Abstract
Background/Aim
Uremic cardiomyopathy (UCM) is a characteristic cardiac pathology that is commonly found in patients with chronic kidney disease. This study dissected the mechanism of SPI1 in myocardial fibrosis and inflammation induced by UCM through S100A8/A9.
Methods
An UCM rat model was established, followed by qRT-PCR and western blot analyses of SPI1 and S100A8/A9 expression in myocardial tissues. After alterations of SPI1 and S100A8/A9 expression in UCM rats, the blood specimens were harvested from the cardiac apex of rats. The levels of creatine phosphokinase-MB (CK-MB), blood creatinine, blood urea nitrogen (BUN), and inflammatory cytokines (interleukin [IL]-6, IL-1β, and tumor necrosis factor-α [TNF-α]) were examined in the collected blood. Collagen fibrosis was assessed by Masson staining. The expression of fibrosis markers [transforming growth factor (TGF)-β1, α-smooth muscle actin (SMA), Collagen 4a1, and Fibronectin], IL-6, IL-1β, and TNF-α was measured in myocardial tissues. Chromatin immunoprecipitation and dual-luciferase reporter gene assays were conducted to test the binding relationship between SPI1 and S100A8/A9.
Results
S100A8/A9 and SPI1 were highly expressed in the myocardial tissues of UCM rats. Mechanistically, SPI1 bound to the promoter of S100A8/A9 to facilitate S100A8/A9 transcription. S100A8/A9 or SPI1 knockdown reduced myocardial fibrosis and inflammation and the levels of CK-MB, blood creatinine, and BUN, as well as the expression of TGF-β1, α-SMA, Collagen 4a1, Fibronectin, IL-6, TNF-α, and IL-1β in UCM rats.
Conclusion
SPI1 knockdown diminished S100A8/A9 transcription, thus suppressing myocardial fibrosis and inflammation caused by UCM.
Keywords: Uremic cardiomyopathy, SPI1, S100A8, S100A9, myocardial fibrosis, inflammation, transcription factor
Introduction
Uremic cardiomyopathy (UCM) is a definition of the cardiac abnormalities present in patients diagnosed with chronic kidney disease (CKD) [1]. As a highly prevalent disorder, CKD afflicts 10–15% of the population worldwide, and its progression is commonly accompanied by the emergence of UCM symptoms [2,3]. Prevalent in patients with end-stage renal disease [4], UCM is featured by obvious myocardial fibrosis, left ventricular hypertrophy, diastolic dysfunction, intramyocardial arteriolar wall thickening, and subsequent abnormalities in systolic function at the advanced stage of overt heart failure [5]. Likewise, inflammation in the myocardium is implicated in the development of UCM [6]. Thereby, the understanding of UCM pathogenesis can be aided by research on molecular mechanisms underpinning myocardial fibrosis and inflammation.
S100 calcium binding protein A8 (S100A8) and S100A9, Ca2+-binding proteins belonging to the S100 family, have been documented to play a decisive role in the development of inflammation-related disorders [7]. As a heterodimer, S100A8/S100A9 is implicated in the process of neutrophil-related inflammation [8]. S100A8/A9 also irreversibly damages tubular epithelial cell contacts to orchestrate kidney fibrosis [9]. S100A8/A9 acts as a pro-inflammatory cytokine in acute hypertension and promotes the production of some cytokines in myocardial fibrosis [10]. Besides, S100A8/A9 plays a negative role in the early period of myocardial infarction [11]. Salmonella pathogenicity island 1 (SPI1, also called PU.1) pivotally orchestrates signaling in the immune system and plays an important part in the growth of lymphocytes and myeloid cells [12]. The inhibition of SPI1 may be a new and effective treatment approach for various fibrotic diseases [13]. The knockdown of SPI1 reduces the expression of pro-inflammatory genes to restrict microglial inflammation [14]. A previous study suggested that SPI1 repression ameliorates angiotensin-II-induced atrial fibrosis [15]. Of note, SPI1 is a transcription factor of the E26 transformation specific family, a large family of transcription factors that contains 28 family members and takes part in various cell processes, which was reported to be associated with the co-expression of S100A9 [16,17].
Based on previous studies, we proposed a hypothesis that SPI1 promoted UCM–induced myocardial fibrosis and inflammation through the regulation of S100A8/A9. This study was conducted to validate this hypothesis with the focus on myocardial fibrosis and inflammation.
Materials and methods
Establishment of the UCM rat model [18–21]
Thirty-six specific pathogen-free male Sprague-Dawley (SD) rats (aged 6 weeks; weighing 200 ± 20 g) were provided by the Experimental Animal Center of the Southern Medical University [Guangzhou, China; license: SCXK (Guangzhou) 2011-0015]. The rats were housed under a constant temperature of 20 ± 2 °C and constant humidity of 50 ± 5% with 12-h artificial lighting and free access to water for one week of acclimation before the experiments. The 36 SD rats were randomly arranged into six groups with six rats per group: sham (normal control), UCM (the UCM rat model), UCM + adeno-associated virus (AAV)-short hairpin RNA negative control (shNC) [UCM rats was infected with NC AAV9 encoding green fluorescent proteins (AAV9-GFP)], UCM + AAV-shS100A8 (UCM rats was infected with S100A8-silencing virus AAV9-GFP-shS100A8), UCM + AAV-shS100A9 (UCM rats was infected with S100A9-silencing virus AAV9-GFP-shS100A9), and UCM + AAV-shSPI1 (UCM rats was infected with SPI1-silencing virus AAV9-GFP-shSPI1) groups. The 5/6-nephrectomy was performed after the rats had been acclimatized for one week. Water and food were deprived 1 day before the operation. The rats were injected intraperitoneally with 1% pentobarbital (30 mg/kg). After the muscles of rats were relaxed, the abdomen was shaved and disinfected, with a sterile sheet paved. Next, the skin was incised along the midline of the abdomen, with an incision of about 2 cm. The subcutaneous tissues of rats were bluntly separated to expose abdominal muscles, and the organs were separated. Thereafter, the right kidney was found and excised after the right renal pedicle was ligated. After one week, the same operation was performed to ligate the left kidney and excise 1/3 of each of the upper and lower ends of the left kidney. The rats in the sham group were only subjected to laparotomy, without renal incision. The abdominal muscles and skin of the rats were sutured layer by layer interruptedly, and then the abdominal wound was disinfected with ethanol. These surgeries were performed under strict aseptic conditions. The rats were intraperitoneally injected with ampicillin sodium (200,000 U/rat) after the operation for 3 days to prevent infection. Eight weeks later, the right carotid artery was cannulated for blood pressure measurement and blood sample collection. Next, the rats were euthanized by overdose anesthesia, and the hearts were attained for subsequent experimentations.
AAV9-GFP carrying either knockdown viruses or NC viruses was purchased from GeneChem (Shanghai, China). The knockdown sequences were as follows: shNC (5′-GGTACGCAATAGGAGTGTGTG-3′), shS100A8 (5′-GCAACGTCATTGAAGTCTACC-3′), shS100A9 (5′-GCAGCATAAGCACCATCATCA-3′), and shSPI1 (5′-GCTTCCCTTATCCACCCTTGT-3′). AAV9-GFP (1 × 109 TU/mL in 200 µL saline) was delivered by a single caudal injection after the second operation.
All animal experiments complied with the regulations and codes of practice for laboratory animal management and ethical requirements related to laboratory animals.
Measurement of biochemical indexes
On week 8, the body and hearts of the rats were weighed. Then, the rats were sedated through intraperitoneal injection of pentobarbital (1%). The blood specimens were harvested from the cardiac apex of rats, put into 2.0 mL heparinized Eppendorf (EP) tubes, mixed completely, and centrifuged at 8000 r/min and 4 °C for 6 min. The supernatant was stored at −80 °C. The creatine phosphokinase-MB (CK-MB) kit (H197-1-1, NanJing JianCheng Bioengineering Institute, Nanjing, China) was utilized to detect CK-MB levels. Sarcosine oxidase and urease methods were employed to measure blood creatinine (ab65340, Abcam, Cambridge, UK) and blood urea nitrogen (BUN; ab83362, Abcam), respectively. Inflammatory cytokines, interleukin-6 [(IL-6), ab234570], tumor necrosis factor-alpha [(TNF-α), ab236712], and interleukin-1 beta [(IL-1β), ab255730] were tested by corresponding enzyme-linked immunosorbent assay (ELISA) kits (Abcam). The above relevant biochemical indexes were measured strictly as per the specific operation steps and precautions of the corresponding kits, and the absorbance was calculated on a microplate reader. The sample concentration was calculated according to the formula or the standard curve regression equation corresponding to the biochemical indexes.
Blood pressure measurement
Before the rats were euthanasia, blood pressure was measured with a 24 G polytetrafluoroethylene heparinized catheter (Descarpack, Sao Paulo, Brazil) connected to a pressure transducer (PowerLab® 4/35; ADInstruments Co., Ltd., New South Wales, Australia) through a carotid cannula. Five min later, pulse waves were recorded to stabilize blood pressure. The mean systolic blood pressure (SBP) was calculated.
Cardiac function test in rats
One day before euthanasia, rats underwent echocardiographic examination on a Philips sonos7500 ultrasound machine (Philips Technologies, Amsterdam, NY). The 2% pentobarbital was injected intraperitoneally into rats (0.2 mL/100 g) for 10–15 min to successfully anesthetize the rats. Afterwards, a small ultrasound probe was placed on the left anterior thoracic wall of the rats. The ultrasound testing was conducted from long-axis views, apical four-chamber views, and two-chamber views of the left ventricle at 10 MHz. Left ventricular (LV) end-systolic volume (ESV), LV internal diameter in systole (LVIDs), interventricular septum (IVS), end-diastolic volume (EDV), LV internal diameter in diastole (LVIDd), and LV posterior wall (LVPW) were measured. All measurements were averaged over three consecutive cardiac cycles. The LV ejection fraction (LVEF) and LV fractional shortening (LVFS) were calculated with the following formula: LVEF = (EDV − ESV)/EDV ×100%. All echocardiographic examinations were performed by specialists with extensive experience in echocardiography through a blind method.
Histological staining
Rat myocardial tissues were stained with hematoxylin and eosin (HE) staining kits (C0105M, Beyotime, Shanghai, China) and Masson staining kits (G1340, Solarbio, Beijing, China). The tissues were fixed in neutral formalin solution (10%), paraffin-embedded, and sectioned into 3 µm on an ultramicrotome. The sections were then dewaxed in xylene and hydrated in gradient ethanol. Next, the sections were washed with tap water, followed by HE staining and Masson staining as per the instructions. The sections were washed in tap water, dehydrated in the order of 70%, 80%, 90%, and anhydrous ethanol (5 min for each) at room temperature, and cleared with xylene twice for 5 min each. Then, the sections were sealed with neutral gum or other sealer and examined under a microscope (Zeiss, Oberkochen, Germany) to observe the arrangement of cardiomyocyte nuclei, cardiomyocyte area, and collagen fiber area. The cardiac myocyte cross-sectional area analysis was performed on HE staining images with Image J 6.0 image analysis software. The collagen fiber area was analyzed in Masson staining images.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay
A TUNEL kit (C1089, Beyotime) was employed to detect apoptosis in rat myocardial tissues in the light of the manuals. Heart paraffin sections were dewaxed with xylene for 5–10 min and with fresh xylene for another 5–10 min successively, reacted with gradient ethanol (100% for 5 min, 90% for 2 min, and 70% for 2 min), and immersed in distilled water for 2 min. Afterwards, the sections were reacted with 20 μg/mL proteinase K without DNase (ST532, Beyotime) at 20–37 °C for 15–30 min. Next, the sections were washed with phosphate-buffered saline (PBS) three times, followed by 20 min of incubation at room temperature in 3% hydrogen peroxide solution prepared with PBS. Subsequent to three washes with PBS, the sections were incubated for 60 min at 37 °C in the dark with 50 μL TUNEL solution and then washed three times with PBS. After 5 min of 4′,6-diamidino-2-phenylindole (C1006, Beyotime) counterstaining and PBS washing, the sections were blocked with anti-fluorescence quenching sealing liquid. Next, five random fields of view were selected under the FV-1000/ES confocal microscope (Olympus, Tokyo, Japan) for observation and photography.
Isolation and culture of rat primary cardiomyocytes
The SD rats aged 1–2 days were purchased from Xiangya Laboratory Animal Center (Hunan, China). The heart of rats was dissected, placed in cold PBS, and then digested with a combination of collagenase type II (Sigma-Aldrich, St. Louis, MO) and trypsin for 5 min. The supernatant was obtained and placed into centrifuge tubes (15 mL) containing 8 mL complete medium. The above process was repeated until the tissues were completely dissolved. Then, the suspensions were combined. The supernatant was removed after 8 min of centrifugation at 1000 rpm. Next, the cells were resuspended and subjected to differential adhesion at 37 °C for 1.5 h to remove fibroblasts. Thereafter, cells were cultured in Dulbecco’s Modified Eagle Medium (GIBCO BRL, Grand Island, NY) encompassing 10% fetal bovine serum (GIBCO BRL) and 1% penicillin-streptomycin (GIBCO BRL), with medium renewed every 3 days. Primary cultures of cardiomyocytes were positively identified by cardiac troponin I (cTnI; ab47003, Abcam) staining, and isolation was considered successful if the positive rate exceeded 90%. The cells in the first passage were used for further studies. Thereafter, the cells were cultured in a 37 °C incubator with 5% CO2 and transfected with Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA). After 48 h of transfection, the subsequent experiments were performed. Small interfering RNA (si)-NC (5′-GATGAAGAGCACCAACTC-3′) and si-SPI1-1 (5′-UCGGAUGAGCUGGUUACUUAU-3′) were synthesized by Sangon Biotech (Shanghai, China).
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total tissue or cellular RNA was extracted with TRIZOL (Invitrogen) and reversely transcribed as instructed in the protocols of a reverse transcription kit (TaKaRa, Tokyo, Japan). Gene expression was detected on a LightCycler 480 (Roche, Indianapolis, IN) fluorescence quantitative PCR instrument. The reaction conditions were set based on the manuals of the fluorescence quantitative PCR kit (SYBR Green Mix, Roche Diagnostics). The thermal cycling parameters were as follows: 95 °C for 10 s, 45 cycles of 95 °C for 5 s, 60 °C for 10 s, and 72 °C for 10 s, and extension at 72 °C for 5 min. Three replicates were set for each PCR. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was adopted as an internal reference. The 2−ΔΔCt method was applied for data analysis: ΔΔCT = ΔCt experimental group – ΔCt control group, where ΔCt = Ct target gene – Ct internal reference gene. Primers are detailed in Table 1.
Table 1.
Primer sequence.
| Name of primer | Sequences (5’–3’) |
|---|---|
| SPI1-F | GAGCAGATGCACGTCCTTGA |
| SPI1-R | GGATAAGGGAAGCACACCCG |
| S100A8-F | GTGCCCTCAGTTTGTGCAG |
| S100A8-R | TCTTTATGAGCTGCCACGCC |
| S100A9-F | AGATGGCTGCCAAAACAGGA |
| S100A9-R | TCAGGGTGTCAGGATGTCCA |
| TNF-α-F | AACACACGAGACGCTGAAGT |
| TNF-α-R | TCCAGTGAGTTCCGAAAGCC |
| IL-6-F | CATTCTGTCTCGAGCCCACC |
| IL-6-R | GCAACTGGCTGGAAGTCTCT |
| GAPDH-F | TGCACCACCAACTGCTTAG |
| GAPDH-R | GATGCAGGGATGATGTTC |
| IL-1β-F | CTCGTGGGATGATGACGACC |
| IL-1β-R | AGCTTTCAGCTCACATGGGT |
| α-SMA-F | CTCCATTGTCCACCGCAAATG |
| α-SMA-R | GGAAAAGAACTGAAGGCGCTG |
| collagen 4a1-F | ATTCCTTTGTGATGCACACCAG |
| collagen 4a1-R | AAGCTGTAAGCATTCGCGTAGTA |
| fibronectin-F | TGGCCACTTCCGAATCTGTC |
| fibronectin-R | CCTCGCTCAGTTCGTACTCC |
| TGF-β1-F | AGGGCTACCATGCCAACTTC |
| TGF-β1-R | CCACGTAGTAGACGATGGGC |
Notes. F: forward; R: reverse; SPI: salmonella pathogenicity island; IL: interleukin; TNF: tumor necrosis factor; SMA: smooth muscle actin; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; TGF: transforming growth factor.
Western blot
Protein samples were obtained by lysing cells or myocardial tissues with Radio-Immunoprecipitation Assay lysis (Beyotime). After measurement of the protein concentration with the bicinchoninic acid kit (Beyotime), the corresponding volume of protein was mixed with the loading buffer (Beyotime) and heated in a boiling-water bath for 3 min for denaturation. Then, electrophoresis was carried out at 80 V for 30 min and then at 120 V for 1–2 h after the bromophenol blue entered the separation gel. The protein was transferred to a membrane in an ice bath with a current of 300 mA for 60 min. The membrane was then rinsed in washing solution for 1–2 min and sealed in sealing solution at room temperature for 60 min or 4 °C overnight. The primary antibodies against S100A8 (PA5-79948, 1:500, Invitrogen), S100A9 (73425, 1:1000, Cell Signaling Technologies, Beverly, MA), SPI1 (PA5-115807, 1:500, Invitrogen), transforming growth factor (TGF)-β1 (ab215715, 1:1000, Abcam), α-smooth muscle actin (α-SMA, ab7817, 1:1000, Abcam), Collagen 4a1 (ab6586, 1:1000, Abcam), and Fibronectin (ab268020, 1:1000, Abcam), and GAPDH (ab8245, 1:5000, Abcam) were incubated with the membrane on a shaker for 1 h at room temperature and the membrane was washed three times with washing solution for 10 min each. The membrane was transferred to secondary goat anti-rabbit (ab6702) or goat anti-mouse (ab6708) immunoglobulin G (IgG) antibodies (1:5000, Abcam) for 1-h incubation at room temperature and then washed three times for 10 min each. Thereafter, an equal quantity of liquid A and liquid B from the electrogenerated chemiluminescence fluorescence detection kit (BB-3501, Amersham, Buckinghamshire, UK) was mixed evenly in a dark chamber and then dropwise added to the membrane. Finally, the membrane was exposed and imaged in a Gel Imager, followed by detection on the chemiluminescence imaging system (Bio-Rad, Hercules, CA).
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed with an EZ-Magna ChIP TMA kit (17-10086, Millipore, Billerica, MA). Rat primary cardiomyocytes at the logarithmic growth phase were cross-linked with 1% formaldehyde for 10 min, which was terminated with 125 mM glycine for 5 min at room temperature. The cells were then washed twice with pre-cooled PBS and harvested after 5 min of centrifugation at 2000 g. Next, the cells were resuspended in cell lysis buffer to a final density of 2 × 106 cells per 200 μL. The cells were added with the protease inhibitor mixture, centrifuged at 5000 g for 5 min, and then resuspended in nuclear separation buffer, followed by an ice-water bath for 10 min. The cells were sonicated to obtain 200–1000 bp chromatin fragments. The fragments were centrifuged at 14,000 g and 4 °C for 10 min. Then, the supernatant was acquired. Next, 100 μL supernatant (DNA fragments) in each group was added with 900 μL ChIP Dilution Buffer and 20 μL 50 × protease inhibitor cocktail. Next, 60 μL Protein A Agarose/Salmon Sperm DNA was added to each group. Later, the tubes were inverted at 4 °C for 1 h, left to stand for 10 min at 4 °C, and then centrifuged at 700 g for 1 min. Thereafter, 20 μL supernatant was used as Input. In the experiment groups, 1 μL rabbit anti-SPI1 antibodies were added to the supernatant, and 1 μL rabbit anti-IgG (ab172730, Abcam) was added to the NC group. Next, 60 μL Protein A was added to each tube that was then inverted at 4 °C for 2 h. After standing for 10 min, the tubes underwent 1 min of centrifugation at 700 g, followed by the removal of the supernatant. The precipitation was washed with l mL low salt wash buffer, high salt wash buffer, LiCl wash buffer, and Tris + ethylenediaminetetraacetic acid (2 times), respectively. The samples in each tube were eluted twice with 250 μL ChIP Wash Buffer. Afterwards, 20 μL of 5 M NaCl was utilized for de-cross-linking to recover DNA. Fluorescence quantitative PCR was implemented to detect the enriched chromatin fragments. In addition, S100A8 promoter primers were utilized, including forward: 5′-GCCTCCTTTGAGAGGGCAAG-3′ and reverse: 5′-GCTTGGAGCTTGTTTGGGAG-3′.
Dual-luciferase reporter gene assay
We constructed dual-luciferase reporter plasmids containing wild-type S100A8 promoter (S100A8-WT), mutant S100A8 promoter (S100A8-Mut, sites 663–669 were changed from AGGAAGT to CTCTCTC and sites 411-416 were changed from GGGAAG to CTCTCT, Supplementary Table 1), S100A9-WT, and S100A9-Mut (sites 769–774 were changed from GGGAAG to CTCTCT, Supplementary Table 1). Thereafter, the reporter plasmids were co-transfected into primary cardiomyocytes with si-NC and si-SPI1. The cells were lysed subsequent to 48 h of transfection and 1 min of centrifugation at 13,000 g. Next, the supernatant was attained for the determination of the luciferase activity with the dual-luciferase reporter gene assay kit (16185, Thermo Scientific, Fremont, CA). With Renilla luciferase activity as an internal reference, the relative activity of luciferase was calculated as the ratio of Firefly luciferase activity to Renilla luciferase activity.
Statistical analysis
GraphPad 9.0 was employed for statistical analysis, and all data were summarized as mean ± standard deviation. The T-test was applied for comparisons between two groups, and the one-way analysis of variance test was utilized for comparisons among multiple groups, with Tukey’s test for post hoc multiple comparisons. p < 0.05 indicated that the difference was statistically significant.
Results
S100A8/A9 was highly expressed in myocardial tissues of UCM rats
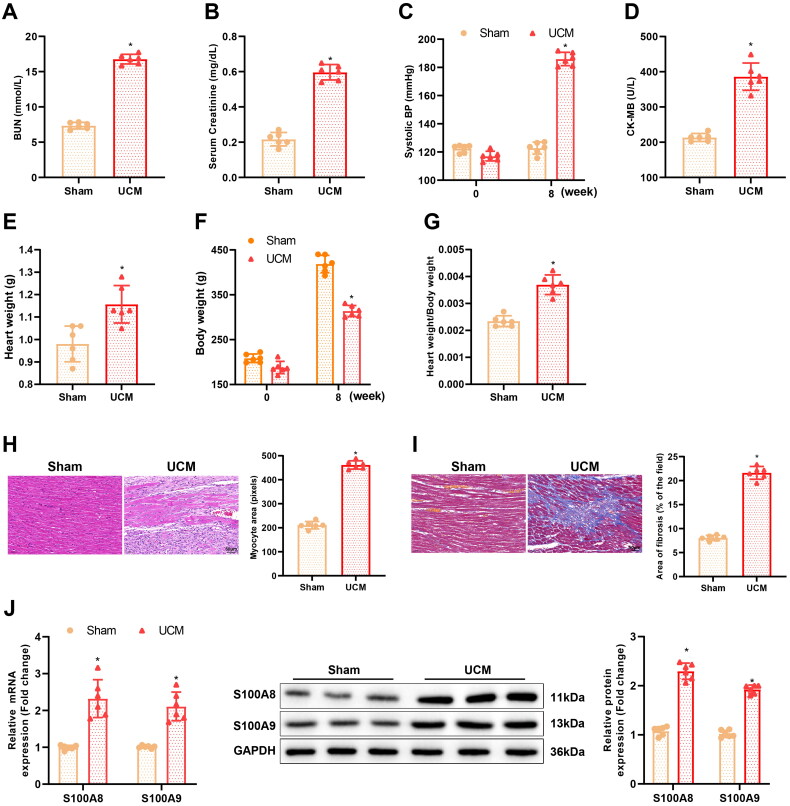
To investigate whether S100A8/A9 was involved in the onset and development of UCM, the UCM rat model was established to detect S100A8 and S100A9 expression. As discovered in Figure 1(A–G), compared to the sham group, there was a significant reduction in body weight but an elevation in BUN levels, blood creatinine levels, SBP, CK-MB levels, heart weight, and cardiac index (heart weight/body weight ratio) in rats of the UCM group. Ultrasound results further displayed that compared to the sham group, rats in the UCM group had an elevation in LV interventricular septum in diastole (LVIVSd) and in systole (LVIVSs), LV posterior wall thickness at diastole (LVPWd) and at systole (LVPWs), and LVEF, as well as a decrease in LVEDV and LVESV (Table 2). In addition, HE staining and Masson staining manifested that cardiomyocytes in the UCM group were hypertrophic and had an increased cell area, with enlarged nuclei, irregular shape, disorganized arrangement, deepened cytoplasmic staining, and memorably enhanced myocardial fibrosis (Figure 1(H,I)), indicating the successful establishment of the UCM rat model.
Figure 1.
S100A8 and S100A9 are upregulated in the myocardial tissues of UCM rats. (A) The levels of BUN in rats of each group; (B) Levels of blood creatinine in rats of each group; (C) SBP in rats of each group; (D) The content of CK-MB in rats of each group; (E–G) The heart weight (E), body weight (F), and the ratio of heart weight to body weight (G) in rats of each group; (H) Representative images of HE staining and quantitation of cardiomyocyte area in rats of each group (scale bar: 50 µm); (I) Masson staining observation of myocardial fibrosis in each group (scale bar: 50 µm); (J) qRT-PCR and western blot detection of mRNA and protein expression of S100A8 and S100A9 in myocardial tissues of rats in each group. *p < 0.05 compared with the sham group. N = 6 rats/group. UCM: uremic cardiomyopathy; BUN: blood urea nitrogen; SBP: systolic blood pressure; CK-MB: creatine phosphokinase-MB; qRT-PCR: quantitative real-time polymerase chain reaction.
Table 2.
Changes in cardiac function (x ± s, N = 6).
| Groups | Sham | UCM | UCM+ | UCM+ | UCM+ | UCM+ |
|---|---|---|---|---|---|---|
| AAV-shNC | AAV-shS100A8 | AAV-shS100A9 | AAV-shSPI1 | |||
| HR (beats/min) | 312 ± 22 | 310 ± 28 | 304 ± 30 | 305 ± 26 | 310 ± 18 | 305 ± 16 |
| IVSs (cm) | 0.14 ± 0.02 | 0.35 ± 0.04* | 0.37 ± 0.10 | 0.20 ± 0.04$ | 0.18 ± 0.03$ | 0.19 ± 0.02$ |
| IVSd (cm) | 0.18 ± 0.03 | 0.27 ± 0.05* | 0.31 ± 0.04 | 0.19 ± 0.05$ | 0.20 ± 0.05$ | 0.19 ± 0. 02$ |
| EDV (µL) | 194 ± 10 | 149 ± 9* | 157 ± 7 | 190 ± 12$ | 183 ± 11$ | 176 ± 12$ |
| ESV (µL) | 69 ± 4 | 34 ± 3* | 38 ± 4 | 58 ± 6$ | 62 ± 7$ | 61 ± 5$ |
| LVPWs (cm) | 0.25 ± 0.06 | 0.37 ± 0.05* | 0.4 ± 0.05 | 0.28 ± 0.05$ | 0.31 ± 0.06$ | 0.29 ± 0.05$ |
| LVPWd (cm) | 0.18 ± 0.02 | 0.35 ± 0.02* | 0.32 ± 0.03 | 0.21 ± 0.02$ | 0.22 ± 0.03$ | 0.20 ± 0.03$ |
| EF (%) | 63 ± 1 | 75 ± 2* | 77 ± 2 | 69 ± 1$ | 66 ± 2$ | 64 ± 2$ |
Notes. UCM: uremic cardiomyopathy; AAV: adeno-associated virus; Sh: short hairpin; NC: negative control; SPI: salmonella pathogenicity island; HR: heart rate; IVSs: interventricular septum in systole; IVSd: interventricular septum in diastole; EDV: end-diastolic volume; ESV: end-systolic volume; LVPWs: left ventricular posterior wall thickness at systole; LVPWd: left ventricular posterior wall thickness at diastole; EF: ejection fraction.
Next, qRT-PCR and western blot demonstrated that S100A8 and S100A9 were expressed highly in the myocardial tissues of UCM rats in contrast to sham-operated rats (Figure 1(J)).
Knockdown of S100A8/A9 repressed myocardial fibrosis and inflammation caused by UCM in rats
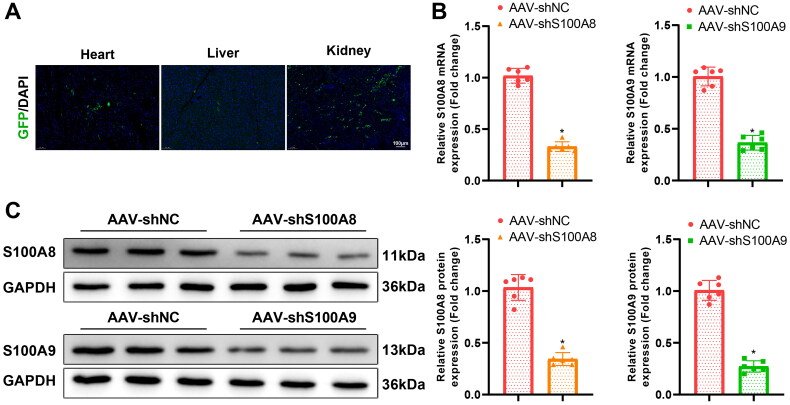
To investigate the role of S100A8 and S100A9 in UCM, the recombination AAV-GFP was constructed to knock down S100A8/A9 in rat myocardial tissues. As manifested in Figure 2(A), GFP expression was clearly detected in the heart, kidney, and liver of rats 2 weeks after transduction with AAV-GFP. Moreover, AAV-shS100A8 or AAV-shS100A9 remarkably reduced S100A8 or S100A9 expression in rat myocardial tissues (Figure 2(B,C)).
Figure 2.
AAV9-GFP expression in rats. (A) Distribution of GFP fluorescence in heart, kidney, and liver tissues after AAV9-GFP infection in rats (scale bar: 100 µm); (B) qRT-PCR examination of the expression of S100A8 and S100A9 in myocardial tissues after AAV-shS100A8 and AAV-shS100A9 infection in rats; (C) The expression of S100A8 and S100A9 in myocardial tissues measured by western blot after AAV-shS100A8 and AAV-shS100A9 infection in rats. *p < 0.05 compared with UCM + AAV-shNC group. N = 6 rats/group. AAV: adeno-associated virus; GFP: green fluorescent protein; qRT-PCR: quantitative real-time polymerase chain reaction; Sh: short hairpin.
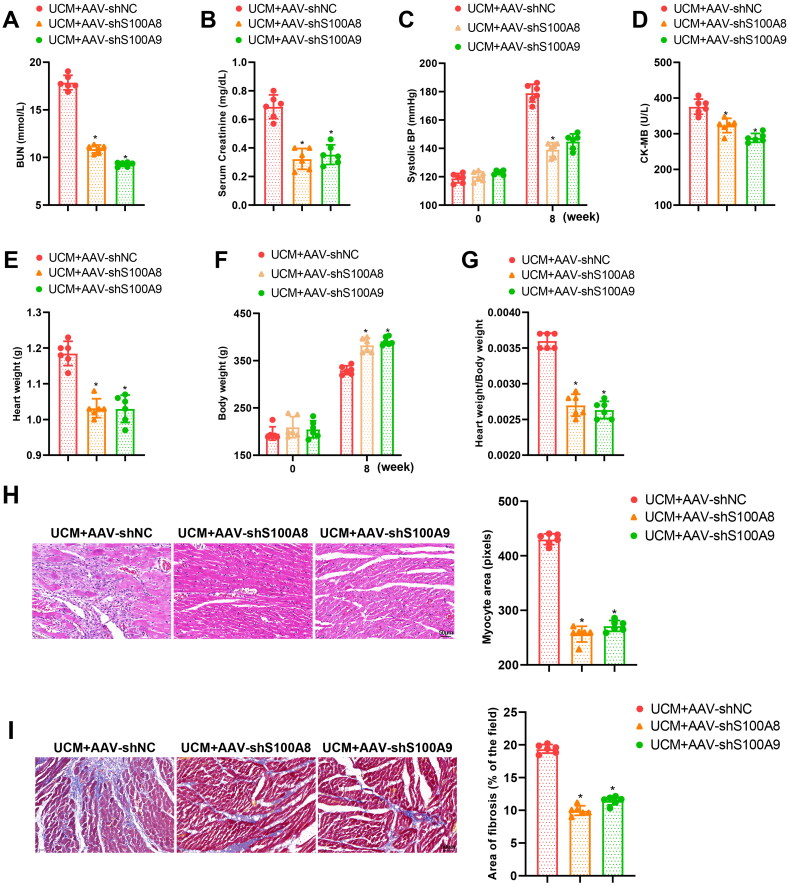
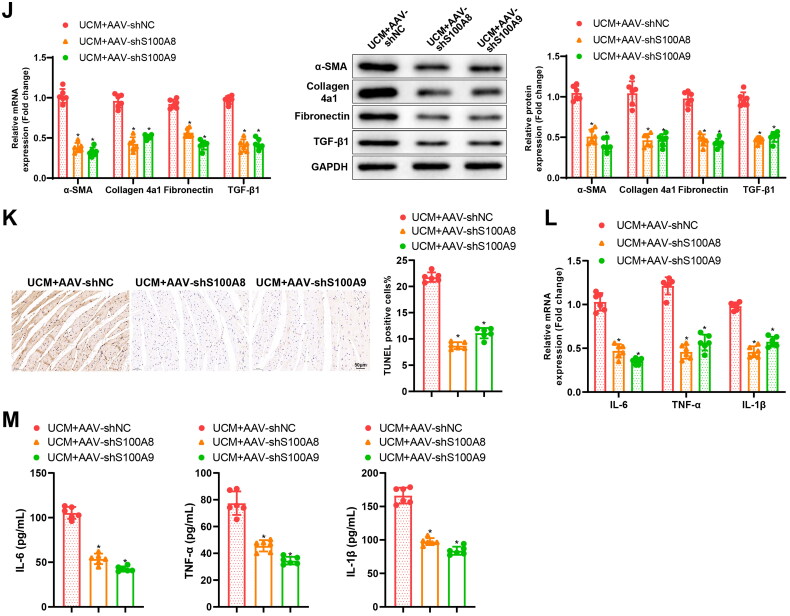
Moreover, the body weight of UCM rats was prominently increased but BUN levels, blood creatinine levels, SBP, CK-MB levels, heart weight, and cardiac index (Figure 3(A–G)) in UCM rats were reduced by AAV-shS100A8 and AAV-shS100A9. As reflected by ultrasound results, AAV-shS100A8 and AAV-shS100A9 decreased LVIVSs, LVIVSd, LVPWs, LVPWd, and LVEF but augmented LVEDV and LVESV in UCM rats (Table 2). The results of HE staining and Masson staining depicted that AAV-shS100A8 and AAV-shS100A9 ameliorated cardiomyocyte hypertrophy and myocardial fibrosis in the myocardial tissues of UCM rats (Figure 3(H,I)). The results of qRT-PCR and western blot also documented that AAV-shS100A8 and AAV-shS100A9 diminished the mRNA and protein expression of fibrosis markers (TGF-β1, α-SMA, Collagen 4a1, and Fibronectin) in the myocardial tissues of UCM rats (Figure 3(J)). In addition, the TUNEL assay presented that apoptosis was evidently reduced in the myocardial tissues of rats after transduction with AAV-shS100A8 and AAV-shS100A9 (Figure 3(K)). Moreover, AAV-shS100A8 and AAV-shS100A9 reduced the expression of inflammatory cytokines, IL-6, TNF-α, and IL-1β, in the myocardial tissues and blood of UCM rats (Figure 3(L,M)). These results suggested that knockdown of S100A8 or S100A9 subdued myocardial fibrosis and inflammation in UCM rats.
Figure 3.
Knockdown of S100A8 or S100A9 restrains myocardial inflammation and fibrosis in rats with UCM. (A) The levels of BUN in rats of each group; (B) Levels of blood creatinine in rats of each group; (C) SBP in rats of each group; (D) CK-MB contents in rats of each group; (E-G) The heart weight (E), body weight (F), and the ratio of heart weight to body weight (G) in rats of each group; (H) Representative images of HE staining and quantitation of cardiomyocyte area in rats of each group (scale bar: 50 µm); (I) Masson staining observation of myocardial fibrosis in rats of each group (scale bar: 50 µm); (J) qRT-PCR and western blot determination of the expression of fibrosis markers (TGF-β1, α-SMA, Collagen 4a1, and Fibronectin) in myocardial tissues of rats; (K) TUNEL staining to test apoptosis in myocardial tissues of rats (scale bar: 50 µm); (L) qRT-PCR to assess mRNA expression of inflammatory cytokines (IL-6, TNF-α, and IL-1β) in myocardial tissues of rats in each group; (M) ELISA detection of the levels of inflammatory cytokines (IL-6, TNF-α, and IL-1β) in blood. * p < 0.05 compared with the UCM + AAV-shNC group. N = 6 rats/group. UCM: uremic cardiomyopathy; BUN: blood urea nitrogen; SBP: systolic blood pressure; CK-MB: creatine phosphokinase-MB; qRT-PCR: quantitative real-time polymerase chain reaction; SMA: smooth muscle actin; mRNA: messenger RNA; IL: interleukin; TNF: tumor necrosis factor; ELISA: enzyme-linked immunosorbent assay; AAV: adeno-associated virus; Sh: short hairpin; NC: negative control; HE: hematoxylin and eosin; TUNEL: terminal deoxynucleotidyl transferase nick-end labeling.
SPI1 bound to the promoter of S100A8 and S100A9 and promoted the transcription of S100A8 and S100A9
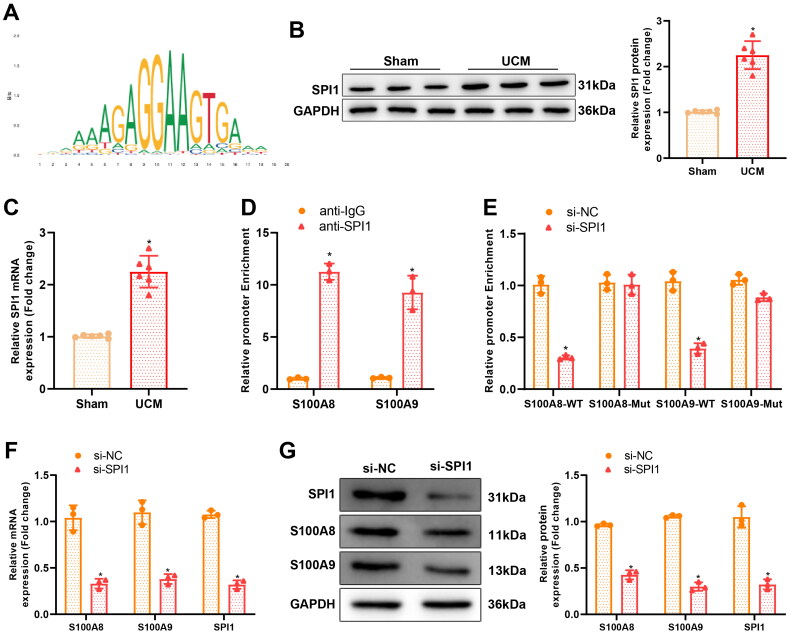
hTFtarget (http://bioinfo.life.hust.edu.cn/hTFtarget#!/) and Jaspar (https://jaspar.genereg.net/) databases predicted that the transcription factor SPI1 manipulated both S100A8 and S100A9 transcription (Figure 4(A).
Figure 4.
The transcription factor SPI1 regulates S100A8 and S100A9 transcription. (A) Binding sites of SPI1 on the promoter of S100A8/A9; (B–C) Western blot and qRT-PCR detection of protein and mRNA expression of SPI1 in rat myocardial tissues, N = 6 rats/group, *p < 0.05 compared with the sham group; (D) ChIP assay determination of SPI1 enrichment in the promoter of S100A8/A9, * p < 0.05 compared with the anti-IgG group; (E) Dual-luciferase reporter gene assay to evaluate the binding relationship between SPI1 and S100A8/A9, * p < 0.05 compared with the si-NC group; (F–G) qRT-PCR and western blot assessment of the expression of related genes in primary rat cardiomyocytes, * p < 0.05 compared with the si-NC group. Cell experiments were repeated three times. SPI: salmonella pathogenicity island; qRT-PCR: quantitative real-time polymerase chain reaction; ChIP: chromatin immunoprecipitation; IgG: Immunoglobulin G; Si: silencing; NC: negative control.
To investigate the relationship between SPI1 and S100A8/A9, SPI1 expression in myocardial tissues of UCM rats was first examined. Western blot and qRT-PCR demonstrated that SPI1 expression was high in the myocardial tissues of UCM rats (Figure 4(B,C)). Next, the Jaspar database predicted the binding sites of SPI1 on the promoter of S100A8/A9 (Supplementary Table 1). The ChIP assay displayed that the SPI1 antibody precipitated the promoter of S100A8 and S100A9 (Figure 4(D)). Moreover, the results of the dual-luciferase reporter gene assay demonstrated that knockdown of SPI1 diminished the luciferase activity of S100A8-WT and S100A9-WT but did not afflict the luciferase activity of S100A8-Mut and S100A9-Mut (Figure 4(E)). With regard to the results of qRT-PCR and western blot, knockdown of SPI1 observably reduced S100A8 and S100A9 expression in rat primary cardiomyocytes (Figure 4(F,G)).
In conclusion, SPI1 was upregulated in the myocardial tissues of UCM rats and bound to the promoter of S100A8/A9 to increase S100A8/A9 transcription.
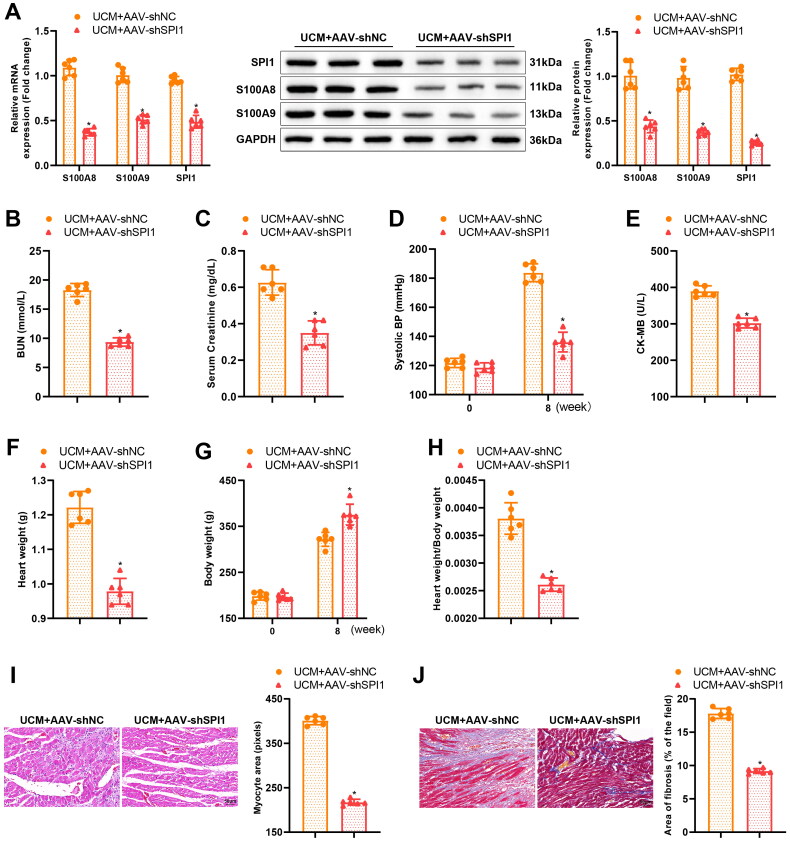
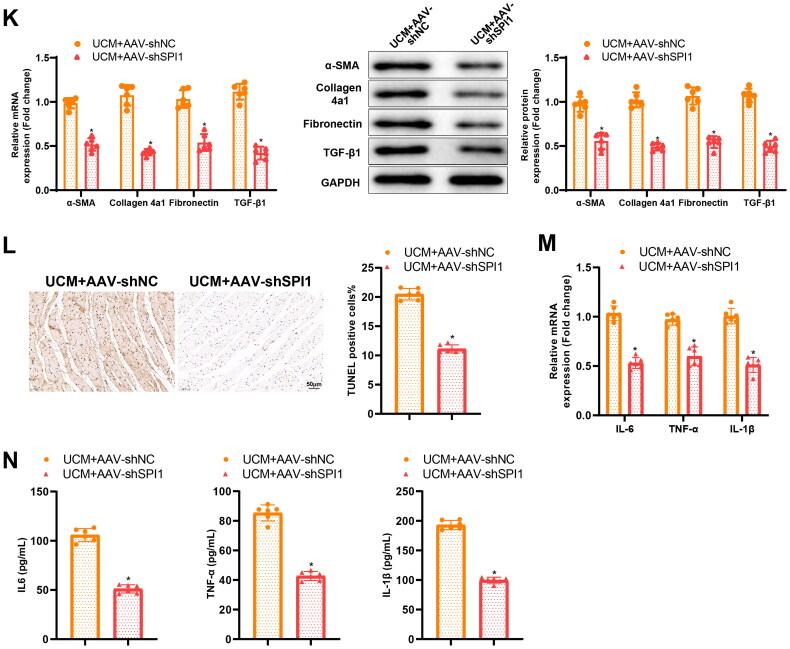
Knockdown of SPI1 restricted UCM-caused myocardial fibrosis and inflammation in rats
Interference of UCM rats with AAV-shSPI1 resulted in a reduction in SPI1, S100A8, and S100A9 expression in rat myocardial tissues (Figure 5(A)). AAV-shSPI1 was also found to substantially increase body weight and decrease BUN levels, blood creatinine levels, SBP, CK-MB levels, heart weight, and cardiac index (Figure 5(B–H)) in UCM rats. Ultrasound results documented that AAV-shSPI1 reduced LVIVSs, LVIVSd, LVPWs, LVPWd, and LVEF in UCM rats, accompanied by elevated LVEDV and LVESV (Table 2). HE staining and Masson staining results manifested that cardiomyocyte hypertrophy and myocardial fibrosis were curbed in myocardial tissues of UCM rats by AAV-shSPI1 (Figure 5(I,J)). qRT-PCR and western blot exhibited that the mRNA and protein expression of TGF-β1, α-SMA, Collagen 4a1, and Fibronectin was diminished in the myocardial tissues of UCM rats subsequent to AAV-shSPI1 transduction (Figure 5(K)). In addition, the TUNEL assay indicated a remarkable decline in apoptosis in myocardial tissues of UCM rats after AAV-shSPI1 treatment (Figure 5(L))). Meanwhile, reduced IL-6, TNF-α, and IL-1β expression was observed in the myocardial tissues and blood of UCM rats transduced with AAV-shSPI1 (Figure 5(M,N)).
Figure 5.
Knockdown of SPI1 alleviates myocardial inflammation and fibrosis in UCM rats. (A) qRT-PCR and western blot detection of the expression of related genes in the myocardial tissues of rats from each group; (B) The level of BUN in each group; (C) The levels of blood creatinine; (D) SBP in rats of each group; (E) Contents of CK-MB in rats of each group; (F–H) The heart weight (F), body weight (G), and ratio of heart weight to body weight (H) of rats in each group; (I) Representative images of HE staining and quantification of cardiomyocyte area in rats of each group (scale bar: 50 µm); (J) Masson staining of myocardial fibrosis in rats of each group (scale bar: 50 µm); (K) qRT-PCR and western blot detection of the expression of SPI1, S100A8, S100A9, and the fibrosis markers (TGF-β1, α-SMA, Collagen 4a1, and Fibronectin) in rats of each group; (L) TUNEL staining determination of myocardial cell apoptosis in rats of each group (scale bar: 50 µm); (M) qRT-PCR to measure the mRNA expression of inflammatory cytokines (IL-6, TNF-α, and IL-1β) in myocardial tissues of rats in each group; (N) ELISA examination of inflammatory cytokines (IL-6, TNF-α, and IL-1β) in the blood of rats in each group. *p < 0.05 compared with UCM + AAV-shNC group. N = 6 rats/group. SPI: salmonella pathogenicity island; UCM: uremic cardiomyopathy; BUN: blood urea nitrogen; SBP: systolic blood pressure; CK-MB: creatine phosphokinase-MB; HE: hematoxylin and eosin; TUNEL: terminal deoxynucleotidyl transferase nick-end labeling; qRT-PCR: quantitative real-time polymerase chain reaction; SMA: smooth muscle actin; mRNA: messenger RNA; ELISA: enzyme-linked immunosorbent assay; AAV: adeno-associated virus; IL: interleukin; TNF: tumor necrosis factor; Sh: short hairpin; NC: negative control.
In summary, the knockdown of SPI1 curbed myocardial fibrosis and inflammation in UCM rats.
Discussion
UCM associated with CKD leads to the adverse cardiac outcome of left ventricular hypertrophy [22]. It is believed that UCM has a linkage with heart failure-induced death of patients with a later stage of CKD [2]. Moreover, the causes of UCM are problematic and the therapeutic strategy remains suboptimal [23]. Consequently, the molecular mechanisms of UCM are well worth discussing. In this context, our study explored the role of SPI1 in the development of myocardial fibrosis and inflammation induced by UCM. The experiment results revealed that SPI1 silencing ameliorated myocardial fibrosis and inflammation in UCM by down-regulating S100A8/A9 transcription.
SPI1 is a transcription factor playing a pivotal role in the differentiation of immune cells, such as T lymphocytes, macrophages, dendritic cells, B lymphocytes, and neutrophils [24]. Evidence has revealed that SPI1 expression of resident macrophages in situ is evidently higher in inflamed colonic mucosa than in healthy mucosa [25]. SPI1 overexpression is also correlated with tendon inflammation [26]. Likewise, SPI1 upregulation results in microglial inflammation in Alzheimer’s disease [14]. In addition, SPI1 upregulation can accelerate the transition of resting fibroblasts in healthy donors to a highly activated and pro-fibrotic phenotype, accompanied by elevated α-SMA and F-actin expression, and SPI1 knockout in fibroblasts mitigated fibrosis in various models [13]. For example, Hu et al. concluded that SPI1 suppression reduces angiotensin-II-induced atrial fibrosis, indicating the potential therapeutic value of SPI1 [15]. Besides, inhibition of SPI1 distinctly reduces inflammation and fibrosis in a dietary mouse model with nonalcoholic steatohepatitis [27]. More importantly, SPI1 inhibition improves cardiac function and suppresses myocardial infarction-induced fibrosis and inflammation in mice [28]. A prior study unveiled that the lowly expressed SPI1 is associated with reduced myocardial injury during acute myocarditis [29]. Moreover, SPI1 expression in heart tissues of rats with diabetic coronary heart disease is higher than that in normal rat tissues [30]. All of these observations contribute to the speculation of the potential promoting role of SPI1 in UCM-induced fibrosis and inflammation. Therefore, a series of assays were carried out in our research to verify this speculation. Of note, our study elucidated that SPI1 was highly expressed in the myocardial tissues of UCM rats. Further analyses revealed that the knockdown of SPI1 inhibited myocardial fibrosis and inflammation induced by UCM in rats, accompanied by downregulated TGF-β1, α-SMA, Collagen 4a1, Fibronectin, IL-6, TNF-α, and IL-1β. In addition, knockdown of SPI1 reduced LVIVSs, LVIVSd, LVPWs, LVPWd, and LVEF, as well as enhanced LVEDV and LVESV, in UCM rats.
It was previously reported that SPI1 transcriptionally elevated S100A8 and S100A9 expression by binding to the promoter of S100A8 and S100A9 during scar formation post-burn [31]. Of note, corroborating trends that SPI1knockdown reduced S100A8 and S100A9 expression were observed in UCM rats and primary cardiomyocytes in our research. S100A8/A9 is abundantly expressed in neutrophils and is rapidly released as a pro-inflammatory alarmin in the circulation and myocardium following myocardial ischemia [32]. S100A8/A9 facilitates inflammation and resolution in myocardial infarction [33]. Previous research revealed that S100A8/A9 is expressed highly in patients with pulmonary fibrosis [34]. Down-regulation of S100A8 causes significant apoptosis acceleration and growth reduction in fibroblasts from hypertrophic scars [35]. S100A8/A9 inhibition can be utilized to prevent renal fibrosis in patients with CKD [9]. In addition, S100A8/A9 expression is elevated in myocarditis and a reduction of S100A8/A9 expression has a connection with decreased cardiac inflammation [36]. However, no research conducted to directly investigate the role of S100A8/A9 in UCM. Our data unraveled the upregulation of S100A8/A9 in the myocardial tissues of UCM rats. Furthermore, we also found that S100A8/A9 knockdown inhibited myocardial fibrosis and inflammation, LVIVSs, LVIVSd, LVPWs, and LVPWd in UCM rats, as well as diminished TGF-β1, α-SMA, Collagen 4a1, Fibronectin, IL-6, TNF-α, and IL-1β expression in myocardial tissues of UCM rats.
All in all, our study collectively demonstrated that SPI1 silencing ameliorated myocardial fibrosis and inflammation in UCM rats by reducing S100A8/A9 transcription. Therefore, the present study revealed the mechanism underlying myocardial fibrosis and inflammation in UCM, providing the scientific basis for the development of new therapeutic modalities for UCM treatment. Nevertheless, in addition to myocardial fibrosis and inflammation, the pathogenesis of UCM also is associated with other mechanisms, such as diastolic dysfunction, left ventricular hypertrophy, volume overload, and renin-angiotensin-aldosterone system, and so on [1,37]. Therefore, further research is warranted to explore other mechanisms of UCM.
Supplementary Material
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Wang X, Shapiro JI.. Evolving concepts in the pathogenesis of uraemic cardiomyopathy. Nat Rev Nephrol. 2019;15(3):159–175. [DOI] [PubMed] [Google Scholar]
- 2.Law JP, Price AM, Pickup L, et al. Clinical potential of targeting fibroblast growth factor-23 and alphaKlotho in the treatment of uremic cardiomyopathy. J Am Heart Assoc. 2020;9(7):e016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levin A, Tonelli M, Bonventre J, et al. Global kidney health 2017 and beyond: a roadmap for closing gaps in care, research, and policy. Lancet. 2017;390(10105):1888–1917. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Liu J, Drummond CA, et al. Sodium potassium adenosine triphosphatase (Na/K-ATPase) as a therapeutic target for uremic cardiomyopathy. Expert Opin Ther Targets. 2017;21(5):531–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lekawanvijit S. Cardiotoxicity of uremic toxins: a driver of cardiorenal syndrome. Toxins (Basel). 2018;10(9):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakano T, Onoue K, Seno A, et al. Involvement of chronic inflammation via monocyte chemoattractant protein-1 in uraemic cardiomyopathy: a human biopsy study. ESC Heart Fail. 2021;8(4):3156–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang S, Song R, Wang Z, et al. S100A8/A9 in inflammation. Front Immunol. 2018;9:1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahler M, Meroni PL, Infantino M, et al. Circulating calprotectin as a biomarker of COVID-19 severity. Expert Rev Clin Immunol. 2021;17(5):431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tammaro A, Florquin S, Brok M, et al. S100A8/A9 promotes parenchymal damage and renal fibrosis in obstructive nephropathy. Clin Exp Immunol. 2018;193(3):361–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Li Y, Zhang C, et al. S100a8/a9 released by CD11b + Gr1+ neutrophils activates cardiac fibroblasts to initiate angiotensin II-Induced cardiac inflammation and injury. Hypertension. 2014;63(6):1241–1250. [DOI] [PubMed] [Google Scholar]
- 11.Cai Z, Xie Q, Hu T, et al. S100A8/A9 in myocardial infarction: a promising biomarker and therapeutic target. Front Cell Dev Biol. 2020;8:603902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang J, Chen W, Jie Z, et al. Comprehensive analysis of immune implications and prognostic value of SPI1 in gastric cancer. Front Oncol. 2022;12:820568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wohlfahrt T, Rauber S, Uebe S, et al. PU.1 controls fibroblast polarization and tissue fibrosis. Nature. 2019;566(7744):344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pimenova AA, Herbinet M, Gupta I, et al. Alzheimer’s-associated PU.1 expression levels regulate microglial inflammatory response. Neurobiol Dis. 2021;148:105217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Zhang JJ, Li L, et al. PU.1 inhibition attenuates atrial fibrosis and atrial fibrillation vulnerability induced by angiotensin-II by reducing TGF-beta1/Smads pathway activation. J Cell Mol Med. 2021;25(14):6746–6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swindell WR, Johnston A, Xing X, et al. Robust shifts in S100a9 expression with aging: a novel mechanism for chronic inflammation. Sci Rep. 2013;3:1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Zhang C, Zhang Z, et al. E26 transformation-specific transcription factor ETS2 as an oncogene promotes the progression of hypopharyngeal cancer. Cancer Biother Radiopharm. 2017;32(9):327–334. [DOI] [PubMed] [Google Scholar]
- 18.Kramann R, Erpenbeck J, Schneider RK, et al. Speckle tracking echocardiography detects uremic cardiomyopathy early and predicts cardiovascular mortality in ESRD. J Am Soc Nephrol. 2014;25(10):2351–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai J, Wu Y, Hang L, et al. [Zhenwu decoction delays ventricular hypertrophy in rats with uremic cardiomyopathy]. Nan Fang Yi Ke Da Xue Xue Bao. 2019;39(1):113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian J, Shidyak A, Periyasamy SM, et al. Spironolactone attenuates experimental uremic cardiomyopathy by antagonizing marinobufagenin. Hypertension. 2009;54(6):1313–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B, Wang ZM, Ji JL, et al. Macrophage-derived exosomal mir-155 regulating cardiomyocyte pyroptosis and hypertrophy in uremic cardiomyopathy. JACC Basic Transl Sci. 2020;5(2):148–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Albuquerque Suassuna PG, Sanders-Pinheiro H, de Paula RB.. Uremic cardiomyopathy: a new piece in the chronic kidney disease-mineral and bone disorder puzzle. Front Med (Lausanne). 2018;5:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semple D, Smith K, Bhandari S, et al. Uremic cardiomyopathy and insulin resistance: a critical role for akt? J Am Soc Nephrol. 2011;22(2):207–215. [DOI] [PubMed] [Google Scholar]
- 24.Li G, Hao W, Hu W.. Transcription factor PU.1 and immune cell differentiation. Int J Mol Med. 2020;46(6):1943–1950. [DOI] [PubMed] [Google Scholar]
- 25.Lasitschka F, Giese T, Paparella M, et al. Human monocytes downregulate innate response receptors following exposure to the microbial metabolite n-butyrate. Immun Inflamm Dis. 2017;5(4):480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freedman BR, Adu-Berchie K, Barnum C, et al. Nonsurgical treatment reduces tendon inflammation and elevates tendon markers in early healing. J Orthop Res. 2022;40(10):2308–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Yu J, Wang L, et al. Inhibition of PU.1 ameliorates metabolic dysfunction and non-alcoholic steatohepatitis. J Hepatol. 2020;73(2):361–370. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z, Huang S.. Upregulation of SPI1 during myocardial infarction aggravates cardiac tissue injury and disease progression through activation of the TLR4/NFkappaB axis. Am J Transl Res. 2022;14(4):2709–2727. [PMC free article] [PubMed] [Google Scholar]
- 29.Corsten MF, Papageorgiou A, Verhesen W, et al. MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ Res. 2012;111(4):415–425. [DOI] [PubMed] [Google Scholar]
- 30.Chen HY, Bao Y, Zou JJ, et al. Activated Th9 cells in diabetic coronary heart disease. J Biol Regul Homeost Agents. 2021;35(3):1137–1144. [DOI] [PubMed] [Google Scholar]
- 31.Xu Z, Cheng C, Kong R, et al. S100A8 and S100A9, both transcriptionally regulated by PU.1, promote epithelial-mesenchymal transformation (EMT) and invasive growth of dermal keratinocytes during scar formation post burn. Aging (Albany NY). 2021;13(11):15523–15537. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinkovic G, Grauen Larsen H, Yndigegn T, et al. Inhibition of pro-inflammatory myeloid cell responses by short-term S100A9 blockade improves cardiac function after myocardial infarction. Eur Heart J. 2019;40(32):2713–2723. [DOI] [PubMed] [Google Scholar]
- 33.Sreejit G, Abdel Latif A, Murphy AJ, et al. Emerging roles of neutrophil-borne S100A8/A9 in cardiovascular inflammation. Pharmacol Res. 2020;161:105212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki K, Kinoshita R, Tomonobu N, et al. The heterodimer S100A8/A9 is a potent therapeutic target for idiopathic pulmonary fibrosis. J Mol Med (Berl). 2021;99(1):131–145. [DOI] [PubMed] [Google Scholar]
- 35.Yaundong L, Dongyan W, Lijun H, et al. Effects of downregulation of S100A8 protein expression on cell cycle and apoptosis of fibroblasts derived from hypertrophic scars. Aesthet Surg J. 2014;34(1):160–167. [DOI] [PubMed] [Google Scholar]
- 36.Muller I, Vogl T, Kuhl U, et al. Serum alarmin S100A8/S100A9 levels and its potential role as biomarker in myocarditis. ESC Heart Fail. 2020;7(4):1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovács ZZA, Szűcs G, Freiwan M, et al. Comparison of the antiremodeling effects of losartan and mirabegron in a rat model of uremic cardiomyopathy. Sci Rep. 2021;11(1):17495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.