Abstract
Naltrexone, an opioid antagonist primarily metabolized by aldo-keto reductase 1C4 (AKR1C4), treats pediatric conditions involving compulsiveness (e.g., autism spectrum, Prader-Willi, eating disorders, non-suicidal self-injury). Pharmacokinetic variability is apparent in adults, yet no data are available for children. This study aimed to examine the impact of age and genetic variation on naltrexone biotransformation. Human liver cytosol (HLC) samples (n = 158) isolated from children and adult organ donors were incubated with therapeutically relevant concentrations of naltrexone (0.1, 1 µM). Naltrexone biotransformation was determined by ultraperformance mass spectrometry quantification of the primary metabolite, 6-beta-naltrexol (6βN), and 6βN formation rates (pmol/mg protein/min) were calculated. HLCs from organ donors, age range 0–79 y (mean 16.0 ± 18.2 y), 37% (n = 60) female, 20% (n = 33) heterozygous and 1.2% (n = 2) homozygous for co-occurring AKR1C4 variants (S145C/L311V) showed >200-fold range in 6βN formation (0.37–76.5 pmol/mg protein/min). Source of donor samples was found to be a substantial contributor to variability. Model estimates for a trimmed data set of source-adjusted pediatric samples (aged 0–18 y) suggested that AKR1C4 genetic variation, age, and sex explained 36% of the variability in 6βN formation. Although activity increased steadily from birth and peaked in middle childhood (2–5 years), genetic variation (S145C/L311V) demonstrated a greater effect on activity than did age. Naltrexone biotransformation is highly variable in pediatric and adult livers and can be partly accounted for by individual factors feasible to obtain (e.g., genetic variability, age, sex). These data may inform a precision therapeutics approach (e.g., exposure optimization) to further study Naltrexone responsiveness in children and adults.
SIGNIFICANCE STATEMENT
Biotransformation of the commonly used opioid antagonist naltrexone is highly variable and may contribute to reduced therapeutic response. Age, sex, and genetic variation in the drug-metabolizing enzyme, AKR1C4, are potential factors contributing to this variability. In pediatric samples, genetic variation (S145C/L311V) demonstrates a greater impact on activity than age. Additionally, the source of donor samples was identified as an important contributor and must be accounted for to confidently elucidate the biological variables most impactful to drug biotransformation.
Introduction
Naltrexone is an opioid antagonist with increasing use for a variety of indications in children, adolescents and adults. Naltrexone is approved by the United States Federal Drug Administration (FDA) for the treatment of opioid and alcohol use disorder in adults (https://www.accessdata.fda.gov/drugsatfda_docs/label/2013/018932s017lbl.pdf). Naltrexone is widely used off-label for conditions that involve compulsiveness and impulsiveness such as eating disorders, obesity, autism spectrum disorder (ASD), Prader-Willi Syndrome, sexual addiction, gambling and non-suicidal self-injury in patients <2 years to >65 years (Stancil, et al., 2021). These conditions are associated with significant morbidity, decreased quality of life, increased health care expenditures, and mortality.
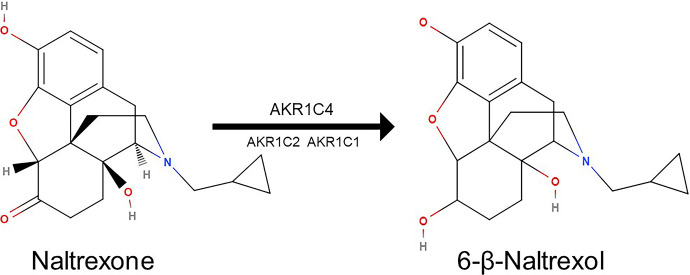
Despite wide use, response to naltrexone is variable, and the optimal dose is not clear. In pediatric conditions, such as autism spectrum disorder, eating disorders, and Prader-Willi Syndrome, response ranges from 40–80% (Kolmen et al., 1995; Willemsen-Swinkels et al., 1996; Kolmen et al., 1997; Feldman et al., 1999; Stancil et al., 2019). Clinical daily doses range from 50 mg to 400 mg (Stancil, et al., 2021). Variability in systemic exposure can contribute to variable responses, and optimizing exposure to achieve response is a core feature of precision therapeutics. Adults treated with oral naltrexone demonstrate >10-fold variability in systemic exposure (e.g., Cmax and Area Under the Curve) (Meyer et al., 1984; McCaul et al., 2000; Mason et al., 2002; Dunbar et al., 2006); however, there are no pharmacokinetic data available in children. Developmental differences in protein expression (i.e., ontogeny) leading to age-dependent alterations in drug clearance and systemic exposure (Kearns et al., 2003) have been well-documented for various drug-metabolizing enzymes (e.g., CYP2D6, CYP2A6, CYP3A). Naltrexone is metabolized primarily by the cytosolic enzyme aldo-keto reductase 1C4 (AKR1C4), with minor contributions from AKR1C2 and AKR1C1 (Fig. 1) (Breyer-Pfaff and Nill, 2004; Stancil et al., 2021). No published data exist regarding the impact of ontogeny on the AKR1C family. Thus, it remains unknown if developmental differences in naltrexone biotransformation exist and the potential consequences for the dose-exposure relationship of naltrexone in children and adolescents.
Fig. 1.
Biotransformation of naltrexone to 6-β-naltrexol. The enzymes known to catalyze the reaction are depicted based on relative affinity (AKR1C4 Km 0.34 µM, AKR1C2 Km 130 µM, AKR1C1 Km 1.4 mM). Molecular structures adapted from molview.org (MolView v2.4). The MolView Project is Open-Source and the source code is released under the GNU AGPL license.
In addition to ontogeny, other factors, such as genetic variability, may contribute to altered naltrexone biotransformation and must be considered. Recombinant enzymes expressing two co-occurring AKR1C4 missense mutations, S145C and L311V, showed a 5-fold reduction in catalytic activity compared with wild type (Kume et al., 1999). The impact of these variants has yet to be investigated in an intact human system; however, they may be a relevant contributor to variability given their mean allele frequencies (MAF) of 10–50% (Sherry et al., 1999).
Understanding the role of ontogeny and genetic variation in naltrexone biotransformation may elucidate variability in exposure, a vital step in untangling the contributing factors to variable response. To fill this gap, this study aims to examine the impact of age and genetics on naltrexone biotransformation across the lifespan using samples from human liver donors. Findings from this study will inform a precision therapeutics approach that facilitates exposure optimization and further our understanding of naltrexone response versus non-response for conditions with high morbidity, mortality, and societal impact.
Materials and Methods
Liver Samples and Tissue Fractions.
Human liver cytosols were isolated from liver samples obtained from pediatric and adult donors (n = 158) and were acquired from five sources: University of Minnesota (n = 34), The Liver, Tissue, and Cell Distribution System at the University of Pittsburg (n = 40), Vitron (Tucson, AZ; n = 4), Xenotech (now Sekisui Xenotech, Kansas City, KS; n = 23), and University of Maryland-Baltimore Brain and Tissue Bank (UMB, n = 57). Samples obtained from Minnesota were flash frozen within 1 hour of donor death/ischemia. Samples from Pittsburg, Vitron, and Xenotech were from livers harvested for transplantation, placed on ice, and perfused with preservation solution. UMB samples were obtained upon autopsy with various post-mortem intervals up to 15 hours.
Chemicals and Reagents.
Naltrexone, 6-beta-naltrexol, naltrexone-d3 and 6-beta-naltrexol-d3 were purchased from Sigma Aldrich (St. Louis, MO). Methanol and formic acid were also purchased from Fisher Scientific (Waltham, MA). All other chemicals were reagent or higher grade and purchased from either Sigma Aldrich or Fisher Scientific.
Human Liver Cytosol In Vitro Incubation Reactions.
Cytosolic fractions were isolated from pediatric and adult human liver samples according to the method of Lu and Levin (Lu and Levin, 1978). Pooled human liver cytosol (n = 200, mixed gender, lot #1410229) was purchased from Xenotech (Kansas City, KS) and was used as an internal assay control (minimal interday variability %CV = 5.5). Incubations were performed in triplicate at 37°C in a shaking water bath for 30 minutes. In a total volume of 400 µl, incubations contained 100 mM phosphate buffer (pH 7.4), nicotinamide adenine dinucleotide phosphate generating system [consisting of nicotinamide adenine dinucleotide phosphate (NADP+ 1 mM), glucose-6- phosphate (G-6-P 5 mM) and glucose-6- phosphate dehydrogenase (G-6-PDH 1 unit/ml)], therapeutically relevant concentrations of naltrexone (0.1, 1 µM) and 0.05 mg/ml of cytosolic protein. Incubation conditions were chosen based on linear product formation achieved between 0 and 45 minutes at 0.05 mg/ml of protein. Reactions were stopped by the addition of an equal volume (400 µl) of ice-cold methanol containing the internal standards, 6-β-naltrexol-d3 and naltrexone-d3 final concentration 10 nM). The samples were gently vortexed and then subjected to centrifugation at 10,000 g × 10 minutes. An aliquot of the supernatant (600 µl) was transferred to a collection plate and evaporated to dryness using heated nitrogen displacement. Samples were reconstituted in 150 µl of 1:1 methanol-water and 10µl was injected onto ultra-performance liquid chromatography tandem mass spectrometry (UPLC-MS/MS).
Naltrexone biotransformation was determined by UPLC-MS/MS quantification of the primary metabolite, 6-beta-naltrexol (6βN), and 6βN formation rates (pmol/mg protein/min) were calculated. All incubations were performed in triplicate and accepted if %CV <15. Results are reported as the mean of triplicate incubations.
Human Microsome In Vitro Incubation Reactions.
Pooled human liver microsomes (n = 200, mixed gender, lot #1710084) were purchased from Xenotech (Kansas City, KS). To screen for potential contribution to naltrexone biotransformation, incubations were performed in triplicate at 37°C in a shaking water bath for 30 minutes. In a total volume of 400 µl, incubations contained 100 mM of phosphate buffer (pH 7.4), nicotinamide adenine dinucleotide phosphate generating system (NADP+ 1 mM, G-6-P 5 mM, G-6-PDH 1 unit/ml), concentrations of naltrexone (1, 10 µM) and 0.5 mg/ml of microsomal protein. Reactions were stopped by the addition of an equal volume (400 µl) of ice-cold methanol containing the internal standard, 6-β-naltrexol-d3 (final concentration 10 nM). The samples were gently vortexed and then subjected to centrifugation at 10,000 g × 10 minutes. An aliquot of the supernatant (600 µl) was transferred to a collection plate and evaporated to dryness using heated nitrogen displacement. Samples were reconstituted in 150 µl of 1:1 methanol-water, and 10µl was injected onto UPLC-MS/MS. naltrexone biotransformation was determined as above.
UPLC-MS/MS Analysis and Analytical Method Validation.
Analytes were detected using a Waters TQD triple quadrupole mass spectrometer coupled to a Waters H-class Acquity UPLC system. Naltrexone was monitored to ensure baseline resolution of structurally similar naltrexone and 6βN and to ensure there was no bleed through. 6βN concentrations were quantified as the focus on this work was metabolite formation. Analytes were separated on a Raptor Biphenyl analytical column (1.8µm, 2.1 × 100 mm) with a matching precolumn held at 60°C. The mobile phase was composed of 0.1% formic acid in water (A) and 0.1% formic acid in methanol (B). The liquid chromatography gradient proceeded from 20–40% B over 3 minutes at a flowrate of 0.3 mL/min. The total chromatographic method time was 6 minutes. Samples were ionized in positive mode with electrospray ionization parameters as follows: capillary voltage, 1.25 kV; sampling cone voltage, 40 V; source temperature, 150°C; desolvation temperature, 400°C; desolvation gas flow, 800 L/h; cone gas flow, 10 L/h The following analyte transitions were monitored: m/z 342.3 → 324.1 for naltrexone, m/z 344.3 → 326.1 for 6β-naltrexol, m/z 345.3 → 327.1 for naltrexone-D3, m/z 347.3 → 329.1 for 6β-naltrexol-D3. The collision energy for all transitions was 20 V. Deuterium‐labeled analytes served as internal standards. Calibration curves were constructed by plotting analyte peak area divided by the area of the internal standard versus the nominal concentration of the analyte. Calibration curves were linear over the range 0.1–100 nM. Quality control (QC) samples were prepared at four concentration levels: 0.1, 0.3, 3, and 80 nM. Method accuracy and precision were within acceptable limits according to the Food and Drug Administration guidelines for a bioanalytical assay (accuracy 85–115% of nominal and coefficient of variation [CV] <15%; lower limit of quantitation accuracy 80–120% of nominal and coefficient of variation [CV] <20%).
Genotype Analysis.
The presence of non-synonymous AKR1C4 coding region single-nucleotide variants (SNPs) (rs3829125 and rs17134592) was determined by Taq-Man genotype assays in all liver samples (n = 158). AKR1C4 SNPs were selected if they met the following search criteria: dbSNP mean allele frequency >5% and evidence of impact on drug biotransformation. AKR1C4 (Kcat/Km 660 ±113 mL/µmol/min) demonstrates ∼20-fold and ∼800-fold higher catalytic efficiency than AKR1C2 (Kcat/Km 34 ± 3 mL/µmol/min) and AKR1C1 (Kcat/Km 0.84 ± 0.08 mL/µmol/min), respectively, in the biotransformation of naltrexone (Breyer-Pfaff and Nill, 2004). DNA samples obtained from Coriell were used as positive controls. All assays were performed in triplicate.
Statistical Analysis.
6βN rates of formation were log transformed to reduce skew. Descriptive statistics and Spearman’s rho were calculated in SPSS version 24 (IBM, Armonk, NY), and statistical modeling was carried out in R (R Core Team, Vienna, Austria).
We initially assessed the potential for source-dependent differences in activity. UMB samples were noted to have consistently lower cytosolic enzyme activity assessed by 6βN formation rate compared with non-UMB samples, and recently published data by our group found reduced microsomal enzyme activity in UMB samples along with reduced tissue quality based on metrics like RNA quality and homogenate POR activity (Leeder et al., In Press). Values for these quality metrics were comparable among the non-UMB samples. Because of these source-specific differences between UMB and non-UMB samples, these cohorts were analyzed separately.
We examined the effects of age and S145C/L311V genotype on naltrexone biotransformation separately, using descriptive statistics and plots and together in a multiple regression model. Statistical modeling was limited to samples from donors through age 18 (n = 73 non-UMB, n = 51 UMB) because all non-UMB samples from donors aged 19 and above were from a single source (Pittsburgh). Source and age effects in the non-UMB data were disentangled for modeling as follows. First, we trimmed the data from each source to create a smaller data set (n = 59) in which the mean age was nearly the same (9.8–10.0 years) for each source. Next, we estimated source effects by fitting a linear model to these trimmed data, modeling log-6βN formation rate as a function of source, controlling for S145C/L311V genotype and a linear age effect. Finally, the resulting source effect estimates were used to adjust each log-6βN value in the untrimmed data set to remove, as much as possible, variability due to source-specific factors.
Using this source-adjusted data set, we used weighted ordinary least squares regression to model log-6βN as a function of age, sex, and S145C/L311V genotype. To allow for a non-linear effect of age, age was centered at 9 years (near its mean) and squared, and this quadratic term was included as an additional explanatory variable. Observations were weighted by the precision of their source effect estimate in the trimmed model. We explored natural spline models for age with 2–3 knots, but these provided negligible additional explanatory value. The same model (allowing for a quadratic effect of age) was fit to the UMB data for comparison.
Results
Donor Demographics.
Human liver donors (n = 158), age range 0-79 y (mean 16.0 ± 18.2 y), 37% (n = 60) female, 20% (n = 33) heterozygous, and 1.2% (n = 2) homozygous for co-occurring AKR1C4 variants (S145C/L311V) demonstrated >200-fold range in 6βN formation (0.37–76.5 pmol/mg protein/min). A 10-fold increase in activity was observed when donor samples were treated with 1 µM of naltrexone, compared with 0.1 µM of naltrexone. All data presented are from incubations containing 1 µM of naltrexone, as this closely approximates maximum concentrations in human plasma (Cmax).
When UMB samples were omitted from the analysis, the fold range in activity for the remaining n = 101 decreased from >200-fold to 21.7-fold. Notably, this non-UMB dataset did not include any samples from donors <1 month of age as these were all obtained from UMB.
Impact of Age on Naltrexone Biotransformation.
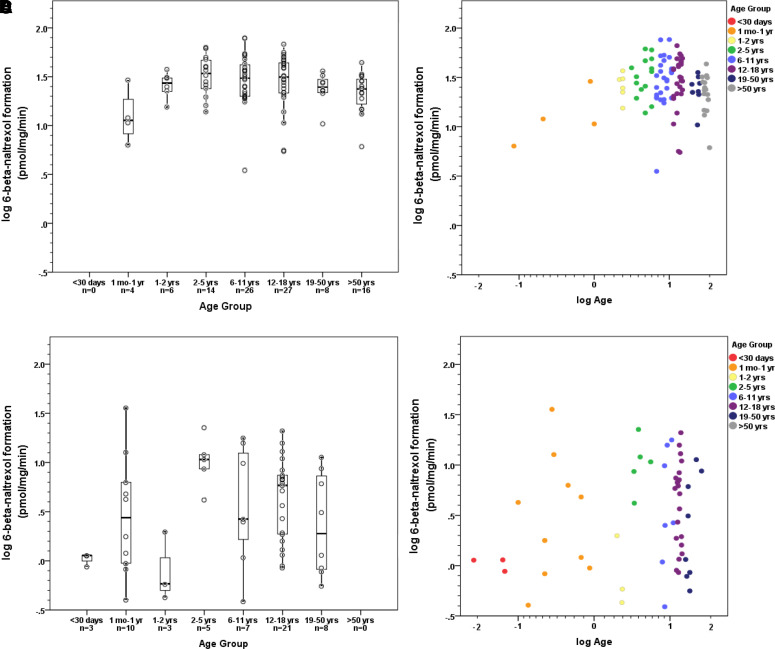
In the non-UMB samples, age appeared to exhibit a non-linear relationship with activity, peaking in early childhood and declining somewhat during adulthood when age is presented as a categorical variable, although it should be noted that all samples from donors >18 years of age were from a single source (Figs. 2A and 1B); this age relationship is less apparent when age is presented as a continuous variable, in part because of the limited number of samples younger than 2 years of age. Within each age group, wide interindividual variability is present. This pattern appeared to be less consistent in the UMB samples (Figs. 2C and 1D).
Fig. 2.
Impact of age on naltrexone biotransformation by source group. (A and B) Liver donors from four sources (non-UMB), n = 101. (C and D) Liver donors from single source (UMB), n = 57.
Impact of Genetics on Naltrexone Biotransformation.
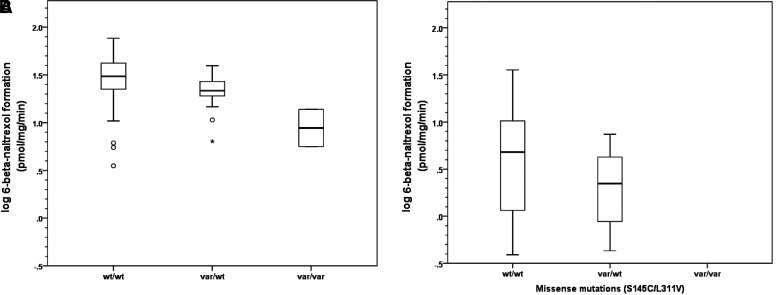
Liver samples were genotyped for two AKR1C4 nonsynonymous SNPs (rs3829125 and rs17134592), also referred to by their amino acid substitutions, S145C and L311V, respectively (Fig. 3). These two SNPs were in complete linkage disequilibrium in our samples.
Fig. 3.
Impact of co-occurring missense mutations on naltrexone biotransformation. (A) Liver donors from four sources (non-UMB), n = 101. (B) Liver donors from single source (UMB), n = 57.
For non-UMB samples, mean activity in homozygous wild-type individuals was 32.6 ± 15.3 pmol/mg/min. Activity was reduced by 31% in heterozygous variant individuals. In the two homozygous variant individuals, activity was reduced by 71% compared with wild-type (32.6 versus 9.7 pmol/mg/min) and 57% compared with heterozygous variants (22.5 ± 7.7 versus 9.7 ± 5.7 pmol/mg/min). In UMB samples, mean activity in homozygous wild-type individuals UMB was 6.8 ± 7.2 pmol/mg/min; activity was reduced by 58% in heterozygous variant individuals.
Statistical Modeling.
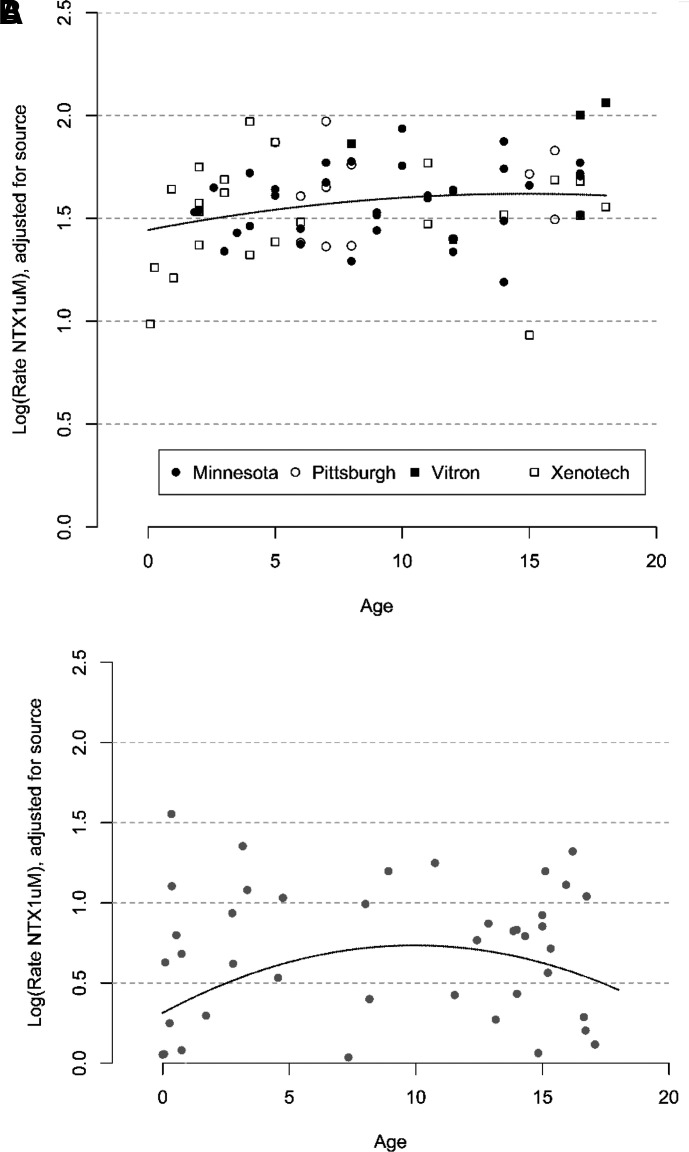
In the source-adjusted non-UMB samples, age, sex, and genetic variation accounted for 36% of the variability in naltrexone biotransformation (Table 1, Fig. 4A). Presence of one variant allele was associated with a standardized beta of -0.73, meaning source-adjusted samples having one variant allele are estimated to have log-transformed naltrexone biotransformation 0.73 s.d. lower, on average, than samples with no variant alleles, holding other model variables constant. The presence of two variant alleles displayed an even larger, supra-additive effect on 6βN formation (standardized beta = -2.66). Age displayed a modest effect in the model as shown in Fig. 4A. Given the variability observed within each age group, the impact of the apparent increase in activity in the first 10-12 years of life may be limited relative to the impact of other factors, such as genetic variation. In addition to this model with a quadratic effect for age, we considered a model with only a linear effect for age. This simpler model fit nearly as well (R2 = 0.35), but we chose to keep the quadratic model, primarily for consistency with the analysis of the UMB samples, in which there was a more pronounced non-linear trend.
TABLE 1.
Model estimates for log transformed naltrexone biotransformation in non-UMB samples, adjusted for source (R2 = 0.36).
| Variable | Beta | 95% CI | Beta (in SD Units) | 95% CI | P Value |
|---|---|---|---|---|---|
| Age (in decades) | 0.09 | 0.01 to 0.17 | 0.42 | 0.06 to 0.78 | 0.024 |
| Age squared | −0.08 | −0.26 to 0.10 | −0.36 | −1.15 to 0.44 | 0.370 |
| Female | −0.05 | −0.13 to 0.04 | −0.21 | −0.6 to 0.18 | 0.282 |
| Var/wta | −0.16 | −0.27 to -0.05 | −0.73 | −1.23 to -0.24 | 0.004 |
| Var/vara | −0.59 | −0.83 to -0.35 | −2.66 | −3.76 to -1.56 | <0.001 |
*Var: co-occurring rs3829125 and rs17134592
Fig. 4.
Source-adjusted model of naltrexone biotransformation by age. (A) Pediatric samples, 0–18 years, from four sources (non-UMB), n = 73. (B) Pediatric samples, 0–18 years, from a single source (UMB), n = 51.
For the UMB samples, the quadratic model had less explanatory power with wide confidence intervals (Table 2, Fig. 4B), making interpretation more difficult. Genetic variation demonstrated a similar directional trend to that seen with non-UMB samples.
TABLE 2.
Model estimates for log transformed naltrexone biotransformation in UMB samples, adjusted for source (R2 = 0.14).
| Variable | Beta | 95% CI | Beta (in SD Units) | 95% CI | P Value |
|---|---|---|---|---|---|
| Age (in decades) | 0.08 | −0.18 to 0.34 | 0.36 | −0.82 to 1.55 | 0.542 |
| Age squared | −0.43 | −1.08 to 0.22 | −1.92 | −4.86 to 1.01 | 0.194 |
| Female | 0.08 | −0.23 to 0.39 | 0.36 | −1.05 to 1.76 | 0.611 |
| Var/wta | −0.30 | −0.67 to 0.06 | −1.37 | −3.02 to 0.28 | 0.101 |
| Var/vara | NA |
aVar: co-occurring rs3829125 and rs17134592
Discussion
Our data describe for the first time the impact of age on naltrexone biotransformation. Given the broad therapeutic use of naltrexone, understanding how age impacts exposure is imperative to begin to untangle questions related to response versus non-response. Other individual variables, specifically genetic variation, also contribute to variability and appear to play a more important role than age. Source effects may be confounding and must be accounted for to appreciate the impact of variables of interest on activity. These findings further our understanding and support the role of precision therapeutics in designing a dosing regimen for opioid antagonism able to achieve exposure optimization.
We observed some evidence that age may impact naltrexone biotransformation in the first 2 years of life in the non-autopsy samples. The major age breakpoint appears to be 2–5 years, with sustained elevation through adolescence before potentially decreasing slightly in adulthood. This relationship was not evident in the autopsy samples. Despite the relationship between age and activity, significant variability exists within each age group; thus, additional variables impacting activity are at play.
AKR1C4 genotype appears to have a large effect on biotransformation of naltrexone. AKR1C4 is expressed almost exclusively in the liver, and endogenous functions include biosynthesis of bile acids and the biotransformation of endogenous steroids like testosterone and progesterone to their inactive 3-α/β-hydroxysteroid metabolites. Testosterone and dihydrotestosterone have been reported to inhibit 6βN formation with Ki values (0.28 ± 0.14 µM and 0.73 ± 0.38 µM, respectively) about 7-fold higher than the upper limit of normal physiologic values (0.04 µM, based on Mayo Clinic Laboratories reference range) of circulating total testosterone in adult males suggesting potential relevance. Additionally, AKR1C4 is involved in detoxification of carcinogens, such as nicotine-derived nitrosamine ketone (NNK) and contributes to the metabolism of chemotherapeutic agents daunorubicin and doxorubicin (readers are directed to Barski et al., 2008 for a comprehensive review of AKR1C family). With respect to naltrexone, AKR1C4 demonstrates ∼20-fold and ∼800-fold higher catalytic efficiency (Kcat/Km) than AKR1C2 and AKR1C1, respectively (Breyer-Pfaff and Nill, 2004), implying that 6βN formation may be a marker of AKR1C4 activity. However, we cannot rule out the possibility that other AKR1C enzymes (or other AKR1 family enzymes) may contribute to formation. We targeted two AKR1C4 SNPs suspected of altering naltrexone biotransformation based on data derived from heterologously expressed enzymes. Our human donor data are consistent with activity in heterologously expressed enzymes that showed a 5-fold reduction in catalytic activity with the two co-occurring AKR1C4 SNPs (Kume et al., 1999). Compared with wild-type, a relative reduction in activity of 31–58% was seen with each variant allele ‘dose’. It is unclear if reduced activity associated with the genetic variants is due to loss of protein function or reduction in protein expression, although reduction in expression is not anticipated given the location of SNPs. Binding site studies, such as the quantitative sudomotor axon reflex test, have yet to be done but may improve our understanding regarding the impact of these variants on substrate binding and function. The rs3829125 and rs17134592 allele variants were in complete linkage disequilibrium in our study (co-occurring in 100% of donors) with an allele frequency of 11%. These two SNPs are approximately 12 kb apart, occurring on exon 4 and exon 9, respectively (GRCh38.p12). Complete linkage disequilibrium with an allele frequency of 4% was shown in another study with nearly 1300 individuals (Multigner et al., 2010). According to dbSNP, rs3829125 and rs17134592 allele frequency is 13% and 11%, respectively, but varies by population (12–14% in European, 1–15% in Asian, 1–5% in African) (Sherry et al., 1999). Additional AKR1C variants identified through sequencing and predicted to have functional relevance may be explored in future studies of naltrexone biotransformation but were outside the scope of this work.
Although our model accounted for over a third of the variability in activity, there are additional factors yet to be identified. Our donors were obtained from five sources with various post-mortem intervals and preservation procedures that could affect sample quality and thus enzyme activity (Leeder et al., Drug Metabolism and Disposition, In Press). Previous studies did not detect a relationship between post-mortem interval, as a proxy for tissue quality, and activity (Stevens et al., 2003; Koukouritaki et al., 2004; Stevens et al., 2008); however, their cohort was from a single source, UMB, that obtained samples acquired after autopsy (Koukouritaki et al., 2002). Our investigation effectively compares UMB with non-UMB samples (e.g., flash frozen, perfused) and demonstrates that these are indeed two distinct cohorts (differences in 6βN formation among the non-UMB sources were minor compared with the difference between the non-UMB cohort and the UMB samples). Although this source effect involves differences in procurement potentially affecting tissue quality, quality may not be the only factor involved. Additional activity data for other cytosolic enzymes would be needed to infer a quality issue should all activities be consistently lower in the UMB samples. Our model attempted to account for source effects and suggested the importance of separately evaluating the data from the lowest performing source, i.e., UMB, demonstrated by activity across multiple substrates, e.g., POR activity and naltrexone.
With UMB samples excluded, we were able to assess the potential tissue source effect by creating a trimmed data set with comparable average age across the four non-UMB sources, thereby reducing any potential confounding effect of age. This exercise resulted in a reduction of inter-individual variability from 200-fold to 20-fold. Because it is not possible at this time to understand all factors associated with in vitro donor activity, simply not reporting the UMB group did not seem justified. Rather, we chose to present the data in two cohorts to allow for more careful interpretation, which may be aided by future investigations. Additional inquiry, such as expanded genomic analysis and proteomic analysis, may elucidate additional factors associated with variability.
Rates of naltrexone biotransformation are highly variable among pediatric and adult liver samples and can be partially explained by individual factors feasible to obtain (e.g., genetic polymorphism, age, sex). Ontogeny may play a role in naltrexone biotransformation during the first two years of life. However, AKR1C4 genetic variation demonstrates a greater impact on activity than age. These data suggest the importance of further inquiry into the impact of AKR1C4 genetic variability on naltrexone biotransformation in patients and will inform a precision therapeutics approach (e.g., exposure optimization) to further study naltrexone responsiveness in a variety of conditions across a wide range of ages.
Abbreviations
- AKR1C4
aldo-ketoreductase 1C4
- 6βN
6-beta-naltrexol
- SNP
single-nucleotide polymorphism
- UMB
University of Maryland-Baltimore Brain and Tissue Bank
- UPLC-MS/MS
ultra-performance liquid chromatography tandem mass spectrometry
Authorship Contributions
Participated in research design: Stancil, Nolte, Pearce, Leeder.
Conducted experiments: Stancil, Nolte.
Performed data analysis: Stancil, Staggs, Leeder.
Wrote or contributed to the writing of the manuscript: Stancil, Nolte, Pearce, Staggs, Leeder.
Footnotes
This work was supported by grants T32 HD069038 (S.L.S.) and U54 HD090258 (J.S.L.) from the Eunice Kennedy Shriver National Institute of Child Health and Development.
The authors declare that they have no conflicts of interest with the contents of the article.
References
- Database of Single Nucleotide Polymorphisms (dbSNP). Bethesda (MD): National Center for Biotechnology Information, National Library of Medicine. dbSNP accession:{rs3829125, rs17134592}, (dbSNP Build ID: {GRCh38.p12}). Available from: http://www.ncbi.nlm.nih.gov/SNP/. [Google Scholar]
- Barski OA, Tipparaju SM, Bhatnagar A (2008) The aldo-keto reductase superfamily and its role in drug metabolism and detoxification. Drug Metab Rev 40:553–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breyer-Pfaff U, Nill K (2004) Carbonyl reduction of naltrexone and dolasetron by oxidoreductases isolated from human liver cytosol. J Pharm Pharmacol 56:1601–1606. [DOI] [PubMed] [Google Scholar]
- Dunbar JL, Turncliff RZ, Dong Q, Silverman BL, Ehrich EW, Lasseter KC (2006) Single- and multiple-dose pharmacokinetics of long-acting injectable naltrexone. Alcohol Clin Exp Res 30:480–490. [DOI] [PubMed] [Google Scholar]
- Feldman HM, Kolmen BK, Gonzaga AM (1999) Naltrexone and communication skills in young children with autism. J Am Acad Child Adolesc Psychiatry 38:587–593. [DOI] [PubMed] [Google Scholar]
- Kearns GL, Abdel-Rahman SM, Alander SW, Blowey DL, Leeder JS, Kauffman RE (2003) Developmental pharmacology--drug disposition, action, and therapy in infants and children. N Engl J Med 349:1157–1167. [DOI] [PubMed] [Google Scholar]
- Kolmen BK, Feldman HM, Handen BL, Janosky JE (1995) Naltrexone in young autistic children: a double-blind, placebo-controlled crossover study. J Am Acad Child Adolesc Psychiatry 34:223–231. [DOI] [PubMed] [Google Scholar]
- Kolmen BK, Feldman HM, Handen BL, Janosky JE (1997) Naltrexone in young autistic children: replication study and learning measures. J Am Acad Child Adolesc Psychiatry 36:1570–1578. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Manro JR, Marsh SA, Stevens JC, Rettie AE, McCarver DG, Hines RN (2004) Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther 308:965–974. [DOI] [PubMed] [Google Scholar]
- Koukouritaki SB, Simpson P, Yeung CK, Rettie AE, Hines RN (2002) Human hepatic flavin-containing monooxygenases 1 (FMO1) and 3 (FMO3) developmental expression. Pediatr Res 51:236–243. [DOI] [PubMed] [Google Scholar]
- Kume T, Iwasa H, Shiraishi H, Yokoi T, Nagashima K, Otsuka M, Terada T, Takagi T, Hara A, Kamataki T (1999) Characterization of a novel variant (S145C/L311V) of 3alpha-hydroxysteroid/dihydrodiol dehydrogenase in human liver. Pharmacogenetics 9:763–771. [PubMed] [Google Scholar]
- Lu AY, Levin W (1978) Purification and assay of liver microsomal epoxide hydrase. Methods Enzymol 52:193–200. [DOI] [PubMed] [Google Scholar]
- Mason BJ, Goodman AM, Dixon RM, Hameed MH, Hulot T, Wesnes K, Hunter JA, Boyeson MG (2002) A pharmacokinetic and pharmacodynamic drug interaction study of acamprosate and naltrexone. Neuropsychopharmacology 27:596–606. [DOI] [PubMed] [Google Scholar]
- McCaul ME, Wand GS, Rohde C, Lee SM (2000) Serum 6-beta-naltrexol levels are related to alcohol responses in heavy drinkers. Alcohol Clin Exp Res 24:1385–1391. [PubMed] [Google Scholar]
- Meyer MC, Straughn AB, Lo MW, Schary WL, Whitney CC (1984) Bioequivalence, dose-proportionality, and pharmacokinetics of naltrexone after oral administration. J Clin Psychiatry 45:15–19. [PubMed] [Google Scholar]
- Multigner L, Ndong JR, Giusti A, Romana M, Delacroix-Maillard H, Cordier S, Jégou B, Thome JP, Blanchet P (2010) Chlordecone exposure and risk of prostate cancer. J Clin Oncol 28:3457–3462. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward M, Sirotkin K (1999) dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res 9:677–679. [PubMed] [Google Scholar]
- Stancil SL , Abdel-Rahman S, Wagner J (2021) Developmental considerations for the use of naltrexone in children and adolescents. J Pediatr Pharmacol Ther 26:675–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancil SL, Adelman W, Dietz A, Abdel-Rahman S (2019) Naltrexone reduces binge eating and purging in adolescents in an eating disorder program. J Child Adolesc Psychopharmacol 29:721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC, Hines RN, Gu C, Koukouritaki SB, Manro JR, Tandler PJ, Zaya MJ (2003) Developmental expression of the major human hepatic CYP3A enzymes. J Pharmacol Exp Ther 307:573–582. [DOI] [PubMed] [Google Scholar]
- Stevens JC, Marsh SA, Zaya MJ, Regina KJ, Divakaran K, Le M, Hines RN (2008) Developmental changes in human liver CYP2D6 expression. Drug Metab Dispos 36:1587–1593. [DOI] [PubMed] [Google Scholar]
- Willemsen-Swinkels SH, Buitelaar JK, van Engeland H (1996) The effects of chronic naltrexone treatment in young autistic children: a double-blind placebo-controlled crossover study. Biol Psychiatry 39:1023–1031. [DOI] [PubMed] [Google Scholar]