Abstract
Klebsiella pneumoniae is the causative agent of a variety of severe infections. Many K. pneumoniae strains are resistant to multiple antibiotics, and this situation creates a need for new antibacterial molecules. K. pneumoniae pathogenicity relies largely on its ability to escape phagocytosis and intracellular killing by phagocytic cells. Interfering with these escape mechanisms may allow to decrease bacterial virulence and to combat infections. In this study, we used Dictyostelium discoideum as a model phagocyte to screen a collection of 1,099 chemical compounds. Phg1A KO D. discoideum cells cannot feed upon K. pneumoniae bacteria, unless bacteria bear mutations decreasing their virulence. We identified 3 non-antibiotic compounds that restored growth of phg1A KO cells on K. pneumoniae, and we characterized the mode of action of one of them, 5-ethyl-2’-deoxyuridine (K2). K2-treated bacteria were more rapidly killed in D. discoideum phagosomes than non-treated bacteria. They were more sensitive to polymyxin and their outer membrane was more accessible to a hydrophobic fluorescent probe. These results suggest that K2 acts by rendering the membrane of K. pneumoniae accessible to antibacterial effectors. K2 was effective on three different K. pneumoniae strains, and acted at concentrations as low as 3 μM. K2 has previously been used to treat viral infections but its precise molecular mechanism of action in K. pneumoniae remains to be determined.
Introduction
Klebsiella pneumoniae is a Gram-negative bacterium responsible for a variety of infections such as pneumonia and urinary tract infections. This opportunistic pathogen is frequently acquired in hospitals by patients with impaired immunity, but community-acquired infections are also common. The emergence of antibiotic-resistant strains represents a life-threatening risk for infected patients, and urgently requires the development of new anti-bacterial therapies [1]. However, development of new antibiotics with novel targets has proved arduous [2], and new screening strategies should be explored. Compounds that inhibit bacterial virulence could in principle be a potential alternative to classical antibiotics [3], although none have yet been fully developed.
Bacterial virulence is broadly defined as the ability of a specific bacterial strain to cause a disease in a given host. It reflects the equilibrium between the pathogenic potential of a bacterium and the host defense systems. A variety of model hosts can be used to assess the virulence of bacteria, ranging from mice to Drosophila melanogaster flies, Caenorhabditis elegans nematodes, Tetrahymena pyriformis ciliate protozoans and Dictyostelium discoideum amoebae [4–7]. For both practical and ethical reasons, non-mammalian hosts are more amenable to phenotypic screening, and have been used to identify new bacterial virulence genes [8] or inhibitors of bacterial virulence [9].
More specifically, D. discoideum has been used as a host to study the virulence of Klebsiella pneumoniae as well as the defense mechanisms of this model phagocytic cell [10–14]. In our laboratory we used a simple assay to monitor interaction between D. discoideum and K. pneumoniae: D. discoideum ingests non-pathogenic KpGE K. pneumoniae bacteria and can efficiently use them as a food source [15]. On the contrary, phg1a KO D. discoideum mutants kill ingested K. pneumoniae inefficiently. Consequently, when K. pneumoniae is the only food source available, phg1a KO D. discoideum grow very slowly [11]. This can be easily visualized by monitoring the ability of D. discoideum cells to grow and to form a phagocytic plaque devoid of live bacteria on a lawn of K. pneumoniae. This assay is robust and reproducible in middle-throughput format. It thus allows to identify D. discoideum mutants with poor antibacterial defense, K. pneumoniae mutants with decreased virulence, or chemical compounds affecting the host-pathogen interaction. Using this assay, the kil2 KO D. discoideum strain was initially identified as a mutant exhibing poor growth in the presence of K. pneumoniae, and Kil2 was later shown to be essential for efficient intracellular killing of K. pneumoniae [16]. Similarly, phg1A KO cells grow more efficiently on a lawn of K. pneumoniae in which waaQ or wbbM are genetically inactivated, perturbing the synthesis of bacterial lipopolysaccharides (LPS) and reducing bacterial virulence [11]. Genetic inactivation of waaQ results in the production of LPS with an altered core [17]. When wbbM is genetically inactivated, the LPS is devoid of its O-antigen polysaccharide [18].
In this study, we used phg1A KO D. discoideum cells to identify chemical inhibitors of K. pneumoniae virulence. We identified and characterized 5-ethyl-2’-deoxyuridine (K2), as a compound facilitating intracellular killing of ingested K. pneumoniae. K2 did not exert an antibiotic activity, but it increased the permeability of the bacterial cell envelope and rendered the bacteria more susceptible to intracellular killing in phagosomes.
Results
Identification of putative inhibitors of K. pneumoniae virulence
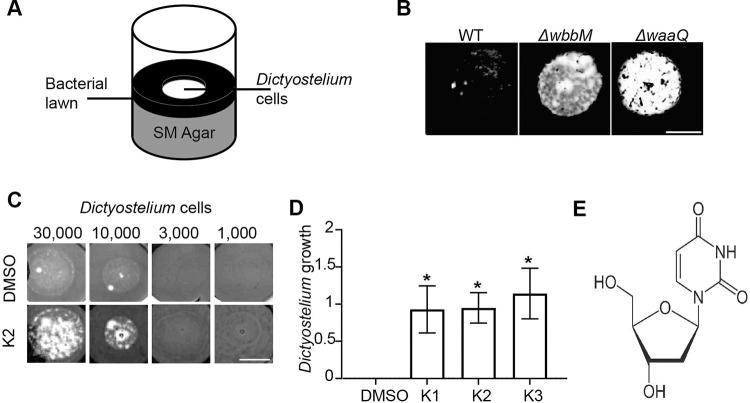
In order to identify compounds that perturb the interaction between K. pneumoniae and host cells, we used a simple assay to visualize growth of phagocytic D. discoideum cells on a lawn of K. pneumoniae. K. pneumoniae bacteria were deposited on Standard Medium nutrient agar, then 10,000 D. discoideum cells were deposited in the center of the well, and allowed to grow for 10 days (Fig 1A). As previously described, wild-type (WT) D. discoideum created a phagocytic plaque in the bacterial lawn [11]. On the contrary, phg1A KO cells were virtually unable to grow on a lawn of WT K. pneumoniae (Fig 1B). Genetic inactivation of LPS synthesis genes (waaQ or wbbM) decreased the virulence of K. pneumoniae and concomitantly restored growth of D. discoideum cells (Fig 1B). This observation indicates that this assay can detect a decrease in the virulence of K. pneumoniae bacteria. The same assay was then repeated adding to each well a test compound at a final concentration of 30 μM, to screen a collection of 1,099 mostly FDA-approved compounds. On the first round of screening 14 compounds were selected, and after re-testing, 3 confirmed hits, named K1 to K3, were selected for further studies (S2 Fig in S1 File). The three compounds were reordered and their effect was confirmed.
Fig 1. Three compounds affect the interaction between K. pneumoniae and phagocytic amoebae.
A. D. discoideum cells were deposited on a lawn of K. pneumoniae and allowed to form a phagocytic plaque for 10 days. B. Phg1A KO cells failed to grow on WT bacteria, but they grew readily on K. pneumoniae mutants with decreased virulence (ΔwaaQ, ΔwbbM) (scale bar: 4mm). C. K2 increased the ability of phg1a KO cells to create phagocytic plaques in comparison with the negative control (DMSO) (scale bar: 4mm). D. The effect of each compound was scored from 4 (visible growth of 1,000 cells) to 0 (no growth of 30,000 cells) and the score of the negative control (DMSO) substracted. In this scale, the result shown in Fig 1C would score as a 0 for DMSO, and 2 for K2. Repeated experiments showed a high variability, but a significant effect for all three selected compounds (mean ± SEM; *: p<0.05; Kruskal-Wallis test, Dunn’s test. DMSO, K2: N = 10; K1, K3: N = 7 independent experiments). The original uncontrasted pictures are shown in S1 Fig in S1 File. E. Chemical structure of the K2 compound.
We then retested each of the confirmed hits in a more quantitative manner, depositing on a lawn of K. pneumoniae an increasing number of phg1A KO D. discoideum cells, from 1,000 to 30,000, and the growth of D. discoideum was scored in each experiment (Fig 1C). A score of 0 indicates that no growth of D. discoideum was observed, suggesting that the K. pneumoniae bacteria are virulent. On the contrary, a score of 4 indicates that even 1,000 cells were sufficient to create a phagocytic plaque in the bacterial lawn, suggesting that the virulence of K. pneumoniae bacteria was strongly decresased. In repeated experiments, all three compounds increased reproducibly and comparably the ability of D. discoideum cells to grow in the presence of K. pneumoniae (Fig 1D). The chemical structure of the K2 compound, on which this study is focused, in shown (Fig 1E).
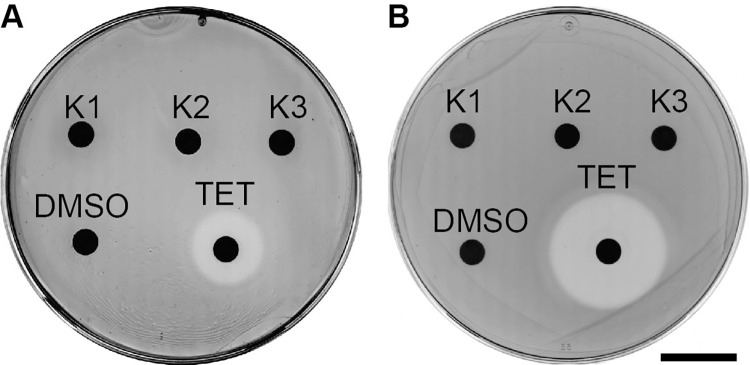
One trivial possibility would be that the selected compounds inhibit the growth of K. pneumoniae, and that this would account for the increased ability of D. discoideum cells to create a plaque in the thinner bacterial lawn. To assess this possibility, we tested directly the ability of each compound to inhibit growth of K. pneumoniae. For this, each compound was deposited on a disc at the surface of an LB-agar plate seeded with K. pneumoniae bacteria. Tetracycline was used as a positive control: it inhibited growth of K. pneumoniae as revealed by a halo of growth inhibition around the site of deposition (Fig 2). None of the selected compounds inhibited bacterial growth in this assay neither in LB (Fig 2A) nor in Standard Medium (Fig 2B).
Fig 2. Selected compounds exhibit no antibiotic activity against K. pneumoniae.
K. pneumoniae were plated on LB- (A) or SM- (B) agar plates. Paper discs with 20 μl of a 10 mM DMSO stock solution of each compound were then placed on the agar and the bacteria allowed to grow. After an overnight incubation at 25°C, a halo of bacterial growth inhibition was observed around a disk containing tetracycline (TET), but none of the selected compounds showed a similar effect (scale bar: 2 cm).
K2 facilitates the intracellular killing of K. pneumoniae by D. discoideum cells
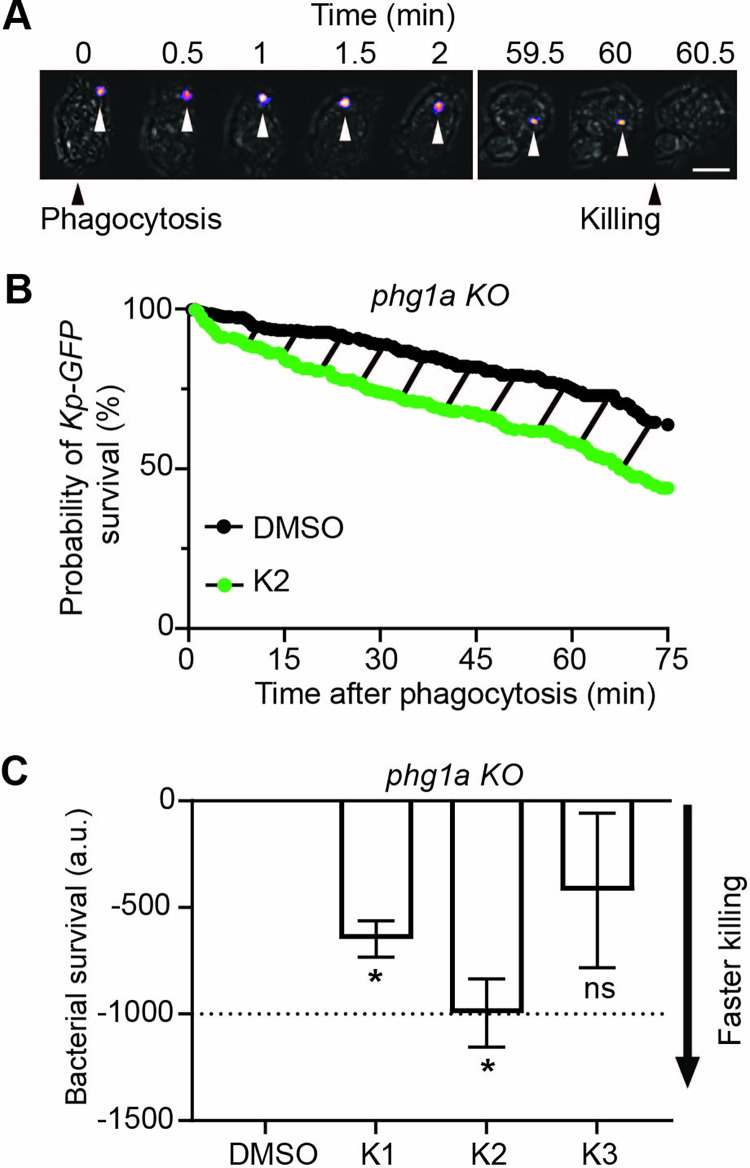
The ability of D. discoideum cells to grow on a lawn of bacteria can be modulated in a number of ways, such as an change in phagocytic uptake, intracellular killing, or phagocyte motility. We first tested if selected compounds restored at least partially the ability of phg1A KO cells to kill ingested K. pneumoniae. For this, we measured intracellular killing of GFP-expressing K. pneumoniae by assessing fluorescence extinction following phagocytosis. Previous experiment have shown that the abrupt extinction of GFP fluorescence provides a good estimate of the moment at which ingested bacteria are killed [19]. Using this method, bacterial fluorescence is extinguished within a few minutes after phagocytosis in WT cells [19]. On the contrary, and in agreement with previous observations [11], ingested bacteria remained alive for a long time in phg1A KO cells: a typical example is shown, where bacterial killing occurred 60 min after ingestion in phg1A KO cells (Fig 3A). Multiple ingestion/killing events were recorded (30 per independent experiment), and the survival of bacteria following ingestion was determined: less than 50% of bacteria were killed within the first hour following their ingestion in phg1A KO cells (Fig 3B; N = 6 independent experiments, n = 180 ingested bacteria). When bacteria and cells were treated with K2, ingested bacteria were killed faster than in the absence of compound. To determine the statistical relevance of these observations, in each independent experiment, the area under the bacterial survival curve was determined, and the control value (DMSO) was substracted (dashed area in Fig 3B). A value below zero corresponds to a killing that is faster in the presence of the compound than in the control condition. The K1 and K2 compounds accelerated intracellular killing of K. pneumoniae, and the effect was strongest for K2 (Fig 3C). Consequently the rest of this study was focused on the K2 compound.
Fig 3. K2 increases the intracellular killing of K. pneumoniae by phg1a KO cells.
A. Time-lapse images showing one representative example of a fluorescent K. pneumoniae ingested by a D. discoideum phg1a KO cell. The phase contrast and fluorescence pictures were superimposed, and the position of the fluorescent bacteria indicated with an arrowhead. Time 0 is defined as the time when the D. discoideum cell engulfs the bacteria (Phagocytosis). In this example, extinction of fluorescence was observed 60 minutes after ingestion (Killing) (scale bar: 6μm). Only the essential time points are shown, showing the moment when the bacteria was phagocytosed (t = 0), and when their fluorescence was lost (t = 60min). B. Survival of K. pneumoniae (%, Kaplan-Meier estimator) ingested by phg1a KO cells was decreased in the presence of the compound K2 (green) compared with the DMSO control (black). These two curves were obtained by combining the results of 6 independent experiments (total 180 bacteria for each condition). The dashed area represents the difference between the two survival curves. C. The Kaplan-Meier survival curves were determined in multiple independent experiments (30 bacteria for each condition) for the three tested compounds (30μM). The area under the curve (AUC) for each compound (K1-K3) and the control (DMSO) was determined for each experiment over 75 min, and the difference (corresponding to the dashed area in Fig 3B) was calculated. A figure inferior to zero indicates that the killing was faster in the presence of the compound than in the control (DMSO) condition. Two compounds, K1 and K2, increased significantly the intracellular killing of K. pneumoniae by phg1a KO cells (mean ± SEM; *: p<0.05; Kruskal-Wallis test, Dunn’s test. K1, K3: N = 3; DMSO; K2: N = 11 independent experiments).
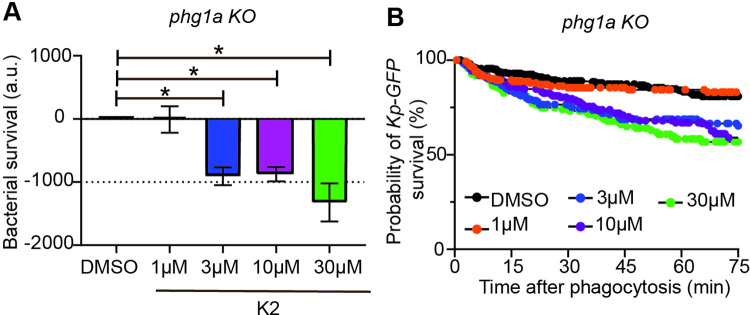
In order to characterize the mode of action of K2, we next tested its effect on intracellular killing over a range of concentrations. K2 stimulated intracellular killing at concentrations of 30 μM, 10 μM and 3 μM, while no effect was observed at 1 μM (Fig 4A). This result was also apparent when the cumulative survival curves of ingested bacteria were compared (Fig 4B).
Fig 4. K2 is active at a concentration of 3 μM and above.
A. The effect of K2 on the intracellular killing of K. pneumoniae was determined as described in the legend to Fig 3 at concentrations of K2 ranging from 1 to 30 μM. K2 increased intracellular killing of bacteria at 3 μM (blue), 10 μM (purple) and 30 μM (green) (mean ± SEM; *: p<0.05; Kruskal-Wallis test, Dunn’s test. DMSO, 3 μM and 10 μM: N = 6; 1 μM: N = 5; 30 μM: N = 4 independent experiments). B. The corresponding survival curves of ingested K. pneumoniae are shown.
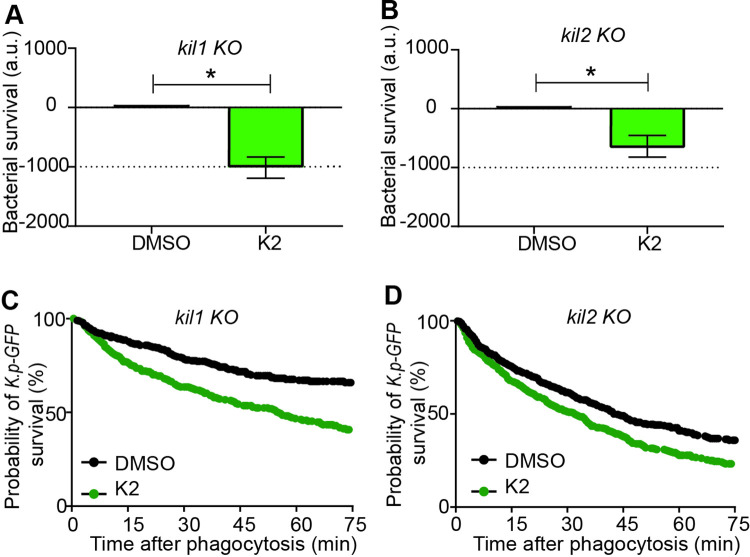
We also tested whether K2 facilitates intracellular killing of K. pneumoniae in other D. discoideum killing-deficient mutants. Indeed, K2 facilitated killing of K. pneumoniae in kil1 KO cells (Fig 5A and 5B) and, to a lesser extent, in kil2 KO cells (Fig 5C and 5D).
Fig 5. K2 increases intracellular killing of K. pneumoniae by kil1 KO and kil2 KO cells.
The effect of the K2 compound on K. pneumoniae intracellular killing was assessed after ingestion by kil1 KO cells (A) or by kil2 KO cells (B). K2 (green) increased significantly bacterial killing in both mutant cells (mean ± SEM; * p<0.05 Mann-whitney test, kil1 KO: N = 9; kil2 KO: N = 13 independent experiments). C, D. The corresponding survival curves of ingested K. pneumoniae in kil1 KO (C) and kil2 KO cells (D) are shown.
K. pneumoniae exposed to K2 are easier to kill
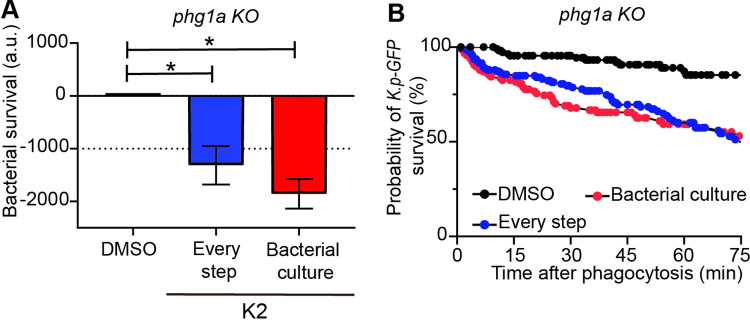
In all experiments up to this point, the compounds were added 16 hours before the experiment both to the K. pneumoniae culture, and to the D. discoideum culture. The compounds were also present when cells and bacteria were mixed and phagocytosis and killing recorded. K2 could in principle act either on bacteria by fragilizing K. pneumoniae, or on D. discoideum cells by stimulating intracellular killing mechanisms. To distinguish between these two possibilities, we measured intracellular killing of bacteria that had been grown in the presence of K2, but omitting K2 in the D. discoideum preculture and during the ingestion and killing of bacteria. In this setting, bacteria were washed twice before use and D. discoideum cells were not exposed to the K2 compound. Exposing bacteria to K2 during their growth was sufficient to facilitate their killing by D. discoideum cells (Fig 6). This result suggests that K2 acts primarily on K. pneumoniae, making it more susceptible to intracellular killing in D. discoideum phagosomes.
Fig 6. K2 treatment renders K. pneumoniae more susceptible to intracellular killing.
A. The effect of K2 on intracellular killing was assessed by adding it at different steps of the experimental process. K2 was added either at every step (bacterial overnight culture, D. discoideum overnight culture and during the ingestion and killing of bacteria; blue), or only in the overnight bacterial culture (red), and compared to a treatment with DMSO (mean ± SEM; *: p<0.05; Kruskal-Wallis test, Dunn’s test. N = 5 independent experiments). B. The corresponding survival curves with K2 added at every step (blue) or only during the bacterial preculture (red) are shown.
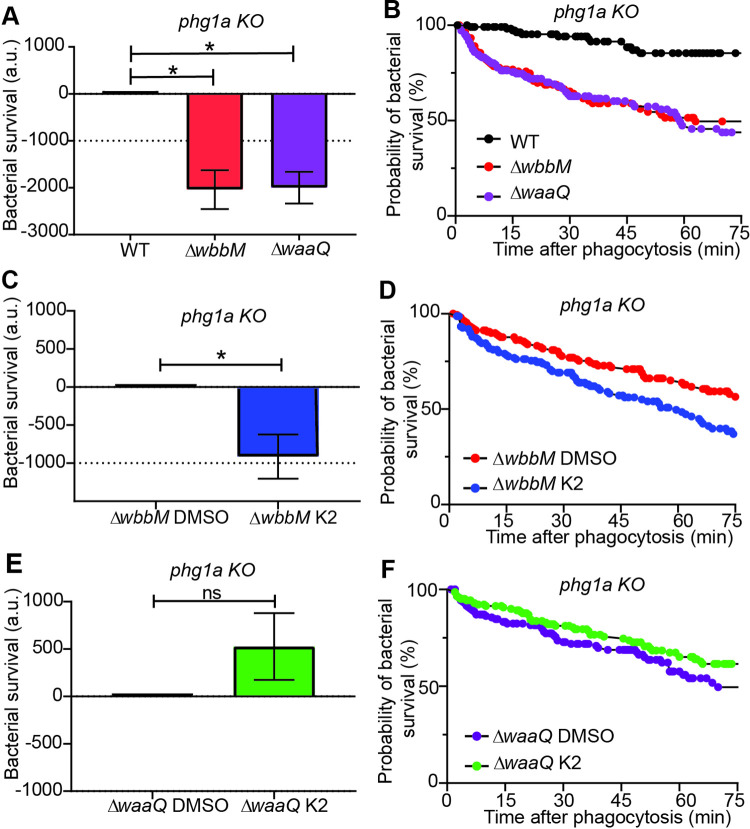
In order to better delineate the mode of action of the K2 compound, we tested its effect on K. pneumoniae mutants with altered LPS synthesis. The wbbM gene is necessary for O-antigen coupling to bacterial LPS, while waaQ participates in the inner core biosynthesis. Genetic inactivation of wbbM prevents the addition of O-antigens to the LPS core. Genetic inactivation of waaQ results in loss of the heptose III side branch of the LPS core, a structure critical for outer membrane stability [17]. Intracellular killing of ΔwaaQ or ΔwbbM K. pneumoniae by phg1A KO D. discoideum was greatly facilitated compared to killing of WT K. pneumoniae (Fig 7A, 7B). K2 further stimulated killing of ΔwbbM bacteria (Fig 7C, 7D), but not that of ΔwaaQ bacteria (Fig 7E, 7F). This observation indicates that when the LPS layer protecting the bacteria is altered by a ΔwaaQ genetic inactivation, no additional effect of K2 on bacterial survival is visible. As detailed below, this suggests that K2 may act by perturbing the protective effect of the outer bacterial layer which includes LPS, but may also include other elements.
Fig 7. Intracellular killing of K. pneumoniae mutants.
A. Intracellular killing of K. pneumoniae (WT: black, ΔwbbM: red, ΔwaaQ: purple) in phg1A KO D. discoideum was assessed as described in the legend to Fig 3 (mean ± SEM; *: p < 0.05; Mann-whitney test. ΔwbbM: N = 4; ΔwaaQ; N = 5 independent experiments) B. The corresponding survival curves of ΔwbbM (red), ΔwaaQ (purple) and WT K. pneumoniae (black) are shown. C. Exposure to K2 further increased the intracellular killing of ΔwbbM K.pneumoniae in phg1a KO cells (mean ± SEM; *: p < 0.05; Mann-whitney test. N = 5 independent experiments). D. The corresponding survival curves of ΔwbbM K. pneumoniae treated with DMSO (red) or K2 (blue) are shown. E. Exposure to K2 did not further increase intracellular killing of ΔwaaQ K. pneumoniae in phg1a KO cells (mean ± SEM; Mann-whitney test. N = 5 independent experiments). F. The corresponding survival curves of ΔwaaQ K. pneumoniae treated with DMSO (purple) or K2 (green) are shown.
Intracellular killing of bacteria can be at least partly reproduced in vitro by exposing bacteria to extracts from D. discoideum cells and following bacterial lysis [20]. Extracts from phg1A KO cells lysed bacteria less rapidly than extracts from WT D. discoideum (S3 Fig in S1 File). When bacteria were grown in the presence of K2, bacterial lysis by D. discoideum extracts was accelerated (S3 Fig in S1 File). This result reinforces the notion that the main effect of K2 is to make K. pneumoniae bacteria more succeptible to attack by cellular antibacterial mechanisms.
K2 treatment disrupts the protective effect of the LPS layer in K. pneumoniae
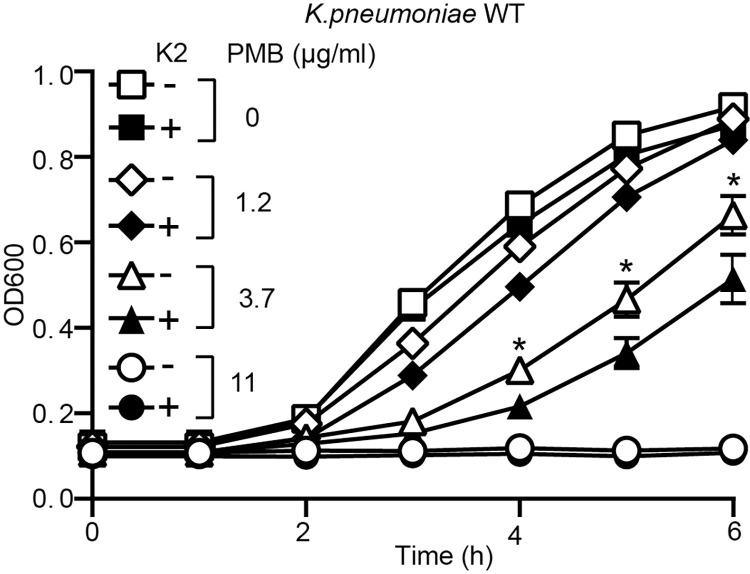
Polymyxin B kills bacteria by disrupting their membranes [21, 22]. To be effective polymyxins need first to cross the protective layer formed by bacterial LPS. Consequently, polymyxins inhibit more efficiently growth of bacteria exhibiting a disorganized LPS layer. A standard bacterial growth inhibition assay failed to reveal any synergistic effect between K2 and polymyxin B or other antibiotics (S4 Fig in S1 File), however this assay may fail to detect subtle effects of K2 on bacterial growth kinetics. We thus measured growth of K. pneumoniae in LB containing increasing concentrations of polymyxin B (Fig 8). In the absence of K2, a concentration of 11 μg/ml of polymyxin B fully inhibited growth of K. pneumoniae, and polymyxin B at 3.7 μg/ml and 1.2 μg/ml partially inhibited bacterial growth. K2 alone (30μM) failed to inhibit bacterial growth. However, when both polymyxin B (3.7 μg/ml) and K2 were added to bacteria, the bacterial growth was slower than when only polymyxin B was used (Fig 8). When a similar experiment was performed with tetracyclin instead of polymyxin B, K2 did not potentiate the effect of tetracycline on bacterial growth (S5 Fig in S1 File). These results suggest that exposure to K2 disorganizes the LPS layer of K. pneumoniae bacteria, making them more sensitive to polymyxin B.
Fig 8. K2 increases the sensitivity of K. pneumoniae to polymyxin B.
WT K. pneumoniae were grown overnight in the presence or absence of K2. The bacteria were then diluted and their growth was assessed for 6 h in the continued presence or absence of K2 and in the presence of increasing concentrations of polymyxin B (PMB: 0-11 μg/ml). While exposure to K2 did not alter the growth of K. pneumoniae, it increased its sensitivity to polymyxin B as evidenced by a slower growth in the presence of 1.2 or 3.7 μg/ml of PMB (mean ± SEM; *: p < 0.05; Mann-whitney test. PMB 3.7 μg/ml: N = 12 independent experiments).
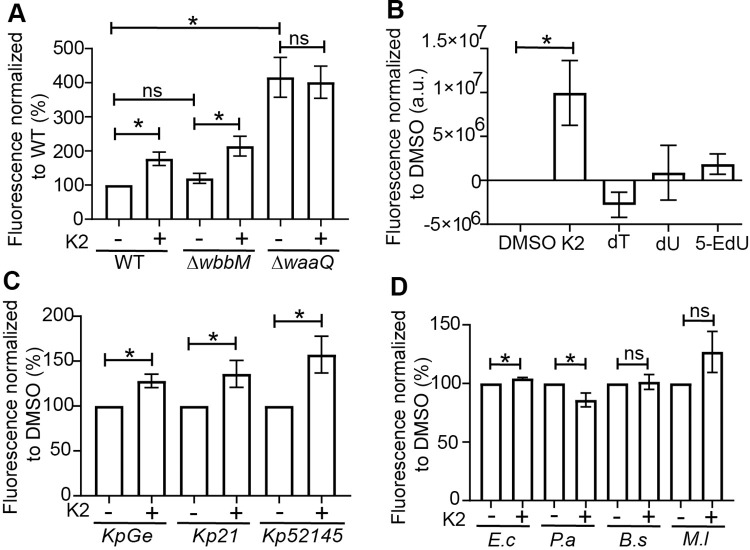
In order to evaluate directly the protective effect of the LPS layer in different strains and conditions, we measured the accessibility of the bacterial membrane using the hydrophobic fluorescent probe 1-N-phenylnaphthylamine (NPN). When NPN inserts in the outer lipidic membrane of bacteria, its fluorescence strongly increases [23]. In WT K. pneumoniae, the LPS layer limits the accessibility of the outer membrane and largely excludes NPN (Fig 9A). Genetic inactivation of waaQ significantly increased access of NPN to the outer membrane, while genetic inactivation of wbbM did not (Fig 9A). K2 increased outer membrane accessibility in WT and ΔwbbM, but not in ΔwaaQ K. pneumoniae (Fig 9A). This effect was seen at concentrations of K2 (3μM and above) similar to those required to render K. pneumoniae easier to kill (S6A Fig in S1 File), and was detectable after 8 h of growth of bacteria in the presence of K2 (S6B Fig in S1 File). Three close chemical analogs of K2 were also tested in this assay (S2 Fig in S1 File) and did not modify outer membrane accessibility (9b Fig), indicating that the action of K2 is highly specific. An increased accumulation of NPN can in principle be caused by an increase in bacteria outer membrane accessibility, or by an inhibition of multi-drug resistance pumps [24]. Indeed outer membrane accessibility was increased by CCCP, an inhibitor of multi-drug resistance pumps, but the effects of CCCP and NPN were additive (S7 Fig in S1 File), suggesting that NPN does not act by inhibiting multi-drug resistance pumps.
Fig 9. K2 treatment increases access of a hydrophobic probe to the bacterial membrane of K. pneumoniae.
A. In order to assess the efficacy with which the LPS shielded the bacterial membrane, bacteria were exposed to the fluorescent probe 1-N-phenylnaphthylamine (NPN), and fluorescence was recorded, providing a measure of the insertion of NPN in the bacterial membrane. The membrane of ΔwaaQ mutant K. pneumoniae was more accessible to NPN than that of WT and ΔwbbM mutant. In K2-treated bacteria, access of NPN increased in WT and in ΔwbbM but not in ΔwaaQ bacteria (mean ± SEM; *: p<0.05; Kruskal-Wallis test; N = 5 independent experiments). B. Three chemical analogs of K2 (dT = deoxythymidine, dU = deoxyuridine and 5-EdU = 5-Ethynyl-2’-deoxyuridine) were tested for their ability to increase the membrane accessibility of NPN. K2 was the only compound that increased significantly the accessibility of the outer membrane of K. pneumoniae to NPN (mean ± SEM; *: p<0.05; Kruskal-Wallis test; DMSO, K2: N = 6; and N = 5 independent experiments for the three analogs of K2). C, D. The effect of K2 was tested on three different strains of K. pneumoniae (KpGE, Kp21 and Kp52145) as well as Escherichia coli (E.c), Pseudomonas aeruginosa (P.a), Bacillus subtilis (B.s) and Microccocus luteus (M.l). K2 increased NPN incorporation in K. pneumoniae, but exhibited little or no effect on other bacteria. (mean ± SEM; *: p<0.05; Mann-whitney test. N = 8 independent experiments).
In order to characterize the spectrum of action of K2, we also determined whether it affected outer membrane accessibility in other strains of bacteria. In addition to the non-virulent KpGE strain of K. pneumoniae used in this study, we tested two virulent strains of K. pneumoniae (Kp21and Kp52145), as well as two other Gram-negative (Escherichia coli, Pseudomonas aeruginosa) and two Gram-positive (Micrococcus luteus, Bacillus subtilis) bacterial strains. K2 increased the accessibility of the outer membrane for the three K. pneumoniae strains tested (Fig 9C), but did not show a significant effect on the other bacteria tested (Fig 9D).
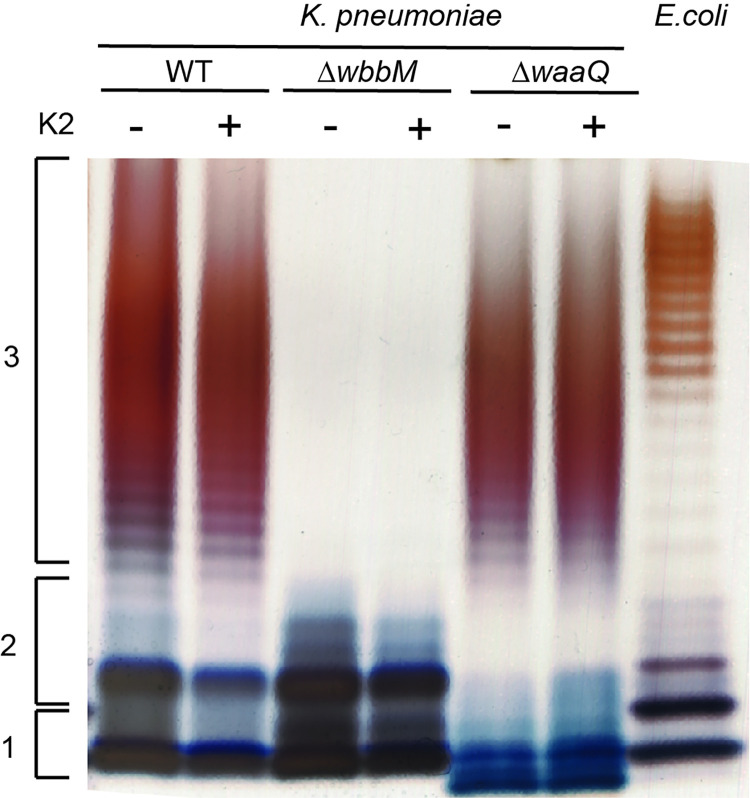
Finally, we assessed directly the effect of K2 on the structure of bacterial LPS. For this, we purified LPS from K. pneumoniae and analyzed its structure by SDS-polyacrylamide gel electrophoresis (Fig 10). LPS purified from WT K. pneumoniae exhibited a lipid A anchor, coupled to an oligosaccharide core, and to a repetitive glycan polymer forming the O-antigen. Incomplete structures composed of only lipid A, or lipid A coupled to the oligosaccharide core, were visible (Fig 10). As expected, LPS from ΔwbbM K. pneumoniae was devoid of the O-antigen, and LPS from ΔwaaQ exhibited an incomplete (smaller) oligosaccharide core that migrated faster in the gel (Fig 10). Upon treatment of bacteria with K2, these LPS profiles remained unchanged (Fig 10). We also visualized the bacterial envelope by electron microscopy, and exposure to K2 did not visibly modify the structure of the bacterial envelope (S8 Fig in S1 File). These observations indicate that K2 does not affect the structure of the bacterial envelope in a manner detectable with these techniques.
Fig 10. K2 treatment does not visibly affect LPS structure.
Bacteria were grown overnight in the presence or absence of K2. LPS from K. pneumoniae WT and mutant strains (ΔwbbM, ΔwaaQ) were then purified and analyzed by SDS-PAGE electrophoresis and silver staining. LPS extracted from E. coli (serotype O111:B4) was used for comparison. The LPS of WT K. pneumoniae showed the three main forms of the LPS: lipid A (1), lipid A+ oligosaccharide core (2), lipid A + core + O-antigen [27]. As expected, the LPS from ΔwbbM bacteria lacked the O-antigen, and ΔwaaQ displayed a smaller oligosaccharide core that migrated further in the gel. No visible alteration of LPS structure was observed in K2-treated bacteria.
Discussion
In this study, we identified three compounds that modify the interaction between K. pneumoniae bacteria and phagocytic D. discoideum cells. We then characterized in more detail the mode of action of one of them, K2, which increases the intracellular killing of K. pneumoniae. There are in principle two ways by which intracellular killing can be facilitated: either by stimulating the intracellular killing mechanisms, or by rendering bacteria more susceptible to intracellular killing. K2 clearly falls into the second category, since bacteria grown in the presence of K2 are more easily killed when they are ingested by D. discoideum, as well as when they are exposed to D. discoideum extracts in vitro. In bacteria treated with K2, the outer membrane is less protected from outer agents as revealed by their increased sensitivity to polymyxin B and by the observation that their outer membrane is more accessible to a hydrophobic fluorescent probe. On these four counts (faster intracellular killing in vivo and in vitro, increased sensitivity to polymyxin B, increased accessibility of bacterial membrane) the phenotype of K2-treated bacteria is similar to that produced by genetic inactivation of waaQ, which alters the structure and function of the LPS layer. The structure of LPS was however not visibly altered in K2-treated bacteria. We conclude that K2 acts by rendering the bacterial membrane more accessible to antibacterial agents, but our results do not indicate that K2 alters the structure of LPS, and do not identify the primary target of the K2 compound. Many antibacterial agents bind to LPS and increase the accessibility of the outer membrane, notably cationic agents, ion chelators, or polymyxins [25, 26]. This is however unlikely to be the case for K2, since its effect only appeared after prolonged (8h) exposure of bacteria to K2. K2 is an analog of thymidine. It may affect genome replication or gene expression and indirectly impact the organization of the bacterial envelope. Further studies will be necessary to determine by which molecular mechanism K2 modifies the properties of bacterial envelope.
In principle a compound affecting the resistance of bacteria to intracellular killing could limit the ability of K. pneumoniae to create virulent infections in patients. In this respect, it is worth noting that ΔwaaQ and ΔwbbM mutant K. pneumoniae strains completely lost the ability to mount a lung infection in mice [11], although their intracellular killing was only partially restored in phg1A KO D. discoideum cells. This observation suggests that even a small increase in the sensitivity of K. pneumoniae bacteria to intracellular killing can strongly diminish their capacity to infect patients.
K2 is 5-ethyl-2’-deoxyuridine, also known as edoxudine. It was used until 1998 as a topical antiviral drug, to treat genital herpes simplex infections [27] and its properties have been carefully studied. It inhibits viral replication in vitro at concentrations ranging from 5 to 50 μM [28]. When administered orally or intravenously, concentrations readily reach values higher than 10 μM in the blood as well as in the lungs [29, 30]. Since K2 decreases K. pneumoniae virulence at a concentration of 3 μM, we speculate that it could be used to treat K. pneumoniae infections. This would be particularly useful when faced with bacteria resistant to multiple antibiotics against which therapeutic options are currently very limited. K. pneumoniae infections can cause severe necrotizing skin infections, often resistant to classical antibiotics, and sometimes fatal [31]. Because K2 was initially approved as a topical drug, it may be used in such situations, possibly as a complement to classical antibiotic treatments. Repurposing of approved drugs to treat bacterial infections is an interesting strategy because information is already available about pharmacological and toxicological profiles in preclinical and clinical studies [32]. In this perspective it is encouraging to find that K2 increases the outer membrane accessibility of three different strains of K. pneumoniae, including two well-characterized pathogenic strains. Detailed studies will however be necessary to determine if K2 acts on pathogenic K. pneumoniae strains in infected patients.
Experimental procedures
Cells and reagents
All D. discoideum cells used in this study were derived from the parental DH1-10 strain [33], referred to as wild-type (WT). Phg1a KO, kil1 KO and kil2 KO cells were described previously [11, 16]. D. discoideum cells were grown in HL5 medium [34] at 21°C. When indicated, D. discoideum cells were grown on a lawn of K. pneumoniae, as previously detailed [34].
The parental K. pneumoniae strain used in this study is the sequenced non-pathogenic laboratory strain KpGE strain [15]. It was grown in LB (lysogeny broth) at 37°C. ΔwaaQ and ΔwbbM mutants of KpGE were described previously [11]. We used for screening a previously described collection of 1,099 compounds [35]. This collection comprises the 1,040 bioactive compounds of the NINDS custom collection 2 (Microsource Inc., Gaylordsville, CT), completed with 59 locally selected compounds. Three quarters of the compounds in the collection are FDA-approved. The three compounds selected (K1: CAS 138-14-7; K2: CAS 15176-29-1; K3: CAS 58-22-0) were re-ordered from Merck (Darmstadt, Germany).
Screening for inhibitors of bacterial virulence
Inhibitors of bacterial virulence were tested as previously described [34]. Briefly, in a 24-well plate containing 2 ml Standard Medium-agar (for 1 L: 10 g bacteriological peptone, 1 g bacto yeast extract, 2.2 g KH2PO4, 1 g K2HPO4, 1 g MgSO4:7H2O, 20% glucose), a 20 μl droplet of each compound was deposited in each well to a final concentration of 30 μM. KpGE bacteria (50 μl of overnight culture) were then added in each well and allowed to dry in a sterile cell culture hood for 2 h. D. discoideum phg1a KO cells (30,000, 10,000, 3,000, or 1,000 cells) were applied onto the bacterial lawn. After ten days, the plates were scanned with an Epson Perfection V850 Pro scanner. phg1a KO cells created phagocytic plaques (white) on the bacterial lawn (black) only when a compound facilitated their growth (Fig 1A). The growth of phg1a KO cells was scored from 4 (efficient growth of 1,000 cells) to 0 (no growth of 30,000 cells).
Antibiotic activity of compounds
An overnight bacterial culture of K. pneumoniae (300 μl) was spread on a Petri dish containing LB-agar. Paper discs with 20 μl of a 10 mM DMSO stock solution of each compound were then placed on the plates and the bacteria allowed to grow at 25°C overnight. Inhibition of bacterial growth around the disc reveals the antibiotic activity of compounds. DMSO was used as a negative control and tetracyclin (12 mg/ml) as a positive control.
To detect a putative additive effect of compounds when combined with antibiotics, bacteria were grown overnight with or without 30 μM of K2. K. pneumoniae bacteria (500 μl culture, 109 cfu / mL) were then spread on a Petri dish containing LB-agar (with or without 30 μM of K2 added in the LB-Agar). Antibiotic discs were then deposited on the plates and bacteria were allowed to grow at 37°C overnight. Inhibition of bacterial growth around the disc revealed antibiotic activities.
To detect synergistic effect of polymyxin B and K2 on bacterial growth, bacteria were grown 16 h at 37°C in a shaken suspension of 2 ml LB containing 6 μl of DMSO with or without K2 (final concentration 30 μM). To test if K2 increases the susceptibility of K. pneumoniae to the antibiotic activity of polymyxin B or tetracycline, 50 μl of DMSO/K2-treated bacteria were transferred to 2 ml of LB supplemented with DMSO or K2. A range of concentrations of polymyxin B or tetracycline was also added to the culture. The cultures were grown at 37°C in a shaken suspension. An aliquot of 150 μl was taken every hour for 6 hours and placed in a 96-well plate (cell culture microplate, PS, F-Bottom, black, Greiner bio-one). The bacterial growth was determined by measuring the OD600nm with a plate reader (SpectraMax Paradigm from Molecular Devices, SoftMaxPro 7.0.)
Intracellular killing of bacteria
To measure intracellular killing of ingested bacteria, GFP-expressing K. pneumoniae were grown 16 h at 37°C, as previsouly described [19]. The culture was washed twice in phosphate buffer (PB)-sorbitol (2 mM Na2HPO4, 14.7 mM KH2PO4, 100 mM sorbitol, pH 6.0) then resuspended in 1 ml PBS. The bacterial suspension was diluted 200 times in PBS, and 150 μL were mixed with 230,000 D. discoideum cells in a final volume of 250 μL PB-sorbitol, and deposited on a glass slide (μ-slide 8 well glass bottom, Ibidi GmbH). When indicated, 30 μM of compound or DMSO (0.3%) was added to each well. The cells were allowed to settle for 10 min, then imaged every 30 sec for 2 h with a Nikon Eclipse Ti2. At each time point, one picture (phase contrast and GFP fluorescence) was taken in five successive focal planes (step size 3 μm) to image the whole cell volume. ImageJ was used to analyze movies. Bacterial fluorescence decreased abruptly (typically a >90% drop within 30 sec), and this event indicated the time when the bacteria was killed. For each bacteria analysed (30 per independent experiment) the time of phagocytosis and the time of killing were recorded. Survival of phagocytosed fluorescent bacteria was computed using the Kaplan–Meier estimator. Statistical analysis was done using GraphPad Prism (V8.1.0). For each condition, at least three independent experiments were performed.
Bacterial lysis in vitro
Bacterial lysis in vitro was assessed as previously described [20]. Briefly, D. discoideum cells were washed twice in phosphate buffer (PB: 2 mM of Na2HPO4 and 14.7 mM of KH2PO4, pH 2.0) and lysed in 800 μL of Lysis buffer (50 mM of sodium phosphate buffer, pH2, 0.5% Triton X‐100) containing protease inhibitors (20 μg/mL of leupeptin, 10 μg/mL of aprotinin, 18 μg/mL of phenylmethylsulfonyl fluoride (PMSF) and 1.8 mg/mL of iodoacetamide (IAA)). The suspension was centrifuged (30,000 g for 60 min at 4°C), and the supernatant was collected and serially in Lysis buffer. Bacteriolytic activity was assessed by mixing in a microtiter plate 100 μL of cell extract with 100 μL of an overnight bacterial culture (grown in the presence or absence of 30 μM K2) washed once in sodium phosphate buffer (50 mM sodium phosphate buffer, pH2) and resuspended in the same buffer to a final optical density (450 nm) of 0.5. The decrease in turbidity (optical density at 450 nm) for 40 min at 21°C was measured with a plate reader (SpectraMax Paradigm from Molecular Devices, SoftMaxPro 7.0.).
Bacterial outer membrane accessibility
The fluorescent probe 1-N-phenylnaphthylamine (NPN; Sigma-Aldrich) was used to assess the accessibility of the outer membrane of bacteria as described previously [23]. The stock solution of NPN was prepared in acetone (0.5 mM), kept at room temperature and used within a week. The stock solution was diluted in HEPES buffer (5 mM, pH 7.2), to a concentration of 40 μM. Bacteria were grown overnight at 37°C in 2 ml LB with or without 30 μM of K2 and reached an OD600nm of 5. The culture was diluted in 9 volumes of HEPES buffer, then 1ml was centrifuged 10 min at 10,000 rpm. Finally, the bacterial pellet was resuspended in 300 μl HEPES buffer and used as described below. For a standard experiment, a 96-well plate (cell culture microplate, PS, F-Bottom, black, Greiner bio-one) was filled with (a) HEPES buffer (200 μl), (b) HEPES buffer (150 μl) + NPN 40 μM (50 μl), (c) HEPES buffer (100 μl) + DMSO/K2-treated bacterial suspension (100 μl), or (d) HEPES buffer (50 μl) + NPN 40 μM (50 μl) + DMSO/K2-treated bacterial suspension (100 μl). The bacterial suspensions were added last, just before measuring fluorescence (360nm excitation, 405nm emission) with a microplate reader. For each well, the flurorescence (arbitrary units; a.u.) was divided by the OD600nm to obtain the corrected fluorescence value.
To assess when the effect of K2 on the bacterial outer membrane accessibility appeared, bacteria were grown 16 h at 37°C in a shaken suspension of 3 ml LB. Bacteria (500 μl) were transferred in LB (20 ml supplemented with 60 μl DMSO or K2). The cultures were grown at 37°C in a shaken suspension for 24 h and aliquots were taken at the indicated times. Each aliquot was transferred in a well (OD600nm of 0.2) and NPN incorporation measured as described above.
The efflux pump inhibitor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) was obtained from Sigma-Aldrich. The experimental procedure to measure the accessibility of the outer membrane was slightly adapted: bacteria resuspended in 300 μl HEPES were treated (or not) with 100 μM of CCCP for 10 min at room temperature, centrifuged 10 min at 10,000 rpm and the pellet resuspended in 300 μl HEPES. The suspension was then used as described above in a 96-well plate to measure NPN incorporation.
LPS purification
K. pneumoniae (KpGE) bacteria (WT, ΔwbbM, ΔwaaQ) were grown for 16 h at 37°C in a shaken suspension of 2 ml LB. As indicated 6 μl of DMSO or K2 (final concentration 30 μM) was added. The Intron Biotechnology kit was used to extract the LPS following manufacturer’s instructions. Briefly, a first lysis step with an organic solution containing phenol was used to disrupt the phospholipid and protein components of the cell membrane. Chloroform was then applied afterward to isolate RNA and genomic DNA/protein. LPS were precipitated at a high salt concentration, washed with 70% EtOH, and resuspended in 10mM Tris-HCl buffer (pH 8.0). Finally, LPS were separated in a 4-15% Mini-PROTEAN® TGX™ Precast protein gel from BioRad then silver-stained. First, the gel was fixed with a fixation solution (40% ethanol, 10% acetic acid, 50% water) for 30 minutes. Then, the gel was incubated overnight with a solution containing 30% ethanol, 260 μl glutaraldehyde (50%), 20% incubation solution (sodium acetate trihydrate 34% and sodium thiosulfate pentahydrate 1%) and 50% water. After the overnight incubation step, the gel was washed three times 5 min in water and exposed 40 min to the silver nitrate solution with 92 μl formaldehyde (36%). Finally, the development solution was applied (sodium carbonate solution and 10 μl formaldehyde 36%) until the appearance of the LPS profiles. A stop solution containing disodium EDTA dehydrate was added 15 s later.
Electron microscopy
To study bacterial morphology by electron microscopy, bacteria were treated overnight with DMSO or 30 μM K2 and then were fixed in 2% glutaraldehyde (1 h at room temperature) and post-fixed in 2% osmium tetroxide (1 h at 4°C), dehydrated and embedded in Epon resin and processed for conventional electron microscopy as previously described [36].
Data availability
All data is available within the manuscript. Unprocessed experimental data is available online on a dedicated server at the following address: https://doi.org/10.26037/yareta:oqmmzyw6crhqtmxfsjhoe65cwa.
Supporting information
(ZIP)
(DOCX)
Abbreviations
- PMB
Polymyxin B
- K2
5-ethyl-2’-deoxyuridine = Edoxudine
- 5-EdU
5-ethynyl-2’-deoxyuridine
- dT
deoxythymidine
- dU
deoxyuridine
- DMSO
Dimethyl sulfoxide
- NPN
1-N-phenylnaphthylamine
- LPS
lipopolysaccharides
Data Availability
**PA at Accept: please follow up with author for experimental data** All processed data is available in the manuscript. All unprocessed experimental data will be made available online on a new dedicated server of our University upon publication.
Funding Statement
This work was supported by Swiss National Science Foundation (SNF) (“Sinergia” grant CRSI33_130016 to PC, TS, HH, LS; 31003A_172951 to PC). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Agyeman AA, Bergen PJ, Rao GG, Nation RL, Landersdorfer CB. 2019. A systematic review and meta-analysis of treatment outcomes following antibiotic therapy among patients with carbapenem-resistant Klebsiella pneumoniae infections. Int J Antimicrob Agents 55:105833. doi: 10.1016/j.ijantimicag.2019.10.014 [DOI] [PubMed] [Google Scholar]
- 2.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. 2007. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov 6:29–40. doi: 10.1038/nrd2201 [DOI] [PubMed] [Google Scholar]
- 3.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27. doi: 10.1038/nrmicro1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cosson P, Zulianello L, Join-Lambert O, Faurisson F, Gebbie L, Benghezal M,. 2002. Pseudomonas aeruginosa virulence analyzed in a Dictyostelium discoideum host system. J Bacteriol 184:3027–33. doi: 10.1128/JB.184.11.3027-3033.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swart AL, Harrison CF, Eichinger L, Steinert M, Hilbi H. 2018. Acanthamoeba and Dictyostelium as Cellular Models for Legionella Infection. Front Cell Infect Microbiol 8:61. doi: 10.3389/fcimb.2018.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cardenal-Munoz E, Barisch C, Lefrancois LH, Lopez-Jimenez AT, Soldati T. 2017. When Dicty Met Myco, a (Not So) Romantic Story about One Amoeba and Its Intracellular Pathogen. Front Cell Infect Microbiol 7:529. doi: 10.3389/fcimb.2017.00529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bozzaro S. 2019. The past, present and future of Dictyostelium as a model system. Int J Dev Biol 63:321–331. doi: 10.1387/ijdb.190128sb [DOI] [PubMed] [Google Scholar]
- 8.Alibaud L, Kohler T, Coudray A, Prigent-Combaret C, Bergeret E, Perrin J, et al. 2008. Pseudomonas aeruginosa virulence genes identified in a Dictyostelium host model. Cell Microbiol 10:729–40. doi: 10.1111/j.1462-5822.2007.01080.x [DOI] [PubMed] [Google Scholar]
- 9.Benghezal M, Adam E, Lucas A, Burn C, Orchard MG, Deuschel C, et al. 2007. Inhibitors of bacterial virulence identified in a surrogate host model. Cell Microbiol 9:1336–42. doi: 10.1111/j.1462-5822.2006.00877.x [DOI] [PubMed] [Google Scholar]
- 10.March C, Cano V, Moranta D, Llobet E, Perez-Gutierrez C, Tomas JM, et al. 2013. Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS One 8:e56847. doi: 10.1371/journal.pone.0056847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, et al. 2006. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol 8:139–48. doi: 10.1111/j.1462-5822.2005.00607.x [DOI] [PubMed] [Google Scholar]
- 12.Sanders D, Borys KD, Kisa F, Rakowski SA, Lozano M, Filutowicz M. 2017. Multiple Dictyostelid Species Destroy Biofilms of Klebsiella oxytoca and Other Gram Negative Species. Protist 168:311–325. doi: 10.1016/j.protis.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcoleta AE, Varas MA, Ortiz-Severin J, Vasquez L, Berrios-Pasten C, Sabag AV, et al. 2018. Evaluating Different Virulence Traits of Klebsiella pneumoniae Using Dictyostelium discoideum and Zebrafish Larvae as Host Models. Front Cell Infect Microbiol 8:30. doi: 10.3389/fcimb.2018.00030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bravo-Toncio C, Alvarez JA, Campos F, Ortiz-Severin J, Varas M, Cabrera R, et al. 2016. Dictyostelium discoideum as a surrogate host-microbe model for antivirulence screening in Pseudomonas aeruginosa PAO1. Int J Antimicrob Agents 47:403–9. doi: 10.1016/j.ijantimicag.2016.02.005 [DOI] [PubMed] [Google Scholar]
- 15.Lima WC, Pillonel T, Bertelli C, Ifrid E, Greub G, Cosson P. 2018. Genome sequencing and functional characterization of the non-pathogenic Klebsiella pneumoniae KpGe bacteria. Microbes Infect 20:293–301. doi: 10.1016/j.micinf.2018.04.001 [DOI] [PubMed] [Google Scholar]
- 16.Lelong E, Marchetti A, Gueho A, Lima WC, Sattler N, Molmeret M, et al. 2011. Role of magnesium and a phagosomal P-type ATPase in intracellular bacterial killing. Cell Microbiol 13:246–58. doi: 10.1111/j.1462-5822.2010.01532.x [DOI] [PubMed] [Google Scholar]
- 17.Yethon JA, Heinrichs DE, Monteiro MA, Perry MB, Whitfield C. 1998. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J Biol Chem 273:26310–6. doi: 10.1074/jbc.273.41.26310 [DOI] [PubMed] [Google Scholar]
- 18.Guan S, Clarke AJ, Whitfield C. 2001. Functional analysis of the galactosyltransferases required for biosynthesis of D-galactan I, a component of the lipopolysaccharide O1 antigen of Klebsiella pneumoniae. J Bacteriol 183:3318–27. doi: 10.1128/JB.183.11.3318-3327.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jauslin T, Lamrabet O, Crespo-Yanez X, Marchetti A, Ayadi I, Ifrid E, et al. 2021. How Phagocytic Cells Kill Different Bacteria: a Quantitative Analysis Using Dictyostelium discoideum. mBio 12:e03169–20. doi: 10.1128/mBio.03169-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guilhen C, Lima WC, Ifrid E, Crespo-Yanez X, Lamrabet O, Cosson P. 2021. A new family of bacteriolytic proteins in Dictyostelium discoideum. Front Cell Infect Microbiol 10: 617310. doi: 10.3389/fcimb.2020.617310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock RE. 1997. Peptide antibiotics. Lancet 349:418–22. doi: 10.1016/S0140-6736(97)80051-7 [DOI] [PubMed] [Google Scholar]
- 22.Velkov T, Thompson PE, Nation RL, Li J. 2010. Structure--activity relationships of polymyxin antibiotics. J Med Chem 53:1898–916. doi: 10.1021/jm900999h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helander IM, Mattila-Sandholm T. 2000. Fluorometric assessment of gram-negative bacterial permeabilization. J Appl Microbiol 88:213–9. doi: 10.1046/j.1365-2672.2000.00971.x [DOI] [PubMed] [Google Scholar]
- 24.Misra R, Morrison KD, Cho HJ, Khuu T. 2015. Importance of Real-Time Assays To Distinguish Multidrug Efflux Pump-Inhibiting and Outer Membrane-Destabilizing Activities in Escherichia coli. J Bacteriol 197:2479–88. doi: 10.1128/JB.02456-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delcour AH. 2009. Outer membrane permeability and antibiotic resistance. Biochim Biophys Acta 1794:808–16. doi: 10.1016/j.bbapap.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaara M. 1992. Agents that increase the permeability of the outer membrane. Microbiol Rev 56:395–411. doi: 10.1128/mr.56.3.395-411.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacks SL, Tyrrell LD, Lawee D, Schlech W 3rd, Gill MJ, Aoki FY, et al. 1991. Randomized, double-blind, placebo-controlled, clinic-initiated, Canadian multicenter trial of topical edoxudine 3.0% cream in the treatment of recurrent genital herpes. Canadian Cooperative Study Group. J Infect Dis 164:665–72. [DOI] [PubMed] [Google Scholar]
- 28.Davis WB, Oakes JE, Taylor JA. 1978. Effect of treatment with 5-ethyl-2’-deoxyuridine on herpes simplex virus encephalitis in normal and immunosuppressed mice. Antimicrob Agents Chemother 14:743–8. doi: 10.1128/AAC.14.5.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheraghali AM, Morin KW, Kumar R, Knaus EE, Wiebe LI. 1994. Synthesis and biodistribution of [4-14C]-5-bromo-6-methoxy-5,6-dihydro-prodrug derivatives of 5-ethyl-2’-deoxyuridine. Drug Des Discov 12:53–61. [PubMed] [Google Scholar]
- 30.Cheraghali AM, Morin KW, Kumar R, Knaus EE, Wiebe LI. 1995. Accumulation of 5-ethyl-2’-deoxyuridine and its 5,6-dihydro prodrugs in murine lung and its potential clinical application. J Pharm Pharmacol 47:595–600. doi: 10.1111/j.2042-7158.1995.tb06721.x [DOI] [PubMed] [Google Scholar]
- 31.Krapp F, Morris AR, Ozer EA, Hauser AR. 2017. Virulence Characteristics of Carbapenem-Resistant Klebsiella pneumoniae Strains from Patients with Necrotizing Skin and Soft Tissue Infections. Sci Rep 7:13533. doi: 10.1038/s41598-017-13524-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miro-Canturri A, Ayerbe-Algaba R, Smani Y. 2019. Drug Repurposing for the Treatment of Bacterial and Fungal Infections. Frontiers in Microbiology 10:41. doi: 10.3389/fmicb.2019.00041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, et al. 2000. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem 275:34287–92. doi: 10.1074/jbc.M006725200 [DOI] [PubMed] [Google Scholar]
- 34.Froquet R, Lelong E, Marchetti A, Cosson P. 2009. Dictyostelium discoideum: a model host to measure bacterial virulence. Nat Protoc 4:25–30. doi: 10.1038/nprot.2008.212 [DOI] [PubMed] [Google Scholar]
- 35.Ouertatani-Sakouhi H, Kicka S, Chiriano G, Harrison CF, Hilbi H, Scapozza L, et al. 2017. Inhibitors of Mycobacterium marinum virulence identified in a Dictyostelium discoideum host model. PLoS One 12:e0181121. doi: 10.1371/journal.pone.0181121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchetti A, Mercanti V, Cornillon S, Alibaud L, Charette SJ, Cosson P. 2004. Formation of multivesicular endosomes in Dictyostelium. J Cell Sci 117:6053–9. doi: 10.1242/jcs.01524 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(ZIP)
(DOCX)
Data Availability Statement
**PA at Accept: please follow up with author for experimental data** All processed data is available in the manuscript. All unprocessed experimental data will be made available online on a new dedicated server of our University upon publication.
All data is available within the manuscript. Unprocessed experimental data is available online on a dedicated server at the following address: https://doi.org/10.26037/yareta:oqmmzyw6crhqtmxfsjhoe65cwa.