Abstract
Imaginal discs are simple epithelial sacs found in Drosophila larvae, which generate adult structures including wings and legs. The first studies of imaginal disc regeneration involved technically challenging transplantation experiments. Yet despite the difficulty, many aspects of regeneration including wound healing, blastema formation, and the repatterning of regenerated tissue were characterized. An important discovery was the phenomenon of transdetermination, where a small group of cells in regenerating tissue collectively switch fate (“collective cell reprogramming”). The development of genetic tissue-ablation systems over the last 12 years has energized this field, by making experiments less technically challenging, more reproducible, and by incorporating additional genetic analysis. Recent progress includes defining mechanistic links between early responses to wounding and the signaling pathways that drive proliferation, uncovering a role for localized silencing of damage-responsive enhancers to limit regenerative capacity as tissues mature, and identifying genes that maintain cellular plasticity within acceptable limits during regeneration.
The phenomenon of appendage regeneration has been studied for more than 250 years; crustacean leg regeneration was described by Reaumur in 1712 and salamander limb regeneration by Spallanzani in 1769 (for reviews, see Morgan 1901; Carlson 2007). Despite years of study in many different organisms, we still do not have definitive answers to some of the fundamental questions pertaining to appendage regeneration: What are the molecular mechanisms that regulate regenerative growth? In what ways do regenerative growth and patterning differ from that which occurs during normal development? Why can some organisms, but not others, regenerate their limbs? And finally, why does the capacity for regeneration in some organisms diminish with age?
For many years, the study of regeneration in both vertebrates and invertebrates was restricted to approaches used by experimental embryologists. These studies, sometimes involving experiments of great ingenuity, uncovered phenomena that are being addressed by contemporary studies of regeneration. These include the timing and morphology of the initial closure of a wound, the formation of an area with localized cell proliferation (referred to as a blastema) that drives regenerative growth, the respecification of cell fates to give rise to a patterned regenerated structure, and, ultimately, the cessation of regeneration when a structure of the appropriate size has been reformed.
Indeed, studies on the regeneration of insect appendages have contributed to our understanding of mechanisms that regulate regeneration. Pioneering studies were conducted on legs of hemimetabolous insects, such as cockroaches and crickets, which develop through a series of nymph stages that can regenerate prior to the final molt (Bohn 1970; French et al. 1976). These studies uncovered some of the mechanisms that regulate tissue patterning during regeneration. They showed that limb amputations result, in most cases, in the generation of structures distal to the site of amputation. Their experiments also demonstrated that regenerative growth is triggered by an intercalation mechanism, such that when tissues of disparate positional identities are in apposition after wound healing regenerative growth generates the missing structures.
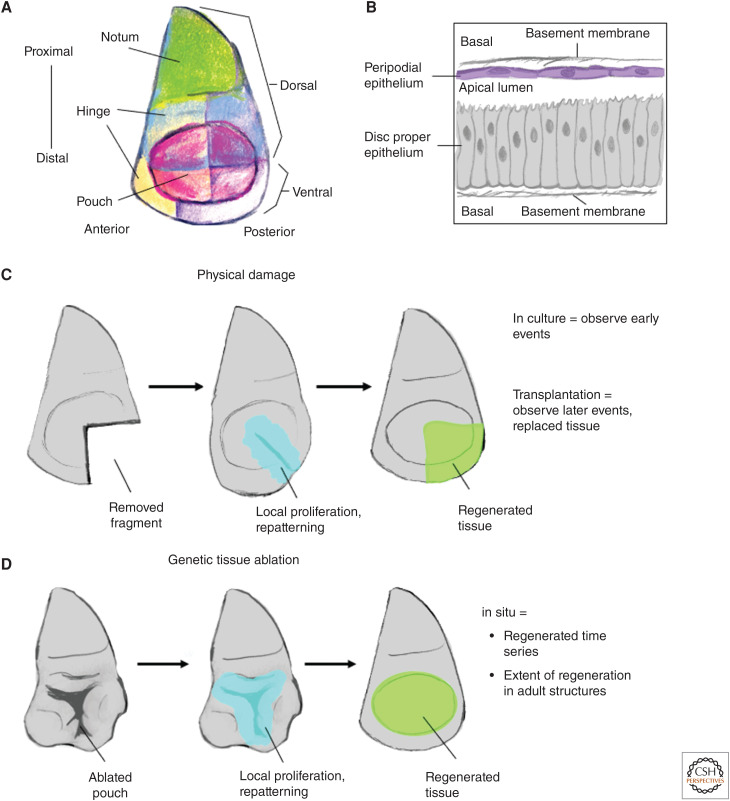
In this review, we focus on the regeneration of imaginal discs (Fig. 1A,B), which are the larval precursors of adult structures such as the legs, eyes, and wing in holometabolous insects, which are insects that undergo a complete metamorphosis. Many of these studies were first conducted using approaches similar to those used by developmental biologists studying regeneration in other organisms (surgical ablation, transplantation) (Fig. 1C). However, the sophisticated genetic tools available to Drosophila geneticists have facilitated the development of ablation systems that are not dependent upon surgery (Fig. 1D), which can be conducted on a large scale, and therefore incorporated into genetic screens. As a result, there has been a renaissance in the field of imaginal disc regeneration resulting in considerable progress toward understanding fundamental issues that pertain to regeneration that have implications well beyond Drosophila.
Figure 1.
Overview of imaginal disc regeneration. (A) Cartoon of a wing imaginal disc showing the different territories of the disc. The notum generates the thoracic wall while the hinge and pouch generate progressively more distal structures of the wing, respectively. (B) Imaginal disc cellular architecture: the disc resembles a flattened sac with the two layers, the disc proper and the peripodial epithelium being continuous around the edges. The apical domain of both epithelial layers face inward toward the lumen. The cells of the disc proper are columnar, while the cells of the peripodial epithelium are squamous. (C) Physical damage of imaginal discs: early responses are best observed in culture while later responses are observed following transplantation into adult abdomens and subsequent and recovery. (D) Genetic tissue ablation models allow more precisely defined ablation regions and for the study of disc regeneration in its natural environment.
In the last 10 years, many review articles have discussed imaginal disc regeneration (Repiso et al. 2011; Worley et al. 2012; Kashio et al. 2014; Sun and Irvine 2014; Beira and Paro 2016; Jaszczak and Halme 2016; Hariharan and Serras 2017; Ahmed-de-Prado and Baonza 2018; Pinal et al. 2019; Fox et al. 2020). Here, we will first briefly review the pioneering research that kick-started the study of regeneration within imaginal discs, and then focus on those aspects where significant progress in understanding the molecular and cellular mechanisms that regulate regeneration have been made in the last decade.
EARLY STUDIES OF IMAGINAL DISC REGENERATION
Imaginal discs (Fig. 1A) are simple epithelia that are formed during embryogenesis by invagination of the epidermal layer (Cohen 1993; Held 2002). They have the appearance of flattened sacs that maintain continuity with the surface of the embryo or larva via a narrow stalk. Different imaginal discs are generated along different positions along the anteroposterior axis of the embryo. Each of the three thoracic segments (T1–T3) of the embryo and larva has a pair of dorsal and ventral imaginal discs. The ventral discs eventually generate the legs that emanate from each of the thoracic segments of the adult body. The dorsal imaginal discs of T2 are the wing discs that generate the adult wings and most of the adult thorax. Whereas most of the modern studies of regeneration use the leg, wing, and eye-antennal discs, many of the earlier studies also used the genital disc, which develops at the posterior end of the embryo. During the larval stages, imaginal discs increase in size as a result of cell proliferation and arrest their growth when they reach a predictable size and shape. Indeed, immature discs transplanted into adult abdomens seem to stop growing at approximately the correct size providing evidence that their eventual size is determined by mechanisms that are intrinsic to the disc itself (Bryant and Levinson 1985). During the pupal stage, additional cell divisions occur as do dramatic morphogenetic events that transform the discs into their cognate adult structures and, by the mechanism of eversion, move these structures externally. Thus far, there is no evidence for specialized stem cells in imaginal discs. Rather, disc cells are similar to committed progenitors that have the capacity to proliferate but eventually undergo terminal differentiation.
The study of imaginal disc regeneration was pioneered by the group of Ernst Hadorn (Hadorn et al. 1949; Ursprung 1959; Hadorn 1965). These early studies were aimed at addressing the stability of the determined state; imaginal discs are initially specified during embryogenesis but their cells terminally differentiate many days later during metamorphosis. By implanting imaginal disc fragments into young larvae or adult abdomens, the Hadorn group found that imaginal discs could indeed regenerate (for review, see Worley et al. 2012). This process could, in principle, be continued indefinitely, thus prolonging the period when these cells were still undifferentiated. By implanting the cultured discs into larvae prior to metamorphosis, the developmental potential of discs could be assessed. In most cases, transplanted discs generated structures that were appropriate for that kind of disc (autotypic structures). However, occasionally, especially after prolonged culture, discs generated structures normally derived from a different kind of disc (allotypic structures), for example, wing structures from a portion of a cultured leg disc. This indicated a change in the determination state of the disc—Hadorn named this phenomenon transdetermination (Hadorn 1965). Intriguingly, transdetermination did not seem to be the result of a stochastic fate change in a single cell but rather the result of a small number of cells collectively changing fate (Gehring 1966; Sustar et al. 2011). Additionally, transdetermination seemed to occur as an aberrant form of regeneration suggesting that tissue damage and regeneration promoted cell plasticity. Even within imaginal discs, specific regions termed “weak points” were more prone to transdetermination (Gehring 1966; Schubiger 1971). These weak points within the tissue often occur in locations with high levels of Wg and Dpp morphogens (Maves and Schubiger 1995, 1998; Johnston and Schubiger 1996; reviewed by Maves and Schubiger 2003; Johnston 2005; McClure and Schubiger 2007).
These studies of imaginal disc regeneration also provided important descriptions of the initial events that occur during regeneration. Imaginal discs are topologically like flattened sacs; the two epithelial layers, one mostly columnar and the other squamous (Fig. 1B), are continuous around the edges. When imaginal discs are cut surgically, the wounds are initially healed by heterotypic contacts between the two epithelial layers that are then resolved when continuity is reestablished in each of the epithelial layers (Reinhardt et al. 1977; Reinhardt and Bryant 1981; Álvarez-Fernández et al. 2015). A zone of localized cell proliferation is observed immediately adjacent to the region of the cut (Abbott et al. 1981; O'Brochta and Bryant 1987). It had been observed that this proliferation precedes the closing of the wound edges (Dale and Bownes 1981) leading to the conclusion that tissue injury rather than the apposition of cells with disparate positional identities (French et al. 1976; Bryant et al. 1981) triggered the cell proliferation. The new tissue is generated from this zone of localized proliferation, the blastema. It was shown that transplantation of the blastema alone into adult abdomens could result in the generation of the missing portion of the disc (Karpen and Schubiger 1981). Also, when a blastema was present, cell proliferation in nondamaged portions of the disc was decreased (Kiehle and Schubiger 1985), a finding confirmed by more recent studies using a genetic ablation system (Smith-Bolton et al. 2009), raising the possibility that regenerative proliferation could, in some unknown way, suppress cell proliferation associated with developmental growth. More recently, lineage-tracing techniques have reemphasized that local cell proliferation generates the regenerated tissue (Bosch et al. 2008; Smith-Bolton et al. 2009; Sustar et al. 2011; Herrera et al. 2013; Worley et al. 2013).
The classical studies of imaginal disc regeneration also provided some insights into how discs are repatterned after damage (Bryant 1971; Schubiger 1971). Many of the studies of regeneration were termed circumferential regeneration where a sector of the disc was removed (Fig. 1C). Typically, in the larger piece, the missing portion was regenerated. Remarkably, the smaller piece often duplicated itself. An intriguing exception to this rule was the T1 leg disc where the smaller piece could regenerate to form a complete disc while the larger piece duplicated itself (Schubiger 1971). These results could be explained by invoking a polar-coordinate model where the missing portions are regenerated by intercalation following the apposition of the two edges with disparate positional identities. Similar results had been obtained by investigators studying cockroach limbs (for review, see Bryant et al. 1981). Intriguingly, subsequent studies on growth and patterning of imaginal discs have still not provided a mechanistic explanation for the polar coordinate model. Less frequently, investigators studied distal regeneration by making a hole in the central portion of the disc that generates the most distal portion of the adult appendage. These studies were consistent with the idea of a gradation of positional identities where proximal structures could generate more distal structures but not vice versa (Haynie and Schubiger 1979).
The classical studies of imaginal disc regeneration described a number of phenomena that appear similar to appendage regeneration in other organisms (e.g., axolotl limb regeneration). These include the formation of a blastema and the idea that regeneration can generate distal structures from proximal structures as well as by intercalation of positional values. Importantly, imaginal disc regeneration, when compared to appendage regeneration in vertebrates, is less complex, since it is a simple epithelial structure that mostly lacks mesodermal elements and is far more amenable to genetic manipulation. As a result, in the last decade, we have witnessed much progress in understanding the molecular events that underlie regeneration that we will discuss here.
DEVELOPMENT OF GENETIC TISSUE-ABLATION SYSTEMS
Since the study of imaginal disc regeneration was technically difficult and laborious, requiring difficult dissections and transplantations, relatively few laboratories worked in this field. As will be discussed later, despite these hurdles, these groups continued to make progress in linking events in regeneration to specific molecular pathways such as the JNK pathway (Bosch et al. 2005; Lee et al. 2005; Mattila et al. 2005) and the JAK/STAT pathway (Katsuyama et al. 2015). The study of imaginal disc regeneration was given fresh energy by the development of genetic tissue ablation systems (Smith-Bolton et al. 2009; Bergantiños et al. 2010). These systems used the Gal4/UAS system (Fischer et al. 1988; Brand and Perrimon 1993) to express a proapoptotic gene in a clearly defined region of the imaginal disc (Fig. 1D). Temporal control of Gal4 activity was provided by concurrent expression of a temperature-sensitive version of the Gal80 repressor (Gal80ts) (McGuire et al. 2003). Thus, embryonic and the early stages of larval development were allowed to occur at 18°C when Gal80ts was active. A temperature shift to 30°C of 24–40 h during the third larval instar rendered Gal80ts inactive and resulted in the expression of a proapoptotic gene in the territory where Gal4 was expressed. Expression of the proapoptotic gene was shut off by shifting the cultures back down to 18°C, which allowed regeneration to occur. Most studies have used the Gal4 drivers that are expressed in the wing pouch and hence the extent of regeneration can be assessed simply by examining the adult wings. When the rotund-Gal4 driver is used (Smith-Bolton et al. 2009), most of the wing pouch, which generates the wing blade, is ablated. Regeneration occurs by the proliferation of more proximally fated cells that normally give rise to the hinge (the structure that joins the wing blade to the body wall), which are respecified to generate more distal structures (Smith-Bolton et al. 2009; Herrera et al. 2013). This genetic model of tissue ablation and regeneration is therefore conceptually similar to experiments where a limb regenerates after amputation where distal structures are generated from the remaining proximal structures. Other Gal4 drivers that are commonly used ablate the medial portion of the wing blade, which is then regenerated from more laterally located cells (Bergantiños et al. 2010).
These first-generation genetic tissue-ablation systems have facilitated many discoveries pertaining to the genetic regulation of regeneration that we discuss below. However, a significant shortcoming in these systems is that the Gal4/UAS system is used to express the proapoptotic gene. The fly community has generated a number of lines expressing RNAi transgenes and cDNAs all under the control of UAS elements. When used in conjunction with these systems, those transgenes are also expressed during the ablation phase and in the same territory as the proapoptotic gene. This has led to the development of a variety of second-generation genetic tissue-ablation systems. One way to express the proapoptotic gene and another transgene in different but overlapping territories is to express a proapoptotic gene under the control of the lexA operator using the LHG driver, thus enabling collections of UAS lines to be expressed under the control of a Gal4 driver (Yagi et al. 2010; Santabárbara-Ruiz et al. 2015; Vizcaya-Molina et al. 2018). However, even in this situation, both transgenes are expressed only during the ablation phase since both Gal4 and LHG are repressed by active Gal80.
More recently, second-generation systems have been developed that specifically allow the expression of UAS-driven transgenes in the region of the blastema during the entire regeneration phase. One such system is DUAL Control (Harris et al. 2020), which uses a bipartite transactivator to express a proapoptotic transgene in the wing pouch following a heat shock. The same heat shock activates a recombination event that turns on Gal4-driven expression of UAS-regulated transgenes in the surrounding region, which, importantly, includes the blastema. The DEMISE system uses FLP-mediated recombination to excise a stop cassette upstream of a UAS-driven proapoptotic gene stochastically in a subset of cells (Cohen et al. 2018). As a result, the cells that are eliminated are surrounded by Gal4-expressing cells. Other ways to uncouple the tissue ablation from genetic manipulation during regeneration is to use the QUAS system (Potter et al. 2010), which has been used to drive a temperature-sensitive toxin for the study of regeneration (Kashio et al. 2016), or ablation activated using an optogenetic method (Makhijani et al. 2017). The latter approach could potentially be used in screens where one disc of each pair is damaged, and the contralateral disc is used as the control. These systems have been reviewed recently by Fox et al. (2020).
EARLY EVENTS IN REGENERATION
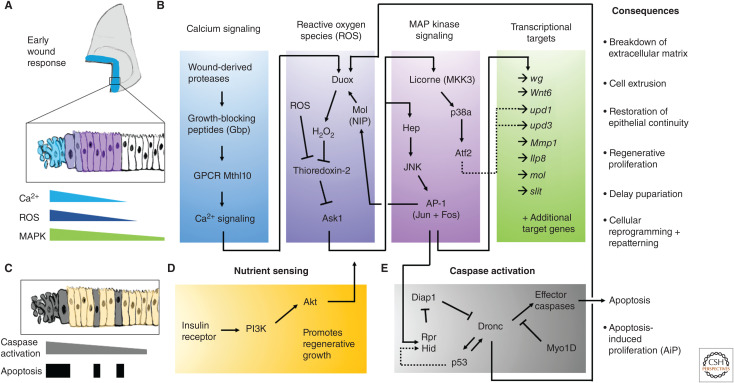
The earliest events observed following tissue damage are waves of cytoplasmic Ca2+ signaling (Narciso et al. 2015), which can persist for 5–10 min after physically damaging the discs (Restrepo and Basler 2016; Yoo et al. 2016). When studied by laser-ablation injury to larval imaginal discs or pupal imaginal disc derivatives, there is an influx of extracellular Ca2+ within milliseconds triggered by microtears in the plasma membrane of damaged cells (Shannon et al. 2017). Spikes in intracellular calcium spread to adjacent cells via gap junctions. A second slower wave of increased intracellular Ca2+ spreads much further (up to 150 m) and peaks in intensity around 120 sec (Shannon et al. 2017; O'Connor et al. 2021). Recent work has defined a mechanism for the propagation of Ca2+ waves within damaged tissues; proteases liberated into the extracellular environment from ruptured cells cleave and activate the Growth-blocking peptides (Gbp) from inactive precursors. These peptides diffuse in the apical extracellular region and function as ligands for the G-protein-coupled receptor Methuselah-like 10 (Mthl 10). Mthl-10 activates phospholipase C via a Gq-α protein and generates inositol-phosphate 3, which results in the release of Ca2+ from the endoplasmic reticulum (O'Connor et al. 2021). The distance of the response is likely determined by the diffusion range and stability of the released proteases and the active peptides that they generate (Fig. 2A,B).
Figure 2.
Early events in imaginal disc regeneration. Recent progress in unraveling the chain of events that link imaginal disc damage to transcriptional responses and physiological changes are summarized. (A) Spatial localization of early wound responses. (B) Molecular pathways connecting several early wound responses, including Ca2+ signaling, reactive oxygen species, and MAP kinase signaling and transcriptional responses. These early signaling events kick-start the regenerative process, including the listed consequences. (C) Deciding whether to live or die: more cells show caspase activation than become apoptotic cells. (D) The sensing of nutrition through the insulin-like signaling pathway is important for promoting cell survival and promoting regeneration. Akt signaling is important for mounting a regenerative response and for cells to survive caspase activation. (E) Caspase activation can lead to various downstream consequences, including promoting the generation of reactive oxygen species via stimulating Duox activity.
Pioneering studies in zebrafish demonstrated, following wounding, a gradient of reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), by using HyPer, a GFP protein that is sensitive to the levels of H2O2. (Niethammer et al. 2009). In that study, the levels of ROS peaked 20 min after wounding and persisted up to 1 h later. The H2O2 was generated by Duox, a membrane-bound NADPH oxidase that generates extracellular H2O2 and can be activated by increased cytoplasmic Ca2+. H2O2 is thought to be able to cross cell membranes. H2O2 was also found to be generated in laser-induced wounds in the Drosophila epidermis (Moreira et al. 2010) and during regeneration within the wing disc (Santabárbara-Ruiz et al. 2015; Brock et al. 2017; Khan et al. 2017). Genetic experiments demonstrated that the generation of H2O2 at wound edges was downstream of an increase in intracellular Ca2+ (Razzell et al. 2013) and that the production of H2O2 by the propagation of the calcium wave could be blocked by reducing the function of the gap-junction component Innexin 2 or the cation channel Trpm (Razzell et al. 2013). Other types of ROS (e.g., O−2) could be generated intracellularly and move between cells via gap junctions or even between ring-canal-like structures that have been observed in cells of the wing disc (McLean and Cooley 2013). Manipulations that enhanced ROS scavenging impaired regeneration thus demonstrating the importance of ROS production in regeneration (Santabárbara-Ruiz et al. 2015). Furthermore, ROS production resulted in the activation of two MAP kinase family members, JNK and p38, and each is activated independently of the other. Of the two p38 paralogs, p38a appears more important (Santabárbara-Ruiz et al. 2015). Regeneration is impaired when the function of either JNK (Bosch et al. 2005; Mattila et al. 2005; Bergantiños et al. 2010) or p38a (Santabárbara-Ruiz et al. 2015) is reduced. ROS levels need to be tightly regulated, as too much ROS prevents regenerative growth (Brock et al. 2017) and excess JNK signaling causes patterning defects (Schuster and Smith-Bolton 2015; Tian and Smith-Bolton 2021). The level and duration of JNK and p38 activity are likely important for coordinating a regenerative response. The MAP kinase kinase kinase Apoptosis Sensing Kinase-1 (Ask1), which functions upstream of both JNK and p38, is necessary for regeneration (Santabárbara-Ruiz et al. 2019) and is activated by ROS; ROS promote the release of the inhibitory function of Thioredoxin on Ask1 (Fig. 2B). However, unlike in dying cells where Ask1 appears to be maximally activated, surrounding cells that will survive have a form of Ask1 that is phosphorylated by Akt on a site outside the kinase domain (Ser83), which reduces its activity (Santabárbara-Ruiz et al. 2019). It has been proposed that this moderate level of Ask1 activation results in an intermediate level of JNK and p38 activity, which is sufficient to promote regeneration but insufficient to activate apoptosis (Santabárbara-Ruiz et al. 2019). Intriguingly, this form of pro-regenerative Ask1/p38 activity is favored in cells that are experiencing signaling via the PI3K/Akt pathway in response to nutrients, highlighting that the nutrient status of a given cell affects how cells respond during regeneration (Esteban-Collado et al. 2021). In addition, the activation of the Akt pathway has been shown to be important for cells to survive temporary caspase activation (Fig. 2C,D; Sun et al. 2020). Other stress-response proteins, such as Growth arrest and DNA damage-inducible 45 (Gadd45) have recently been shown to be important for effective regeneration (Camilleri-Robles et al. 2019), likely through activating the MAPK pathway.
As described above, ROS can activate JNK. JNK signaling is, in turn, able to promote the expression of moladietz (mol), which encodes for a maturation factor for the H2O2-generating enzyme Duox (Fig. 2; Khan et al. 2017). This positive feedback loop might explain the prolonged JNK activity that is necessary to maintain regenerative growth and, thus, highlights a role of ROS in maintaining MAPK signaling.
LINKING EARLY EVENTS TO REGENERATIVE GROWTH
How does wounding and tissue ablation activate cell proliferation? One possible explanation is that the damage itself causes the release of signals that promote proliferation in the surrounding cells. Consistent with this model was the observation made in early studies that the onset of proliferation preceded wound closure (Dale and Bownes 1981). An alternative possibility is that once wound healing has occurred, the differences in positional identities of cells from the two wound edges would trigger proliferation and also direct the extent of proliferation (French et al. 1976). These two types of mechanisms are not mutually exclusive; it is possible that proliferation could be initially triggered by damage and subsequent maintenance of proliferation could depend on differences in positional identity between the edges. Such a mechanism would not delay proliferation until wound healing has completed but would also ensure that the extent of proliferation that occurs would match the amount of tissue that has been lost.
Much progress has been made in linking the early changes that follow damage to pathways that promote cell proliferation. Whereas many of the signaling pathways that regulate regenerative growth are also used during normal development, a key difference appears to be the way in which those signaling pathways are activated during regeneration. Key promoters of regenerative cell proliferation include the Wnt ligand Wingless (Wg) (Gibson and Schubiger 1999; McClure et al. 2008; Smith-Bolton et al. 2009; Harris et al. 2016), Myc (Smith-Bolton et al. 2009), the JAK/STAT pathway (Katsuyama et al. 2015; Santabárbara-Ruiz et al. 2015; La Fortezza et al. 2016), the EGFR/Ras pathway (Fan et al. 2014), and Yorkie, the transcriptional coactivator downstream of the Hippo pathway (Grusche et al. 2011; Sun and Irvine 2011; Repiso et al. 2013; Meserve and Duronio 2015). JNK has been shown to activate the expression of wg (Harris et al. 2016), the unpaired family of ligands (Upd, Upd2, and Upd3) (Pastor-Pareja et al. 2008; Bunker et al. 2015; Katsuyama et al. 2015; Santabárbara-Ruiz et al. 2015; La Fortezza et al. 2016), which are ligands upstream of the JAK/STAT pathway, and spitz (spi), which encodes a ligand for EGFR (Fan et al. 2014). Indeed, investigations of the transcriptional changes during regeneration predict that JNK/AP1 regulates the expression of many of these genes (Blanco et al. 2010; Katsuyama et al. 2015; Khan et al. 2017; Harris et al. 2020). Increased Yki activity likely results from a combination of increased JNK activity (Sun and Irvine 2011) and changes in mechanical forces (Dupont et al. 2011) caused by tissue damage. In addition to regulating proliferation, components of the Hippo pathway regulate the orientation of cell division of the proliferating cells so as to position new cells to most effectively replace those that have been lost (Repiso et al. 2013). Myc can be activated by Wg via a double-repression mechanism involving Notch (Smith-Bolton et al. 2009) and by increased Yki activity (Neto-Silva et al. 2010). Thus, JNK appears to be a key player in activating multiple pathways that promote regeneration, and cells of the regenerated tissue are derived from cells that activated the JNK pathway (Bosch et al. 2008). Importantly, damage to the same tissue in different ways can involve different pathways during regeneration (Diaz-Garcia and Baonza 2013; Herrera et al. 2013), suggesting that different tissues might also use distinct mechanisms.
There are hints from studies in many organisms that apoptosis is necessary for optimal regeneration (Tseng et al. 2007; for review, see Guerin et al. 2021). Cell death is observed near the cut edges of damaged discs and extensive cell death occurs when genetic tissue ablation systems are used (Álvarez-Fernández et al. 2015; Diaz-Garcia et al. 2016; Yoo et al. 2016; Klemm et al. 2021). Apoptosis itself has been shown to induce additional apoptosis within the tissues (via apoptosis-induced apoptosis) (Pérez-Garijo et al. 2013) and also promotes proliferation (via apoptosis-induced proliferation) (for review, see Fogarty and Bergmann 2017). Recent work has discovered that inducing necrosis in a section of the wing disc also prompts apoptosis in the adjacent tissue, termed “necrosis-induced apoptosis,” and that this apoptosis is necessary for efficient regeneration (Klemm et al. 2021). Thus, dying cells or those that have undergone a near-death experience appear to promote regeneration, likely by secreting factors that promote regeneration (reviewed by Morata et al. 2011; Ryoo and Bergmann 2012; Fogarty and Bergmann 2017). As cells initiate apoptosis, many studies have shown that dying cells release morphogens, including Wg and Dpp to promote other cells to be their replacement (Huh et al. 2004; Pérez-Garijo et al. 2004; Ryoo et al. 2004). The initiator caspase DRONC was discovered to function in an amplification loop that activates JNK signaling and the stress-responsive p53 transcription factor (Shlevkov and Morata 2012), which is known to be important for promoting both compensatory proliferation and regeneration (Wells et al. 2006; Wells and Johnston 2012). This amplification loop promotes ROS production and JNK activation, and, if left unchecked, can promote tissue overgrowth (Pérez-Garijo et al. 2009; Pinal et al. 2018). Recent work has uncovered a role for the unconventional Myosin1D (Myo1D) in sequestering DRONC to the plasma membrane and specifically activating Duox to produce ROS (Fig. 2E; Amcheslavsky et al. 2018).
There has been recent progress in understanding the mechanisms that allow cells to survive high ROS and JNK levels caused by tissue damage and regeneration. The activation of the JAK/STAT pathway (La Fortezza et al. 2016; Verghese and Su 2016) and arresting cells in the G2 phase of the cell cycle (Cosolo et al. 2019) both promote cell survival. Ways of marking cells that survive caspase activation and their descendants have been developed, and these studies show that they can make extensive contributions to regenerated tissue (Sun et al. 2020).
Ultimately, the clearance of dying cells from the epithelium is vital for proper wound healing and regeneration (Yoo et al. 2016; Iida et al. 2019). Two axon guidance pathways, Semaphorin-PlexA (Yoo et al. 2016) and Slit/Robo (Iida et al. 2019) are required for promoting the extrusion of dying cells during regeneration. The ligand Slit is up-regulated in a JNK-dependent manner within the dying cells and is critical for the regeneration of damaged discs (Iida et al. 2019). Damaged tissues are known to recruit circulating immune cells, called hemocytes, via JAK/STAT signaling (Pastor-Pareja et al. 2008) and ROS activity (Fogarty et al. 2016). These hemocytes likely play a role in cleaning up cellular debris (Fig. 3A–C). However, the importance of hemocytes is unclear, because tissues were still able to regenerate without help from hemocytes (Katsuyama and Paro 2013). In this case, the adjacent epithelial cells may engulf and clean up the cellular debris, as has been observed for cell competition (for reviews, see Amoyel and Bach 2014; Ohsawa et al. 2018). A better understanding of these non-cell-autonomous pro-regenerative mechanisms is likely to emerge in the coming years. Links between the earliest changes in damaged imaginal discs and the activation of pro-regenerative pathways are summarized in Figure 2. Many of the same molecular pathways that function in imaginal disc regeneration have also been found to regulate stem-cell-based replacement and regeneration of the adult fly gut (for review, see Zhang and Edgar 2021).
Figure 3.
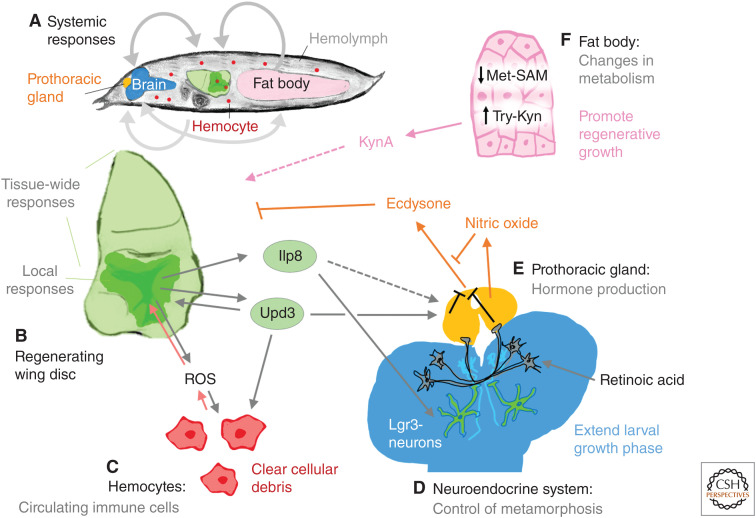
Systemic regenerative response. (A) Inter-organ signaling events between the damaged imaginal disc and other tissues. (B) Cells of the blastema in regenerating imaginal discs secrete a number of ligands that are known to impact distant tissues. (C) Hemocytes are recruited to the damaged tissue. (D,E) The brain (D) and prothoracic gland (E) make up the neuroendocrine system. Neurons in the brain respond to secreted signals (such as Ilp8) and prevent the prothoracic gland from producing the hormone ecdysone. (F) The larval fat body shows several changes in metabolism during regeneration that are necessary for regeneration to occur.
Repairing tissue damage in a developing organism poses some additional challenges. Tissues in general seem to become less capable of regeneration as they mature (see below). Additionally, in Drosophila, the onset of metamorphosis would prevent further intake of nutrients that are likely required to fuel regeneration. Damage to imaginal discs induces a delay in metamorphosis (Smith-Bolton et al. 2009; Halme et al. 2010; Hackney et al. 2012), indicating that mechanisms have evolved that delay pupariation following damage. The best characterized mechanism involves the production of the insulin/relaxin family member Ilp8 by damaged tissue (Fig. 3B; Colombani et al. 2012; Garelli et al. 2012). Ilp8 binds to the Lgr3 receptor in the brain and prothoracic gland to suppress the production of the steroid hormone ecdysone, which drives metamorphosis (Fig. 3D,E; Colombani et al. 2015; Garelli et al. 2015; Vallejo et al. 2015; Jaszczak et al. 2016). Ilp8 also activates nitric oxide synthase in the prothoracic gland, which inhibits ecdysone production (Fig. 3E; Jaszczak et al. 2015). Indeed, Ilp8 levels are crucial for effective regeneration (Garelli et al. 2012; Jaszczak et al. 2015; Skinner et al. 2015; Kashio et al. 2016). Other signaling molecules are known to delay the production of ecdysone including retinoic acid (Halme et al. 2010). Recent work has shown that the Unpaired3 (Upd3) protein that activates the JAK/STAT pathway can also inhibit ecdysone production by the PG (Romão et al. 2021). Other proteins that are involved in tissue patterning such as Hedgehog (Rodenfels et al. 2014) and the BMP 2/4 ortholog Dpp can also function as inter-organ signals to delay pupariation (Setiawan et al. 2018). In addition, inter-organ communication, either directly or indirectly, between the regenerating tissue and the larval fat body is critical for promoting regeneration (Kashio et al. 2016; Kashio and Miura 2020). Recent work has identified that a metabolite of Tryptophan, kynurenic acid (KynA), which is produced at a distance in the fat body, plays a role in promoting regenerative growth (Fig. 3F; Kashio and Miura 2020). Thus, it is likely that more humoral mechanisms will be discovered in the coming years that alter organismal physiology in ways that are conducive to regeneration.
REGENERATIVE CAPACITY REGULATED BY DAMAGE-RESPONSIVE ENHANCERS
A common feature of regeneration in many organisms is that the capacity for regeneration decreases as organisms mature. Notable examples include the response to amputation of limbs in tadpoles as they progress through metamorphosis (Dent 1962). Even more dramatic is the ability of the mouse heart to regenerate following an experimentally induced infarct. In the first week of life, mice can regenerate damaged cardiac tissue, but this ability is rapidly lost and infarcts after this period result in fibrotic scars (Porrello et al. 2011). As in these other situations, Drosophila imaginal discs lose the capacity for regeneration following damage in situ as they progress through the third larval instar (L3). This is observed following genetic ablation (Smith-Bolton et al. 2009) and when whole larvae are irradiated (Halme et al. 2010). Examination of the response to discs to damage at different points in L3 show clear differences. Whereas the JNK/AP1 pathway appears to be activated to similar levels at both time points, other JNK-responsive genes that are expressed near the site of damage including wg and Ilp8 are expressed at much reduced levels in late L3 (Smith-Bolton et al. 2009; Harris et al. 2016).
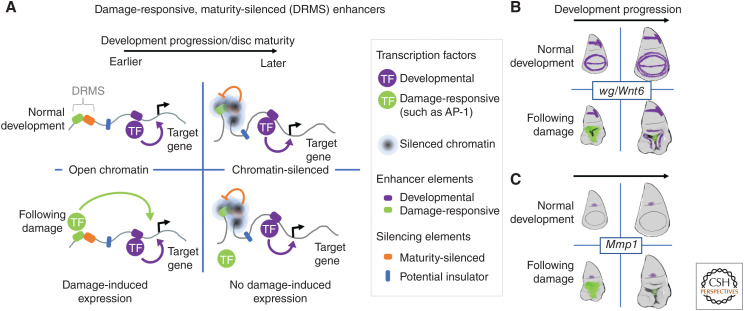
A damage-responsive enhancer is found between the wg and Wnt6 genes (Fig. 4A,B; Schubiger et al. 2010; Harris et al. 2016). Expression of both genes following damage requires this enhancer and a deletion that removes this enhancer significantly reduces regeneration (Harris et al. 2016). Molecular dissection of this enhancer (Harris et al. 2016) revealed a bipartite structure—it is composed of two separable elements. The first element is a damage-responsive element, contains binding sites for the JNK-responsive transcription factor AP-1, and is equally active in both immature and mature larvae. The second adjacent element has no enhancer activity on its own, but it can attenuate the damage-responsive enhancer in cis. This silencing activity becomes more effective as the larva matures and correlates with highly localized Polycomb-mediated silencing of the damage-responsive enhancer as characterized by increased H3K27 trimethylation. The increase in H3K27 trimethylation was found to be confined to the enhancer and did not spread to the adjacent genes. This provides an explanation for how damage-responsive expression of both genes could be curtailed without compromising their expression using other enhancers that function at later stages of development or for homeostatic processes. The expression of other genes that promote regeneration including Matrix metalloproteinase 1 (Mmp1) (Fig. 4C; McClure et al. 2008; Harris et al. 2020) and asperous (aspr), which encodes a protein with multiple EGF repeats that appears to be secreted (Harris et al. 2020), are regulated by damage-responsive maturity-silenced (DRMS) enhancers (Harris et al. 2020). Thus, the reduction in the capacity for regeneration as larvae mature can be accounted for, in significant part, by the silencing of damage-responsive enhancers at multiple loci. Damage-responsive enhancers have also been identified in vertebrates (for example, see Guenther et al. 2015; Kang et al. 2016; Wang et al. 2020). It will be of interest to know whether the loss of regenerative capacity that accompanies maturity in vertebrates, such as the loss of heart regeneration in newborn mice over the first week (Porrello et al. 2011), is characterized by epigenetic silencing of such enhancers.
Figure 4.
Mechanism for reducing regenerative capacity with organismal maturation. (A) The localized silencing of damage-responsive enhancers at multiple loci appears to be an important mechanism for reducing regeneration as the larva matures. Sparing the coding regions of the genes from silencing allows these genes to continue to be used for development and homeostasis under the control of other unsilenced enhancers. (B,C) Examples of when and where these damage-responsive, maturity-silenced enhancers are active are shown in green. Gene expression from developmental enhancers is shown in purple. Note that the damage-responsive enhancer can be silenced without interfering with normal developmental expression. (TF) Transcription factor.
Can this decline in regenerative capacity be reversed? Given that silencing at multiple loci might have a major role in curtailing regeneration, it is not unexpected that restoring the expression of individual genes such as wg cannot enable imaginal disc regeneration in mature larvae. Some improvement in regeneration in mature discs has been observed by increasing levels of the growth-promoting protein Myc (Smith-Bolton et al. 2009; Harris et al. 2016), by reducing function of the extra sex combs (esc) gene that encodes a component of the PRC2 complex (Harris et al. 2020) or by reducing the level of the Broad (Br) transcription factor (Narbonne-Reveau and Maurange 2019). Chinmo and Broad are transcription factors that are expressed in immature and mature discs, respectively, and each appears to antagonize the function of the other (Narbonne-Reveau and Maurange 2019). The switch from chinmo expression to expression of the br isoform br-Z1 coincides with the increase of ecdysone levels and passage through “critical weight” (Beadle et al. 1938; Mirth et al. 2005) when larvae proceed to pupariation even if they are deprived of food. Thus far, a mechanistic link between the onset of br-Z1 expression and the silencing of damage-responsive enhancers has not been established.
The increase in ecdysone levels that occurs as imaginal discs mature could also impact regeneration by its effect on the architecture of the imaginal disc. Cells in damaged imaginal discs secrete Ilp8 via their apical surfaces. The released Ilp8 protein acts on receptors in the brain and prothoracic gland to delay pupariation (Fig. 3). Once epithelial continuity is reestablished in discs, Ilp8 would accumulate in the lumen between the disc proper and the peripodial epithelium and therefore it would need to pass between epithelial cells of the disc to reach these distant locations. It has been found that the epithelial barrier becomes less permeable as discs mature, which could potentially limit the ability of mature discs to sufficiently delay pupariation to regenerate (DaCrema et al. 2021). However, other mechanisms (e.g., silencing of DRMS enhancers) must operate in parallel, since even massive tissue damage in mature discs that would be expected to disrupt epithelial barrier function does not elicit a developmental delay (Smith-Bolton et al. 2009; Harris et al. 2016).
PLASTICITY AND REPATTERNING DURING REGENERATION
When a contiguous group of cells is lost as a result of damage, adjacent cells often need to either respecify their fates or generate cells with altered fates so as to replace the lost cells, Thus, regeneration requires some degree of cell plasticity. Indeed lineage-tracing experiments in regenerating wing discs have shown plasticity between multiple different cell fates including vein–intervein (Repiso et al. 2013) and anterior–posterior (Szabad et al. 1979; Herrera and Morata 2014) fates. What are the molecular and cellular mechanisms that regulate cellular plasticity during regeneration? This fundamental question of how cells properly regulate the ability to change fate has been approached in several ways, and genetic screens have uncovered genes that are important for both maintaining proper fate in the face of damage and for promoting cellular plasticity.
Clear examples of tissue injury promoting such cellular reprogramming in imaginal discs comes from the pioneering work on transdetermination by the Hadorn group (Hadorn 1965), which preceded the work on reprogramming fibroblasts into pluripotent stem cells (Takahashi and Yamanaka 2006) by decades. This work demonstrated that cells with seemingly established cell fates could be reprogrammed to very different fates. Their work showed that depending on the region of the disc damaged, the regenerating tissue would occasionally adopt a new determined state (for review, see Worley et al. 2012). Thus, while some plasticity is necessary during regeneration, excessive plasticity can be deleterious and hence plasticity needs to be tightly regulated. Even more intriguing was the observation that transdetermination resulted not from a stochastic fate change in a single cell, but from a coordinated fate change in a small group of cells (∼3–5 cells) (Gehring 1966; Sustar et al. 2011), a phenomenon that can be described as “collective cell reprogramming.” The mechanism by which multiple cells undergo rare cell fates changes together remains mysterious.
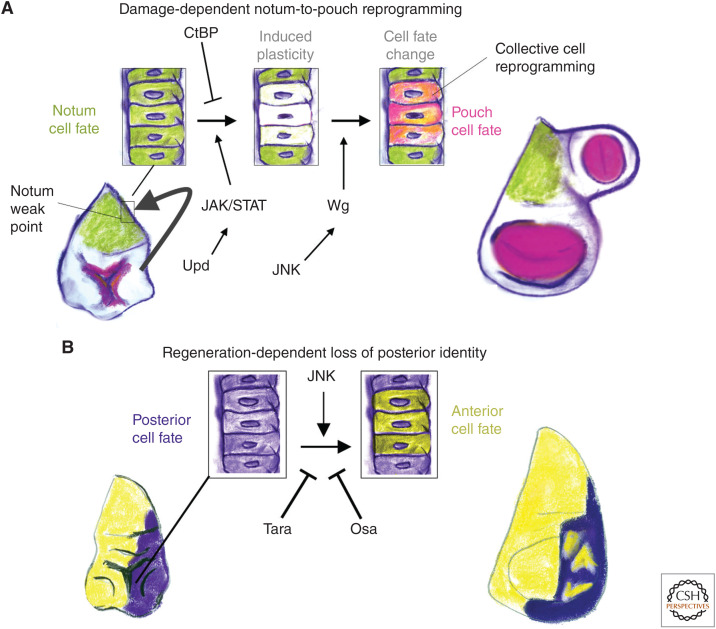
A damage-induced cell fate change from notum to wing pouch is observed at varying frequencies when the imaginal discs are damaged by either X-rays (Verghese and Su 2017) or with a genetic tissue-ablation system (Worley et al. 2018). This damage-induced cell fate change and the subsequent growth of an ectopic wing pouch results in the presence of an additional wing blade from a secondary location on the dorsal thorax (Worley et al. 2018). This phenotype was used to screen for genes that are important for maintaining identity during the regenerative process and identified the transcriptional corepressor carboxy-terminal binding protein (CtBP), CtBP as being critical for notum cells, specifically at a “weak point,” to maintain their identity in the face of tissue damage (Fig. 5A; Worley et al. 2018). Interestingly, this form of transdetermination also involved collective cell reprogramming, as shown by lineage tracing (Worley et al. 2018). It is not yet known whether multiple cells are simultaneously reprogrammed or whether a leader cell takes on a new identity and recruits others. In addition, there is some indication that cellular reprogramming and repatterning during regeneration can occur in the absence of cell division (Diaz-Garcia and Baonza 2013; Verghese and Su 2017), highlighting the importance of remodeling during the regenerative process. Many of the same molecular pathways that promote collective cell reprogramming (Verghese and Su 2017; Worley et al. 2018), namely, JNK and JAK/STAT, also promote cellular plasticity during regeneration (Lee et al. 2005; Herrera et al. 2013; Herrera and Morata 2014; Ahmed-de-Prado et al. 2018).
Figure 5.
Regulation of plasticity during regeneration. Two types of cellular reprogramming during regeneration are shown. (A) Damage-induced notum-to-pouch transformations are promoted by JNK-induced Wg and by JAK/STAT signaling. The transcriptional co-repressor carboxy-terminal binding protein (CtBP) acts to prevent collective cell reprogramming and maintain proper notum identity during regeneration. (B) High JNK signaling can promote cells to switch their compartment identities at relatively late stages of regeneration. This inappropriate cell fate change is resisted by the chromatin regulators Taranis and Osa. Together, these chromatin regulators maintain proper levels of the posterior transcription factor Engrailed (En) to prevent posterior cells adopting an anterior cell fate.
There are chromatin modifiers that are critical for proper initiation of the regenerative program, potentially by regulating the accessibility of damage-responsive enhancers that allow for the expression of pro-regenerative genetic programs (Harris et al. 2016, 2020; Vizcaya-Molina et al. 2018; Tian and Smith-Bolton 2021). There are also chromatin modifiers that act at a later point during regeneration, to ensure that the cells that have contributed to the regenerate are able to maintain their proper identity (Fig. 5B). These include Taranis (Tara) and Osa (a subunit of the BAP chromatin remodeling complex), which have been shown to be important for regulating the expression levels of the gene engrailed (en) that encodes a transcription factor that is necessary for establishing and maintaining the identity of cells in the posterior compartment (Schuster and Smith-Bolton 2015; Tian and Smith-Bolton 2021). When Engrailed levels are not properly maintained, cells can switch to expressing anterior compartment markers. Interestingly, Tara also modulates cell-fate changes induced by the overexpression of the constitutively active transcription factor Yorkie (Bairzin et al. 2020).
Genes and molecular pathways implicated in transdetermination include JNK, chromatin modifiers, such as Polycomb, as well as Wingless and the JAK/STAT signaling pathways (McClure and Schubiger 2008; Worley et al. 2018). Transdetermination events were observed to occur in regions of discs that showed increased JNK activity. Increased JNK activity resulted in decreased expression of several PcG group genes (Lee et al. 2005) that could normally serve to reduce expression of genes that might direct alternate fates. Increased expression of wg promotes leg-to-wing transdetermination (Maves and Schubiger 1995, 1998; Johnston and Schubiger 1996), although the mechanism for this is still not clear. This system was used to search for genes whose expression was increased during transdetermination using microarrays (Klebes et al. 2005). Interestingly, the most up-regulated gene was later identified as the pupariation-delaying factor Ilp8 (Colombani et al. 2012; Garelli et al. 2012). The screens for genes that regulate transdetermination are far from saturation and future screens are likely to provide more mechanistic insights into the phenomenon of collective cell reprogramming.
CONCLUDING REMARKS
The study of imaginal disc regeneration offers a wonderful opportunity to address fundamental questions in the biology of regeneration both because of its simplicity and because of the availability of many sophisticated genetic tools. The development of genetic tissue ablation systems has transformed a field that previously required technically difficult and laborious experimentation to one that can be conducted in most laboratories. In the coming years, we expect single-cell multiomics-based approaches to have a major impact, as we are already learning more about the unique cell types within developing imaginal discs (Bageritz et al. 2019; Deng et al. 2019; Zappia et al. 2020; Everetts et al. 2021), and that regeneration is driven by interactions between small heterogeneous populations of cells. Single-cell transcriptomics and single-cell chromatin profiling (Chappell et al. 2018) will provide important insights into gene-regulatory networks that drive regeneration, and also inform about cellular states in different regions of the disc—those actively participating in regeneration and those that appear to be bystanders. Techniques such as lattice-light sheet microscopy (Lemon and McDole 2020) and expansion microscopy (Chen et al. 2015) will provide us with detailed information regarding the architecture of the blastema and might reveal an important role for cellular processes that have thus far escaped our attention. These technological innovations coupled with experimental genetics should provide many exciting insights into regeneration in the coming years.
ACKNOWLEDGMENTS
We apologize to those whose work we have neglected to cover in this brief review that summarizes 70 years of research. We thank Donald Fox, Robin Harris, Rachel Smith-Bolton, and Nicholas Everetts for comments on the manuscript. We welcome feedback that brings important work that we might have overlooked to our attention.
Footnotes
Editors: Kenneth D. Poss and Donald T. Fox
Additional Perspectives on Regeneration available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abbott LC, Karpen GH, Schubiger G. 1981. Compartmental restrictions and blastema formation during pattern regulation in Drosophila imaginal leg discs. Dev Biol 87: 64–75. 10.1016/0012-1606(81)90061-0 [DOI] [PubMed] [Google Scholar]
- Ahmed-de-Prado S, Baonza A. 2018. Drosophila as a model system to study cell signaling in organ regeneration. Biomed Res Int 2018: 7359267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed-de-Prado S, Diaz-Garcia S, Baonza A. 2018. JNK and JAK/STAT signalling are required for inducing loss of cell fate specification during imaginal wing discs regeneration in Drosophila melanogaster. Dev Biol 441: 31–41. 10.1016/j.ydbio.2018.05.021 [DOI] [PubMed] [Google Scholar]
- Álvarez-Fernández C, Tamirisa S, Prada F, Chernomoretz A, Podhajcer O, Blanco E, Martín-Blanco E. 2015. Identification and functional analysis of healing regulators in Drosophila. PLoS Genet 11: e1004965. 10.1371/journal.pgen.1004965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A, Wang S, Fogarty CE, Lindblad JL, Fan Y, Bergmann A. 2018. Plasma membrane localization of apoptotic caspases for non-apoptotic functions. Dev Cell 45: 450–464.e3. 10.1016/j.devcel.2018.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amoyel M, Bach EA. 2014. Cell competition: how to eliminate your neighbours. Development 141: 988–1000. 10.1242/dev.079129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bageritz J, Willnow P, Valentini E, Leible S, Boutros M, Teleman AA. 2019. Gene expression atlas of a developing tissue by single cell expression correlation analysis. Nat Methods 16: 750–756. 10.1038/s41592-019-0492-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairzin JCD, Emmons-Bell M, Hariharan IK. 2020. The Hippo pathway coactivator Yorkie can reprogram cell fates and create compartment-boundary-like interactions at clone margins. Sci Adv 6: eabe8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G, Tatum E, Clancy C. 1938. Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Bio Bull Mar Biol Lab, Woods Hole 75: 447–462. 10.2307/1537573 [DOI] [Google Scholar]
- Beira JV, Paro R. 2016. The legacy of Drosophila imaginal discs. Chromosoma 125: 573–592. 10.1007/s00412-016-0595-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergantiños C, Corominas M, Serras F. 2010. Cell death-induced regeneration in wing imaginal discs requires JNK signalling. Development 137: 1169–1179. 10.1242/dev.045559 [DOI] [PubMed] [Google Scholar]
- Blanco E, Ruiz-Romero M, Beltran S, Bosch M, Punset A, Serras F, Corominas M. 2010. Gene expression following induction of regeneration in Drosophila wing imaginal discs. Expression profile of regenerating wing discs. BMC Dev Biol 10: 94. 10.1186/1471-213X-10-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohn H. 1970. Intercalary regeneration and segmental gradients in the extremities of Leucophaea (Blattaria) larvae. II: Coxa and tarsus. Dev Biol 23: 355–379. 10.1016/0012-1606(70)90104-1 [DOI] [PubMed] [Google Scholar]
- Bosch M, Serras F, Martín-Blanco E, Baguñà J. 2005. JNK signaling pathway required for wound healing in regenerating Drosophila wing imaginal discs. Dev Biol 280: 73–86. 10.1016/j.ydbio.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Bosch M, Bagun J, Serras F. 2008. Origin and proliferation of blastema cells during regeneration of Drosophila wing imaginal discs. Int J Dev Biol 52: 1043–1050. 10.1387/ijdb.082608mb [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415. 10.1242/dev.118.2.401 [DOI] [PubMed] [Google Scholar]
- Brock AR, Seto M, Smith-Bolton RK. 2017. Cap-n-Collar promotes tissue regeneration by regulating ROS and JNK signaling in the Drosophila melanogaster wing imaginal disc. Genetics 206: 1505–1520. 10.1534/genetics.116.196832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant PJ. 1971. Regeneration and duplication following operations in situ on the imaginal discs of Drosophila melanogaster. Dev Biol 26: 637–651. 10.1016/0012-1606(71)90146-1 [DOI] [PubMed] [Google Scholar]
- Bryant PJ, Levinson P. 1985. Intrinsic growth control in the imaginal primordia of Drosophila, and the autonomous action of a lethal mutation causing overgrowth. Dev Biol 107: 355–363. 10.1016/0012-1606(85)90317-3 [DOI] [PubMed] [Google Scholar]
- Bryant SV, French V, Bryant PJ. 1981. Distal regeneration and symmetry. Science 212: 993–1002. 10.1126/science.212.4498.993 [DOI] [PubMed] [Google Scholar]
- Bunker BD, Nellimoottil TT, Boileau RM, Classen AK, Bilder D. 2015. The transcriptional response to tumorigenic polarity loss in Drosophila. eLife 4: e03189. 10.7554/eLife.03189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri-Robles C, Serras F, Corominas M. 2019. Role of D-GADD45 in JNK-dependent apoptosis and regeneration in Drosophila. Genes (Basel) 10: 378. 10.3390/genes10050378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson BM. 2007. Principles of regenerative biology. Academic, Cambridge, MA. [Google Scholar]
- Chappell L, Russell AJC, Voet T. 2018. Single-cell (multi)omics technologies. Annu Rev Genomics Hum Genet 19: 15–41. 10.1146/annurev-genom-091416-035324 [DOI] [PubMed] [Google Scholar]
- Chen F, Tillberg PW, Boyden ES. 2015. Optical imaging. Expansion microscopy. Science 347: 543–548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. 1993. Imaginal disc development. In The development of Drosophila melanogaster (ed. Bate M). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Cohen E, Allen SR, Sawyer JK, Fox DT. 2018. Fizzy-related dictates A cell cycle switch during organ repair and tissue growth responses in the Drosophila hindgut. eLife 7: e38327. 10.7554/eLife.38327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Leopold P. 2012. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336: 582–585. 10.1126/science.1216689 [DOI] [PubMed] [Google Scholar]
- Colombani J, Andersen DS, Boulan L, Boone E, Romero N, Virolle V, Texada M, Léopold P. 2015. Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr Biol 25: 2723–2729. 10.1016/j.cub.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Cosolo A, Jaiswal J, Csordás G, Grass I, Uhlirova M, Classen AK. 2019. JNK-dependent cell cycle stalling in G2 promotes survival and senescence-like phenotypes in tissue stress. eLife 8: e41036. 10.7554/eLife.41036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaCrema D, Bhandari R, Karanja F, Yano R, Halme A. 2021. Ecdysone regulates the Drosophila imaginal disc epithelial barrier, determining the length of regeneration checkpoint delay. Development 148: dev195057. 10.1242/dev.195057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale L, Bownes M. 1981. Wound healing and regeneration in the imaginal wing disc of Drosophila. Wilehm Roux Arch Dev Biol 190: 185–190. 10.1007/BF00848301 [DOI] [PubMed] [Google Scholar]
- Deng M, Wang Y, Zhang L, Yang Y, Huang S, Wang J, Ge H, Ishibashi T, Yan Y. 2019. Single cell transcriptomic landscapes of pattern formation, proliferation and growth in Drosophila wing imaginal discs. Development 146: dev179754. 10.1242/dev.179754 [DOI] [PubMed] [Google Scholar]
- Dent JN. 1962. Limb regeneration in larvae and metamorphosing individuals of the South African clawed toad. J Morphol 110: 61–77. 10.1002/jmor.1051100105 [DOI] [PubMed] [Google Scholar]
- Diaz-Garcia S, Baonza A. 2013. Pattern reorganization occurs independently of cell division during Drosophila wing disc regeneration in situ. Proc Natl Acad Sci 110: 13032–13037. 10.1073/pnas.1220543110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Garcia S, Ahmed S, Baonza A. 2016. Analysis of the function of apoptosis during imaginal wing disc regeneration in Drosophila melanogaster. PLoS ONE 11: e0165554. 10.1371/journal.pone.0165554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. 2011. Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183. 10.1038/nature10137 [DOI] [PubMed] [Google Scholar]
- Esteban-Collado J, Corominas M, Serras F. 2021. Nutrition and PI3K/Akt signaling are required for p38-dependent regeneration. Development 148: dev197087. 10.1242/dev.197087 [DOI] [PubMed] [Google Scholar]
- Everetts NJ, Worley MI, Yasutomi R, Yosef N, Hariharan IK. 2021. Single-cell transcriptomics of the Drosophila wing disc reveals instructive epithelium-to-myoblast interactions. eLife 10: e61276. 10.7554/eLife.61276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Wang S, Hernandez J, Yenigun VB, Hertlein G, Fogarty CE, Lindblad JL, Bergmann A. 2014. Genetic models of apoptosis-induced proliferation decipher activation of JNK and identify a requirement of EGFR signaling for tissue regenerative responses in Drosophila. PLoS Genet 10: e1004131. 10.1371/journal.pgen.1004131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer JA, Giniger E, Maniatis T, Ptashne M. 1988. GAL4 activates transcription in Drosophila. Nature 332: 853–856. 10.1038/332853a0 [DOI] [PubMed] [Google Scholar]
- Fogarty CE, Bergmann A. 2017. Killers creating new life: caspases drive apoptosis-induced proliferation in tissue repair and disease. Cell Death Differ 24: 1390–1400. 10.1038/cdd.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty CE, Diwanji N, Lindblad JL, Tare M, Amcheslavsky A, Makhijani K, Brückner K, Fan Y, Bergmann A. 2016. Extracellular reactive oxygen species drive apoptosis-induced proliferation via Drosophila macrophages. Curr Biol 26: 575–584. 10.1016/j.cub.2015.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Cohen E, Smith-Bolton R. 2020. Model systems for regeneration: Drosophila. Development 147: dev173781. 10.1242/dev.173781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French V, Bryant PJ, Bryant SV. 1976. Pattern regulation in epimorphic fields. Science 193: 969–981. 10.1126/science.948762 [DOI] [PubMed] [Google Scholar]
- Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. 2012. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336: 579–582. 10.1126/science.1216735 [DOI] [PubMed] [Google Scholar]
- Garelli A, Heredia F, Casimiro AP, Macedo A, Nunes C, Garcez M, Dias ARM, Volonte YA, Uhlmann T, Caparros E, et al. 2015. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat Commun 6: 8732. 10.1038/ncomms9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring W. 1966. Cell heredity and changes of determination in cultures of imaginal discs in Drosophila melanogaster. J Embryol Exp Morphol 15: 77–111. [PubMed] [Google Scholar]
- Gibson MC, Schubiger G. 1999. Hedgehog is required for activation of engrailed during regeneration of fragmented Drosophila imaginal discs. Development 126: 1591–1599. 10.1242/dev.126.8.1591 [DOI] [PubMed] [Google Scholar]
- Grusche FA, Degoutin JL, Richardson HE, Harvey KF. 2011. The Salvador/Warts/Hippo pathway controls regenerative tissue growth in Drosophila melanogaster. Dev Biol 350: 255–266. 10.1016/j.ydbio.2010.11.020 [DOI] [PubMed] [Google Scholar]
- Guenther CA, Wang Z, Li E, Tran MC, Logan CY, Nusse R, Pantalena-Filho L, Yang GP, Kingsley DM. 2015. A distinct regulatory region of the Bmp5 locus activates gene expression following adult bone fracture or soft tissue injury. Bone 77: 31–41. 10.1016/j.bone.2015.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerin DJ, Kha CX, Tseng KA. 2021. From cell death to regeneration: rebuilding after injury. Front Cell Dev Biol 9: 655048. 10.3389/fcell.2021.655048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney JF, Zolali-Meybodi O, Cherbas P. 2012. Tissue damage disrupts developmental progression and ecdysteroid biosynthesis in Drosophila. PLoS ONE 7: e49105. 10.1371/journal.pone.0049105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn E. 1965. Problems of determination and transdetermination. Brookhaven Symp Biol 18: 148–161. [Google Scholar]
- Hadorn E, Bertani G, Gallera J. 1949. Regulative capacity and field organization of male genital discs in Drosophila melanogaster. Roux's Arch Dev Biol 144: 31–70. 10.1007/BF00575293 [DOI] [PubMed] [Google Scholar]
- Halme A, Cheng M, Hariharan IK. 2010. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr Biol 20: 458–463. 10.1016/j.cub.2010.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariharan IK, Serras F. 2017. Imaginal disc regeneration takes flight. Curr Opin Cell Biol 48: 10–16. 10.1016/j.ceb.2017.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, Setiawan L, Saul J, Hariharan IK. 2016. Localized epigenetic silencing of a damage-activated WNT enhancer limits regeneration in mature Drosophila imaginal discs. eLife 5: e11588. 10.7554/eLife.11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RE, Stinchfield MJ, Nystrom SL, McKay DJ, Hariharan IK. 2020. Damage-responsive, maturity-silenced enhancers regulate multiple genes that direct regeneration in Drosophila. eLife 9: e58305. 10.7554/eLife.58305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynie J, Schubiger G. 1979. Absence of distal to proximal intercalary regeneration in imaginal wing discs of Drosophila melanogaster. Dev Biol 68: 151–161. 10.1016/0012-1606(79)90250-1 [DOI] [PubMed] [Google Scholar]
- Held LI Jr. 2002. Imaginal discs: The genetic and cellular logic of pattern formation. Cambridge University Press, Cambridge. [Google Scholar]
- Herrera SC, Morata G. 2014. Transgressions of compartment boundaries and cell reprogramming during regeneration in Drosophila. eLife 3: e01831. 10.7554/eLife.01831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera SC, Martín R, Morata G. 2013. Tissue homeostasis in the wing disc of Drosophila melanogaster: immediate response to massive damage during development. PLoS Genet 9: e1003446. 10.1371/journal.pgen.1003446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. 2004. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol 14: 1262–1266. 10.1016/j.cub.2004.06.015 [DOI] [PubMed] [Google Scholar]
- Iida C, Ohsawa S, Taniguchi K, Yamamoto M, Morata G, Igaki T. 2019. JNK-mediated Slit-Robo signaling facilitates epithelial wound repair by extruding dying cells. Sci Rep 9: 19549. 10.1038/s41598-019-56137-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczak JS, Halme A. 2016. Arrested development: coordinating regeneration with development and growth in Drosophila melanogaster. Curr Opin Genet Dev 40: 87–94. 10.1016/j.gde.2016.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczak JS, Wolpe JB, Dao AQ, Halme A. 2015. Nitric oxide synthase regulates growth coordination during Drosophila melanogaster imaginal disc regeneration. Genetics 200: 1219–1228. 10.1534/genetics.115.178053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczak JS, Wolpe JB, Bhandari R, Jaszczak RG, Halme A. 2016. Growth coordination during Drosophila melanogaster imaginal disc regeneration is mediated by signaling through the relaxin receptor Lgr3 in the prothoracic gland. Genetics 204: 703–709. 10.1534/genetics.116.193706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LA. 2005. Regeneration and transdetermination: new tricks from old cells. Cell 120: 288–290. 10.1016/j.cell.2005.01.022 [DOI] [PubMed] [Google Scholar]
- Johnston LA, Schubiger G. 1996. Ectopic expression of wingless in imaginal discs interferes with decapentaplegic expression and alters cell determination. Development 122: 3519–3529. 10.1242/dev.122.11.3519 [DOI] [PubMed] [Google Scholar]
- Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD. 2016. Modulation of tissue repair by regeneration enhancer elements. Nature 532: 201–206. 10.1038/nature17644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Schubiger G. 1981. Extensive regulatory capabilities of a Drosophila imaginal disk blastema. Nature 294: 744–747. 10.1038/294744a0 [DOI] [PubMed] [Google Scholar]
- Kashio S, Miura M. 2020. Kynurenine metabolism in the fat body non-autonomously regulates imaginal disc repair in Drosophila. iScience 23: 101738. 10.1016/j.isci.2020.101738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashio S, Obata F, Miura M. 2014. Interplay of cell proliferation and cell death in Drosophila tissue regeneration. Dev Growth Differ 56: 368–375. 10.1111/dgd.12139 [DOI] [PubMed] [Google Scholar]
- Kashio S, Obata F, Zhang L, Katsuyama T, Chihara T, Miura M. 2016. Tissue nonautonomous effects of fat body methionine metabolism on imaginal disc repair in Drosophila. Proc Natl Acad Sci 113: 1835–1840. 10.1073/pnas.1523681113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T, Paro R. 2013. Innate immune cells are dispensable for regenerative growth of imaginal discs. Mech Dev 130: 112–121. 10.1016/j.mod.2012.11.005 [DOI] [PubMed] [Google Scholar]
- Katsuyama T, Comoglio F, Seimiya M, Cabuy E, Paro R. 2015. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc Natl Acad Sci 112: E2327–E2336. 10.1073/pnas.1423074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SJ, Abidi SNF, Skinner A, Tian Y, Smith-Bolton RK. 2017. The Drosophila Duox maturation factor is a key component of a positive feedback loop that sustains regeneration signaling. PLoS Genet 13: e1006937. 10.1371/journal.pgen.1006937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiehle CP, Schubiger G. 1985. Cell proliferation changes during pattern regulation in imaginal leg discs of Drosophila melanogaster. Dev Biol 109: 336–346. 10.1016/0012-1606(85)90460-9 [DOI] [PubMed] [Google Scholar]
- Klebes A, Sustar A, Kechris K, Li H, Schubiger G, Kornberg TB. 2005. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development 132: 3753–3765. 10.1242/dev.01927 [DOI] [PubMed] [Google Scholar]
- Klemm J, Stinchfield MJ, Harris RE. 2021. Necrosis-induced apoptosis promotes regeneration in Drosophila wing imaginal discs. Genetics iyab144. 10.1093/genetics/iyab144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Fortezza M, Schenk M, Cosolo A, Kolybaba A, Grass I, Classen AK. 2016. JAK/STAT signalling mediates cell survival in response to tissue stress. Development 143: 2907–2919. [DOI] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R. 2005. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature 438: 234–237. 10.1038/nature04120 [DOI] [PubMed] [Google Scholar]
- Lemon WC, McDole K. 2020. Live-cell imaging in the era of too many microscopes. Curr Opin Cell Biol 66: 34–42. 10.1016/j.ceb.2020.04.008 [DOI] [PubMed] [Google Scholar]
- Makhijani K, To TL, Ruiz-González R, Lafaye C, Royant A, Shu X. 2017. Precision optogenetic tool for selective single- and multiple-cell ablation in a live animal model system. Cell Chem Biol 24: 110–119. 10.1016/j.chembiol.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J, Omelyanchuk L, Kyttala S, Turunen H, Nokkala S. 2005. Role of Jun N-terminal Kinase (JNK) signaling in the wound healing and regeneration of a Drosophila melanogaster wing imaginal disc. Int J Dev Biol 49: 391–399. 10.1387/ijdb.052006jm [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. 1995. Wingless induces transdetermination in developing Drosophila imaginal discs. Development 121: 1263–1272. 10.1242/dev.121.5.1263 [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. 1998. A molecular basis for transdetermination in Drosophila imaginal discs: interactions between wingless and decapentaplegic signaling. Development 125: 115–124. 10.1242/dev.125.1.115 [DOI] [PubMed] [Google Scholar]
- Maves L, Schubiger G. 2003. Transdetermination in Drosophila imaginal discs: a model for understanding pluripotency and selector gene maintenance. Curr Opin Genet Dev 13: 472–479. 10.1016/j.gde.2003.08.006 [DOI] [PubMed] [Google Scholar]
- McClure KD, Schubiger G. 2007. Transdetermination: Drosophila imaginal disc cells exhibit stem cell-like potency. Int J Biochem Cell Biol 39: 1105–1118. 10.1016/j.biocel.2007.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure KD, Schubiger G. 2008. A screen for genes that function in leg disc regeneration in Drosophila melanogaster. Mech Dev 125: 67–80. 10.1016/j.mod.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure KD, Sustar A, Schubiger G. 2008. Three genes control the timing, the site and the size of blastema formation in Drosophila. Dev Biol 319: 68–77. 10.1016/j.ydbio.2008.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- McLean PF, Cooley L. 2013. Protein equilibration through somatic ring canals in Drosophila. Science 340: 1445–1447. 10.1126/science.1234887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meserve JH, Duronio RJ. 2015. Scalloped and Yorkie are required for cell cycle re-entry of quiescent cells after tissue damage. Development 142: 2740–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C, Truman JW, Riddiford LM. 2005. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr Biol 15: 1796–1807. 10.1016/j.cub.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Morata G, Shlevkov E, Pérez-Garijo A. 2011. Mitogenic signaling from apoptotic cells in Drosophila. Dev Growth Differ 53: 168–176. 10.1111/j.1440-169X.2010.01225.x [DOI] [PubMed] [Google Scholar]
- Moreira S, Stramer B, Evans I, Wood W, Martin P. 2010. Prioritization of competing damage and developmental signals by migrating macrophages in the Drosophila embryo. Curr Biol 20: 464–470. 10.1016/j.cub.2010.01.047 [DOI] [PubMed] [Google Scholar]
- Morgan TH. 1901. Regeneration. Macmillan, New York. [Google Scholar]
- Narbonne-Reveau K, Maurange C. 2019. Developmental regulation of regenerative potential in Drosophila by ecdysone through a bistable loop of ZBTB transcription factors. PLoS Biol 17: e3000149. 10.1371/journal.pbio.3000149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso C, Wu Q, Brodskiy P, Garston G, Baker R, Fletcher A, Zartman J. 2015. Patterning of wound-induced intercellular Ca2+ flashes in a developing epithelium. Phys Biol 12: 056005. 10.1088/1478-3975/12/5/056005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva RM, de Beco S, Johnston LA. 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev Cell 19: 507–520. 10.1016/j.devcel.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niethammer P, Grabher C, Look AT, Mitchison TJ. 2009. A tissue-scale gradient of hydrogen peroxide mediates rapid wound detection in zebrafish. Nature 459: 996–999. 10.1038/nature08119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brochta DA, Bryant PJ. 1987. Distribution of S-phase cells during the regeneration of Drosophila imaginal wing discs. Dev Biol 119: 137–142. 10.1016/0012-1606(87)90215-6 [DOI] [PubMed] [Google Scholar]
- O'Connor JT, Stevens AC, Shannon EK, Akbar FB, LaFever KS, Narayanan NP, Gailey CD, Hutson MS, Page-McCaw A. 2021. Proteolytic activation of growth-blocking peptides triggers calcium responses through the GPCR Mthl10 during epithelial wound detection. Dev Cell 56: 2160–2175.e5. 10.1016/j.devcel.2021.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa S, Vaughen J, Igaki T. 2018. Cell extrusion: a stress-responsive force for good or evil in epithelial homeostasis. Dev Cell 44: 284–296. 10.1016/j.devcel.2018.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor-Pareja JC, Wu M, Xu T. 2008. An innate immune response of blood cells to tumors and tissue damage in Drosophila. Dis Model Mech 1: 144–154. 10.1242/dmm.000950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Garijo A, Martín FA, Morata G. 2004. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development 131: 5591–5598. 10.1242/dev.01432 [DOI] [PubMed] [Google Scholar]
- Pérez-Garijo A, Shlevkov E, Morata G. 2009. The role of Dpp and Wg in compensatory proliferation and in the formation of hyperplastic overgrowths caused by apoptotic cells in the Drosophila wing disc. Development 136: 1169–1177. 10.1242/dev.034017 [DOI] [PubMed] [Google Scholar]
- Pérez-Garijo A, Fuchs Y, Steller H. 2013. Apoptotic cells can induce non-autonomous apoptosis through the TNF pathway. eLife 2: e01004. 10.7554/eLife.01004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal N, Martín M, Medina I, Morata G. 2018. Short-term activation of the Jun N-terminal kinase pathway in apoptosis-deficient cells of Drosophila induces tumorigenesis. Nat Commun 9: 1541. 10.1038/s41467-018-04000-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinal N, Calleja M, Morata G. 2019. Pro-apoptotic and pro-proliferation functions of the JNK pathway of Drosophila: roles in cell competition, tumorigenesis and regeneration. Open Biol 9: 180256. 10.1098/rsob.180256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. 2011. Transient regenerative potential of the neonatal mouse heart. Science 331: 1078–1080. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter CJ, Tasic B, Russler EV, Liang L, Luo L. 2010. The Q system: a repressible binary system for transgene expression, lineage tracing, and mosaic analysis. Cell 141: 536–548. 10.1016/j.cell.2010.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzell W, Evans IR, Martin P, Wood W. 2013. Calcium flashes orchestrate the wound inflammatory response through DUOX activation and hydrogen peroxide release. Curr Biol 23: 424–429. 10.1016/j.cub.2013.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt CA, Bryant PJ. 1981. Wound healing in the imaginal discs of Drosophila. II: Transmission electron microscopy of normal and healing wing discs. J Exp Zool 216: 45–61. 10.1002/jez.1402160107 [DOI] [PubMed] [Google Scholar]
- Reinhardt CA, Hodgkin NM, Bryant PJ. 1977. Wound healing in the imaginal discs of Drosophila. I: Scanning electron microscopy of normal and healing wing discs. Dev Biol 60: 238–257. 10.1016/0012-1606(77)90122-1 [DOI] [PubMed] [Google Scholar]
- Repiso A, Bergantiños C, Corominas M, Serras F. 2011. Tissue repair and regeneration in Drosophila imaginal discs. Dev Growth Differ 53: 177–185. 10.1111/j.1440-169X.2010.01247.x [DOI] [PubMed] [Google Scholar]
- Repiso A, Bergantiños C, Serras F. 2013. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development 140: 3541–3551. 10.1242/dev.095760 [DOI] [PubMed] [Google Scholar]
- Restrepo S, Basler K. 2016. Drosophila wing imaginal discs respond to mechanical injury via slow InsP3R-mediated intercellular calcium waves. Nat Commun 7: 12450. 10.1038/ncomms12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenfels J, Lavrynenko O, Ayciriex S, Sampaio JL, Carvalho M, Shevchenko A, Eaton S. 2014. Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev 28: 2636–2651. 10.1101/gad.249763.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romão D, Muzzopappa M, Barrio L, Milán M. 2021. The Upd3 cytokine couples inflammation to maturation defects in Drosophila. Curr Biol 31: 1780–1787.e6. 10.1016/j.cub.2021.01.080 [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Bergmann A. 2012. The role of apoptosis-induced proliferation for regeneration and cancer. Cold Spring Harb Perspect Biol 4: a008797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. 2004. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell 7: 491–501. 10.1016/j.devcel.2004.08.019 [DOI] [PubMed] [Google Scholar]
- Santabárbara-Ruiz P, López-Santillán M, Martínez-Rodríguez I, Binagui-Casas A, Pérez L, Milán M, Corominas M, Serras F. 2015. ROS-induced JNK and p38 signaling is required for unpaired cytokine activation during Drosophila regeneration. PLoS Genet 11: e1005595. 10.1371/journal.pgen.1005595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santabárbara-Ruiz P, Esteban-Collado J, Pérez L, Viola G, Abril JF, Milán M, Corominas M, Serras F. 2019. Ask1 and Akt act synergistically to promote ROS-dependent regeneration in Drosophila. PLoS Genet 15: e1007926. 10.1371/journal.pgen.1007926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger G. 1971. Regeneration, duplication and transdetermination in fragments of the leg disc of Drosophila melanogaster. Dev Biol 26: 277–295. 10.1016/0012-1606(71)90127-8 [DOI] [PubMed] [Google Scholar]
- Schubiger M, Sustar A, Schubiger G. 2010. Regeneration and transdetermination: the role of wingless and its regulation. Dev Biol 347: 315–324. 10.1016/j.ydbio.2010.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster KJ, Smith-Bolton RK. 2015. Taranis protects regenerating tissue from fate changes induced by the wound response in Drosophila. Dev Cell 34: 119–128. 10.1016/j.devcel.2015.04.017 [DOI] [PubMed] [Google Scholar]
- Setiawan L, Pan X, Woods AL, O'Connor MB, Hariharan IK. 2018. The BMP2/4 ortholog Dpp can function as an inter-organ signal that regulates developmental timing. Life Sci Alliance 1: e201800216. 10.26508/lsa.201800216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon EK, Stevens A, Edrington W, Zhao Y, Jayasinghe AK, Page-McCaw A, Hutson MS. 2017. Multiple mechanisms drive calcium signal dynamics around laser-induced epithelial wounds. Biophys J 113: 1623–1635. 10.1016/j.bpj.2017.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlevkov E, Morata G. 2012. A dp53/JNK-dependant feedback amplification loop is essential for the apoptotic response to stress in Drosophila. Cell Death Differ 19: 451–460. 10.1038/cdd.2011.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner A, Khan SJ, Smith-Bolton RK. 2015. Trithorax regulates systemic signaling during Drosophila imaginal disc regeneration. Development 142: 3500–3511. 10.1242/dev.122564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Bolton RK, Worley MI, Kanda H, Hariharan IK. 2009. Regenerative growth in Drosophila imaginal discs is regulated by Wingless and Myc. Dev Cell 16: 797–809. 10.1016/j.devcel.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD. 2011. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol 350: 139–151. 10.1016/j.ydbio.2010.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Irvine KD. 2014. Control of growth during regeneration. Curr Top Dev Biol 108: 95–120. 10.1016/B978-0-12-391498-9.00003-6 [DOI] [PubMed] [Google Scholar]
- Sun G, Ding XA, Argaw Y, Guo X, Montell DJ. 2020. Akt1 and dCIZ1 promote cell survival from apoptotic caspase activation during regeneration and oncogenic overgrowth. Nat Commun 11: 5726. 10.1038/s41467-020-19068-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sustar A, Bonvin M, Schubiger M, Schubiger G. 2011. Drosophila twin spot clones reveal cell division dynamics in regenerating imaginal discs. Dev Biol 356: 576–587. 10.1016/j.ydbio.2011.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabad J, Simpson P, Nothiger R. 1979. Regeneration and compartments in Drosophila. J Embryol Exp Morphol 49: 229–241. [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126: 663–676. 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Tian Y, Smith-Bolton RK. 2021. Regulation of growth and cell fate during tissue regeneration by the two SWI/SNF chromatin-remodeling complexes of Drosophila. Genetics 217: 1–16. 10.1093/genetics/iyaa028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng AS, Adams DS, Qiu D, Koustubhan P, Levin M. 2007. Apoptosis is required during early stages of tail regeneration in Xenopus laevis. Dev Biol 301: 62–69. 10.1016/j.ydbio.2006.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursprung H. 1959. Fragmentation and radiation experiments to determine the determination and fate map of the Drosophila genital disc. Roux's Arch Dev Biol 151: 501–558. [Google Scholar]
- Vallejo DM, Juarez-Carreño S, Bolivar J, Morante J, Dominguez M. 2015. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science 350: aac6767. 10.1126/science.aac6767 [DOI] [PubMed] [Google Scholar]
- Verghese S, Su TT. 2016. Drosophila Wnt and STAT define apoptosis-resistant epithelial cells for tissue regeneration after irradiation. PLoS Biol 14: e1002536. 10.1371/journal.pbio.1002536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese S, Su TT. 2017. STAT, Wingless, and Nurf-38 determine the accuracy of regeneration after radiation damage in Drosophila. PLoS Genet 13: e1007055. 10.1371/journal.pgen.1007055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaya-Molina E, Klein CC, Serras F, Mishra RK, Guigó R, Corominas M. 2018. Damage-responsive elements in Drosophila regeneration. Genome Res 28: 1852–1866. 10.1101/gr.233098.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hu CK, Zeng A, Alegre D, Hu D, Gotting K, Ortega Granillo A, Wang Y, Robb S, Schnittker R, et al. 2020. Changes in regeneration-responsive enhancers shape regenerative capacities in vertebrates. Science 369: eaaz3090. 10.1126/science.aaz3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Johnston LA. 2012. Maintenance of imaginal disc plasticity and regenerative potential in Drosophila by p53. Dev Biol 361: 263–276. 10.1016/j.ydbio.2011.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells BS, Yoshida E, Johnston LA. 2006. Compensatory proliferation in Drosophila imaginal discs requires Dronc-dependent p53 activity. Curr Biol 16: 1606–1615. 10.1016/j.cub.2006.07.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley MI, Setiawan L, Hariharan IK. 2012. Regeneration and transdetermination in Drosophila imaginal discs. Annu Rev Genet 46: 289–310. 10.1146/annurev-genet-110711-155637 [DOI] [PubMed] [Google Scholar]
- Worley MI, Setiawan L, Hariharan IK. 2013. TIE-DYE: a combinatorial marking system to visualize and genetically manipulate clones during development in Drosophila melanogaster. Development 140: 3275–3284. 10.1242/dev.096057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley MI, Alexander LA, Hariharan IK. 2018. CtBP impedes JNK- and Upd/STAT-driven cell fate misspecifications in regenerating Drosophila imaginal discs. eLife 7: e30391. 10.7554/eLife.30391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Mayer F, Basler K. 2010. Refined LexA transactivators and their use in combination with the Drosophila Gal4 system. Proc Natl Acad Sci 107: 16166–16171. 10.1073/pnas.1005957107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Pascoe HG, Pereira T, Kondo S, Jacinto A, Zhang X, Hariharan IK. 2016. Plexins function in epithelial repair in both Drosophila and zebrafish. Nat Commun 7: 12282. 10.1038/ncomms12282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia MP, de Castro L, Ariss MM, Jefferson H, Islam AB, Frolov MV. 2020. A cell atlas of adult muscle precursors uncovers early events in fibre-type divergence in Drosophila. EMBO Rep 21: e49555. 10.15252/embr.201949555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zhang P, Edgar BA. 2021. Insect gut regeneration. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a040915 [DOI] [PMC free article] [PubMed] [Google Scholar]