Abstract
Acute myeloid leukemia (AML) is a type of leukemia with a poor prognosis and survival characterized by abnormal cell proliferation and differentiation. Despite advances in treatment, AML still has a low complete remission rate, particularly in elderly patients, and recurrences are frequently seen even after complete remissions. The major challenge in treating AML is the resistance of leukemia cells to chemotherapy drugs. Thus, to overcome this issue, it can be crucial to conduct new investigations to explore the mechanisms of chemo-resistance in AML and target them. In this review, the potential role of autophagy induced by FLT3-ITD and acid ceramidase in chemo-resistance in AML patients are analyzed. With regard to the high prevalence of FLT3-ITD mutation (about 25% of AML cases) and high level of acid ceramidase in these patients, we hypothesized that both of these factors could lead to chemo-resistance by inducing autophagy. Therefore, pharmacological targeting of autophagy, FLT3-ITD, and acid ceramidase production could be a promising therapeutic approach for such AML patients to overcome chemo-resistance.
Video abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-022-00956-7.
Keywords: Acute myeloid leukemia, FLT3-ITD, Autophagy, Acid ceramidase, Sphingosine, Chemo-resistance
Introduction
Chemo-resistance in AML and its challenges
Acute myeloid leukemia (AML) is the most common acute leukemia in adults [1]. It is characterized by clonal proliferation of undifferentiated myeloid precursors, thereby impairing hematopoiesis and giving rise to bone marrow failure [2]. Several genetic mutations provide diagnostic and prognostic data, including mutations in FMS-like tyrosine kinase-3 (FLT3), Nucleophosmin 1 (NPM1), KIT, CCAAT/enhancer-binding protein alpha (CEBPA), and ten-eleven-translocation 2 (TET2) [3]. While many patients respond well to induction chemotherapy, chemo-resistance is a significant challenge in the treatment of AML patients particularly older patients are more resistant to treatment making the cure more challenging and only 5–25% of them are likely to achieve a cure [4]. Therefore, drug resistance is a major barrier to cancer chemotherapy [5]. Some gene mutations like FLT3, autophagy signal pathway, drug resistance-related protein and enzyme-like MDR (multiple drug resistance), and abnormal expression of microRNA (miRNA) will cause drug resistance in AML patients [6]. Although patients undergoing initial induction chemotherapy can attain a complete response rate as high as 80%, most AML patients will eventually be diagnosed with recurrent or refractory disease [6]. A therapeutic strategy for AML patients is an intensive induction regimen including Cytarabine and Anthracycline infusion that is usually used to induce complete remission. A complete remission is usually achieved in about 60–80% of younger adults and 40–60% older than 65 years. In addition, allogeneic stem cell transplants (alloSCT) are recommended for AML patients with poor prognosis, and the post-remission regimen consists of consolidation chemotherapy, autologous and allogeneic stem cell transplantation [7].
FLT3-ITD and acid ceramidase levels and related association with drug resistance in AML patients
Around 25% of AML cases have mutations in the FLT3 receptor [8]. FLT3 is an oncogene in AML, which exhibits two types of mutations; internal tandem duplication (ITD) in 20–25% of patients, and tyrosine kinase domain (TKD) mutations in 5–10% of patients [6, 9]. FLT3 is a tyrosine kinase receptor that contributes to the differentiation, proliferation, and apoptosis of hematopoietic stem cells [7]. Mutation in FLT3-ITD has been identified as a relapse-associated genetic marker [6]. Myelodysplastic syndromes (MDS) and AML are hematological malignancies associated with aberrant splicing patterns [10] and FLT3 is one of the most mis-spliced genes in AML, dysregulation of the splicing process can also disturb apoptosis and cause resistance to therapies [4]. FLT3 mutation carriers, especially those with ITD mutations, are more likely to have poor prognoses, with fewer chances of complete remission and overall survival [11].
Furthermore, in AML, acid ceramide expression is elevated. Overexpression of acid ceramidase increases ceramide breakdown and increases S1P production. In healthy cells, a stable balance exists between ceramides and S1Ps levels, but disruption of this equilibrium contributes to the progression of several diseases including multiple cancers [12]. Thus, there is an elevated level of acid ceramidase in several types of solid tumors and leukemia, such as AML, and patients with elevated acid ceramidase have a poor prognosis and lower overall survival [13]. As ceramide has a pro-apoptotic role and sphingosine has a pro-survival role, acid ceramidase promotes cellular proliferation and increases the growth rate of tumor cells. In addition, upregulation of acid ceramidase contributes to chemo-resistance in solid tumors such as prostate cancer and hepatoma cancer cell lines [14]. It is also shown that a high acid ceramidase level can increase the survival of AML cells [12].
Autophagy and drug resistance in AML and other malignancies
In recent years, there have been an increase in autophagy research. By definition, autophagy is the process of phagocytosing its own cytoplasmic organelles or proteins. Autophagy originates from a Greek word that means eating oneself. During autophagy, old proteins or organelles are broken down by lysosomes within the cells, nutrients and new synthetic materials are supplied to maintain cellular homeostasis [15, 16]. As its package is transferred into vesicles, it will be fused with lysosomes, creating autolysosomes, and eventually degrading its contents. During this process, organelles and cell metabolism are renewed. A chemotherapeutic drug can induce autophagy, resulting in drug resistance in cancer cells [6]. A study by Shang et al. has shown that cirpan3 (a kind of circular RNA) is involved in drug resistance in AML by promoting autophagy through the AMPK/mTOR pathway [17]. In other leukemias, such as chronic lymphocytic leukemia (CLL), bone marrow stromal cells stimulate autophagy which contributes to drug resistance in CLL cells [18]. As well, autophagy has a pro-survival role in Multiple Myeloma (MM) and it promotes the resistance of MM cells to proteasome inhibitors [19]. Moreover, autophagy can lead to chemo-resistance in solid tumors such as prostate and renal cancer cells [20]. In glioblastoma, hypoxia induces autophagy, which contributes to tumor cell survival and resistance to antiangiogenic treatments [21]. Therefore, autophagy is a significant process that can lead to chemo-resistance in AML and other malignancies [22–24]. This suggests that it is a potential target in cancer treatments for preventing or reducing chemo-resistance.
The role of FLT3-ITD in induction of autophagy
Based on studies in xenografted mice, FLT3-ITD activity increased basal autophagy in AML cells, which is needed for cell proliferation in vitro. FLT3-ITD-induced autophagy was found to be mediated by ATF4 (activating transcription factor 4). Downregulation of autophagy and ATF4 inhibits the proliferation of AML cells and improves survival in mice. It appears that autophagy is involved in the proliferation and degradation of the FLT3-ITD receptor [25]. The RET receptor, a tyrosine kinase receptor often activated in AML, has been demonstrated to suppress autophagy via mTORC1 (mammalian target of rapamycin complex1), leading to the stabilization of mutant FLT3 receptors [26].
Recent studies have found that SHP2, which interacts with FLT3-ITD phosphorylated Gab2, is involved in activating the MEK/ERK pathway, as well as RSK's negative feedback regulation of that pathway in FLT3-ITD-positive AML cells. Activation of the MEK/ERK or phosphatidylinositol 3-kinase (PI3K)/AKT pathways can occur through the interaction of SHP2 or p85 with tyrosine-phosphorylated Gab2 via their SH2 domains, respectively, downstream of several tyrosine kinases or cytokine receptors [27, 28]. FLT3-ITD is capable of triggering the transcription factors signal transducer and activator of transcription (STAT) 5. STAT5 to translocate into the nucleus, triggering the production of oncogenic proteins such as (proviral insertion site) PIM kinases and Bcl-xL [29]. Through the STAT5/PIM and PI3K/AKT pathways, FLT3-ITD cooperatively activates the mTORC1/S6K/4EBP1 pathway [30].
By activating mTORC1, several cellular processes can be regulated that affect the metabolic state of the cell. A number of mechanisms underlie mTORC1 signaling, including inhibition of autophagy and stimulation of biosynthesis pathways [31].
The mTORC1 pathway inhibits autophagy at a number of stages. The mechanism through which mTORC1 inhibits autophagy is best understood as direct control of unc-51-like autophagy activating kinase 1 (ULK1), but it is also associated with the human class III PI3K (Vps34) complex containing autophagy-related protein 14 (ATG14) and transcription factor EB (TFEB). The autophagy initiators ULK1 and ATG13, which form a complex with the focal adhesion kinase family interacting protein of 200KDa (FIP200) and ATG101, are inactivated by mTORC1 in amino acid-rich conditions which binds, phosphorylates, and thus inactivates ULK1 [31–33].
A second mechanism involves mTORC1 phosphorylating TFEB and TFE3 and this event allows them to interact with the cytosolic chaperone 14-3-3 to remain in the cytoplasm [34, 35].
Lack of activating stimuli, in turn, induces autophagy through dissociation between mTORC1 and the ULK1 complex, thereby reducing the inhibition of ULK1, which is then phosphorylated along with ATG13, FIP200, and Raptor [33].
ULK1 can then activate the PI3K complex and induce autophagosome synthesis. Furthermore, mTORC1 inactivation leads to the re-localization of TFEB and TFE3 to the nucleus, where they induce the expression of multiple autophagy-related genes. As a result, the cell maintains a critical level of energy and metabolites in order to survive the starvation state (Fig. 1) [36].
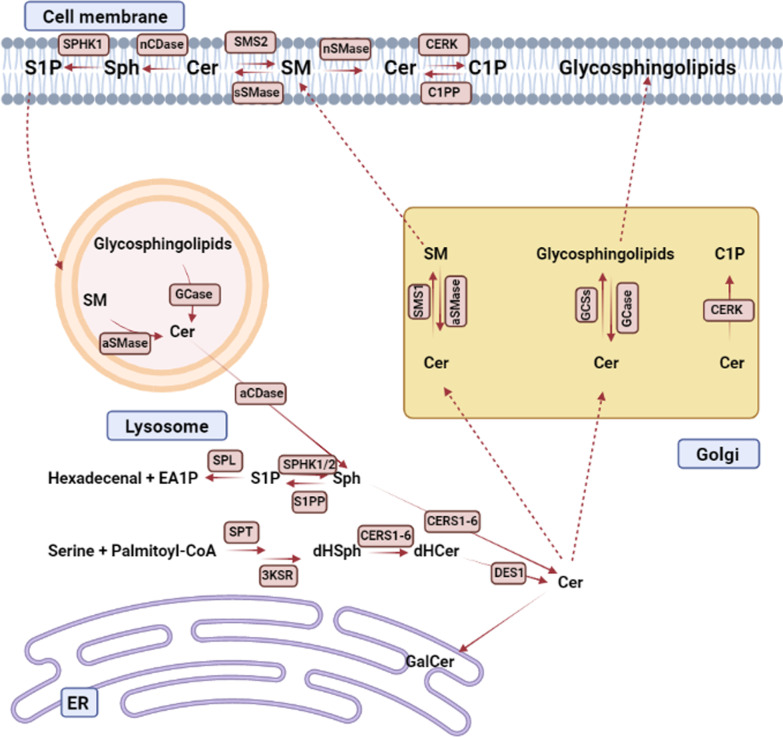
Fig. 1.
Sphingolipid metabolism in a summary. Ceramide is a key component of sphingolipid metabolism. Ceramide is made from the condensation of serine and palmitoyl-CoA in the endoplasmatic reticulum (ER) by serine palmitoyltransferase (SPT). After further reduction and acylation by a ceramide synthase (CERS1-6), dihydrosphingosine (dHSph) is produced, which is then desaturated to generate ceramide. Ceramide can be altered to galactosylceramide (GalCer) in the ER, although most ceramide change occurs at the Golgi in a manner that is dependent on their following use. Ceramide is used in the Golgi to produce sphingomyelin (SM) and glycosphingolipids in processes mediated by sphingomyelin synthase 1 (SMS1) and glycosphingolipid synthases (GCSs). SM and glycosphingolipids are delivered to the plasma membrane from the Golgi. By the actions of secretory and neutral sphingomyelinases, SM can be converted back to ceramide (sSMase and nSMase). Ceramide can then be converted to ceramide-1-phosphate (C1P), sphingosine-1-phosphate (S1P), or SM. By accessing the endolysosomal process, complex sphingolipids in the membrane can be utilized as a pool for ceramide recycling. Acid SMases (aSMase) and glycosidases (GCase) create ceramide, which can then be hydrolyzed into sphingosine and utilized in ceramide production or degraded by phosphorylation into S1P followed by degradation to hexadecenal and ethanolamine-1-phosphate (EA1P). Ceramide kinase (CERK) can phosphorylate ceramide in the Golgi, resulting in ceramide-1-phosphate (C1P)
Nevertheless, these signaling pathways connected to mTOR and ULK1 are controversial, and it is also possible for FLT3-ITD to promote autophagy through some other mechanisms and signaling pathways. According to the above evidence, for the first time, in a study by Heydt et al. [25], ATF4 (a transcription factor) was recognized as a critical factor of FLT3-ITD-induced autophagy (Fig. 2). The level of ATF4 in cells was highly dependent on FLT3-ITD activity, and ATF4 downregulation inhibited autophagy and cell proliferation in AML cells and increased survival of mice in a similar manner to autophagy inhibition.
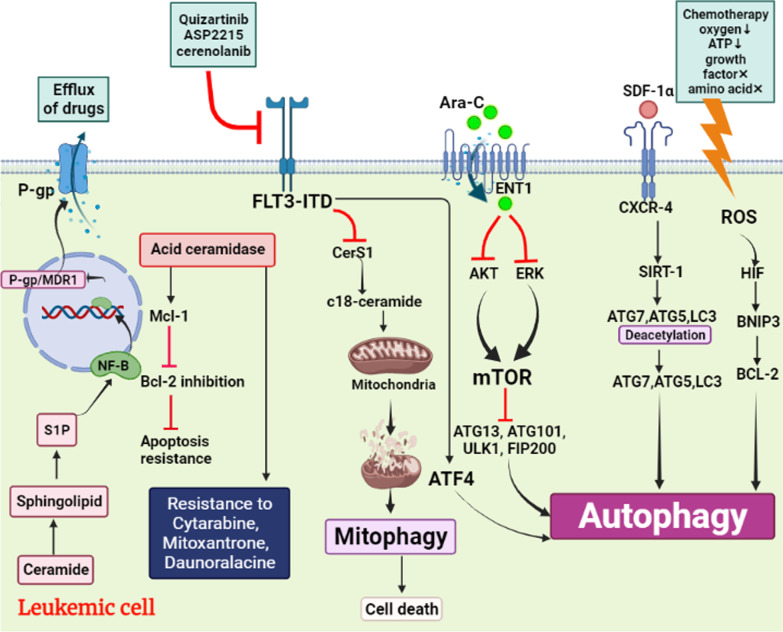
Fig. 2.
Illustration shows that leukemic cells design mechanisms against a variety of therapies and ultimately achieve chemotherapy-resistance through autophagy: 1—Chemotherapy increases reactive oxygen species (ROS) and induces autophagy through HIF/ BNIP3/BCL-2 signaling pathway. 2—Binding of SDF-1α as a drug to CXCR-4 activates SIRT-1 and SIRT-1 by deacetylation of ATG7, ATG5, LC3 induces autophagy. 3—Ara-C modulates AKT and ERK signaling pathways and induces mTOR-dependent autophagy. 4—By activating ATF4, it directly induces autophagy and inhibits mitophagy by inhibiting CerS1. 5—S1P production as a result of increased ceramidase acid in AML cells activates NF-κB and increases P-gp expression as a channel that causes drug efflux
Based on available evidence, targeting autophagy or ATF4 in patients expressing FLT3 mutations could be a novel potential promising therapeutic approach for AML patients with or even without significant chemo-resistance. In order to provide other meaningful insights into chemo-resistance in AML patients, more studies are needed to figure out the exact mechanism of correlations between FLT3-ITD and autophagy.
The role of acid ceramidase and ceramide in induction of autophagy
Acid ceramidase is a hydrolase that converts ceramide into sphingosine and free fatty acids, located in the lysosome. By degrading ceramide into its metabolites, ceramide decreases in the cell while sphingosine increases [37]. Sphingosine is then phosphorylated and converted into sphingosine-1-phosphate (S1P) by sphingosine kinase [38]. S1P and ceramide play roles in determining whether cells undergo apoptosis or proliferation [39]. The accumulation of ceramide in the cell causes apoptosis. In contrast, S1P stimulates angiogenesis through G protein-coupled receptors [37]. Ceramide and S1P have been identified as bioactive signaling molecules that regulate cell growth, differentiation, senescence, apoptosis, and autophagy [40].
Numerous studies have suggested sphingolipids (SLs), as well as ceramide, are autophagosome membrane components [41, 42]. In recent years, the role of ceramide in autophagy has become known. The compound ceramide inhibits the transport of nutrients and initiates autophagy [15]. Ceramide stimulates autophagy by regulating classic or atypical autophagic pathways. By virtue of these signals, class I PI3Ks and AKT negatively regulate autophagy, whereas class III PI3Ks stimulate autophagy. Ceramide was determined to act as an inhibitor of AKT, by inducing phosphoprotein phosphatase 2A, and to promote interaction between PI3K class III and other autophagy regulators [43, 44]. Studies have shown that ceramide interacts directly with microtubule-associated protein light chain 3 (LC3) on mitochondrial membranes to induce deadly autophagy via an increase in intracellular mitophagy [45]. It has been reported that ceramide-induced autophagy can result in cell death under high growth factor conditions [15]. In addition, ceramides activate the transcription factor c-Jun, which is involved in the increase of the autophagic protein Beclin-1 [46]. The chemotherapeutic agents may induce the biosynthesis of intracellular ceramide that can result in autophagy-mediated cell death; however, an increase in S1P levels results from nutrition starvation and intercedes cytoprotective autophagy. Therefore, during the conversion of ceramide into S1P as a result of overexpression of acid ceramidase, the survival effects accumulate as well as the death signals are removed and it is suggested that acid ceramidase can influence the response to chemotherapeutics [46, 47].
Autophagy may support cellular survival and help cancer cells to resist metabolic and therapeutic stress [48]. Three types of autophagy are identified so far: macroautophagy (also called 'autophagy'), microautophagy, and chaperone-mediated autophagy (CMA) [45, 49]. During macroautophagy (autophagy), a double membrane structure called a phagophore engulfs intracellular components such as proteins and organelles. An autophagosome is formed when this transient compartment matures. Autophagosomes and lysosomes fuse to form autolysosomes, which digest vesicle contents for recycling. Macroautophagy has been linked to the development of AML in various studies. There is a correlation between the high expression of key genes involved in autophagic processes such as SIRT1 (sirtuin 1), BECN1 (beclin 1), STK11/LKB1 (serine/threonine kinase 11), and ATG7 (autophagy-related 7) and poor clinical outcome, short remission duration and drug resistance in patients with AML [50, 51].
Cancers such as prostate cancer, liver cancer, melanoma, and AML exhibit high expression of acid ceramidase [12, 52, 53]. Cancer cells expressing a high level of acid ceramidase may resist chemotherapy and radiation therapy since acid ceramidase inhibits apoptosis caused by TNF-α [45, 46, 54]. It is worth noting, that AC is not only found in lysosomes in cancer cells but is also present in the cytoplasm [46]. In a study by Turner et al. prostate cancer cell lines overexpressing acid ceramidase showed an increased lysosomal density and an increased autophagic activity [54]. In melanoma cells treated with doxorubicin, Michele Lai et al. found that acid ceramidase controlled apoptosis and increased autophagy [55]. Su-Fern Tan et al. demonstrated for the first time how targeting acid ceramidase reverses the aberrant lipid profile of AML, characterized by low ceramide levels and elevated S1P, which ultimately leads to cell death [12]. According to collected evidence, the upregulation of acid ceramidase in cancers can boost cytoprotective autophagy, which is associated with drug resistance in many types of cancers, especially AML.
Sphingolipid metabolism
Sphingolipids are a large group of lipids that varies structurally from other lipids in that they have a sphingoid group as their structural backbone. Sphingolipid metabolism is a complex network in which the balance of sphingolipid formation, degradation, and recycling are carefully controlled in terms of cell response and fate.
Serine palmitoyl-transferase (SPT) catalyzes the condensation of serine and palmitoyl-CoA to produce 3-keto-dihydro sphinganine at the cytosolic side of the ER membrane, where de novo formation of sphingolipids occurs. The sphingoid base dihydrosphingosine (sphinganine) is generated from 3-keto-dihydro sphinganine, which, along with sphingosine, forms the backbone of sphingolipids.
Ceramide synthases (CERS1-6) catalyze the N-acylation of sphingoid bases, which leads to ceramide formation. The CERSs have variable chain length unique qualities [56], which adds to the sphingolipid's diversity. CERS1 favors C18-CoAs, whereas CERS2 uses acyl-CoAs in the range of C20 to C26. CERS3 has an affinity for ultra-long-chain acyl-CoAs (C26–36), whereas CERS4 has a preference for C18- and C20-CoAs. C16-CoAs are largely present in CERS5 and CERS6. Ceramides can be phosphorylated to produce ceramide-1-phosphate (C1P), modified with phosphocholine to generate sphingomyelin, or glycosylated to produce a wide range of glycosyl ceramides (Fig. 1).
Ceramides can be produced from sphingosine, recycled from glycosyl ceramides and sphingomyelin, and dephosphorylated from C1P [57]. Furthermore, while de novo ceramide formation by CERSs takes hours [58], ceramide generation through recycling pathways, such as sphingomyelin degradation, occurs within minutes after activation [59]. Therefore, ceramides and other sphingolipids potentially regulate cellular functions in the short term via salvaging pathways, while de novo sphingolipid synthesis might affect long-term cellular processes.
Given the critical roles of sphingolipids in autophagy, it is important to note that autophagy affects sphingolipid levels, particularly ceramide levels [60], and also the mobilization and storage of glycerolipids in lipid droplets [61–63]. Overall, research demonstrates that autophagy and lipid metabolism are coordinated, highlighting the relevance of sphingolipids in metabolic regulation. Some of the effects of sphingolipids have been documented, however, they are based on short-chain ceramides, which may have very different effects than ceramides generated in vivo. Instead of utilizing such non-natural ceramides, the involvement of sphingolipids in autophagy must be investigated further using genetically amenable model systems using loss-of-function and overexpression approaches.
The role of autophagy in AML chemo-resistance
For decades, the use of chemotherapy to control and treat many cancers, such as AML has been common. However, this method has been along with several challenges, such as chemotherapy resistance. Importantly, autophagy is one of the most important mechanisms in chemotherapy resistance. A variety of studies revealed that multiple signaling pathways are related to energy metabolism and gene mutations can lead to autophagy [64]. Interestingly, reactive oxygen species (ROS), causes autophagy in AML cells via HIF/BNIP3/Bcl-2 [65]. It is noticeable to state that the level of ROS is increased in chemotherapy [66]. Sometimes it is possible that environmental conditions cause increase levels of ROS that are named stress signaling in general, such as the low level of oxygen, growth factors deprivation, acid amine deprivation, the low level of ATP, and lack of glucose [67]. In addition, temozolomide as a drug used in AML initiates autophagy and blockade of the apoptotic pathway via ERK/ROS signaling pathway [6]. To place more emphasis on cellular stress signaling, it is noticeable that these signaling lead to activation of JNK as a signaling factor and JNK causes activation of C-Jun, C-Fos, and FOXO1/3 as transcription factors and leads to an increased level of transcription of some autophagy factors (i.g Beclin 1, VPS34, and LC3) [65]. Also, stress pressure on AML cells activates heat shock transcription factor 1 (HSTF1) that launches transcription of ATG7 as an autophagy factor and chemo-resistance inducer [6]. Interestingly, AML treatment with some common drugs like Ara-C can induce autophagy and lead to drug resistance in AML cells. For instance, according to X Hu et al.'s studies, AML treatment with Ara-C leads to an increase expression of CXCR4 on AML cells. In this study, authors have found that CXCR4/SDF-1α axis can induce upregulation of SIRT1 in these cell lines [68]. Importantly, SIRT1 stimulates autophagy via deacetylation of ATG5, ATG7, ATG8, and LC3 [69]. On the other hand, Ara-C modulates several signaling pathways, such as AKT and ERK that lead to the activation of autophagy via mTOR pathway [70]. With this in mind, in addition to chemotherapy, recent treatments such as epigenetic therapies are no exception. Circular RNAs (circRNAs), are several types of non-coding RNAs used in some cancers, like AML. Accordingly, J. shang et al. have found that treatment with circRNAs in AML cells is associated with increased circPAN3 that induces autophagy through the AMPK/mTOR signaling pathway [17]. In a study by Jin Shang et al. [17] it has been demonstrated that circPAN3 may increase AML drug resistance by inducing autophagy. Also, Xiaojia Hu et al. [68] have found that the autophagy-related protein SIRT1 expression is associated with stimulation of SDF-1a-CXCR4 signaling, interacting with autophagy proteins (e.g. ATG5 and LC3). In addition, they have revealed that among primary human AML samples, a high CXCR4 expression level is correlated with SIRT1 and other autophagy-related proteins' elevated expression levels. Overall, their investigation proposed new roles for SDF-1a-CXCR4 signaling in the induction of autophagy in AML cells, which consequently facilitated their survival and drug resistance under stress. Specifically, in thisreview, we described that acid ceramidase and FLT3-ITD mutations can lead to autophagy in AML patients. In addition, multiple studies have revealed autophagy as a major cause of drug resistance in AML patients and other malignancies [17]. Therefore, acid ceramidase overexpression and FLT3-ITD mutation can increase drug resistance in these patients.
The role of FLT3-ITD mutation in AML drug resistance
Midostaurin is a multi-targeted tyrosine kinase inhibitor that inhibits FLT3 [71]. The presence of FLT3-ITD and FLT3-TKD mutations in the same subclone of an AML patient may induce primary resistance to FLT3 inhibitors [72]. Because of the wide range of FLT3-ITD mutations, the position and amino acid sequence of ITD may also be involved in primary resistance by modifying protein structure, which activates alternate downstream signaling pathways [73]. It is unclear how mutant FLT3-internal tandem duplication (ITD) regulates cellular signaling pathways that contribute to AML cell death resistance. However, studies showed that FLT3-ITD targeting increased ceramide accumulation on outer mitochondrial membranes, which in turn resulted in the binding of autophagy-inducing light chain 3 (LC3), which is involved its I35 and F52 residues, to recruit autophagosomes to execute lethal mitophagy [74].
Quizartinib is a small molecule inhibitor with activity against FLT3-ITD. It is a very efficient FLT3 inhibitor that was designed specifically for this purpose. Unfortunately, when AML patients are treated with quizartinib, resistance develops fast, typically owing to secondary mutations in the FLT3 gene. Pexidartinib has been proven to be effective against FLT3-ITD, but the tumor is also resistant to pexidartinib due to additional secondary mutations [71, 75, 76]. Sorafenib (Nexavar) is a small molecule multitargeted kinase inhibitor that targets FLT3 [71]. Smith et al. identified secondary point FLT3 mutations in eight relapsed patients with ITD mutated AML. As well, these mutations have shown resistance to sorafenib [75].
Mutation in FLT3 is associated with poor prognosis in AML patients. Interestingly, mutations in FLT3 causes a kind of drug resistance. As part of a study on this issue, Dany et al. have found that the cause of drug resistance in FLT3 mutant AML cells is FLT3-ITD signaling that leads to the reduction of C18-ceramide. In this regard, CerS1 activation is stopped and we have stopped producing C18-ceramide [74]. On the other hand, when the activity of FLT3-ITD is suppressed with some drugs such as ASP2215, quizortinib, sorafenib, and cerenolanib, the percentage of cancerous cells death increases through mitophagy resulting from CerS1 activity that causes an increased level of C18-ceramide [10, 11]. Importantly, this mutation causes resistance to some drugs like Doxorabicin that target mitochondria via mitophagy, leading to AML cell death [12]. Furthermore, according to studies, mutation in FLT3 causes Ara-C resistance in AML cells due to a decreased uptake of Ara-C. To that end, the expression level of Ara-C transport which is named equilibrative nucleoside transport 1 (ENT1) is decreased fallowing HIF-1α activity [13]. According to Jin et al. increased HIF-1α activity originated from PI3K/AKT and MAPK/ERK signaling pathways that are activated directly by FLT3 [14]. Interestingly, increased activity of HIF-1α causes decreased expression of ENT1 [15].
The role of acid ceramidase overexpression in AML drug resistance
Acid ceramidase inhibition has also significantly improved survival in mice with AML. In AML, acid ceramidase plays a crucial role in blast survival and drug resistance [77].
Bcl-2 is a crucial factor in the apoptosis signaling pathways. Researchers find that Bcl-2 inhibition overcomes apoptosis resistance and significantly increases survival in models of resistant AML [78]. In addition, it was shown that acid ceramidase activity promotes the production of pro-survival Mcl-1 protein and confers resistance to Bcl-2 inhibition [12].
The combination of daunorubicin (dnr) and cytarabine (Ara-C) is a critical component of AML treatment and resistance to these drugs greatly contributes to the failure of treatments [79]. Using parental HL-60 cells and drug-resistant derivatives as the model, researchers found that acid ceramidase overexpression in HL-60 induced resistance to the AML chemotherapy drugs such as cytarabine, mitoxantrone, and daunorubicin [80]. Turner et al. [81] have concluded that acid ceramidase overexpression enhances autophagy in prostate cancer and that enhances autophagy increases resistance to ceramide. The findings imply that prostate cancer cells overexpressing acid ceramidase have a significantly higher amount of autophagy than parental cell lines, thus, resulting in an 'insult-ready' phenotype in which cells are more resistant to initial insult and can quickly metabolize any ceramide generated. Therefore, they believe that inhibition of autophagy improves treatment response. According to studies, acid ceramidase is significantly increased in AML blasts. Importantly, when sphingosine (SPH) is made by acid ceramidase, it is phosphorylated by sphingosine kinase (SPHK), and sphingosine 1 phosphate (S1P) levels rise in the AML blasts. This series of events causes upregulation of NF-κB (Fig. 1) [16]. NF-κB in AML blasts actives P-glycoprotein (P-gp)/multidrug resistance protein 1 (MDR1) promoter. Interestingly P-gp is a protein with a flexible structure that causes leaking out of the drugs that are used for AML treatment [17].
Conclusion and future directions
Chemo-resistance as the major challenge in AML therapy, reduces the survival rate and treatment failure in multiple malignancies such as AML. In order to overcome this serious issue, the mechanisms of chemo-resistance in AML must be uncovered. One of the mechanisms of chemo-resistance in AML is autophagy. In this study, we suggested that FLT3-ITD and high levels of acid ceramidase could lead to chemo-resistance by inducing autophagy. In patients with AML, some types of gene mutations such as FLT3-ITD, overexpression of some chemo-resistance-related enzymes such as acid ceramidase, and autophagy signaling pathways, lead to relapse and chemo-resistance. By targeting these adverse factors, chemo-resistance could be alleviated. Patients with AML must be evaluated to determine whether they harbor high-risk factors for drug resistance, as we suggested in this study FLT3-ITD and overexpression of acid ceramidase could be considered as adverse factors that lead to autophagy and chemo-resistance in these patients. Therefore, the combination of drugs targeting FLT3-ITD and its downstream signaling pathways, acid ceramidase, and autophagy with common chemotherapies might be a promising therapeutic approach for these AML patients in order to overcome chemo-resistance and improve long-term survival rate. However, further investigations are recommended to evaluate the interplay between FLT3-ITD and acid ceramidase in acute myeloid leukemia chemo-resistance and subsequently, more clinical, in-vitro, and in-vivo studies are needed to better understand these mechanisms and prepare therapeutic strategies for them.
Acknowledgements
Not applicable.
Abbreviations
- AML
Acute myeloid leukemia
- FLT3-ITD
Fms-like tyrosine kinase 3-internal tandem duplication
- ATG
Autophagy-related gene
- Ara-C
Cytarabine or cytosine arabinoside
- MDR
Multidrug resistance
- HIF-1α
Hypoxia-inducible factor-1 alpha
- ROS
Reactive oxygen species
- NF-κB
Nuclear factor kappa B
- PI3K
Phosphatidylinositol-3-kinase
- mTOR
Mammalian target of rapamycin
- MAPK
Mitogen-activated protein kinase
- ERK
Extracellular signal-regulated kinase
- Cer
Ceramide
- Sph
Sphingosine
- dHCer
Dihydroceramide
- dHSph
Dihydrosphingosine
- C1P
Ceramide-1-phosphate
- S1P
Sphingosine-1-phosphate
- 3KSR
3-Ketosphinganine reductase
- CPP
Ceramide phosphatase
- DES1
Dihydroceramide desaturase 1
- aCDase
Acid ceramidase
- nCDase
Neutral ceramidase
- SPHK
Sphingosine kinase
- aSMase
Acid sphingomyelinase
- SPL
S1P lyase
- S1PP
Sphingosine phosphate phosphatase
Author contributions
HZ conceived the study and designed the headings and study. HZ, MB, AA, FA, HS, ML, ZZ, and SY, wrote the manuscript text. MB, FA, and HZ created the figures. MGH, VT, and HZ supervised the study. HZ, MB, HS and MGH revised the study. All authors read and approved the final manuscript.
Funding
There is no foundation for this study.
Availability of data and materials
Not applicable.
Declarations
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors have read the manuscript and given their consent for publication.
Competing interests
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hamidreza Zalpoor, Email: hamidreza.zlpr1998@gmail.com.
Vahideh Tarhriz, Email: tarhrizv@tbzmed.ac.ir.
Mazdak Ganjalikhani-Hakemi, Email: mghakemi@med.mui.ac.ir.
References
- 1.Ameri M, Alipour M, Madihi M, Nezafat N. Identification of intrinsically disordered regions in hub genes of acute myeloid leukemia: a bioinformatics approach. Biotechnol Appl Biochem. 2021. [DOI] [PubMed]
- 2.Papaemmanuil E, Doehner H, Campbell PJ. Genomic classification in acute myeloid leukemia. N Engl J Med. 2016;375(9):900–901. doi: 10.1056/NEJMc1608739. [DOI] [PubMed] [Google Scholar]
- 3.Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Necochea-Campion R, Shouse GP, Zhou Q, Mirshahidi S, Chen C-S. Aberrant splicing and drug resistance in AML. J Hematol Oncol. 2016;9(1):1–9. doi: 10.1186/s13045-016-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuczynski EA, Sargent DJ, Grothey A, Kerbel RS. Drug rechallenge and treatment beyond progression—implications for drug resistance. Nat Rev Clin Oncol. 2013;10(10):571–587. doi: 10.1038/nrclinonc.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Gu Y, Chen B. Mechanisms of drug resistance in acute myeloid leukemia. Onco Targets Ther. 2019;12:1937. doi: 10.2147/OTT.S191621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luppi M, Fabbiano F, Visani G, Martinelli G, Venditti A. Novel agents for acute myeloid leukemia. Cancers. 2018;10(11):429. doi: 10.3390/cancers10110429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapoorian H, Zalpoor H, Ganjalikhani-Hakemi M. The correlation between Flt3-ITD mutation in dendritic cells with TIM-3 expression in acute myeloid leukemia. Blood Sci. 2021;3(04):132–135. doi: 10.1097/BS9.0000000000000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Short NJ, Konopleva M, Kadia TM, Borthakur G, Ravandi F, DiNardo CD, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506–525. doi: 10.1158/2159-8290.CD-19-1011. [DOI] [PubMed] [Google Scholar]
- 10.Larsson CA, Cote G, Quintás-Cardama A. The changing mutational landscape of acute myeloid leukemia and myelodysplastic syndrome. Mol Cancer Res. 2013;11(8):815–827. doi: 10.1158/1541-7786.MCR-12-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stone RM, DeAngelo DJ, Klimek V, Galinsky I, Estey E, Nimer SD, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105(1):54–60. doi: 10.1182/blood-2004-03-0891. [DOI] [PubMed] [Google Scholar]
- 12.Tan S-F, Liu X, Fox TE, Barth BM, Sharma A, Turner SD, et al. Acid ceramidase is upregulated in AML and represents a novel therapeutic target. Oncotarget. 2016;7(50):83208. doi: 10.18632/oncotarget.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan S-F, Pearson JM, Feith DJ, Loughran TP., Jr The emergence of acid ceramidase as a therapeutic target for acute myeloid leukemia. Expert Opin Ther Targets. 2017;21(6):583–590. doi: 10.1080/14728222.2017.1322065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeidan YH, Jenkins RW, Korman JB, Liu X, Obeid LM, Norris JS, et al. Molecular targeting of acid ceramidase: implications to cancer therapy. Curr Drug Targets. 2008;9(8):653–661. doi: 10.2174/138945008785132358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther GG, Peralta ER, Rosales KR, Wong SY, Siskind LJ, Edinger AL. Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc Natl Acad Sci. 2008;105(45):17402–17407. doi: 10.1073/pnas.0802781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang J, Chen W-M, Liu S, Wang Z-H, Wei T-N, Chen Z-Z, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. 2019;85:106198. doi: 10.1016/j.leukres.2019.106198. [DOI] [PubMed] [Google Scholar]
- 18.Ding L, Zhang W, Yang L, Pelicano H, Zhou K, Yin R, et al. Targeting the autophagy in bone marrow stromal cells overcomes resistance to vorinostat in chronic lymphocytic leukemia. Onco Targets Ther. 2018;11:5151. doi: 10.2147/OTT.S170392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desantis V, Saltarella I, Lamanuzzi A, Mariggiò M, Racanelli V, Vacca A, et al. Autophagy: a new mechanism of prosurvival and drug resistance in multiple myeloma. Transl Oncol. 2018;11(6):1350–1357. doi: 10.1016/j.tranon.2018.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, et al. Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis. 2013;4(10):e838-e. doi: 10.1038/cddis.2013.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y-L, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, et al. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Can Res. 2012;72(7):1773–1783. doi: 10.1158/0008-5472.CAN-11-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zalpoor H, Rezaei M, Yahyazadeh S, Ganjalikhani-Hakemi M. Flt3-ITD mutated acute myeloid leukemia patients and COVID-19: potential roles of autophagy and HIF-1α in leukemia progression and mortality. Hum Cell. 2022;35:1304–1305. doi: 10.1007/s13577-022-00718-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zalpoor H, Akbari A, Nayerain Jazi N, Liaghat M, Bakhtiyari M. Possible role of autophagy induced by COVID-19 in cancer progression, chemo-resistance, and tumor recurrence. Infect Agents Cancer. 2022;17(1):1–4. doi: 10.1186/s13027-022-00450-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu D, Chen Y, Yang Y, Yin Z, Huang C, Wang Q, et al. Autophagy activation mediates resistance to FLT3 inhibitors in acute myeloid leukemia with FLT3-ITD mutation. J Transl Med. 2022;20(1):1–12. doi: 10.1186/s12967-022-03498-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heydt Q, Larrue C, Saland E, Bertoli S, Sarry J, Besson A, et al. Oncogenic FLT3-ITD supports autophagy via ATF4 in acute myeloid leukemia. Oncogene. 2018;37(6):787–797. doi: 10.1038/onc.2017.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudat S, Pfaus A, Cheng Y, Holtmann J, Ellegast JM, Buehler C, et al. RET-mediated autophagy suppression as targetable co-dependence in acute myeloid leukemia. Leukemia. 2018;32(10):2189–2202. doi: 10.1038/s41375-018-0102-4. [DOI] [PubMed] [Google Scholar]
- 27.Wöhrle FU, Daly RJ, Brummer T. Function, regulation and pathological roles of the Gab/DOS docking proteins. Cell Commun Signal. 2009;7(1):1–28. doi: 10.1186/1478-811X-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma S, Vaughan T, Bunting KD. Gab adapter proteins as therapeutic targets for hematologic disease. Adv Hematol. 2012;2012:380635. doi: 10.1155/2012/380635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hospital M-A, Green AS, Maciel TT, Moura IC, Leung AY, Bouscary D, et al. FLT3 inhibitors: clinical potential in acute myeloid leukemia. Onco Targets Ther. 2017;10:607. doi: 10.2147/OTT.S103790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada K, Nogami A, Ishida S, Akiyama H, Chen C, Umezawa Y, et al. FLT3-ITD induces expression of Pim kinases through STAT5 to confer resistance to the PI3K/Akt pathway inhibitors on leukemic cells by enhancing the mTORC1/Mcl-1 pathway. Oncotarget. 2018;9(10):8870. doi: 10.18632/oncotarget.22926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalle Pezze P, Ruf S, Sonntag AG, Langelaar-Makkinje M, Hall P, Heberle AM, et al. A systems study reveals concurrent activation of AMPK and mTOR by amino acids. Nat Commun. 2016;7(1):1–19. doi: 10.1038/ncomms13254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Ganley IG, Lam DH, Wang J, Ding X, Chen S, Jiang X. ULK1· ATG13· FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284(18):12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. 2012;8(6):903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puertollano R. mTOR and lysosome regulation. F1000prime Rep. 2014;6:52. doi: 10.12703/P6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao C, Obeid LM. Ceramidases: regulators of cellular responses mediated by ceramide, sphingosine, and sphingosine-1-phosphate. Biochim Biophys Acta. 2008;1781(9):424–434. doi: 10.1016/j.bbalip.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olivera A, Kohama T, Edsall L, Nava V, Cuvillier O, Poulton S, et al. Sphingosine kinase expression increases intracellular sphingosine-1-phosphate and promotes cell growth and survival. J Cell Biol. 1999;147(3):545–558. doi: 10.1083/jcb.147.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee O-H, Kim Y-M, Lee YM, Moon E-J, Lee D-J, Kim J-H, et al. Sphingosine 1-phosphate induces angiogenesis: its angiogenic action and signaling mechanism in human umbilical vein endothelial cells. Biochem Biophys Res Commun. 1999;264(3):743–750. doi: 10.1006/bbrc.1999.1586. [DOI] [PubMed] [Google Scholar]
- 40.Bedia C, Levade T, Codogno P. Regulation of autophagy by sphingolipids. Anticancer Agents Med Chem. 2011;11(9):844–853. doi: 10.2174/187152011797655131. [DOI] [PubMed] [Google Scholar]
- 41.Yamagata M, Obara K, Kihara A. Sphingolipid synthesis is involved in autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2011;410(4):786–791. doi: 10.1016/j.bbrc.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 42.Sims K, Haynes CA, Kelly S, Allegood JC, Wang E, Momin A, et al. Kdo2-lipid A, a TLR4-specific agonist, induces de novo sphingolipid biosynthesis in RAW264. 7 macrophages, which is essential for induction of autophagy. J Biol Chem. 2010;285(49):38568–38579. doi: 10.1074/jbc.M110.170621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng X, Overmeyer JH, Maltese WA. Functional specificity of the mammalian Beclin-Vps34 PI 3-kinase complex in macroautophagy versus endocytosis and lysosomal enzyme trafficking. J Cell Sci. 2006;119(2):259–270. doi: 10.1242/jcs.02735. [DOI] [PubMed] [Google Scholar]
- 44.Scarlatti F, Bauvy C, Ventruti A, Sala G, Cluzeaud F, Vandewalle A, et al. Ceramide-mediated macroautophagy involves inhibition of protein kinase B and up-regulation of beclin 1. J Biol Chem. 2004;279(18):18384–18391. doi: 10.1074/jbc.M313561200. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Li S, Qin X, Hou W, Dong H, Yao L, et al. The pleiotropic roles of sphingolipid signaling in autophagy. Cell Death Dis. 2014;5(5):e1245-e. doi: 10.1038/cddis.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai M, La Rocca V, Amato R, Freer G, Pistello M. Sphingolipid/ceramide pathways and autophagy in the onset and progression of melanoma: novel therapeutic targets and opportunities. Int J Mol Sci. 2019;20(14):3436. doi: 10.3390/ijms20143436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Brocklyn JR, Williams JB. The control of the balance between ceramide and sphingosine-1-phosphate by sphingosine kinase: oxidative stress and the seesaw of cell survival and death. Comp Biochem Physiol B: Biochem Mol Biol. 2012;163(1):26–36. doi: 10.1016/j.cbpb.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 48.Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. 2019;9(9):1167–1181. doi: 10.1158/2159-8290.CD-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du W, Xu A, Huang Y, Cao J, Zhu H, Yang B, et al. The role of autophagy in targeted therapy for acute myeloid leukemia. Autophagy. 2020;17:2665–2679. doi: 10.1080/15548627.2020.1822628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piya S, Kornblau SM, Ruvolo VR, Mu H, Ruvolo PP, McQueen T, et al. Atg7 suppression enhances chemotherapeutic agent sensitivity and overcomes stroma-mediated chemoresistance in acute myeloid leukemia. Blood. 2016;128(9):1260–1269. doi: 10.1182/blood-2016-01-692244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auberger P, Puissant A. Autophagy, a key mechanism of oncogenesis and resistance in leukemia. Blood. 2017;129(5):547–552. doi: 10.1182/blood-2016-07-692707. [DOI] [PubMed] [Google Scholar]
- 52.Liu X, Cheng JC, Turner LS, Elojeimy S, Beckham TH, Bielawska A, et al. Acid ceramidase upregulation in prostate cancer: role in tumor development and implications for therapy. Expert Opin Ther Targets. 2009;13(12):1449–1458. doi: 10.1517/14728220903357512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Realini N, Palese F, Pizzirani D, Pontis S, Basit A, Bach A, et al. Acid ceramidase in melanoma: expression, localization, and effects of pharmacological inhibition. J Biol Chem. 2016;291(5):2422–2434. doi: 10.1074/jbc.M115.666909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turner LS, Cheng JC, Beckham TH, Keane T, Norris J, Liu X. Autophagy is increased in prostate cancer cells overexpressing acid ceramidase and enhances resistance to C 6 ceramide. Prostate Cancer Prostatic Dis. 2011;14(1):30–37. doi: 10.1038/pcan.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lai M, Amato R, La Rocca V, Bilgin M, Freer G, Spezia P, et al. Acid ceramidase controls apoptosis and increases autophagy in human melanoma cells treated with doxorubicin. Sci Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-90219-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stiban J, Tidhar R, Futerman AH. Ceramide synthases: roles in cell physiology and signaling. In: Chalfant C, Poeta MD, editors. Sphingolipids as signaling and regulatory molecules. New York: Springer; 2010. pp. 60–71. [DOI] [PubMed] [Google Scholar]
- 57.Gault CR, Obeid LM, Hannun YA. An overview of sphingolipid metabolism: from synthesis to breakdown. In: Chalfant C, Poeta MD, editors. Sphingolipids as signaling and regulatory molecules. New York: Springer; 2010. pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huwiler A, Zangemeister-Wittke U. Targeting the conversion of ceramide to sphingosine 1-phosphate as a novel strategy for cancer therapy. Crit Rev Oncol Hematol. 2007;63(2):150–159. doi: 10.1016/j.critrevonc.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Kolesnick RN, Krönke M. Regulation of ceramide production and apoptosis. Annu Rev Physiol. 1998;60(1):643–665. doi: 10.1146/annurev.physiol.60.1.643. [DOI] [PubMed] [Google Scholar]
- 60.Alexaki A, Gupta SD, Majumder S, Kono M, Tuymetova G, Harmon JM, et al. Autophagy regulates sphingolipid levels in the liver [S] J Lipid Res. 2014;55(12):2521–2531. doi: 10.1194/jlr.M051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu K, Czaja M. Regulation of lipid stores and metabolism by lipophagy. Cell Death Differ. 2013;20(1):3–11. doi: 10.1038/cdd.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, et al. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, et al. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Curr Biol. 2014;24(6):609–620. doi: 10.1016/j.cub.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Du W, Xu A, Huang Y, Cao J, Zhu H, Yang B, et al. The role of autophagy in targeted therapy for acute myeloid leukemia. Autophagy. 2021;17(10):2665–2679. doi: 10.1080/15548627.2020.1822628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chang H, Zou Z. Targeting autophagy to overcome drug resistance: further developments. J Hematol Oncol. 2020;13(1):1–18. doi: 10.1186/s13045-020-01000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu S-Y, Wen Y-C, Ku C-C, Yang Y-C, Chow J-M, Yang S-F, et al. Penfluridol triggers cytoprotective autophagy and cellular apoptosis through ROS induction and activation of the PP2A-modulated MAPK pathway in acute myeloid leukemia with different FLT3 statuses. J Biomed Sci. 2019;26(1):1–13. doi: 10.1186/s12929-019-0557-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ravanan P, Srikumar IF, Talwar P. Autophagy: The spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 68.Hu X, Mei S, Meng W, Xue S, Jiang L, Yang Y, et al. CXCR4-mediated signaling regulates autophagy and influences acute myeloid leukemia cell survival and drug resistance. Cancer Lett. 2018;425:1–12. doi: 10.1016/j.canlet.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 69.Huang F-T, Sun J, Zhang L, He X, Zhu Y-H, Dong H-J, et al. Role of SIRT1 in hematologic malignancies. J Zhejiang Univ Sci B. 2019;20(5):391–398. doi: 10.1631/jzus.B1900148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva VR, Neves SP, Santos LS, Dias RB, Bezerra DP. Challenges and therapeutic opportunities of autophagy in cancer therapy. Cancers. 2020;12(11):3461. doi: 10.3390/cancers12113461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Konig H, Levis M. Targeting FLT3 to treat leukemia. Expert Opin Ther Targets. 2015;19(1):37–54. doi: 10.1517/14728222.2014.960843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bagrintseva K, Geisenhof S, Kern R, Eichenlaub S, Reindl C, Ellwart JW, et al. FLT3-ITD-TKD dual mutants associated with AML confer resistance to FLT3 PTK inhibitors and cytotoxic agents by overexpression of Bcl-x (L) Blood. 2005;105(9):3679–3685. doi: 10.1182/blood-2004-06-2459. [DOI] [PubMed] [Google Scholar]
- 73.Breitenbuecher F, Markova B, Kasper S, Carius B, Stauder T, Böhmer FD, et al. A novel molecular mechanism of primary resistance to FLT3-kinase inhibitors in AML. Blood. 2009;113(17):4063–4073. doi: 10.1182/blood-2007-11-126664. [DOI] [PubMed] [Google Scholar]
- 74.Dany M, Gencer S, Nganga R, Thomas RJ, Oleinik N, Baron KD, et al. Targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML. Blood. 2016;128(15):1944–1958. doi: 10.1182/blood-2016-04-708750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith CC, Wang Q, Chin C-S, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 2012;485(7397):260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang J, Lindström HJG, Friedman R. Combating drug resistance in acute myeloid leukaemia by drug rotations: the effects of quizartinib and pexidartinib. Cancer Cell Int. 2021;21(1):1–14. doi: 10.1186/s12935-021-01856-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan S-F. Role of acid ceramidase in regulating survival and drug resistance in acute myeloid leukemia. 2013.
- 78.Pan R, Ruvolo V, Mu H, Leverson JD, Nichols G, Reed JC, et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell. 2017;32(6):748–760.e6. doi: 10.1016/j.ccell.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kao L-P, Morad SA, Davis TS, MacDougall MR, Kassai M, Abdelmageed N, et al. Chemotherapy selection pressure alters sphingolipid composition and mitochondrial bioenergetics in resistant HL-60 cells. J Lipid Res. 2019;60(9):1590–1602. doi: 10.1194/jlr.RA119000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan S-F, Dunton W, Liu X, Fox TE, Morad SA, Desai D, et al. Acid ceramidase promotes drug resistance in acute myeloid leukemia through NF-κB-dependent P-glycoprotein upregulation. J Lipid Res. 2019;60(6):1078–1086. doi: 10.1194/jlr.M091876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Turner LS, Cheng JC, Beckham TH, Keane T, Norris J, Liu X. Autophagy is increased in prostate cancer cells overexpressing acid ceramidase and enhances resistance to C6 ceramide. Prostate Cancer Prostatic Dis. 2011;14(1):30–37. doi: 10.1038/pcan.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.