Abstract
The monkeypox virus is excreted in the feces of infected individuals. Therefore, there is an interest in using viral load detection in wastewater for sentinel early surveillance at a community level and as a complementary approach to syndromic surveillance. We collected wastewater from 63 sewered and non-sewered locations in Bangkok city center between May and August 2022. Monkeypox viral DNA copy numbers were quantified using real-time polymerase chain reaction (PCR) and confirmed positive by Sanger sequencing. Monkeypox viral DNA was first detected in wastewater from the second week of June 2022, with a mean copy number of 16.4 copies/ml (n = 3). From the first week of July, the number of viral DNA copies increased to a mean copy number of 45.92 copies/ml. Positive samples were Sanger sequenced and confirmed the presence of the monkeypox virus. Our study is the first to detect monkeypox viral DNA in wastewater from various locations within Thailand. Results suggest that this could be a complementary source for detecting viral DNA and predicting upcoming outbreaks.
Keywords: Monkeypox, WBE, Monkeypox viral DNA, Wastewater surveillance, Monkeypox virus in wastewater, Monkeypox Asia
Graphical abstract
Human monkeypox is a zoonotic viral disease that occurs mainly in the rainforests of Central and Western Africa (Peiró-Mestres et al., 2022; Thornhill et al., 2022; Wannigama et al., 2022). Since early May 2022, over 60,000 monkeypox virus infections have been reported across 75 countries, prompting the World Health Organization to declare this outbreak a public health emergency of international concern. The monkeypox virus is a double-stranded DNA virus belonging to a member of the genus Orthopoxvirus (family Poxviridae, subfamily Chordopoxvirinae) (Thornhill et al., 2022). It has been found to infect various tissues, including the heart, brain, ovaries, and lymphoid tissue (Thornhill et al., 2022). The primary transmission mode is via close physical contact, including skin-to-skin contact, exchange of body fluids, and respiratory droplets at close range (Miura et al., 2022; Peiró-Mestres et al., 2022; Thornhill et al., 2022; Weinstein et al., 2005).
Monkeypox viral shedding via feces has been previously reported in the stool of infected individuals (Adler et al., 2022; Peiró-Mestres et al., 2022; Sklenovská and Van Ranst, 2018; Tiwari et al., 2023). Therefore it follows that the circulation of the virus within a population may increase the viral load in sewer systems (Wolfe et al., 2022). Wastewater surveillance could be a powerful tool to monitor the virus in communities. It can estimate asymptomatic or pre-symptomatic and symptomatic transmission and predict future waves (Ahmed et al., 2021; Wannigama et al., 2021). However, up to now, little information has been available on monkeypox viral DNA in wastewater. Detection of monkeypox, specifically in Thailand, has not been investigated before, and only seven cases have been reported at the time of writing this manuscript (https://ddc.moph.go.th/en).
To assess the presence of monkeypox DNA in wastewater, we retrieved samples from an ongoing SARS-CoV-2 wastewater surveillance program (Wannigama et al., 2021) collected between May and August 2022. Samples were collected twice a month (every two weeks) from 63 locations (shopping centers, condominium complexes, office complexes, food markets, wastewater treatment plants, and entertainment venues) in Bangkok province, covering 51 sub-districts. The monkeypox DNA copy number in samples was quantified by real-time PCR. E5R/ OPG117 and N3R/OPG016 genes were amplified by PCR from the DNA of RT-PCR positive samples collected on the 4th week of August, and Sanger sequencing was conducted to validate the RT-PCR results (Amarasiri et al., 2020; Covarrubias et al., 2022; Kumar et al., 2018; Lum et al., 2022; Shu and McCauley, 2017) (Details are available in the supporting information). All those locations have closed wastewater storage systems.
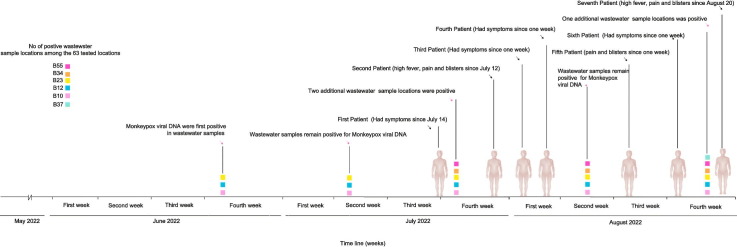
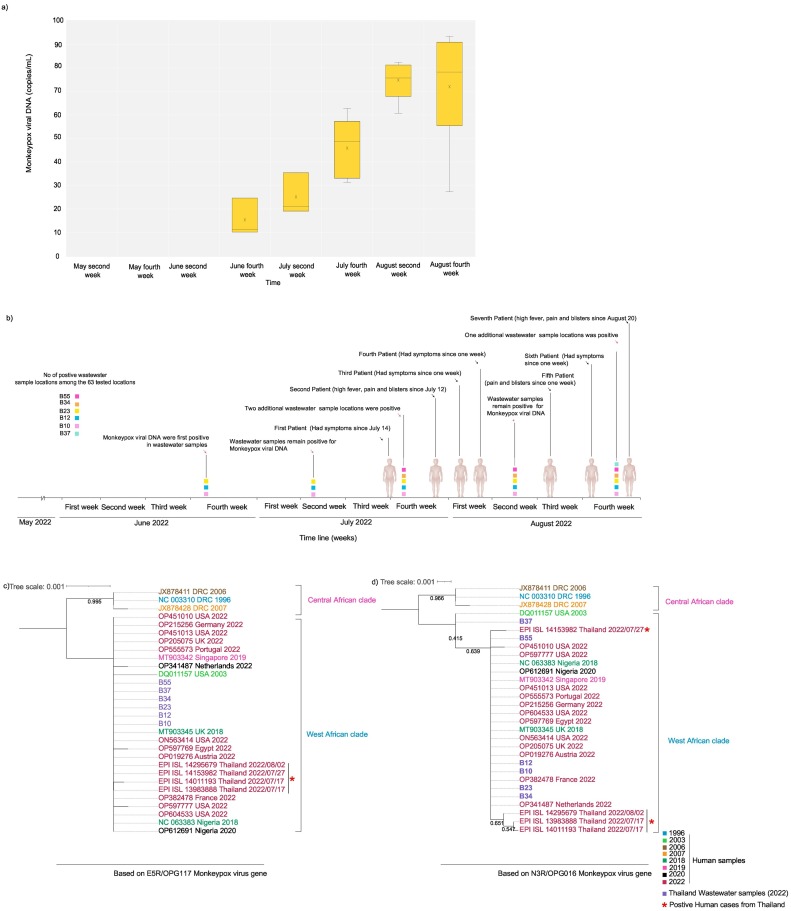
Monkeypox viral DNA was detected in wastewater starting the last week of June 2022. The positivity rate for monkeypox DNA increased from 4.76 % (n = 3) to 9.52 % (n = 6) between the 4th week of June and the 4th week of August (Table 1 ). The average copy number of monkeypox DNA detected in the 4th week of June was 16.4 copies/ml (n = 3). Average viral DNA concentration increased to 46 copies/ml by the 4th week of July (n = 5) and to 80 copies/ml by the 4th week of August (n = 6) (Fig. 1 and Table 1). Quantification of monkeypox viral DNA in samples taken a year ago confirmed the absence of the monkeypox virus in those samples. The Sanger sequencing of E5R/ OPG117 and N3R/OPG016 gene fragments confirmed the monkeypox virus's identification and validated the PCR findings. Preliminary analysis (Figs. 1c and d) shows that the monkeypox virus DNA from wastewater samples belongs to the West African clade and is closely related to the genomes from the 2022 outbreak, including genomes from Thailand monkeypox cases. All the monkeypox DNA-positive locations were commercial or public venues with closed non-sewered systems. Thailand's first monkeypox case was reported on July 21, 2022, in a 27-year-old male who had been sick for a week, while the second case was reported on July 28, 2022: a 47-year-old male, who had symptoms since July 12, 2022. Subsequently, third (August 3, 2022), fourth (August 5, 2022), fifth (August 15, 2022), sixth (August 21, 2022), and seventh (August 28, 2022) cases were reported. According to the press releases, most patients had symptoms a week preceding positive diagnoses. They had been to entertainment venues or crowded places with tourists and locals, which may correspond to the wastewater findings. Unfortunately, due to the personal data protection act, we cannot access complete contact tracing details of the positive cases.
Table 1.
Monkeypox viral DNA copies in wastewater samples from six positive locations among the 63 tested locations in Bangkok city from June second week to August fourth week.
| Month | Sampling Locations | Monkeypox viral DNA (copies/mL)a | Standard deviation | Cumulative cases national (total Monkeypox cases for each month)b | Monkeypox viral DNA positivity rate (%) |
|---|---|---|---|---|---|
| June fourth week | B10 | 11.35 | 3.15 | 0 | 4.76 |
| B12 | 13.22 | 2.56 | |||
| B23 | 24.68 | 2.50 | |||
| July Second week | B10 | 35.48 | 1.50 | 2 | 4.76 |
| B12 | 19.14 | 3.52 | |||
| B23 | 21.0 | 2.31 | |||
| July fourth week | B10 | 31.30 | 1.01 | 7.93 | |
| B12 | 35.67 | 1.00 | |||
| B23 | 62.89 | 2.25 | |||
| B34 | 48.71 | 4.00 | |||
| B55 | 51.84 | 3.74 | |||
| August second week | B10 | 51.8 | 2.00 | 5 | 7.93 |
| B12 | 60.78 | 2.24 | |||
| B23 | 75.74 | 3.52 | |||
| B34 | 80.09 | 4.10 | |||
| B55 | 82.43 | 2.10 | |||
| August fourth week | B10 | 75.12 | 3.78 | 9.52 | |
| B12 | 68.44 | 5.69 | |||
| B23 | 88.07 | 5.82 | |||
| B34 | 90.16 | 3.67 | |||
| B55 | 93.43 | 4.75 | |||
| B37 | 64.87 | 4.63 |
Average copy number of monkeypox viral DNA at positive locations.
Total confirmed monkeypox cases in Thailand obtained form Department of Disease Control, Ministry of Public Health, Thailand. https://ddc.moph.go.th/en.
Fig. 1.
a) Viral loads of monkeypox DNA copies in wastewater at six positive locations among the 63 tested locations in Bangkok from June to August 2022. The lower and upper boundaries of the box (interquartile) represent the 25th and the 75th percentile, respectively. The line within the box corresponds to the median and the cross to the mean of the distribution, while the whiskers indicate the highest and the lowest monkeypox DNA copy values. b) Timeline of the monkeypox wastewater viral DNA detection and monkeypox-positive patients reported. Evolutionary history was inferred using Maximum Likelihood method and Hasegawa-Kishino-Yano model for E5R/OPG117 (c) and Tamura 3-parameter model for N3R/OPG016 (d). The trees with highest log likelihood (−6078.89 and −1778.21) are shown.
Detected amounts of monkeypox DNA have increased regarding the quantity detected at positive sampling sites and the number of positive sampling sites. These results suggest the growing transmission of monkeypox and the exact time of monkeypox virus 2022 circulating in Thailand was far earlier than July 2022 (The first positive human case). Unfortunately, we could not match these results to case or contact tracing data. However, phylogenetic analysis of the sequence of E5R/ OPG117 and N3R/OPG016 gene reflect that wastewater samples are clustering with positive clinical cases from Thailand and other geographical origins connected to the monkeypox 2022 outbreak (Xiong et al., 2022). Thailand opened its borders in late May 2022 and is a major tourist destination for travelers from Europe and North America, where many monkeypox cases are currently reported. Thailand is a major risk area for emerging infectious diseases because it is a key destination for the sex tourism industry, where infections can appear and spread easily and quickly across the region (Kueakulpattana et al., 2021). Therefore, we believe that number of individuals carrying the monkeypox virus may be higher in the community than in reported cases. We observed low monkeypox viral DNA concentrations during the sampling period, and studies from US and Europe reported similar findings (de Jonge et al., 2022; Girón-Guzmán et al., 2022; La Rosa et al., 2022; Wolfe et al., 2022). However, all the positive samples in our study are from closed, non-sewered locations collected in sparse sampling events compared to frequent sampling from centralized wastewater treatment plants as in reported studies (de Jonge et al., 2022; Girón-Guzmán et al., 2022; La Rosa et al., 2022; Wolfe et al., 2022). Therefore, our findings strongly emphasize that tracing monkeypox viral DNA in wastewater is feasible even with sparse sampling, including non-sewered wastewater, which allows the application of wastewater-based surveillance to countries/regions with lower resources without established sewer systems. Since Bangkok province does not have an organized wastewater infrastructure like developed countries and it's difficult to track emerging infectious diseases based entirely on centralized wastewater treatment plants (Wannigama et al., 2021). To overcome this challenge, we selected 90 % of sampling locations in commercial or public venues based on population density, accessibility via the transportation system, and frequency of visitation. This comes with its limitation: we cannot attribute most of these locations to actual catchment numbers or flow due to the nature of non-sewered (Wannigama et al., 2021). However, this approach helped us to successfully identify the potential clusters and transmission in urban and rural communities during the SARS-CoV-2 pandemic (Wannigama et al., 2021). Moreover, our detection methodology is entirely based on standard qPCR techniques, which may be easily adaptable to resource-limited settings compared to droplet digital PCR or whole genome sequencing (de Jonge et al., 2022; Girón-Guzmán et al., 2022). Therefore, we believe that our findings might also advance health equity and provide strong real-world evidence for wastewater-based epidemiology in densely packed urban populations with poor sewer infrastructure.
The fewer number of reported positive cases in Thailand may be attributed to treatment-seeking behavior, LGBTQ people facing stigma and bias over mislabeling monkeypox as a sexually transmitted disease (STD), misdiagnosis with secondary syphilis, and unable to locate blisters in internal organs (rectum or throat cavity) (Girometti et al., 2022; Miura et al., 2022; Tarín-Vicente et al., 2022; Thornhill et al., 2022). Also, the lack of access to testing and the unavailability of diagnostics could steer people away from getting tested and seeking treatments (Girometti et al., 2022; Miura et al., 2022; Tarín-Vicente et al., 2022; Thornhill et al., 2022). According to a recent report, many of those infected individuals experience mild symptoms, including singular lesions or mild rectal pain; for example, a single lesion or sore in the oral cavity or on the genitals (Miura et al., 2022; Peiró-Mestres et al., 2022; Thornhill et al., 2022). Also, some people with monkeypox may not develop any symptoms (Ferré et al., 2022; Miura et al., 2022). Despite the mildness or absence of symptoms, infected people are likely to shed the virus into wastewater via routes connected to the location of the lesion; those shed through urine and feces are most likely to end up in wastewater (Peiró-Mestres et al., 2022). We also determined the absence of monkeypox viral DNA in historical samples (such as those taken a year ago as part of a study into SARS-CoV-2 in wastewater), which may suggest the monkeypox DNA detected in this study is excreted to the wastewater by the infected individuals from the current outbreak.
However, 15.3 % of the country's total population resides in Bangkok. Our results based on biweekly wastewater sampling frequency may not provide a clear view of the overall picture of monkeypox transmission in Thailand. The samples in this study were characterized according to the United States Centers for Disease Control and Prevention (CDC) Monkeypox virus-specific real-time PCR assay (CDC, 2022) like other studies (de Jonge et al., 2022; La Rosa et al., 2022; Wolfe et al., 2022). However, further characterization with Sanger sequencing assured the strength of our findings and the credibility of the CDC PCR assay. In addition, the inability to easily correlate virions in wastewater to the number of infected people makes it challenging to interpret the results. Furthermore, there is limited information regarding the intensity and duration of monkeypox viral DNA shedding in feces throughout infection (Miura et al., 2022). However, our findings provide an opportunity for ensuring an equitable approach to wastewater monitoring even with less frequent sampling, including non-sewered wastewater in countries/regions with lower resources.
CRediT authorship contribution statement
Dhammika Leshan Wannigama conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
Mohan Amarasiri conception, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
Parichart Hongsing conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
Cameron Hurst conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
Charin Modchang data curation, formal analysis, supervision, writing the original draft of the manuscript.
Sudarat Chadsuthi data curation, formal analysis, supervision, writing the original draft of the manuscript.
Suparinthon Anupong data curation, formal analysis, supervision, writing the original draft of the manuscript.
Phatthranit Phattharapornjaroen supervision, critical review and editing of the manuscript.
Ali Hosseini Rad S. M. supervision, critical review and editing of the manuscript.
Stefan Fernandez. critical review and editing of the manuscript.
Angkana T. Huang. critical review and editing of the manuscript.
Porames Vatanaprasan data curation, formal analysis.
Thammakorn Saethang data curation, formal analysis.
Sirirat Luk-in supervision, critical review and editing of the manuscript.
Robin James Storer supervision, critical review and editing of the manuscript.
Puey Ounjai supervision, critical review and editing of the manuscript.
Naveen Kumar Devanga Ragupathi supervision, critical review and editing of the manuscript.
Phitsanuruk Kanthawee supervision, critical review and editing of the manuscript.
Daisuke Sano supervision, critical review and editing of the manuscript.
Takashi Furukawa supervision, critical review and editing of the manuscript.
Kazunari Sei supervision, critical review and editing of the manuscript.
Asada Leelahavanichkul supervision, critical review and editing of the manuscript.
Talerngsak Kanjanabuch supervision, critical review and editing of the manuscript.
Nattiya Hirankarn supervision, critical review and editing of the manuscript.
Paul G. Higgins supervision, critical review and editing of the manuscript.
Anthony Kicic supervision, critical review and editing of the manuscript.
Tanittha Chatsuwan supervision, critical review and editing of the manuscript.
Alexander D McLellan supervision, critical review and editing of the manuscript.
Shuichi Abe supervision, critical review and editing of the manuscript.
Ethical approval
Ethics approval was not required for this type of environmental wastewater surveillance study.
Funding sources
Dhammika Leshan Wannigama was supported by Balvi Filantropic Fund and Chulalongkorn University Second Century Fund-C2F Postdoctoral Fellowship, University of Western Australia (Overseas Research Experience Fellowship) and Yamagata Prefectural Central Hospital, Yamagata, Japan (Clinical Residency Fellowship). Charin Modchang was supported by the Centre of Excellence in Mathematics, Ministry of Higher Education, Science, Research and Innovation, Thailand, Center of Excellence on Medical Biotechnology (CEMB), and Thailand Center of Excellence in Physics (ThEP). Anthony Kicic is a Rothwell Family Fellow.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all the volunteers who kindly supported with sample collection. Also, thanks to Dr. Ong-orn Prasarnphanich at Centers for Disease Control and Prevention (CDC) Thailand for the technical support. We gratefully acknowledge all data contributors, i.e., the Authors and their Originating laboratories responsible for obtaining the specimens, and their Submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative and NCBI genbank.
Editor: Kevin V. Thomas
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.159816.
Appendix A. Supplementary data
Supplementary material
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its additional information
References
- Adler H., Gould S., Hine P., Snell L.B., Wong W., Houlihan C.F., et al. Clinical features and management of human monkeypox: a retrospective observational study in the UK. Lancet Infect. Dis. 2022;22:1153–1162. doi: 10.1016/S1473-3099(22)00228-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasiri M., Utagawa E., Sano D., Katayama K. Identification of novel norovirus polymerase genotypes from pediatric fecal samples collected between the year 1997 and 2000 in Japan. Infect. Genet. Evol. 2020;82 doi: 10.1016/j.meegid.2020.104313. [DOI] [PubMed] [Google Scholar]
- CDC . Test Procedure: Monkeypox virus Generic Real-Time PCR Test. Centers for Disease Control & Prevention; 2022. pp. 1–7. [Google Scholar]
- Ferré V.M., Bachelard A., Zaidi M., Armand-Lefevre L., Descamps D., Charpentier C., et al. Detection of monkeypox virus in anorectal swabs from asymptomatic men who have sex with men in a sexually transmitted infection screening program in Paris, France. Ann. Intern. Med. 2022;175(10):1491–1492. doi: 10.7326/M22-2183. [DOI] [PubMed] [Google Scholar]
- Girometti N., Byrne R., Bracchi M., Heskin J., McOwan A., Tittle V., et al. Demographic and clinical characteristics of confirmed human monkeypox virus cases in individuals attending a sexual health Centre in London, UK: an observational analysis. Lancet Infect. Dis. 2022;22(9):1321–1328. doi: 10.1016/S1473-3099(22)00411-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girón-Guzmán I., Díaz-Reolid A., Truchado P., Carcereny A., Garcia-Pedemonte D., Hernaez B. Wastewater Based Epidemiology Beyond SARS-CoV-2: Spanish Wastewater Reveals the Current Spread of Monkeypox Virus. medRxiv. 2022 doi: 10.1101/2022.09.19.22280084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias P., Castillo A.E., Gomez V, Campano C, Guajardo M, Rojas M, et al. 2022. Draft Genome of the First Case of the Monkeypox Virus in Chile Associated With the 2022 Outbreak.https://virological.org/t/draft-genome-of-the-first-case-of-the-monkeypox-virus-in-chile-associated-with-the-2022-outbreak/887 https://virological.org/ [Google Scholar]
- de Jonge E.F., Peterse C.M., Koelewijn J.M., van der Drift A.-M.R., van der Beek R.F.H.J., Nagelkerke E., et al. The detection of monkeypox virus DNA in wastewater samples in the Netherlands. Sci. Total Environ. 2022;852 doi: 10.1016/j.scitotenv.2022.158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kueakulpattana N., Wannigama D.L., Luk-in S., Hongsing P., Hurst C., Badavath V.N., et al. Multidrug-resistant neisseria gonorrhoeae infection in heterosexual men with reduced susceptibility to ceftriaxone, first report in Thailand. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-00675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Mancini P., Veneri C., Bonanno Ferraro G., Lucentini L., Iaconelli M. Detection of Monkeypox Virus DNA in the Wastewater of an Airport in Rome, Italy: Expanding Environmental Surveillance to Emerging Threats. medRxiv. 2022 doi: 10.1101/2022.08.18.22278932. [DOI] [Google Scholar]
- Lum F.-M., Torres-Ruesta A., Tay M.Z., Lin R.T.P., Lye D.C., Rénia L., et al. Monkeypox: disease epidemiology, host immunity and clinical interventions. Nat. Rev. Immunol. 2022;22:597–613. doi: 10.1038/s41577-022-00775-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F., van Ewijk C.E., Backer J.A., Xiridou M., Franz E., Op de Coul E., et al. Estimated incubation period for monkeypox cases confirmed in the Netherlands, May 2022. Eurosurveillance. 2022;27(24) doi: 10.2807/1560-7917.ES.2022.27.24.2200448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiró-Mestres A., Fuertes I., Camprubí-Ferrer D., Marcos M.Á., Vilella A., Navarro M., et al. Frequent detection of monkeypox virus DNA in saliva, semen, and other clinical samples from 12 patients, Barcelona, Spain, May to June 2022. Eurosurveillance. 2022;27(28) doi: 10.2807/1560-7917.ES.2022.27.28.2200503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklenovská N., Van Ranst M. Emergence of monkeypox as the Most important orthopoxvirus infection in humans. Front. Public Health. 2018;6 doi: 10.3389/fpubh.2018.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarín-Vicente E.J., Alemany A., Agud-Dios M., Ubals M., Suñer C., Antón A., et al. Clinical presentation and virological assessment of confirmed human monkeypox virus cases in Spain: a prospective observational cohort study. Lancet. 2022;400:661–669. doi: 10.1016/S0140-6736(22)01436-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill J.P., Barkati S., Walmsley S., Rockstroh J., Antinori A., Harrison L.B., et al. Monkeypox virus infection in humans across 16 countries — April–June 2022. N. Engl. J. Med. 2022;387:679–691. doi: 10.1056/NEJMoa2207323. [DOI] [PubMed] [Google Scholar]
- Tiwari A., Adhikari S., Kaya D., Islam M.A., Malla B., Sherchan S.P., et al. Monkeypox outbreak: wastewater and environmental surveillance perspective. Sci. Total Environ. 2023;856(2) doi: 10.1016/j.scitotenv.2022.159166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., et al. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021;108:296–299. doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wannigama D.L., Brown J., Hongsing P., Phattharapornjaroen P., Amarasiri M., Abe S., et al. Monkeypox: prevalence, diagnostics, and prevention. Greater Mekong Sub-region Med. J. 2022;2(3):185–196. [Google Scholar]
- Weinstein R.A., Nalca A., Rimoin A.W., Bavari S., Whitehouse C.A. Reemergence of monkeypox: prevalence, diagnostics, and countermeasures. Clin. Infect. Dis. 2005;41:1765–1771. doi: 10.1086/498155. [DOI] [PubMed] [Google Scholar]
- Wolfe M.K., Duong D., Hughes B., Chan-Herur V., White B.J., Boehm A.B. Detection of Monkeypox Viral DNA in a Routine Wastewater Monitoring Program. medRxiv. 2022 doi: 10.1101/2022.07.25.22278043. [DOI] [Google Scholar]
- Xiong C, Li Y, Hou J, Sun Z, Han W, Thilakavathy K, Chen W, Wang Y, Liu X, Gao Q, Lu S, Shao Z, Lu Y, Wang W, Hu J. Monkeypox Virus 2022, Gene Heterogeneity and Protein Polymorphism. Research Square. 2022 doi: 10.21203/rs.3.rs-2162648/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its additional information