SARS-CoV-2, the causative agent of the COVID-19 pandemic, continues to evolve. A subvariant of SARS-CoV-2 omicron (B.1.1.529), known as BA.4.6, emerged in March, 2022, and it appears to be expanding its coverage even in the presence of BA.5, the globally dominant subvariant in recent months (appendix p 2).1, 2 Compared with subvariants BA.4 and BA.5 (hereafter referred to as BA.4/5), BA.4.6 contains two additional mutations, R346T and N658S, in the spike protein (appendix p 2). Three other nascent omicron subvariants with similar spike mutations, BA.4.7 with R346S, BA.5.9 with R346I, and BF.7 with R346T, have also been detected, although at very low frequencies (appendix p 2). The fact that these four new subvariants all have mutations at the R346 residue raises concerns for further antibody evasion, because R346K in a previous subvariant of omicron (BA.1.1) impaired the potency of several therapeutic monoclonal antibodies (mAbs).3, 4
We aimed to characterise viral receptor affinities and antibody evasion properties of the newly emerging subvariants of BA.4/5. First, we examined whether the transmission advantage of BA.4.6 could be due to a higher affinity for the viral receptor. We measured the affinity of the binding of purified spike trimers of D614G, BA.2, BA.4/5, BA.4.6, BA.4.7, BA.5.9, and BF.7 to dimeric human ACE2 by surface plasmon resonance (appendix p 3). All the spike proteins from BA.4/5 sublineages, and those of BA.4/5 carrying point mutations of R346S and N658S, showed similar binding affinities to ACE2, with dissociation constant values ranging 0·39–0·49 nM. Therefore, the expansion of BA.4.6 cannot be explained by a higher affinity for human ACE2.
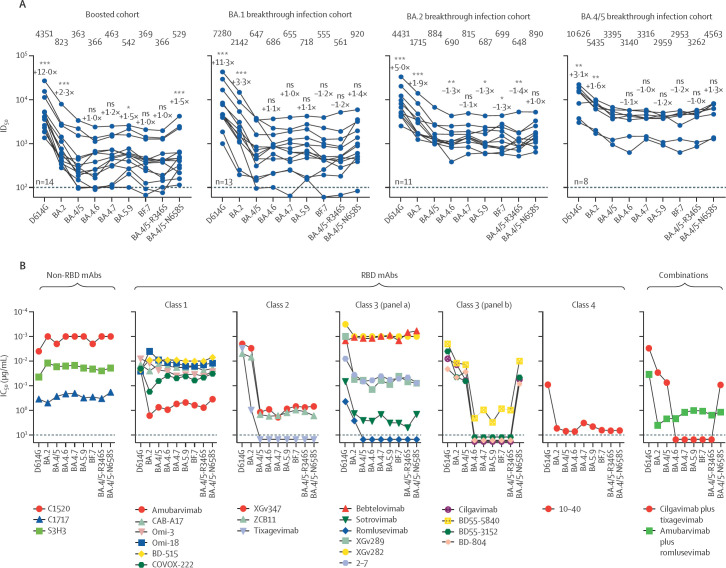
Next, to investigate the antibody evasion properties of BA.4.6, BA.4.7, BA.5.9, and BF.7, we assessed the sensitivity of their corresponding pseudoviruses to neutralisation with serum samples from healthy individuals who had received three doses of a COVID-19 mRNA vaccine BNT162b2 (Pfizer-BioNTech) or mRNA-1273 (Moderna; ie, who had received booster doses) and patients with either BA.1, BA.2, or BA.4/5 breakthrough infection after vaccination (figure ; appendix p 1). The 50% inhibitory dose (ID50) titres of the boosted samples against BA.4.6, BA.4.7, BA.5.9, and BF.7 were similar to that against BA.4/5, with no more than 1·5-fold deviation in the geometric mean values (figure). Likewise, the individual mutations R346S and N658S in the background of BA.4/5 had little effect on the neutralisation profiles. A similar trend in serum neutralisation was also observed for BA.1 and BA.4/5 breakthrough samples, but for the BA.2 breakthrough samples, BA.4.6 was slightly (1·3-fold) but significantly (p<0·01) more resistant than BA.4/5; although whether this marginal difference could explain the recent expansion of BA.4.6 worldwide remains unclear. Notably, in BA.4/5 breakthrough cohorts, neutralising titres against new emerging omicron subvariants were higher than those of the serum samples from BA.1 and BA.2 breakthrough cohorts.
Figure.
Antibody neutralisation profiles of new omicron subvariants
(A) Neutralisation ID50 titres of serum samples from cohorts who were healthy and had received a booster vaccination and who have been vaccinated (some having received a booster) and had BA.1 breakthrough infections, BA.2 breakthrough infections, and BA.4/5 breakthrough infections. Numbers along the top of the graph are the geometric mean ID50 values, the values closest to the datapoints are the fold-change in geometric mean ID50 from that of BA.4/5, and values on the lower left of each plot indicate the sample size (n). The limit of detection is 100 (dotted line). Comparisons were made against BA.4/5 using the two-tailed Wilcoxon matched-pairs signed-rank tests. (B) Neutralisation by mAbs of pseudotyped D614G, omicron subvariants, and point mutants in the background of BA.4/5. Datapoints above the maximum antibody concentration tested (10 μg/mL, indicated by the dotted line) are arbitrarily plotted to allow for visualisation of each sample. Preclinical mAbs are denoted by their laboratory designations, and clinical mAbs are denoted by their generic names. The combination of cilgavimab and tixagevimab is marketed as Evusheld. IC50=50% inhibitory concentration. ID50=50% inhibitory dose. mAbs=monoclonal antibodies. NS=not significant. RBD=recptor binding domain. *p<0·05. **p<0·01. ***p<0·001.
To further characterise the antigenic properties of BA.4.6, along with BA.4.7, BA.5.9, and BF.7, we measured the sensitivity of each subvariant pseudovirus to neutralisation by a panel of 23 mAbs that retained potency against earlier omicron subvariants, including some that targeted different epitope clusters (classes 1, 2, 3, and 4) of the receptor-binding domain (RBD) of the viral spike and others that target non-RBD epitopes (figure; appendix p 4). In general, the neutralisation profiles of BA.4.6, BA.4.7, BA.5.9, and BF.7 did not differ much from that of BA.4/5. The only exceptions were mAbs in RBD class 3 (figure B), which showed substantial reduction in their neutralisation potency against the new subvariants. This loss of neutralising activity was due to mutation R346T, R346S, or R346I, but not due to N658S. Structural analyses revealed that R346T, R346S, or R346I mutations eliminated or weakened hydrogen bonds or salt bridges, or both, between R346 and some RBD class 3 mAbs (appendix p 5), explaining why these mutations led to substantial neutralisation resistance. These findings suggest that BA.4.6, BA.4.7, BA.5.9, and BF.7 probably emerged under the selective pressure of RBD class 3 antibodies in infected individuals.
Importantly, several mAbs in clinical use were also included in the neutralisation assays against the new omicron subvariants (figure; appendix p 6). The combination of cilgavimab and tixagevimab, which had received emergency use authorisation for the prevention of COVID-19,5 could not neutralise BA.4.6, BA.4.7, BA.5.9, or BF.7, nor the authentic BA.4.6 (appendix p 6). The loss of this antibody combination against BA.4.6 leaves bebtelovimab as the only therapeutic mAb that retained potent activity against all circulating forms of SARS-CoV-2.
As the COVID-19 pandemic and SARS-CoV-2 continue to evolve, our arsenal of authorised monoclonal antibodies might soon be depleted, thereby jeopardising the wellbeing of millions of immunocompromised individuals who cannot robustly respond to COVID-19 vaccines.
QW and ZL contributed equally to this Correspondence. LL and DDH contributed equally to this work as joint senior authors. JY, LL, and DDH are inventors on patent applications (WO2021236998) or provisional patent applications (63/271,627) filed by Columbia University for a number of SARS-CoV-2 neutralising antibodies described in this Correspondence; both sets of applications are under review. DDH is a cofounder of TaiMed Biologics and RenBio, consultant to WuXi Biologics and Brii Biosciences, and board director for Vicarious Surgical. All other authors declare no competing interests.
Acknowledgments
Editorial note: The Lancet Group takes a neutral position with respect to territorial claims in institutional affiliations.
Supplementary Material
References
- 1.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.US Centers for Disease Control and Prevention COVID data tracker. Daily updates for the United States. Aug 31, 2022. https://covid.cdc.gov/covid-data-tracker
- 3.Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Guo Y, Iketani S, et al. Antibody evasion by SARS-CoV-2 omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608:603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.US Food and Drug Administration FDA releases important information about risk of COVID-19 due to certain variants not neutralized by Evusheld. June 29, 2022. https://www.fda.gov/drugs/drug-safety-and-availability/fda-authorizes-revisions-evusheld-dosing
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.