ABSTRACT
Exogenous oestrogen 17β-oestradiol (E2) has been shown to effectively induce feminization in teleosts. However, the molecular mechanisms underlying the process remain unclear. Here, we determined global DNA methylation and gene expression profiles of channel catfish (Ictalurus punctatus) during early sex differentiation after E2 treatment. Overall, the levels of global DNA methylation after E2 treatment were not significantly different from those of controls. However, a specific set of genes were differentially methylated, which included many sex differentiation-related pathways, such as MARK signalling, adrenergic signalling, Wnt signalling, GnRH signalling, ErbB signalling, and ECM–receptor interactions. Many genes involved in these pathways were also differentially expressed after E2 treatment. Specifically, E2 treatments resulted in upregulation of female-related genes and downregulation of male-related genes in genetic males during sex reversal. However, E2-induced sex reversal did not cause sex-specific changes in methylation profiles or gene expression within the sex determination region (SDR) on chromosome 4, suggesting that E2-induced sex reversal was a downstream process independent of the sex determination process that was regulated by sex-specific methylation within the SDR.
KEYWORDS: Sex reversal, methylation, oestrogen, transcription, sex differentiation, teleost
Introduction
Fish account for more than half of all vertebrate species and exhibit very diverse reproductive strategies [1,2]. Among them, gonochoristic species have only ovarian or testicular tissues after sex determination and differentiation, while sequential hermaphrodites can change from male to female or from female to male in their life cycle. For instance, zebrafish (Danio rerio) is a gonochoristic species without sex switch as adults [3], whereas bluehead wrasse (Thalassoma bifasciatum) exhibits dramatic and complete female-to-male sex reversal following social stresses [4]. This diversity and plasticity in fish provide excellent models to study sex determination and differentiation.
Sex determination leads to the binary fate of ovary or testis. In species where sex is determined by genetic factors, the potential of an individual to become specific sex with either an ovary (female) or a testis (male) is determined by their chromosomal composition. However, in many species, especially those with lower teleost fish, sex differentiation may be delayed and altered by environment or sex hormones [5]. Sex steroids are involved in the natural process of sex differentiation and maintenance [6,7], and the administration of exogenous sex steroids can affect the process [8]. In many fish species, oestrogens cause feminization, whereas androgens cause masculinization [8–10].
17β-oestradiol (E2), one of the exogenous oestrogens, has been reported to effectively induce feminization in more than 50 fish species, including, but not limited to the taxonomic families of Cyprinidae, Salmonidae, Cichlidae, and Ictaluridae [8,11], but the underlying mechanisms of the steroid-induced sex reversal remain elusive. In model organisms, oestrogen is involved in the regulation of methylation and transcription [12–14]. After E2 exposure of zebrafish, genes related to ovarian development were significantly upregulated, and they were enriched with genes involved in ECM–receptor interaction, gap junction, and cell adhesion molecules [12].
Previous studies demonstrated that exogenous application of sex steroids, either oestrogens or androgens, caused feminization of channel catfish [15,16]. In the latter case, exogenously applied androgens were likely converted into oestrogens after application [17], but the molecular mechanisms of the hormone-induced sex reversal are unknown. Sex hormones were known to cause changes in genome methylation profiles of several fish species, including zebrafish and three-spine stickleback [13,18]. The objective of this study was to determine the effect of E2 on global methylation and genome expression profiles, in relation to its phenotypic effect on sex reversal. Here, we report the dynamic changes in genome expression of a large set of genes involved in female gonadal sex differentiation, whereas whole-genome methylation and the sex-specific methylation profiles within the sex determination region (SDR) on chromosome 4 were not significantly modified after E2 treatment.
Materials and methods
Ethical statement
This study was performed according to the Guide for the Care and Use of Laboratory Animals and the Animal Welfare Act in the USA. All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at Auburn University.
Experimental design and sample collection
All fish work was performed at the Fish Genetics Laboratory, Auburn University. Fertilized eggs of channel catfish were incubated in wheel troughs to avoid fungal growth. It is difficult to control the concentration of oestrogen in the flow-through trough system. Therefore, starting within the first 2 hours post fertilization, the embryos were taken out of the hatching trough and moved to a tub with static water containing E2 (E8875, Sigma-Aldrich, Saint Louis, USA) at 400 μg/L for 4 hours per day (2 hours in the morning and 2 hours in the afternoon) until 10 days post fertilization (dpf); the embryos were incubated in the hatching troughs between each of the treatment in the morning and afternoon. Starting from 10 dpf, the fry were fed with diets containing E2 at a dose of 200 mg/kg, three times per day to 110 dpf. This concentration in the feed was used in previous studies, e.g., with zebra cichlid Cichlasoma nigrofasciatum, which resulted in 100% feminization [19]. Control fish were reared in the same way as the treatment, except that E2 was not added during the whole process.
Samples from treatment and control groups were collected at 3, 9, 12, 16, and 110 dpf. Embryos or fish were euthanized with MS222, then placed in a 1.5 ml tube and plunged into liquid nitrogen immediately. At 3, 9, 12, and 16 dpf, gonadal differentiation was morphologically invisible, and, therefore, the whole fish samples were collected. Then, the head and tail parts were dissected for DNA extraction to identify genetic sex via PCR analysis using a sex-linked microsatellite marker AUEST0678 [20]. The remaining part was divided longitudinally into two equal parts from the spine, with one-half for DNA isolation for whole-genome bisulphite sequencing (WGBS), and the other half for RNA isolation for RNA sequencing. For samples at 110 dpf, gonads were visible, and the phenotypic sex was determined by gonad morphology; genetic sex was determined via PCR analysis. Gonads were dissected and collected for DNA and RNA isolation.
DNA and RNA extraction
Genomic DNA was extracted using DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer’s protocol. Total RNA was extracted by RNeasy Plus Universal Tissue Mini Kit (Qiagen) according to the manufacturer’s instruction. Genomic DNA and RNA were quantified using Nanodrop 2000, and the quality was assessed by 1% agarose gel electrophoresis.
Whole-genome bisulphite sequencing (WGBS)
For time points at 3, 9, 12, 16, and 110 dpf, equal amounts of genomic DNA from five fish samples of genetic females or males in the treatment and control groups were pooled as one replicate for WGBS libraries construction. At 110 dpf, five DNA samples from pseudo-female gonads were also pooled for library preparation. Three replicates were used for each time point, each sex, and each treatment. A total of 63 libraries (2 sexes × 5 time points × 2 treatment × 3 replicates + 3 pseudo-females) were prepared and sequenced by Illumina NovaSeq6000 S4 platform with paired-end (PE) 150 (CD Genomics, Shirley, NY, USA).
WGBS data analysis
Bisulphite conversion efficiency was assessed by adding unmethylated lambda phage DNA, and over 99% conversion rates were achieved for all samples. Quality control of raw WGBS reads was performed by FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Trimmomatic v0.37 [21] was used to remove adaptor sequences, ambiguous nucleotides, short reads (< 36 bp), and low-quality reads (quality score < 20). Before mapping, the trimmed reads were transformed into fully bisulphite-converted forward (C to T) and reverse read (G to A conversion of the forward strand) versions by the bisulphite alignment programme Bismark v0.22.1 [22]. Channel catfish reference genome IpCoco_1.2 [23] was also converted (C to T and G to A converted) by ‘bismark_genome_preparation’ tool in the Bismark v0.22.1. Then, sequence reads were aligned to converted versions of the genome using Bowtie 2 [24]. Methylation level on each site was extracted using ‘bismark_methylation_extractor’ tool. Upon completion, a run report including percentage methylation of cytosines in CpG, CHG and CHH context, number of methylated and unmethylated cytosines, and the per cent methylation value (context) was produced.
Methylation levels were analysed using SeqMonk v1.45.2 (https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). The methylation sites located on the 29 chromosomes of channel catfish were imported to SeqMonk, and the minimum read count per position was set to 10 when defining the probe. Methylation percentages were calculated using the ‘bisulfite methylation over feature’ pipeline in SeqMonk with default parameters (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/). A statistical test between the two groups was performed by logistic regression of proportion-based statistics in SeqMonk with p value < 0.01 with a minimum observation of 10. The determination of differentially methylated sites (DMSs) between the two groups also required a percentage difference greater than 25%.
Differentially methylated genes (DMGs) were defined as DMSs located in their promoter [the 2 kb region upstream of the transcription start site (TSS)] and/or gene body. Gene ontology (GO) and Kyoto Encyclopaedia of Genes and Genomes (KEGG) enrichment analyses of the DMGs were performed using the R package clusterProfiler [25].
Principal component analysis (PCA) was conducted using methyl Kit [26] and plotted by ggplot2 in R [27]. CpG methylation levels of features in the channel catfish genome and average CpG methylation percentage near TSS of different groups were calculated with the results from Bismark.
RNA-seq samples and data analysis
For RNA-seq libraries, RNA from three samples of the same sex (genetic female or genetic male) and the same group (treatment and control) were pooled equalling as a replicate at 3, 16, and 110 dpf. Three replicates were prepared for each group. Additionally, RNA from sex-reversed fish at 110 dpf were also extracted. A total of 39 libraries were generated and sequenced by Illumina NovaSeq6000 S4 platform with PE150 (CD Genomics, Shirley, NY, USA).
Raw reads were trimmed by Trimmomatic v0.37 with length > 36 bp and quality score > 20 [21]. The quality of each sample was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The analysis was conducted by mapping reads to channel catfish reference genome IpCoco_1.2 [23] with HISAT2 v2.1.0 [28]. The alignments were saved in SAM format, which were converted and sorted to bam files with SAMtools [29]. HTSeq v0.11.0 was used to quantify the number of expressed transcripts [30]. Differentially expressed genes (DEGs) were identified with DESeq2 with | log2(fold change) | > 1 and adjusted p value < 0.05 [31]. Functional enrichment analysis of DEGs was conducted in R using clusterProfiler [25].
Results
Feminization by E2 exposure
The genetic and phenotypic sex of experimental fish were determined at 110 dpf. Of the 233 fish that were treated with E2, 101 were genetic females and 132 were genetic males. All genetic females were phenotypic females. Among the 132 genetic males, 116 were sex-reversed to phenotypic females (pseudo-females), with a sex reversal rate of 87.9%. In total, 217 phenotypic females (91.8%) were identified among the E2-treated fish (Figure 1). The sex ratio of control was nearly 1:1, including 81 females and 78 males, with no fish being sex-reversed (Figure 1).
Figure 1.
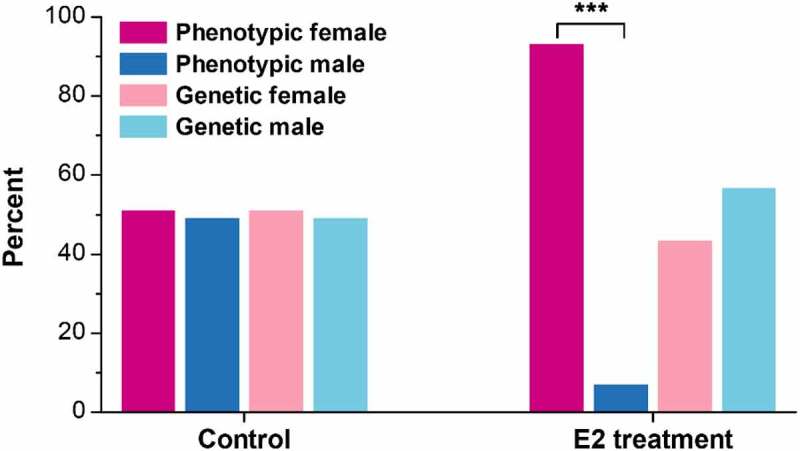
The percent of genetic and phenotypic female and male channel catfish (Ictalurus punctatus) in the control and 17β-oestradiol (E2) treatment at 110 days post fertilization (dpf). ***, p < 0.001 by the Chi-square goodness-of-fit test.
Genome-wide DNA methylation profiles after E2 exposure
To determine the effect of E2 on genome methylation, WGBS was conducted with samples of females and males at 3, 9, 12, 16, and 110 dpf, with and without E2 treatment. Samples from pseudo-females at 110 dpf after E2 exposure were also analysed. A total of 1,794 Gb methylome data with 12 billion sequence reads were produced, which yielded an average depth of 35× per sample (Supplementary Table 1). The raw data has been submitted to the NCBI SRA with the BioProject ID PRJNA826119. Of these datasets, an average of 163 million methylated cytosines (mCs) per sample were identified, accounting for 8% of the total sequenced cytosines. Among them, 95% of mCs were in the CpG context, and nearly 80% of the total CpG were methylated (Figure 2(a), Supplementary Table 2). No significant difference in genome methylation levels was observed between E2 treatment and control in both females and males (Figure 2(a)). Likewise, the percentage of mCpG between E2 treatment and control at each time point was also similar (Figure 2(b)). The overall mCpG percentage declined with gonadal differentiation over time from 3 to 110 dpf (Figure 2(b)). Additionally, slight increases in mCpG percentage were observed after E2 treatment at 3, 9, and 16 dpf in both females and males, but a slight decrease was observed at 110 dpf in females; all these differences were not statistically significant (Figure 2(b)). The methylation level of pseudo-females was most similar to that of E2-treated females at 110 dpf (Figure 2(b)).
Figure 2.
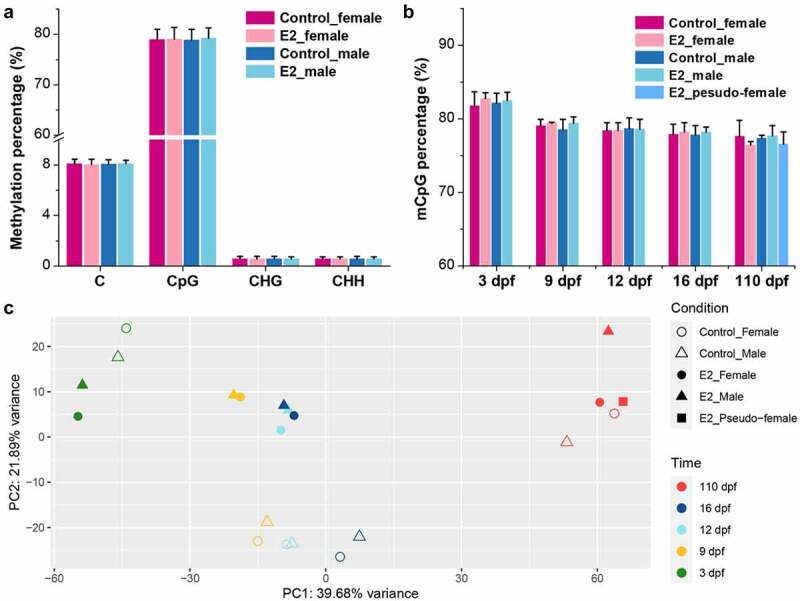
Genome-wide methylation levels of the control and 17β-oestradiol (E2) treated channel catfish, Ictalurus punctatus. (a) Methylation percentage in the contexts of C, CpG, CHG, and CHH. (b) Relative percentage of CpG methylation at different time points; dpf: days post fertilization. (c) PCA of all WGBS samples based on methylated CpG sites. PCA plots of replicate samples are shown in Supplemental Figure 1, showing variations of samples.
PCA revealed that the methylation patterns were initially clustered by development, but then by treatment (Figure 2(c)). Samples of E2 treatment and control were effectively separated at 9, 12, and 16 dpf. At 110 dpf, pseudo-females clustered together with both control females and E2-treated females, while control males and E2-treated males were more different.
DMGs and involved gene pathways after E2 exposure
Although the overall levels of methylation after E2 treatment were not significantly different from those of controls, a large number of genes were differentially methylated after E2 treatment (Figure 3(a), Supplementary Table 3). The number of DMGs in males was greater than that in females at both 3 and 9 dpf. At 12 dpf, the numbers of DMGs in females and males were similar between females and males, but became smaller than those at 3 and 9 dpf (Figure 3(a)). At 16 dpf, the number of DMGs in females and males remained low, and they were even smaller in males than in females. At 110 dpf, the number of DMGs became very large, with females harbouring more DMGs than males (Figure 3(a)).
Figure 3.
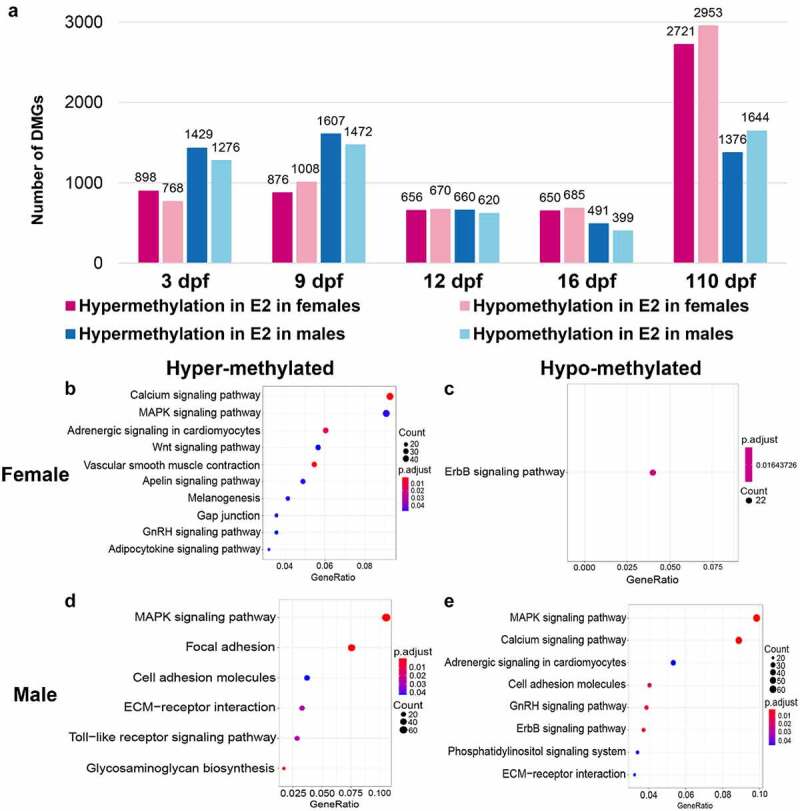
Numbers of differentially methylated genes (DMGs) and their enriched gene pathways after treatment of 17β-oestradiol (E2). (a) Comparison of DMGs after E2 treatment in females and males at 3, 9, 12, 16, and 110 days post fertilization (dpf). B-E KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway enrichment analysis of hypermethylated methylated genes (DMGs) in females (b), hypomethylated DMGs in females (c), hypermethylated DMGs in males (d), and hypomethylated DMGs in males (e) at 3 and 16 dpf after E2 treatment.
To gain initial insight into the nature of DMGs after E2 treatment during early gonadal differentiation (3, 9, 12, and 16 dpf), the involved pathways were determined through a KEGG pathway enrichment analysis. As shown in Figure 3(b), hypermethylated genes in females were enriched in 10 pathways, including calcium signalling, MARK signalling, adrenergic signalling, Wnt signalling, vascular smooth muscle contraction, apelin signalling, melanogenesis, gap junction, GnRH signalling, and adipocytokine signalling. Hypomethylated genes in females were involved in ErbB signalling (Figure 3(c)).
There were remarkable similarities in pathways involving genes that were hypermethylated in females but hypomethylated in males. The top three pathways involving the largest number of genes had the same pathways hypermethylated in females but hypomethylated in males (compare Figure 3(b) with Figure 3(e)). Genes involved in the ErbB signalling pathway were hypomethylated in both females and males (Figure 3(c,e)). Interestingly, genes involved in certain pathways were both hypermethylated and hypomethylated in males, including cell adhesion molecules and ECM–receptor interaction (Figure 3(d,e)).
Differential expression after E2 exposure
RNA-seq was conducted to determine genomic expression after E2 treatment. A total of 3,950 million raw reads were produced, and 3,689 million clean reads were generated. They have been submitted to NCBI SRA with the BioProject ID PRJNA826119. The average mapping rate was 93.4% (Supplementary Table 4). Expression levels were compared between treatment and control in both females and males, and DEGs were listed in Supplementary Table 5. At 3 dpf, 208 and 283 genes were upregulated in females and males, respectively; 610 and 99 genes were down-regulated in females and males, respectively (Table 1). At 16 dpf, many more genes were differentially expressed in both females (3,636 up and 1,875 down) and males (2,575 up and 1,492 down), but at 110 dpf, the number of DEGs in females was relatively small, with 876 up-regulated genes and 213 down-regulated genes. However, the number of DEGs in males was extremely large, with 5,665 up-regulated genes and 4,609 down-regulated genes (Table 1), presumably because of the transition from genetic males to phenotypic females.
Table 1.
The numbers of differentially expressed genes (DEGs) after 17β-oestradiol (E2) treatment at 3, 16, and 110 days post fertilization (dpf).
| Time point (dpf) | Up-DEGs in females | Down-DEGs in females | Up-DEGs in males | Down-DEGs in males |
|---|---|---|---|---|
| 3 | 208 | 610 | 283 | 99 |
| 16 | 3,636 | 1,875 | 2,507 | 1,492 |
| 110 | 876 | 213 | 5,665 | 4,609 |
At 110 dpf, gonads were differentiated such that phenotypic sex could be determined. The KEGG enrichment analysis was conducted using RNA-seq datasets of 3 and 16 dpf, to reveal gene pathways involved in E2-induced sex reversal. As expected, enriched gene pathways among up-regulated genes in females and males were very similar, with each enriched for gene pathways of neuroactive ligand–receptor interaction, calcium signalling, MARK signalling, adrenergic signalling in cardiomyocytes, cell adhesion molecules, and cardiac muscle contraction. The enrichment of these pathways apparently reflected significant advances towards female sex differentiation with both genetic females and genetic males, although the number of genes involved in these enriched pathways in genetic females was larger (compare Figure 4(a) with Figure 4(c)). In contrast, a much larger number of pathways were enriched among down-regulated genes in both females and males. Most of the down-regulated gene pathways were shared between females and males, including DNA replication, focal adhesion, cardiac muscle contraction, cell cycle, ECM–receptor interaction, fatty acid metabolism, glycolysis-pyruvate metabolism, amino acid metabolism, and terpenoid backbone biosynthesis (Figure 4(b,d)). Most of these enriched pathways were involved in growth rather than differentiation, turning the course of prolonged cellular growth as naturally occurring in males, to gonadal sex differentiation as naturally occurring in females.
Figure 4.
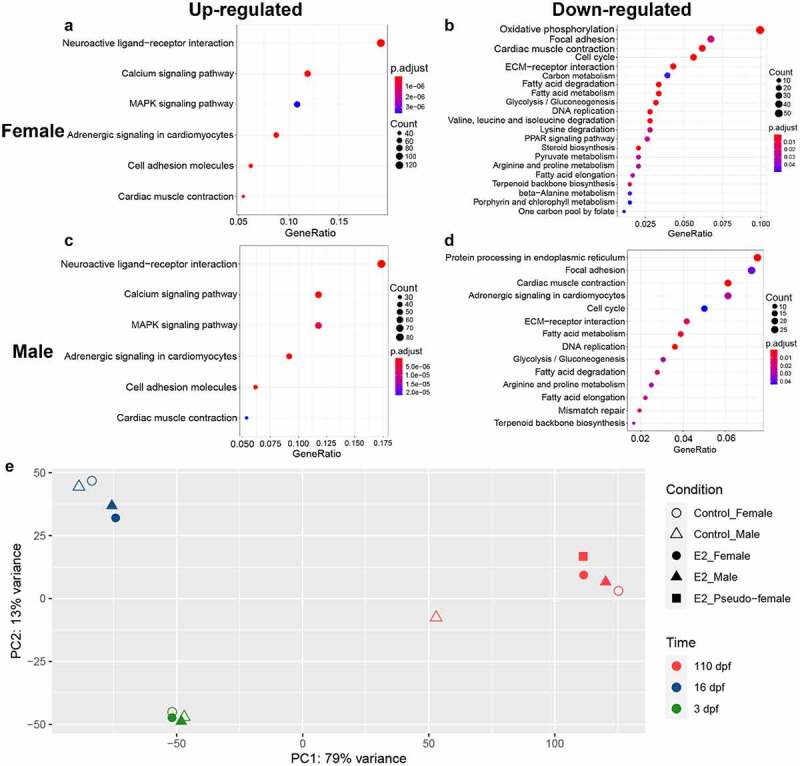
Enriched gene pathways of differentially expressed genes (DEGs) after 17β-oestradiol (E2) treatment. A-D KEGG (Kyoto Encyclopaedia of Genes and Genomes) pathway enrichment analysis of up-regulated DEGs in females (a), down-regulated DEGs in females (b), up-regulated DEGs in males (c), and down-regulated DEGs in males (d) at 3 and 16 days post fertilization (dpf) after E2 treatment. (e) PCA of RNA-seq samples using regularized-logarithm transformation of normalized expression data in DESeq2. PCA plots of replicate RNA-Seq samples are shown in Supplemental Figure 2, showing variations of samples.
Principal component analysis was conducted using genomic expression datasets of RNA-seq at 3, 16, and 110 dpf. As shown in Figure 4(e), the developmental time accounted for the largest proportion of variance of gene expression. As such, samples of 3, 16, and 110 dpf were mostly clustered within their own group at the same time point after fertilization, especially at 3 dpf. Over time, the treatment effect accounted for increasing amount of variance in expression. At 16 dpf, the control females and males clustered together, while the treated females and males clustered together. This pattern, however, did not hold true for samples at 110 dpf when control females, treated males, treated females and pseudo-females clustered together, whereas control males were very distant (Figure 4(e)), suggesting all treated fish were most similar to females with regard to genome expression patterns at 110 dpf.
Methylation and gene expression in the SDR after E2 treatment
As with the whole genome, treatment of E2 did not have a specific effect on methylation within the SDR between females and males. As shown in Figure 5, hypermethylation was observed with females in both control and E2-treated fish at 3, 9, 12, and 16 dpf. The overall DMSs between females and males were strikingly similar, with or without treatment, suggesting that E2-induced sex reversal was independent of methylation within the SDR.
Figure 5.
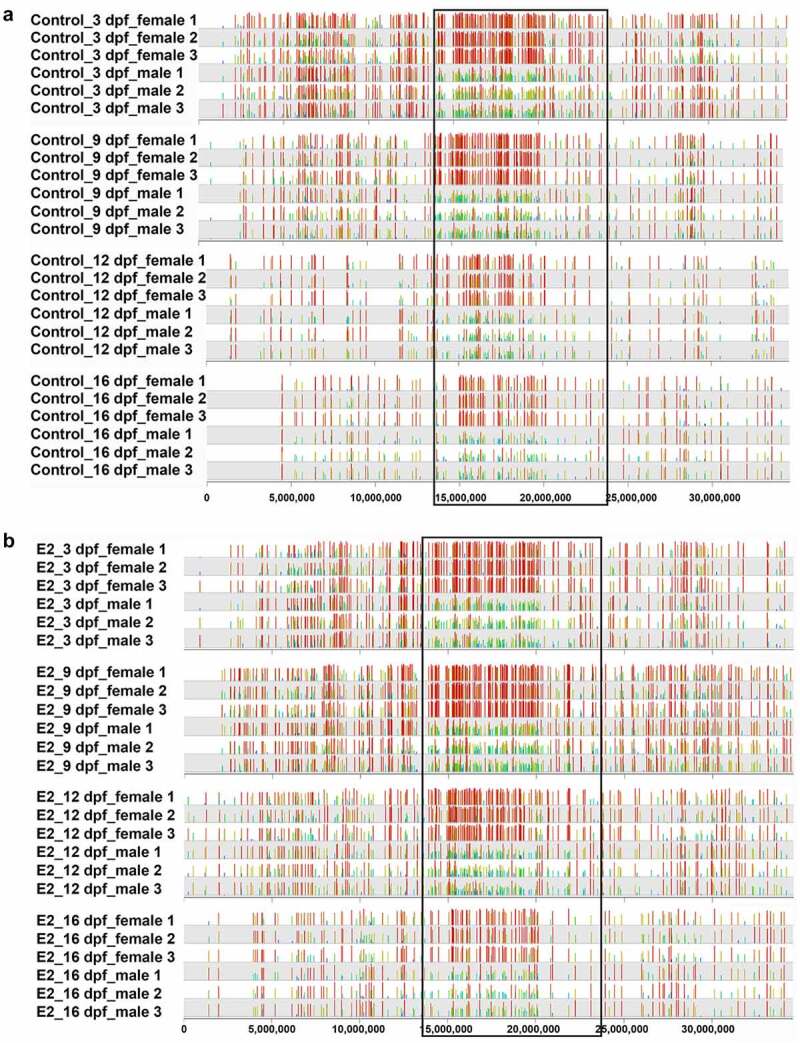
The distribution of differentially methylated CpG sites between females and males of channel catfish (Ictalurus punctatus) on chromosome 4 in control (a) and 17β-oestradiol (E2) treatment (b). The sex determination region (SDR) is located within the black box.
The DMGs and DEGs within the SDR after the E2 treatment are summarized in Table 2. A total of 13 genes within the SDR were found to be differentially methylated at least at one time point at 3 or 16 dpf. The patterns of methylation, however, were more dynamic than trending. For example, hydin gene was hypermethylated in females at 3 dpf but not in 16 dpf; while in males it was hypomethylated at 3 dpf but hypermethylated at 16 dpf. There was no clear relationship between methylation and expression of these 13 genes, only six genes, hydin, spred3, rasgrf1, reln, essrrg, and sphkap, of the 13 genes were differentially expressed in at least one sex or at one time point (Table 2). An additional 21 genes were differentially expressed but were not differentially methylated, again suggesting that methylation and expression regulation within the SDR region was independent after E2 treatment.
Table 2.
Differentially methylated genes (DMGs) and differentially expressed genes (DEGs) within the sex determination region (SDR) after 17β-oestradiol (E2) treatment. Hyper- and hypo-indicated hyper- and hypo-methylated genes, respectively, and up and down indicated up- and down-regulated genes, respectively, as compared to the control. The numbers in the parenthesis indicated percentage of difference in methylation or fold change in expression. ‘-’ indicates no significant difference after E2 treatment.
| Gene | Female |
Male |
||||||
|---|---|---|---|---|---|---|---|---|
| 3 dpf |
16 dpf |
3 dpf |
16 dpf |
|||||
| Methylation | Expression | Methylation | Expression | Methylated | Expression | Methylation | Expression | |
| hydin | Hyper (31) | - | - | Up (15.2) | Hypo (−25) | - | Hyper (26) | Up (3.7) |
| aplp2 | Hypo (−32) | - | - | - | Hyper (28) | - | - | - |
| rasgrf1 | - | - | - | Up (8.0) | Hyper (31) | - | - | Up (4.9) |
| alpk3a | Hyper (−31) | - | - | Down (2.2) | - | - | - | - |
| nectin1 | Hypo (−27) | - | - | - | - | - | - | - |
| reln | - | - | - | Up (8.9) | - | - | Hyper (26) | Up (7.3) |
| ltbp4 | - | - | - | - | Hypo (−25) | - | - | - |
| golgb1 | - | - | - | - | Hypo (−33) | - | - | - |
| ppfia1 | Hypo (−38) | - | - | - | - | - | - | - |
| zbbx | - | - | Hypo (−27) | - | - | - | - | Up (2.6) |
| mbtps1 | - | - | Hypo (−29) | - | Hyper (32) | - | Hyper (39) | - |
| bcar1 | Hypo (−37) | Hyper (25) | - | - | - | - | - | |
| pard6a | Hyper (29) | Down (2.0) | - | - | Hypo (−37) | - | - | - |
| kcnj1 | - | - | - | Up (2.9) | - | - | - | - |
| ehd2b | - | Down (2.1) | - | - | - | - | - | - |
| idh2 | - | - | - | Down (4.1) | - | - | - | Down (2.8) |
| ttc36 | - | - | - | Up (2.3) | - | - | - | - |
| slc28a1 | - | - | - | - | - | - | - | Down (2.1) |
| treh | - | - | - | - | - | - | - | Down (2.4) |
| spred3 | - | - | - | Up (11.0) | - | - | - | Up (4.0) |
| dner | - | - | - | Up (15.2) | - | - | - | Up (6.0) |
| sphkap | - | - | - | Up (9.4) | - | - | - | Up (5.5) |
| sptbn4 | - | - | - | Up (2.9) | - | - | - | Up (2.7) |
| esrrd | - | - | - | Up (6.6) | - | - | - | Up (4.0) |
| shank2 | - | - | - | Up (7.6) | - | - | - | Up (7.9) |
| serpini1 | - | - | - | Up (2.9) | - | - | - | Up (2.1) |
| wdr49 | - | - | - | - | - | - | - | Up (2.2) |
| necab2 | - | - | - | Up (5.0) | - | - | - | Up (3.0) |
| slc38a8 | - | - | - | Up (7.9) | - | - | - | - |
| nutf2 | - | - | - | Down (2.2) | - | - | - | - |
| tsnaxip1 | - | - | - | Up (4.7) | - | - | - | Up (4.9) |
| carmil2 | - | - | - | Up (6.2) | - | - | - | Up (3.2) |
| gnao1a | - | - | - | Up (3.6) | - | - | - | Up (2.5) |
| nr2f2 | - | - | - | Up (2.7) | - | - | - | Up (3.3) |
Methylation and gene expression in pseudo-females
As described above, E2 treatment caused sex reversal of 87.9% of the genetic males to phenotypic females. To elucidate methylation and gene expression in pseudo-females, patterns of DNA methylation, both within the SDR and the whole genome, and profiles of gene expression were compared between females, males, and pseudo-females. As shown in Figure 6, methylation profile within the SDR of pseudo-females at 110 dpf was more similar to those of males than to those of females. Thus, for most of the DMSs, hypermethylation was observed within females in the control (Figure 6(a)). Similarly, many of the hypermethylated sites within females as compared with males remain hypermethylated when compared with pseudo-females (Figure 6(b)). In contrast, there were few DMSs between control males and pseudo-females at 110 dpf (Figure 6(c)), suggesting a high level of similarity of methylation between pseudo-females and males within SDR.
Figure 6.
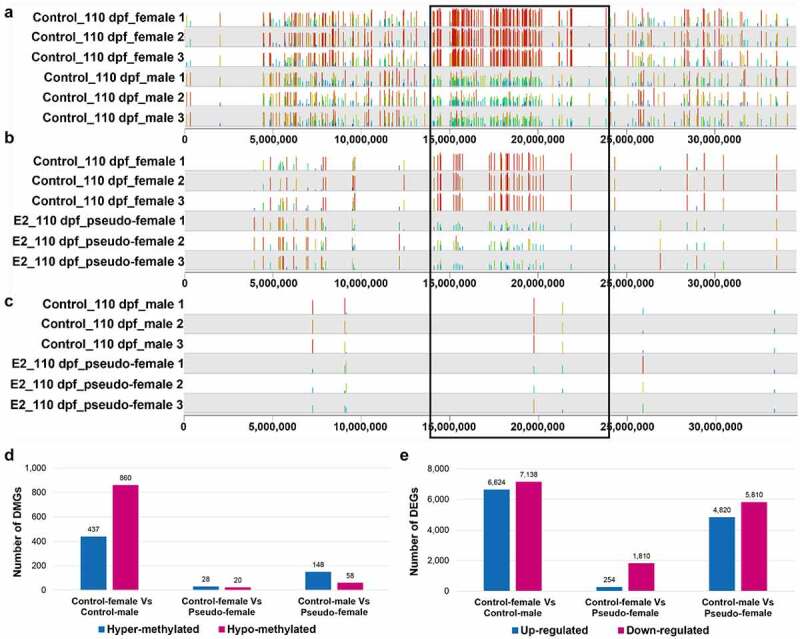
Methylation and gene expression in pseudo-females of channel catfish, Ictalurus punctatus. A-C The distribution of differentially methylated CpG sites (DMSs) on chromosome 4 between control females and control males (a), between control females and pseudo-females (b), and between control males and pseudo-females (c) at 110 days post fertilization (dpf). The black box indicates the sex determination region (SDR). E2: 17β-oestradiol. (d) The number of differentially methylated genes (DMGs) in the genome between different gonads at 110 dpf. (e) The number of differentially expressed genes (DEGs) between different gonads at 110 dpf.
The methylation profile in the whole genome was different from those of the SDR. As shown in Figure 6(d), when the whole-genome DMGs were compared, the largest difference was found to be between control females and control males. Thus, 437 DMGs were hypermethylated in control females, and 860 DMGs were hypomethylated in control females when compared with control males. The difference between control females and pseudo-females was very small, with only 28 DMGs being hypermethylated in control females and 20 DMGs being hypomethylated in control females. When pseudo-females were compared with control males, the difference was larger than the difference between control females and pseudo-females, but much smaller than the difference between control females and males (Figure 6(d)). Taken together, the whole-genome methylation profiles of pseudo-females were most similar to those of control females.
Whole-genome expression of pseudo-females at 110 dpf was also much more similar to that of females. As shown in Figure 6(e), a total of 13,762 and 10,630 genes were differentially expressed between control females and control males and between control males and pseudo-females at 110 dpf, respectively. However, the number of DEGs between control females and pseudo-females was much smaller, only 2,064 (254 higher in control females and 1,810 higher in pseudo-females).
Methylation and expression of some known sex-related genes
Methylation and expression of a set of 12 female sex-related genes and 15 male sex-related genes were examined in E2-treated genetic males. A detailed description of these genes is provided in Supplementary Table 6. Methylation and expression profiles of these sex-related genes between E2-treated genetic males and control males were compared. As shown in Figure 7, in most cases, female sex-related genes were up-regulated following E2 treatment, especially at 16 dpf, while the expression of most male sex-related genes was down-regulated, especially at 110 dpf.
Figure 7.
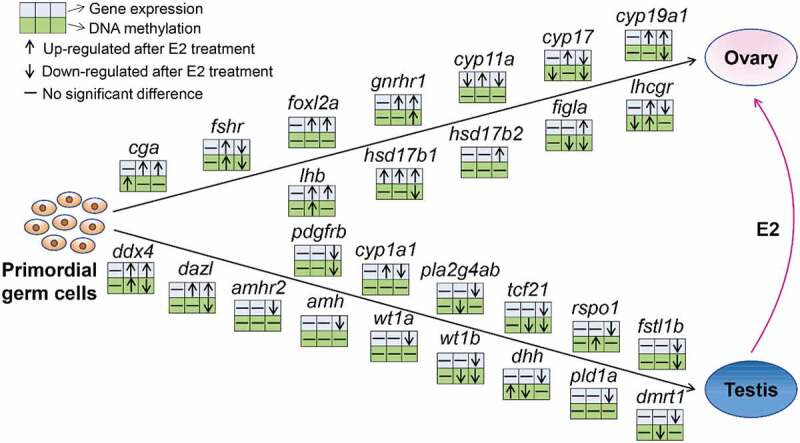
Changes in methylation and expression of a selected set of sex-related genes in the process of sex reversal from genetic males to phenotypic females after E2 exposure in channel catfish, Ictalurus punctatus. For each gene, the three upper blue squares and the three lower green squares represent gene expression and methylation, from the comparison of: between E2-treated males and control males at 3 dpf (first square), between E2-treated males and control males at 16 dpf (second square), and between E2-induced pseudo-females and control males at 110 dpf (third square). At 3 dpf, the samples were from the fertilized eggs; at 16 dpf, the samples were taken from the middle part of the fry with head and tail removed; at 110 dpf, the samples were from gonads. The details of these genes are listed in the Supplementary Table 6.
The effect of E2 treatment on methylation profiles with these genes did not show a specific pattern. Diverse relationships between methylation and gene expression were observed (Figure 7). For example, female-related genes, cyp19a1, figla, and hsd17b1 exhibited an inverse correlation between methylation and gene expression, while no obvious correlation was found in some male-related genes (dmrt1, pld1a, amh, amhr2, cyp1a1, pla2g4ab, and rspo1) and in some female-related genes (foxl2a, cyp11a, and hsd17b2). Additionally, the methylation of some genes was positively correlated with gene expression, including fshr, gnrhr1, cyp17, tcf21, fstl1b, pdgfrb, and wt1b.
Discussion
It is well known that the sex of many fish can be reversed at the undifferentiated stage of gonad by the administration of high doses of exogenous 17β-oestradiol (E2) [32–35]. However, the molecular mechanisms underlying sex reversal are still not fully understood. In this study, whole-genome DNA methylation and gene expression profiles were analysed in females and males of channel catfish during the time of gonadal sex differentiation after E2 exposure. The treatment of E2 caused sex reversal of over 87% of the genetic males into phenotypic females (pseudo-females), but the global methylation profiles were not significantly modified in either females or males. However, a specific set of genes, especially those that were normally highly expressed in females, were differentially expressed after treatment, with or without differential methylation, suggesting that epigenetic regulation may be involved, but were not particularly directional in relation to sex reversal.
DNA methylation is highly dynamic, playing important roles in response to environmental cues [36,37]. Increasing numbers of reports indicated that external stimuli may cause epigenetic changes in various organisms [37–39]. For instance, female-to-male sex reversal occurred under high temperature in Nile tilapia (Oreochromis niloticus), and both females and males showed increased methylation levels [37]. The hatchery-reared and seawater brown trout (Salmo trutta) showed significant differences in genome-wide methylation patterns, and salt-rich diets affected genome-wide methylation in hatchery-reared brown trout [40]. E2 exposure caused global genomic hypermethylation in male gonads of stickleback [15].
In the current study, whole-genome methylation level was not significantly different after E2 treatment in either females or males. However, a large number of DMSs and DMGs were identified in both females and males. DMGs after E2 exposure were significantly enriched in pathways involved in gonadal sex differentiation, such as calcium signalling pathway, MAPK signalling pathway, adrenergic signalling pathway, Wnt signalling pathway, GnRH signalling pathway, ErbB signalling pathway, and enriched in differentiation such as melanogenesis, gap junctions, adipokine signalling pathway, and ECM–receptor interactions. GnRH signalling pathway is a key regulator of the reproductive system [41]. The binding of GnRH to the receptor triggers a series of signal transduction events and leads to the synthesis and release of the gonadotropins, luteinizing hormone (LH), and follicle stimulating hormone (FSH) [42]. The ErbB receptors signal through Akt, MAPK, and many other pathways to regulate cell proliferation, migration, differentiation, apoptosis, and cell motility [43]. Studies have revealed that erbB1 and erbB4 receptors signalling in glial cells controlled female sexual development and the onset of female puberty in mammals [44,45]. ECM–receptor interaction is involved in a variety of cellular activities such as adhesion, migration, differentiation, proliferation, and apoptosis [46] and can regulate the synthesis of steroid hormone [47]. ECM–receptor interaction was associated with the primordial follicle activation and follicular development in mice [48], which was also implicated in ovarian function in pigs [47], goats [49], cattle [50], and tilapia [51]. The MAPK signalling pathway integrated with the GnRH signalling pathway is necessary for normal fertility [42]. The MAPK signalling pathway mediates the balance between ovary- and testis-promoting genes and is related to the gonadal sex reversal in mice and humans [52–56].
It was interesting to note that most of the enriched pathways with genes hypermethylated in E2-treated females overlapped with those with genes hypomethylated in E2-treated males (Figure 3(b,e)), suggesting sex-specific methylation and regulated expression of the same set of genes were important for female gonadal sex differentiation, leading to sex reversal from genetic males to phenotypic females.
Up-regulated DEGs after E2 exposure were enriched in ligand–receptor interaction, calcium signalling, MARK signalling, and cell adhesion molecules pathways, while down-regulated DEGs were involved in growth rather than differentiation (Figure 4(a-d)). Given that female gonads differentiate well ahead of male gonadal sex differentiation in channel catfish (female differentiation at around 19 dpf, male at over 90 dpf) [57], these results were expected because increased expression of genes leading to female gonadal sex differentiation should have promoted sex differentiation into females, rather than the normal situation in genetic males that continued with prolonged cell growth and proliferation.
Despite regulated methylation and expression of a specific set of genes, E2-induced sex reversal appeared to be independent of methylation and regulated expression in the sex determination region (SDR). The methylation profiles within the SDR remained similar before and after treatment with E2, in both females and males (Figure 5). Although over a dozen genes within the SDR were differentially expressed after the treatment of E2, their expression also appeared to be independent of the methylation within the SDR (Table 2). These results suggested that the E2-treated males, even the pseudo-females at 110 dpf, exhibited a methylation profile within the SDR similar to that of normal males. However, in terms of whole-genome methylation profiles, the pseudo-females exhibited a pattern that was extremely similar to that of normal females at 110 dpf. Similarly, for gene expression, the E2-treated fish were increasingly more like normal females such that at 110 dpf, the number of DEGs between control females and pseudo-females was the smallest of the three pairs of comparisons (Figure 6).
Oestrogen is likely at the downstream of the sex determination process [58,59]. In channel catfish, sex is determined genetically by inheritance, with XY being males. Through genetic linkage mapping and genome-wide association studies, the sex determination locus of channel catfish was mapped to chromosome 4 within the 8.9 Mb SDR [60,61]. This region was shown to have sex-specific DNA methylation, and the sex-specific methylation in turn regulates expression of a set of genes within the SDR [62]. In this study, DMGs and DEGs within the SDR after E2 treatment exhibited dynamic changes at different time points (Table 2) without specific correlations with sex. Additionally, the DMSs within the SDR between females and males were strikingly similar in E2 treatment and control (Figure 5), suggesting that E2-induced sex reversal was independent of the SDR and was the downstream of the sex determination pathway.
Sex development is regulated by a network of genes that generate gene cascade leading to establishment of sex phenotypes [63,64]. The functions of these sex-related genes are highly conserved across species. Our results revealed that the majority of genes documented to be associated with sex differentiation and ovarian steroidogenesis in other vertebrates had similar overall expression patterns in channel catfish. Most of the genes known to be highly expressed during female sex differentiation in other species were up-regulated in E2-treated males, whereas most of the genes known to be highly expressed during male sex differentiation in other species were down-regulated in E2-treated channel catfish males (Figure 7).
The E2 treatment-induced expression of a specific set of genes important for female sex differentiation, including cyp19a1, cyp17, cyp11a, gnrhr1, foxl2a, cga, lhb, hsd17b1, hsd17b2, and figla. These findings are consistent with those reported in the Asian seabass (Lates calcarifer) implanted with E2 [39]. Cyp19a1 is a key gene in the conversion of endogenous androgens to oestrogen and ovarian differentiation in various teleosts. It is regulated by sex-biased methylation and involved in DNA methylation mediated sex reversal following heat induction in the European sea bass [65]. In this study, DNA methylation in promoter of cyp19a1 was reduced, and gene expression was increased in pseudo-females compared with control males. Foxl2 is a conserved gene with function of ovarian differentiation in vertebrates [66,67]. Studies have demonstrated that foxl2 was a direct transcriptional activator of the cyp19a1 gene, which was observed in goat [68] and chicken [69]. The expression pattern of foxl2 was also found to be synchronized with cyp19a1 in teleosts, such as air-breathing catfish (Clarias gariepinus) [70,71], zebrafish [72], Nile tilapia [73], medaka (Oryzias latipes) [74], rainbow trout [75], spotted scat (Scatophagus argus) [76], and black porgy (Acanthopagrus schlegeli) [77]. In the present study, foxl2a was up-regulated after E2 exposure since 16 dpf, without significant changes in DNA methylation, which was similar to the study in Asian seabass [39]. The results implied that exogenous oestrogen-induced upregulation of foxl2 in genetic males of channel catfish, which resulted in increased expression of cyp19a1, leading to the conversion of endogenous androgens to oestrogens. The accumulation of endogenous oestrogen and the reduction of androgens were considered to be the determinant of sex differentiation in teleost fish [11,78], explaining the conversion of genetic males to phenotypic females after E2 exposure. Furthermore, figla is another female-specific gene which plays important roles in folliculogenesis, fertilization, and early development [79,80]. Disruption of figla gene led to an all-male phenotype in zebrafish [80]. In this study, the promoter methylation of figla was decreased, and its expression was significantly increased after E2 treatment.
Phenotypic sex is governed by an antagonistic interaction between feminizing and masculinizing gene networks [81]. E2 treatment also suppressed a specific set of genes important for male sex differentiation, leading to sex reversal from genetic males to phenotypic females. These genes included amhr2, amh, wt1a, wt1b, dhh, pld1a, dmrt1, pdgfrb, cyp1a1, pla2g4ab, tcf21, rspo1, and fstl1b (Figure 7). Dmrt1 is essential to testis determination in birds, reptiles, amphibians, and teleost fish [81–85]. Exogenous oestrogen exposure has been observed to reduce dmrt1 expression in red‐eared slider turtle (Trachemys scripta), pejerrey (Odontesthes bonariensis) and rainbow trout [86–88]. In addition, amh is also involved in testicular development and function [89]. Exposure to 17α-ethinylestradiol suppressed the expression of amh in zebrafish [90]. Taken together, increased expression of female sex-related genes and reduced expression of male sex-related genes worked together for sex reversal of genetic males to phenotypic females.
DNA methylation was generally thought to be negatively correlated with gene expression, while recent studies revealed a more dynamic and complex association between DNA methylation and expression than previously known [91–93]. In this study, we observed negative relationships between methylation and gene expression in female-related genes (cyp19a1, figla, and hsd17b1), but most male-related genes did not show obvious correlation between methylation and gene expression (Figure 7), suggesting that the regulation of genes involved in sex reversal may be more complex, with some being epigenetically regulated, while many others may be regulated by other factors including being a secondary response after the primary response to the oestrogen treatment.
Supplementary Material
Acknowledgments
This project was supported by the Agriculture and Food Research Initiative (AFRI) Competitive Grants 2019-67015-29409 from the USDA National Institute of Food and Agriculture (NIFA) Animal Reproduction Program.
Funding Statement
This work was supported by the National Institute of Food and Agriculture [2019-67015-29409].
Data availability statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. The short sequence reads were deposited to NCBI SRA https://www.ncbi.nlm.nih.gov/sra/docs/ with the BioProject ID PRJNA826119.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethical approval
Experimental protocols of animal care and tissue collection were approved by the Auburn University Institutional Animal Care and Use Committee (AU-IACUC).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15592294.2022.2086725
References
- [1].Kobayashi Y, Nagahama Y, Nakamura M.. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 2013;7(1–3):115–125. [DOI] [PubMed] [Google Scholar]
- [2].Devlin RH, Nagahama Y. Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences. Aquaculture. 2002;208(3–4):191–364. [Google Scholar]
- [3].Kossack ME, Draper BW. Genetic regulation of sex determination and maintenance in zebrafish (Danio rerio). Curr Top Dev Biol. 2019;134:119–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Todd EV, Ortega-Recalde O, Liu H, et al. Stress, novel sex genes, and epigenetic reprogramming orchestrate socially controlled sex change. Sci Adv. 2019;5(7):eaaw7006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].MacLaughlin DT, Donahoe PK. Sex determination and differentiation. N Engl J Med. 2004;350(4):367–378. [DOI] [PubMed] [Google Scholar]
- [6].Li M, Sun L, Wang D. Roles of estrogens in fish sexual plasticity and sex differentiation. Gen Comp Endocrinol. 2019;277:9–16. [DOI] [PubMed] [Google Scholar]
- [7].Nakamura M, Bhandari RK, Higa M. The role estrogens play in sex differentiation and sex changes of fish. Fish Physiol Biochem. 2003;28(1):113–117. [Google Scholar]
- [8].Pandian TJ, Sheela SG. Hormonal induction of sex reversal in fish. Recent Results in Cancer Research. Fortschritte der Krebsforschung. Progres Dans Les Recherches Sur le Cancer. 1995;138(1–4):1–22.7899686 [Google Scholar]
- [9].Bhandari RK, Alam MA, Higa M, et al. Evidence that estrogen regulates the sex change of honeycomb grouper (Epinephelus merra), a protogynous hermaphrodite fish. J Exp Zool Part A Comp Exp Biol. 2005;303(6):497–503. [DOI] [PubMed] [Google Scholar]
- [10].Pandian TJ, Kirankumar S. Recent advances in hormonal induction of sex-reversal in fish. J Appl Aquac. 2003;13(3–4):205–230. [Google Scholar]
- [11].Piferrer F. Endocrine sex control strategies for the feminization of teleost fish. Reprod Biotechnol Finfish Aquac. 2001;197(1–4):229–281. [Google Scholar]
- [12].Zheng Y, Yuan J, Meng S, et al. Testicular transcriptome alterations in zebrafish (Danio rerio) exposure to 17β-estradiol. Chemosphere. 2019;218:14–25. [DOI] [PubMed] [Google Scholar]
- [13].Zhao F, Wei P, Wang J, et al. Estrogenic effects associated with bisphenol a exposure in male zebrafish (Danio rerio) is associated with changes of endogenous 17β-estradiol and gene specific DNA methylation levels. Gen Comp Endocrinol. 2017;252:27–35. [DOI] [PubMed] [Google Scholar]
- [14].Kovács T, Szabó-Meleg E, Ábrahám IM. Estradiol-induced epigenetically mediated mechanisms and regulation of gene expression. Int J Mol Sci. 2020;21(9):3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Goudie CA, Redner BD, Simco BA, et al. Feminization of channel catfish by oral administration of steroid sex hormones. Trans Am Fish Soc. 1983;112(5):670–672. [Google Scholar]
- [16].Davis KB, Simco BA, Goudie CA, et al. Hormonal sex manipulation and evidence for female homogamety in channel catfish. Gen Comp Endocrinol. 1990;78(2):218–223. [DOI] [PubMed] [Google Scholar]
- [17].Lou Z, Johnson JV, James MO. Intestinal and hepatic microsomal metabolism of testosterone and progesterone by a 3α-hydroxysteroid dehydrogenase to the 3α-hydroxy derivatives in the channel catfish, Ictalurus punctatus. J Steroid Biochem Mol Biol. 2002;82(4–5):413. [DOI] [PubMed] [Google Scholar]
- [18].Aniagu SO, Williams TD, Allen Y, et al. Global genomic methylation levels in the liver and gonads of the three-spine stickleback (Gasterosteus aculeatus) after exposure to hexabromocyclododecane and 17-β oestradiol. Environ Int. 2008;34(3):310–317. [DOI] [PubMed] [Google Scholar]
- [19].George T, Pandian TJ. Hormonal induction of sex reversal and progeny testing in the zebra cichlid Cichlasoma nigrofasciatum. J Exp Zool. 1996;275(5):374–382. [Google Scholar]
- [20].Ninwichian P, Peatman E, Perera D, et al. Identification of a sex-linked marker for channel catfish. Anim Genet. 2012;439(4):476–477. [DOI] [PubMed] [Google Scholar]
- [21].Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics (Oxford, England). 2011;27(11):1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Liu Z, Liu S, Yao J, et al. The channel catfish genome sequence provides insights into the evolution of scale formation in teleosts. Nat Commun. 2016;7(1):11757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yu G, Wang LG, Han Y, et al. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omi A J Integr Biol. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Akalin A, Kormaksson M, Li S, et al. MethylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wickham H. ggplot2. Wiley Interdiscip Rev Comput Stat. 2011;3(2):180–185. [Google Scholar]
- [28].Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Karsli Z, Aral O, Yeşilayer N. The effects of different proportions of the 17β-estradiol and 17α-methyltestosterone on growth, sex reversal and skin colouration of the electric blue hap (Sciaenochromis ahli Trewavas, 1935). Aquac Res. 2016;47(2):640–648. [Google Scholar]
- [33].Lin S, Benfey TJ, Martin-Robichaud DJ. Hormonal sex reversal in Atlantic cod, Gadus morhua. Aquaculture. 2012;364:192–197. [Google Scholar]
- [34].Imai S, Koyama J, Fujii K. Effects of 17β-estradiol on the reproduction of Java-medaka (Oryzias javanicus), a new test fish species. Mar Pollut Bull. 2005;51(8–12):708–714. [DOI] [PubMed] [Google Scholar]
- [35].Seki M, Fujishima S, Nozaka T, et al. Comparison of response to 17β‐estradiol and 17β‐trenbolone among three small fish species. Environ Toxicol Chem. 2006;25(10):2742–2752. [DOI] [PubMed] [Google Scholar]
- [36].Manolakou P, Lavranos G, Angelopoulou R. Molecular patterns of sex determination in the animal kingdom: a comparative study of the biology of reproduction. Reprod Biol Endocrinol. 2006;4(1):59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sun LX, Wang YY, Zhao Y, et al. Global DNA methylation changes in Nile tilapia gonads during high temperature-induced masculinization. PLoS One. 2016;11(8):e0158483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Strömqvist M, Tooke N, Brunström B. DNA methylation levels in the 5’ flanking region of the vitellogenin I gene in liver and brain of adult zebrafish (Danio rerio)-Sex and tissue differences and effects of 17α-ethinylestradiol exposure. Aquat Toxicol. 2010;98(3):275–281. [DOI] [PubMed] [Google Scholar]
- [39].Banh QQT, Guppy JL, Domingos JA, et al. Induction of precocious females in the protandrous barramundi (Lates calcarifer) through implants containing 17β-estradiol - effects on gonadal morphology, gene expression and DNA methylation of key sex genes. Aquaculture. 2021;539:736601. [Google Scholar]
- [40].Morán P, Marco-Rius F, Megías M, et al. Environmental induced methylation changes associated with seawater adaptation in brown trout. Aquaculture. 2013;392:77–83. [Google Scholar]
- [41].Kraus S, Naor Z, Seger R. Intracellular signaling pathways mediated by the gonadotropin-releasing hormone (GnRH) receptor. Arch Med Res. 2001;32(6):499–509. [DOI] [PubMed] [Google Scholar]
- [42].Bliss SP, Navratil AM, Xie J, et al. GnRH signaling, the gonadotrope and endocrine control of fertility. Front Neuroendocrinol. 2010;31(3):322–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Marmor MD, Skaria KB, Yarden Y. Signal transduction and oncogenesis by ErbB/HER receptors. Int J Radiat Oncol Biol Phys. 2004;58(3):903–913. [DOI] [PubMed] [Google Scholar]
- [44].Prevot V, Rio C, Cho GJ, et al. Normal female sexual development requires neuregulin-erbB receptor signaling in hypothalamic astrocytes. J Neurosci. 2003;23(1):230–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Prevot V, Lomniczi A, Corfas G, et al. erbB-1 and erbB-4 receptors act in concert to facilitate female sexual development and mature reproductive function. Endocrinology. 2005;146(3):1465–1472. [DOI] [PubMed] [Google Scholar]
- [46].Zhang HJ, Tao J, Sheng L, et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang X, Otsu K, Saito H, et al. Sandwich configuration of type I collagen suppresses progesterone production in primary cultured porcine granulosa cells by reducing gene expression of cytochrome P450 cholesterol side-chain cleavage enzyme. Arch Biochem Biophys. 2000;376(1):117–123. [DOI] [PubMed] [Google Scholar]
- [48].Xiong J, Wu M, Zhang Q, et al. Proteomic analysis of mouse ovaries during the prepubertal stages. Exp Cell Res. 2019;377(1–2):36–46. [DOI] [PubMed] [Google Scholar]
- [49].Wang L, Sun X, Guo F, et al. Transcriptome analysis of the uniparous and multiparous goats ovaries. Reprod Domest Anim. 2016;51(6):877–885. [DOI] [PubMed] [Google Scholar]
- [50].Lan D, Xiong X, Huang C, et al. Toward understanding the genetic basis of yak ovary reproduction: a characterization and comparative analyses of estrus ovary transcriptiome in yak and cattle. PLoS One. 2016;11(4):e0152675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang R, Wen L, Ma H, et al. Effects of gonadotropin-releasing hormone analog (GnRHa) immunization on the gonadal transcriptome and proteome of tilapia (Oreochromis niloticus). Comp Biochem Physiol - Part D Genomics Proteomics. 2021;37:100780. [DOI] [PubMed] [Google Scholar]
- [52].Warr N, Carre GA, Siggers P, et al. Gadd45γ and Map3k4 interactions regulate mouse testis determination via p38 MAPK-mediated control of Sry expression. Dev Cell. 2012;23(5):1020–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Gierl MS, Gruhn WH, von Seggern A, et al. GADD45G functions in male sex determination by promoting p38 signaling and Sry expression. Dev Cell. 2012;23(5):1032–1042. [DOI] [PubMed] [Google Scholar]
- [54].Loke J, Pearlman A, Radi O, et al. Mutations in MAP3K1 tilt the balance from SOX9/FGF9 to WNT/β-catenin signaling. Hum Mol Genet. 2014;23(4):1073–1083. [DOI] [PubMed] [Google Scholar]
- [55].Pearlman A, Loke J, Le Caignec C, et al. Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am J Hum Genet. 2010;87(6):898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bogani D, Siggers P, Brixey R, et al. Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol. 2009;7(9):1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Patiño R, Davis KB, Schoore JE, et al. Sex differentiation of channel catfish gonads: normal development and effects of temperature. J Exp Zool. 1996;276(3):209–218. [Google Scholar]
- [58].Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet. 2017;18(11):675–689. [DOI] [PubMed] [Google Scholar]
- [59].Purcell CM, Seetharam AS, Snodgrass O, et al. Insights into teleost sex determination from the Seriola dorsalis genome assembly. BMC Genomics. 2018;19(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Li Y, Liu S, Qin Z, et al. Construction of a high-density, high-resolution genetic map and its integration with BAC-based physical map in channel catfish. DNA Res. 2015;22(1):39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bao L, Tian C, Liu S, et al. The Y chromosome sequence of the channel catfish suggests novel sex determination mechanisms in teleost fish. BMC Biol. 2019;17(1):1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang Y, Zhou T, Liu Y, et al. Identification of an epigenetically marked locus within the sex determination region of channel catfish (Ictalurus punctatus). Intl J Mol Sci. 2022;23(10):5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Munger SC, Capel B. Sex and the circuitry: progress toward a systems-level understanding of vertebrate sex determination. Wiley Interdiscip Rev Syst Biol Med. 2012;4(4):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Angelopoulou R, Lavranos G, Manolakou P. Sex determination strategies in 2012: towards a common regulatory model? Reprod Biol Endocrinol. 2012;10(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Navarro-Martín L, Viñas J, Ribas L, et al. DNA methylation of the gonadal aromatase (cyp19a) promoter is involved in temperature-dependent sex ratio shifts in the European sea bass. PLoS Genet. 2011;7(12):e1002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ottolenghi C, Omari S, Garcia-Ortiz JE, et al. Foxl2 is required for commitment to ovary differentiation. Hum Mol Genet. 2005;14(4):2053–2062. [DOI] [PubMed] [Google Scholar]
- [67].Yao HHC. The pathway to femaleness: current knowledge on embryonic development of the ovary. Mol Cell Endocrinol. 2005;230(1–2):87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Pannetier M, Fabre S, Batista F, et al. FOXL2 activates P450 aromatase gene transcription: towards a better characterization of the early steps of mammalian ovarian development. J Mol Endocrinol. 2006;36(3):399–413. [DOI] [PubMed] [Google Scholar]
- [69].Govoroun MS, Pannetier M, Pailhoux E, et al. Isolation of chicken homolog of the FOXL2 gene and comparison of its expression patterns with those of aromatase during ovarian development. Dev Dyn. 2004;231(4):859–870. [DOI] [PubMed] [Google Scholar]
- [70].Sridevi P, Senthilkumaran B. Cloning and differential expression of FOXL2 during ovarian development and recrudescence of the catfish, Clarias gariepinus. Gen Comp Endocrinol. 2011;174(3):259–268. [DOI] [PubMed] [Google Scholar]
- [71].Sridevi P, Chaitanya RK, Dutta-Gupta A, et al. FTZ-F1 and FOXL2 up-regulate catfish brain aromatase gene transcription by specific binding to the promoter motifs. Biochim Biophys Acta - Gene Regul Mech. 2012;1819(1):57–66. [DOI] [PubMed] [Google Scholar]
- [72].Caulier M, Brion F, Chadili E, et al. Localization of steroidogenic enzymes and Foxl2a in the gonads of mature zebrafish (Danio rerio). Comp Biochem Physiol -Part A Mol Integr Physiol. 2015;188:96–106. [DOI] [PubMed] [Google Scholar]
- [73].Wang DS, Kobayashi T, Zhou LY, et al. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor. Mol Endocrinol. 2007;21(3):712–725. [DOI] [PubMed] [Google Scholar]
- [74].Nakamoto M, Matsuda M, Wang DS, et al. Molecular cloning and analysis of gonadal expression of Foxl2 in the medaka, Oryzias latipes. Biochem Biophys Res Commun. 2006;344(1):353–361. [DOI] [PubMed] [Google Scholar]
- [75].Baron D, Cocquet J, Xia X, et al. An evolutionary and functional analysis of FoxL2 in rainbow trout gonad differentiation. J Mol Endocrinol. 2004;33(3):705–715. [DOI] [PubMed] [Google Scholar]
- [76].Liu H, Mu X, Gui L, et al. Characterization and gonadal expression of FOXL2 relative to Cyp19a genes in spotted scat Scatophagus argus. Gene. 2015;561(1):6–14. [DOI] [PubMed] [Google Scholar]
- [77].Wu GC, Tomy S, Nakamura M, et al. Dual roles of cyp19a1a in gonadal sex differentiation and development in the protandrous black porgy, Acanthopagrus schlegeli. Biol Reprod. 2008;79(6):1111–1120. [DOI] [PubMed] [Google Scholar]
- [78].Piferrer F. Epigenetics of sex determination and gonadogenesis. Dev Dyn. 2013;242(4):360–370. [DOI] [PubMed] [Google Scholar]
- [79].Joshi S, Davies H, Sims LP, et al. Ovarian gene expression in the absence of FIGLA, an oocyte-specific transcription factor. BMC Dev Biol. 2007;7(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Qin M, Zhang Z, Song W, et al. Roles of Figla/figla in juvenile ovary development and follicle formation during zebrafish gonadogenesis. Endocrinology. 2018;159(11):3699–3722. [DOI] [PubMed] [Google Scholar]
- [81].Capel B. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat Rev Genet. 2017;18(11):675–689. [DOI] [PubMed] [Google Scholar]
- [82].Ferguson-Smith M. The evolution of sex chromosomes and sex determination in vertebrates and the key role of DMRT1. Sex Dev. 2006;1(1):2–11. [DOI] [PubMed] [Google Scholar]
- [83].Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat Rev Genet. 2012;13(3):163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Smith CA, Roeszler KN, Ohnesorg T, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461(7261):267–271. [DOI] [PubMed] [Google Scholar]
- [85].Cui Z, Liu Y, Wang W, et al. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis). Sci Rep. 2017;7(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Marchand O, Govoroun M, D’Cotta H, et al. DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochim Biophys Acta - Gene Struct Expr. 2000;1493(1–2):180–187. [DOI] [PubMed] [Google Scholar]
- [87].Murdock C, Wibbels T. Dmrt1 expression in response to estrogen treatment in a reptile with temperature-dependent sex determination. J Exp Zool Part B Mol Dev Evol. 2006;306(2):134–139. [DOI] [PubMed] [Google Scholar]
- [88].Fernandino JI, Hattori RS, Shinoda T, et al. Dimorphic expression of dmrt1 and cyp19a1 (ovarian aromatase) during early gonadal development in pejerrey, Odontesthes bonariensis. Sex Dev. 2009;2(6):316–324. [DOI] [PubMed] [Google Scholar]
- [89].Matuszczak E, Hermanowicz A, Komarowska M, et al. Serum AMH in physiology and pathology of male gonads. Int J Endocrinol. 2013;2013:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Schulz R, Bogerd J, Male R, et al. Estrogen-induced alterations in amh and dmrt1 expression signal for disruption in male sexual development in the zebrafish. Environ Sci Technol. 2007;41(17):6305–6310. [DOI] [PubMed] [Google Scholar]
- [91].Siegfried Z, Simon I. DNA methylation and gene expression. Wiley Interdiscip Rev Syst Biol Med. 2010;2(3):362–371. [DOI] [PubMed] [Google Scholar]
- [92].Wagner JR, Busche S, Ge B, et al. The relationship between DNA methylation, genetic and expression inter-individual variation in untransformed human fibroblasts. Genome Biol. 2014;15(2):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Moarii M, Boeva V, Vert JP, et al. Changes in correlation between promoter methylation and gene expression in cancer. BMC Genomics. 2015;16(1):873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. The short sequence reads were deposited to NCBI SRA https://www.ncbi.nlm.nih.gov/sra/docs/ with the BioProject ID PRJNA826119.


