Figure 2.
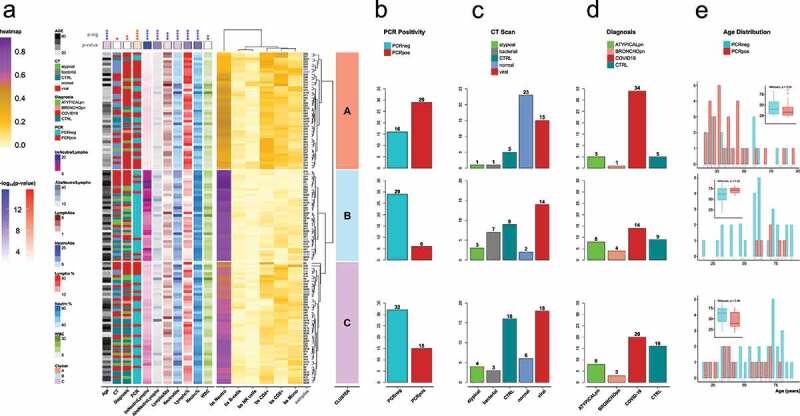
a) Heatmap of the individual samples based on the values of 5meC-calculated cell types: neutrophils (bs Neutro); B cells (bs B-cells); NK cells (bs NK cells); CD4 T cells (bs CD4+); CD8 T cells (bs CD8+); monocytes (bs Mono). Samples are divided in three groups based on the hierarchical clustering: cluster A (salmon); cluster B (light blue); cluster C (orchid). Technical replicates are labelled in blue. The annotations on the left refers to: age (Age); CT scan classification (CT): atypical, bacterial, non-pneumonia control (CTRL), pneumonia normal (normal), viral; diagnosis classification (Diagnosis): atypical pneumonia (ATYPICALpn), bronchopneumonia (BRONCHOpn), COVID-19 pneumonia (COVID-19), non-pneumonia control (CTRL); PCR test positivity (PCR): negative (PCRneg), positive (PCRpos); ratio between 5meC-calculated Neutrophils and Lymphocytes (bsNeutroLympho); ratio between Neutrophil and Lymphocyte counts (AbsNeutroLympho); Lymphocyte counts (LymphoAbs); Neutrophil counts (NeutroAbs); Lymphocyte percentage (Lympho %); Neutrophil percentage (Neutro %); white blood cell counts (WBC). Statistical analysis of the distribution of the annotation features among the three clusters: blue for continuous values (Kruskal-Wallis); red for categorical values (Fisher’s exact test): Diagnosis (controls, COVID-19 pneumonia, non-COVID-19 pneumonia); CT (control, viral, non-viral); PCR (positive, negative); statistical significance (p-sig; ns = p-val >0.05; * = p-val <0.05; ** = p-val <0.01; *** = p-val <0.001; **** = p-val <0.0001). The p-value coloured boxes below the p-sig, and above the annotations represents the -log10(p-value). b,c,d) Bar Plot of counts for each category of PCR test positivity (b), CT scan results (c), and Diagnosis (d) in each cluster. (e) Age distribution grouped by PCR test positivity for each cluster. Positive: groups II, and III vs. negative: groups I, IV, and IV (refer to Supplementary Figure 1).
