ABSTRACT
Background
Comanagement of glycemia and adiposity is the cornerstone of cardiometabolic risk reduction in type 1 diabetes (T1D), but targets are often not met. The intestinal microbiota and microbiota-derived short-chain fatty acids (SCFAs) influence glycemia and adiposity but have not been sufficiently investigated in longstanding T1D.
Objectives
We evaluated the hypothesis that an increased abundance of SCFA-producing gut microbes, fecal SCFAs, and intestinal microbial diversity were associated with improved glycemia but increased adiposity in young adults with longstanding T1D.
Methods
Participants provided stool samples at ≤4 time points (NCT03651622: https://clinicaltrials.gov/ct2/show/NCT03651622). Sequencing of the 16S ribosomal RNA gene measured abundances of SCFA-producing intestinal microbes. GC-MS measured total and specific SCFAs (acetate, butyrate, propionate). DXA (body fat percentage and percentage lean mass) and anthropometrics (BMI) measured adiposity. Continuous glucose monitoring [percentage of time in range (70–180 mg/dL), above range (>180 mg/dL), and below range (54–69 mg/dL)] and glycated hemoglobin (i.e., HbA1c) assessed glycemia. Adjusted and Bonferroni-corrected generalized estimating equations modeled the associations of SCFA-producing gut microbes, fecal SCFAs, and intestinal microbial diversity with glycemia and adiposity. COVID-19 interrupted data collection, so models were repeated restricted to pre-COVID-19 visits.
Results
Data were available for ≤45 participants at 101 visits (including 40 participants at 54 visits pre-COVID-19). Abundance of Eubacterium hallii was associated inversely with BMI (all data). Pre-COVID-19, increased fecal propionate was associated with increased percentage of time above range and reduced percentage of time in target and below range; and abundances of 3 SCFA-producing taxa (Ruminococcus gnavus, Eubacterium ventriosum, and Lachnospira) were associated inversely with body fat percentage, of which two microbes were positively associated with percentage lean mass. Abundance of Anaerostipes was associated with reduced percentage of time in range (all data) and with increased body fat percentage and reduced percentage lean mass (pre-COVID-19).
Conclusions
Unexpectedly, fecal propionate was associated with detriment to glycemia, whereas most SCFA-producing intestinal microbes were associated with benefit to adiposity. Future studies should confirm these associations and determine their potential causal linkages in T1D.
This study is registered at clinical.trials.gov (NCT03651622; https://clinicaltrials.gov/ct2/show/NCT03651622).
Keywords: type 1 diabetes, gut microbiota, short-chain fatty acids, continuous glucose monitoring, dual-energy X-ray absorptiometry, hemoglobin A1c, glycemia, adiposity, body mass index
Fecal propionate and SCFA-producing gut microbes were associated with impaired glycemia but reduced adiposity, respectively, in young adults with type 1 diabetes who were overweight.
Introduction
Individuals with type 1 diabetes (T1D) are at a 3–7 times higher risk of micro- and macrovascular (cardiovascular) complications than those without diabetes (1). Comanagement of glycemia and adiposity is the cornerstone of cardiometabolic risk reduction for people with T1D. However, glycemic and weight targets are often not met, particularly in young adults, in whom glycated hemoglobin (HbA1c) peaks between the ages of 19 and 30 (2). Glycemia has not improved in the United States whereas the prevalence of obesity has increased in people with T1D in recent decades (3–5) despite numerous advances in and uptake of diabetes self-management technologies [e.g., continuous glucose monitoring (CGM) and insulin pump therapy] (6). There is a pressing need to identify novel factors that ease the burden of comanaging glycemia and adiposity in T1D.
One potential disruptor of comanagement of glycemia and adiposity is an altered composition of the intestinal microbiota—the vast community of microbes residing in the intestinal tract—and its effector metabolites, short-chain fatty acids (SCFAs), produced through gut microbial fermentation of nondigestible carbohydrates such as fiber (7). Individuals with T1D may have a reduced abundance of SCFA-producing gut microbes and fecal SCFAs compared with controls without T1D (8–10). This altered microbial ecosystem could impact glycemia and adiposity via metabolic regulation. Specifically, SCFAs have been shown to bind to receptors in the liver and adipose tissue leading to improved hepatic and peripheral insulin sensitivity, enhanced neurologically mediated satiety, and glucagon-like peptide 1 (GLP-1) production by intestinal epithelial L-cells—which can enhance satiety through afferent signals to the brain's appetite regulatory centers (7, 11, 12). However, salient to weight management, specific gut microbes can increase energy harvest by fermenting fiber to SCFAs, which invites speculation that SCFA-producing intestinal microbes could contribute to positive energy balance and therefore weight gain (13–15). The notion that SCFAs can contribute to adiposity is supported by the finding that mice lacking the G-protein coupled SCFA receptor GPR41 are leaner than their wild-type littermates (16), although another preclinical study found a protective effect of SCFAs on adiposity through their signals to a different SCFA receptor, GPR43 (17).
Investigations of the role of the gut microbiota in T1D etiology have revealed differences in the abundances of major gut microbial taxa and their functional genomic potential (e.g., increases or decreases in facets of carbohydrate metabolism) in individuals with T1D compared with controls without T1D (8, 9, 18). However, prior associations of gut microbes and SCFAs with glucose metabolism and adiposity in animals, and in humans with metabolic syndrome (19, 20) have not been effectively translated to the metabolically unique setting of longstanding T1D. Therefore, in this hypothesis-generating study, we assessed whether an increased abundance of SCFA-producing gut microbes, fecal SCFAs, and intestinal microbial diversity were associated with improved glycemia, but also with increased adiposity, in young adults with longstanding T1D and overweight or obesity.
Methods
Study sample
Participants were young adults with T1D aged 19–30 y (T1D duration ≥1 y), literate in English, HbA1c <13.0% (<119 mmol/mol), and BMI 27–39.9 kg/m2 who enrolled in the NIH-funded Advancing Care for Type 1 Diabetes and Obesity Network (ACT1ON) Sequential Multiple Assignment Randomized Trial (SMART) pilot for weight and glycemic management (1DP3DK113358-01, NCT03651622). Those included in the present analysis participated in an ancillary gut microbiome pilot study under the umbrella of ACT1ON. The study design of ACT1ON has been described elsewhere (21). Briefly, the ACT1ON SMART pilot was a 9-mo feasibility intervention conducted at the University of North Carolina at Chapel Hill (UNC) and Stanford University. Its objective was to identify acceptable dietary strategies (hypocaloric low carbohydrate, hypocaloric moderate low fat, or Mediterranean diet without calorie restriction) to co-optimize weight and glycemia in young adults with T1D. Per the SMART design, participants were assigned to an initial diet arm at study enrollment, after which the trial adapted dynamically to participant responses by rerandomizing those who rated the diet as being unacceptable or who did not achieve a minimum weight loss, or whose glycemic parameters deteriorated according to HbA1c or self-reported hypoglycemia. Diets were assigned using permuted block randomization stratified by site at 3 and 6 mo of the intervention (22–24). The primary parent study outcomes were weight, HbA1c, and percentage of time in clinical hypoglycemia (54–69 mg/dL) (25) assessed by CGM at the end of each of 3 diet periods. Secondary outcomes were percentage body fat assessed by DXA, and percentage of time in target glucose range (CGM, 70–180 mg/dL). (25) UNC coordinated the study.
We identified eligible young adults according to medical record data for participation in the parent ACT1ON SMART using a 2-step recruitment process (26). Participants completed 4 measurement visits. All study visits were completed between November 12, 2018 and February 2, 2021. As of April 27, 2020, the study moved to a virtual format via a Health Insurance Portability and Accountability Act–secure Zoom account in continued response to COVID-19. Dietary counseling and data collection were both done virtually, and recruitment ceased (we enrolled 68 participants whereas the target was 72). We conducted the first virtual visit during COVID-19 on June 17, 2020. Using standardized protocols with support from study staff, participants collected HbA1c samples and inserted CGM sensors at home. DXA was discontinued and measures of body composition were therefore only available pre-COVID-19.
Ancillary gut microbiome pilot study
We invited ACT1ON study participants who had not taken antibiotics in the prior month to provide stool samples for an ancillary hypothesis-generating gut microbiome study via a home collection during the 2 weeks in which all other measurement visit data were collected (i.e., before the beginning of each diet period). We originally planned to collect samples only at baseline and measurement visit 2, which we did pre-COVID-19. During COVID-19, we added voluntary stool collection at measurement visits 3 and 4 due to participant dropout and diminished sample size resulting in part from the COVID-19 pandemic. We invited participants who provided samples at the baseline visit to provide additional samples at follow-up visits if they reported no antibiotic use in the month prior to collection.
Sixty-eight parent ACT1ON participants completed 200 visits across the 4 study timepoints. Forty-five participants voluntarily provided stool for the ancillary gut microbiome pilot study, including 112 stool samples across the 4 study timepoints (we excluded 6 participants for antibiotic use, 2 who did not return the stool sample before diet randomization due to shipment issues, 2 who had difficulty with producing a sample, 4 who declined participation, and 9 who initially agreed to participate but did not return the sample prior to randomization). We restricted the analysis to visits with concurrently available 24-h dietary recall data, because it was necessary to adjust for fiber intake as an important potential confounder. Thus, we excluded 11 samples from study analysis due to missing diet data. SCFA and diet data were available for 101 samples, of which all had HbA1c and BMI data, 43 were missing DXA data (due to the post-COVID-19 virtual format), and 24 had missing (n = 9) or insufficient (n = 15) CGM data. An additional 4–5 samples (dependent on the outcome) did not pass quality controls [filtering and denoising in the Quantitative Insights Into Microbial Ecology 2 (QIIME2) analytic pipeline] and were therefore excluded from analysis of gut microbial taxonomy and diversity but still provided SCFA data. We show a Consolidated Standards of Reporting Trials diagram with the derived sample size for each outcome, including after restriction to pre-COVID-19 data (Supplemental Figure 1).
Measures
Gut microbiota characterization
We isolated genomic microbial DNA from human fecal samples using a phenol-chloroform extraction combined with a bead beating step using 0.1-mm glass beads (Bio Spec products) to physically disrupt bacterial cells, and a DNA clean-up kit (Qiagen DNeasy Blood and Tissue extraction kit), as previously described (27). We characterized fecal microbiotas using the variable 4 region of the 16S rRNA gene to create sequencing libraries via PCR and sequencing on the Illumina MiSeq platform at the High-Throughput Sequencing Facility in the Carolina Center for Genome Sciences at the UNC School of Medicine, as previously described (28).
We managed 16S rRNA gene sequences generated by the Illumina MiSeq platform via the QIIME2 pipeline, which included demultiplexing and denoising reads via the Divisive Amplicon Denoising Algorithm (DADA2) (29). We generated sequence variants at 100% identity threshold using DADA2. The total number of sequence reads was 11,105,926 [98,558.5 (IQR: quartile 1, 78,072.0; quartile 3, 129,325.8) per sample] and there were 2339 generated sequence variants. We considered several approaches to normalize read counts (30, 31) and ultimately selected a previously published method using the following formula (32):
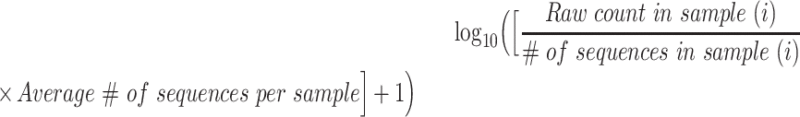 |
We performed taxonomic classification using the DADA2-formatted reference database Silva (33). We used QIIME2 to derive rarefied intestinal microbial diversity—which we report as the number of unique sequence variants (i.e., the number of unique taxa) per sample. To reduce potential bias stemming from imbalanced replication of sequence reads during PCR steps, we rarefied our measure of within-sample intestinal microbial diversity (i.e., we normalized sequencing depth to 3000 sequence reads per sample) (34). Per published methods, we retained only “non-rare” taxa (i.e., those that were present in ≥25% of samples) (35).
We conducted a rigorous literature review to identify genus- and species-level SCFA-producing taxa. (13, 17, 36–38). We detected the following SCFA-producing taxa in the stool of our study participants: Akkermansia, Alistipes, Anaerostipes, Bacteroides, Bifidobacterium, two members of the Clostridium genus (Clostridium sensu stricto cluster 1 and Clostridium innocuum), Dialister, three members of the Eubacterium genus (E. eligens, E. hallii, and E. ventriosum), Faecalibacterium, Intestinimonas, Lachnospira, two members of the Prevotella genus (Prevotella clusters 7 and 9), Roseburia, four members of the Ruminococcus genus (Ruminococcus gnavus, Ruminococcus torques, and Ruminococcus clusters 1 and 2), Sutterella, Streptococcus, and Veillonella. We removed three taxa that were present in <25% of samples (Sutterella and Prevotella clusters 7 and 9).
SCFA analysis
We analyzed total and specific fecal SCFAs using GC-MS (Agilent 7820), as previously described. Values were expressed in micromoles per gram (39).
CGM
Study participants wore a blinded CGM (Freestyle Libre Pro; Abbott Diabetes Care Inc) for 2 weeks following each measurement visit. We computed percentage of time in target glucose range (70–180 mg/dL), percentage of time above target range (i.e., hyperglycemia, >180 mg/dL), and percentage of time below range (i.e., clinical hypoglycemia, 54–69 mg/dL) for use as outcome variables in the present analysis. We did not use CGM values in the range of clinically serious hypoglycemia (<54 mg/dL) (25) because factors related to insulin dosing are likely to cause clinically serious hypoglycemic events, and because the amount of time in clinical hypoglycemia was limited (1.8%; IQR, 0.73%; 4.1%). We included observations with ≥1 wk of CGM data (i.e., ≥168 h regardless of gaps in readings) based on a recent consensus statement (25).
DXA
We quantified body fat percentage and percentage lean mass using a DXA scan (UNC: GE Lunar iDXA, GE Medical Systems Ultrasound & Primary Care Diagnostics; Stanford: Horizon Model A, Hologic).
HbA1c
We collected venous blood samples in person prior to COVID-19 and sent them to the Northwest Lipid Metabolism and Diabetes Research Laboratories at the University of Washington School of Medicine for determination of HbA1c. During COVID-19, participants obtained capillary blood samples at home using provided kits (BIO-RAD Hemoglobin Capillary Collection System for HbA1c Testing) with live instruction from study staff via Zoom. Participants mailed home kits to the Diabetes Diagnostic Lab at the University of Missouri, Columbia for determination of HbA1c. In a prior study of 122 participants with T1D or type 2 diabetes at 22 clinical centers, venous HbA1c was highly correlated with capillary HbA1c (R2 = 0.993), and 96.7% of measurements differed by ≤0.2% (2.2 mmol/mol) (40).
Anthropometrics
Weight (to the nearest 0.1 kilograms) was measured in person pre-COVID-19 at each measurement visit following standard procedures. During COVID-19, participants measured weights at home using BodyTrace Bluetooth scales, which were provided at study enrollment for voluntary weight tracking. We used baseline height measurements (to the nearest 0.1 cm) and the weight at each measurement visit to calculate BMI (kg/m²).
Demographic and clinical covariates
Participants self-reported demographic data including age, gender, race and ethnicity, and insulin regimen (twice daily, 3 times daily, >3 times daily injections, or insulin pump) using standardized questionnaires. Self-reported race categories included African American, American Indian/Alaska Native, Asian, Native Hawaiian/Other Pacific Islander, Other race, or white. Ethnicity was classified as Spanish/Hispanic/Latino or not. Given sample size limitations, we collapsed race and ethnicity into a single indicator variable: Other race and ethnicity or non-Hispanic White. We provide information about raw race and ethnicity in all relevant table legends. We imputed 3 missing observations for insulin regimen forwards or backwards from the closest visit in time. Insulin regimen was dichotomized as insulin pump or injections due to sample size limitations.
Dietary intake
Trained UNC NIH/National Institute of Diabetes and Digestive and Kidney Diseases Nutrition Obesity Research Center (NORC) staff administered 24-h dietary recalls via telephone at each measurement visit using a multipass method (41, 42). Staff collected recalls following a standard script on nonconsecutive days, ideally including 1 weekday and 1 weekend day. If 2 dietary recalls were available for a participant at a given measurement visit, the nutrient values were averaged across the 2 d. We removed unreliable dietary recalls according to the interviewer or that participants described as being “a lot more” or “a lot less” than they usually ate from analysis. NORC used the Nutrition Data System for Research (NDSR, version 2019; Nutrition Coordinating Center, University of Minnesota) (43) to derive nutrients associated with recalled foods and beverages.
Design covariates
We constructed an indicator variable denoting whether each visit was completed during COVID-19; the duration (months) of each diet period given increased variability during COVID-19; the diet period (1, 2, or 3) to account for a likely greater impact of the intervention in the first diet period; diet assignment; and study site.
Statistical analysis
We compared the baseline demographic and clinical characteristics of ACT1ON study participants included and excluded from the analysis to assess representativeness. We conducted sensitivity analyses for outlier observations that were ≥2 SDs from the mean by rerunning the models without the outlier. If the outlier had undue influence on the results, the outlying value was truncated (winsorized) to 10% above or below than the next highest or lowest absolute value (44). We chose this process because even when an outlier was determined not to be the result of measurement error, we did not want a single value to have undue influence on the results. We winsorized outlier values for fecal butyrate and the normalized abundance of Anaerostipes. An additional sensitivity analysis tested whether exclusion of 2 outlier observations for fecal acetate influenced results. They did not, so fecal acetate outliers were not winsorized.
Effect size and power
After correction for multiple comparisons, we were powered to detect an R² of 0.07 with 80% power and an R² of 0.10 with 90% power given a sample size of n = 101 (the sample size for all available HbA1c or weight data when SCFA data were also available). Using the sample size of n = 58 for available DXA data (similar to the sample size for pre-COVID-19 data), we were powered to detect an R² of 0.13 with 80% power and an R² of 0.16 with 90% power. The magnitude of these effect sizes is smaller than those found in prior studies of the intestinal microbiota and adiposity (Spearman ρ = 0.28–0.6) with smaller sample sizes than ours (n = 30–39), suggesting that we were powered to detect observable effects (45, 46).
Modeled analysis
We fit separate generalized estimating equation (GEE) models predicting outcomes (percentage of time in target glucose range, percentage of time above range, percentage of time below range, body fat percentage, percentage lean mass, BMI, and HbA1c) from each exposure variable [abundance of each SCFA-producing taxon, fecal SCFA (butyrate, propionate, acetate, and total) concentrations, and intestinal microbial diversity (number of unique taxa per sample)] using data from the 4 measurement timepoints (time 0 and roughly at 3, 6, and 9 mo).
We elected to use GEEs because they account for nonindependence of repeated measures. Although linear mixed models also have this capability, GEEs can better handle zero-inflated gut microbiome data (47). Because the adult fecal microbiome has high interindividual variability and temporal stability (48–50), substantial changes to diet are necessary to observe changes in the fecal microbiome; therefore, because ACT1ON was a free-living diet study, we designed this analysis as a repeated measures interindividual comparison rather than an intraindividual longitudinal analysis of how changes in the gut microbiome predict changes in glycemia and adiposity. We computed standardized β coefficients by dividing each β estimate from GEE models by its SE to allow for comparability across estimates and report these unitless standardized coefficients in the figures (51).
We repeated all modeled analyses restricted to pre-COVID-19 data, given changes in the mode of intervention delivery, in the assessment methods for the primary ACT1ON parent study outcomes of glycemia and weight, and reduced study retention and adherence to diet assignments during COVID-19. Analysis of the larger parent study outcomes revealed that the statistically significant ∼5-lb (∼2.27-kg) mean weight loss at the end of the first diet period pre-COVID-19 was attenuated, although not to nonsignificance, when including participants who completed the first diet period during COVID-19 (D Igudesman, J Crandell, KD Corbin, DP Zaharieva, A Addala, JM Thomas, A Casu, MS Kirkman, T Pokaprakarn, MC Riddell, K Burger, RE Pratley, MR Kosorok, DM Maahs, EJ Mayer-Davis, unpublished results, 2022).
Model 1 was unadjusted but accounted for within-subject correlations of repeated measures. To maximize utility of this pilot study sample, we used stepdown approaches to evaluate which nondesign potential confounders (age, gender, race and ethnicity, BMI, insulin regimen, and dietary fiber intake) to retain in statistical models (52). We retained age, gender, race and ethnicity, diabetes duration, and fiber intake in Model 2 because these were the informative variables (P < 0.1). Model 2 also included the design covariates of diet assignment, study site, the COVID-19 indicator, diet duration, and diet period. We did not adjust Model 2 for the COVID-19 indicator or diet duration when restricting to pre-COVID-19 visits.
Given the hypothesis-generating nature of this study and our conservative method of correction for multiple comparisons, we considered Bonferroni-corrected (53) P values to be statistically significant at an α level <0.1. We estimated power calculations with R software version 4.1.1. We conducted all other analyses using SAS version 9.4.
Human participants
This study was approved by the UNC and Stanford Institutional Review Boards (IRBs) and study participation did not begin until participants signed informed consent. The procedures followed were in accordance with the ethical standards of the UNC and Stanford IRBs. Participants signed an informed consent form stating that their data would be used anonymously in future publications. Participants were compensated ≤ 780.00 for taking part in the parent ACT1ON study. Additionally, participants received
780.00 for taking part in the parent ACT1ON study. Additionally, participants received  30,
30,  50,
50,  70, and
70, and  90 for voluntarily providing a stool sample at the baseline visit and measurement visits 2, 3, and 4, respectively.
90 for voluntarily providing a stool sample at the baseline visit and measurement visits 2, 3, and 4, respectively.
Study physicians reviewed all laboratory findings in a timely fashion. Research staff received formalized guidance (“alert values”) for when to proactively contact the study physician. The interventionists (Registered Dietitians) and study physicians communicated regularly (verbally and in writing per protocol) regarding blood glucose values so that any changes in medical management were made in a timely fashion. Study coordinators immediately reported any adverse events to the study physician at the local site and to the study investigators, who kept a log of adverse events and serious adverse events.
All collected data were anonymized by using a participant identification number, which was stored in a secure location and could only be linked to participant identifiers by select study staff. The study project manager oversaw data security. Potential benefits of participation included (but did not guarantee) improved blood glucose management and weight loss. Participants were provided with ample time to review and ask questions about the informed consent form. Study coordinators administered an informed consent comprehension checklist to each study participant as part of enrollment.
Results
At enrollment, the 45 participants included in the present analysis had a mean age of 25.4 ± 3.3 y, a median BMI of 30.8 (IQR: quartile 1, 28.2; quartile 3, 34.0), mean HbA1c of 7.8 ± 1.4%, and a mean diabetes duration of 15.1 ± 6.4 y (Table 1). Over two-thirds of included participants identified with a female gender (68.9%), 62.8% were enrolled at UNC, and 58.1% used insulin pump therapy for their diabetes management. ACT1ON study participants who were included (n = 45) or excluded (n = 23) from analysis did not differ with respect to age, gender, enrollment across study sites, BMI, insulin pump use, dietary fiber intake, or any of the study outcomes related to glycemia or adiposity at the baseline visit (all P > 0.05). ACT1ON participants included in the analytic sample had a longer diabetes duration (15.1 ± 6.4 y) and were less racially and ethnically diverse (75.6% had a non-Hispanic white race and ethnicity) than those excluded (diabetes duration 11.8 ± 5.7 y, P = 0.03; 47.8% had a non-Hispanic white race and ethnicity, P = 0.02).
TABLE 1.
Baseline characteristics among ACT1ON study participants included or excluded from the analytic sample (total n = 68)1
| Included (n = 45) | Excluded (n = 23) | P value | |
|---|---|---|---|
| Age, mean ± SD, y | 25.4 ± 3.3 | 25.6 ± 2.8 | 0.73 |
| Female gender, n (%) | 31 (68.9) | 18 (78.3) | 0.42 |
| Non-Hispanic White race and ethnicity,2n (%) | 34 (75.6) | 11 (47.8) | 0.02 |
| UNC site, n (%) | 27 (62.8) | 12 (48.0) | 0.23 |
| Diabetes duration, mean ± SD, y | 15.1 ± 6.4 | 11.8 ± 5.7 | 0.03 |
| Insulin pump use, n (%) | 25 (58.1) | 15 (60.0) | 0.88 |
| BMI, median (Q1, Q3) | 30.8 (28.2, 34.0) | 29.7 (27.1, 33.2) | 0.33 |
| HbA1c, mean ± SD, % | 7.8 ± 1.4 | 8.0 ± 1.3 | 0.52 |
| Body fat percentage, median (Q1, Q3) | 41.6 (34.7, 45.1) | 39.2 (32.6, 43.8) | 0.46 |
| Percentage lean mass, median (Q1, Q3) | 55.2 (52.3, 62.1) | 57.6 (54.0, 64.2) | 0.40 |
| Included (n = 39) 3 | Excluded (n = 19) 3 | ||
| Total fiber intake, grams | 12.9 (9.3, 22.1) | 14.2 (10.5, 19.3) | 0.76 |
| Included (n = 38) 3 | Excluded (n = 19) 3 | ||
| Time in range (70–180 mg/dL), mean ± SD, % | 52.6 ± 22.0 | 44.1 ± 20.5 | 0.16 |
| Time above range (>180 mg/dL), mean ± SD, % | 40.2 ± 25.2 | 50.4 ± 22.8 | 0.14 |
| Time below range (54–70 mg/dL), median (Q1, Q3), % | 3.7 (1.7, 7.4) | 3.3 (1.0, 5.5) | 0.39 |
Group differences in continuous variables were tested using independent t-tests for normally distributed variables or Mann–Whitney U (Wilcoxon rank sum test) for nonnormally distributed continuous variables. Group differences in categorical variables were tested using the χ2 test for independence or the Fisher exact test if any cell sizes were less than n = 5. Note: Body composition measures were only available pre-COVID-19 due to discontinuation of DXA after the transition to a virtual protocol during COVID-19. ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network; HbA1c, glycated hemoglobin; Q1, quartile 1; Q3, quartile 3; UNC, University of North Carolina at Chapel Hill.
Race and ethnicity were collapsed into non-Hispanic white and Other due to sample size limitations. To avoid the possibility of participant identification, we express frequencies with <3 individuals as “n <3” rather than disclosing the absolute number: of those included in the analytic sample, n = 4 (8.9%) identified as African American, n < 3 identified as Asian, n < 3 identified with >1 race; n = 36 (80.0%) identified as White; n = 6 (13.3%) identified as Hispanic or Latino. Of those excluded from the analytic sample, n <3 identified as African American, n <3 identified as Native Hawaiian/Other Pacific Islander, n <3 identified as Asian, n <3 identified as Other race, n = 4 (17.4%) identified with >1 race; n = 13 (56.5%) identified as White; n = 5 (21.7%) identified as Hispanic or Latino.
Six and 7 participants included in the analysis were missing data for diet and continuous glucose monitoring, respectively, at the baseline visit.
Mean or median values for SCFAs and the outcomes of glycemia and adiposity across all timepoints of the ACT1ON gut microbiome pilot study are displayed in Table 2. Collapsing across visits, the mean ± SD values for fecal SCFAs were 54.7 ± 20.3 μmol/g for acetate, 16.7 ± 6.6 μmol/g for propionate, and 6.9 ± 1.9 μmol/g for butyrate.
TABLE 2.
SCFA and study outcome data over time for participants included in the analytic sample1
| Measurement visit 2 | |||||
|---|---|---|---|---|---|
| SCFA, μmol/g | Baseline | All data | Pre-COVID-19 | Measurement visit 3 | Measurement visit 4 |
| n | 39 | 25 | 19 | 15 | 22 |
| Acetate | 52.9 ± 19.3 | 58.0 ± 24.1 | 56.4 ± 26.5 | 50.8 ± 16.9 | 56.9 ± 20.1 |
| Butyrate | 7.0 ± 1.8 | 6.6 ± 1.9 | 6.5 ± 2.1 | 7.4 ± 2.2 | 6.6 ± 2.0 |
| Propionate | 16.4 ± 6.1 | 16.6 ± 7.8 | 14.8 ± 7.5 | 17.7 ± 7.7 | 16.8 ± 5.6 |
| Total SCFA | 76.3 ± 23.6 | 81.1 ± 29.7 | 77.6 ± 32.1 | 75.9 ± 16.0 | 80.3 ± 24.4 |
| CGM metrics | |||||
| n | 26 | 19 | 14 | 13 | 19 |
| Time in range, mean ± SD, % | 52.4 ± 20.5 | 60.1 ± 19.2 | 61.1 ± 20.9 | 59.2 ± 17.9 | 53.6 ± 18.9 |
| Time above range (>180 mg/dL), mean ± SD, % | 40.8 ± 23.0 | 32.0 ± 21.8 | 32.1 ± 23.8 | 28.3 ± 23.0 | 36.9 ± 24.5 |
| Time below range (54–70 mg/dL), median (Q1, Q3), % | 3.7 (1.9, 7.2) | 3.6 (2.2, 10.7) | 3.6 (2.2, 6.2) | 7.0 (4.8, 10.7) | 4.0 (1.5, 9.1) |
| DXA metrics | Baseline | Measurement visit 2 | Measurement visit 3 | Measurement visit 4 | |
| n | 39 | 19 | 0 | 0 | |
| Body fat percentage, median (Q1, Q3) | 42.8 (34.3, 45.2) | 36.6 (31.0, 43.9) | — | — | |
| Percentage lean mass, median (Q1, Q3) | 54.7 (51.8, 62.8) | 60.5 (54.5, 65.5) | — | — | |
| HbA1c, % (mmol/mol) | |||||
| n | 39 | 25 | 19 | 15 | 22 |
| Mean ± SD | 8.0 ± 1.4% (64 ± 15. mmol/mol) | 7.3 ± 1.2 (56 ± 13.1) | 7.2 ± 1.2 (55 ± 13.1) | 7.1 ± 1.2 (54 ± 13.1) | 7.4 ± 1.3 (57 ± 14.2) |
| BMI, kg/m² | |||||
| n | 39 | 25 | 19 | 15 | 22 |
| Median (Q1, Q3) | 30.3 (28.0, 34.0) | 28.7 (27.3, 33.1) | 28.6 (27.0, 32.4) | 29.3 (27.6, 34.1) | 28.9 (26.8, 35.1) |
Data are from participants who had both SCFA and outcome data at the corresponding timepoint. Thirty-seven participants had CGM and SCFA data at 77 visits (32 participants with 40 visits pre-COVID-19); 45 participants had DXA and SCFA data at 58 visits pre-COVID-19; 45 participants had BMI and SCFA data at 101 visits (43 participants with 58 visits pre-COVID-19); 45 participants had HbA1c and SCFA data at 101 visits (40 participants with 54 visits pre-COVID-19). Stool collection was added to Measurement visits 3 and 4 only after COVID-19 began, and DXA was removed from the protocol when COVID-19 began; therefore, no participants have both DXA and gut microbiome data at Measurement visits 3 and 4. Note that baseline sample sizes may be smaller in Table 2 than in Table 1 because fecal samples were not available for all included participants at baseline. CGM, continuous glucose monitoring; HbA1c, glycated hemoglobin; Q1, quartile 1; Q3, quartile 3.
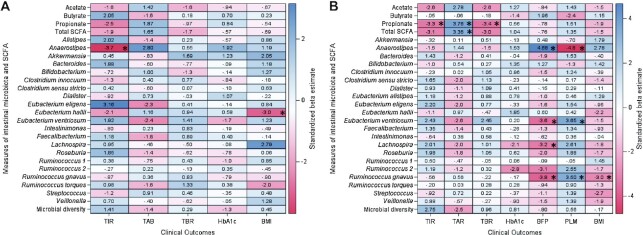
Using all available data, the only associations among SCFA-producing microbes, fecal SCFA, and α-diversity with glycemia and adiposity that remained statistically significant after adjustment for potential confounders and Bonferroni correction were: 1) a negative association of the abundance ofEubacterium hallii with BMI (unstandardized β estimate −0.70 [95% CI:−1.2,−0.24] P=0.07); and 2) a negative association of the abundance ofAnaerostipes with percentage of time in target glucose range (unstandardized β estimate −8.9% [95% CI −13.1,−4.0;P=0.01).
Using pre-COVID-19 data, 4 covariate-adjusted and Bonferroni-corrected associations remained statistically significant for outcomes of glycemia, and 8 associations remained statistically significant for outcomes of adiposity. All statistically significant associations for glycemia used CGM data and had the following unstandardized β estimates: a 1 SD increase in fecal propionate or in total fecal SCFA was associated with a 11.7% (95% CI 5.6, 17.7) and an 11.7% (95% CI 4.9, 18.5) increase in the percentage of time spent in hyperglycemia (>180 mg/dL), respectively; and a 1 SD increase in fecal propionate was associated with a 1.6% (95% CI 2.6, 0.69) reduction in the percentage of time spent in clinical hypoglycemia (54–69 mg/dL) and a 9.3% (95% CI 14.8, 3.8) reduction in the percentage of time spent in target glucose range (70–180 mg/dL), respectively. The increased abundance of 3 SCFA-producing intestinal taxa (Ruminococcus gnavus, Eubacterium ventriosum, and Lachnospira) was associated with reduced body fat percentage; of these, increased Ruminococcus gnavus and Eubacterium ventriosum were also associated with increased percentage lean mass, and increased Ruminococcus gnavus was associated with reduced BMI. The normalized abundance of the SCFA producer Anaerostipes was associated with increased BFP, and with reduced percentage lean mass. The largest unstandardized β estimates corresponding to a 1 unit increase in the normalized abundance of the intestinal microbiota were a 1.7 (95% CI 0.92, 2.5) percentage point increase in body fat percentage with increasing Anaerostipes and a −1.6 (95% CI −2.4, −0.78) percentage point reduction in body fat percentage with increasing Ruminococcus gnavus. Heatmaps with standardized β coefficients for covariate-adjusted and Bonferroni-corrected estimates are shown in Figure 1A (all data) and Figure 1B (pre-COVID-19 data). Crude unadjusted estimates are not presented due to major confounding by dietary fiber intake, whose inclusion in the adjusted models changed some point estimates by ∼5-fold or more.
FIGURE 1.
Heatmaps with standardized β estimates from covariate-adjusted GEE models for associations of SCFA-producing microbes, fecal SCFAs, and α-diversity with glycemia and adiposity. (A) Results using all available data; (B) Results restricted to pre-COVID-19 data.P values were Bonferroni corrected and statistically significant at P < 0.1 (denoted by asterisks). Units are micromoles per gram for SCFAs, normalized abundance for gen us-level intestinal microbes, and the number of unique taxa from the phylum to the genus level for intestinal microbial diversity. DXA was only performed prior to COVID-19. BFP, body fat percentage; GEE, generalized estimating equation; HbA1c, glycated hemoglobin; PLM, percentage lean mass; TAB, percentage of time above target glucose range (>180 mg/dL); TBR, percentage of time below target glucose range (54–69 mg/dL); TIR, percentage of time in target glucose range (70–180 mg/dL).
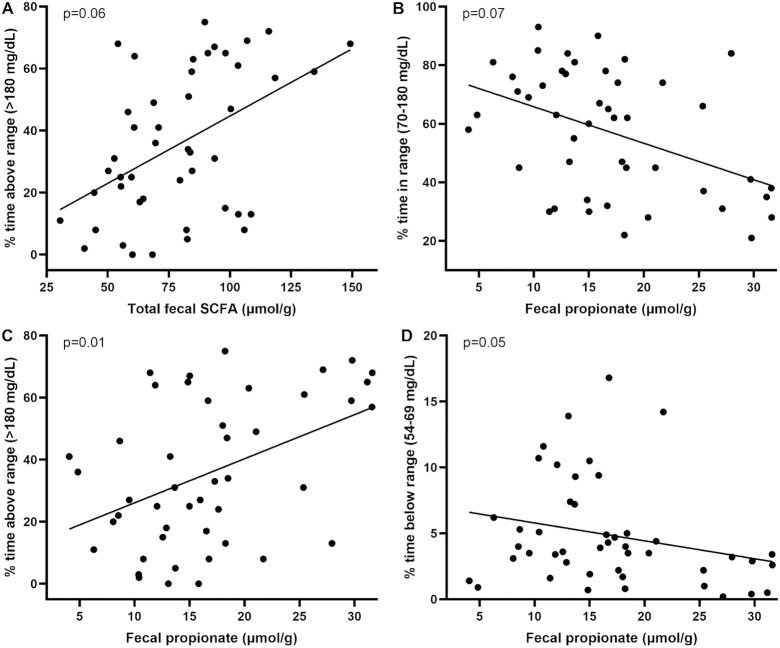
Scatterplots of raw data for statistically significant associations of fecal SCFA with measures of glycemia are shown in Figure 2.
FIGURE 2.
Scatterplots of raw pre-COVID-19 data for associations of fecal SCFAs with measures of glycemia that were statistically significant after adjustment for potential confounders and correction for multiple hypothesis testing. (A, B) Increased total fecal SCFAs and fecal propionate were associated with reduced percentage of time in target glucose range. (C, D) Increased fecal propionate was associated with reduced percentage of time above target glucose range (C) and percentage of time in clinical hypoglycemia (D).
Discussion
To our knowledge, our hypothesis-generating study is the first to rigorously assess advanced metrics of glycemia and adiposity in association with measures of the intestinal microbiota and SCFAs in individuals with longstanding T1D. Whereas we surmised from preclinical evidence and from limited evidence in individuals without T1D that an increased SCFA-producing capacity of the intestinal microbiota is associated with improved glycemia but with increased adiposity, the results of our covariate-adjusted models suggested the opposite: increased fecal propionate, total fecal SCFAs and the abundance of the butyrogenic genus Anaerostipes were associated with more percentage of time in hyperglycemia and less percentage of time in target glucose range, whereas the abundance of several SCFA-producing enteric microbial genera that are known SCFA producers was inversely associated with percentage body fat or BMI or positively with percentage lean mass. There was one exception: the abundance of the SCFA producer Anaerostipes was associated with increased adiposity.
Our results suggesting that propionate may have a net negative impact on glycemia are unanticipated, given evidence from preclinical models and humans with metabolic syndrome that SCFAs, including propionate, can improve blood glucose homeostasis and help to resolve inflammation (54–59). Of note, although propionate was associated with a modest reduction in percentage of time in hypoglycemia, this was likely due to increased percentage of time in hyperglycemia. Therefore, we do not consider propionate to be protective from hypoglycemia. One preclinical study found that provision of propionate to healthy human-derived hepatocytes activated 5′-activated AMP kinase, which downregulated the expression of gluconeogenic enzymes and reduced gluconeogenesis, suggesting a potential benefit to glycemia (60). In a rat model, propionate decreased hepatic gluconeogenesis purportedly through stimulation of intestinal gluconeogenesis (61). However, several glucoregulatory mechanisms are disrupted in T1D, including deficient insulin, glucagon, and amylin production (62). This raises the possibility that propionate's ability to regulate blood glucose is blunted in T1D.
The observed estimates for the association of propionate with hyperglycemia are clinically relevant if it is possible to manipulate the diet or the SCFA-producing capacity of the intestinal microbiota such that propionate is reduced by 1–2 SDs (i.e., a ∼10–20% reduction in hyperglycemia). However, these associational data must be interpreted with caution and require confirmation in proof-of-mechanism studies to assess the direct biological effect of propionate administration on blood glucose excursions in individuals with T1D. Of note, calcium propionate is sometimes added to foods as a mold inhibitor (e.g., in certain bread products) (63) but is not calculated by the NDSR software, so we were unable to determine its dietary contribution. We assume that the major source of propionate is microbe-derived, although this requires further investigation (64).
Our results suggest that SCFAs may function differently in people with T1D than in those with metabolic syndrome. Prior use of fecal microbiota transplantation (FMT), or transfer of healthy donor stool to individuals with metabolic syndrome, suggests the potential to improve insulin sensitivity or energy expenditure through FMT, presumably through the action of the intestinal microbiota or its effector metabolites including SCFAs (20, 55, 57, 65). It is surprising that we did not find associations of butyrate (57, 66) with measures of glycemia or adiposity in our study, given its prior associations with metabolic benefits in preclinical models and humans with obesity or type 2 diabetes (67–69). Butyrate might indeed function uniquely in people with T1D: a recent randomized crossover study of 30 participants with longstanding T1D aged 18–65 y (BMI 18.5–25.0), did not find a benefit of daily oral supplementation with 4 g butyrate for 1 mo on immune or glycemic parameters (70). In people with non-alcoholic fatty liver disease, the abundance of butyrogenic Anaerostipes was associated inversely with fasting blood glucose (PMID: 31177662); however, in line with our results, Anaerostipes was associated positively with blood glucose among children with T1D from China (PMID: 34040330). Longer-term studies, including those that enroll adults with T1D and overweight or obesity, are required to fully understand the influence of SCFAs in the clinical management of T1D.
Although our results should be interpreted with caution, they suggest that SCFA-producing gut microbes may uniquely protect against adiposity in people with T1D. However, these observational results should be interpreted with caution, especially given that the SCFA producer Anaerostipes was associated with increased adiposity in our study. Contrary to our results, a preclinical study in mice and a clinical study in people without T1D both found an increased abundance of Ruminococcus gnavus in a group with overweight or obesity compared with their lean counterparts, which the authors attributed to the microbe's SCFA-producing (i.e., energy-harvesting) abilities (71, 72). In another preclinical mouse model of malnutrition, Ruminococcus gnavus ameliorated an impaired growth phenotype, potentially owing to its production of acylcarnitines—an energy source—from fermentation of branched-chain amino acids (73). Eubacterium ventriosum was similarly elevated in Japanese individuals with obesity compared with those who were lean (74), and Lachnospira decreased with decreasing body weight during a 3-mo weight loss intervention (75). Nonetheless, a well-established function of SCFAs is their ability to stimulate satiety hormone secretion (e.g., peptide YY and GLP-1) from intestinal epithelial L-cells, which signal to central homeostatic satiety regions (16, 76–78). It is possible that in people with T1D, the SCFA-stimulated increase in satiety hormones predominates over energy-accumulating pathways. Ultimately, novel strategies are needed to determine the net ecosystem-wide effects of the gut microbiota and its metabolites on host physiology and energy balance, particularly in people with T1D.
The results of our study should be interpreted in the context of its limitations. Given the hypothesis-generating nature of this study, its observational design, our conservative method of correction for multiple comparisons, modest sample size, and the small effect sizes that characterize associations with the intestinal microbiota, we cannot rule out the possibility of type I or type II error (79) or reverse causality. We focused on the SCFA-producing capacity of the intestinal microbiota, but other metabolic capabilities of the gut microbes we studied might have contributed to their inverse associations with adiposity—which can be investigated using whole-genome sequencing in future research.
We included individuals if they had not taken antibiotics in the prior month, which is long enough for many, but not all, intestinal microbes to reconstitute the intestinal tract (80). Thus, we cannot exclude the possibility that antibiotic use >1 mo prior to stool collection influenced the gut microbial composition such that it was not “usual.” Generalizability of our findings to individuals with T1D who do not have overweight or obesity, who are racially or ethnically more diverse than our study participants, who are middle-aged or older adults, or who were recently diagnosed with T1D could be limited. We lacked sufficient statistical power to adjust for race and ethnicity in their raw form and therefore used a combined binary specification of these variables.
The greater number of statistically significant findings when using pre-COVID-19 data compared with all available data might be due to changes in the composition of the intestinal microbiota and SCFAs during COVID-19 due to reduced exposure to the external environment, changes to diet, and other lifestyle factors such as physical activity, use of antibiotics, inflammatory responses to the COVID-19 virus or other infections, and changes to hygiene practices (81). It is also possible that participants’ management of glycemia and weight changed during COVID-19 (82), or that there was variability in study dropout according to variability in success with managing glycemia or adiposity. We used fecal SCFAs as a proxy for production, which is common in other studies but is not a direct measure (83, 84). SCFAs are absorbed across the intestinal epithelium with high efficiency and therefore represent ∼5–10% of total SCFAs produced (85). Future in vivo tracer studies can directly measure SCFA production by the intestinal microbiota (86).
Our study also includes several strengths. We addressed a substantial gap in the literature by assessing the links among the intestinal microbiota, SCFAs, and clinical outcomes in people with longstanding T1D. We used novel and rigorous methods of analyzing the intestinal microbiota in association with advanced metrics of glycemia and adiposity, and we carefully adjusted models for potential confounding and design covariates. All statistically significant associations of the intestinal microbiota and SCFAs with glycemia were detected using CGM-based metrics, which highlights the utility of parsing hyper-, hypo-, and euglycemia from HbA1c—a 3-mo average that is less informative for day-to-day management (87). The majority of statistically significant associations with adiposity used DXA-based metrics—which measure body composition directly and better predict metabolic risk than BMI (88)—so the contribution of intestinal microbes and SCFAs to adiposity may be better approximated using DXA.
Given that overweight status and glycemia have not improved or have worsened in people with T1D in the United States over the past several decades (3, 4, 89, 90), our hypothesis-generating study assessed whether the intestinal microbiota and SCFAs warrant further research in the clinical management of T1D. We identified 4 candidate microbes whose abundance was inversely associated with adiposity; and 1 SCFA (i.e., propionate) whose abundance was associated with a potential harm to glycemia. Additionally, the increased abundance of butyrogenic Anaerostipes was associated with more hyperglycemia and a higher body fat percentage, so additional studies should examine this gut microbe in people with T1D and overweight or obesity specifically. These findings require confirmation in additional observational and mechanistic research, which could ultimately determine whether the intestinal microbiota or SCFAs are worthy of investigation in interventional trials to reduce the cardiometabolic risk factors of dysglycemia and adiposity in people with T1D.
Supplementary Material
ACKNOWLEDGEMENTS
The ACT1ON Study is indebted to the young adults whose participation made this study possible. Twenty-four-hour recalls were obtained by the UNC Nutrition Obesity Research Center staff, funded by the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH under Award Number 1DP3DK113358; Multiple Principle Investigators Mayer-Davis, Maahs, Pratley. Analysis of SCFAs was performed by Yifei Yang and Dr Yunjia Lai under the supervision of Dr Kun Lu at the University of North Carolina at Chapel Hill. We thank Dr Anthony Fodor for his provision of technical advice related to analysis of the intestinal microbiota.
The authors’ responsibilities were as follows—DI, IMC, and EJM-D: designed research; DI, IMC, EJM-D, JMT, DMM, DPZ, FM, KDC, and REP: conducted research; IMC: provided essential materials; DI: had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis; DI, IMC, MRK, JC, and EJM-D: designed the analyses; DI: conducted the analyses with the oversight of IMC, JC, and MRK; DI: wrote the initial manuscript; and all authors: provided critical review and read and approved the final manuscript.
Notes
This study was supported by the NIH/National Institute of Diabetes and Digestive and Kidney Diseases (1DP3DK113358-01, PNC DK056350, and P30 DK034987). DPZ is supported by an International Society for Pediatric and Adolescent Diabetes-Juvenile Diabetes Research Foundation (JDRF) Research Fellowship and by the Leona M. and Harry B. Charitable Trust. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author disclosures: DMM has had research support from the NIH, JDRF, National Science Foundation, and the Helmsley Charitable Trust, and his institution has had research support from Medtronic, Dexcom, Insulet, Bigfoot Biomedical, Tandem, and Roche; and has consulted for Abbott, Aditxt, the Helmsley Charitable Trust, Lifescan, Mannkind, Sanofi, Novo Nordisk, Eli Lilly, Medtronic, Insulet, Dompe, and Biospex. EJM-D has consulted for Helmsley Charitable Trust. CMB reports: Shire (grant recipient, Scientific Advisory Board member); Idorsia (consultant); Lundbeckfonden (grant recipient); Pearson (author, royalty recipient); Equip Health Inc (Clinical Advisory Board). IMC is a consultant for Vivilex; former consultant for Salix Pharmaceuticals and receives funding from NIH (R21-AI125800–01-02) and National Institute of Mental Health (R01-MN105684–03).
DI was supported by the Global Cardiometabolic Disease training grant (National Heart, Lung, and Blood Institute of the NIH) awarded to the Department of Nutrition at the University of North Carolina at Chapel Hill under Award Number HL129969. All other authors report no conflicts of interest.
Supplemental Figure 1 is available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/cdn/.
Abbreviations used: ACT1ON, Advancing Care for Type 1 Diabetes and Obesity Network; CGM, continuous glucose monitoring; DADA2, Divisive Amplicon Denoising Algorithm; FMT, fecal microbiota transplantation; GEE, generalized estimating equation; GLP-1, glucagon-like peptide 1; GPR, G-protein coupled receptor; HbA1c, glycated hemoglobin; IRB, Institutional Review Board; NDSR, Nutrition Data System for Research; NORC, Nutrition Obesity Research Center; QIIME2, Quantitative Insights Into Microbial Ecology 2; rRNA, ribosomal RNA; SMART, Sequential Multiple Assignment Randomized Trial; T1D, type 1 diabetes; UNC, University of North Carolina at Chapel Hill.
Contributor Information
Daria Igudesman, Email: daria.igudesman@adventhealth.com, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Jamie Crandell, Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Karen D Corbin, AdventHealth Translational Research Institute, Orlando, FL, USA.
Franklin Muntis, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Dessi P Zaharieva, Department of Pediatrics, Division of Endocrinology, Stanford University, Stanford, CA, USA.
Anna Casu, AdventHealth Translational Research Institute, Orlando, FL, USA.
Joan M Thomas, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Cynthia M Bulik, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Psychiatry, University of North Carolina at Chapel Hill, Chapel Hill, CA, USA; Department of Medical Epidemiology and Biostatistics, Karolinska Institutet, Stockholm, Sweden.
Ian M Carroll, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
Brian W Pence, Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, CA, USA.
Richard E Pratley, AdventHealth Translational Research Institute, Orlando, FL, USA.
Michael R Kosorok, Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA.
David M Maahs, Department of Pediatrics, Division of Endocrinology, Stanford University, Stanford, CA, USA.
Elizabeth J Mayer-Davis, Department of Nutrition, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, CA, USA.
Data Availability
Data described in the manuscript, code book, and analytic code will be made available upon request pending application and approval by the ACT1ON Publications and Presentations Committee. If necessary, a data use or other data sharing agreement will be established.
References
- 1. De Ferranti SD, De Boer IH, Fonseca Vet al. . Type 1 diabetes mellitus and cardiovascular disease: a scientific statement from the American Heart Association and American Diabetes Association. Circulation. 2014;130(13):1110–30. [DOI] [PubMed] [Google Scholar]
- 2. Miller KM, Foster NC, Beck RWet al. . Current state of type 1 diabetes treatment in the US: updated data from the T1D Exchange clinic registry. Diabetes Care. 2015;38(6):971–8. [DOI] [PubMed] [Google Scholar]
- 3. Liu LL, Lawrence JM, Davis Cet al. . Prevalence of overweight and obesity in youth with diabetes in USA: the SEARCH for Diabetes in Youth study. Pediatr Diabetes. 2010;11(1):4–11. [DOI] [PubMed] [Google Scholar]
- 4. Malik FS, Sauder KA, Isom Set al. . Trends in glycemic control among youth and young adults with diabetes: the SEARCH for Diabetes in Youth study. Diabetes Care. 2022;45(2):285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wallace AS, Chang AR, Shin J-Iet al. . Obesity and chronic kidney disease in US adults with type 1 and type 2 diabetes mellitus. J Clin Endocrinol Metab. 2022;107(5):1247–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tauschmann M, Hovorka R. Technology in the management of type 1 diabetes mellitus—current status and future prospects. Nat Rev Endocrinol. 2018;14(8):464–75. [DOI] [PubMed] [Google Scholar]
- 7. Chambers ES, Morrison DJ, Frost G. Control of appetite and energy intake by SCFA: what are the potential underlying mechanisms?. Proc Nutr Soc. 2015;74(3):328–36. [DOI] [PubMed] [Google Scholar]
- 8. Murri M, Leiva I, Gomez-Zumaquero JMet al. . Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11(1):46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown CT, Davis-Richardson AG, Giongo Aet al. . Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS One. 2011;6(10):e25792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Goffau MC, Luopajärvi K, Knip Met al. . Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62(4):1238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577. [DOI] [PubMed] [Google Scholar]
- 12. De Groot PF, Belzer C, Aydin Öet al. . Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One. 2017;12(12):e0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Koh A, De Vadder F, Kovatcheva-Datchary P, Bäckhed F. From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites. Cell. 2016;165(6):1332–45. [DOI] [PubMed] [Google Scholar]
- 14. David LA, Maurice CF, Carmody RNet al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27(2):201–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Samuel BS, Shaito A, Motoike Tet al. . Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, GPR41. Proc Natl Acad Sci U S A. 2008;105(43):16767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kimura I, Ozawa K, Inoue Det al. . The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. de Groot P, Nikolic T, Pellegrini Set al. . Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut. 2021;70(1):92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. den Besten G, Bleeker A, Gerding Aet al. . Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 2015;64(7):2398–408. [DOI] [PubMed] [Google Scholar]
- 20. Kootte RS, Levin E, Salojärvi Jet al. . Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab. 2017;26(4):611–9.e6. [DOI] [PubMed] [Google Scholar]
- 21. Corbin K. D., Igudesman D., Addala A., Casu A., Crandell J., Kosorok M. R.et al. . Design of the Advancing Care for Type 1 Diabetes and Obesity Network energy metabolism and sequential multiple assignment randomized trial nutrition pilot studies: An integrated approach to develop weight management solutions for individuals with type 1 diabetes. Contemp Clin Trials. 2022;117:106765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kidwell KM. SMART designs in cancer research: past, present, and future. Clin Trials. 2014;11(4):445–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lavori PW, Dawson R. Dynamic treatment regimes: practical design considerations. Clin Trials. 2004;1(1):9–20. [DOI] [PubMed] [Google Scholar]
- 24. Murphy SA. An experimental design for the development of adaptive treatment strategies. Stat Med. 2005;24(10):1455–81. [DOI] [PubMed] [Google Scholar]
- 25. Battelino T, Danne T, Bergenstal RMet al. . Clinical targets for continuous glucose monitoring data interpretation: recommendations from the International Consensus on Time in Range. Diabetes Care. 2019;42(8):1593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Standiford DA, Morwessel N, Bishop FKet al. . Two-step recruitment process optimizes retention in FLEX clinical trial. Contemp Clin Trials Commun. 2018;12:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fouladi F, Glenny EM, Bulik-Sullivan ECet al. . Sequence variant analysis reveals poor cor in microbial taxonomic abundance between humans and mice after gnotobiotic transfer. ISME J. 2020;14(7):1809–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kleiman SC, Glenny EM, Bulik-Sullivan ECet al. . Daily changes in composition and diversity of the intestinal microbiota in patients with anorexia nervosa: a series of three cases. Eur Eat Disord Rev. 2017;25(5):423–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lin H, Peddada SD. Analysis of microbial compositions: a review of normalization and differential abundance analysis. NPJ Biofilms Microbiomes. 2020;6:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Van den Berge K, Perraudeau F, Soneson Cet al. . Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol. 2018;19(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Noble EE, Hsu TM, Jones RB, Fodor AA, Goran MI, Kanoski SE. Early-life sugar consumption affects the rat microbiome independently of obesity. J Nutr. 2017;147(1):20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quast C, Pruesse E, Yilmaz Pet al. . The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012;41(D1):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Willis AD. Rarefaction, alpha diversity, and statistics. Front Microbiol. 2019;10:2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Noble EE, Hsu TM, Jones RB, Fodor AA, Goran MI, Kanoski SE. Early-life sugar consumption affects the rat microbiome independently of obesity. J Nutr. 2017;147(1):20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y, Wang H, Howard AGet al. . Circulating short-chain fatty acids are positively associated with adiposity measures in Chinese adults. Nutrients. 2020;12(7):2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Remely M, Aumueller E, Merold Cet al. . Effects of short chain fatty acid producing bacteria on epigenetic regulation of FFAR3 in type 2 diabetes and obesity. Gene. 2014;537(1):85–92. [DOI] [PubMed] [Google Scholar]
- 38. Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12(2):304–14. [DOI] [PubMed] [Google Scholar]
- 39. van der Lelie D, Oka A, Taghavi Set al. . Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat Commun. 2021;12(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nathan DM, Krause-Steinrauf H, Braffett BHet al. . Comparison of central laboratory HbA1c measurements obtained from a capillary collection versus a standard venous whole blood collection in the GRADE and EDIC studies. PLoS One. 2021;16(11):e0257154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Posner BM, Smigelski C, Duggal A, Morgan J, Cobb J, Cupples L. Validation of two-dimensional models for estimation of portion size in nutrition research. J Am Diet Assoc. 1992;92((6):):738–41. [PubMed] [Google Scholar]
- 42. Beaton GH, Milner J, Corey Pet al. . Sources of variance in 24-hour dietary recall data: implications for nutrition study design and interpretation. Am J Clin Nutr. 1979;32(12):2546–59. [DOI] [PubMed] [Google Scholar]
- 43. University of Minnesota . Nutrition Data System for Research. 2014. [Google Scholar]
- 44. Tukey JW. The future of data analysis. Ann Math Stat. 1962;33(1):1–67. [Google Scholar]
- 45. Dewulf EM, Cani PD, Claus SPet al. . Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62(8):1112–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nadal I, Santacruz A, Marcos Aet al. . Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes. 2009;33(7):758–67. [DOI] [PubMed] [Google Scholar]
- 47. Weiss S, Xu ZZ, Peddada Set al. . Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Turnbaugh PJ, Hamady M, Yatsunenko Tet al. . A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Faith JJ, Guruge JL, Charbonneau Met al. . The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hanley JA, Negassa A, MDd E, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157(4):364–75. [DOI] [PubMed] [Google Scholar]
- 52. Mantel N. Why stepdown procedures in variable selection. Technometrics. 1970;12(3):621–5. [Google Scholar]
- 53. Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. BMJ. 1995;310(6973):170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Robertson MD, Bickerton AS, Dennis AL, Vidal H, Frayn KN. Insulin-sensitizing effects of dietary resistant starch and effects on skeletal muscle and adipose tissue metabolism. Am J Clin Nutr. 2005;82(3):559–67. [DOI] [PubMed] [Google Scholar]
- 55. De Groot P, Scheithauer T, Bakker GJet al. . Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut. 2020;69(3):502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Canfora EE, van der Beek CM, Jocken JWet al. . Colonic infusions of short-chain fatty acid mixtures promote energy metabolism in overweight/obese men: a randomized crossover trial. Sci Rep. 2017;7(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Vrieze A, Van Nood E, Holleman Fet al. . Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143(4):913–6.e7. [DOI] [PubMed] [Google Scholar]
- 58. Singh N, Gurav A, Sivaprakasam Set al. . Activation of GPR109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13(20):2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yoshida H, Ishii M, Akagawa M. Propionate suppresses hepatic gluconeogenesis via GPR43/AMPK signaling pathway. Arch Biochem Biophys. 2019;672:108057. [DOI] [PubMed] [Google Scholar]
- 61. De Vadder F, Kovatcheva-Datchary P, Goncalves Det al. . Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1-2):84–96. [DOI] [PubMed] [Google Scholar]
- 62. Gerich JE, Langlois M, Noacco C, Karam JH, Forsham PH. Lack of glucagon response to hypoglycemia in diabetes: evidence for an intrinsic pancreatic alpha cell defect. Science. 1973;182(4108):171–3. [DOI] [PubMed] [Google Scholar]
- 63. Darzi J, Frost GS, Robertson MD. Effects of a novel propionate-rich sourdough bread on appetite and food intake. Eur J Clin Nutr. 2012;66(7):789–94. [DOI] [PubMed] [Google Scholar]
- 64. Hoyles L, Snelling T, Umlai U-Ket al. . Microbiome–host systems interactions: protective effects of propionate upon the blood–brain barrier. Microbiome. 2018;6(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yu EW, Gao L, Stastka Pet al. . Fecal microbiota transplantation for the improvement of metabolism in obesity: the FMT-TRIM double-blind placebo-controlled pilot trial. PLoS Med. 2020;17(3):e1003051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Engels C, Ruscheweyh H-J, Beerenwinkel N, Lacroix C, Schwab C. The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol. 2016;7:713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Roshanravan N, Mahdavi R, Alizadeh Eet al. . Effect of butyrate and inulin supplementation on glycemic status, lipid profile and glucagon-like peptide 1 level in patients with type 2 diabetes: a randomized double-blind, placebo-controlled trial. Horm Metab Res. 2017;49(11):886–91. [DOI] [PubMed] [Google Scholar]
- 68. Jia L, Li D, Feng Net al. . Anti-diabetic effects of Clostridium butyricum CGMCC0313.1 through promoting the growth of gut butyrate-producing bacteria in type 2 diabetic mice. Sci Rep. 2017;7(1):7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lin HV, Frassetto A, Kowalik EJ Jret al. . Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS One. 2012;7(4):e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. De Groot PF, Nikolic T, Imangaliyev Set al. . Oral butyrate does not affect innate immunity and islet autoimmunity in individuals with longstanding type 1 diabetes: a randomised controlled trial. Diabetologia. 2020;63(3):597–610. [DOI] [PubMed] [Google Scholar]
- 71. Petriz BA, Castro AP, Almeida JAet al. . Exercise induction of gut microbiota modifications in obese, non-obese and hypertensive rats. BMC Genomics. 2014;15(1):511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Palmas V, Pisanu S, Madau Vet al. . Gut microbiota markers associated with obesity and overweight in Italian adults. Sci Rep. 2021;11(1):5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Blanton LV, Charbonneau MR, Salih Tet al. . Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science. 2016;351(6275):aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kasai C, Sugimoto K, Moritani Iet al. . Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterol. 2015;15(1):100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mayengbam S, Lambert JE, Parnell JAet al. . Impact of dietary fiber supplementation on modulating microbiota–host–metabolic axes in obesity. J Nutr Biochem. 2019;64:228–36. [DOI] [PubMed] [Google Scholar]
- 76. Tolhurst G, Heffron H, Lam YSet al. . Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein–coupled receptor FFAR2. Diabetes. 2012;61(2):364–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ceriello A, Novials A, Ortega Eet al. . Glucagon-like peptide 1 reduces endothelial dysfunction, inflammation, and oxidative stress induced by both hyperglycemia and hypoglycemia in type 1 diabetes. Diabetes Care. 2013;36(8):2346–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Frost G, Sleeth ML, Sahuri-arisoylu Met al. . The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5:3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ma Z, Li L, Gotelli NJ. Diversity-disease relationships and shared species analyses for human microbiome-associated diseases. ISME J. 2019;13(8):1911–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Suez J, Zmora N, Zilberman-Schapira Get al. . Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell. 2018;174(6):1406–23.e16. [DOI] [PubMed] [Google Scholar]
- 81. Finlay BB, Amato KR, Azad Met al. . The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc Natl Acad Sci U S A. 2021;118(6):e2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tornese G, Ceconi V, Monasta L, Carletti C, Faleschini E, Barbi E. Glycemic control in type 1 diabetes mellitus during COVID-19 quarantine and the role of in-home physical activity. Diabetes Technol Ther. 2020;22(6):462–7. [DOI] [PubMed] [Google Scholar]
- 83. De la Cuesta-Zuluaga J, Mueller NT, Álvarez-Quintero Ret al. . Higher fecal short-chain fatty acid levels are associated with gut microbiome dysbiosis, obesity, hypertension and cardiometabolic disease risk factors. Nutrients. 2019;11(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kim KN, Yao Y, Ju SY. Short chain fatty acids and fecal microbiota abundance in humans with obesity: a systematic review and meta-analysis. Nutrients. 2019;11(10):2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Boets E, Gomand SV, Deroover Let al. . Systemic availability and metabolism of colonic-derived short-chain fatty acids in healthy subjects: a stable isotope study. J Physiol. 2017;595(2):541–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Beyond A1C Writing Group . Need for regulatory change to incorporate beyond A1C glycemic metrics. Diabetes Care. 2018;41(6):e92–4. [DOI] [PubMed] [Google Scholar]
- 88. Wang Y, Rimm EB, Stampfer MJ, Willett WC, Hu FB. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am J Clin Nutr. 2005;81(3):555–63. [DOI] [PubMed] [Google Scholar]
- 89. DuBose SN, Hermann JM, Tamborlane WVet al. . Obesity in youth with type 1 diabetes in Germany, Austria, and the United States. J Pediatr. 2015;167(3):627–32.e4. [DOI] [PubMed] [Google Scholar]
- 90. Conway B, Miller RG, Costacou Tet al. . Temporal patterns in overweight and obesity in type 1 diabetes. Diabetes Med. 2010;27(4):398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data described in the manuscript, code book, and analytic code will be made available upon request pending application and approval by the ACT1ON Publications and Presentations Committee. If necessary, a data use or other data sharing agreement will be established.