Abstract
Autophagy is an intracellular degradation system involving de novo generation of autophagosomes, which function as a transporting vesicle of cytoplasmic components to lysosomes for degradation. Isolation membranes (IMs) or phagophores, the precursor membranes of autophagosomes, require millions of phospholipids to expand and transform into autophagosomes, with the endoplasmic reticulum (ER) being the primary lipid source. Recent advances in structural and biochemical studies of autophagy-related proteins have revealed their lipid transport activities: Atg2 at the interface between IM and ER possesses intermembrane lipid transfer activities, while Atg9 at IM and VMP1 and TMEM41B at ER possess lipid scrambling activities. In this review, we summarize recent advances in the establishment of the lipid transport activities of these proteins and their collaboration mechanisms for lipid transport between the ER and IM, and further discuss how unidirectional lipid transport from the ER to IM occurs during autophagosome formation.
Autophagy is a lysosomal degradation mechanism for intracellular components. For lysosomal degradation, cytoplasmic components are delivered to lysosomes by carriers known as autophagosomes during macroautophagy (referred to as autophagy hereafter). Autophagosomes are double-membrane-bound organelles as large as ∼1 µm that are de novo generated upon autophagy induction. During autophagosome formation, cytoplasmic components, including proteins, organelles, and invasive microbes, are sequestered into the lumen of autophagosomes, which are eventually delivered to the lysosomal lumen upon the fusion of autophagosome with lysosome and degraded by lysosomal hydrolases (Yang and Klionsky 2010; Mizushima et al. 2011; Nakatogawa 2020). Autophagosome formation occurs at the proximity of the endoplasmic reticulum (ER), especially the ER exit site (ERES) in the case of budding yeast (Graef et al. 2013; Suzuki et al. 2013). Autophagosome formation requires a large amount of phospholipids, which are obtained from various organelles (Lamb et al. 2013; Melia et al. 2020; Gómez-Sánchez et al. 2021). In particular, ER is believed to be the primary phospholipid source for autophagosome formation because the autophagosome intermediate termed isolation membrane (IM) or phagophore is tightly linked to the ER membrane (Axe et al. 2008; Hayashi-Nishino et al. 2009; Ylä-Anttila et al. 2009). The mechanism by which phospholipids at the ER are delivered to the IM has long been a mystery.
Proteins responsible for autophagosome formation were initially identified in budding yeast, called autophagy-related (Atg), and assigned numbers (ATG is used for mammalian counterparts) (Nakatogawa 2020). Among them, 16 Atg proteins are evolutionarily conserved from yeast to humans, including the Atg1 kinase complex that undergoes phase separation to concentrate Atg proteins, the phosphatidylinositol (PI) kinase complex that produces PI-3 phosphate (PI3P), the PI3P-binding protein Atg18, the Atg8 and Atg12 conjugation systems that decorate autophagic membranes, and the less characterized Atg2 and Atg9 proteins, involved in lipid transport from the ER to the autophagic membranes (Fig. 1; Nakatogawa 2020; Matoba and Noda 2021).
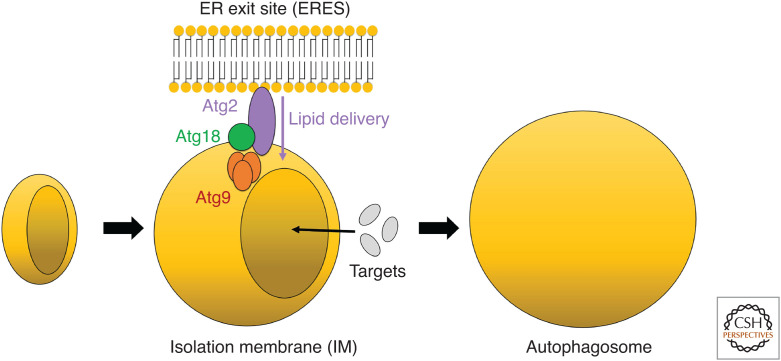
Figure 1.
Schematic drawing of autophagosomal membrane expansion in budding yeast. The Atg2-Atg18 complex and Atg9 at the expanding edge of the isolation membrane (IM) contact the endoplasmic reticulum exit site (ERES) and are considered to be involved in lipid transfer from the ERES to the IM.
In this review, we describe the current model of autophagosomal membrane expansion mediated by Atg2, Atg9, and ER-resident factors, especially focusing on their structures, and we discuss the molecular mechanisms of unidirectional lipid transport from the ER to IM. For detailed biological aspects of these proteins, please refer to other recent reviews (Osawa and Noda 2019; Lees and Reinisch 2020; Noda 2021).
A NEW MECHANISM OF BULK LIPID DELIVERY BETWEEN ORGANELLES
In autophagy, IM is thought to expand using phospholipids primarily derived from the ER to mature into the autophagosome, which contains little membrane proteins (Baba et al. 1995; Fengsrud et al. 2000). Autophagosomes are formed within 10 min, necessitating the rapid transfer of a large amount of phospholipids from the ER to IM. High-speed bulk transfer of lipids was thought to be mediated by direct connection between organelles or vesicular transport. However, it is unclear whether these mechanisms apply to bulk lipid transfer in autophagosome formation because they would transfer membrane proteins in addition to lipids. The Atg2-mediated lipid transfer has recently been proposed as a third mechanism for lipid delivery from ER to IM (Maeda et al. 2019; Osawa et al. 2019, 2020; Valverde et al. 2019). Atg2 is a large protein (composed of 1592 residues for yeast Atg2) and one of the evolutionarily conserved essential factors for autophagosome formation. During autophagosome formation, Atg2 forms a complex with Atg18 that binds PI3P and interacts with a transmembrane protein Atg9 embedded in the IM, enabling Atg2 to localize at the edge of the expanding IM (Obara et al. 2008; Gómez-Sánchez et al. 2018). Moreover, Atg2 contacts with the ERES, a subdomain of the ER responsible for COPII vesicle generation, by an unknown mechanism (Graef et al. 2013; Suzuki et al. 2013). Atg2 was first reported to mediate tethering between the ER and the IM at IM-ERES contact sites (Chowdhury et al. 2018; Kotani et al. 2018), but recent breakthroughs in structural biology of Atg2 revealed that Atg2 is a novel type of a lipid transfer protein possessing both membrane tethering and lipid transfer activities, proposing a model in which Atg2 delivers bulk lipids from the ER to the IM while inhibiting the flow of membrane proteins to the IM, enabling IM expansion (Fig. 1; Maeda et al. 2019; Osawa et al. 2019; Valverde et al. 2019).
Canonical lipid transfer proteins or their lipid-binding domains are considered to shuttle between membranes to transfer lipids (Lev 2010; Wong et al. 2019). Contrary to this principle, Atg2 tethers membranes instead of shuttling between membranes for efficient lipid transfer. The membrane tethering activity of Atg2 significantly depends on membrane curvature. In vitro, Atg2 efficiently tethers small liposomes (<100 nm in diameter) but exhibits little membrane tethering activity for larger liposomes (Osawa et al. 2019). Atg18 and PI3P enhance the membrane tethering activity of Atg2. Membrane tethering analysis of the ATG2A-WIPI4 complex, a human counterpart of the Atg2-Atg18 complex, revealed that ATG2A can tether PI3P-containing large liposomes through an interaction between WIPI4 and PI3P (Chowdhury et al. 2018). The membrane tethering activity of Atg2 is closely associated with the lipid transfer activity of Atg2. Atg2 can mediate efficient lipid transfer only between membranes tethered to each other by Atg2 (Maeda et al. 2019; Osawa et al. 2019). Therefore, the IM-ERES contact site is a suitable place for Atg2-mediated lipid transfer because both the ERES and edge of the IM could be highly curved. The amino- and carboxy-terminal regions of Atg2 (residues 2–22 and residues 1347–1592) encompass membrane-binding sites, which are crucial for the membrane tethering activity of Atg2; however, either is adequate for the membrane-binding activity (Kotani et al. 2018). The substitution of amino-terminal conserved basic residues (residues 10–12) with aspartate significantly reduced membrane tethering and lipid transfer activities of Atg2 and impaired autophagic activity in vivo, demonstrating a direct relationship between in vitro activities and in vivo autophagic activities of Atg2 (Kotani et al. 2018; Osawa et al. 2019).
LIPID SPECIFICITY AND TRANSFER RATE OF ATG2
Lipid-binding analyses of Atg2 and ATG2A revealed that Atg2 efficiently binds to phospholipids with minimal specificity to their groups and ∼20 phospholipids can be loaded into one Atg2 molecule at once (Osawa et al. 2019; Valverde et al. 2019). On the other hand, the binding affinity of Atg2 for cholesterol and fatty acid is significantly weaker than for phospholipids (Osawa et al. 2019). These observations indicate that Atg2 mainly transfers phospholipids, which is reasonable because autophagic membranes are primarily composed of phospholipids (Schütter et al. 2020). The lipid transfer rate of Atg2 must be fast enough to achieve autophagosome formation. The autophagosome with a diameter of 1 µm is estimated to contain ∼25 million phospholipids. To form such autophagosomes within 10 min, one Atg2 molecule needs to transfer approximately 42 phospholipids per second, assuming that 100 Atg2 molecules work at the ERES-IM contact site and other pathways for lipid delivery are not considered. Unfortunately, no accurate lipid transfer rate of Atg2 has been experimentally determined. The lipid transfer activity of Atg2 in vitro is detected by a lipid transfer assay based on fluorescence resonance energy transfer (FRET). The FRET-based lipid transfer assay was originally developed for lipid transfer proteins shuttling between membranes (Connerth et al. 2012) and is therefore unsuitable for the correct evaluation of the lipid transfer activity of Atg2. Thus, the Atg2-mediated lipid transfer rate was indirectly estimated by applying a special kinetic model in which phospholipids bidirectionally move between membranes using Atg2 as a bridge. According to the kinetic analysis, Atg2 and ATG2A transfer at a rate of approximately 750 and 115 phospholipids per second and protein, respectively, which are sufficient for autophagosome formation (von Bulow and Hummer 2020).
THE STRUCTURAL BASIS OF ATG2-MEDIATED LIPID TRANSFER
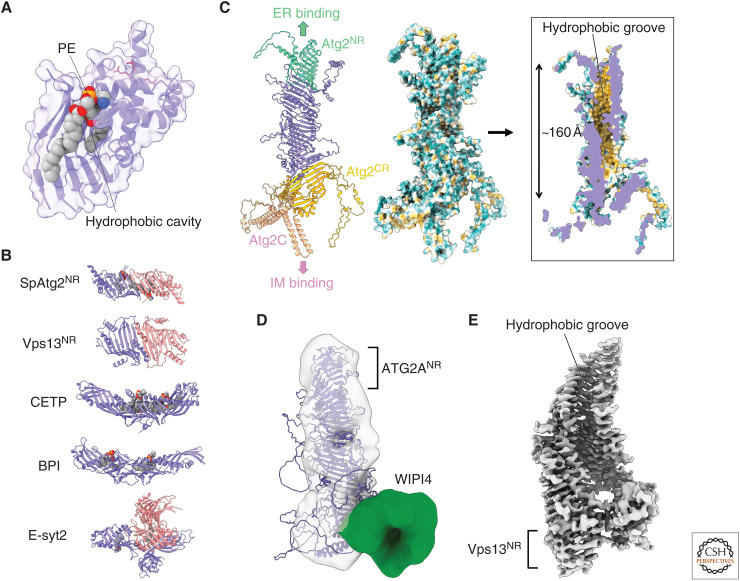
The amino-terminal region of Atg2 (Atg2NR), which contains about 240 amino acid residues, can extract phospholipids from membranes (Osawa et al. 2019). Additionally, the GFP-tagged amino-terminal 46 residues alone localize to the ER (Kotani et al. 2018). These findings imply that Atg2-mediated lipid transfer from ER to IM starts at Atg2NR. This critical function of Atg2NR was initially discovered by a crystallographic study on Schizosaccharomyces pombe Atg2NR (SpAtg2NR) (Osawa et al. 2019). To date, available atomic resolution structures related to Atg2 have been limited to SpAtg2NR and its phosphatidylethanolamine (PE)-bound form. The PE-bound SpAtg2NR structure demonstrated the phospholipid-binding manner of Atg2 (Fig. 2A). SpAtg2NR adopts a vase-like fold harboring a hydrophobic cavity largely composed of β-sheets. Mutational analysis on human ATG2A revealed that hydrophobic residues constructing the cavity are important for phospholipid transfer activity in vitro and autophagy activity in vivo (Valverde et al. 2019). The basic residues responsible for membrane binding in budding yeast Atg2 are located at the first α-helix and form the outer surface of the vase. SpAtg2NR enwraps acyl chains of PE through its hydrophobic cavity but has little interactions with the PE head group, except for an electrostatic interaction between the phosphoryl group and arginine residues. For these structural reasons, Atg2 exhibits broad binding specificity to phospholipids. The phospholipid-binding mode of SpAtg2NR is quite similar to that of the synaptotagmin-like mitochondrial-lipid-binding (SMP) domains, which act as a direct lipid carrier in lipid transfer proteins that localize at organelle contact sites (Jeong et al. 2016; Reinisch and De Camilli 2016; Kawano et al. 2018). However, compared to SMP domains, SpAtg2NR exhibits little lipid transfer activity. As a result, Atg2NR cannot unload lipids into membranes, and additional regions other than Atg2NR are required for lipid unloading and efficient lipid transfer. Conversely, the amino-terminal region of ATG2A (ATG2ANR), consisting of 345 residues, possesses the lipid transfer activity, and ATG2ANR overexpression restored autophagic activity in ATG2A/ATG2B double knock out HEK293 cells (Valverde et al. 2019). Thus, ATG2ANR is regarded as a minimum region essential for lipid delivery in mammalian autophagy. However, we must carefully interpret this ATG2ANR analysis for the following reasons. In the lipid transfer assay, ATG2ANR was engineered to bind to membranes through a membrane-binding tag. Moreover, ATG2ANR might form a capsule-like dimer in which two hydrophobic cavities are directly connected to sequester themselves from the water phase, as SpAtg2NR and Atg2-related lipid transfer protein Vps13 form capsule-like dimers in crystals (Fig. 2B; Kumar et al. 2018; Osawa et al. 2019). In various lipid transfer proteins, capsule-like structures formed by two SMP domains function as a lipid carrier (Fig. 2B). Thus, ATG2ANR might form functional SMP-domain-like dimers with lipid transfer activity.
Figure 2.
Structural basis of Atg2-mediated lipid transfer. (A) Crystal structure of phosphatidylethanolamine (PE)-bound SpAtg2NR (PDB 6A9J). Basic residues responsible for the membrane tethering activity of Atg2 are shown with red stick models. All structural models in this manuscript were prepared using PyMOL. (B) Capsule-like structures found in various lipid transfer proteins: SpAtg2NR, the amino-terminal region of Vps13 (Vps13NR; PDB 6CBC), cholesteryl ester transfer protein (CETP; PDB 2OBD), bactericidal permeability-increasing protein (BPI; PDB 1BP1), and extended synaptotagmin 2 (E-syt2; PDB 4P42). Monomers in capsule-like structures are colored in slate or salmon. Lipids bound to capsule-like structures are displayed as a sphere model. (C) AlphaFold structure of Atg2. (Left) Ribbon model of Atg2 whose domains are colored differently. (Middle and right) Outer and inner surface model of Atg2 in which hydrophobicity is shown in Kyte–Doolittle scale. The most hydrophobic region is colored in orange-red. (D) Fitting of the AlphaFold structure of ATG2A to the electron microscopy (EM) density map of the ATG2A-WIPI4 complex (EMD-8899). (E) Cryo-EM density map of the amino-terminal fragment of Chaetomium thermophilum VPS13 (EMD-21113).
OVERALL STRUCTURE OF ATG2 PREDICTED BY AlphaFold
AlphaFold is an artificial intelligence system that predicts protein structures based on amino acid sequences (Jumper et al. 2021). Its high prediction accuracy is revolutionizing structural biology. We can easily access AlphaFold structures of full-length Atg2 from various species on AlphaFold Protein Structure Database. The AlphaFold structure of Atg2 has a rod-like shape, which is consistent with the low-resolution electron microscopy (EM) density map of the ATG2A-WIPI4 complex (Fig. 2C,D; Chowdhury et al. 2018). A major portion of the Atg2 structure consists of β-sheets, meanwhile α-helices are locally found at both ends of the rod. A striking structural feature of Atg2 is a ∼160 Å-long groove with the inner surface mostly composed of hydrophobic residues (Fig. 2C, right). The hydrophobic groove exists along the long axis of Atg2 with its slit forming a spiral. Atg2NR forms one end of the hydrophobic groove and its hydrophobic cavity directly connects to the downstream groove. The inner diameter of the hydrophobic cavity of Atg2NR (∼15 Å) is almost similar to that of the downstream groove, indicating that lipids loaded into Atg2NR can move within the hydrophobic groove. Cryo-electron microscopy analysis revealed that the amino-terminal fragment of VPS13 (residues 1–1390), which contains the Atg2NR-like region, also harbors a long hydrophobic groove (Fig. 2E; Li et al. 2020). Atg2NR would essentially function as a component of the long hydrophobic groove. The carboxy-terminal region of Atg2 (Atg2CR), which structurally resembles Atg2NR, forms the other end of the hydrophobic groove (Fig. 2C). Atg2CR contains the Atg9-interacting region. In AlphaFold structures of ATG2, the WIPI4-interacting region known as the Y/HF motif or the WIR motif (Zheng et al. 2017; Ren et al. 2020) is located at the region corresponding to Atg2CR. The relative location of ATG2ANR and WIPI4-interacting region in the AlphaFold structure is consistent with that in EM analysis of the ATG2A-WIPI4 complex (Fig. 2D; Chowdhury et al. 2018). Since Atg18 directly recognizes PI3P in IM and Atg9 is a transmembrane protein embedded in IM, Atg2CR would contact the IM and unload lipids into it. Atg2CR is followed by the carboxy-terminal membrane-binding region of Atg2 (Atg2C) (Fig. 2C). Atg2C folds into a crescent shape and contains a helix bundle composed of four amphipathic α-helices forming a hydrophobic concave surface. Substitution of hydrophobic residues of the concave surface with acidic residues impaired the membrane tethering activity of Atg2 and hindered Atg2 from localizing to the initiation site for autophagosome formation (Kotani et al. 2018). In addition to the Atg18- and Atg9-mediated indirect interactions with IM, Atg2 may directly interact with the highly curved edge of IMs using the concave surface of Atg2C. All regions essential for interaction with IM are located at the opposite side of Atg2NR within the rod-like architecture, enabling Atg2 to bridge between the ERES and IM in the determined orientation. The AlphaFold structure suggests that after lipids are extracted from the ER by Atg2NR, they travel to the IM through the 160 Å-long groove. Mutational studies based on the AlphaFold structure of Atg2 are required to validate this hypothesis.
MECHANISM OF DISTRIBUTING PHOSPHOLIPIDS TO BOTH LEAFLETS OF THE IM FOR EXPANSION
Since Atg2 is not a transmembrane protein, it is considered to transfer phospholipids between the cytosolic leaflets of the ER and IM. Therefore, phospholipids transferred from the ER to the IM by Atg2 would accumulate at the cytosolic leaflet of the IM if they were not transferred to the luminal leaflet of the IM, destabilizing rather than expanding the membrane structure. To achieve IM expansion, delivered phospholipids must be distributed to both leaflets of the IM. The efficient movement of phospholipids between membrane leaflets, known as a flip-flop, requires transmembrane proteins, such as lipid flippases, floppases, and scramblases (Sharom 2011; Nagata et al. 2020). Flippases and floppases mediate unidirectional transport of specific lipids between membrane leaflets using ATP, whereas scramblases mediate bidirectional transport of lipids with low specificity without an ATP requirement. Recent in vitro analyses revealed that Atg9, the sole core Atg transmembrane protein, can transfer fluorescently labeled phospholipids across leaflets using dithionite and BSA extraction assays, which is routinely employed for detecting lipid scrambling activities (Matoba et al. 2020). This assay was also applied to human ATG9A, revealing that ATG9A possesses the same activity (Maeda et al. 2020). These activities did not require ATP and showed little specificity to the head group of phospholipids for both yeast Atg9 and human ATG9A, indicating that they are phospholipid scramblases. Moreover, the PI3P transport assay was designed to detect the activity to transfer PI3P, a natural phospholipid known to exist at the IM, between membrane leaflets, demonstrating that both yeast Atg9 and human ATG9A possess the activity to transport PI3P between membrane leaflets (Matoba et al. 2020). These data strongly indicate that Atg9-family proteins are phospholipid scramblases, although the transport efficiency between the two directions (cytosolic-to-luminal leaflet vs. luminal-to-cytosolic leaflet) has not been characterized experimentally.
THE STRUCTURAL BASIS OF Atg9-MEDIATED LIPID SCRAMBLING
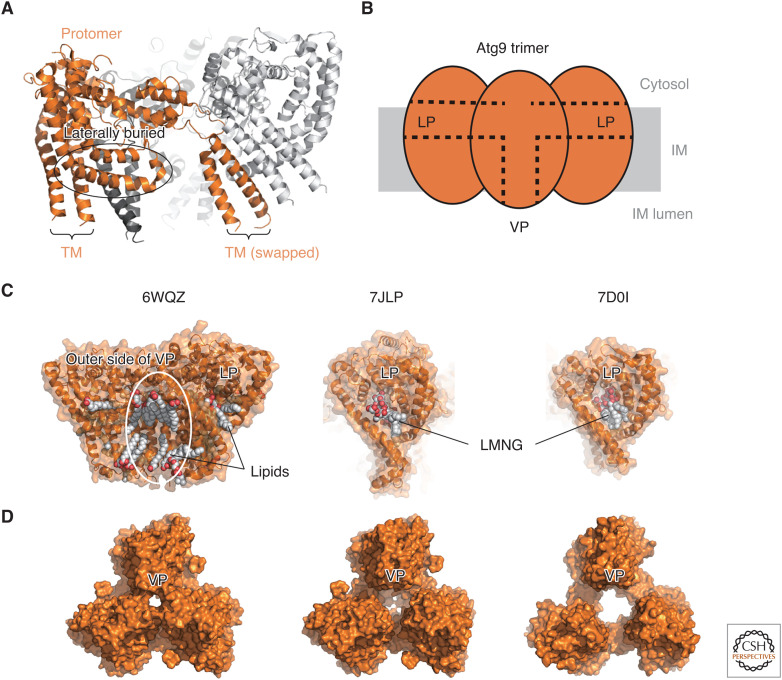
Until recently, the structure of Atg9 has been unknown. From the amino-acid sequence, Atg9 was proposed to possess six to eight (most probably six) transmembrane helices, with large intrinsically disordered regions at both termini (Noda et al. 2000; Young et al. 2006). Furthermore, it was proposed that Atg9 functions as a multimer, although the exact number of assemblies is unknown (He et al. 2008). Cryo-EM analysis was first applied to plant ATG9, revealing that it forms a trimer with C3 symmetry (Lai et al. 2020). However, due to the low resolution, structural models were not built, and the number of transmembrane helices was estimated to be six. Later, cryo-EM analysis of human ATG9A and fission yeast Atg9 yielded a high-resolution density map and a structural model, revealing the conserved unique architecture of Atg9-family proteins (Fig. 3A; Guardia et al. 2020; Maeda et al. 2020; Matoba et al. 2020). Mainly three unexpected structural features have been unveiled. First, Atg9-family proteins possess four, not six, transmembrane helices. In addition to four transmembrane helices, two helices are laterally buried in the cytosolic leaflet, which do not penetrate the membrane. Second, Atg9-family proteins form a domain-swapped trimer, with two transmembrane helices bound to another Atg9 molecule within the trimer. Third, Atg9-family proteins possess two unique pores, one is located at the center of each protomer and lateral to the cytosolic leaflet of the membrane (termed lateral pore [LP]), whereas another is located at the trimer center and penetrating the membrane (termed vertical pore [VP]) (Fig. 3B). Three LPs are connected to the VP at the center of the trimer. These structural features are totally distinct from known lipid scramblases. However, careful structural inspection revealed that the amino-terminal and carboxy-terminal halves of ATG9A exhibited weak similarity to each other, as did the amino-terminal half of the type I ABC transporter fold, including the lipid flippase MsbA (Maeda et al. 2020). Interestingly, structural superimposition revealed that the substrate lipopolysaccharide bound to MsbA is located close to the VP of the superimposed ATG9A, indicating that the VP may accommodate lipids as a substrate. Moreover, phospholipids within a nanodisc used for the cryo-EM analysis of ATG9A were observed to be bound to the LP and VP, and detergents used for the cryo-EM analysis of Atg9 and ATG9A were observed to be bound to the LP, suggesting that both the LP and VP accommodate lipids (Fig. 3C; Guardia et al. 2020; Maeda et al. 2020; Matoba et al. 2020).
Figure 3.
Structural basis of Atg9-mediated lipid scrambling. (A) Ribbon model of fission yeast Atg9 (PDB 7D0I). One protomer is colored orange, whereas the other two protomers within a trimer are colored light and dark gray, respectively. (TM) Transmembrane helices. (B) Schematic drawing of Atg9 trimer. (C) Lipids and detergents bound to the lateral pore (LP) and vertical pore (VP) of Atg9/ATG9A. (Left) Lipids bound to the outer side of VP and LP of human ATG9A (PDB 6WQZ). (Middle) LMNG bound to the LP of human ATG9A (PDB 7JLP). (Right) LMNG bound to the LP of fission yeast Atg9. (D) Various-sized VPs observed at the center of the Atg9/ATG9A trimers.
Structure-based mutational analyses revealed that residues constituting VP and LP are essential for the lipid scrambling activity of Atg9. Consistently, the VP of ATG9A is crucial for lipid scrambling activity. These data suggest that the phospholipid scrambling activities of these proteins are mediated by two pores. Importantly, the mutations at the VP and LP of Atg9 that impaired the lipid scrambling activities significantly decreased the number and size of autophagic bodies (inner membranes of autophagosomes accumulated at the vacuole by protease inhibition) and severely impaired autophagy activity in yeast (Matoba et al. 2020). In the case of ATG9A, mutations at the VP of ATG9A that reduced the lipid scrambling activities significantly reduced the size, but not the number, of autophagosomes (Maeda et al. 2020). Moreover, mutations at the LP of ATG9A were shown to inhibit autophagy activity although their effects on the lipid scrambling activities were not analyzed (Guardia et al. 2020). These data strongly indicate that the phospholipid scrambling activities of Atg9-family proteins, which are mediated by VP and LP, are crucial for IM expansion.
Then, how do Atg9-family proteins exert their phospholipid scrambling activities and how are these activities regulated in cells? When atomic molecular dynamics (MDs) simulation was applied to human ATG9A embedded in the membrane, the protomer−protomer interface of the VP gradually opened, and phospholipids inserted their head group into the VP through the opened interface, reorienting themselves laterally (Maeda et al. 2020). This MD result implies that ATG9A promotes phospholipid flip-flop using the VP. Structural comparison between cryo-EM structures of Atg9 and ATG9A indicates that the VP size can be influenced by the environment, although the LP size is almost invariant (Fig. 3D; Guardia et al. 2020; Maeda et al. 2020; Matoba et al. 2020). MD simulation of Atg9 in a hydrophobic plane mimicking the lipid bilayer showed that the largely opened VP structure of Atg9, which was observed by cryo-EM as a detergent-solubilized state, was closed when embedded in the membrane (Matoba et al. 2020). Another MD simulation of ATG9A revealed that clustering ATG9A trimers in the membrane induce positive membrane curvature, which is similar to that of ATG9A vesicles observed in cells (Guardia et al. 2020). These findings imply that the lipid scrambling activity of Atg9-family proteins could be regulated by the membrane environment through VP size regulation. Although the role and regulation of the VP are gradually becoming clear, little is known for the LP, another critical pore for the scrambling activity. Further studies are required to unveil the molecular mechanisms and regulation of phospholipid scrambling activities of Atg9-family proteins.
MECHANISM OF PHOSPHOLIPID DISTRIBUTION TO BOTH LEAFLETS OF THE ER
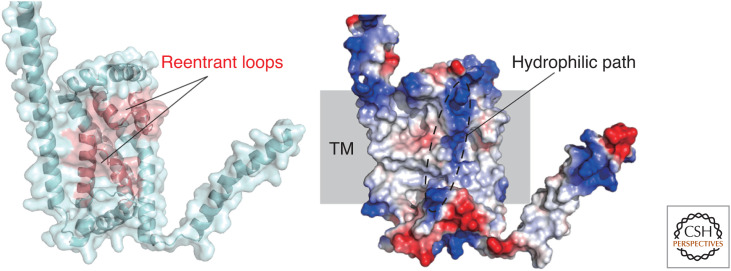
As described above, advances in structure-based analyses revealed that phospholipids delivered to the cytosolic leaflet of IM by Atg2-family proteins would be distributed to the luminal leaflet by the scrambling activity of IM-resident Atg9-family proteins. Then, what about the ER membrane? Because Atg2 is suggested to extract phospholipids from the cytosolic leaflet of the ER and deliver them to the IM, the integrity of the ER membrane would be severely impaired if the phospholipid imbalance between the cytosolic and luminal leaflets of the ER is not resolved. In higher eukaryotes, ER-resident transmembrane proteins, VMP1 and TMEM41B, have been reported to be essential for autophagosome formation in addition to their roles in lipid droplet homeostasis and lipoprotein secretion (Ropolo et al. 2007; Moretti et al. 2018; Morita et al. 2018, 2019; Shoemaker et al. 2019). They share the VMP1, TMEM41, and Tvp38 (VTT) domains and belong to the DedA superfamily (Okawa et al. 2021). According to coevolution-based structural prediction, the VTT/DedA domain possesses two reentrant loops facing each other in the membrane and is topologically similar to ion-coupled transporters (Mesdaghi et al. 2020; Okawa et al. 2021). These predictions are consistent with the AlphaFold structural model of TMEM41B (Fig. 4).
Figure 4.
AlphaFold structure of TMEM41B. (Left) Two reentrant loops are colored red. (Right) Red and blue shading represents negatively and positively charged regions, respectively. TM indicates predicted transmembrane region.
In 2021, several groups independently performed in vitro lipid scrambling assays on VMP1 and TMEM41B using fluorescently labeled lipids and revealed that they are phospholipid scramblases (Ghanbarpour et al. 2021; Huang et al. 2021; Li et al. 2021). Moreover, ATG2A was shown to interact with VMP1 and TMEM41B in addition to ATG9A by co-immunoprecipitation and floatation assays (Ghanbarpour et al. 2021). Based on these data, TMEM41B and VMP1 were proposed to transport phospholipids from the luminal to the cytosolic leaflet of the ER, resolving the phospholipid imbalance between the leaflets caused by phospholipid extraction at the cytosolic leaflet by ATG2 during IM expansion. Consistent with these observations, the amount of phosphatidylserine at the luminal leaflet of the ER, which was similar to that at the cytosolic leaflet, significantly decreased by deleting VMP1 or TMEM41B (Li et al. 2021). Furthermore, TMEM41B was shown to mediate bulk phospholipid transport from the cytosolic to the luminal leaflet of the ER for lipoprotein biogenesis at the ER lumen (Huang et al. 2021). Together, these recent advances indicate that VMP1 and TMEM41B are ER-resident phospholipid scramblases that would promote autophagosome formation by resolving the phospholipid imbalance between the ER leaflets. The AlphaFold structure of TMEM41B predicts the existence of a hydrophilic path across the transmembrane region (Fig. 4, right), which might function in phospholipid scrambling as is observed for canonical scramblases. To further confirm these attractive models, it is crucial to discover mutations that specifically impair the scramblase activities of VMP1/TMEM41B and study the effect of these mutations on autophagosome formation. Also, it is vital to reveal the mechanism by which budding yeast maintains ER integrity during IM expansion since they do not require DedA proteins for autophagosome formation.
POSSIBLE MECHANISMS OF UNIDIRECTIONAL LIPID TRANSFER FROM THE ER TO THE IM
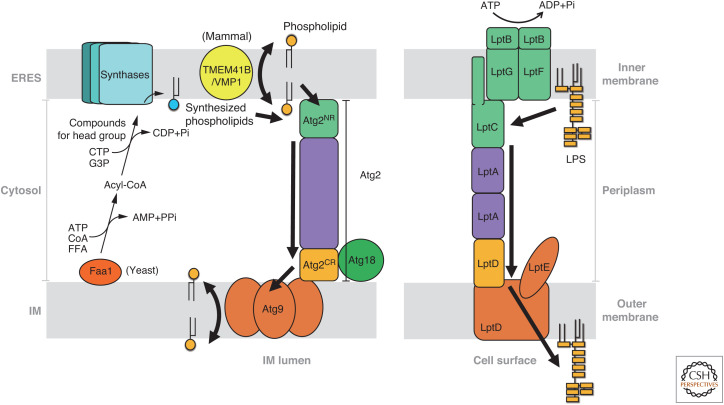
The lipid transfer and scrambling activities of Atg2 and Atg9, as well as VMP1/TMEM41B, described above, enable lipid movement between the ER and IM, which is analogous to that of lipopolysaccharide (LPS) transport of gram-negative bacteria (Fig. 5; Ruiz et al. 2009; Sherman et al. 2018). The cytoplasm of gram-negative bacteria is surrounded by the inner and outer membranes. The outer membrane constituting the cell surface contains LPS in its outer leaflet. LPS is synthesized in the inner membranes and is subsequently transferred to the outer membranes by seven different Lpt proteins (LptA-LptG). The Lpt proteins form an extended complex that bridges the periplasm between the inner and outer membranes. In the Lpt complex, the LptB2FG tetramer, which is an ATP-binding cassette transporter, extracts LPS from inner membranes using ATP hydrolysis. LPS extracted by the LptB2FG tetramer is transferred to LptC anchored to inner membranes. The repeat of LPS loading into LptC is thought to induce LPS movement within the Lpt complex. This mechanism known as the PEZ dispenser model (Okuda et al. 2016; Sherman et al. 2018) would apply to Atg2-mediated lipid transfer in which Atg2NR plays a role corresponding to the LptB2FG-LptC complex. However, unlike the LptB2FG-LptC complex, Atg2NR does not require ATP hydrolysis to extract lipids from membranes. None of the factors described above (Atg2, Atg9, VMP1, TMEM41B) can utilize ATP. Although it has not been discovered, some sources of free energy are required for unidirectional lipid flow from the ER to the IM in the Atg2-mediated lipid transfer. LptC is stacked with the LptA oligomer, which further binds to a periplasmic region of the outer membrane protein LptD. LptA, LptC, and the periplasmic region of LptD fold into a β-jellyroll structure with a hydrophobic cavity that forms a path of LPS flow to outer membranes (Suits et al. 2008; Dong et al. 2014; Qiao et al. 2014; Li et al. 2019). The β-jellyroll region of LptD is followed by a β-barrel transmembrane region of LptD, which forms a complex with LptE. This outer membrane protein complex translocates LPS to the outer leaflet of outer membranes. Therefore, the β-jellyroll region of LptD and β-barrel region of LptD bound to LptE can be considered to correlate with Atg2CR and Atg9, respectively. Notably, the β-jellyroll region of LptD directly connects to the β-barrel pore through which a hydrophilic polysaccharide moiety of LPS probably passes to the outer leaflet of outer membranes (Dong et al. 2014; Qiao et al. 2014). The domain arrangement of LptD is consistent with the possibility that Atg2 directly passes lipids to Atg9 for efficient collaboration between lipid transfer and lipid scrambling, which is supported by the findings that Atg2 and ATG2A physically interact with Atg9 and ATG9A, respectively. Similarly, it is proposed that VMP1 and TMEM41B, which are ER-resident lipid scramblases, directly pass lipids to ATG2A by the observation that ATG2A physically interacts with VMP1 and TMEM41B.
Figure 5.
Mechanisms of the ER-to-IM phospholipid transfer (left) and LPS transport (right). Regions responsible for similar functions are displayed in the same colors. Arrows indicate the lipid flow. For clarity, peptidoglycan layers are not shown in LPS transport. (G3P) Glycerol 3-phosphate, (FFA) free fatty acid, (Pi) inorganic phosphate, (PPi) inorganic pyrophosphate.
Then, what is the source of free energy for unidirectional bulk lipid transfer from the ER to the IM? Although still in the hypothesis stage, de novo phospholipid biosynthesis at the cytosolic leaflet of the ER is a promising candidate for the free energy source. In budding yeast, an acyl-coA synthetase Faa1 was shown to localize at the IM and play an essential role in autophagosome formation (Schütter et al. 2020). It was proposed that Faa1 at IM produces acyl-CoA using ATP and channels it to the phospholipid synthesizing enzymes at the ER, promoting de novo phospholipid synthesis. Phospholipid generation at the cytosolic leaflet of the ER will increase, at least transiently, the chemical potentials of phospholipids at the cytosolic leaflet, which could be a driving force for unidirectional transfer to IM. De novo synthesized phosphatidylcholine was shown to be preferentially transferred to IM under starvation in both budding yeast and mammals, indicating the existence of some mechanisms that channel de novo synthesized phospholipids to Atg2 (and thus to the IM) (Ogasawara et al. 2020; Orii et al. 2021). When mammalian cells were subjected to long-term starvation, β3 isoforms of CTP: phosphocholine cytidylyltransferase (CCT), the rate-limiting enzyme in the Kennedy pathway, were shown to localize at the autophagosome formation site and play critical roles in autophagosome formation, supporting the idea that de novo phospholipid synthesis is important for autophagosome formation (Ogasawara et al. 2020). Moreover, PI synthase was concentrated at the autophagosome formation site on the ER, and PI depletion there impaired autophagosome formation in mammals (Nishimura et al. 2017). These observations suggest that de novo phospholipid synthesis at the ER plays a crucial role in autophagosome formation. It should be noted that the increased local crowding of lipids would result in higher chemical potentials for both de novo synthesized phospholipids and preexisting phospholipids surrounding them, both of which might be unidirectionally transferred to the IM via Atg2. However, phospholipid scrambling at the ER by VMP1 and TMEM41B would reduce such locally increased chemical potentials of lipids at the cytosolic leaflet of the ER and could be detrimental to the unidirectional lipid transfer from the ER to the IM. Further mechanistic studies are required to unveil the relationship between de novo phospholipid synthesis, ER scramblases, and Atg2 to understand the mechanism of unidirectional phospholipid transport to IM.
CONCLUDING REMARKS
In recent years, our understanding of the mechanisms of phospholipid transfer from ER to IM has advanced significantly and appears to have succeeded in establishing the fundamental principles of autophagosomal membrane expansion. However, many questions remain unanswered, including how Atg2 recognizes a specific subdomain of the ER (ERES in the case of budding yeast), what drives the unidirectional lipid transfer through Atg2, how lipid scramblases at the ER (VMP1/TMEM41B) and IM (Atg9) collaborate with Atg2 for membrane expansion, and to what extent de novo phospholipid synthesis contributes to the autophagosomal membrane expansion. Furthermore, it is crucial to determine the proportion of phospholipids that make up the autophagosome provided by Atg2-mediated transfer from the ER. Understanding the mechanisms of autophagosome formation would contribute not only to the autophagy field but also to the entire field of cell biology by establishing fundamental principles of de novo organelle biogenesis.
ACKNOWLEDGMENTS
This work was supported in part by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Nos. 18H03989 and 19H05707 (to N.N.N.), 20K06532 (to T.O.), 21K06055 (to K.M.), CREST, Japan Science and Technology Agency Grant No. JPMJCR20E3 (to N.N.N.), and grants from the Takeda Science Foundation (to N.N.N., T.O., and K.M.).
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182: 685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Osumi M, Ohsumi Y. 1995. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct 20: 465–471. 10.1247/csf.20.465 [DOI] [PubMed] [Google Scholar]
- Chowdhury S, Otomo C, Leitner A, Ohashi K, Aebersold R, Lander GC, Otomo T. 2018. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc Natl Acad Sci 115: E9792–E9801. 10.1073/pnas.1811874115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. 2012. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science 338: 815–818. 10.1126/science.1225625 [DOI] [PubMed] [Google Scholar]
- Dong H, Xiang Q, Gu Y, Wang Z, Paterson NG, Stansfeld PJ, He C, Zhang Y, Wang W, Dong C. 2014. Structural basis for outer membrane lipopolysaccharide insertion. Nature 511: 52–56. 10.1038/nature13464 [DOI] [PubMed] [Google Scholar]
- Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO. 2000. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur J Cell Biol 79: 871–882. 10.1078/0171-9335-00125 [DOI] [PubMed] [Google Scholar]
- Ghanbarpour A, Valverde DP, Melia TJ, Reinisch KM. 2021. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc Natl Acad Sci 118: e2101562118. 10.1073/pnas.2101562118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez R, Rose J, Guimarães R, Mari M, Papinski D, Rieter E, Geerts WJ, Hardenberg R, Kraft C, Ungermann C, et al. 2018. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol 217: 2743–2763. 10.1083/jcb.201710116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Sánchez R, Tooze SA, Reggiori F. 2021. Membrane supply and remodeling during autophagosome biogenesis. Curr Opin Cell Biol 71: 112–119. 10.1016/j.ceb.2021.02.001 [DOI] [PubMed] [Google Scholar]
- Graef M, Friedman JR, Graham C, Babu M, Nunnari J. 2013. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell 24: 2918–2931. 10.1091/mbc.e13-07-0381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guardia CM, Tan XF, Lian T, Rana MS, Zhou W, Christenson ET, Lowry AJ, Faraldo-Gómez JD, Bonifacino JS, Jiang J, et al. 2020. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep 31: 107837. 10.1016/j.celrep.2020.107837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. 2009. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 11: 1433–1437. 10.1038/ncb1991 [DOI] [PubMed] [Google Scholar]
- He C, Baba M, Cao Y, Klionsky DJ. 2008. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell 19: 5506–5516. 10.1091/mbc.e08-05-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Xu B, Liu L, Wu L, Zhu Y, Ghanbarpour A, Wang Y, Chen FJ, Lyu J, Hu Y, et al. 2021. TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab 33: 1655–1670.e8. 10.1016/j.cmet.2021.05.006 [DOI] [PubMed] [Google Scholar]
- Jeong H, Park J, Lee C. 2016. Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex. EMBO Rep 17: 1857–1871. 10.15252/embr.201642706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Židek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with alphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Tamura Y, Kojima R, Bala S, Asai E, Michel AH, Kornmann B, Riezman I, Riezman H, Sakae Y, et al. 2018. Structure-function insights into direct lipid transfer between membranes by Mmm1-Mdm12 of ERMES. J Cell Biol 217: 959–974. 10.1083/jcb.201704119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Kirisako H, Koizumi M, Ohsumi Y, Nakatogawa H. 2018. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci 115: 10363–10368. 10.1073/pnas.1806727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, De Camilli P. 2018. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J Cell Biol 217: 3625–3639. 10.1083/jcb.201807019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai LTF, Yu C, Wong JSK, Lo HS, Benlekbir S, Jiang L, Lau WCY. 2020. Subnanometer resolution cryo-EM structure of Arabidopsis thaliana ATG9. Autophagy 16: 575–583. 10.1080/15548627.2019.1639300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. 2013. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol 14: 759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Lees JA, Reinisch KM. 2020. Inter-organelle lipid transfer: a channel model for Vps13 and chorein-N motif proteins. Curr Opin Cell Biol 65: 66–71. 10.1016/j.ceb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S. 2010. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol 11: 739–750. 10.1038/nrm2971 [DOI] [PubMed] [Google Scholar]
- Li Y, Orlando BJ, Liao M. 2019. Structural basis of lipopolysaccharide extraction by the LptB2FGC complex. Nature 567: 486–490. 10.1038/s41586-019-1025-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lees JA, Lusk CP, Reinisch KM. 2020. Cryo-EM reconstruction of a VPS13 fragment reveals a long groove to channel lipids between membranes. J Cell Biol 219: e202001161. doi: 10.1083/jcb.202001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YE, Wang Y, Du X, Zhang T, Mak HY, Hancock SE, McEwen H, Pandzic E, Whan RM, Aw YC, et al. 2021. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J Cell Biol 220: e202103105. doi: 10.1083/jcb.202103105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Otomo C, Otomo T. 2019. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 8: e45777. 10.7554/eLife.45777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, Grishin NV, Forli S, Mizushima N, Otomo T. 2020. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol 27: 1194–1201. 10.1038/s41594-020-00520-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba K, Noda NN. 2021. Structural catalog of core Atg proteins opens new era of autophagy research. J Biochem 169: 517–525. 10.1093/jb/mvab017 [DOI] [PubMed] [Google Scholar]
- Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, Sugita Y, Nomura N, Iwata S, Ohsumi Y, et al. 2020. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol 27: 1185–1193. 10.1038/s41594-020-00518-w [DOI] [PubMed] [Google Scholar]
- Melia TJ, Lystad AH, Simonsen A. 2020. Autophagosome biogenesis: from membrane growth to closure. J Cell Biol 219: e202002085. 10.1083/jcb.202002085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesdaghi S, Murphy DL, Sánchez Rodríguez F, Burgos-Mármol JJ, Rigden DJ. 2020. In silico prediction of structure and function for a large family of transmembrane proteins that includes human Tmem41b. F1000Res 9: 1395. 10.12688/f1000research.27676.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. 2011. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol 27: 107–132. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- Moretti F, Bergman P, Dodgson S, Marcellin D, Claerr I, Goodwin JM, DeJesus R, Kang Z, Antczak C, Begue D, et al. 2018. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep 19: e45889. 10.15252/embr.201845889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hama Y, Izume T, Tamura N, Ueno T, Yamashita Y, Sakamaki Y, Mimura K, Morishita H, Shihoya W, et al. 2018. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol 217: 3817–3828. 10.1083/jcb.201804132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita K, Hama Y, Mizushima N. 2019. TMEM41B functions with VMP1 in autophagosome formation. Autophagy 15: 922–923. 10.1080/15548627.2019.1582952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata S, Sakuragi T, Segawa K. 2020. Flippase and scramblase for phosphatidylserine exposure. Curr Opin Immunol 62: 31–38. 10.1016/j.coi.2019.11.009 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H. 2020. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol 21: 439–458. 10.1038/s41580-020-0241-0 [DOI] [PubMed] [Google Scholar]
- Nishimura T, Tamura N, Kono N, Shimanaka Y, Arai H, Yamamoto H, Mizushima N. 2017. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J 36: 1719–1735. 10.15252/embj.201695189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN. 2021. Atg2 and Atg9: intermembrane and interleaflet lipid transporters driving autophagy. Biochim Biophys Acta Mol Cell Biol Lipids 1866: 158956. 10.1016/j.bbalip.2021.158956 [DOI] [PubMed] [Google Scholar]
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. 2000. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 148: 465–480. 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Sekito T, Niimi K, Ohsumi Y. 2008. The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function. J Biol Chem 283: 23972–23980. 10.1074/jbc.M803180200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara Y, Cheng J, Tatematsu T, Uchida M, Murase O, Yoshikawa S, Ohsaki Y, Fujimoto T. 2020. Long-term autophagy is sustained by activation of CCTβ3 on lipid droplets. Nat Commun 11: 4480. 10.1038/s41467-020-18153-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okawa F, Hama Y, Zhang S, Morishita H, Yamamoto H, Levine TP, Mizushima N. 2021. Evolution and insights into the structure and function of the dedA superfamily containing TMEM41B and VMP1. J Cell Sci 134: jcs255877. 10.1242/jcs.255877 [DOI] [PubMed] [Google Scholar]
- Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14: 337–345. 10.1038/nrmicro.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orii M, Tsuji T, Ogasawara Y, Fujimoto T. 2021. Transmembrane phospholipid translocation mediated by Atg9 is involved in autophagosome formation. J Cell Biol 220: e202009194. 10.1083/jcb.202009194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Noda NN. 2019. Atg2: a novel phospholipid transfer protein that mediates de novo autophagosome biogenesis. Protein Sci 28: 1005–1012. 10.1002/pro.3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, Ohsumi Y, Noda NN. 2019. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol 26: 281–288. 10.1038/s41594-019-0203-4 [DOI] [PubMed] [Google Scholar]
- Osawa T, Ishii Y, Noda NN. 2020. Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells 25: 65–70. 10.1111/gtc.12733 [DOI] [PubMed] [Google Scholar]
- Qiao S, Luo Q, Zhao Y, Zhang XC, Huang Y. 2014. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 511: 108–111. 10.1038/nature13484 [DOI] [PubMed] [Google Scholar]
- Reinisch KM, De Camilli P. 2016. SMP-domain proteins at membrane contact sites: structure and function. Biochim Biophys Acta 1861: 924–927. 10.1016/j.bbalip.2015.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren JQ, Liang RB, Wang WJ, Zhang DC, Yu L, Feng W. 2020. Multi-site-mediated entwining of the linear WIR-motif around WIPI β-propellers for autophagy. Nat Commun 11: 2702. 10.1038/s41467-020-16523-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropolo A, Grasso D, Pardo R, Sacchetti ML, Archange C, Lo Re A, Seux M, Nowak J, Gonzalez CD, Iovanna JL, et al. 2007. The pancreatitis-induced vacuole membrane protein 1 triggers autophagy in mammalian cells. J Biol Chem 282: 37124–37133. 10.1074/jbc.M706956200 [DOI] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. 2009. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol 7: 677–683. 10.1038/nrmicro2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schütter M, Giavalisco P, Brodesser S, Graef M. 2020. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell 180: 135–149.e14. 10.1016/j.cell.2019.12.005 [DOI] [PubMed] [Google Scholar]
- Sharom FJ. 2011. Flipping and flopping—lipids on the move. IUBMB Life 63: 736–746. [DOI] [PubMed] [Google Scholar]
- Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D. 2018. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359: 798–801. 10.1126/science.aar1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Huang TQ, Weir NR, Polyakov NJ, Schultz SW, Denic V. 2019. CRISPR screening using an expanded toolkit of autophagy reporters identifies TMEM41B as a novel autophagy factor. PLoS Biol 17: e2007044. 10.1371/journal.pbio.2007044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suits MD, Sperandeo P, Dehò G, Polissi A, Jia Z. 2008. Novel structure of the conserved gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. J Mol Biol 380: 476–488. 10.1016/j.jmb.2008.04.045 [DOI] [PubMed] [Google Scholar]
- Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. 2013. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 126: 2534–2544. doi: 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ. 2019. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol 218: 1787–1798. 10.1083/jcb.201811139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bulow S, Hummer G. 2020. Kinetics of Atg2-mediated lipid transfer from the ER can account for phagophore expansion. bioRxiv doi 10.1101/2020.05.12.090977 [DOI] [Google Scholar]
- Wong LH, Gatta AT, Levine TP. 2019. Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes. Nat Rev Mol Cell Biol 20: 85–101. 10.1038/s41580-018-0071-5 [DOI] [PubMed] [Google Scholar]
- Yang Z, Klionsky DJ. 2010. Eaten alive: a history of macroautophagy. Nat Cell Biol 12: 814–822. 10.1038/ncb0910-814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 2009. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 5: 1180–1185. 10.4161/auto.5.8.10274 [DOI] [PubMed] [Google Scholar]
- Young AR, Chan EY, Hu XW, Köchl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 119: 3888–3900. 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- Zheng JX, Li Y, Ding YH, Liu JJ, Zhang MJ, Dong MQ, Wang HW, Yu L. 2017. Architecture of the ATG2B-WDR45 complex and an aromatic Y/HF motif crucial for complex formation. Autophagy 13: 1870–1883. 10.1080/15548627.2017.1359381 [DOI] [PMC free article] [PubMed] [Google Scholar]