ABSTRACT
Low-cost, refrigerator-stable COVID-19 vaccines will facilitate global access and improve vaccine coverage in low- and middle-income countries. To this end, subunit-based approaches targeting the receptor-binding domain (RBD) of SARS-CoV-2 Spike protein remain attractive. Antibodies against RBD neutralize SARS-CoV-2 by blocking viral attachment to the host cell receptor, ACE2. Here, a yeast-produced recombinant RBD antigen (RBD-L452K-F490W or RBD-J) was formulated with various combinations of aluminum-salt (Alhydrogel®, AH; AdjuPhos®, AP) and CpG 1018 adjuvants. We assessed the effect of antigen-adjuvant interactions on the stability and mouse immunogenicity of various RBD-J preparations. While RBD-J was 50% adsorbed to AH and <15% to AP, addition of CpG resulted in complete AH binding, yet no improvement in AP adsorption. ACE2 competition ELISA analyses of formulated RBD-J stored at varying temperatures (4, 25, 37°C) revealed that RBD-J was destabilized by AH, an effect exacerbated by CpG. DSC studies demonstrated that aluminum-salt and CpG adjuvants decrease the conformational stability of RBD-J and suggest a direct CpG-RBD-J interaction. Although AH+CpG-adjuvanted RBD-J was the least stable in vitro, the formulation was most potent at eliciting SARS-CoV-2 pseudovirus neutralizing antibodies in mice. In contrast, RBD-J formulated with AP+CpG showed minimal antigen-adjuvant interactions, a better stability profile, but suboptimal immune responses. Interestingly, the loss of in vivo potency associated with heat-stressed RBD-J formulated with AH+CpG after one dose was abrogated by a booster. Our findings highlight the importance of elucidating the key interrelationships between antigen-adjuvant interactions, storage stability, and in vivo performance to enable successful formulation development of stable and efficacious subunit vaccines.
KEYWORDS: COVID-19, vaccine, RBD, alum, CpG, adjuvant, formulation, stability, immunogenicity
Introduction
Monumental efforts over the past 2 years have led to the successful approval and use of first-generation COVID-19 vaccines in record time.1 Nevertheless, global vaccine access and vaccination coverage in Low- and Middle-Income countries (LMICs) remains well behind the rest of the globe.2 In fact, according to the World Health Organization (WHO), nearly 85% of the world’s current COVID-19 vaccine supply has gone to the well-to-do high- and upper-middle-income countries.3 Moreover, the two widely administered mRNA vaccines—Comirnaty (Pfizer-BioNTech) and Spikevax (Moderna)—are expensive and have ultra-low frozen storage temperature requirements, which present cold chain and delivery challenges in resource-limited LMICs.4–6 A next-generation, low-cost, and easily manufactured subunit COVID-19 vaccine stable at refrigerated (or ambient) temperatures is thus urgently needed to facilitate improved vaccine coverage in LMICs.7
SARS-CoV-2 is the etiologic agent of COVID-19. The surface of SARS-CoV-2 is studded with 40 copies of a trimeric club-shaped glycoprotein called spike. The receptor-binding domain (RBD) of spike protein promotes attachment to angiotensin-converting enzyme 2 (ACE2) and thereby promotes viral entry into host cells.8,9 Antibodies against multiple conformation-dependent epitopes on RBD are capable of blocking interactions with ACE2, thereby neutralizing SARS-CoV-2.10,11 Due to RBD’s important functional biological properties (e.g., ACE2 binding, key neutralizing epitopes, and independent folding),12 as well as its attractive developability properties (e.g., high thermal stability,13 low-cost manufacturing, and ease in scalability),14,15 there are numerous efforts to develop RBD-based subunit vaccine candidates.16,17 Due to limited immunogenicity, however, recombinant subunit vaccines invariably require the use of adjuvants, and RBD is no exception.18,19 Aluminum salts are the most commonly used adjuvants with well-established safety and efficacy records over the past 70 years and are in use in numerous approved vaccines, including DTaP, Tdap, Hep B, Men B, HPV, and anthrax.20–22 Aluminum salt adjuvants induce primarily humoral Th-2 type immune response and are widely available at low cost.23,24 In contrast, CpG oligodeoxynucleotide adjuvant is a TLR-9 agonist currently widely used in pre-clinical and clinical testing25,26 but commercially only in the hepatitis B subunit vaccine (HeplisavTM).27,28 CpG induces a Th1-type immune response and, therefore, can be used in synergy with aluminum salt adjuvants to elicit a robust and balanced cellular and humoral immune response.29–31
There are not only several reports of aluminum salt and/or CpG-adjuvanted RBD-based subunit vaccine candidates in pre-clinical and clinical development but there has also been a recent emergency use authorization in India of an RBD-based vaccine containing both aluminum salt and CpG adjuvants.32–44 In general, adjuvanted RBD-based candidates have been reported to induce robust neutralizing antibody responses and protection in SARS-CoV-2 challenges studies. Nevertheless, the key pharmaceutical attributes of candidate RBD-based vaccine formulations, such as the nature of antigen-adjuvant interactions, have only been reported for one RBD candidate formulated with aluminum salt adjuvant,45 and the effect of such interactions on the storage stability of RBD antigen has not been reported for any candidate. A careful evaluation and optimization of these pharmaceutical attributes is critical to optimize vaccine formulation in terms of preserving structural integrity and physicochemical properties of the antigen during manufacturing, long-term storage, and administration. Additionally, the development and use of appropriate analytical methods that can characterize the RBD on aluminum salt and indicate its stability are critical. Due to interference from aluminum salt adjuvants, the antigen often needs to be desorbed from the surface of the adjuvant prior to analysis.46 A desorbed antigen may have different conformational properties than an adjuvant-bound antigen, and therefore, the measured properties may not give a true picture of the antigen when adsorbed to an adjuvant.
In this work, we report on a combination of mouse immunogenicity and storage stability results with a recombinant RBD variant referred to as RBD-J (RBD-L452K-F490W) expressed in a low-cost Pichia pastoris expression system,36 and formulated in the presence of aluminum salt (Alhydrogel®; AH and AdjuPhos®; AP) and CpG adjuvants. Compared to wild-type RBD, RBD-J demonstrates enhanced production titers and ACE2 binding affinity, and is highly immunogenic in mice.36 We established analytical methods to characterize antigen-adjuvant interactions (SDS-PAGE, UV-Visible spectroscopy, and intact protein mass spectrometry) and determined their effects on RBD-J conformational stability (DSC). To monitor the storage stability of antigen adsorbed to aluminum salt adjuvant, we developed a competitive ELISA that measures the ACE2 binding activity of RBD-J without desorption. Next, we prepared various formulations of RBD-J with AH or AP in the presence and absence of CpG. We also assessed the effect of varying levels of antigen-adjuvant interactions on in vivo mouse immunogenicity and in vitro storage stability of RBD-J. Finally, we investigated the relationship between the stability of RBD-J formulations and their immunogenicity in mice.
Materials and methods
Materials
RBD-J was produced by secretion from Pichia pastoris, purified, and stored at −80°C in 20 mM NaPO4, 100 mM NaCl buffer (pH 8.0) as described.36 Frozen RBD-J was thawed at ambient temperature for 30 min, 0.02% PS80 (Thermo Scientific, 28329) was spiked in, and then the protein solution was dialyzed into a formulation buffer (20 mM histidine, 100 mM NaCl, 0.02% PS80, pH 6.5) using 3.5 K MWCO dialysis device (Thermo Scientific, 88400) as per manufacturer’s protocol and stored at 4°C until sample preparation on the same day. AH (10 mg/mL Al content, # vac-alu-250) and AP (5 mg/mL Al content, # vac-phos-250) were purchased from InvivoGen. CpG 1018 was obtained from Dynavax Technologies in the form of a lyophilized powder containing the oligonucleotide along with some residual NaCl. The freeze-dried material was reconstituted to ~12 mg/mL in 20 mM Tris, 100 mM NaCl buffer at pH 7.5 per the manufacturer’s protocol. Sodium chloride and sodium phosphate dibasic heptahydrate were purchased from Thermo Fisher, L-histidine, and sodium phosphate monobasic monohydrate were purchased from Sigma-Aldrich.
Methods
Experimental details of sample preparation and setup of antigen-adjuvant binding studies, as well as preparation of formulations used in mouse immunogenicity and storage stability studies, are provided in the Supplementary methods. Descriptions of the various physicochemical techniques (including SDS-PAGE, UV-Visible spectroscopy, intact protein mass spectrometry, and DSC) used in this work have been described previously (see ref.47) and are also described in the Supplementary methods. The competitive ELISA used to monitor binding of adjuvanted RBD-J to ACE2 (obtained through Global Health Vaccine Accelerator Platforms portal, GH-VAP) was adapted from methodologies described for other antigens.48 Experimental procedures and materials used for mouse immunogenicity studies including endpoint titer ELISA, multiplex immune assay, and pseudovirus neutralization assay are also provided in the Supplementary methods.
Results
Interaction of RBD-J antigen with aluminum salt adjuvants with and without the addition of CpG
The extent of RBD-J binding to AH and AP in the presence and absence of CpG was determined using quantitative SDS-PAGE analysis under reducing conditions (Figure 1a). The in-solution protein control for the standard curve (Lanes #1–5) resolved into a main band at ~30 kDa and higher molecular weight (HMW) species ranging up to ~160 kDa. PNGase-F treatment combined with SDS-PAGE and intact protein mass spectrometry analysis revealed that RBD-J is (1) glycosylated (hyper-mannosylated forms ranging from 9 to 23 units), and (2) has N-terminal heterogeneity (Pro-RBD-J) due to inefficient clipping of the [pro] region of the alpha-factor signal peptide (Suppl. Fig. S1, see discussion). These various species of RBD-J are functional and can bind ACE2 as observed by competitive ELISA (see below).
Figure 1.
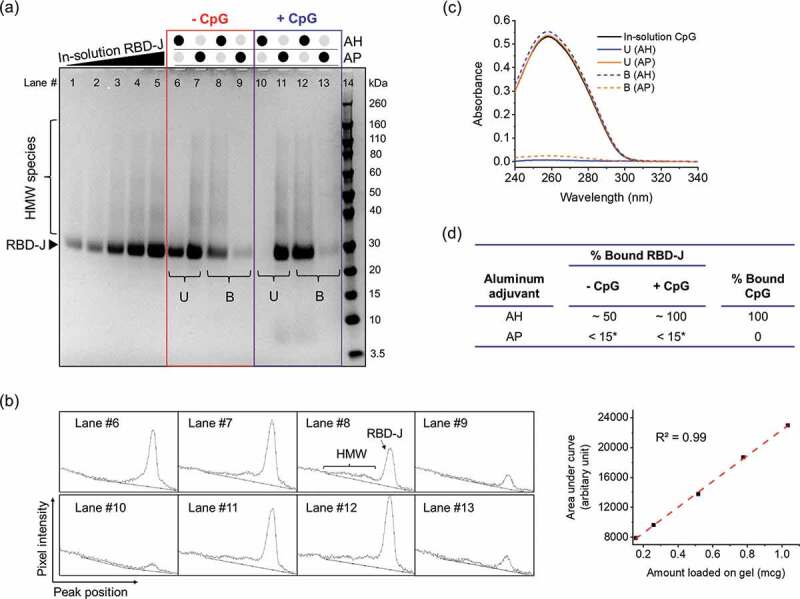
Interaction and binding of RBD-J and CpG to aluminum salt adjuvants (AH and AP). (a) Reducing SDS-PAGE to determine binding of RBD-J to AH and AP in the absence (highlighted in the red box, Lanes #6–9) and presence (highlighted in the blue box, Lanes #10–13) of CpG 1018. U= Unbound RBD-J, B = Bound RBD-J. Lanes #1–5 are increasing amounts of unadjuvanted in-solution RBD-J to generate standard curve for RBD-J quantitation. Lane #14 is a molecular weight standard. (b) Representative spectra from densitometric analysis of gel using ImageJ software along with the standard curve generated to quantify amount of unbound and bound protein (c) Representative UV-Visible spectroscopy spectra for CpG in unbound (U) and bound fractions (B) after adsorption to AH and AP, and (d) summary table with compiled results for percentage of RBD-J and CpG bound to AH and AP. *LOQ = 15%.
The total amount of RBD-J (main band + HMW species) bound (or unbound) to aluminum salt adjuvant in the presence and absence of CpG was determined by densitometric analysis of the gel (Figure 1b). As summarized in Figure 1d, RBD-J binds only partially to AH (~50%), and as shown in Figure 1b, the HMW species preferentially bind, the main band species partially bind, and thus the unbound fraction consists of the main band species only. Interestingly, when CpG is included, RBD-J binds 100% to AH (Figure 1d). In the case of AP, which has the opposite surface charge of AH under these solution conditions, much less RBD-J binds (< LOQ = 15%), and CpG addition does not affect antigen-adjuvant binding (see discussion).
The binding of CpG to both AH and AP in the presence of RBD-J was determined by measuring the A260 of supernatant and pellet fraction using UV-Visible spectroscopy (Figure 1c). In the presence of RBD-J, CpG (a negatively charged oligodeoxynucleotide) binds completely to the positively charged AH and does not bind to the negatively charged AP (Figure 1d). Similar results were observed without RBD-J (data not shown). In summary, in the case of AH, CpG increases the levels of AH-bound RBD-J (from ~50% to ~100%), but RBD-J has no effect on AH-bound CpG (100% bound). In the case of AP, both CpG and RBD-J separately or in combination remain mostly unbound (<15% bound).
Effect of antigen-adjuvant interactions on mouse immunogenicity of RBD-J formulations
Based on our understanding of RBD-J-adjuvant interactions described above, we prepared seven different formulations with varying degrees of RBD-J binding to aluminum salt adjuvants, with and without the addition of CpG (Table 1). Formulations F1 and F2 with AH contained ~50% bound and ~100% AH-bound RBD-J, respectively (the 100% AH-bound RBD-J sample was generated using 2X antigen levels followed by removal of supernatant upon ~50% AH binding) (see Supplementary methods). The F2 formulation contained a greater degree of HMW species of RBD-J due to their preferential binding to AH (see Discussion). In formulation F3, RBD-J was combined with both AH and CpG (AH+CpG) such that both RBD-J and CpG were completely (100%) bound to AH. In contrast, formulations F4 and F5 contained AP, without and with CpG, respectively, with <15% RBD-J bound to AP in both formulations. Formulation F6 contained RBD-J in solution with CpG, while formulation F7 was an unadjuvanted RBD-J control.
Table 1.
RBD-J formulations prepared to assess the effect of antigen-adjuvant interactions on mouse immunogenicity and storage stability. Seven formulations (F1-F7) of RBD-J were prepared in various combinations with AH, AP, and CpG. The composition of each formulation and the percentage of RBD-J and CpG bound to AH or AP is displayed along with the concentration of antigen and adjuvants in each formulation and the dose subcutaneously injected in mice (Balb/c, 50 µl injection volume). Note that the target concentration of RBD-J in each formulation was the same (100 mcg/ml). *LOQ = 15%.
| |
|
% Bound to aluminum adjuvant |
Concentration (mcg/mL) |
Dose injected in mice (mcg) Injection volume = 50 µL |
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Formulation No. | Adjuvant | RBD-J | CpG 1018 | RBD-J | Aluminum adjuvant | CpG 1018 | RBD-J | Aluminum adjuvant | CpG 1018 | No. of mice |
| F1 | AH | 50 | - | 100 | 1500 | - | 5 | 75 | - | 9 |
| F2 | AH | 100 | - | 100 | 1500 | - | 5 | 75 | - | 9 |
| F3 | AH+CpG | 100 | 100 | 100 | 1500 | 600 | 5 | 75 | 30 | 9 |
| F4 | AP | <15* | - | 100 | 1500 | - | 5 | 75 | - | 9 |
| F5 | AP+CpG | <15* | 0 | 100 | 1500 | 600 | 5 | 75 | 30 | 9 |
| F6 | CpG | - | - | 100 | - | 600 | 5 | - | 30 | 9 |
| F7 | No adjuvant | - | - | 100 | - | - | 5 | - | - | 9 |
The impact of varying extent of antigen-adjuvant interactions on immunogenicity was assessed in a mouse model, and a summary of the prepared formulations is shown in Table 1. Groups of BALB/c mice (n = 9 per group) received 5 mcg RBD-J on day 0, followed by a booster of the same dose and formulation on day 21. The degree of adsorption of RBD-J samples injected into mice on days 0 and 21 in each aluminum-adjuvanted formulation was confirmed by SDS-PAGE and no differences over time were observed. Serum samples were collected on days 21, 35, and 65 and tested for RBD and full-length trimeric spike-binding IgG, IgG subclass distributions, and SARS-CoV-2 neutralizing titers using a lentivirus-based pseudovirus (Figure 2a). On day 21, 8 out of 9 mice that received AH+CpG-adjuvanted RBD-J had robust anti-RBD-J serum IgG titers (mean titer >105) (Figure 2b). In the other groups of animals, only an occasional mouse seroconverted by day 21. After a boost on day 21, virtually all mice seroconverted by day 35 or 65, although there was a range of titers dependent on the formulation. Serum IgG reactivity to wild-type (WT) RBD and full-length trimeric spike mirrored that of RBD-J with the highest titers elicited by AH+CpG group (Suppl. Fig. S2a). Analysis of day 65 sera demonstrated that RBD-J formulations containing AH (50% bound), AH+CpG, and AP+CpG had higher WT RBD-specific titers as compared to other formulations (Suppl. Fig. S2a). Finally, the IgG response across all formulations consisted predominantly of IgG1, with detectable levels of IgG2a and IgG2b induced in AH+CpG and AP+CpG formulations (Suppl. Fig. S2b).
Figure 2.
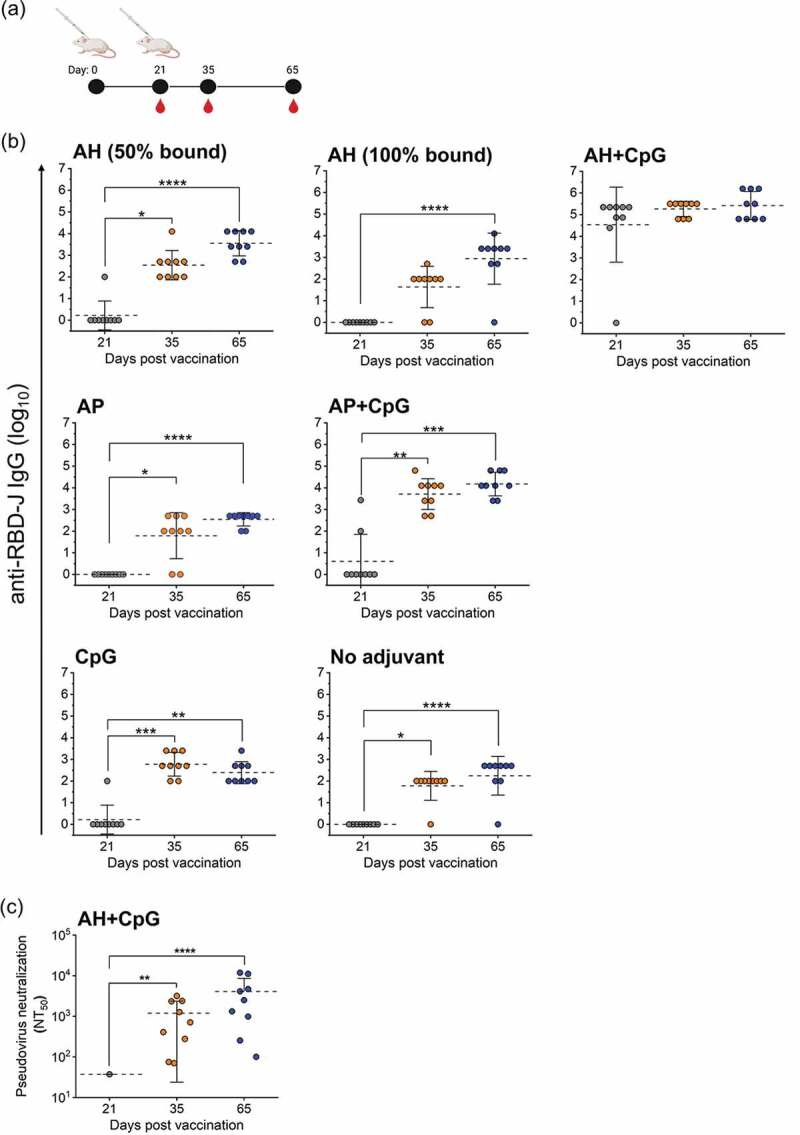
In vivo mouse immunogenicity of RBD-J adjuvanted with aluminum salt and CpG adjuvants. (a) BALB/c mice (n = 9) were given a subcutaneous primary vaccination on day 0 and boosted on day 21 with 5 mcg RBD-J in various combinations with aluminum salt and CpG adjuvants. Sera were collected on day 21, 35, and 65. (b) anti-RBD-J IgG titer for each formulation group and (c) Pseudovirus neutralization titers (NT50) for AH+CpG group on day 21, 35, and 65. All NT50 values were determined using a best-fit nonlinear regression on serum dilution curves. The dashed lines represent group mean and the errors bars indicate standard deviation. Significance between time-points was determined using Kruskal–Wallis test and post hoc Dunn’s multiple comparisons test (*p ≤ 0.05; **p ≤ 0.0; ***p ≤ 0.001; ****p ≤ 0.0001). Illustration in (a) was created with Biorender.com.
We used a lentivirus-based pseudovirus assay to assess SARS-CoV-2 neutralizing titers in mice that were vaccinated with the RBD-J adjuvanted with AH+CpG (Figure 2c). On day 21, only a single mouse had detectable neutralization titers (NT50). However, by day 35 (2 weeks after boost), all nine mice in this group achieved NT50 titers and those titers increased further by day 65. In contrast, despite all other RBD-J formulations yielding elevated endpoint titers at days 35 and 65, only one mouse given AH (50% bound) and 2 mice given AP+CpG formulations had detectable neutralizing responses on day 65 (data not shown). In summary, only the AH+CpG-containing formulation of RBD-J induced a robust virus-neutralizing antibody response, while the other adjuvant combinations primarily elicited non-neutralizing antibodies.
Development of competitive ELISA method to determine storage stability of adjuvanted RBD-J formulations by measuring antigen binding to ACE2
We developed a competitive ELISA that measures the binding of RBD-J to ACE2 in the presence or absence of adjuvants without the need to desorb the RBD-J bound to AH (Figure 3). This assay was used to monitor the storage stability of the seven RBD-J formulations evaluated in the mouse immunogenicity studies described above. In this competitive ELISA format, ACE2 was pre-incubated with formulated RBD-J samples and then transferred to an ELISA plate coated with RBD-J antigen, followed by detection of ACE2 binding to the plate (see Supplementary methods). This assay results in an inverse dose–response curve where increased ACE2 - RBD-J binding in the test samples leads to decreased binding of ACE2 to the RBD-J antigen bound to the ELISA plate.
Figure 3.
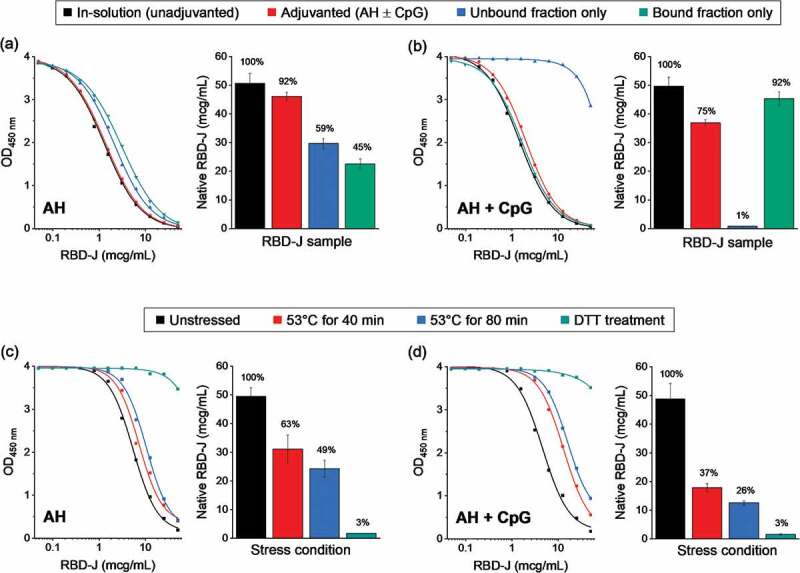
Competitive ELISA method development to measure the ACE2 binding of RBD-J in the presence and absence of adjuvants. Representative dose–response curves along with measured native RBD-J concentration in (a) AH-adjuvanted and (b) AH+CpG-adjuvanted formulations. Adjuvanted drug product, along with unbound and bound fractions of the drug product (generated by centrifugation of the sample) were compared to unadjuvanted in-solution RBD-J reference. Representative dose response curves along with measured native RBD-J concentration from forced degradation studies on (c) AH and (d) AH+CpG-adjuvanted formulation. Partially and completely denatured RBD-J samples (heat and DTT treatment, respectively) were adjuvanted and then compared to the corresponding unstressed adjuvanted RBD-J reference. Bars represent mean and error bars represent range of values with n = 2 (2 independent runs). For each test condition, the percentage of native RBD-J relative to reference is indicated above the bars.
Competitive ELISA on AH-adjuvanted and in-solution RBD-J gave similar concentrations of native RBD-J (i.e., RBD-J that binds ACE2) in both samples (Figure 3a), indicating that the AH-adjuvanted RBD-J bound similar amounts of ACE2 as RBD-J in solution. Upon separation of AH-bound and unbound (in-solution) RBD-J, the measured concentration of native RBD-J in each fraction was nearly similar, indicating partial (~50%) binding of RBD-J to AH, a result consistent with the previously described SDS-PAGE analysis (see Figure 1). For RBD-J formulated with AH+CpG, the native RBD-J concentrations approached that of the unadjuvanted in-solution reference (ranging from ~75% to ~92%) (Figure 3b). Upon partitioning of AH-bound and unbound (in solution) fractions, it was confirmed that essentially 100% RBD-J was in the bound fraction, indicating complete binding of RBD-J to AH in the presence of CpG (again consistent with SDS-PAGE analysis; Figure 1). In summary, minimal to no losses were observed in the ability of RBD-J to bind ACE2 upon the addition of adjuvants. Using this competitive ELISA method, RBD-J samples were successfully evaluated for ACE2 binding without the need for desorption from adjuvants, and the percent of AH-bound RBD-J (± CpG) obtained by this method correlated well with SDS-PAGE analysis.
Next, we determined the stability-indicating nature of the above competitive ELISA method. Forced-degraded AH- or AH+CpG-adjuvanted RBD-J samples prepared by either partially or completely denaturing the protein (by heating at 53°C for up to 80 min or by incubating with 20 mM dithiothreitol (DTT) at 37°C for 30 min) were compared to the unstressed adjuvanted RBD-J control. The heat-stressed samples displayed a gradual time-dependent decrease in RBD-J-ACE2 binding, indicating loss of native RBD-J (Figure 3c,d). Interestingly, following heat stress, AH+CpG-adjuvanted samples of RBD-J displayed a more pronounced loss of ACE2 binding compared to RBD-J adjuvanted with AH alone. Additionally, reduction of all four disulfide bonds of RBD-J upon treatment with DTT (as confirmed by intact protein mass spectrometry; data not shown) resulted in a complete loss of ACE2 binding-native RBD-J. Overall, these results established the stability-indicating nature of this competitive ELISA method, which was then used to monitor the storage stability of adjuvanted RBD-J formulations.
Storage stability profiles of RBD-J formulated with various combinations of aluminum salt and CpG adjuvants
Formulations F1-F7 shown in Table 1 were stored at 4°C for 3 months, 25°C for 2 months, and 37°C for 1 week and analyzed for ACE2 binding using the competitive ELISA (Figure 4). The measured concentration (ranging from 90 to 110 mcg/mL) of native RBD-J in each formulation at time zero (t0) was close to the target concentration of 100 mcg/mL (Figure 4a). The relative percent of native RBD-J remaining at subsequent stability time-points at the three temperatures was determined by normalizing the results to the t0 value. The average slope values (i.e., percent loss in native RBD-J per month) along with the corresponding 95% confidence intervals for 4°C and 25°C samples were calculated by linear regression analysis.
Figure 4.
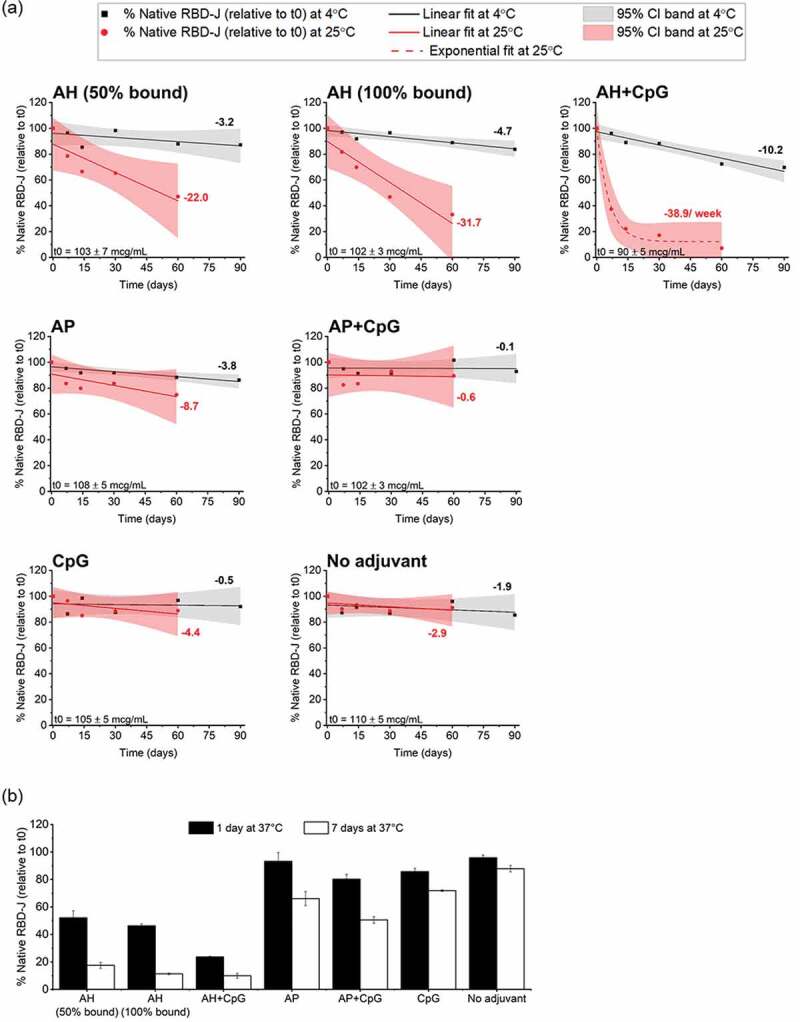
Stability profile of RBD-J in formulations F1-F7 at 4, 25 and 37°C as measured by competitive ELISA. (a) Stability at 4°C and 25°C up to 90 days and 60 days, respectively. Concentration of ACE2-binding native RBD-J at each time point was normalized against values at time zero (t0) and then plotted as a function of time. Solid lines at 4°C (black) and 25°C (red) represent linear regression fit of data points and the corresponding slope values displayed alongside represent percentage loss per month. The red dotted line at 25°C for AH+CpG formulation represents best exponential fit and the indicated slope for percentage loss per week was determined by fitting t0, 7 days, 14 days data points to linear model. The shaded bands indicate the 95% confidence interval. The measured concentration of RBD-J at t0 in each formulation is displayed at the bottom right as mean ± SD (n = 4). (b) Percentage of native RBD-J, relative to t0, remaining in each formulation after 1 day and 7 days at 37°C is presented as mean ± SD for n = 4 measurements.
At 4°C (Figure 4a), the AH+CpG formulation lost ~30% native RBD-J at a rate of ~10% loss per month, whereas all the other formulations retained >85% native protein over 3 months. Notable differences in the stability profiles of adjuvanted RBD-J across various formulations were observed over 2 months of storage at 25°C (Figure 4a). For example, the rate of loss in 100% AH-bound RBD-J formulation was greater than the loss in 50% AH-bound RBD-J formulation (32% vs. 22% loss per month, respectively). The instability of RBD-J accelerated exponentially (with biphasic kinetics) when CpG was present along with AH, with an initial native RBD-J loss rate of ~39% per week at 25°C (determined by linear regression analysis of the first three data points).
In contrast, AP-containing formulations with <15% AP-bound RBD-J were much more stable even in combination with CpG when stored at 4 and 25°C. The monthly loss rate of native RBD-J in AP and AP+CpG formulations at 4°C was 4% and 0.1% over 3 months, respectively, and at 25°C was 9% and ~1% over 2 months, respectively (Figure 4a). Interestingly, unadjuvanted RBD-J and RBD-J formulated with CpG (no aluminum salt adjuvant) showed virtually no loss in RBD-J -ACE2 binding and maintained a similar stability profile when stored at either 4°C (0.5% vs. 2% loss per month, respectively) or 25°C (4% vs. 3% loss per month, respectively).
Finally, under more aggressive storage conditions (37°C for up to 1 week) (Figure 4b), the unadjuvanted RBD-J maintained excellent stability. In contrast, all adjuvanted formulations resulted in a substantial loss of native RBD-J under these stress conditions. The observed loss in 100% AH-bound formulation trended slightly higher than 50% AH-bound formulation with nearly 50% and 80–90% native RBD-J lost in both formulations over 24 h and 7 days at 37°C, respectively. The AH+CpG formulation was the most destabilized with ~80% native RBD-J protein lost in 24 h and only minimal (10%) native protein remaining after 7 days at 37°C. Finally, formulations with AP ± CpG, or CpG alone, showed 30–50% loss of native RBD-J protein over 7 days at 37°C. Overall, at all three storage temperatures, the RBD-J formulation containing AH ± CpG demonstrated notably higher losses of native RBD-J over time, compared to RBD-J formulated with AP ± CpG or CpG alone.
Effect of aluminum salt and CpG adjuvants on the conformational stability of RBD-J
To better understand differences in the storage stability of RBD-J in various adjuvanted formulations, the overall conformational stability of RBD-J in each freshly prepared formulation was measured using DSC. Unadjuvanted RBD-J displayed a single major endothermic peak with a thermal unfolding temperature (Tm) value of 53°C (Figure 5a). This value was used as a “baseline value” for comparison to the adjuvanted RBD-J formulations by determining the corresponding ΔTm values (Figure 5b). A similar relative stability profile for RBD-J in the adjuvanted formulations was determined when comparing the thermal onset temperature (Tonset) (Figure 5b).
Figure 5.
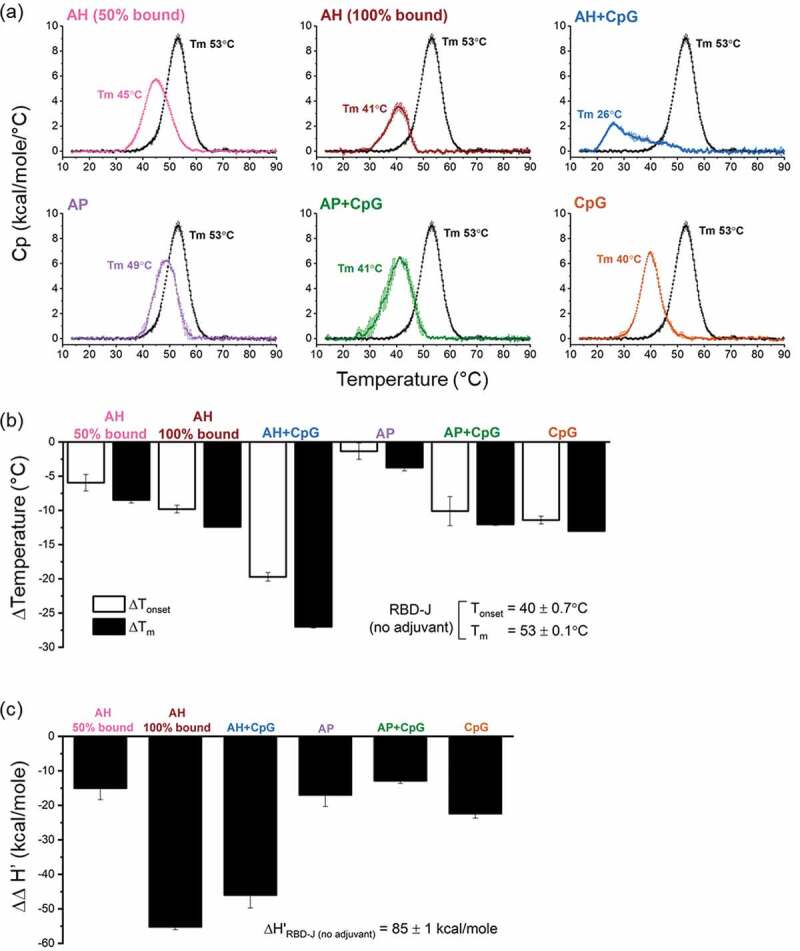
Conformational stability of RBD-J in formulations F1-F7 as measured by DSC. (a) Representative DSC thermograms of RBD-J in each formulation. DSC thermogram of unadjuvanted in-solution RBD-J (black) is plotted alongside each adjuvanted formulation for comparison. Thermal unfolding temperature (Tm) values are shown as the mean of n = 2 measurements with range = ± 0.1–0.6°C. Effect of adjuvants on (b) ΔTonset and ΔTm values and (c) Δapparent enthalpy (∆∆H’) values of RBD-J in F1-F6 as determined by subtracting the results for each formulation from respective values of the unadjuvanted in-solution RBD-J control. Bars indicate mean of n = 2 measurements and the errors bars represent the data range.
Each adjuvanted RBD-J formulation had a notably different conformational stability profile when compared to the unadjuvanted protein (Figure 5b). For example, the Tm value of RBD-J when partially or completely AH-bound decreased from 53°C to 45°C and 41°C, respectively. A further major drop in the Tm value of RBD-J to ~26°C was observed in the AH+CpG formulation. Interestingly, RBD-J formulations containing AP, in the absence and presence of CpG, demonstrated intermediate conformational stability profiles with Tm values of 49°C and 41°C, respectively. Finally, the CpG-containing formulation with no aluminum salt resulted in a Tm value of RBD-J of 40°C. A similar rank ordering of the stability profile was observed for RBD-J in different formulations when comparing the thermal onset temperature (Tonset) values. The relative ΔTm and ΔTonset values for RBD-J in the presence of different adjuvants, compared to the unadjuvanted control, are displayed in Figure 5b. Overall, the largest destabilization of RBD-J was observed in the AH+CpG formulation, while the AP formulation (where RBD-J was essentially all unbound and no CpG was present) was the most stable.
To further estimate the degree of RBD-J destabilization in various adjuvanted formulations as measured by DSC, we also compared the area under the curve (apparent enthalpy of unfolding, ∆H’) of formulated RBD-J samples vs unadjuvanted control. Each adjuvanted formulation destabilized RBD-J to varying extents, as indicated by negative ∆∆H’ values (Figure 5c). Using these criteria, the highest destabilization of RBD-J was observed where RBD-J was completely AH-bound with and without CpG.
Comparison of in vitro stability profile (ability to bind ACE2) of RBD-J with in mouse immunogenicity using stressed AH+CpG-adjuvanted RBD-J
To evaluate the effect of loss of in vitro ACE2-binding activity of RBD-J formulated with AH+CpG adjuvants on its in vivo immunogenicity profile, we performed two different mouse immunization studies. We first tested the adjuvanted RBD-J samples stored at 4°C or 25°C for 2 months. The 4°C sample (~80% native RBD-J) was used as a control and compared to the 25°C sample (~10% native RBD-J). Mice (BALB/c, n = 6) were vaccinated with 5 mcg of each adjuvanted RBD-J samples on days 0 (prime) and 21 (boost) (Figure 6a). Sera were collected on days 21, 35, and 65 and analyzed for total antibody titers and SARS-CoV-2 neutralization titers. All the vaccinated mice in both temperature groups seroconverted by day 21 and demonstrated a robust anti-RBD-J IgG response that persisted to day 65 (Figure 6b).
Figure 6.
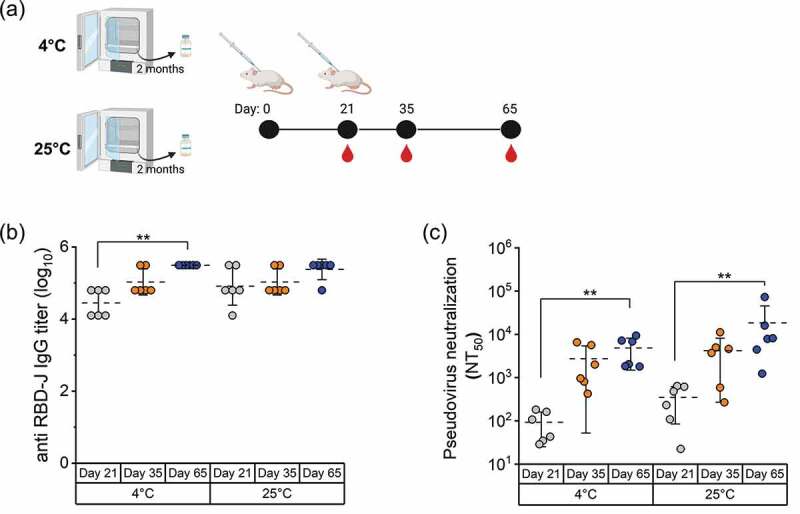
In vivo mouse immunogenicity of AH+CpG-adjuvanted RBD-J formulation stored at 4°C and 25°C for two months. (a) Mice (n = 6) were immunized by subcutaneous route with two 5 mcg doses (days 0 and 21) of AH+CpG-adjuvanted RBD-J stored at 4 and 25°C for 2 months. Serum was collected on days 21, 35, and 65. (b) Sera anti-RBD-J IgG titer as determined by ELISA and (c) Individual NT50 values as determined using a best-fit nonlinear regression on serum dilution curves on day 21, 35, and 65. The dashed lines represent group mean, and the errors bars indicate standard deviation. Statistical significance between time-points in each group was determined by Kruskal-Wallis test and post hoc Dunn’s multiple comparisons test (**p ≤ 0.01). Illustration in (a) was created with Biorender.com.
In terms of neutralizing responses (Figure 6c), 6 of 6 mice that received 4°C control sample had NT50 values on day 21 and those values increased by day 65. Surprisingly, a similar neutralizing response was observed in mice immunized with 25°C sample (~10% native RBD-J). In fact, on day 21, NT50 values for mice treated with 25°C sample trended somewhat higher (albeit not statistically significant, using two-tailed Mann–Whitney test) than for those vaccinated with 4°C samples (Figure 6c). Moreover, sera collected on day 65 from the 4°C and 25°C sample groups demonstrated comparable neutralizing activity against pseudovirus expressing the spike protein from SARS-CoV-2 Delta variant and the WT virus (p > 0.05, Wilcoxon matched-pairs signed rank test) (Suppl. Fig. S3). These results indicate that the loss of in vitro ACE2 binding activity of AH+CpG-adjuvanted RBD-J, observed during storage at 25°C for 2 months, did not adversely affect the ability of vaccine antigen to induce SARS-CoV-2 neutralizing antibodies.
To further investigate the apparent inconsistency between loss of native RBD-J in vitro and vaccine immunogenicity in vivo, we performed an additional mouse study with formulated RBD-J samples that had been subjected to various forced degradation conditions (see Table 2; Study I). RBD-J samples were adjuvanted with AH+CpG and heated at 37°C for 1 week (group #2) or 70°C for 1 h (group #3), respectively, resulting in 80–90% loss of ACE2-binding native RBD-J, respectively (data not shown). Another sample was treated with DTT (group #4), resulting in a reduction of all four disulfide bonds and complete loss of ACE2 binding activity (data not shown). Unstressed AH+CpG-adjuvanted RBD-J formulation prepared at 4°C (group #1) was used as control. Similar to the previous study, mice were subcutaneously immunized at day 0 (prime) and 21 (boost), and serum was collected on days 21, 35, and 65 (Figure 7a). Native RBD-J content in control, heat-stressed, and DTT-treated formulations remained unchanged between day 0 and day 21 (confirmed by ACE2 competition ELISA, data not shown). In terms of total antibody responses (Figure 7b), all immunized mice were seroconverted after a single dose (anti-RBD-J IgG titer >104), and strong antibody responses were observed in the control and forced degraded formulations after single or two doses.
Table 2.
Summary of RBD-J formulations prepared to evaluate the correlation of in vitro stability of RBD-J with in vivo mouse immunogenicity. Study I (groups #1–4) included unstressed and forced degraded (37 or 70°C heat-stressed and DTT-treated) RBD-J formulated with AH and CpG. The stress condition for each formulation is displayed along with the concentration of antigen and adjuvants. RBD-J (5 mcg) was administered to mice subcutaneously (50 µl). Study II (groups #5–6) included varying doses (5, 1, 0.5 mcg) of unstressed RBD-J adjuvanted with AH+CpG and administered as noted above. Note that the doses of AH and CpG in each group in study I and II were same (75 mcg and 30 mcg, respectively).
| |
|
|
Concentration (mcg/mL) |
Dose injected in mice (mcg) Injection volume = 50 µL |
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Study arm | Group No. | Condition | Total RBD-J | Aluminum adjuvant | CpG | Total RBD-J | Aluminum adjuvant | CpG | No. of mice |
| I | 1 | Unstressed (4°C) | 100 | 1500 | 600 | 5 | 75 | 30 | 9 |
| 2 | 37°C for 1 week | 100 | 1500 | 600 | 5 | 75 | 30 | 9 | |
| 3 | 70°C for 1 h | 100 | 1500 | 600 | 5 | 75 | 30 | 5 | |
| 4 | DTT | 100 | 1500 | 600 | 5 | 75 | 30 | 5 | |
| II | 5 | Unstressed (4°C) | 20 | 1500 | 600 | 1 | 75 | 30 | 9 |
| 6 | Unstressed (4°C) | 10 | 1500 | 600 | 0.5 | 75 | 30 | 9 | |
Figure 7.
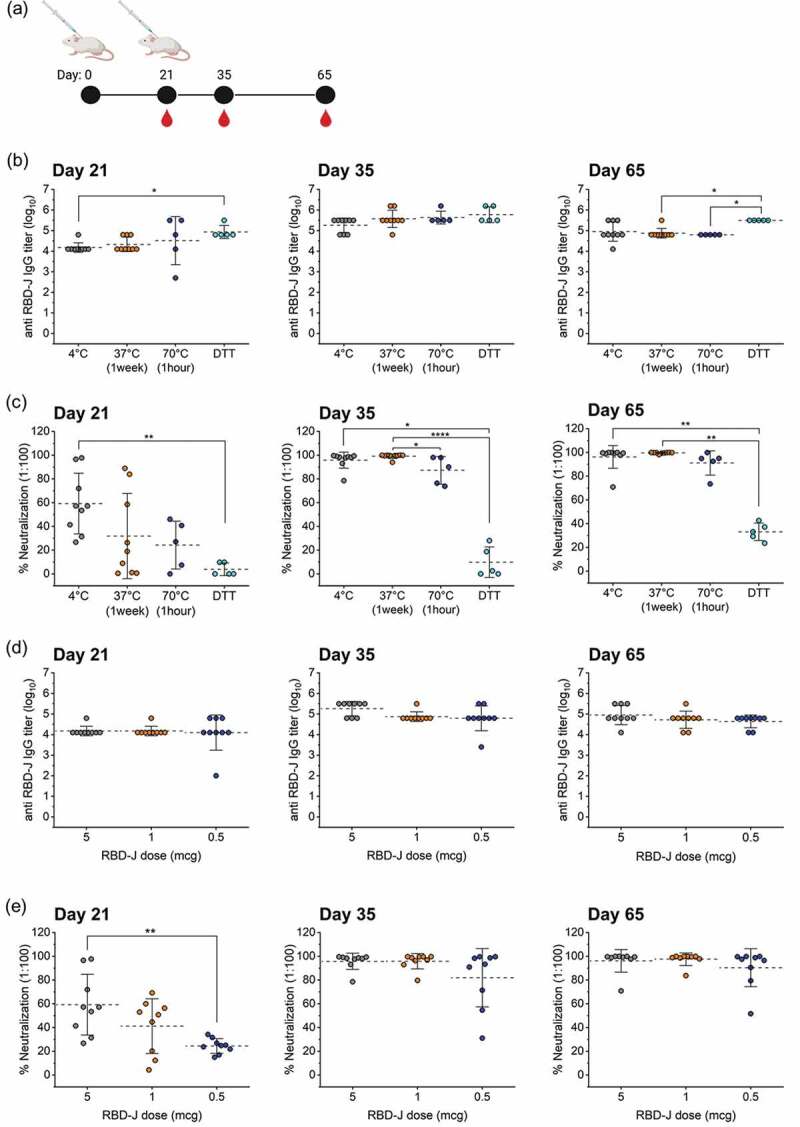
Effect of forced degradation and dose-ranging of AH+CpG–adjuvanted RBD-J on in vivo mouse immunogenicity. (a) Mice were subcutaneously immunized at day 0 and 21 and serum was collected on days 21, 35, and 65. (b) anti- RBD-J IgG titer as determined by ELISA and (c) % Neutralization at 1:100 serum dilution on day 21, 35, and 65 for mice immunized with unstressed (4°C) and stressed (37°C, 70°C and DTT-treated) AH+CpG- adjuvanted RBD-J formulations. (d) anti- RBD-J IgG titer and (e) % Neutralization at 1:100 serum dilution on day 21, 35, and 65 for mice immunized with decreasing doses of RBD-J (5, 1 and 0.5 mcg) formulated with AH+CpG adjuvants. The dashed lines represent group mean and the errors bars indicate standard deviation. P-values were determined using Kruskal–Wallis test and post hoc Dunn’s multiple comparisons test (*p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001). Illustration in (a) was created with Biorender.com.
In contrast, the control and forced degraded RBD-J sample groups demonstrated differences in pseudovirus neutralization activities at day 21 (Figure 7c). As expected, the heat-stressed formulation groups (37°C for 1 week and 70°C for 1 h) had lower neutralizing antibody levels (~30% and ~20% neutralization, respectively), as compared to the 4°C control (~60% neutralization). No virus-neutralizing activity was detected in sera from mice that received the DTT-treated sample, even though those same animals had anti-RBD-J antibody titers equivalent to (and even greater than) the control. This observation is consistent with the complete loss of native RBD-J in this formulation as reflected by the ACE2 competition ELISA. After the booster dose on day 21, virus-neutralizing activity improved significantly in mice that were administered the heat-stressed RBD-J samples (37°C for 1 week and 70°C for 1 h), while neutralizing activity in mice that received DTT-treated RBD-J remained near or at baseline. In summary, the endpoint titers for native (4°C) versus structurally altered RBD-J formulated with AH+CpG (after treatment at 37°C, 70°C or with DTT) were similar after one or two doses, but the neutralizing antibody response on day 21 correlated with the loss of ACE2 binding activity in vitro. In addition, the 10–20% ACE2 binding activity remaining in the structurally altered heat-treated AH+CpG-adjuvanted RBD-J was sufficient to elicit a robust neutralizing response after two doses.
To better understand the above results, as part of this same mouse study, we also investigated the effect of varying the dose of native RBD-J (within the AH+CpG-adjuvanted formulation) on total antibody responses and pseudovirus neutralization activity in mice (see Table 2, Study II). Mice (BALB/c, n = 9) were administered 1 mcg or 0.5 mcg adjuvanted RBD-J (group #5 and group #6, respectively) and compared to mice that received 5 mcg RBD-J formulated with the same adjuvants (group #1, Study I) (Figure 7d,e). Mice in all three groups were seroconverted to similar anti-RBD-J IgG levels after a single dose (>104) and the titers improved further after the second dose (Figure 7d). There was, however, no evidence of a dose-dependent reduction in anti-RBD-J antibody titers at any time-point, suggesting that even the lowest RBD-J dose tested (0.5 mcg) was sufficient to induce maximal antibody response. In contrast, SARS-CoV-2 pseudovirus neutralization activity in serum collected on day 21 reflected a clear dose–response (Figure 7e). For example, the neutralizing activity in the mice that received 5 mcg RBD-J was significantly higher than in mice that received 0.5 mcg RBD-J (p ≤ 0.01). After the second dose, however, differences in neutralization activity diminished and all three groups achieved >80% neutralization at a 1:100 dilution, albeit more variability was seen across the mice given the lowest dose (0.5 mcg).
In summary, after two doses, a 10-fold decrease in the RBD-J dose (5 vs. 0.5 mcg) could still elicit a neutralizing response in the presence of AH+CpG in this mouse model. After one dose, however, the pseudovirus neutralization response on day 21 showed a dose–response relationship. Combining the results from study I (stressed RBD-J samples) and study II (RBD-J dose ranging), pseudovirus neutralization results at day 21 (after one dose) were identified as the most reliable stability indicator for mouse immunogenicity testing of RBD-J formulated with AH+CpG.
Discussion
Recombinant subunit vaccines have the potential to enable the rapid deployment of low-cost COVID-19 vaccines to LMICs and improve global vaccine coverage due to (1) ease of manufacturing and scale-up at low-cost using existing recombinant protein production technologies, and (2) the ability to formulate and distribute them under refrigerator storage conditions.49,50 Compared to other vaccine types (i.e., live-attenuated vaccines), recombinant protein-based vaccines are generally formulated with adjuvants to accentuate the magnitude and breadth of the immune response.51 The widely available, inexpensive, and safe ‘classical’ aluminum salt adjuvants are being tested (as single adjuvant or in combination with other immunostimulatory molecules like CpG ODN) in several recombinant protein-based COVID-19 vaccine candidates, but several critical attributes of vaccine formulation such as antigen-adjuvant interaction(s), and its impact on both storage stability and in vivo potency of the vaccine have not been reported. Previous vaccine formulation development studies have exemplified the necessity to examine these attributes carefully for each antigen-adjuvant combination to optimize vaccine stability and immunogenicity.52,53 The purpose of our work, therefore, was to evaluate the effect of antigen-adjuvant interactions on in vivo performance (mouse immunogenicity studies) and storage stability (using a competitive ELISA to monitor ACE2 binding in vitro) of a recombinant RBD variant (RBD-J) formulated with aluminum salt adjuvants with and without CpG. In addition, we evaluated in vitro-in vivo correlation of storage stability of adjuvanted RBD-J formulations with their immunogenicity in mice and performed several different biophysical analyses to better understand the nature of the RBD-J interaction with these adjuvant combinations.
Effect of antigen-adjuvant interactions on mouse immunogenicity of RBD-J
We systematically prepared formulations with varying levels of RBD-J adsorption to AH (50% or 100% bound RBD-J) and AP (< LOQ = 15% bound RBD-J) with or without CpG and examined their effect on immunogenicity in mice. RBD-J formulations with aluminum salt adjuvant alone (AH or AP) elicited either greater (in case of AH) or similar (in case of AP) levels of antigen-specific total antibody response compared to unadjuvanted RBD-J, but these formulations generated little to no neutralizing antibodies. Similar results have been previously reported with a minimally characterized AH-adjuvanted RBD-J sample prepared shortly before administration.36 When comparing 0%, 50%, and 100% aluminum-adsorbed RBD-J formulations (i.e., formulated with AP, AH, and AH with settle-decant concentration procedure, respectively), increasing RBD-J adsorption from unbound to the 50% bound state showed modest improvements in immunogenicity, while further adsorption to 100% did not further enhance the RBD-J immunogenicity. Contrary to our observations, Pollet et al. have reported strong neutralizing antibody responses in mice after two injections of their AH-adsorbed RBD vaccine at dose 25 mcg RBD and 100–500 mcg AH.45 The differences in the observed immune responses may be due to a different RBD variant (see below), considerably higher doses of antigen used in their study, and/or differences in the aluminum salt adjuvant dose used. In fact, in another study with a lower dose of the same RBD-based antigen (7 mcg) formulated with 200 mcg AH the authors did not observe notable neutralizing antibody responses.38
We then examined the effect of a second adjuvant, CpG, either alone or in combination with aluminum salt adjuvants. The CpG 1018 adjuvant used in this study is a well-known activator of TLR9.27 Both RBD-J ± CpG formulations produced low levels of total antigen-specific antibodies (mean titer: 102 – 103) and undetectable neutralizing antibody titers, suggesting that CpG alone is not sufficient as an adjuvant. RBD-J formulated with CpG in combination with aluminum salt adjuvants (AH or AP), however, evoked a strong antibody response, especially under conditions where CpG and RBD-J were completely bound to AH (mean titer >105). Interestingly, there was a clear difference between the unbound and bound RBD-J (and CpG) formulations in their ability to elicit neutralizing antibodies, and only the bound RBD-J (and CpG) formulation displayed high pseudovirus neutralization titers (NT50 > 103). Similar results have been reported previously with a minimally characterized AH+CpG-adjuvanted RBD-J prepared shortly before injection.36 In comparison, optimization of antigen-adjuvant interactions has been demonstrated to improve the immunogenicity of other vaccine candidates targeting various pathogens; in the case of the recombinant poxvirus protein L1, the inclusion of CpG with AH as well as binding of the antigen to AH were critical for optimal protection of mice from vaccinia infection.54,55 Likewise, with the P. falciparum vaccine antigen AMA1, the physical association of CpG and antigen (by adsorption to AH) produced an optimal immune response.56 Interestingly, it has also been reported that lack of interaction between protein antigens and aluminum salt adjuvants has potentiation effects on immune responses,57 thus indicating the need to assess the effects of such antigen-adjuvant interactions on immune responses for each protein antigen candidate during vaccine formulation development.52
In this work, we focused on humoral immune responses to compare different adjuvanted formulations. Although T-cell responses were outside the scope of this work, T-cell activation is also critical for protection against SARS-CoV-2.58 It would therefore be of interest in future to evaluate cell-mediated immune responses to these formulations. Furthermore, non-neutralizing antibodies, through their Fc-dependent effector functions, can protect against viral infections.59,60 Future work, including mouse challenge studies, could help to elucidate the correlation between pseudovirus neutralizing activity of elicited antibodies in bound vs. unbound RBD-J (and CpG) groups and their ability to protect against live SARS-CoV-2 infection.
Effect of antigen-adjuvant interactions on the storage stability of RBD-J
Next, we evaluated the effect of antigen-adjuvant interactions on the stability of the same RBD-J vaccine formulations after storage at 4, 25, and 37°C using ACE2 competition ELISA. The unadjuvanted in-solution RBD-J antigen demonstrated excellent stability at all three storage temperatures. In contrast, RBD-J adjuvanted with aluminum salt adjuvant alone (AH or AP) displayed a good storage stability profile at 4°C up to 3 months, but a loss in ACE2 binding was observed at higher temperatures. Our results indicate that RBD-J stability at 25 and 37°C is dependent upon the extent of antigen-adjuvant interactions and declines with greater adsorption to the aluminum adjuvant. For example, at 25 and 37°C up to 2 months and 1 week, respectively, AH formulations (partially or completely adsorbed RBD-J) were notably more de-stabilized than AP formulations (minimally adsorbed RBD-J), and the observed losses were greater in fully adsorbed than partially adsorbed RBD-J formulation. Recent publications on RBD-based subunit vaccine candidates have evaluated the stability of antigen in solution, but not in the presence of adjuvants.42,61 Nonetheless, the de-stabilizing effect of adsorption on aluminum salt adjuvant has been noted for other protein antigens including diphtheria toxoid as well as the recombinant NRRV subunit and Streptococcus pneumoniae subunit vaccine candidates.47,62–65 Based on these results, the effects of aluminum adjuvant on vaccine storage stability should be carefully evaluated for each RBD vaccine candidate.
We also assessed the stability of RBD-J formulations with CpG (alone or in combination with aluminum salt adjuvants). The addition of CpG by itself did not have any prominent effect on the stability of antigen in solution, but its presence along with AH (such that both CpG and RBD-J were completely adsorbed to AH) resulted in enhanced loss of ACE2 binding at 4, 25, and 37°C within 2 months, 1 week and 1 day, respectively. In contrast, the destabilizing effect of CpG on RBD-J minimally bound to AP was not observed at 4 or 25°C and was only seen at 37°C after 7 days. The rapid destabilization of RBD-J in the AH+CpG formulation at 25 and 37°C over a period of weeks is potentially concerning because it may be an early indicator of instability of RBD-J during real-world storage conditions including (1) storage at refrigerated temperatures over months and years, or (2) accidental exposure to elevated temperature conditions during storage at and transportation to remote vaccine administration sites in LMICs. At the same time, this observation is specific to RBD-J, and other RBD antigens of different sequences or lengths may have different stability profiles. Possible approaches to overcome this observed de-stabilization of RBD-J formulated with AH+CpG could include (1) limiting the shelf-life, (2) optimizing the solution conditions using excipients to improve antigen stability, or (3) packaging the antigens and adjuvants in separate containers and extemporaneously combining them before administration. However, this latter approach (i.e., “bedside mix”) may not be practical for widespread use due to financial and logistical limitations.
Nature of molecular interactions of RBD-J with aluminum salt and CpG adjuvants
During RBD-J binding studies, we observed partial and minimal adsorption of RBD-J to AH and AP, respectively. This result was surprising for two reasons. First, at the formulation pH 6.5, the RBD-J antigen has a net positive charge (calculated pI ~8.7). Therefore, if the nature of antigen binding to aluminum adjuvants follows electrostatic charge–charge interactions, the RBD-J antigen should bind AP, which has a net negative charge in the pH range of 5–7, and not AH, which has a net positive charge in the pH range of 5–7 (measured zeta potential values of AP and AH in formulation buffer at pH 6.5 were −18 mV and +22 mV, respectively (data not shown)). However, this is not what we observed; thus, the binding of RBD-J to aluminum salt adjuvants cannot be explained by the commonly observed electrostatic charge–charge interactions. Second, only partial RBD-J binding (~50%) to AH was achieved, and we observed that the HMW species in the RBD-J solution were preferentially bound to AH. We also tested higher AH concentrations (up to 3000 mcg/mL) and another source of aluminum hydroxide (Rehydragel-HPA),52 but no increases beyond 50% RBD-J adsorption were observed (data not shown). These results suggest some molecular heterogeneity in the RBD-J preparation in terms of ability to bind aluminum hydroxide-based adjuvants.
Contrary to our observations, a recent study reported >98% binding of an RBD vaccine candidate (with a similarly calculated pI ~8.9) to AH in a Tris-based formulation (20 mM Tris, 150 mM NaCl at pH 7.5) at similar antigen-adjuvant concentrations.45 To investigate if the differences in formulation buffer and pH contributed to the differential binding behavior of RBD-J, we performed RBD-J adsorption in a Tris-based formulation buffer but did not detect improved binding to AH (data not shown). It is likely that the structural differences between RBD-J and the other RBD antigens, the latter containing an additional beta-hairpin motif arising from extended C-terminus (331–549), could contribute to increased binding of this RBD candidate to AH.
Further analysis of RBD-J antigen by de-glycosylation followed by mass spectrometry analysis revealed the presence of glycosylated RBD-J and glycosylated Pro-RBD-J (contains inefficiently clipped [Pro] region of the alpha signal peptide) species. The presence of these Pro-RBD-J species is not unexpected, given the known inefficiency of Kex2 and Ste13-dependent protease cleavage of Pichia pastoris alpha factor “Pre Pro” secretion peptide.66 Other groups have also noted similar inefficient cleavage of the signal peptide in recombinant proteins expressed in Pichia pastoris.38 Interestingly, the additional N-terminus amino acid residues arising from Pro-peptide lower the calculated pI values of RBD-J from 8.7 to 5.3, which likely explains the observed preferential binding of HMW species (containing negatively charged glycosylated Pro-RBD-J species) to the positively charged AH adjuvant. Future adjuvant-binding studies using PNGase-F-treated RBD-J resolved into aglycosylated RBD-J and Pro-RBD-J species will shed light on this hypothesis and provide insights as to the observed partial binding to AH. Furthermore, in support of the above hypothesis, we have recently observed 100% binding to AH when an acidic peptide/linker was recombinantly added to the N- or C-terminus of RBD-J (data not shown).67
When RBD-J binding studies were performed with aluminum salt adjuvants in the presence of CpG, both RBD-J and CpG adsorbed completely to AH. CpG 1018 has a phosphorothioate backbone and can bind to AH via electrostatic interaction or ligand-exchange mechanism.68 In terms of the effect of CpG on antigen-AH binding, Aebig et al. reported reduced or similar binding of four model antigens upon addition of CpG.69 To the best of our knowledge, however, ours is the first report of CpG enhancing the binding of a protein antigen to AH. In our study, CpG was first added to AH, followed by the addition of RBD-J; however, other formulation compounding schemes were also tested (such as adding RBD-J to AH before CpG addition or the simultaneous addition of RBD-J and CpG to AH). Regardless of the mixing order, 100% adsorption of RBD-J and CpG to AH was observed in all cases (data not shown).
Furthermore, DSC analysis revealed major de-stabilization of RBD-J in AH ± CpG formulations, suggesting a direct interaction of RBD-J with CpG. Developing a better understanding of RBD-J–CpG interaction and the mechanism of adjuvant-induced destabilization of RBD-J will be the focus of our future work. There are reports on the electrostatic interaction of RBD with polyanions (e.g., inorganic polyphosphates and glycosaminoglycans, such as heparin and its derivatives) via RBD’s solvent-exposed ‘poly-anion binding site’ composed of a continuous stretch of basic (Arg and Lys) amino acid residues.70,71 We hypothesize that a similar interaction may occur between RBD-J and the negatively charged CpG, which in turn may trigger protein destabilization. To this end, mechanistic studies utilizing hydrogen exchange-mass spectrometry (HX-MS) to elucidate the peptide-level RBD-J–CpG interaction sites are ongoing.
Assessing storage stability of AH+CpG-adjuvanted RBD-J by in vitro vs. in assays
We first evaluated in vitro–in vivo correlations for RBD-J adjuvanted with AH+CpG samples that had been stored at 4 and 25°C for 2 months. Although large stability differences were observed between the two samples using the ACE2 competition ELISA, both formulations elicited a strong neutralizing antibody response. Surprisingly, the notable loss of in vitro ACE2 binding activity in the 25°C sample did not result in a corresponding loss of in vivo immunogenicity in the formulation under these conditions. Interestingly, similar observations have been reported with different vaccines, such as the inactivated poliovirus vaccine (IPV), where the loss of D-antigen content measured by in vitro ELISA potency assay did not have any implications on the immunogenicity of the vaccine in the rat model.72–74 In this particular case, the D-antigen assay is considered a more sensitive method to monitor structural alterations in the IPV antigens during storage. We, therefore, hypothesize that similar to observations made with D-antigen in IPV, the ~10% of native RBD-J remaining in stressed AH+CpG formulation (2 months at 25°C) was sufficient to elicit a robust neutralizing antibody response in mice at these antigen doses and in the presence of these two adjuvants.
To test the above hypothesis, an additional mouse study was performed with partially or completely structurally altered RBD-J antigen (by heat-stress and DTT treatment, respectively) in the presence of AH+CpG. The heat-treated samples contained 10–20% native RBD-J, whereas the DTT-treated sample displayed complete loss of native protein (due to reduction of the four disulfide bonds). All RBD-J samples, irrespective of the nature of stress conditions or the extent of loss of ACE2 binding as measured in vitro by competitive ELISA, generated high levels of total anti-RBD-J antibodies in mice. In contrast, the trends observed in neutralizing responses in mice after a single dose were able to distinguish 4°C vs. stressed RBD-J formulations, a result that correlated with their measured loss of ACE2 binding. However, after two doses, the differences in neutralizing responses in mice in the 4°C vs. heat-stressed RBD-J formulations were notably diminished.
Additionally, a dose ranging study was performed with varying amounts of RBD-J in AH+CpG-adjuvanted formulation. An RBD-J dose–response effect was seen in neutralizing activity after a single dose, but this effect subsided after two doses. In fact, after two vaccine doses, even a 10-fold lower dose (0.5 mcg) of the antigen was sufficient to produce a neutralizing response statistically equal to the highest dose (5 mcg). Taken together, these results agree with the above hypothesis that 10–20% native RBD-J remaining in samples stored at 25°C for 2 months was sufficient to generate a strong neutralizing antibody response in our mice model in the presence of AH+CpG adjuvants, especially after two doses. Moreover, these findings demonstrate that pseudovirus neutralization titer in mice after one dose at day 21 was the best indicator of native RBD-J loss in formulated samples, and these results best correlated with in vitro ACE2 binding results.
Additional work is required to better understand the stability profile of adjuvanted RBD-J formulations when compared by in vivo vs. in vitro assays. On the one hand, the in vitro ACE2 binding assay is a sensitive and convenient method to monitor structural alterations in formulated RBD-J in adjuvanted formulations during storage, but partial loss of structural integrity does not necessarily affect in vivo performance. On the other hand, pseudovirus neutralization assays in mouse immunogenicity studies can be stability-indicating under certain conditions (e.g., antigen-adjuvant dose, injection, bleed time-point, etc.), but they are not as sensitive to partially altered structures within RBD-J. In the future, it will be of interest to explore if other mechanisms may help explain some of these in vitro vs in vivo results including the possible role of (1) linear neutralization epitopes within the receptor-binding motif (RBM) which may be exposed upon structural alterations of RBD-J during stress and may directly block binding of RBD-J to ACE2,75,76 and (2) conformational epitopes outside of the ACE2 binding region that may remain unperturbed in structurally altered RBD-J protein after storage, and thus could elicit antibodies that may sterically hinder ACE2-RBD-J interactions and neutralize the pseudovirus.77
Supplementary Material
Acknowledgments
We gratefully acknowledge the Wadsworth Center’s Veterinary Sciences staff for assistance with animal care, and the Tissue Culture core facility for media preparation. The authors would also like to thank Dr. Dong Yu, Dr. Mattew J. Bottomley, and Dr. Robert L. Coffman at Dynavax Technologies for providing the CpG 1018 adjuvant and reviewing this manuscript.
Funding Statement
This work was supported, in whole or in part, by the Bill & Melinda Gates Foundation [Investment IDs INV-002740 and INV-027417]. Research conducted at the Wadsworth Center was also supported, in part, by the National Cancer Institute of the National Institutes of Health under award number U01CA260508. N.C.D. was supported by the Ludwig Center at MIT’s Koch Institute. RBD and trimeric Spike antigen used for the analysis of mouse sera were kindly provided by MassBiologics (Boston) and produced under a Project Award Agreement from the National Institute for Innovation in Manufacturing Biopharmaceuticals (NIIMBL) and financial assistance award 70NANB20H037 from the U.S. Department of Commerce, National Institutes of Standards and Technologies. We gratefully acknowledge the Wadsworth Center’s Veterinary Sciences staff for assistance with animal care, and the Tissue Culture core facility for media preparation. The authors would also like to thank Dr. Dong Yu, Dr. Mattew J. Bottomley and Dr. Robert L. Coffman at Dynavax Technologies for providing CpG 1018 adjuvant and reviewing this manuscript.
Abbreviations
- AH
Alhydrogel®
- AP
AdjuPhos®
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The dataset generated and/or analyzed during the current study are available in the KU ScholarWorks repository, https://doi.org/10.17161/1808.32758. The data is also available with the corresponding author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2022.2079346.
References
- 1.COVID-19 vaccine tracker and landscape [Internet]. World Health Organization; [accessed 2021 Aug 23]. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 2.Forman R, Shah S, Jeurissen P, Jit M, Mossialos E.. COVID-19 vaccine challenges: what have we learned so far and what remains to be done? Health Policy (New York). 2021;125(5):1–17. doi: 10.1016/j.healthpol.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Director-General’s opening remarks at the media briefing on COVID-19 – 9 April 2021 [Internet]; [accessed 2021 Aug 28]. https://www.who.int/director-general/speeches/detail/director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-9-april-2021.
- 4.Holm MR, Poland GA. Critical aspects of packaging, storage, preparation, and administration of mRNA and adenovirus-vectored COVID-19 vaccines for optimal efficacy. Vaccine. 2021;39(3):457–59. doi: 10.1016/j.vaccine.2020.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson NAC, Kester KE, Casimiro D, Gurunathan S, DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5(1):1–6. doi: 10.1038/s41541-020-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumru OS, Joshi SB, Smith DE, Middaugh CR, Prusik T, Volkin DB. Vaccine instability in the cold chain: mechanisms, analysis and formulation strategies. Biologicals. 2014;42(5):237–59. doi: 10.1016/j.biologicals.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Kavanagh MM, Gostin LO, Sunder M. Sharing technology and vaccine doses to address global vaccine inequity and end the COVID-19 pandemic. Jama. 2021;326(3):219–20. doi: 10.1001/jama.2021.10823. [DOI] [PubMed] [Google Scholar]
- 8.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–48. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang M-Y, Zhao R, Gao L-J, Gao X-F, Wang D-P, Cao J-M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front Cell Infect Microbiol. 2020;10:724. doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piccoli L, Park Y-J, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183(4):1024–42.e21. doi: 10.1016/j.cell.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Law JLM, Logan M, Joyce MA, Landi A, Hockman D, Crawford K, Johnson J, LaChance G, Saffran HA, Shields J, et al. SARS-COV-2 recombinant Receptor-Binding-Domain (RBD) induces neutralizing antibodies against variant strains of SARS-CoV-2 and SARS-CoV-1. Vaccine. 2021;39(40):5769–79. doi: 10.1016/j.vaccine.2021.08.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborti S, Prabakaran P, Xiao X, Dimitrov DS. The SARS coronavirus S Glycoprotein receptor binding domain: fine mapping and functional characterization. Virol J. 2005;2(1):73. doi: 10.1186/1743-422X-2-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malladi SK, Singh R, Pandey S, Gayathri S, Kanjo K, Ahmed S, Khan MS, Kalita P, Girish N, Upadhyaya A, et al. Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J Biol Chem. 2021;296:100025. doi: 10.1074/jbc.RA120.016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalvie NC, Biedermann AM, Rodriguez-Aponte SA, Naranjo CA, Rao HD, Rajurkar MP, Lothe RR, Shaligram US, Johnston RS, Crowell LE, et al. Scalable, methanol-free manufacturing of the SARS-CoV-2 receptor-binding domain in engineered Komagataella phaffii. Biotechnol Bioeng. 2022;119(2):657–62. doi: 10.1002/bit.27979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, Liu Z, Chen W-H, Wei J, Kundu R, Adhikari R, Rivera JA, Gillespie PM, Strych U, Zhan B, et al. Process development and scale-up optimization of the SARS-CoV-2 receptor binding domain–based vaccine candidate, RBD219-N1C1. Appl Microbiol Biotechnol. 2021;105(10):4153–65. doi: 10.1007/s00253-021-11281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleanthous H, Silverman JM, Makar KW, Yoon I-K, Jackson N, Vaughn DW. Scientific rationale for developing potent RBD-based vaccines targeting COVID-19. NPJ Vaccines. 2021;6(1):1–10. doi: 10.1038/s41541-021-00393-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel FR. Improving vaccine performance with adjuvants. Clin Infect Dis. 2000;30(Supplement_3):S266–270. doi: 10.1086/313883. [DOI] [PubMed] [Google Scholar]
- 19.Reed SG, Orr MT, Fox CB. Key roles of adjuvants in modern vaccines. Nat Med. 2013;19(12):1597–608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 20.Baylor NW, Egan W, Richman P. Aluminum salts in vaccines—us perspective. Vaccine. 2002;20:S18–23. doi: 10.1016/S0264-410X(02)00166-4. [DOI] [PubMed] [Google Scholar]
- 21.Gupta RK. Aluminum compounds as vaccine adjuvants. Adv Drug Deliv Rev. 1998;32(3):155–72. doi: 10.1016/S0169-409X(98)00008-8. [DOI] [PubMed] [Google Scholar]
- 22.Lindblad EB. Aluminium compounds for use in vaccines. Immunol Cell Biol. 2004;82(5):497–505. doi: 10.1111/j.0818-9641.2004.01286.x. [DOI] [PubMed] [Google Scholar]
- 23.Awate S, Babiuk LAB, Mutwiri G. Mechanisms of Action of Adjuvants. Front Immunol [Internet]. 2013. [accessed 2021 Jan 18];4. 10.3389/fimmu.2013.00114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogenesch H. Mechanism of immunopotentiation and safety of aluminum adjuvants. Front Immunol. 2012;3:406. doi: 10.3389/fimmu.2012.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheiermann J, Klinman DM. Clinical evaluation of CpG oligonucleotides as adjuvants for vaccines targeting infectious diseases and cancer. Vaccine. 2014;32(48):6377–89. doi: 10.1016/j.vaccine.2014.06.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shirota H, Klinman DM. CpG oligodeoxynucleotides as adjuvants for clinical use [Internet]. In: Immunopotentiators in modern vaccines. Elsevier Inc.; 2017. [accessed 2021 Nov 17]. p. 163–98. doi: 10.1016/B978-0-12-804019-5.00009-8. [DOI] [Google Scholar]
- 27.Campbell JD. Development of the CpG adjuvant 1018: a case study. Methods Mol Biol. 2017;1494:15–27. [DOI] [PubMed] [Google Scholar]
- 28.HeplisavTM: a new hepatitis B vaccine. Future Virology [Internet]; [accessed 2021 Aug 22]. https://www.futuremedicine.com/doi/abs/10.2217/17460794.3.2.109.
- 29.Klinman DM, Currie D, Gursel I, Verthelyi D. Use of CpG oligodeoxynucleotides as immune adjuvants. Immunol Rev. 2004;199(1):201–16. doi: 10.1111/j.0105-2896.2004.00148.x. [DOI] [PubMed] [Google Scholar]
- 30.Mirotti L, Alberca Custódio RW, Gomes E, Rammauro F, de Araujo EF, Garcia Calich VL, Russo M. CpG-ODN shapes alum adjuvant activity signaling via MyD88 and IL-10. Front Immunol. 2017;8:47. doi: 10.3389/fimmu.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugai T, Mori M, Nakazawa M, Ichino M, Naruto T, Kobayashi N, Kobayashi Y, Minami M, Yokota S. A CpG-containing oligodeoxynucleotide as an efficient adjuvant counterbalancing the Th1/Th2 immune response in diphtheria–tetanus–pertussis vaccine. Vaccine. 2005;23(46–47):5450–56. doi: 10.1016/j.vaccine.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 32.Texas children’s hospital and Baylor college of medicine covid-19 vaccine technology secures emergency use authorization in India. Texas Children’s Hospital [Internet]; [accessed 2022 Mar 4]. https://www.texaschildrens.org/texas-children%E2%80%99s-hospital-and-baylor-college-medicine-covid-19-vaccine-technology-secures-emergency
- 33.Thuluva S, Paradkar V, Turaga K, Gunneri S, Yerroju V, Mogulla R, Kyasani M, Manoharan SK, Medigeshi G, Singh J, et al. Selection of optimum formulation of RBD-based protein sub-unit covid19 vaccine (Corbevax) based on safety and immunogenicity in an open-label, randomized Phase-1 and 2 clinical studies Internet. Human Vacc Immunother. 2022. [accessed 2022 Mar 24]:1–11. https://www.medrxiv.org/content/10.1101/2022.03.08.22271822v2 [Google Scholar]
- 34.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–77. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 35.Pollet J, Strych U, Chen W-H, Versteeg L, Keegan B, Zhan B, Wei J, Liu Z, Lee J, Kundu R, et al. Receptor-Binding domain recombinant protein RBD219-N1C1 on alum-CpG induces broad protection against SARS-CoV-2 variants of concern [Internet]. Mol Biol. 2021. [accessed 2021 Nov 16]. http://biorxiv.org/lookup/doi/10.1101/2021.07.06.451353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dalvie NC, Rodriguez-Aponte SA, Hartwell BL, Tostanoski LH, Biedermann AM, Crowell LE, Kaur K, Kumru OS, Carter L, Yu J, et al. Engineered SARS-CoV-2 receptor binding domain improves manufacturability in yeast and immunogenicity in mice. PNAS [Internet] 2021. [accessed 2021 Oct 10]; 118. https://www.pnas.org/content/118/38/e2106845118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang S, Li Y, Dai L, Wang J, He P, Li C, Fang X, Wang C, Zhao X, Huang E, et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107–19. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W-H, Pollet J, Strych U, Lee J, Liu Z, Kundu RT, Versteeg L, Villar MJ, Adhikari R, Wei J, et al. Yeast-Expressed recombinant SARS-CoV-2 receptor binding domain RBD203-N1 as a COVID-19 protein vaccine candidate. Protein Expr Purif. 2022;190:106003. doi: 10.1016/j.pep.2021.106003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adimmune Corporation . A randomized, single center, open-label, dose-finding, phase i study to evaluate the safety and immunogenicity of pandemic virus vaccine, AdimrSC-2f (SARS-CoV-2), in healthy volunteers [Internet]. clinicaltrials.gov; 2021. [accessed 2021 Nov 16]. https://clinicaltrials.gov/ct2/show/NCT04522089.
- 40.United Biomedical Inc., Asia . A Phase I, open-label study to evaluate the safety, tolerability, and immunogenicity of UB-612 vaccine in healthy adult volunteers [Internet]. clinicaltrials.gov; 2021. [accessed 2021 Nov 16]. https://clinicaltrials.gov/ct2/show/NCT04545749
- 41.Sun S, He L, Zhao Z, Gu H, Fang X, Wang T, Yang X, Chen S, Deng Y, Li J, et al. Recombinant vaccine containing an RBD-Fc fusion induced protection against SARS-CoV-2 in nonhuman primates and mice. Cell Mol Immunol. 2021;18:1070–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan X, Shi J, Hu X, Wu Y, Zeng L, Yao Y, Shang W, Liu K, Gao G, Guo W, et al. RBD-Homodimer, a COVID-19 subunit vaccine candidate, elicits immunogenicity and protection in rodents and nonhuman primates. Cell Discov. 2021;7(1):1–15. doi: 10.1038/s41421-021-00320-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.An Y, Li S, Jin X, Han J, Xu K, Xu S, Han Y, Liu C, Zheng T, Liu M, et al. A tandem-repeat dimeric RBD protein-based COVID-19 vaccine ZF2001 protects mice and nonhuman primates [Internet]. Immunology. 2021. [accessed 2021 Nov 12]. http://biorxiv.org/lookup/doi/10.1101/2021.03.11.434928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nanishi E, Borriello F, O’-Meara TR, McGrath ME, Saito Y, Haupt RE, Seo H-S, van HS, Cavazzoni CB, Brook B, et al. An aluminum hydroxide:cpg adjuvant enhances protection elicited by a SARS-CoV-2 receptor-binding domain vaccine in aged mice. Sci Transl Med [Internet]. 2021. [accessed 2022 Jan 21]. https://www.science.org/doi/abs/10.1126/scitranslmed.abj5305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pollet J, Chen W-H, Versteeg L, Keegan B, Zhan B, Wei J, Liu Z, Lee J, Kundu R, Adhikari R, et al. Sars‑cov-2 RBD219-N1C1: a yeast-expressed SARS-CoV-2 recombinant receptor-binding domain candidate vaccine stimulates virus neutralizing antibodies and T-cell immunity in mice. Human Vacc Immunother. 2021;17(8):2356–66. doi: 10.1080/21645515.2021.1901545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamborg M, Foged C. Characterizing the association between antigens and adjuvants [Internet]. In: Foged C; Rades T; Perrie YHook S, editors. Subunit Vaccine Delivery. New York (NY): Springer; 2015. [accessed 2021 Nov 20]. p. 413–26. 10.1007/978-1-4939-1417-3_21 [DOI] [Google Scholar]
- 47.Sawant N, Kaur K, Holland DA, Hickey JM, Agarwal S, Brady JR, Dalvie NC, Tracey MK, Velez-Suberbie ML, Morris SA, et al. Rapid developability assessments to formulate recombinant protein antigens as stable, low-cost, multi-dose vaccine candidates: case-study with Non-Replicating Rotavirus (NRRV) Vaccine antigens. J Pharm Sci. 2021;110(3):1042–53. doi: 10.1016/j.xphs.2020.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAdams D, Lakatos K, Estrada M, Chen D, Plikaytis B, Sitrin R, White JA. Quantification of trivalent non-replicating rotavirus vaccine antigens in the presence of aluminum adjuvant. J Immunol Methods. 2021;494:113056. doi: 10.1016/j.jim.2021.113056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hotez PJ, Bottazzi ME. Whole inactivated virus and protein-based COVID-19 vaccines. Annu Rev Med. 2022;73(1):55–64. doi: 10.1146/annurev-med-042420-113212. [DOI] [PubMed] [Google Scholar]
- 50.Pollet J, Chen W-H, Strych U. Recombinant protein vaccines, a proven approach against coronavirus pandemics. Adv Drug Deliv Rev. 2021;170:71–82. doi: 10.1016/j.addr.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nanishi E, Dowling D, Levy O. Toward precision adjuvants: Optimizing science and safety. Curr Opin Pediatr. 2020;32(1):1. doi: 10.1097/MOP.0000000000000868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.HogenEsch H, O’-Hagan DT, Fox CB. Optimizing the utilization of aluminum adjuvants in vaccines: you might just get what you want. NPJ Vaccines. 2018;3(1):1–11. doi: 10.1038/s41541-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fox CB, Kramer RM, Barnes VL, Dowling QM, Vedvick TS. Working together: interactions between vaccine antigens and adjuvants. Ther Adv Vaccines. 2013;1(1):7–20. doi: 10.1177/2051013613480144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao Y, Zeng Y, Alexander E, Mehta S, Joshi SB, Buchman GW, Volkin DB, Russell Middaugh C, Isaacs SN. Adsorption of recombinant poxvirus L1-protein to aluminum hydroxide/cpg vaccine adjuvants enhances immune responses and protection of mice from vaccinia virus challenge. Vaccine. 2013;31(2):319–26. doi: 10.1016/j.vaccine.2012.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao Y, Zeng Y, Schante C, Joshi SB, Buchman GW, Volkin DB, Middaugh CR, Isaacs SN. Short-Term and longer-term protective immune responses generated by subunit vaccination with smallpox A33, B5, L1 or A27 proteins adjuvanted with aluminum hydroxide and CpG in mice challenged with vaccinia virus. Vaccine. 2020;38(38):6007–18. doi: 10.1016/j.vaccine.2020.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mullen GED, Aebig JA, Dobrescu G, Rausch K, Lambert L, Long CA, Miles AP, Saul A. Enhanced antibody production in mice to the malaria antigen AMA1 by CPG 7909 requires physical association of CpG and antigen. Vaccine. 2007;25(29):5343–47. doi: 10.1016/j.vaccine.2007.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Romero Méndez IZ, Shi Y, HogenEsch H, Hem SL. Potentiation of the immune response to non-adsorbed antigens by aluminum-containing adjuvants. Vaccine. 2007;25(5):825–33. doi: 10.1016/j.vaccine.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 58.Moss P. The T cell immune response against SARS-CoV-2. Nat Immunol. 2022;23(2):186–93. doi: 10.1038/s41590-021-01122-w. [DOI] [PubMed] [Google Scholar]
- 59.Beaudoin-Bussières G, Chen Y, Ullah I, Prévost J, Tolbert WD, Symmes K, Ding S, Benlarbi M, Gong SY, Tauzin A, et al. A Fc-enhanced NTD-binding non-neutralizing antibody delays virus spread and synergizes with a nAb to protect mice from lethal SARS-CoV-2 infection. Cell Rep. 2022;38(7):110368. doi: 10.1016/j.celrep.2022.110368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sedova ES, Scherbinin DN, Lysenko AA, Alekseeva SV, Artemova EA, Shmarov MM. Non-neutralizing antibodies directed at conservative influenza antigens. Acta Naturae. 2019;11(4):22–32. doi: 10.32607/20758251-2019-11-4-22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shrivastava T, Singh B, Rizvi ZA, Verma R, Goswami S, Vishwakarma P, Jakhar K, Sonar S, Mani S, Bhattacharyya S, et al. Comparative immunomodulatory evaluation of the receptor binding domain of the SARS-CoV-2 spike protein; a potential vaccine candidate which imparts potent humoral and Th1 Type immune response in a mouse model. Front Immunol. 2021;12:641447. doi: 10.3389/fimmu.2021.641447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones LS, Peek LJ, Power J, Markham A, Yazzie B, Middaugh CR. Effects of adsorption to aluminum salt adjuvants on the structure and stability of model protein antigens. J Biol Chem. 2005;280(14):13406–14. doi: 10.1074/jbc.M500687200. [DOI] [PubMed] [Google Scholar]
- 63.Iyer V, Hu L, Liyanage MR, Esfandiary R, Reinisch C, Meinke A, Maisonneuve J, Volkin DB, Joshi SB, Middaugh CR. Preformulation characterization of an aluminum salt-adjuvanted trivalent recombinant protein-based vaccine candidate against Streptococcus pneumoniae. J Pharm Sci. 2012;101(9):3078–90. doi: 10.1002/jps.23175. [DOI] [PubMed] [Google Scholar]
- 64.Régnier M, Metz B, Tilstra W, Hendriksen C, Jiskoot W, Norde W, Kersten G. Structural perturbation of diphtheria toxoid upon adsorption to aluminium hydroxide adjuvant. Vaccine. 2012;30(48):6783–88. doi: 10.1016/j.vaccine.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Agarwal S, Hickey JM, McAdams D, White JA, Sitrin R, Khandke L, Cryz S, Joshi SB, Volkin DB. Effect of aluminum adjuvant and preservatives on structural integrity and physicochemical stability profiles of three recombinant subunit rotavirus vaccine antigens. J Pharm Sci. 2020;109(1):476–87. doi: 10.1016/j.xphs.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cereghino JL, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol Rev. 2000;24(1):45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]
- 67.Dalvie NC, Tostanoski LH, Rodriguez-Aponte SA, Kaur K, Bajoria S, Kumru OS, Martinot AJ, Chandrashekar A, McMahan K, Mercado NB, et al. SARS-CoV-2 receptor binding domain displayed on HBsAg virus–like particles elicits protective immunity in macaques. Sci Adv. 2022;8(11). doi: 10.1126/sciadv.abl6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fox C, Dutill T, Chesko J, Anderson R, Reed S, Vedvick T. Characterization of aluminum hydroxide gel and oil-in-water emulsion formulations containing CpG ODNs. BioPharm International. 2010;23:14–19. [Google Scholar]
- 69.Aebig JA, Mullen GED, Dobrescu G, Rausch K, Lambert L, Ajose-Popoola O, Long CA, Saul A, Miles AP. Formulation of vaccines containing CpG oligonucleotides and alum. J Immunol Methods. 2007;323(2):139–46. doi: 10.1016/j.jim.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mycroft-West C, Su D, Elli S, Li Y, Guimond S, Miller G, Turnbull J, Yates E, Guerrini M, Fernig D, et al. The 2019 coronavirus (SARS-CoV-2) surface protein (Spike) S1 Receptor binding domain undergoes conformational change upon heparin binding [Internet]; 2020. [accessed 2022 Jan 21]. https://www.biorxiv.org/content/10.1101/2020.02.29.971093v2.
- 71.Neufurth M, Wang X, Tolba E, Lieberwirth I, Wang S, Schröder HC, Müller WEG. The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem Pharmacol. 2020;182:114215. doi: 10.1016/j.bcp.2020.114215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.White JA, Estrada M, Weldon WC, Chumakov K, Kouiavskaia D, Fournier-Caruana J, Stevens E, Gary HE, Maes EF, Oberste MS, et al. Assessing the potency and immunogenicity of inactivated poliovirus vaccine after exposure to freezing temperatures. Biologicals. 2018;53:30–38. doi: 10.1016/j.biologicals.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cai W, Ping L, Shen W, Liu J, Zhang M, Zhou J, Peng J, Wang M, Zhu Y, Ji G, et al. Potency of the Sabin inactivated poliovirus vaccine (sIPV) after exposure to freezing temperatures in cold chains. Human Vacc Immunother. 2020;16(8):1866–74. doi: 10.1080/21645515.2019.1709352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murakami K, Fujii Y, Someya Y. Effects of the thermal denaturation of Sabin-derived inactivated polio vaccines on the D-antigenicity and the immunogenicity in rats. Vaccine. 2020;38(17):3295–99. doi: 10.1016/j.vaccine.2020.03.027. [DOI] [PubMed] [Google Scholar]
- 75.Nitahara Y, Nakagama Y, Kaku N, Candray K, Michimuko Y, Tshibangu-Kabamba E, Kaneko A, Yamamoto H, Mizobata Y, Kakeya H, et al. High-resolution linear epitope mapping of the receptor binding domain of SARS-CoV-2 spike protein in COVID-19 mRNA vaccine recipients. Microbiol Spectr. 2021;9(3):e0096521. doi: 10.1128/Spectrum.00965-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jiang M, Zhang G, Liu H, Ding P, Liu Y, Tian Y, Wang Y, Wang A. Epitope profiling reveals the critical antigenic determinants in SARS-CoV-2 RBD-based antigen. Front Immunol. 2021;12:707977. doi: 10.3389/fimmu.2021.707977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Finkelstein MT, Mermelstein AG, Parker Miller E, Seth PC, Stancofski E-S, Fera D. Structural analysis of neutralizing epitopes of the SARS-CoV-2 spike to guide therapy and vaccine design strategies. Viruses. 2021;13(1):134. doi: 10.3390/v13010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The dataset generated and/or analyzed during the current study are available in the KU ScholarWorks repository, https://doi.org/10.17161/1808.32758. The data is also available with the corresponding author(s).


