary posttranslational modifications to ensure proper conformation. Conversely, proteins expressed in bacteria do not possess any posttranslational modifications; therefore, such an approach may fail to give clear results when complex antigenic structures are involved. Previously, PRRS virus N protein mapping studies were performed with E. coli-expressed protein fragments (22). A linear epitope was identified between amino acids 50 and 66; however, neither this nor any of the other E. coli-expressed N protein fragments proved efficacious for diagnostic purposes. This can be attributed to the fact that the majority of N-specific MAbs produced during infection are conformation-dependent (6, 18). Consequently, for our mapping studies we produced the N protein deletion mutants in mammalian cells by the T7-based vaccinia virus expression system so as to approximate native conformation.
The purpose of this study was to elucidate the structural requirements for Ab binding by using a collection of 12 MAbs with a total of nine different N protein constructs expressed in mammalian cells. Immunoreactivities of the MAbs with the individual mutants are summarized in Table 2, and the domains of antigenic importance identified in our study are depicted in Fig. 5. In accordance with the experimental results, MAbs were divided into four different groups, each specifying a unique region of antigenic importance on the N protein. Domain I, which was identified by MAb SDOW17 (group 1), was localized to the amino-terminal region between amino acids 18 and 52 (Fig. 4A). During the preparation of this manuscript, an additional deletion mutant, N-30, was constructed. All the MAbs tested reacted with the N-30 product in immunoprecipitation assays (data not shown). The N-30 deletion mutant comprises amino acids 30 to 123, and N-52 comprises amino acids 52 to 123, indicating that a region of antigenic importance is actually located between amino acids 30 and 52 (domain I). All of the other conformation-dependent MAbs retained binding activity upon removal of 52 amino acids from the amino terminus (Table 2), which suggests that this region recognized by SDOW17 contains a relatively unique epitope. The specificity exhibited exclusively by SDOW17 for domain I was not unexpected. It has been demonstrated that SDOW17 is the only MAb capable of recognizing an epitope common to almost all European and North American isolates of PRRS virus. Consequently, SDOW17 represents a valuable diagnostic reagent. Nelson et al. (16) identified an amino acid present on nearly all field isolates of PRRS virus but absent from the Prime Pac PRRS vaccine strain. By successive passages, the Prime Pac PRRS vaccine strain acquired a mutation at position 61 that resulted in a change from aspartic acid to tyrosine. A mutation of this nature is predicted to disrupt α-helix conformation. This amino acid difference impaired SDOW17 recognition of the vaccine strain. Given the fact that SDOW17 recognizes a conformational epitope common to both European and North American genotypes, this epitope is likely to be localized within a highly conserved region of the N protein. Moreover, sequence analysis revealed that although there were amino acid changes scattered throughout the N protein, a central region of well-conserved amino acids existed between residues 47 and 100 (22). Secondary structure prediction using the method of Garnier et al. (8) suggests that this region forms a helical conformation; therefore, it is possible that SDOW17 recognizes this highly conserved hydrophilic loop structure and that removal of 52 amino acids, but not 30 amino acids, alters the local conformation such that SDOW17 is no longer able to recognize the N protein. Similarly, the change from aspartic acid to tyrosine, which is predicted to disrupt α-helix formation, may influence overall conformation, explaining why the vaccine strain is unrecognizable by SDOW17.
TABLE 2.
Summary of immunoreactivities of N-specific MAbs with N protein deletion mutants
| N-specific MAb | Immunoreactivity with:
|
Required domain(s) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WT | N-18 | N-52 | N-69 | C-98 | C-86 | C-66 | C-50 | C-11 | ||
| SDOW17 | + | + | − | − | − | − | − | − | − | I + V |
| VO17 | + | + | + | − | − | − | − | − | − | III + V |
| EP147 | + | + | + | − | − | − | − | − | − | III + V |
| SR30 | + | + | + | + | − | − | − | − | − | IV + V |
| MR40 | + | + | + | − | − | − | − | − | − | III + V |
| JP25 | + | + | + | − | − | − | − | − | − | III + V |
| 1D2 | + | + | + | − | − | − | − | − | − | III + V |
| 2G7 | + | + | + | − | − | − | − | − | − | III + V |
| 7C10 | + | + | + | − | − | − | − | − | − | III + V |
| 5H2.3B12.1C9 | + | + | − | − | − | − | + | + | + | II |
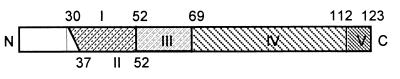
FIG. 5.
Illustration of the five antigenically important domains localized on the N protein. The shaded areas flanked by amino acid positions represent domains localized by the mapping studies.
Domain III, defined by amino acids 52 to 69, lies within a well-conserved region of the N protein. This stretch of amino acids appears to be part of an immunodominant epitope, as 70% of the MAbs on the panel recognized local protein conformation present in the N-52 but not in the N-69 deletion mutant (Table 2), which suggests good immunogenicity for this region. These results are corroborated by a previous report which suggests that a well-conserved linear epitope is located between amino acids 50 and 66 (22). Hydropathic profile indicates that amino acids 40 to 60 are present in a hydrophilic region of the N protein, demonstrating high solvent accessibility. It is therefore likely that the majority of MAbs recognize this portion of the protein because of its conspicuous location on the outer capsid surface.
Domain IV, which extends from amino acids 69 to 123, was defined by the specific binding activity of MAb SR30 with deletion mutants N-18, N-52, and N-69 (Fig. 4C). Deletion of the amino terminus does not appear to affect the local protein conformation forming the SR30 specific epitope. In a survey of over 300 North American PRRS virus isolates, the MAb SR30 reacted with 100% of the field isolates tested, whereas SDOW17 reacted with 99.4% of the isolates (17). This suggests that like SDOW17, SR30 recognizes a highly conserved epitope on the N protein and therefore constitutes a valuable diagnostic tool.
MAb 5H2.3B12.1C9 was able to recognize the carboxy-terminal deletion mutants C-11, C-50, and C-66 (Fig. 4D). It was possible to delete up to 66 amino acids from the C terminus without the C-66 mutant losing the ability to be bound by 5H2.3B12.1C9. However, deletion of more than 30 amino acids from the N terminus disrupted the MAb binding (data not shown). Therefore, the epitope specific for this MAb should be located between amino acids 37 and 57 (domain II). The difference observed between 5H2.3B12.1C9 and other MAbs may be due to the nature of the antigen used for immunization. Unlike the other MAbs, formalin-inactivated PRRS virus was used in the production of 5H2.3B12.1C9. Treatment with formalin, a protein cross-linking agent, promotes conformational changes or denaturation of the antigen; therefore, MAbs made in this fashion are likely to be specific for linear as opposed to conformational epitopes. Thus, it is conceivable that, because 5H2.3B12.1C9 reacted positively in Western blot experiments (Table 1), domain II constitutes a linear epitope.
To facilitate our mapping studies, reactivities of carboxy-terminal deletion mutants were also examined with the panel of N-specific MAbs. Remarkably, all of the MAbs, with the exception of 5H2.3B12.1C9, were unable to recognize the mutants with carboxy terminus deletions (Fig. 4A to C; Table 2). Deletion of as few as 11 amino acids from the carboxy terminus (C-11) of the N protein rendered all of the conformation-dependent MAbs unresponsive. Given the fact that the only MAb on the panel capable of recognizing the carboxy-terminal deletions (5H2.3B12.1C9) was conformation independent, it is likely that the region defined by domain V plays a critical role in maintaining overall protein structure. Carboxy-terminal deletions must, therefore, influence the structure of epitopes recognized by the conformation-dependent N-specific MAbs under consideration. In previous studies, conformation-dependent N-specific MAbs produced against a European strain of PRRS virus failed to recognize N protein fragments whose carboxy termini had been removed (22), which is consistent with our observations. It is possible that the region defined by domain V plays a role in N protein multimerization and nucleocapsid assembly. Mutational analysis to dissect the C-terminal 11 amino acids for their contribution to MAb binding and the studies to delineate multimerization of the N protein are presently in progress.
ACKNOWLEDGMENTS
This research was supported by the Ontario Ministry of Agriculture, Food and Rural Affairs; the Ontario Research Enhancement Program of Agriculture and Agri-Food Canada; Ontario Pork; and USDA-NRI-CGP grant 9602270.
We thank J. Cho (Animal Disease Research Institute) for the anti-PRRS virus swine polyclonal antiserum and the PA-8 strain of PRRS virus; R. Magar for MAb 5H2.3B12.1C9; D. Deregt for MAbs 2G7, 1D2, 2D6, and 7C10; and Eva Varady for her technical help.
REFERENCES
- 1.Benfield D A, Nelson E, Collins J E, Harris L, Goyal S M, Robinson D, Christianson W T, Morrison R B, Gorcyca D E, Chladek D W. Characterization of swine infertility and respiratory syndrome (SIRS) virus (isolate ATCC VR-2332) J Vet Diagn Investig. 1992;4:127–133. doi: 10.1177/104063879200400202. [DOI] [PubMed] [Google Scholar]
- 2.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 3.Collins J E, Benfield D A, Christianson W T, Harris J E, Hennings L, Shaw J C, Goyal D P, McCullough S M, Morrison S, Joo R B, Gorcyca H S, Chladek D W. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Investig. 1992;4:117–126. doi: 10.1177/104063879200400201. [DOI] [PubMed] [Google Scholar]
- 4.Conzelmann K, Visser N, van Woensel P, Thiel H. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the arterivirus group. Virology. 1993;193:329. doi: 10.1006/viro.1993.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denac H, Tratschin J D, Hofmann M A. An indirect ELISA for the detection of antibodies against porcine reproductive and respiratory syndrome virus using recombinant nucleocapsid protein as antigen. J Virol Methods. 1997;65:169–181. doi: 10.1016/s0166-0934(97)02186-1. [DOI] [PubMed] [Google Scholar]
- 5a.Deregt, D., and R. Magar. Personal communication.
- 6.Drew T W, Meulenber J J M, Sands J J, Paton D J. Production, characterization and reactivity of monoclonal antibodies to the porcine reproductive and respiratory syndrome virus. J Gen Virol. 1995;76:1361–1369. doi: 10.1099/0022-1317-76-6-1361. [DOI] [PubMed] [Google Scholar]
- 7.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 9.Kapur V, Elam M R, Pawlovich T M, Murtaugh M P. Genetic variation in porcine reproductive and respiratory syndrome virus isolates in the Midwestern United States. J Gen Virol. 1996;77:1271–1276. doi: 10.1099/0022-1317-77-6-1271. [DOI] [PubMed] [Google Scholar]
- 10.Kim H S, Kwang J, Yoon I J, Joo H S, Frey M L. Enhanced replication of PRRS virus in a homogeneous subpopulation of MA-104 cell line. Arch Virol. 1993;133:477–483. doi: 10.1007/BF01313785. [DOI] [PubMed] [Google Scholar]
- 11.Magar R, Larochelle R, Dea S, Gagnon C A, Nelson E A, Christopher-Hennings J, Benfield D A. Antigenic comparison of Canadian and US isolates of porcine reproductive and respiratory syndrome virus using monoclonal antibodies to the nucleocapsid protein. Can J Vet Res. 1995;59:232–234. [PMC free article] [PubMed] [Google Scholar]
- 12.Meng X J, Paul P S, Halbur P G, Lum M A. Phylogenic analysis of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the U.S.A. and Europe. Arch Virol. 1995;140:745–755. doi: 10.1007/BF01309962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meulenberg J J M, den Besten A, De Kluyver E P, Moormann R J M, Schaaper V M M, Wensvoort G. Characterization of proteins encoded by ORFs 2 to 7 of Lelystad virus. Virology. 1995;206:155–163. doi: 10.1016/S0042-6822(95)80030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meulenberg J J M, Hulst M M, de Meijer E J, Moonen P J L M, den Besten A, De Kluyver E P, Wensvoort G, Moormann R J M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson E, Christopher-Hennings J, Benfield D A. Serum immune responses to the proteins of PRRS virus. J Vet Diagn Investig. 1994;6:410–415. doi: 10.1177/104063879400600402. [DOI] [PubMed] [Google Scholar]
- 16.Nelson E, Nelson J K, Couture L P, Lau M L, Christopher-Hennings J, Chase C C L, Hesse R A. Proceedings of the 78th Conference of Research Workers in Animal Diseases, Chicago, Ill. 1997. A single amino acid substitution allows for the differentiation of the Prime Pac PRRS® vaccine from field isolates of PRRSV, abstr. 202. [Google Scholar]
- 17.Nelson E A, Christopher-Hennings J K, Drew T, Wensvoort G, Collins J, Benfield D A. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J Clin Microbiol. 1993;31:3184–3189. doi: 10.1128/jcm.31.12.3184-3189.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson E A, Nelson J K, Christopher-Hennings J, Yoon K-J, Magar R, Benfield D A. Proceedings of the 14th International Pig Veterinary Society Congress, Bologna, Italy. 1996. Reactivity of North American PRRSV isolates with a monoclonal antibody panel; p. 88. [Google Scholar]
- 19.Plagemann P G W, Moenning V. Lactate dehydrogenase elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive strand RNA viruses. Adv Virus Res. 1992;41:99–192. doi: 10.1016/S0065-3527(08)60036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plana-Duran J, Climent I, Sarraseca J, Uriniza A, Cortes E, Vela C, Casal J I. Baculovirus expression of proteins of porcine reproductive and respiratory syndrome virus strain Olot/91. Involvement of ORF3 and ORF5 proteins in protection. Virus Genes. 1997;14:19–29. doi: 10.1023/a:1007931322271. [DOI] [PubMed] [Google Scholar]
- 21.Raux H, Iseni F, Lafay F, Blondel D. Mapping of monoclonal antibody epitopes of the rabies virus P protein. J Gen Virol. 1997;78:119–124. doi: 10.1099/0022-1317-78-1-119. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez M J, Sarraseca J, Garcia J, Sanz A, Plana-Duran J, Casal J I. Epitope mapping of the nucleocapsid protein of European and North American isolates of porcine reproductive and respiratory syndrome virus. J Gen Virol. 1997;78:2269–2278. doi: 10.1099/0022-1317-78-9-2269. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 24.van Alistine W G, Stevenson G W, Kanitz C L. Diagnosis of PRRS. Swine Health Prod. 1993;4:24–28. [Google Scholar]
- 25.Wensvoort G, de Kluyver E P, Luitjze E A, den Besten A, Harris L, Collins J E, Christianson W T, Chladek D. Antigenic comparison of Lelystad virus and swine infertility and respiratory syndrome (SIRS) virus. J Vet Diagn Investig. 1992;4:134–138. doi: 10.1177/104063879200400203. [DOI] [PubMed] [Google Scholar]
- 26.Wensvoort G, Tepstra C, Pol J M A, ter Laak E A, Bloemraad M, de Kluyver E P, Kragten C, van Buiten L, den Besten A, Wagenaar F, Broekhuijsen J M, Moonen P L J M, Zestra T, de Boer E A, Tibben H J, de Jong M F, van Veld P, Groenland G J R, van Gennep J A, Voets M T, Verheijden J H M, Braamskamp J. Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q. 1991;13:121–130. doi: 10.1080/01652176.1991.9694296. [DOI] [PubMed] [Google Scholar]
- 27.Yoo D, Parker D M, Song J, Cox G J, Deregt D, Babiuk L A. Structural analysis of the conformational domains involved in the neutralization of bovine coronavirus using deletion mutants of the spike glycoprotein S1 subunit expressed by recombinant baculoviruses. Virology. 1991;183:91–98. doi: 10.1016/0042-6822(91)90121-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]