A CRISPR-Cas9 screen reveals that loss of integrin-linked kinase synergizes with SRC inhibition, providing a new opportunity for enhancing the clinical effectiveness of SRC inhibitors in breast cancer.
Abstract
SRC is a nonreceptor tyrosine kinase with key roles in breast cancer development and progression. Despite this, SRC tyrosine kinase inhibitors have so far failed to live up to their promise in clinical trials, with poor overall response rates. We aimed to identify possible synergistic gene–drug interactions to discover new rational combination therapies for SRC inhibitors. An unbiased genome-wide CRISPR-Cas9 knockout screen in a model of triple-negative breast cancer revealed that loss of integrin-linked kinase (ILK) and its binding partners α-Parvin and PINCH-1 sensitizes cells to bosutinib, a clinically approved SRC/ABL kinase inhibitor. Sensitivity to bosutinib did not correlate with ABL dependency; instead, bosutinib likely induces these effects by acting as a SRC tyrosine kinase inhibitor. Furthermore, in vitro and in vivo models showed that loss of ILK enhanced sensitivity to eCF506, a novel and highly selective inhibitor of SRC with a unique mode of action. Whole-genome RNA sequencing following bosutinib treatment in ILK knockout cells identified broad changes in the expression of genes regulating cell adhesion and cell–extracellular matrix. Increased sensitivity to SRC inhibition in ILK knockout cells was associated with defective adhesion, resulting in reduced cell number as well as increased G1 arrest and apoptosis. These findings support the potential of ILK loss as an exploitable therapeutic vulnerability in breast cancer, enhancing the effectiveness of clinical SRC inhibitors.
Significance:
A CRISPR-Cas9 screen reveals that loss of integrin-linked kinase synergizes with SRC inhibition, providing a new opportunity for enhancing the clinical effectiveness of SRC inhibitors in breast cancer.
Introduction
SRC family kinases are group of nine nonreceptor tyrosine kinases, of which, SRC is the prototype. SRC is a key regulator of cell–matrix and cell–cell adhesions and is known to play important roles in a number of cancer-associated phenotypes including proliferation, migration and invasion. Disease-associated upregulation of SRC kinase activity has been demonstrated in many malignancies, including breast, colon and gastric cancer. In breast cancer, elevated SRC expression is associated with poor prognosis (1). Despite the validated etiologic role of this target and several commercial drug discovery programs, there has been limited evidence of clinical efficacy in solid tumors, and no progress in understanding the molecular signatures that predict sensitivity to available inhibitors (2, 3). Bosutinib (SKI-606) is a multi-kinase inhibitor and has activity against all nine SRC family members as well as ABL (4). Bosutinib is currently used as a second-line treatment for patients with chronic myeloid leukemia that carry the BCR-ABL oncogene (5), and has shown activity in preclinical models of breast cancer (6, 7). In a phase II clinical trial of bosutinib monotherapy in heavily pretreated, advanced breast cancer patients, bosutinib presented poor overall response rates of 5.5% (8). However, there was evidence of disease stabilization (37%), and together with the reported disease control rates following treatment with dasatinib, a multi-kinase inhibitor with activity against SRC family members (9), this suggests there is a subset of breast cancer patients that are responsive to dual SRC/ABL inhibition.
One of the reasons that SRC/ABL tyrosine kinase inhibitors (TKI) such as bosutinib have so far failed to live up to their promise in clinical trials is due to limited information on the molecular determinants of SRC inhibitor sensitivity. To address this, we undertook an unbiased genome-wide CRISPR-Cas9 screen in a model of triple-negative breast cancer (TNBC) to identify genes whose depletion alters the cellular sensitivity to bosutinib treatment. We show that loss of the cell-matrix adhesion protein integrin-linked kinase (ILK) and its binding partners α-Parvin and PINCH-1 can potentiate the inhibitory effect of bosutinib.
Materials and Methods
Additional information on all Methods can be found in the Supplementary Information.
Cell culture
All cell lines were grown in a humidified cell culture incubator at 37°C and 5% CO2. MDA-MB-231, MDA-MB-468, MDA-MB-157, MDA-MB-134-VI, SKBR3, and MCF7 were maintained in DMEM +10% FBS. ZR-75–1 and T47D were maintained in RPMI1640 +10% FBS. All cell lines were passaged for no more than 20 passages. Automated cell counts for passage calculations were obtained from the CellCountess II automated cell counter (Thermo Fisher Scientific). Cells were routinely tested for Mycoplasma and were authenticated by STR profiling.
Two-dimensional cell viability assays
Unless stated otherwise, cells were seeded at 1,000 cells/well into flat-bottom 96-well plates (Greiner 655180). Edge wells were excluded and plates were left for 30 min at room temperature before transferring to an incubator. Plates for real-time analysis were transferred to the IncuCyte FLR imaging system (Essen BioScience) as previously described (10). Unless stated otherwise, compounds were added (5 μL/well) at two days post seeding, for five days. Compounds were reconstituted in dimethyl sulfoxide (DMSO). Plates were assayed for cell viability using the resazurin reduction assay and/or cell counts as described previously (10). The Spark 20M (TECAN) was used to quantify resazurin fluorescence following best practice procedures described previously (11). The ImageXpress system (Molecular Devices) was used to image Hoechst 33342 stained plates. CellProfiler (www.cellprofiler.org) was used to enumerate cell counts as previously described (12). Normalized cell viability was calculated as follows: sample/DMSO × 100. Relative half-inhibitory concentrations (EC50) were calculated in Prism from the nonlinear regression algorithm of the normalized cell viability plotted against an 8-point compound dilution.
Three-dimensional spheroid cell viability assays
Cells were seeded at 2,000 cells/well into round-bottom 96-well plates (Corning #7007). Once spheroids had formed over two to three days, plates were imaged at ×4 magnification using the ImageXpress and subsequently compounds were added. On day seven post drugging, spheroids were stained with Calcein-AM (3.75 μmol/L final) to give a “live cell” readout, propidium iodide (1.0 μmol/L final) or Draq7 (3.0 μmol/L final) for a “dead cell” readout and Hoechst 33342 (1.0 μmol/L final) for a “total cells” readout. The ImageXpress was used to create a single flattened maximum intensity projection from an eight-step Z stack with a 50 μmol/L step size. A custom CellProfiler pipeline was used to quantify the spheroid area and intensity values for each spheroid. Data was normalized as follows: sample/DMSO × 100.
Cell adhesion challenge
Cells were seeded at 20,000 cells/well in serum-free media into 96-well plates (Greiner #655180). DMSO or bosutinib was added at the time of seeding. Plates were imaged every 10 min in the IncuCyte FLR for 2 hours and then washed gently twice with 100 μL PBS taking care to not disrupt adhered cells. Plates were then returned to the IncuCyte for a final post-wash scan to determine the adhered cells.
Western blotting
Cell lysates were prepared in RIPA buffer (150 mmol/L NaCl, 50 mmol/L Tris pH 7.2, 0.1% SDS, 1.0% Triton X-100) with protease and phosphatase inhibitors (10 μg/mL aprotinin, 125 mmol/L phenylmethylsulfonyl fluoride, 100 μmol/L sodium orthovanadate, and 0.5 mmol/L sodium fluoride). For three-dimensional (3D) spheroid protein extraction, spheroids were pooled and then solubilised in RIPA using sonication. Protein concentration was determined using the BCA protein assay kit. Whole-cell lysates were fractionated by SDS-PAGE (20 μg per sample) and transferred onto nitrocellulose membranes. Primary antibodies used were: ABL (SCBT, #sc-23 and #sc-56887) and pY245 ABL (CST #2861); Arg (Ab134134); cofillin (CST #5175); CrkL (CST #3182) and pY207 CrkL (CST #3181); FAK (CST #3285) and pY397 FAK (CST #3283); GAPDH (CST #5174); ILK (BD #611803 and Abcam #ab52480); mouse IgG1 (CST #5415); rabbit IgG (CST #2729); α-Parvin (CST #8190 and CST #4026); PINCH-1 (CST #11890); SRC (CST #2109) and pY416 SRC (CST #2101). Protein–antibody complexes were visualized by enhanced chemiluminescence detection (GE Healthcare) with horseradish peroxidase-conjugated secondary antibodies (CST #7076 and #7074) using the ChemiDoc XRS imaging system (Bio-Rad Laboratories). Experiments were repeated at least three times. Blots were cropped for clarity. The volume tool in Image Lab (v5.0) was used to quantify intensity. The same standard rectangle was used for each sample, as well as background correction for each lane.
Genome-wide CRISPR-Cas9 dropout screen in MDA-MB-231 cells
The CRSIPR screen was performed using the pooled lentiviral genome-wide CRISPR-SpCas9 TKOv3 library targeting 18,053 protein-coding genes (4 gRNAs/gene), a gift from Jason Moffat (Addgene #90294), that was amplified and packaged into lentiviral particles (Vector Core, University of Michigan) as described (13). A total of 9 × 107 MDA-MB-231 cells were transduced with the TKOv3 lentiviral library (71,090 gRNAs) at a low MOI (0.36) to ensure each cell receives only one sgRNA and to achieve >300-fold library coverage after selection. To select for transductants, 24 hours after infection, cells were grown in the presence of 2.5 μg/mL puromycin for 48 hours, which was considered the initial time point (T0). Cells were then passaged every 3–4 days and library coverage of ≥300 cells per gRNA was maintained at every step. Seven days after puromycin selection (T7), two technical replicates containing 24 × 106 cells were established for DMSO and bosutinib (provided by Pfizer), respectively. An EC20 concentration of bosutinib (0.9 μmol/L) was added 24 hours postseeding for 3 days and cells were treated with bosutinib for five rounds or for approximately 12 population doubling times over 20 days. To capture the full representation of the screen, 25 × 106 cells were collected at each passage point for genomic DNA extraction using the QIAamp Blood Maxi Kit (#51192). The representation of genome-integrated gRNA sequences was determined using a two-step PCR in which the first step “PCR1” amplifies the lentiviral sequence containing the 20 bp gRNA cassette from genomic DNA and the second step “PCR2” attaches unique barcodes for sample deconvolution and Illumina TruSeq adapters (i5 and i7) for next-generation sequencing (Supplementary Fig. S1; Supplementary Tables S1–S3). The final pooled library was quantified with the Qubit 2.0 (Invitrogen) and Agilent 2100 Bioanalyser platforms. The diluted and denatured NGS library was sequenced on a NextSeq 500/550 High Output Kit v2 (75 cycles) with a custom “dark cycle recipe,” for dual-index, single-read reads. Sequencing was performed by the Edinburgh Welcome Trust Clinical Research Facility. DrugZ (14) was used to calculate a gene-level normalized Z-score for determining significant differences between the DMSO and bosutinib samples at both T19 (three rounds of drugging) and T27 (five rounds of drugging). Gene copy number data was incorporated into the analysis as double-stranded DNA breaks from CRISPR-Cas9 can generate false-positive hits in genes with high copy numbers due to cutting-related genotoxicity (15). There was no correlation between gene copy number and gene-level normalized Z-score (Supplementary Table S4). Supplementary Dataset 1 contains the DrugZ results file.
Interaction network analysis
Composite functional association networks were constructed for putative enhancer and suppressor genes using GeneMANIA (version 3.5.2; human interactions: https://genemania.org/) in Cytoscape (version 3.8.0: https://cytoscape.org/). Network edges were weighted according to evidence of cofunctionality using GeneMANIA. Connected networks were clustered using the force-directed algorithm in the Prefuse toolkit (https://github.com/prefuse/Prefuse).
Patient survival analysis
cBioPortal was used to download the METABRIC data (16) and The Cancer Genome Atlas (TCGA-PanCancer: http://cancergenome.nih.gov/) data. Data were analyzed using the R packages survival (17), survival (18) and their dependencies. In the METABRIC dataset, the TNBC subset was created from the clinical data for estrogen receptor alpha (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2). The TNBC subset had a 75% overlap with the PAM50-basal METABRIC data. The TNBC subtype increased the total number of patients compared to the PAM50-basal subtype, thereby increasing the power of the statistical test for high versus low ILK. The optimal threshold for dichotomization of the datasets into high and low ILK expression was identified using the survivALL R package and “plotALL” function for multi-cut point analysis from the METABRIC dataset. The same dichotomization threshold was then applied to the TCGA dataset to validate the cut-off point.
CRISPR knockout of ILK and ABL1 in mammalian cells
CRISPR-(e)SpCas9 target sites designed against ILK and ABL1 were created using the Broad Institute's gRNA design tool (https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design), which uses the optimal gRNA and off-target design principles of Doench and colleagues (19). The gRNAs target exons 1 and 8 of ILK (deposited with Addgene plasmid #163320 and 163321), within the N-terminal ankyrin repeat domain-1 and the C-terminal protein kinase domain, respectively (Supplementary Table S5). For ABL1, the two designed gRNAs target exon 4 (Deposited with Addgene plasmid #163322 and 163323) within the protein kinase domain of both ABL1 isoforms 1a and 1b (Supplementary Table S6). The CRISPR plasmid constructs PX459v2 and eSpCas9(1.1) were purchased from Addgene (plasmid #62988 and #71814) and were deposited by Prof. Feng Zhang's laboratory (20). The CRISPR-(e)Cas9 constructs were created as described by Ran and colleagues (21). Individual transfected cells [Nucleofector system (Lonza)] were selected for clonal expansion using puromycin selection (PX459v2) and flow cytometry. Monoclonal colonies were screened for loss of the gene of interest using western blotting with the ILK monoclonal antibodies BD #611801 and Abcam #ab52480 or the ABL monoclonal antibodies 8E9 (sc-56887) and 24–11 (sc-23). The plasmids pDONR223-ABL1 (Plasmid #23939) and pLX301 (Plasmid #25895) were purchased from Addgene, and were deposited by William Hahn & David Root (22) and David Root (23), respectively. Using gateway cloning, pDONR223-ABL1 (containing the ABL1 isoform 1a (ABL1–1A)) was cut and inserted into pLX301. 293T cells were used for viral packaging.
Cell-cycle analysis
Cells were seeded in 6-well plates and drugged at either 48 h post seeding (48-hour drug time point) or 72 hours (24-hour drug time point). Cells were harvested using trypsin and fixed in cold 70% ethanol for 48 hours. Pellets were treated with RNAse H (NEB, #M0297L) and stained with 20 μg/mL propidium iodide. Cells were sorted on the BD LSRFortessa and analysed using BD FACSDiva software (v8.0.1).
Apoptosis and cell death assay
Cells were seeded in 6-well plates and drugged for 4 days. Medium was removed and spun down to include floating cells. Cells were then trypsinized and resuspended with 5% FBS in PBS. Cells were sorted on the BD LSRFortessa and analyzed using BD FACSDiva software (v8.0.1). CellEvent Caspase 3/7 (apoptotic cells) and DAPI (dead cells) dyes were used.
Breast cancer cell line datasets
We generated a 16 BC cell line dataset for sensitivity to bosutinib using a PrestoBlue (Resazurin reduction) cell viability assay as described above (24). The cb5 cell line dataset is an integrated compendium of 5 Affymetrix datasets and was downloaded from Moleirinho and colleagues (25). Multiple cell line microarray datasets were combined for ILK expression levels in the 16 breast cancer cell lines. The Cancerxgene dataset was downloaded from: https://www.cancerrxgene.org/. The Cancerxgene dataset included 43 BC cell lines with data for bosutinib treatment. The Depmap interactive website is available at: https://depmap.org/portal/. The Depmap interactive portal of 20 BC cell lines used bosutinib cell viability data compared to ILK RNA sequencing (RNA-seq) expression levels and ILK relative protein expression.
RNA-seq
Bosutinib EC20 (0.9 μmol/L) or DMSO were added to cells at 24 hours postseeding in 10-cm dishes. Three independent experimental replicates were performed. Total RNA was extracted at 72 hours postdrugging using RNAeasy Miniprep Kit (Qiagen) according to the manufacturer's protocol. RNA quality and integrity were assessed on the Fragment Analyser Automated Capillary Electrophoresis System (Agilent Technologies). RNA yield and DNA contamination was quantified using the Qubit 2.0. Library preparation and sequencing was performed by the Edinburgh Welcome Trust Clinical Research Facility. The Lexogen QuantSeq 3′mRNA-Seq Library Prep Kit (FWD) for Illumina was used to generate libraries from 200 ng total RNA. Library quantity and quality were assessed using Qubit 2.0 and the Bioanalyser. Single-read sequencing was performed using the NextSeq 500/550 High-Output v2.5 (75 cycles) Kit on the NextSeq 550 platform (Illumina). Demultiplexed FASTQ files with md5 checksums were imported into the Lexogen Bluebee platform and the Quantseq 2.3.6 FWD pipeline was used. Trimming was performed using bbduk v35.92. STAR v2.5.2a (26) was used to align the read data to human genome build human GRCh38. The expected counts from HTSeq-count v0.6.0 were imported into the statistical software package R (www.r-project.org). Differential expression analysis was undertaken with the EdgeR (v3.26.8) and Limma (v3.40.6) packages. Significantly differentially expressed genes were determined using the FDR (Benjamini and Hochberg) P value adjustment <0.05 and a log2 fold change of ±1. Gene ontology (GO) functional enrichment analysis was carried out using ToppGene (https://toppgene.cchmc.org/). Read count and differential expression analysis data is available in Supplementary Datasets 2–5.
Tumorigenicity assay
Immunodeficient Rag2-Il2rg double knockout mice (R2G2) were obtained from Envigo, UK. Female mice (2–4 months of age) were bilaterally injected subcutaneously with 2.5 × 106 MDA-MB-231 PX45v2 or ILK gRNA 2 cells (100 μL in PBS). Once tumors developed, mice were randomized (using the standard RAND function in Microsoft Excel) to daily oral gavage with either vehicle (50 mmol/L lactic acid with 0.1 mol/L NaOH, pH 6), eCF506 (40 mg/kg body weight, 0.1 mL/20 g) or bosutinib (75 mg/kg body weight, 0.1 mL/20 g). Statisticians were blinded to treatment group allocation. Mice were monitored twice weekly and culled if they became sickly or tumors had reached a maximum size of 15 mm. Tumors were collected at the end of the study. Formalin-fixed tissues were processed into paraffin wax blocks. Sections were cut, dewaxed, and rehydrated before heat-mediated antigen retrieval in sodium citrate buffer. After blocking, sections were incubated with Ki67 antibody at 1:400 (Cell Signaling Technology #9027) followed by 3,3′-diaminobenzidine (DAB) Chromagen (Agilent #K3468) for visualization. Sections were counterstained with hematoxylin and QuPath used to quantify Ki67 positive nuclei (https://qupath.github.io/). All animal studies and procedures were approved by the University of Edinburgh Ethical review committee (Application #116-WGH-20) and conducted in accordance with United Kingdom Home Office regulations.
Data availability
The datasets generated during and/or analyzed during the current study are available as Supplementary Files. Additional data is available from the corresponding author on reasonable request.
Results
Bosutinib demonstrates antiproliferative activity in several breast cancer cell lines
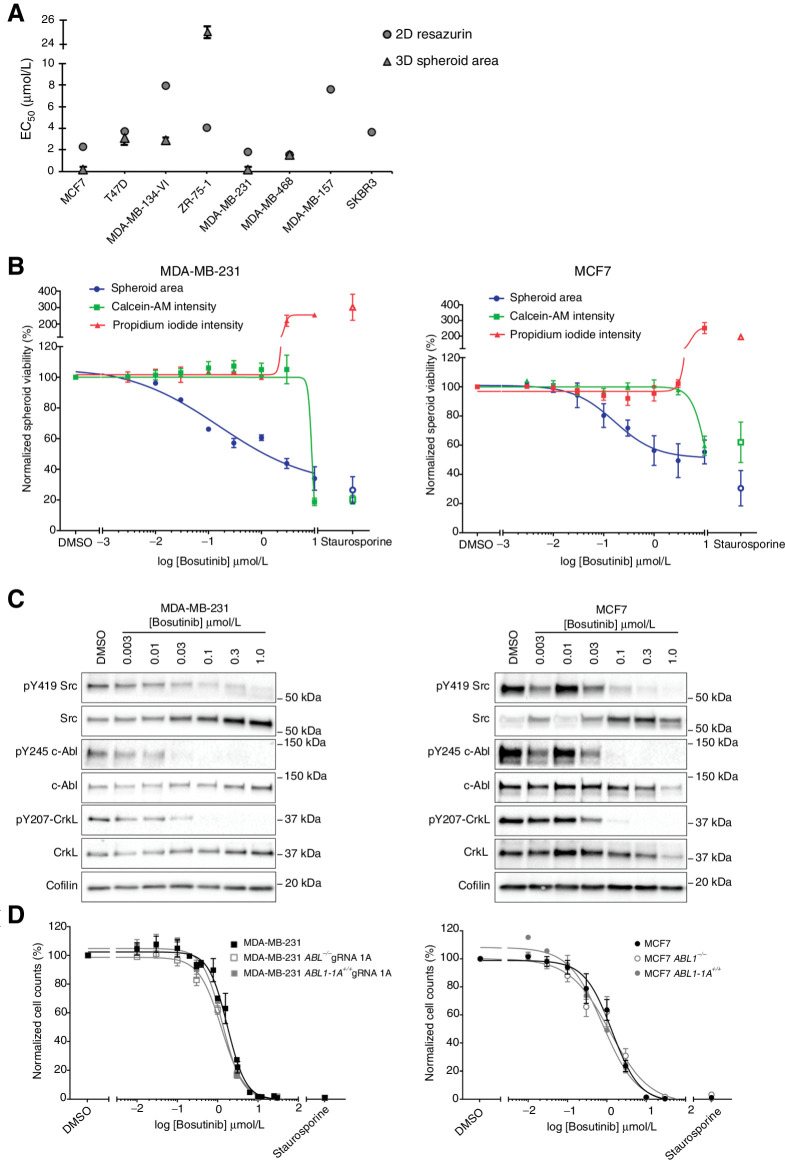
Because of limited published data on sensitivity to bosutinib in breast cancer cell lines (6), we tested a panel of eight cell lines in both two-dimensional (2D) and 3D viability assays. Bosutinib sensitivity did not correlate with breast cancer subtype in our cell line panel (Fig. 1A; Supplementary Table S7); however, the triple-negative MDA-MB-231 and the luminal A MCF7 cell lines were the most sensitive to bosutinib.
Figure 1.
Effect of bosutinib treatment on MDA-MB-231 and MCF7 cell lines. A, Bosutinib 2D and 3D EC50 values in a panel of breast cancer cell lines. MDA-MB-157 and SKBR3 are unable to form spheroids. B, 3D spheroid cell viability. Spheroid area, Calcein-AM intensity, and propidium iodide intensity were quantified using a custom CellProfiler pipeline. Staurosporine (1 μmol/L) was used as a positive control to induce cell death. C, Western blot analysis of cell lines grown as 3D spheroids. Data are representative of two independent experimental replicates. D, Loss of ABL1 does not alter bosutinib sensitivity. 2D bosutinib 8-point dilution for half-inhibitory concentrations (EC50). Additional 2D and 3D data are shown in Supplementary Fig. S4. All data shown are the mean of at least three independent experimental replicates except MDA-MB-134-VI and MDA-MB-468, which were two independent experimental replicates. ***, P < 0.0002; ****, P < 0.0001, as determined by a two-way ANOVA with Bonferroni multiple comparisons correction. All error bars are SEM.
Both cell lines were more sensitive to bosutinib in a 3D spheroid growth assay (Fig. 1A). However, although the half maximal effective concentration (EC50) values for bosutinib were in the submicromolar range for both MDA-MB-231 and MCF7 cells, no significant cell death was seen at these concentrations using a 3D spheroid live/dead assay (Fig. 1B; Supplementary Fig. S2A). In both cell lines, bosutinib treatment caused a dose-dependent decrease in spheroid area (EC50 = 0.151 μmol/L and 0.194 μmol/L, respectively), but no corresponding decrease in Calcein-AM (viable cells) or increase in propidium iodide intensity (dead cells) until concentrations greater than 3 μmol/L.
We confirmed that bosutinib was inhibiting its designed targets SRC and ABL in the spheroid assay by Western blot analysis, where bosutinib treatment resulted in a dose-dependent reduction in pY419-SRC, with almost complete inhibition (∼90% reduction) at 0.1 μmol/L (Fig. 1C; Supplementary Fig. S2B). Total SRC was increased, as has been reported following treatment with SRC inhibitors (27). There was a marked reduction in ABL phosphorylation at Y245, a site required for its kinase activity, following treatment with bosutinib at 30 and 100 nmol/L for the MDA-MB-231 and MCF7 cells, respectively. This correlated with a 90% and 70% reduction in pY207-CRKL at 30 nmol/L in the MDA-MB-231 and MCF7 cell lines, respectively (Fig. 1C; Supplementary Fig. S2C).
To provide mechanistic insight into the molecular pathways underlying bosutinib sensitivity, we performed a reverse-phase protein array (RPPA) analysis using 125 antibodies covering a panel of signaling proteins and phosphoproteins in MDA-MB-231 and MCF7 cells following treatment with bosutinib (Supplementary Fig. S3). This identified 17 and 7 significantly differentially regulated proteins and phosphoproteins in MDA-MB-231 and MCF7 cells, respectively (P < 0.05, log2 fold change ±0.75). As expected, bosutinib treatment inhibited its designed targets SRC and ABL, with a significantly decreased signal intensity in SRC autophosphorylation at tyrosine (pY)419 (70% reduction at 1.25 μmol/L), as well as an 80% and 70% reduction in pY207-CRKL, a key substrate of ABL, at 1.25 μmol/L for the MDA-MB-231 and MCF7 cells, respectively (Supplementary Fig. S3). Other consistent changes that were seen between the cell lines included a decrease in pY15 CDK1 and total CDK1, and a decrease in phosphorylation of phospho-threonine (pThr)183-JNK (Supplementary Fig. S3A).
Loss of ABL does not alter bosutinib effects in MDA-MB-231 and MCF7 cells
As bosutinib is a dual SRC/ABL inhibitor, which effectively inhibited ABL activity in both the MDA-MB-231 and MCF7 cell lines, we asked whether the antiproliferative activity of bosutinib was due to ABL inhibition. We created ABL1 knockout isogenic cell lines using CRISPR-eCas9 (Supplementary Fig. S4A). Using 2D cell viability assays there were no differences between bosutinib EC50 values for the ABL1 knockout MCF7 or MDA-MB-231 cell lines compared with the parental cells or ABL1 knockout cell lines in which ABL1 was reexpressed (Fig. 1D). Similar results were observed with three other MDA-MB-231 ABL1 gRNA cell lines (Supplementary Fig. S4B). Loss of ABL1 also did not alter the sensitivity to bosutinib in the 3D spheroid assay (Supplementary Fig. S4C). Thus, loss of ABL1 does not alter bosutinib activity in MDA-MB-231 and MCF7 cells in both 2D and 3D assays, indicating that bosutinib is unlikely to act via ABL1 inhibition.
A genome-wide CRISPR-Cas9 knockout screen identifies the ILK–Parvin–PINCH complex as a key determinant of bosutinib sensitivity
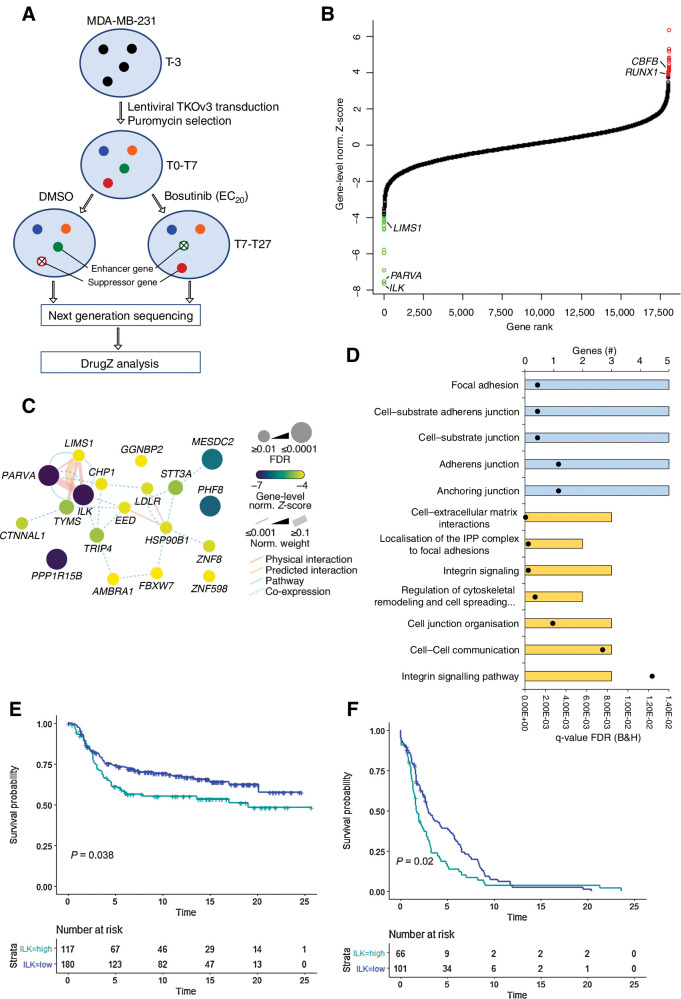
To identify genes that can alter bosutinib sensitivity we performed an unbiased genome-wide CRISPR-Cas9 negative selection screen in the MDA-MB-231 cell line (Fig. 2A). We chose the Toronto knockout version 3 (TKOv3) library (13) that contains sequence-optimized guide RNA (gRNA) following empirically determined on- and off-target prediction enabling increased accuracy and scalability of the CRISPR screen. Cells were transduced with the lentiviral TKOv3 library at a low multiplicity of infection (MOI = 0.36) and selected with puromycin. Since almost complete modification of genomic loci occurs after seven days of CRISPR treatment (28), cells were passaged for seven days after puromycin selection (T7), after which, either an EC20 concentration of bosutinib (0.9 μmol/L) or DMSO as a control were added 24 hours postseeding for 3 days. A 24-hour time point for bosutinib addition was chosen in order for cell attachment to occur, but before log-phase growth began. Cells were treated in this way for around 12 population doubling times (PD) over 20 days. The screen was performed in technical duplicates and genomic DNA was extracted and genome-integrated gRNA sequences were amplified and labeled with barcodes via a 2-step polymerase chain reaction (PCR). Next-generation sequencing was used to quantify the library representation of the gRNA sequences and DrugZ (14) was used to calculate gene-level normalized Z-scores for determining differences between the DMSO and bosutinib samples at the T19 (∼7 PDs) and T27 (∼12 PDs) time points (Fig. 2A). This allowed the identification of enhancer genes whose loss of function enhance sensitivity to bosutinib and suppressor gene interactions whose loss of function reduce bosutinib efficacy.
Figure 2.
The ILK–Parvin–PINCH complex is the top enhancer of bosutinib-induced cell growth inhibition. A, Overview of the genome-wide CRISPR-Cas9 knockout screen in MDA-MB-231 cells. T0, initial time point immediately after puromycin selection; T7, 7 days post puromycin selection; T19, three rounds of drug treatment; T27, five rounds of drug treatment. B, DrugZ analysis of the genome-wide bosutinib CRISPR-Cas9 screen. The gene-level normalized Z-score from the T27 time point is shown. Genes are ranked according to the enhancer genes. The Benjamini and Hochberg FDR was used to calculate significance. Green data points are enhancer genes with FDR < 0.05 and red are suppressor genes with FDR < 0.05. C, Network map for the enhancer genes identified at T27. Network edges were weighted according to evidence of cofunctionality using GeneMANIA in Cytoscape. D, ToppGene gene ontology analysis for the combined T19 and T27 enhancer genes. Blue, cellular component category; yellow, pathway category. Black dots, q values. E, High ILK expression correlates with worse survival. Kaplan Meier plot for the METABRIC TNBC subtype. The optimal threshold for dichotomisation of the datasets into high and low ILK expression was identified using the survivALL R package and “plotALL” function for multicut point analysis using the METABRIC dataset. F, Kaplan–Meier plot for the TCGA-PanCancer basal subtype used as a validation dataset. Breast cancer-specific survival was used.
Hierarchical cluster analysis of the individual gRNAs from the top enhancer and suppressor genes revealed groupings of replicates and treatments across time points, emphasizing consistency in the results between the T19 and T27 time points at the gRNA level (Supplementary Fig. S5A). DrugZ analysis of the T27 time point identified 19 enhancer genes and 23 suppressor genes with a significant gene-level normalized Z-score (FDR<0.05) with 12 enhancer genes and 15 suppressor genes also identified at the T19 time point (Fig. 2B; Supplementary Fig. S5B). Integrin-linked kinase (ILK) and α-Parvin (PARVA) were the highest ranked enhancer genes at both the T27 and T19 time points. PINCH-1 (LIMS1) also had a significantly decreased gene-level normalized Z-score upon bosutinib treatment at both time points (Supplementary Table S4). These three proteins form the ILK–Parvin–PINCH (IPP) complex. The IPP complex tethers cells to their surrounding environment via integrins and is involved in cytoskeleton remodeling, angiogenesis, proliferation, survival, and differentiation (29). Hence, within the pool of CRISPR knockout cells, those cells with loss of ILK or its binding partners α-Parvin and PINCH-1 significantly enhanced the inhibitory effect of bosutinib on MDA-MB-231 cell growth.
Functional network analysis revealed connectivity between many of the top enhancer genes (Fig. 2C; Supplementary Fig. S6A). Interestingly, both the expected targets of bosutinib, SRC and ABL, and many of the enhancer genes, such as those involved in the IPP complex, AMBRA1, EED, and TYMS, converge on the focal adhesion kinase (FAK) network (Supplementary Fig. S6B) and are linked to cytoskeletal organization. Consistent with this, gene ontology (GO) analysis of the top enhancer genes centered around the interactions of the IPP complex. The key enriched molecular pathways were adherens junctions, cell-extracellular matrix (ECM) interactions, focal adhesions and the regulation of cytoskeletal remodeling and cell spreading (Fig. 2D). There were also a number of interesting suppressor genes such as CBFB, RUNX1, EP300 and AHR, but no GO terms were significantly enriched for the top suppressor genes (Supplementary Fig. S6C; Supplementary Table S8).
Loss of the ILK–Parvin–PINCH complex enhances sensitivity to bosutinib
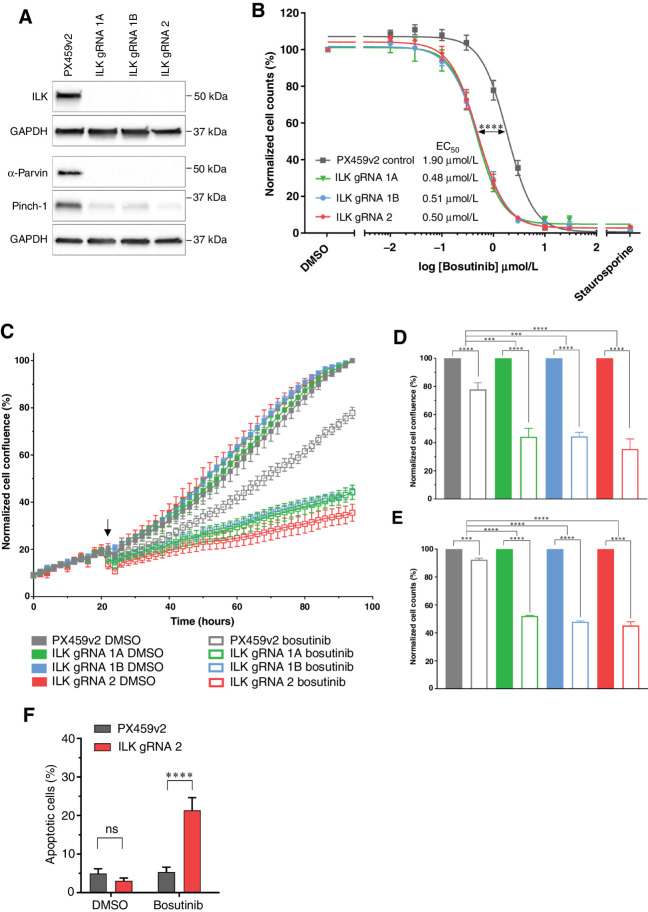
We focused on ILK as the top ranked enhancer gene for validation studies. Analysis of the METABRIC and TCGA-PanCancer breast cancer datasets showed that high ILK expression was significantly associated with poor breast cancer-specific survival for the TNBC and basal subtypes (Kaplan–Meier log-rank test, P < 0.05), but not for the remaining breast cancer patients (Fig. 2E and F; Supplementary Fig. S7), suggesting a functional role for ILK in these subtypes. To validate that loss of ILK, α-Parvin or PINCH-1 can potentiate the inhibitory effect of bosutinib, we generated clonal ILK knockout cell lines. We created MDA-MB-231 ILK knockout cell lines using two gRNAs targeting ILK and a transiently expressed CRISPR-Cas9 system (PX459v2) to generate single-cell–derived cell lines lacking ILK. We generated three cell lines for ILK gRNA 1 and three for ILK gRNA 2 (Supplementary Fig. S8A). The IPP complex members are interdependent for their protein stability (30). Accordingly, our western blot analysis showed that ILK knockout in MDA-MB-231 cells resulted in the downregulation of PINCH-1 and α-Parvin in addition to loss of ILK (Fig. 3A; Supplementary Fig. S8A). Therefore, we did not endeavor to create PINCH-1 or α-Parvin CRISPR knockouts in addition to the ILK knockout cell lines.
Figure 3.
Loss of the ILK–Parvin–PINCH complex potentiates the effects of bosutinib in MDA-MB-231 cells. A, CRISPR-Cas9 targeting ILK results in ILK loss in the MDA-MB-231 cell line as assessed by Western blot. B, Bosutinib 8-point dose response curves using normalized cell counts form Hoechst-stained images. Significance test refers to EC50 values calculated in Prism. C–G) Cells were treated with the EC20 for bosutinib (0.9 μmol/L). C, ILK loss potentiates bosutinib inhibition in real-time assays. Cells were seeded in 96-well plates at 8,000 cells/well and transferred to the IncuCyte Zoom. Arrow shows drug addition at 24 hours post seeding. D, Endpoint quantification using normalized cell confluence from the IncuCyte software. E, Endpoint quantification using normalized cell counts form Hoechst-stained images. F, Apoptosis assay using CellEvent Caspase 3/7 and fluorescence-activated cell sorting. All data shown are the mean of at least three independent experimental replicates. ns, nonsignificant; ***, P < 0.001; ****, P < 0.0001, as determined by a two-way ANOVA with Bonferroni multiple comparisons correction. All error bars are SEM. PX459v2, empty-gRNA CRISPR control cells; gRNA 1A, gRNA 1B and gRNA 2, ILK knockout clones.
We next assessed the impact of bosutinib treatment on cell viability in cells with or without ILK. Loss of ILK resulted in an approximately 4-fold decrease (P < 0.0001) in the EC50 of bosutinib compared with the empty-gRNA CRISPR control (PX459v2; Fig. 3B). ILK loss alone did not impact the growth rate of the cells, as assessed by the change in cell confluence over time (Fig. 3C). At the EC20 concentration of 0.9 μmol/L, bosutinib significantly decreased the real-time growth of all MDA-MB-231 ILK knockout cell lines compared to the PX459v2 control (Fig. 3C; Supplementary Fig. S8B). This resulted in an approximately 3-fold change decrease in cell confluence between cells with ILK loss and the PX459v2 control at the end of the assay (Fig. 3D). Bosutinib treatment also resulted in a significant decrease in cell number in all six MDA-MB-231 ILK knockout cell lines compared to the PX459v2 control (Fig. 3E; Supplementary Fig. S8C). A similar enhancement of bosutinib activity was seen in the T47D breast cancer cell line following knockdown of ILK (Supplementary Fig. S8D and S8E). Cell-cycle analysis revealed no changes in cell-cycle distribution upon ILK loss or bosutinib treatment (EC20, 0.9 μmol/L). However, there was an increased G1 phase arrest (P < 0.05) when ILK knockout cells were treated with bosutinib (Supplementary Fig. S8F). Notably, there was also a 4-fold increase (P < 0.0001) in the percent of apoptotic cells when ILK knockout cells were treated with bosutinib compared to PX459v2 control cells (Fig. 3F). Taken together, these data confirm that loss of ILK in MDA-MB-231 cells enhances sensitivity to bosutinib through an enhanced G1 arrest and increased apoptosis.
SRC but not ABL is responsible for the increased activity of bosutinib in combination with ILK loss
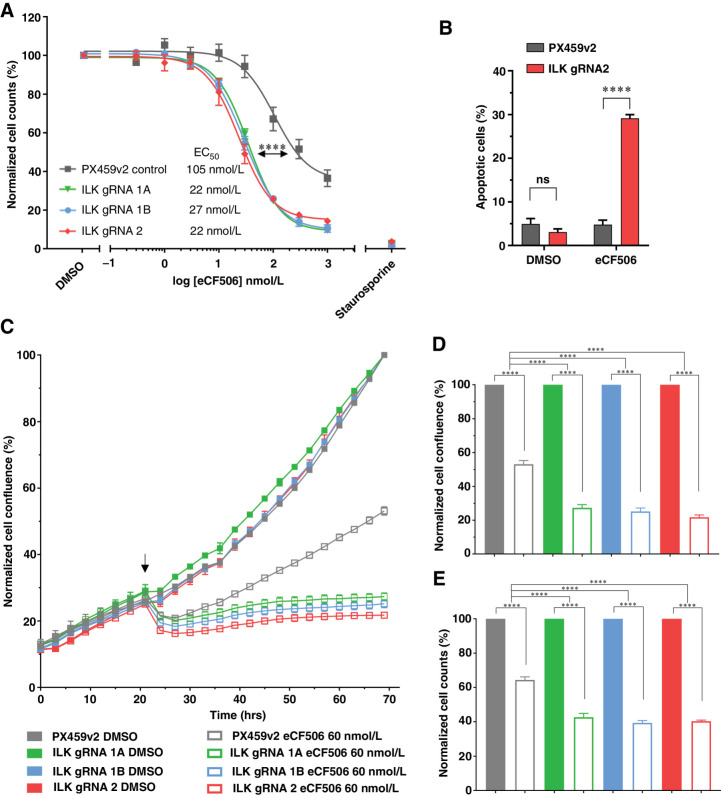
The other designed target of bosutinib is SRC. However, as there are nine SRC family members it is difficult to create suitable knockouts. In this respect, the SRC-specific inhibitor eCF506 is an effective tool compound as it inhibits all nine SRC family members and has been shown to have a 1,000-fold selectivity for SRC over ABL and potent growth inhibitory effects in both MDA-MB-231 and MCF7 cell lines (24, 27), supporting the importance of SRC activity in driving their proliferation. 2D cell viability assays confirmed that there were no differences between eCF506 EC50 values for the ABL1 knockout MDA-MB-231 cell lines compared to the parental cells (Supplementary Fig. S9A). However, in the ILK knockout cells there was an approximately 4-fold decrease (P < 0.0001) in the EC50 for eCF506 compared with the PX459v2 CRISPR control cells (Fig. 4A). Similar to bosutinib, analysis of apoptotic cells revealed a 6-fold increase (P < 0.0001) in caspase-3/7–positive cells upon eCF506 inhibition (40 nmol/L; EC20) in ILK depleted cells compared with the PX459v2 control cells (Fig. 4B). In addition, cell-cycle analysis showed a small but consistent increase in the G1 peak for eCF506 in ILK depleted cells (Supplementary Fig. S8F). Treatment of the MDA-MB-231 ILK knockout isogenic cell lines with the ABL allosteric inhibitor GNF-2 (31) did not change cell viabilities (Supplementary Fig. S9B). As expected, addition of eCF506 to the PX459v2 MDA-MB-231 cells resulted in a significant reduction in cell growth as measured in real-time in the IncuCyte and endpoint cell counts (Fig. 4C). Similar to bosutinib treatment, the combined effect of ILK loss and eCF506 treatment (60 nmol/L; EC30) resulted in a further significant decrease in cell growth as well as cell counts compared with the PX459v2 control (Fig. 4D and E). Thus, loss of ILK in the MDA-MB-231 cell lines enhanced the effects of SRC inhibition in the absence of ABL inhibition. These findings taken together confirm that SRC inhibition is required for the decreased viability in cells with ILK loss following treatment with bosutinib or eCF506.
Figure 4.
Loss of the ILK–Parvin–PINCH complex potentiates the effects of eCF506 in MDA-MB-231 cells. A, eCF506 8-point dilution for half-inhibitory concentrations (EC50). B, Apoptosis assay using CellEvent Caspase 3/7 and fluorescence-activated cell sorting. eCF506 treatment (40 nmol/L; EC20). C, ILK loss potentiates eCF506 inhibition in real time. MDA-MB-231 cells were seeded in 96-well plates at 8,000 cells/well and transferred to the IncuCyte Zoom. Arrow, drug addition. D, Endpoint quantification using normalized cell confluence from the IncuCyte software. E, Endpoint quantification using normalized cell counts form Hoechst-stained images. C–E, An EC30 concentration for eCF506 of 60 nmol/L is shown. Data shown is the mean of two independent experimental replicates. ns, nonsignificant; ****, P < 0.0001, as determined by a two-way ANOVA with Bonferroni multiple comparisons correction. All error bars are SEM. PX459v2, empty-gRNA CRISPR control cells; gRNA 1A, gRNA 1B and gRNA 2, ILK knockout clones.
Low ILK expression moderately correlates with sensitivity to bosutinib
To establish whether ILK expression was associated with bosutinib sensitivity in a larger panel of breast cancer cell lines, Pearson rank correlation coefficients were used as a measure of the strength of association between sensitivity to bosutinib and ILK mRNA expression levels in three different cell line datasets, two publicly available and one in-house (Supplementary Table S9; Supplementary Fig. S10). We generated a dataset from 16 breast cancer cell lines using EC50 cell viability measurements to determine sensitivity to bosutinib (24). For ILK mRNA expression, we used an integrated compendium of 5 Affymetrix cell line datasets (cb5; ref. 25). Decreased ILK expression correlated with a lower bosutinib EC50, although with only a moderate effect size (Pearson: r(61) = 0.358, P = 0.004). The publicly available Cancerxgene dataset allowed us to include 43 breast cancer cell lines with data for bosutinib treatment queried against the cb5 dataset. This also resulted in a weak but positively correlated relationship (Pearson: r(119) = 0.283, P = 0.002). Similar correlations were seen between bosutinib sensitivity and LIMS1 and PARVA in these two datasets (Supplementary Table S9). Additionally, we utilized the Depmap interactive portal of 20 breast cancer cell lines with bosutinib cell viability data compared to ILK RNA-seq and protein expression levels. Although this dataset had a positive correlation for both RNA-seq and protein expression it was not significant (Pearson: r (18) = 0.395, P = 0.085 and r (18) = 0.299, P = 0.199, respectively), while the correlation with LIMS1 and PARVA was significant (Supplementary Table S9). The small number of cell lines in these datasets that have low ILK expression limits the predictivity of these analyses, but taken together suggest that ILK gene expression or protein alone is not sufficient to predict sensitivity to bosutinib.
To look at gene expression of ILK in human breast tumors, we utilized an integrated compendium of 17 Affymetrix datasets (cb17) representing 2,996 breast cancers (25). This dataset has detection calls calculated from perfect and mis-matched probes, which effectively indicate whether genes are detectably “present” or “absent” in each sample. The basal subtype had significantly lower ILK expression (Tukey one-way ANOVA, P < 0.001) compared with all other subtypes (Supplementary Fig. S11A). Interestingly, ILK was significantly called “absent” in 15% of basal breast tumors (χ2 test, P < 0.001) compared with around 6% for other subtypes (Supplementary Fig. S11B and S11C). Although METABRIC and TCGA-PanCancer samples do not have “absent” or “present” detection calls, ILK expression was significantly lower in the basal subtype (Tukey HSD, P < 0.05) compared with all other subtypes except HER2 (Supplementary Fig. S11D). A similar pattern was seen for PARVA but not LIMS1, other components of the IPP complex. Interestingly, in the same datasets SRC was higher in the basal subtype (Supplementary Fig. S11D). It remains to be established whether there is a small subset of patients that would benefit from bosutinib treatment due to the absence of ILK, and the sensitization of the luminal A T47D cell line to SRC inhibitor treatment following loss of ILK, indicates that this will not be subtype specific.
ILK loss in combination with bosutinib regulates extracellular matrix and adhesion protein transcripts
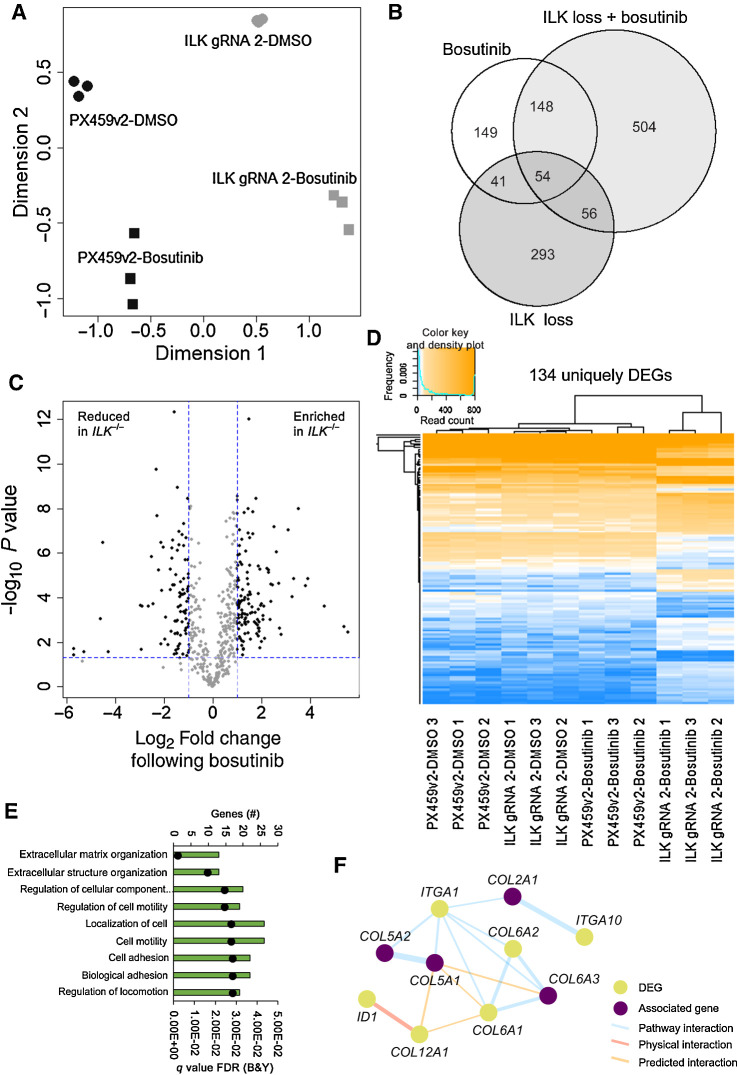
We undertook an unbiased genome-wide RNA sequencing approach to understand and exploit the mechanism(s) at the transciptional level that enabled ILK loss to potentiate SRC inhibition in our TNBC model. Principal component analysis identified good clustering of similar samples and separation between different groups. Hierarchical clustering of log2 counts per million for the 500 most variable genes across all 12 samples also correctly clustered samples (Supplementary Fig. S12A). The largest variation (dimension 1) was seen between the presence or absence of ILK, which resulted in 444 DEGs (Fig. 5A and B). This identified genes associated with the ECM, actin cytoskeleton, membrane structures, integrin and growth factor binding (Supplementary Fig. S12B) consistent with ILKs known role in cell-matrix adhesions. For the MDA-MB-231 PX459v2 CRISPR control, bosutinib treatment resulted in 392 DEGs (DEGs; log2 fold change ±1 and p-value < 0.05) (Fig. 5B). Importantly, there were 504 DEGs unique to the combination of ILK loss and bosutinib addition (Fig. 5B). To determine the magnitude of change in the 504 uniquely DEGs we compared the normalized read counts for bosutinib treatment with or without ILK. This resulted in 134 genes with significant (log2 fold change ±1 and P < 0.05) changes upon bosutinib addition and ILK loss (Fig. 5C). These 134 DEGs are shown across all samples, confirming that the changes are unique to bosutinib treatment of ILK knockout cells (Fig. 5D). These uniquely DEGs were involved in ECM and cell adhesion processes as shown using ToppGene (Fig. 5E). Network analysis of the 134 unique DEGs revealed a number of interactions with the most prominent being a collagen-integrin network of associated genes (Fig. 5F), but also adhesion complex associated, and ECM-related gene expression interactions (Supplementary Fig. S12C). Clearly, cell adhesion and cell-ECM interactions were the core effected processes in the context of ILK knockout-mediated bosutinib sensitivity, suggesting they may be responsible for the increased sensitivity to bosutinib observed in the ILK depleted cells.
Figure 5.
ILK loss in combination with bosutinib treatment results in differentially expressed genes. A, Principal component analysis using the Limma R package. B, Venn diagram of differentially expressed genes. A cutoff of FDR (Benjamini and Hochberg) P value adjustment <0.05 and a log2-fold change of ±1 was used for the differential expression analysis (DEA). Bosutinib EC20 (0.9 μmol/L) used. Bosutinib DEA: PX459v2-DMSO versus PX459v2-bosutinib; ILK loss DEA: PX459v2-DMSO versus ILK gRNA 2-DMSO; ILK loss + bosutinib DEA: ILK gRNA 2-DMSO versus ILK gRNA 2-bosutinib. Venn diagram created using the VennDiagram R package. C, Volcano plot of the 504 differentially expressed genes unique to ILK gRNA 2-bosutinib samples shown in B. The 504 unique genes were subject to a further cut-off of P < 0.05 (horizontal dotted blue line) and a log2-fold change of ±1 (vertical dotted blue lines) for ILK gRNA 2-bosutinib vs. PX459v2-bosutinib. D, Heatmap of the 134 unique and significantly changed genes shown in C, across all 12 samples. Heatmap created using the ggplots R package. Euclidean distance and complete linkage clustering were used. E, ToppGene gene ontology analysis for the 134 unique DEGs. The top hits from the biological process category are shown. Black dots represent q values. F, Integrin-collagen network map from the 134 unique DEGs created using GeneMANIA in Cytoscape. Associated genes were determined in GeneMANIA based on published databases and specific omics publications.
ILK knockout in combination with bosutinib causes cell adhesion defects
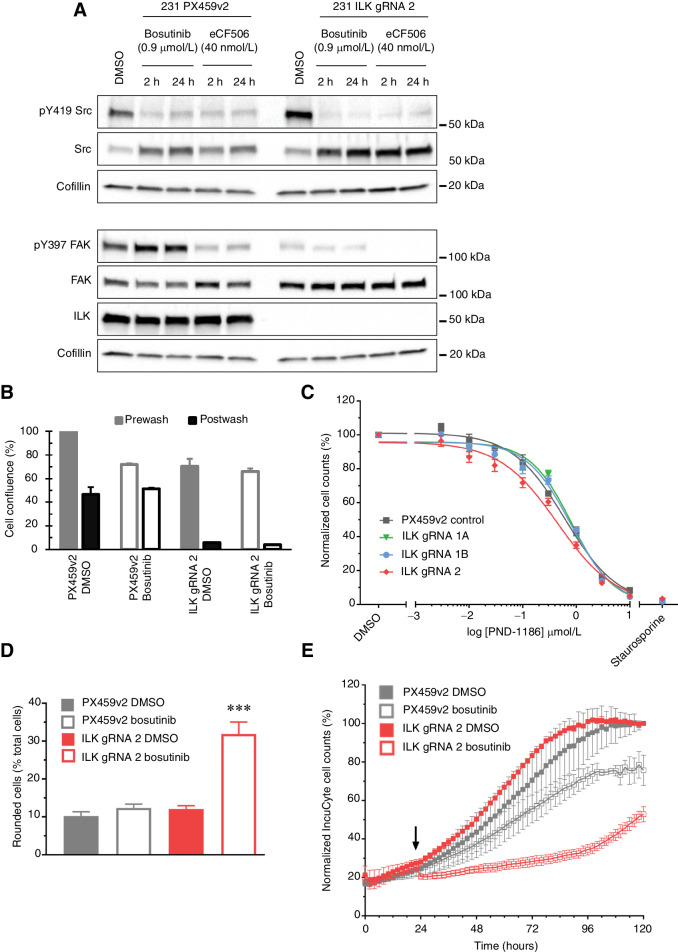
The IPP complex members and SRC are involved in regulating cell matrix adhesion. Taken together with the RNA-seq data, this suggested that defective adhesion may be involved in the increased sensitivity to bosutinib eCF506 treatment in ILK knockout cells. To characterize the adhesive defects in the ILK knockout cells, we first analyzed their focal adhesions by carrying out immunofluorescence using a number of antibodies against adhesion proteins (Supplementary Fig. S13A and S13B). Using paxillin as a marker of focal adhesions, we showed that adhesion length was significantly reduced in the ILK knockout cells. Furthermore, the distribution of adhesions, was also altered: ILK knockout cells lacked fibrillar adhesions located towards the center of a cell. These fibrillar adhesions are distinguished from focal adhesions by their low or absent phosphotyrosine content as was also seen in the ILK knockout cells. Fibrillar adhesions evolve from mature focal adhesions, indicating that the ILK knockout cells have a defect in focal adhesion maturation. This has been described previously in ILK knockout fibroblasts (30). Integrin clustering in focal adhesions results in the activation of FAK at the tyrosine (Y)397 autophosphorylation site. Consistent with decreased integrin activation, ILK loss decreased pY397-FAK by 2.4-fold (p-value = 0.047; Fig. 6A; quantification Supplementary Fig. S13C). We then measured cell–substrate adhesion following an adhesion “challenge” with a gentle PBS wash. In the ILK knockout cells, there was a complete loss of adherent cells following the adhesion challenge (Fig. 6B; data for additional ILK gRNA lines shown in Supplementary Fig. S13D). Interestingly, bosutinib, but not eCF506, caused an increase in cell adhesion after the adhesion challenge in the control cells, but both drugs were not able to rescue the adhesion defect in the ILK knockout cells (Fig. 6B; Supplementary 13D). It is not clear why bosutinib increases adhesion in the control cells as the reduction in adhesion observed with eCF506 is more consistent with the well documented role of SRC family kinases in cell–matrix adhesion (32). eCF506 reduced pY397-FAK levels in both control and ILK knockout cells. This can be attributed to the ability of eCF506 to bind SRC in an inactive conformation, leading to destabilization of the FAK-SRC complex and a loss of FAK phosphorylation. In contrast, bosutinib binds SRC in an active conformation, thus stabilizing FAK-SRC binding and Y397-FAK phosphorylation (24). The loss of ILK did not alter the ability of SRC and FAK to form a complex, although the levels of pY397–FAK complexed with SRC was reduced reflecting the reduced phosphorylation of FAK on Y397 in the ILK knockout cells (Supplementary Fig. S13E).
Figure 6.
ILK knockout in combination with bosutinib treatment causes cell adhesion defects. A, Western blot analysis of cell lines grown in 2D. Data are representative of three independent experimental replicates. B, Adhesion challenge: IncuCyte cell confluence was normalized to DMSO PX459v2. DMSO, solid fill; bosutinib, clear fill. C, 2D 8-point dilution for half-inhibitory concentrations (EC50) for PND-1186. Endpoint quantification using normalized cell counts form Hoechst-stained images. D, Cell-by-cell mask and a “rounded cells” classifier whereby area <600 μm2 and eccentricity <0.65, one hour post drug addition. E, IncuCyte S3 cell-by-cell quantification for MDA-MB-231 cells plated at 8,000 cells per well. Arrow, drug addition. A bosutinib EC20 (0.9 μmol/L) concentration was used in all experiments. ***, P < 0.001.
To address whether inhibition of FAK, could also synergize with ILK loss, ILK knockout cells were treated with the FAK inhibitor PND-1186. There was no difference in EC50 values for PND-1186 between the PX459v2 and ILK knockout cells, although pY397-FAK was effectively inhibited in both (Fig. 6C; Supplementary Fig. S14A). pY419-SRC was unaltered following PND-1186 treatment (Supplementary Fig. S14A and S14B). Previous studies have shown that inhibition of FAK is not sufficient to reduce phosphorylation of SRC and its substrates in integrin adhesion complexes, suggesting that either the residual FAK activity is sufficient to maintain SRC-dependent phosphorylation events, or that SRC is activated in a FAK-independent manner (33). To determine whether downstream signaling pathways were differentially regulated following FAK and SRC inhibitor treatment in the ILK knockout cells, a RPPA was carried out. This identified 24 and 10 uniquely differentially regulated proteins and phosphoproteins following bosutinib and PND-1186 respectively (P < 0.05, log2 fold change), while 13 were common to both drugs (Supplementary Fig. S14C). Taken together, these data suggest that although SRC and FAK have overlapping functions the reduction in Y397-FAK phosphorylation following FAK inhibitor treatment is not sufficient to prevent downstream SRC signaling and/or that they have independent signaling roles.
Initial microscopy observations showed that bosutinib treatment (EC20; 0.9 μmol/L) led to a rapid “rounding up” of MDA-MB-231 ILK knockout cells within one hour of addition (Supplementary Fig. S15A and S15B). We investigated this change by quantifying the number of total and “rounded cells” following bosutinib treatment using the IncuCyte S3 (Fig. 6D). Within the first hour of bosutinib addition, there was an increase in the number of rounded cells (cells with an area <600 μm2 and eccentricity <0.65): in the ILK knockout cells there was a 2.6-fold increase in rounded cells (p-value <0.0001) following bosutinib treatment compared to the ILK gRNA 2 DMSO-treated cells (Fig. 6D). This cell rounding was accompanied by an immediate decrease in total cell number in the MDA-MB-231 ILK knockout cells, but not PX459v2 cells (Fig. 6E), and a 3.0-fold reduction (P < 0.0001) in cell number after 72 hours in the ILK knockout cells as well as a 1.3-fold reduction (P = 0.045) in the PX459v2 cells. Tracking of apoptotic cells using a fluorescent reporter of caspase-3/7 activity showed that apoptotic cell numbers correlated with the rounded phenotype (Supplementary Fig. S15C and S15D). Taken together with the increased apoptosis observed by FACS in the ILK knockout cells following treatment with bosutinib over 4 days (Fig. 3F), this indicates that the cell rounding and loss of adhesion contributes to the cell death and decreased cell numbers observed. Thus, the increased sensitivity to bosutinib following ILK loss can be attributed to reduced adhesiveness when SRC inhibition is combined with ILK loss. Interestingly, siRNA targeting of ILK in MDA-MB-231 cells did not result in enhanced sensitivity to bosutinib or eCF506 despite reducing ILK by 90% (Supplementary Fig. S16), suggesting that complete loss of ILK is required to disrupt adhesion complexes in the MDA-MB-231 cells. In support of this, quantification of focal adhesions in the siRNA ILK-treated cells showed that there was no difference in focal adhesions in contrast to the disruption of adhesions seen in the sgRNA ILK knockout cells (Supplementary Fig. S13B).
ILK loss enhances SRC inhibition in vivo
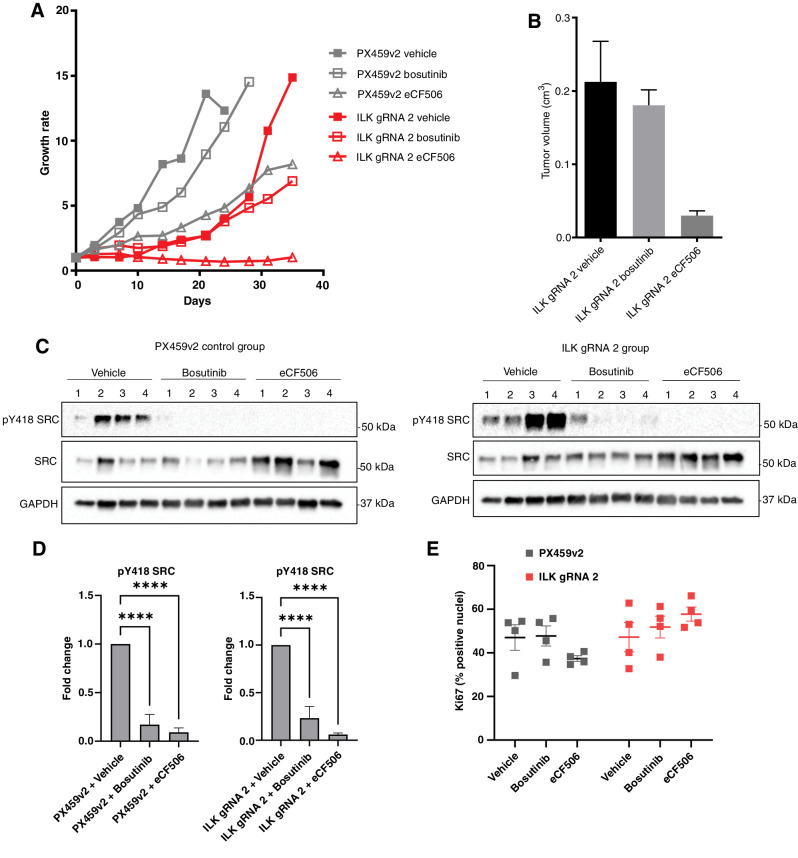
Finally, we characterized the effect of combined SRC inhibition and ILK loss on tumor growth in vivo. Immunodeficient R2G2 mice were subcutaneously injected with MDA-MB-231 cells with or without ILK, and once tumors had established, were treated with either bosutinib or eCF506 at doses previously shown to inhibit SRC activity in tumors (Fig. 7A; ref. 24). The rapid growth of the control tumors resulted in all animals being sacrificed by day 24 of treatment, which was extended to day 28 in the bosutinib treated animals. In contrast eCF506 reduced the growth of the control tumors and all animals were sacrificed at day 35 at the end of the experiment. There was a delay in tumor growth in the ILK gRNA tumors, but once established the tumors also grew rapidly and animals were sacrificed after 35 days of treatment. Treatment with eCF506 completely blocked the growth of the ILK gRNA tumors (Fig. 7A) and analysis of the final tumors showed a significant reduction in tumor volumes (Fig. 7B). Western blot analysis confirmed effective inhibition of SRC in tumors from both the bosutinib and eCF506-treated animals (Fig. 7C and D). Ki67 positivity in the tumors was analyzed as a measure of proliferation and showed that there was no difference in the proliferation of the ILK-deficient tumors compared with the controls at the end point of the study when the tumors were collected (Fig. 7E). Taken together with the lack of effect of ILK loss on proliferation in 2D (Fig. 3C), this indicates that ILK is not required for the intrinsic proliferative capacity of the cells. Bosutinib treatment prevented the rapid growth of the ILK gRNA tumors at the later time points; however, final tumor volumes and Ki67 positivity were not significantly different between the ILK gRNA vehicle and bosutinib-treated animals (Fig. 7). In contrast treatment with eCF506 completely blocked the growth of the ILK knockout tumors. The difference in activity of bosutinib and eCF506 is not fully understood but may reflect the reported ability of eCF506 to inhibit SRC in an inactive conformation, leading to disruption of protein–protein interactions in addition to inhibition of SRC kinase activity (24), as pY418 SRC was reduced in both bosutinib and eCF506-treated tumors (Fig. 7C and D).
Figure 7.
ILK loss enhances bosutinib and eCF506 treatment in vivo. A, Tumor growth rates for PX459v2 and ILK gRNA 2 tumors growing in immunodeficient Rag2-Il2rg double knockout mice (5 mice per group, 2 tumors per mouse) following treatment with vehicle, bosutinib (75 mg/kg), or eCF506 (40 mg/kg) given once daily via oral gavage. B, Final tumor volumes at day 35. C, Bosutinib and eCF506 inhibit pSrc in vivo as shown via Western blot analysis. D, Quantification of Western blots shown in C. E, Ki67 analysis using QuPath. ****, P < 0.0001, as determined by a two-way ANOVA with Bonferroni multiple comparisons correction. All error bars are SEM.
Discussion
SRC/ABL TKIs such as bosutinib and dasatinib have been investigated for treating breast cancer for some time (6, 8, 9, 34, 35). Yet so far these drugs have had limited clinical efficacy in solid tumors, despite attempts to validate potential gene signatures of sensitivity (36, 37). We aimed to identify possible synergistic gene–drug interactions in an attempt to understand the molecular determinants of bosutinib sensitivity and discover new rational combination therapies for SRC inhibitors. Using an unbiased genome-wide CRISPR-Cas9 knockout screen, we identified loss of the ILK–Parvin–PINCH complex as the key enhancer of SRC inhibition in in vitro and in vivo breast cancer models. ILK serves as a key scaffold protein in cell–matrix interactions and focal adhesions, tethering cells to their surrounding environment and interacting with multiple signaling pathways through interactions with its binding partners Parvin and PINCH (29). Furthermore, Horton and colleagues identified a consensus integrin adhesome comprising 60 proteins representing the core cell adhesion machinery, which is centered around four functionally interconnected axes, with the ILK–PINCH–PARVIN complex segregating into one of these (38). SRC is also a key regulator of integrin adhesions and ILK is known to interact with the SH3 domain of SRC demonstrating interconnected signaling pathways (39). However, they also have distinct biological activities and our data support a model in which targeting either SRC or ILK alone is not sufficient to inhibit proliferation and induce apoptosis, but that taking out both of these integrin complex components has a synergistic effect. The RNA-seq analysis identified a number of integrins and ECM proteins to be significantly differentially expressed in the ILK knockout cells, with further changes in adhesion related genes seen following treatment with bosutinib. This supports defective adhesion signaling as being the critical pathway involved in the enhanced sensitivity to bosutinib. Interestingly, there was no synergism between loss of ILK and treatment with the FAK inhibitor PND-1186, suggesting that the decreased activity of FAK in the ILK knockout cells is already sufficient to disrupt adhesion signaling and prime sensitivity to SRC inhibition. Previous studies have shown that SRC and FAK inhibitors can reduce the phosphorylation of different proteins within integrin adhesion complexes, and that FAK inhibition is not sufficient to reduce phosphorylation of SRC substrates (33). Taken together with our data, this suggests that although SRC and FAK have overlapping functions, the reduction in pY397-FAK following FAK inhibitor treatment is not sufficient to prevent downstream SRC signaling and/or that they have independent signaling roles. Indeed, analysis of signaling pathways that were altered following treatment with either bosutinib or PND-1186 revealed a number of differences in the ILK knockout cells. A greater understanding of the interplay between ILK, SRC and other adhesion signaling pathways in tumors will be required to fully exploit this vulnerability.
Upregulation of ILK is frequently observed in cancer tissues such as breast, colorectal and prostate (40). In addition, in prostate cancer, ILK expression is a promising prognostic marker and is correlated with tumor grade, progression, and 5-year survival (41). In breast cancer there is limited data, but one study has reported that ILK overexpression is associated with poor overall survival using immunohistochemistry methods (42), and that mRNA levels are higher in tumor compared with adjacent noncancerous tissues. We utilized the two largest publicly available breast cancer gene expression datasets (METABRIC and TCGA) to show that high ILK expression is associated with poor outcomes for the TNBC and basal subtypes. This is consistent with a number of in vitro and in vivo studies that have reported ILK-dependent effects on breast cancer proliferation, migration, invasion and tumor growth and initiation (43). Whether there is a subtype specific role for ILK in breast cancer remains to be established.
The potential of ILK as an anticancer target has been recognised for a number of years, with a number of putative inhibitors showing significant suppressive effect on cancer development and progression and hence a potential to target ILK for cancer treatment (40). However, although it was originally thought that ILK exerts its oncogenic effects via its kinase domain, mammalian ILK lacks kinase activity (44, 45) and thus off-target effects are anticipated for small molecules designed to target ILK kinase activity [e.g., Cpd 22 (46), QLT0267 (47), and QLT0254 (48)]. These putative “ILK inhibitors” in our hands and others (49) do not act via ILK kinase inhibition and do not mimic ILK knockdown (49). Notably, a recent study in a model of chronic myeloid leukemia has found QLT-0267 synergizes with dasatinib treatment (50); however, they did not demonstrate that it was working via inhibition of ILK kinase activity, and further investigation into the potential mechanism is required. There is therefore a need to create new bona fide ILK inhibitors that could be used in combination with SRC kinase inhibitors such as bosutinib and eCF506. One possibility is the development of drugs that can disrupt the IPP complex, which has shown some promise (51). This work provides the first step towards new rational drug combinations of ILK and SRC inhibitors in tumors expressing ILK.
Authors' Disclosures
O. Murina reports being a full-time employee and shareholder of AstraZeneca. A. Unciti-Broceta reports a patent for EP3298015B1 issued, licensed, and with royalties paid from Nuvectis Pharma, a patent for JP6684831B2 issued, licensed, and with royalties paid from Nuvectis Pharma, a patent for US10294227B2 issued, licensed, and with royalties paid from Nuvectis Pharma, a patent for CN107849050B issued, licensed, and with royalties paid from Nuvectis Pharma, and a patent for CA3021550A1 issued, licensed, and with royalties paid from Nuvectis Pharma. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank Dr John Dawson and Prof. Neil Carragher (Cancer Research UK Edinburgh Centre, University of Edinburgh) for their expertise and the use of the IncuCyte and ImageXpress systems. The authors are grateful to Prof. Andrew Jackson [Medical Research Council Human Genetics Unit (MRC), University of Edinburgh] and Prof. Margaret Frame [Director, Institute of Genetics and Cancer (IGC), University of Edinburgh] for their helpful discussions and comments as well as Richard Clark (Wellcome Trust Clinical Research Facility, Edinburgh) and Dr. Clare Logan (Illumina, UK) for their expertise setting up the dark cycles for the NGS run. They also acknowledge the Vector Core facility (University of Michigan) for the lentiviral production. They thank Elisabeth Freyer and Robbie Pineda (Flow Cytometry Facility, IGC, University of Edinburgh) for their help with the flow cytometry experiments and analysis. Additionally, the authors thank Kenneth Macleod (Microarray Services, IGC, University of Edinburgh) for running the RPPA samples and the associated analysis. The authors also acknowledge Fraser Laing (IGC, University of Edinburgh) for his help with the in vivo experiments.
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funding from Breast Cancer Now (Project reference: 2016NovPCC008). Dr. Olga Murina and Dr. David Parry were supported by the UK Medical Research Council (MRC) Human Genetics Unit core grant (no. U127580972). Professor Valerie Brunton, Dr. Adam Byron, Dr. Billie Griffith and Dr. Alexander Loftus were supported by Cancer Research UK (C157/A24837).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
H. Beetham: Data curation, software, formal analysis, supervision, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. B.G.C. Griffith: Formal analysis, investigation. O. Murina: Resources, validation, methodology, writing–review and editing. A.E.P. Loftus: Formal analysis, investigation, writing–review and editing. D.A. Parry: Resources, software, formal analysis. C. Temps: Resources, data curation, investigation. J. Culley: Investigation. M. Muir: Resources, formal analysis, investigation. A. Unciti-Broceta: Resources, supervision, writing–review and editing. A.H. Sims: Resources, supervision, writing–review and editing. A. Byron: Formal analysis, investigation, writing–review and editing. V.G. Brunton: Conceptualization, supervision, funding acquisition, visualization, writing–original draft, project administration, writing–review and editing.
References
- 1. Ottenhoff AE. Characterization of protein tyrosine kinases from human breast cancer: involvement of the c-src oncogene product. Cancer Res 1992;52:4773–8. [PubMed] [Google Scholar]
- 2. Martellucci S, Clementi L, Sabetta S, Mattei V, Botta L, Angelucci A. Src family kinases as therapeutic targets in advanced solid tumors: what we have learned so far. Cancers 2020;12:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Creedon H, Brunton VG. Src kinase inhibitors: promising cancer therapeutics? Crit Rev Oncog 2012;17:145. [DOI] [PubMed] [Google Scholar]
- 4. Klaeger S, Heinzlmeir S, Wilhelm M, Polzer H, Vick B, Koenig P-A, et al. The target landscape of clinical kinase drugs. Science 2017;358:1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khoury HJ, Cortes JE, Kantarjian HM, Gambacorti-Passerini C, Baccarani M, Kim D-W, et al. Bosutinib is active in chronic phase chronic myeloid leukemia after imatinib and dasatinib and/or nilotinib therapy failure. Blood 2012;119:3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vultur A, Buettner R, Kowolik C, Liang W, Smith D, Boschelli F, et al. SKI-606 (bosutinib), a novel Src kinase inhibitor, suppresses migration and invasion of human breast cancer cells. Mol Cancer Ther 2008;7:1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jallal H, Valentino ML, Chen G, Boschelli F, Ali S, Rabbani SA. A Src/Abl kinase inhibitor, SKI-606, blocks breast cancer invasion, growth, and metastasis in vitro and in vivo. Cancer Res 2007;67:1580. [DOI] [PubMed] [Google Scholar]
- 8. Campone M, Bondarenko I, Brincat S, Hotko Y, Munster PN, Chmielowska E, et al. Phase II study of single-agent bosutinib, a Src/Abl tyrosine kinase inhibitor, in patients with locally advanced or metastatic breast cancer pretreated with chemotherapy. Ann Oncol 2011;23:610. [DOI] [PubMed] [Google Scholar]
- 9. Finn RS, Bengala C, Ibrahim N, Roché H, Sparano J, Strauss LC, et al. Dasatinib as a single agent in triple-negative breast cancer: Results of an open-label phase 2 study. Clin Cancer Res 2011;17:6905. [DOI] [PubMed] [Google Scholar]
- 10. Single A, Beetham H, Telford BJ, Guilford P, Chen A. A Comparison of real-time and endpoint cell viability assays for improved synthetic lethal drug validation. J Biomol Screen 2015;20:1286. [DOI] [PubMed] [Google Scholar]
- 11. Beetham H, Chen A, Telford BJ, Single A, Jarman KE, Lackovic K, et al. A high-throughput screen to identify novel synthetic lethal compounds for the treatment of E-cadherin-deficient cells. Sci Rep 2019;9:12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006;7:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hart T, Tong AHY, Chan K, Van Leeuwen J, Seetharaman A, Aregger M, et al. Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 2017;7:2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colic M, Wang G, Zimmermann M, Mascall K, McLaughlin M, Bertolet L, et al. Identifying chemogenetic interactions from CRISPR screens with drugZ. Genome Med 2019;11:019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ong SH, Li Y, Koike-Yusa H, Yusa K. Optimised metrics for CRISPR-KO screens with second-generation gRNA libraries. Sci Rep 2017;7:7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pearce DA, Nirmal AJ, Freeman TC, Sims AH. Continuous biomarker assessment by exhaustive survival analysis. biorxivorg 2017. [Google Scholar]
- 18. Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. Statistics for Biology and Health 2000;40:978. [Google Scholar]
- 19. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 2016;34:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science 2015;351:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010;468:968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods 2011;8:659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Temps C, Lietha D, Webb ER, Li XF, Dawson JC. A novel mode of inhibiting SRC improves drug efficacy and tolerability. Cancer Res 2021. Online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moleirinho S, Chang N, Sims AH, Tilston-Lünel AM, Angus L, Steele A, et al. KIBRA exhibits MST-independent functional regulation of the Hippo signaling pathway in mammals. Oncogene 2013;32:1821. [DOI] [PubMed] [Google Scholar]
- 26. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fraser C, Dawson JC, Dowling R, Houston DR, Weiss JT, Munro AF, et al. Rapid discovery and structure–activity relationships of pyrazolopyrimidines that potently suppress breast cancer cell growth via SRC kinase inhibition with exceptional selectivity over ABL kinase. J Med Chem 2016;59:4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, et al. Up, down, and out: optimized libraries for CRISPRa, CRISPRi, and CRISPR-knockout genetic screens. Nat Commun 2018;9:5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Widmaier M, Rognoni E, Radovanac K, Azimifar SB, Fassler R. Integrin-linked kinase at a glance. J Cell Sci 2012;125:1839. [DOI] [PubMed] [Google Scholar]
- 30. Stanchi F, Grashoff C, Yonga CFN, Grall D, Fassler R, Van Obberghen-Schilling E. Molecular dissection of the ILK-PINCH-parvin triad reveals a fundamental role for the ILK kinase domain in the late stages of focal-adhesion maturation. J Cell Sci 2009;122:1800. [DOI] [PubMed] [Google Scholar]
- 31. Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, et al. Targeting Bcr–Abl by combining allosteric with ATP-binding-site inhibitors. Nature 2010;463:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeatman TJ. A renaissance for SRC. Nat Rev Cancer 2004;4:470–80. [DOI] [PubMed] [Google Scholar]
- 33. Horton ER, Humphries JD, Stutchbury B. Modulation of FAK and Src adhesion signaling occurs independently of adhesion complex composition. J Cell Biol 2016;212:349–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian J, Raffa Al F, Dai M, Moamer A, Khadang B, Hachim IY, et al. Dasatinib sensitises triple negative breast cancer cells to chemotherapy by targeting breast cancer stem cells. Br J Cancer 2018;119:1495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Colicelli J. ABL tyrosine kinases: Evolution of function, regulation, and specificity. Sci Signal 2010;3:re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pusztai L, Moulder S, Altan M, Kwiatkowski D, Valero V, Ueno NT, et al. Gene signature-guided Dasatinib therapy in metastatic breast cancer. Clin Cancer Res 2014;20:5265. [DOI] [PubMed] [Google Scholar]
- 37. Huang F, Reeves K, Han X, Fairchild C, Platero S, Wong TW, et al. Identification of candidate molecular markers predicting sensitivity in solid tumors to dasatinib: rationale for patient selection. Cancer Res 2007;67:2226. [DOI] [PubMed] [Google Scholar]
- 38. Horton ER, Byron A, Askari JA, Ng D. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 2015;17:1577–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kim Y-B, Choi S, Choi M-C, Oh M-A, Lee S-A, Cho M, et al. Cell adhesion-dependent cofilin serine 3 phosphorylation by the integrin-linked kinase center dot c-Src complex. J Biol Chem 2008;283:10089–96. [DOI] [PubMed] [Google Scholar]
- 40. Zheng CC, Hu HF, Hong P, Zhang QH, Xu WW, He Q, et al. Significance of integrin-linked kinase (ILK) in tumorigenesis and its potential implication as a biomarker and therapeutic target for human cancer. Am J Cancer 2019;9:186–97. [PMC free article] [PubMed] [Google Scholar]
- 41. Graff JR, Deddens JA, Konicek BW, Colligan BM. Integrin-linked kinase expression increases with prostate tumor grade. Clin Cancer Res 2001;7:1987. [PubMed] [Google Scholar]
- 42. Yang H-J, Zheng Y-B, Ji T, Ding X-F, Zhu C, Yu X-F, et al. Overexpression of ILK1 in breast cancer associates with poor prognosis. Tumor Biol 2013;34:3933. [DOI] [PubMed] [Google Scholar]
- 43. Tsirtsaki K, Gkretsi V. The focal adhesion protein Integrin-Linked Kinase (ILK) as an important player in breast cancer pathogenesis. Cell Adh Migr 2020;14:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Arrington J, Xue L, Wang W-H, Geahlen RL, Tao WA. Identification of the direct substrates of the ABL kinase via kinase assay linked phosphoproteomics with multiple drug treatments. J Proteome Res 2019;157:10. [DOI] [PubMed] [Google Scholar]
- 45. Fukuda K, Gupta S, Chen K, Wu C, Qin J. The pseudoactive site of ILK is essential for its binding to α-Parvin and localization to focal adhesions. Mol Cell 2009;36:819–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee S-L, Hsu E-C, Chou C-C, Chuang H-C, Bai L-Y, Kulp SK, et al. Identification and characterization of a novel integrin-linked kinase inhibitor. J Med Chem 2011;54:6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kalra J, Warburton C, Fang K, Edwards L, Daynard T, Waterhouse D, et al. QLT0267, a small molecule inhibitor targeting integrin-linked kinase (ILK), and docetaxel can combine to produce synergistic interactions linked to enhanced cytotoxicity, reductions in P-AKT levels, altered F-actin architecture and improved treatment outcomes in an orthotopic breast cancer model. Nat Biotechnol 2009;11:1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wheeler JJ, Sutton KL, Hedley DW. Inhibition of integrin-linked kinase by a selective small molecule inhibitor, QLT0254, inhibits the PI3K/PKB/mTOR, Stat3, and FKHR pathways and tumor growth, and enhances gemcitabine-induced apoptosis in human orthotopic primary pancreatic cancer xenografts. Cancer Res 2005;65:1497. [DOI] [PubMed] [Google Scholar]
- 49. Fang C-C, Chou T-H, Huang J-W, Lee C-C, Chen S-C. The small molecule inhibitor QLT-0267 decreases the production of Fibrin-induced inflammatory cytokines and prevents post-surgical peritoneal adhesions. Sci Rep 2018;8:9481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rothe K, Babaian A, Nakamichi N, Chen M, Chafe SC, Watanabe A, et al. Integrin-linked kinase mediates therapeutic resistance of Quiescent CML stem cells to tyrosine kinase inhibitors. Cell Stem Cell 2020;27:110. [DOI] [PubMed] [Google Scholar]
- 51. Kim O, Hwangbo C, Kim J, Li D-H, Min B-S, Lee J-H. Chelidonine suppresses migration and invasion of MDA-MB-231 cells by inhibiting formation of the integrin-linked kinase/PINCH/α-parvin complex. Mol Med Rep 2015;12:2161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available as Supplementary Files. Additional data is available from the corresponding author on reasonable request.