Abstract
The development of gene delivery vehicles with high organ specificity when administered systemically is a critical goal for gene therapy. We combine optical and positron emission tomography (PET) imaging of 1) reporter genes and 2) capsid tags to assess the temporal and spatial distribution and transduction of adeno-associated viruses (AAVs). AAV9 and two engineered AAV vectors (PHP.eB and CAP-B10) that are noteworthy for maximizing blood-brain barrier transport were compared. CAP-B10 shares a modification in the 588 loop with PHP.eB, but also has a modification in the 455 loop, added with the goal of reducing off-target transduction. PET and optical imaging revealed that the additional modifications retained brain receptor affinity. In the liver, the accumulation of AAV9 and the engineered AAV capsids was similar (~15% of the injected dose per cc and not significantly different between capsids at 21 h). However, the engineered capsids were primarily internalized by Kupffer cells rather than hepatocytes, and liver transduction was greatly reduced. PET reporter gene imaging after engineered AAV systemic injection provided a non-invasive method to monitor AAV-mediated protein expression over time. Through comparison with capsid tagging, differences between brain localization and transduction were revealed. In summary, AAV capsids bearing imaging tags and reporter gene payloads create a unique and powerful platform to assay the pharmacokinetics, cellular specificity and protein expression kinetics of AAV vectors in vivo, a key enabler for the field of gene therapy.
1. Introduction
With gene therapy now an approved and expanding disease correction modality, current efforts are focused on improving the safety and specificity of delivery vehicles. Of particular interest are gene therapies for neurological disorders where naturally occurring viral vectors have been re-engineered to efficiently transduce the brain with minimal integration in peripheral and clearance tissues. Many vectors have been engineered for tissue tropism and the ability to cross difficult body barriers [1,2]. An unmet need is the ability to non-invasively visualize in vivo the influence of capsid characteristics on delivery pharmacokinetics (PK) and gene transduction efficiency. In this study, we focus on imaging of recently engineered adeno-associated viral (AAV) vectors (single--stranded deoxyribonucleic acid (ssDNA) encapsulated within 25-nm protein capsids) for central nervous system (CNS) gene delivery [1,3,4] and focus on their impact on the brain and liver.
In vivo imaging techniques have great potential to contribute to the understanding of gene delivery. Biochemical techniques have previously included real-time PCR, Southern and Western blotting of the transduced gene and immunohistochemistry (IHC) [5]. However, these invasive methods do not provide the ability to track changes in situ over time and cannot be applied to the measurement of transduction in deep human tissues. Multimodality imaging can answer fundamental questions regarding vector PK and the resulting protein expression in both target and clearance organs across scale in the same subject.
Labeling a virus with trace amounts of radioactivity facilitates non-invasive PK studies via in vivo imaging for direct and non-invasive assessment of receptor binding and comparison between gene therapy vehicles. We have previously shown with positron emission tomography (PET) that radiolabeled engineered AAVs can deliver up to 30% of the injected dose per cubic centimeter (%ID/cc) to regions of the brain [6]. A unique aspect of our current work is the tetrazine modification of the AAV as a conjugation platform, which allows PET or optical tags to be easily incorporated for in vivo or ex vivo PK imaging, respectively.
For the assessment of liver accumulation, PET provides kinetic quantification that cannot be obtained with optical imaging. We then complement organ-level PET imaging with optical fluorescence confocal microscopy and IHC to more precisely characterize the cellular specificity and transduction of AAVs. Specifically, capsids are tagged with the fluorescent Cy5 dye, and the mNeonGreen (mNG) optical reporter gene is incorporated as a payload. Evaluation of the mechanisms behind liver detargeting of AAVs is important as liver toxicity has been a major factor limiting successful gene therapy. Severe liver dysfunction has been reported in recent studies of AAV9-based gene therapy for spinal muscular atrophy [7]. Progressive cholestatic hepatitis, a unique hepatic injury pattern predominantly in zone 3, developed in an AAV trial in patients with X-linked Myotubular Myopathy (XLMTM). The liver is heterogeneous in terms of molecular phenotypes and oxygen concentration, with reduced blood and intracellular oxygen observed in zone 3, near the central vein [8]. Hexagonal-shaped lobules surround the central vein, and the corners of each hexagon are formed by portal triads consisting of branches from the portal vein and hepatic artery and a bile duct. Others have reported zonal variation in AAV liver accumulation [9]. However, the underlying mechanisms were not determined and an improved understanding of the transport of AAVs within the liver is urgently needed. Kupffer cells are liver-resident macrophages adherent to liver vascular endothelial cells [10]. We hypothesize that differential AAV liver transduction across capsids results from differences in Kupffer cell uptake, and therefore we particularly focus on cell specificity of uptake and transduction.
Here, for the first time, we systemically administer AAV variants tagged for PET imaging with 64Cu on the capsid and also carrying a PET reporter gene cargo [11]. This is important as it allows us to spatially map both accumulation and transduction and to measure the timing of AAV transduction. PET reporter gene imaging enables serial assessment of transduction over days to years, through periodic injection of the reporter probe. Pyruvate kinase isoform M2 (PKM2), the PET reporter gene applied here is an isoform of pyruvate kinase involved in the final step of glycolysis with relatively low expression in the brain [11]. PKM2 is imaged by PET with 1-((2-fluoro-6-[18F]fluorophenyl)sulfonyl)-4-((4-methoxyphenyl)sulfonyl)piperazine ([18F]DASA-23) [11,12], a tracer that freely crosses the blood-brain barrier (BBB). [18F]DASA-23 was previously developed to assess tumor glycolysis [13–17] and has been shown to be safe for human imaging in the context of cancer [15, 16]. [18F]DASA-23 is the only currently identified radiotracer for brain reporter gene imaging, crosses the BBB freely and is undergoing a phase 1 clinical trial in patients with intracranial tumors (NCT03539731). Furthermore, in previous work [18], D. Brown et al. indicated that PKM2 was not upregulated in the brain as a result of novel AAV administration. This has previously been understood to be a slow process in which the single stranded genome carried by recombinant AAV (rAAV) is trafficked to the nucleus where particles are uncoated and the cargo converted to double-stranded DNA. Previous work with locally administered AAV9 has indicated that this process requires months to produce significant amounts of protein within the brain [12] and we quantify this here for novel AAVs and systemic administration [19].
We apply these imaging tools to evaluate modified capsids discovered using a directed evolution approach to viral capsid engineering and selection by the Gradinaru laboratory. They demonstrated that specific peptides displayed on the capsid surface enhanced neuronal transduction 40- to 90-fold throughout the brain when compared to conventionally-used AAV serotypes following IV injection in mice [3,4]. Such selections have provided multiple generations of engineered capsids that transduce the mouse brain efficiently following intravenous (IV) injection and reduce expression in the liver, spleen, kidneys, and lungs. Recent work indicates that systemic delivery of engineered AAVs to the brain can be achieved in marmosets [20]. Delivery was identified as the key issue in human clinical trials of AAVs for gene therapies targeting the CNS, where postmortem studies indicated that local/-stereotactic delivery in humans has been inadequate [21,22].
Capsid proteins impact endothelial transcytosis, mediate endosomal escape and nuclear import, and therefore varied capsids can differ in both systemic distribution and intracellular trafficking. A commonly manipulated loop in selected AAV capsids is the AA588 loop [23–25]. AAV-PHP.eB (denoted as PHP.eB hereafter), a variant of AAV9, was previously engineered by manipulation of the AA588 loop, resulting in increased efficiency in crossing the BBB and transducing neurons [3]. The AA455 loop of AAV9 is the furthest protruding loop from the surface of the capsid and has previously been implicated in neutralizing antibody binding [26] and liver transduction [27]. Starting with PHP.eB, two rounds of selection based on substitutions in the AA455 loop (AA452-458) resulted in a small subset of sequences that exhibited high levels of enrichment for the brain and negative enrichment for the liver and other peripheral organs. In mice, AAV.CAP-B10 (denoted as CAP-B10 hereafter) stands out as exhibiting higher transduction in the brain than PHP.eB with negligible liver transduction, and CAP-B10 is one of the capsids that conserves BBB-crossing activity in marmosets [20]. We begin by comparing brain uptake and liver cell uptake differences in this variant. Importantly, the tropism profile of AAV.CAP-B10 in mice is shifted toward neurons and away from astrocytes when compared with AAV-PHP.eB [18].
In summary, the unique tetrazine-modified AAV described here allows both 64Cu and Cy5 tagging of AAVs to assess both their tissue and cellular localization by PET and confocal microscopy, respectively. We combine this approach with [18F]DASA-23 PET and mNG optical reporter gene imaging for tissue and cellular level assessment of transduction. The powerful combination of quantifiable multi-modality imaging of tags and reporter genes allows for zonal and cell-specific evaluation of novel AAV capsid gene delivery.
2. Results
2.1. AAV labeling approach for PET and optical imaging
We previously developed a multichelator approach to increase the molar activity of the PET labeling of AAVs [6]. The multichelator was conjugated to AAV surface lysine and cysteines by bioorthogonal chemistry. Conjugation to surface lysine was previously shown to be feasible in fluorescence and PET imaging [6,28–31]. Here, for the first time, we extend the biorthogonal approach by tagging the AAV with an optical or PET label using the same tetrazine-AAV conjugation methodology to compare the PK of PHP.eB [4], CAP-B10 [20] and AAV9 [32]. Since CAP-B10 has > 98% peptide sequence homology compared to parent AAV9 and > 99% to PHP.eB, our multichelator labeling approach was again applied to compare in vivo pharmacokinetics. Based on surface solvent accessibility in the X-ray structure of the capsid [27], the estimated number of exposed lysine on AAV9, CAP-B10 and PHP.eB ranges from 420 to 480 out of 1185 and 1245 total lysine per AAV particle. This includes 7–8 lysine per viral protein (VP), where each viral particle was composed of 60 units of VP. The surface lysine labeling strategy is based on an inverse electron demand Diels–Alder reaction (IEDDA) [33].
The tetrazine-NHS ester attachment to lysine residues on AAV9 and PHP.eB did not alter transduction efficiency in an in vitro assay [6], and the same modification on CAP-B10 minimally affected transduction efficiency (decreased by 3%) (Figs. S1A–B) in an in vitro assay. For the 64Cu-radiolabeling of AAVs, we used 261 ± 35 equivalents Tz-NHS ester to modify the surface lysine of the AAV viral genome (5 × 1012 ± 6 × 1011 vg), followed by a reaction with 8 ± 6 equivalents of the 64Cu-multichelator-transcyclooctene (64Cu-(NOTA)8-TCO). Two-step radiolabeling of AAVs with 64Cu-(NOTA)8-TCO via dialysis and centrifugal filter purification afforded 51 ± 6% AAV recovery yields, and the average number of multichelators on 64Cu-AAVs was 0.2 ± 0.1 (n = 3) per viral genome. For the Cy5 fluorescent dye labeling, the surface lysine modification was performed with 413 ± 21 equivalents Tz-NHS and 69 ± 4 equivalents Cy5-TCO (n = 3) and yielded 13 ± 3 dye molecules per viral genome. Scaling by 50–80% capsid full/empty ratio, based on quality control data routinely acquired in CLOVER Center at Caltech, the yield was roughly 0.1–0.16 multichelators and 7–10 dye molecules per particle. The recovery yield of AAVs was 19 ± 7% based on viral genome.
2.2. While blood clearance was accelerated, brain accumulation of PHB. eB and CAP-B10 was enhanced upon PET imaging of tagged capsids
The novel capsid CAP-B10, evolved from PHP.eB and AAV9, displayed comparable transduction to PHP.eB in the brain and reduced transduction in the liver compared with AAV9 and PHP.eB [4,20]. In order to assess differences in the rate of target accumulation and blood clearance, we evaluated the PET-tagged capsids. The PK and biodistribution of the 64Cu-AAV9, -PHP.eB and -CAP-B10 capsids were assessed in C57BL/6 mice (n = 3–4/group) with PET/CT as illustrated in Fig. 1A–B. The projection images acquired after AAV systemic injection revealed two distinctions: the high brain uptake of CAP-B10 was similar to PHP.eB and the blood circulation of CAP-B10 was extended compared with PHP.eB (Fig. 1C). Sliced sagittal whole-body images from PET/CT (after 21 h) showed a similar distribution of 64Cu-PHP.eB and -CAP-B10 capsids in the internal organs and brain (Fig. 1D left). The accumulation of viral particles within the brain was largest in the cerebellum, midbrain and cerebral cortex (Fig. 1D right, Fig. S2).
Fig. 1.
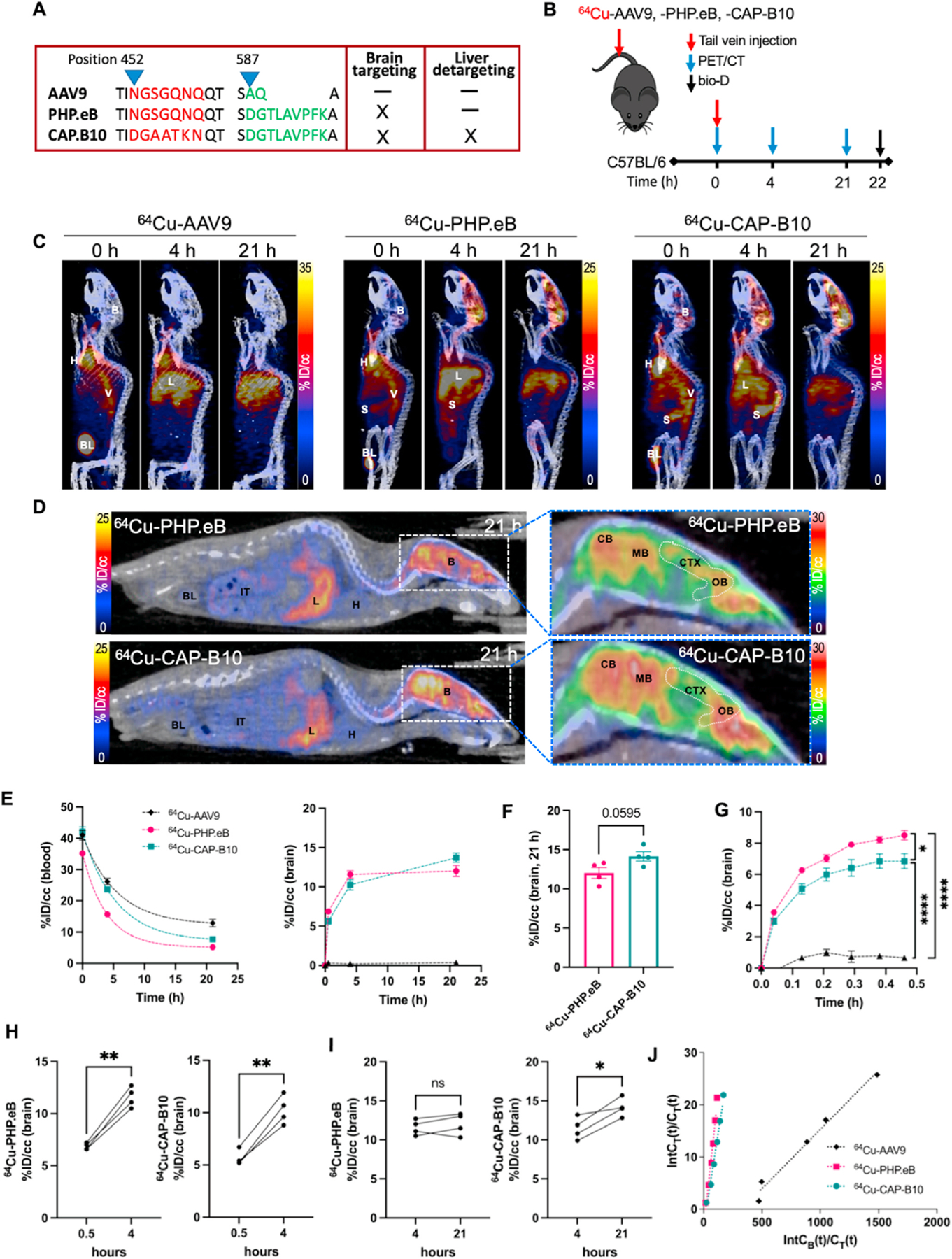
64Cu-AAV pharmacokinetics from PET image analysis of the radiolabeled capsids in the brain and blood (AAV9, PHP.eB, and CAP-B10). A. Capsid protein sequence of AAVs with enhanced brain targeting and reduced liver transduction. B. Experimental scheme of PET imaging and biodistribution of AAV9, PHP.eB, and CAP-B10 in C57BL/6 mice. C. Representative maximum intensity projected (MIP) PET/CT images of mice systemically administered 64Cu-AAV9 (n = 3), -PHP.eB (n = 4) and -CAP-B10 (n = 4). Mice were scanned for 30 min post-injection (p.i.) and rescanned at 4 and 21 h. D. Representative sliced whole body (left) and brain (right) PET/CT images of 64Cu-PHP.eB and -CAP-B10 accumulation. E. Time-activity curve (TAC) (%ID/cc) of AAVs (AAV9: black, PHP.eB: magenta, CAP-B10: green) from blood (left) and blood subtracted radioactivity in the brain (right). F. Blood subtracted radioactivity in the brain (%ID/cc) at 21 h p.i. G. The early binding kinetics of AAVs in the brain over 30 min p.i. H–I. Blood subtracted radioactivity in the brain (%ID/cc) of individual mice from 0.5 to 4 h (H) and 4–21 h (I). Significant changes in radioactivity value represent the gradual accumulation of AAVs. J. Logan plots of brain uptake rate after AAVs administration. Abbreviations: %ID/cc: Percent injected dose per cubic centimeter, B: brain, L: liver, H: heart, V: vena cava, BL: bladder, S: spleen, IT: intestinal track, CB: cerebellum, MB: midbrain, CTX: cerebellar cortex, OB, olfactory bulb. Data and error bars are presented as mean ± SEM. One-way ANOVA formed in Fig. 1G with Tukey’s multiple hypothesis correction. Unpaired two-tailed t-test with Welch’s correction was performed in Fig. 1F, H and I n.s.: not significant, *P < 0.05, **P < 0.01, ****P < 0.0001.
Blood circulation of AAV9 (t1/2 = 3.8 h) was longer than that of either PHP.eB (t1/2 = 2.7 h) or CAP-B10 (t1/2 = 3.7 h) (Fig. 1E left, Table S1). Brain accumulation of PHP.eB (n = 4, P < 0.0001 at 0, 4 and 21 h) was greater at all time points than that of AAV9 (n = 3) (Fig. 1E right). CAP-B10 similarly outperformed AAV9 (P < 0.0001 at 0, 4 and 21 h), accumulating more slowly but ultimately achieving a higher peak than PHP.eB (P = 0.0595, 21 h) (Fig. 1E right, Fig. 1 F). Mean blood subtracted brain uptake (%ID/cc) of 64Cu-PHP.eB and -CAP-B10 was 12–14%ID/cc (Fig. 1F). The blood subtracted accumulation of 64Cu-AAVs in the brain was analyzed in dynamic intervals over the first 30 min with accumulation by 64Cu-PHP.eB > 64Cu-CAP-B10 > 64Cu-AAV9 during this period (Fig. 1G). The accumulation of both 64Cu-PHP.eB and -CAP-B10 capsids was rapid and reached 8 and 6%ID/cc, respectively, after 30 min (Fig. 1G). 64Cu-PHP.eB peak brain accumulation occurred near 4 h (Fig. 1H left, Fig. 1I left); however, 64Cu-CAP-B10 accumulation continued to increase over 21 h (4 h vs 21 h, P = 0.028) (Fig. 1H right, Fig. 1I right). At 4 h post-injection (p.i.) an additional 4 to 5%ID/cc of AAVs (P < 0.01) had bound as compared with the initial 30 min (Fig. 1H). The biodistribution after perfusion was assessed at 22 h after injection of radiolabeled capsids, and the enhanced brain accumulation of the novel capsids was confirmed (Fig. S3A).
A Logan plot is a graphical analysis method for the estimation of the reversible uptake of tracer [34]. Logan plots confirmed that the 30-min accumulation in the brain was greater for 64Cu-PHP.eB than 64Cu-CAP-B10 and much greater than 64Cu-AAV9, with a distribution volume of 0.207, 0.143, and 0.0227, respectively (Fig. 1J). The estimated distribution volume of 64Cu-CAP-B10 was 30% lower than for 64Cu-PHP.eB. Over this 30-min period, we estimate that the increase in the distribution volume results from an increase in the number of bound AAVs.
2.3. On PET reporter gene imaging, brain transduction rapidly increases after systemic PHP.eB injection
We previously incorporated the PKM2 reporter gene within AAV9 viral capsids [19,35], and conducted a study of transduction with intracranial injection of AAVs [19]. The [18F]DASA-23 tracer is known to reversibly bind to PKM2 protein and consequently clears slowly from the brain when PKM2 is expressed. In that study, the mRNA level was correlated to the PET signal and expression of the reporter gene was significantly increased after ~2 months.
Here, we systemically administered AAV9 or PHP.eB and monitored the spatial distribution and longitudinal expression of PKM2 in the brain with [18F]DASA-23 (Fig. 2A). EF1A-PKM2 transgene was packaged in both AAVs, and transduction was monitored at 1, 3, 8, 16 and 34 weeks in C57BL/6 mice after injection of 2 × 1011 vector genome (vg). The subsequent PKM2 protein expression of PHP.eB at 3 weeks p.i. was apparent (Fig. 2B) with [18F]DASA-23 imaging. We directly compared the brain distribution of the tagged AAVs (Fig. 2C1) to the reporter gene image (Fig. 2C2). Interestingly, the [18F]DASA-23 tracer intensity was greater in the cerebral cortex than in the cerebellum (Fig. 2B, Fig. 2C2), and therefore differed from the distribution of the labeled capsid, where accumulation was greater in the cerebellum than in the cortex (Fig. 2C1). The spatial distribution of protein expression on PET reporter gene imaging was confirmed with fluorescence microscopy imaging of the optical reporter gene mNG (Fig. 2C3). Doses to acquire the images of Fig. 2C were 1 × 1011 ~ 1 × 1012 vg per mouse across the studies where we assessed the co-localization of AAV capsid delivery and their transductions. We have confirmed that dose differences in this range do not affect the regional distribution. Here we kept the EF1A and CAG promotor consistent with our previous PET studies (e.g. AAV9:EF1A-PKM2 for reporter gene imaging was consistent with Haywood et al. [19] and CAG used for 64Cu-AAV was consistent with our previous work [6]). In a previous study by Qin et al. [36], the comparison of gene expression in eight mammalian cells with six commonly used mammalian constitutive promotors (including EF1A and CAG) in lentiviral vector demonstrated that EF1A and CAG promoters were consistent in all mammalian cell types. Here, EF1A-PKM2 (Fig. 2C2) and CAG-mNG (Fig. 2C3) within the same capsid should therefore be comparable. Taken together, the results suggest differing affinity for specific cell types.
Fig. 2.
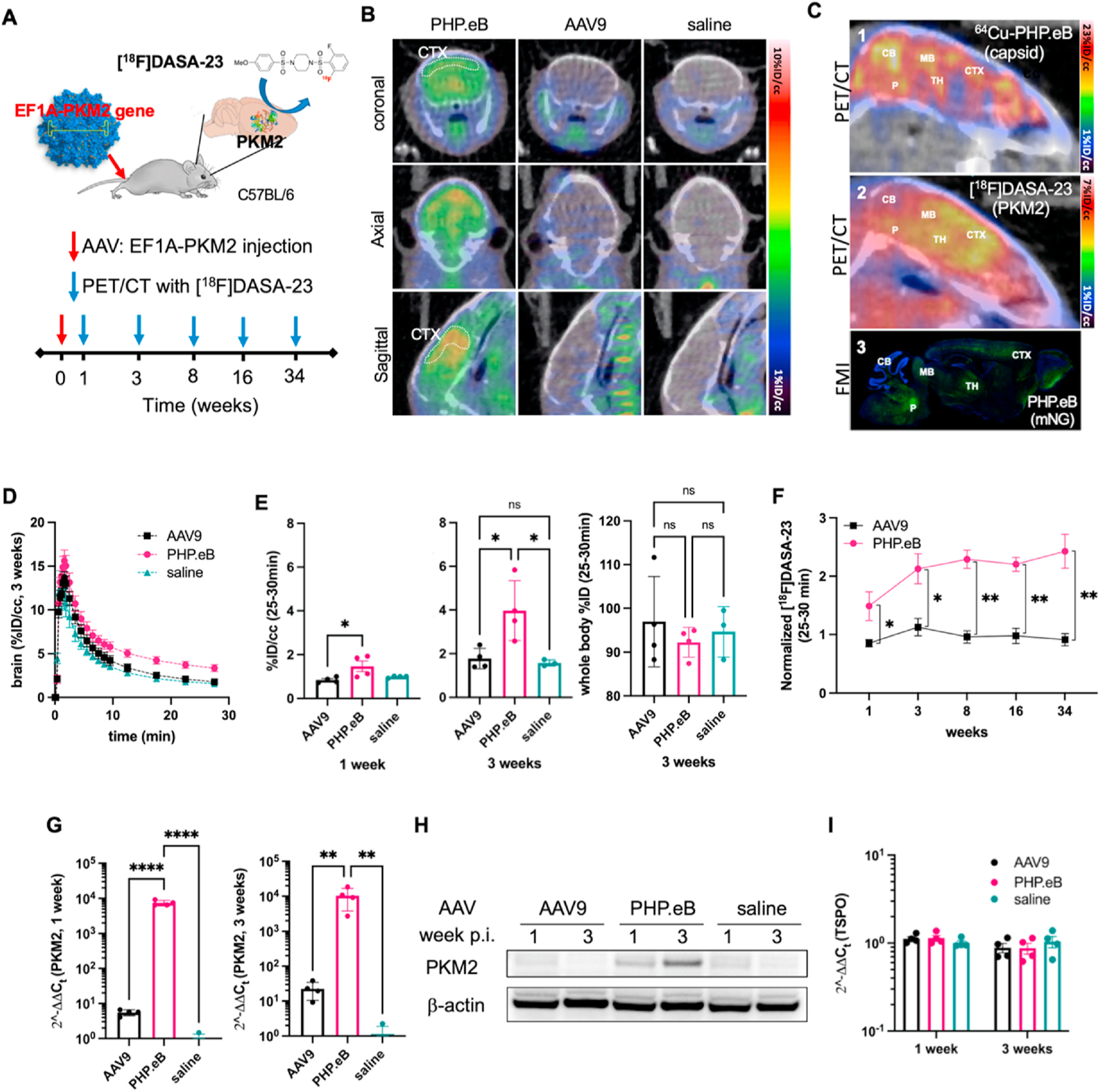
PET reporter gene imaging and quantification, comparing AAV9 and PHP.eB transduction of the PKM2 gene in the brain. A. Schematic illustration of the longitudinal PET reporter gene (PKM2) expression study. AAVs, PHP.eB (n = 4) and AAV9 (n = 4) with EF1A-PKM2, and saline (n = 4) are systemically injected in C57BL/6 mice, and PKM2 transduction was assessed by [18F]DASA-23 at 1, 3, 8, 16, and 34 weeks p.i. B. Representative sliced PET/CT images of brains from mice (at 3 weeks p.i. of AAVs) systemically injected with [18F]DASA-23. C. Comparison of PET/CT and fluorescent microscope images of the mouse brain. 1) PET/CT intensity image represents accumulated 64Cu-PHP.eB (6 × 1011 vg) capsid location at 21 h p.i. 2) PET/CT intensity image represents retained [18F]DASA-23 indicating where the PKM2 protein is overexpressed at 3 weeks after tail-vein administration of PHP.eB:EF1A-PKM2 (2 × 1011 vg). 3) Fluorescence microscopy intensity (FMI) of mouse brain overexpressing mNG at 3 weeks after tail-vein administration of PHP.eB:CAG-mNG (7 × 1011 vg). Green fluorescence represents mNG. Blue represents nuclear (DAPI) staining. CB, cerebellum; CTX, cerebral cortex; MB, midbrain; P, Pons; TH, Thalamus. D. Time-activity curve for the initial 30-min dynamic brain acquisition following [18F]DASA-23 administration. Dynamic data are from mice systemically injected with PHP.eB, AAV9, or saline at 3 weeks p.i. (n = 3–4 per group). [18F]DASA-23 retention increased in PHP.eB-treated mice compared to both AAV9 and saline treated mice. E. [18F]DASA-23 uptake (25–30 min) in the brain from PET/CT images acquired at 1 week (left) and 3 weeks (middle) p.i. of AAVs, and whole body (WB) retention (right) at 3 weeks p.i. F. Normalized fold uptake of [18F]DASA-23 (%ID/cc) over saline control mice (%ID/cc) at 1, 3, 8, 16 and 34 weeks p.i. of PHP.eB (left) and AAV9 (right). G. Gene expression of PKM2 from the mouse brain at 1 and 3 weeks p.i. of AAV9, PHP.eB, and saline (each week, nAAV9 = 4, nPHP.eB = 4, and nsaline = 4). H. Protein bands of PKM2 and β-actin control from the brain of mice at 1 and 3 weeks p.i. of AAV9, PHP.eB, and saline (each week, nAAV9 = 2, nPHP.eB = 2, nsaline = 2). I. Gene expression of TSPO (translocator protein) from the mouse brain at 1 and 3 weeks p.i. of AAV9, PHP.eB, and saline (each week, nAAV9 = 4, nPHP.eB = 4, and nsaline = 4). Data and error bars are presented as mean ± SEM. Ordinary one-way ANOVA with Tukey’s multiple comparison test (E and G) and multiple unpaired t-test corrected for multiple comparisons using the Holm-Sidak method (F) were performed for the statistical analysis. *P < 0.05, **P < 0.01, ****P < 0.0001.
The time-activity curve from the initial 30-min dynamic acquisition of brain confirmed the high specificity and longer retention of [18F] DASA-23 in the brain transfected with PHP.eB (Fig. 2D). PKM2 expression, as measured by [18F]DASA-23 at 1 week p.i., was significantly higher in PHP.eB-dosed mice (P = 0.033) than in AAV9-dosed mice. [18F]DASA-23 uptake was elevated 1.7-fold at 1 week (P = 0.033) (Fig. 2E left) and 2.2-fold at 3 weeks (P = 0.020) p.i. of PHP.eB (Fig. 2E middle). Enhanced [18F]DASA-23 retention was observed in the brain and resulted from the engineered capsid delivery. [18F]DASA-23 clears through the liver resulting in increased radioactivity in this region (Fig. S4). No significant difference in the reporter probe was observed through the whole-body analysis within the 30-min imaging window (Fig. 2E right, Fig. S4), suggesting that the difference in gene delivery between the capsids was limited to the brain. The PKM2 expression in brains of mice dosed with PHP.eB and detected by [18F]DASA-23 gradually increased over 8 weeks (1 week vs 8 weeks, P = 0.0425), remaining enhanced for more than 34 weeks and was significantly higher than in mice dosed with AAV9. (Fig. 2F).
We also assessed the mRNA and protein expression of PKM2 by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and Western blots from the brains of mice injected with AAV9, PHP.eB, or saline (Fig. 2G, Fig. 2H). For AAVs packaging EF1A-PKM2, systemic administration of 2 × 1011 vg of AAV9 and PHP.eB, resulted in a 6- and 7464-fold increase in mRNA, respectively, at 1 week and a 23- and 12,871-fold increase at 3 weeks compared with control mice injected with saline. The increased mRNA for PHP.eB (but not AAV9) was significant at each time point (p < 0.0001 and p < 0.001 at 1 and 3 weeks, respectively) (Fig. 2G). PKM2 mRNA expression following AAV9 and PHP.eB injection increased 17- and 5400-fold respectively from 1 to 3 weeks. The increase in PKM2 resulted in a significant increase in [18F] DASA-23 binding in the brain of mice injected with PHP.eB. Transcribed PKM2 mRNA from PHP.eB in the mouse brain was translated to PKM2 protein at 1 and 3 weeks, detected by Western blot (Fig. 2H). We also evaluated the mRNA expression of translocator protein (TSPO), associated with brain inflammation, to determine whether the expression of exogenously delivered PKM2 increased neuroinflammation. No significant elevation of TSPO mRNA was found in brains transduced with PKM2 by AAV9 and PHP.eB at 1 and 3 weeks (Fig. 2I).
2.4. PET image analysis and biodistribution of tagged AAV capsids provide insight into clearance
We then analyzed the pharmacokinetics of the three tagged capsids as they accumulated in the liver. The distribution of viral particles in bladder, intestinal track and liver was clearly visualized in Fig. 1D. Since the blood clearance of 64Cu-PHP.eB was demonstrated in Fig. 1E to be faster than the other capsids studied, we asked whether rapid liver uptake was a factor. In fact, at 0.5 h after injection, the liver accumulation of 64Cu-PHP.eB (Fig. 3A) was 8.7%ID/cc (P = 0.038), which was greater than 64Cu-CAP-B10 (5.6%ID/cc, P = 0.038) and 64Cu-AAV9 (3.1%ID/cc, P = 0.0026) (Fig. 3B), in part accounting for the faster blood clearance. Similarly, 64Cu-PHP.eB liver accumulation was greater than 64Cu-CAP- B10 and 64Cu-AAV9 at 4 h after injection (Fig. 3C). However, the 21 h accumulation of the three capsids was not significantly different (Fig. 3D).
Fig. 3.
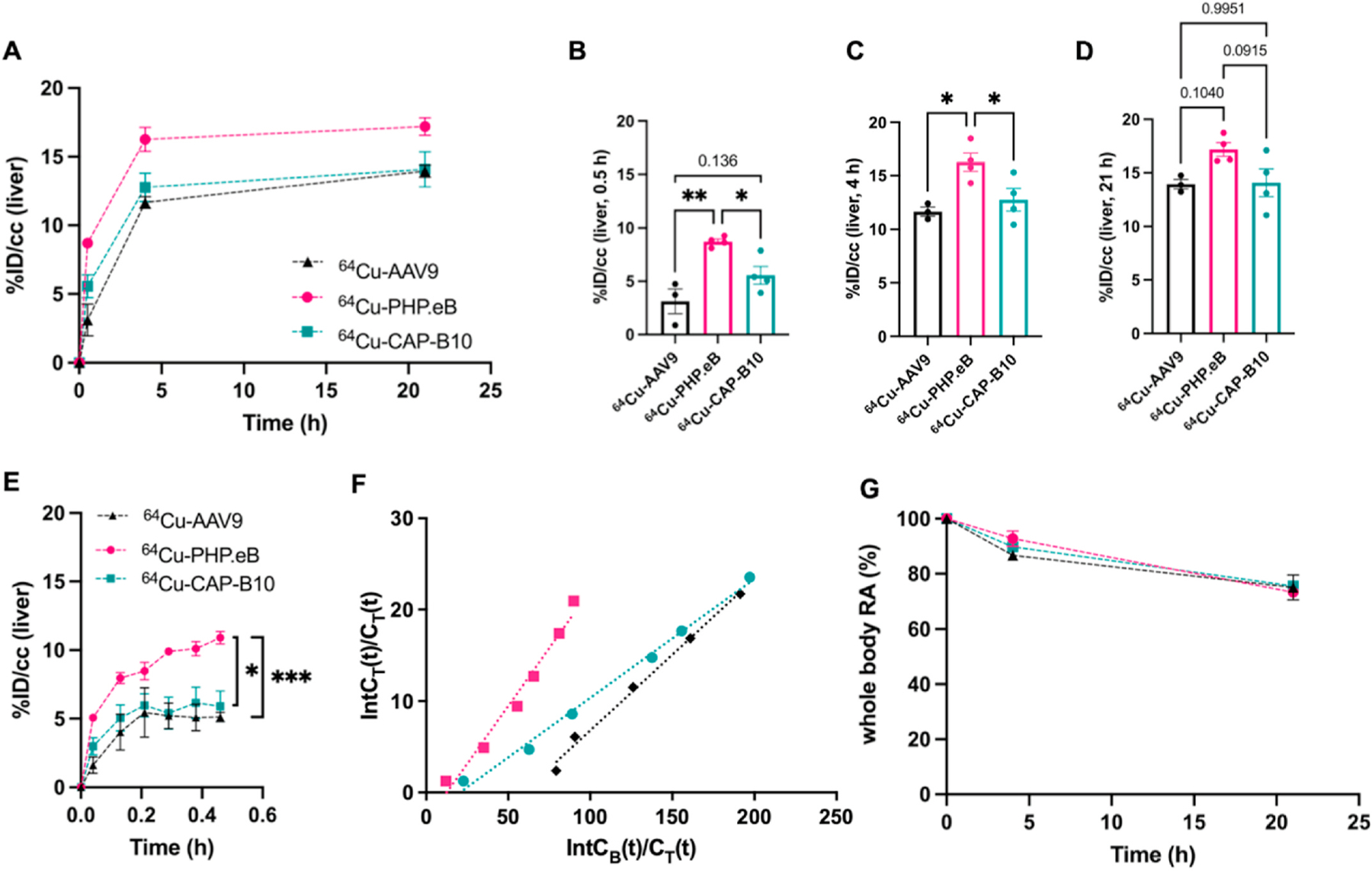
The pharmacokinetics of 64Cu-AAVs in the liver and entire body. A. Blood subtracted accumulation of 64Cu-AAV9 (n = 3), -PHP.eB (n = 4), and –CAP-B10 (n = 4) in the mouse liver (C57BL/6). B-D. Bar graphs displaying statistical significance of liver radioactivity from Fig. 3A at 0.5 h (B) 4 h (C) and 21 h (D) time points. E. The early binding kinetics of AAVs in the liver over 30 min. F. Logan plots of liver uptake rate after AAVs administration. CB(t) and CT(t) are the radioactivity concentration in the blood and target at a given time, and IntCB(t) and IntCT(t) are the accumulated radioactivity in the blood and target, respectively, from the time of injection to 30 min. G. The clearance of 64Cu-AAV9 (n = 3), -PHP.eB (n = 4), and –CAP-B10 (n = 4) from the body. RA: radioactivity. Data and error bars are presented as mean ± SEM. Ordinary one-way ANOVA (B, C, D) and two-way ANOVA (E) with Tukey’s multiple comparison test were performed. *P < 0.05, **P < 0.01, ***P < 0.001.
The accumulation of 64Cu-AAVs in the liver was then plotted in dynamic intervals over the first 30 min. The accumulation of 64Cu-PHP.eB capsids was greater than 64Cu-CAP-B10 (P < 0.05) and 64Cu-AAV9 (P < 0.001) at 0.5 h p.i. (Fig. 3E). Logan plots of liver uptake confirmed that the 30 min accumulation was greater for 64Cu-PHP.eB > 64Cu-AAV9~64Cu-CAP-B10 with a distribution volume of 0.253, 0.166, and 0.130, respectively (Fig. 3F). The early retention of 64Cu-CAP-B10 in the liver was estimated to be 49% lower than for 64Cu-PHP.eB (Fig. 3F). The whole-body clearance of radioactivity was similar for the three capsids (Fig. 3G).
The biodistribution assessed at 22 h after the PET/CT image acquisition and blood perfusion confirmed that the liver, spleen, and brain were the major AAV uptake organs, each with more than 10%ID/g for the novel capsids (Fig. S3B, Table S2). At this time point, the accumulation of 64Cu-PHPeB and -CAP-B10 in the liver was slightly (<2%ID/g) higher than 64Cu-AAV9 (Fig. S3C). In the spleen, the accumulation of 64Cu-PHPeB and -CAP-B10 was 2- and 3-fold higher as compared to 64Cu-AAV9 (Fig. S3D).
2.5. AAV zonal and cellular localization within the liver varies with the capsid
We then applied optical imaging of tagged AAVs to determine the capsid localization in liver tissue on the cellular scale. We employed an optical probe (Cy5-TCO) attached to capsid lysine in a two-step reaction (Fig. 4A). We injected the tagged Cy5-AAV9, -PHP.eB, and -CAP-B10 systemically through the tail vein and investigated the location of viral capsids within the liver at 4 h p.i. and transduction at 3 weeks p.i. (Fig. 4B). Since hepatocytes and Kupffer cells are the major cell populations in the liver, we stained Kupffer cells with the Clec4F antibody to differentiate the location of viral capsids. With a 20× magnification, we found that the fluorescence from PHP.eB and CAP-B10 capsids was spatially correlated with Kupffer cells and enhanced compared to that of AAV9 and saline (Fig. 4C, Fig. S5). The trends observed for the mean fluorescent intensity (Cy5) of liver tissue obtained from mice at 4 h p.i. of Cy5-AAVs (Fig. 4C) were similar to the radiotracer result (Fig. 3C) which showed the accumulation of Cy5-PHP.eB was significantly greater than that of Cy5-CAP-B10 or -AAV9. As shown by the high magnification images (63×), we identified Cy5-PHP.eB and -CAP-B10 capsids within Kupffer cells (Fig. 4D). The overlap coefficient of Cy5-AAVs (red channel) on Kupffer cells (green channel) confirmed the greater colocalization of Cy5-PHP.eB and -CAP-B10 capsids within Kupffer cells compared to Cy5-AAV9 (Fig. 4E), where the overlap was greater for Cy5-CAP-B10 than for Cy5-PHP.eB. High resolution microscopic images of Kupffer cells further demonstrate the cytoplasmic localization of the Cy5-PHP. eB (Fig. 4F) and -CAP-B10 (Fig. S6).
Fig. 4.
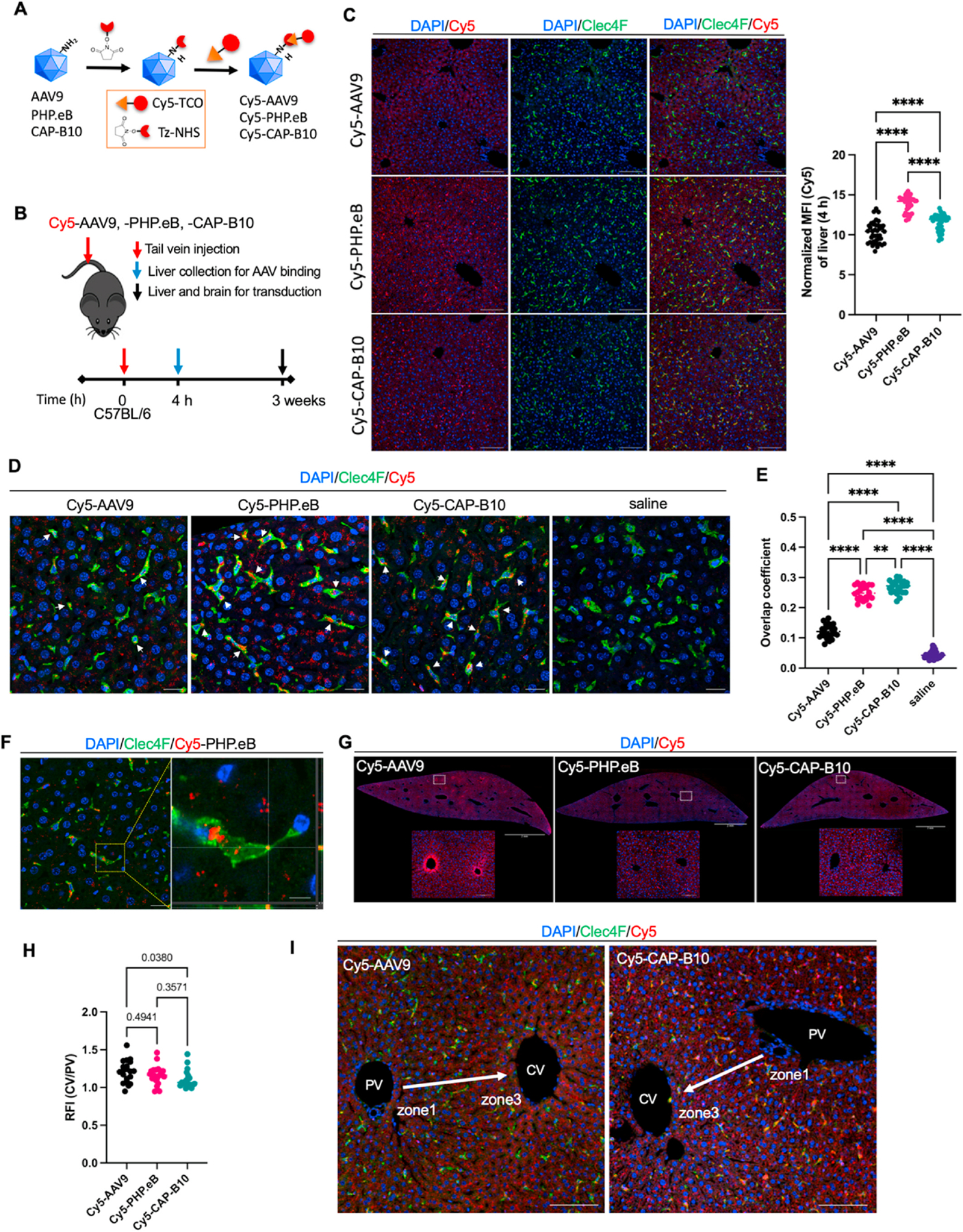
On optical imaging at 4 h after injection, liver distribution of Cy5-labeled AAV9 variants displays a distinct cellular and zonal pattern. A. Labeling scheme for Cy5-AAVs (AAV9, PHP.eB and CAP-B10). B. Schematic of study for identifying viral particle distribution and transduction in liver. C. Representative fluorescence microscope images (FMI) of mice (C57BL/6) liver sections stained with Clec4F (green) and DAPI (blue) and the quantitation of the accumulated Cy5-AAVs in whole liver tissue (right) at 4 h p.i. Red and green channel represent the presence of Cy5-labeled viral particles and Kupffer cells, respectively. Bar graph represents a normalized mean fluorescent inteisity (MFI) of Cy5-AAVs in whole liver tissues. Scale bar: 100 μm. D. Maximum intensity projection (MIP) of liver tissue stained with Clec4F (green) and DAPI (blue). White arrow indicates Cy5-AAVs (red) in Kupffer cells. Scale bar: 20 μm. E. Manders’ overlap coefficient of AAVs. Colocalization of Cy5-AAV (red channel) and Kupffer cells stained with Clec4F (binarized green channel). F. A Z-section of Cy5-PHP.eB in Fig. 4D (left) and magnified square region of interest from left image (right). Cy5-PHP.eB (red) is found on the surface and cytoplasm of Kupffer cell (green). Scale bar: 20 and 5 μm. G. FMIs of mouse (C57BL/6) liver sections collected at 4 h p.i. of Cy5-AAV9, -PHP.eB, and -CAP-B10. Sliced tissues were stained with DAPI (blue). Maximum threshold was adjusted to 10% of raw image to enhance the Cy5 fluorescent channel (red). Scale bar: 2 mm (whole liver image) and 100 μm (magnified image). H. Relative fluorescent intensity of cells in zone 1 portal vein (PV) over zone 3 central vein (CV) in liver tissues. I. Representative FMI of mouse liver sections stained with DAPI (blue) and Clec4F (green) with Cy5 fluorescence (red) around the CV and PV. Arrow indicates blood flow from PV to CV. Scale bar: 100 μm. Data and error bars are presented as mean ± SEM. Ordinary one-way ANOVA with Tukey’s multiple comparison test was performed for the statistical analysis (C, E, and H). **P < 0.01, ****P < 0.0001.
Interestingly, we observed higher fluorescence in the Cy5-AAV channel in hepatocytes surrounding the central vein of the liver in mice administered Cy5-AAV9 as compared to Cy5-PHP.eB and -CAP-B10 (Fig. 4G). To quantify the differences in the fluorescence from zone 1 (portal vein) to zone 3 (central vein), the relative fluorescent intensity (RFI) of cells in zone 3 over zone 1 was measured after administration of Cy5-AAV9, -PHP.eB, and -CAP-B10. Results from region of interest (ROI) analysis (Fig. S7) confirmed the enhanced accumulation of Cy5-AAV9 in zone 3 compared with Cy5-CAP-B10 (P = 0.038) and Cy5-PHP.eB (P = 0.494) (Fig. 4H and I).
2.6. Optical imaging of the spatial and cellular distribution of transduction
Finally, optical imaging of a fluorescent reporter protein was used to assess the spatial and cellular distribution of transduction. To compare the transduction pattern of Cy5-labeled AAVs with the distribution of Cy5 labeled AAVs in Fig. 4, we evaluated transduction of CAG-mNG (mNeonGreen) in the mouse liver at 3 weeks p.i. of 7 × 1011 vg Cy5-AAV9, -PHP.eB, and -CAP-B10. In microscope images of the entire liver, mNG fluorescent protein was highly expressed in mice administered Cy5-AAV9 compared to Cy5-PHP.eB and -CAP-B10 (Fig. 5A). Based on the cellular morphology, we did not observe transduction of Kupffer cells. In this study we did not differentiate endothelial cells and hepatocytes, both of which have previously been observed to accumulate AAV8 and be transduced in a previous optical imaging study [10]. Brain transduction of mNG was greater in mice injected with Cy5-PHP. eB and -CAP-B10 than with Cy5-AAV9 (Fig. 5A) as in our previous studies. Quantification of mean liver green fluorescence intensity revealed that Cy5-AAV9 transduced a significantly greater amount of mNG than Cy5-PHP.eB and -CAP-B10, likely due to the higher uptake of the engineered AAVs, PHP.eB and CAP-B10, in Kupffer cells compared to AAV9 (Fig. 5B).
Fig. 5.
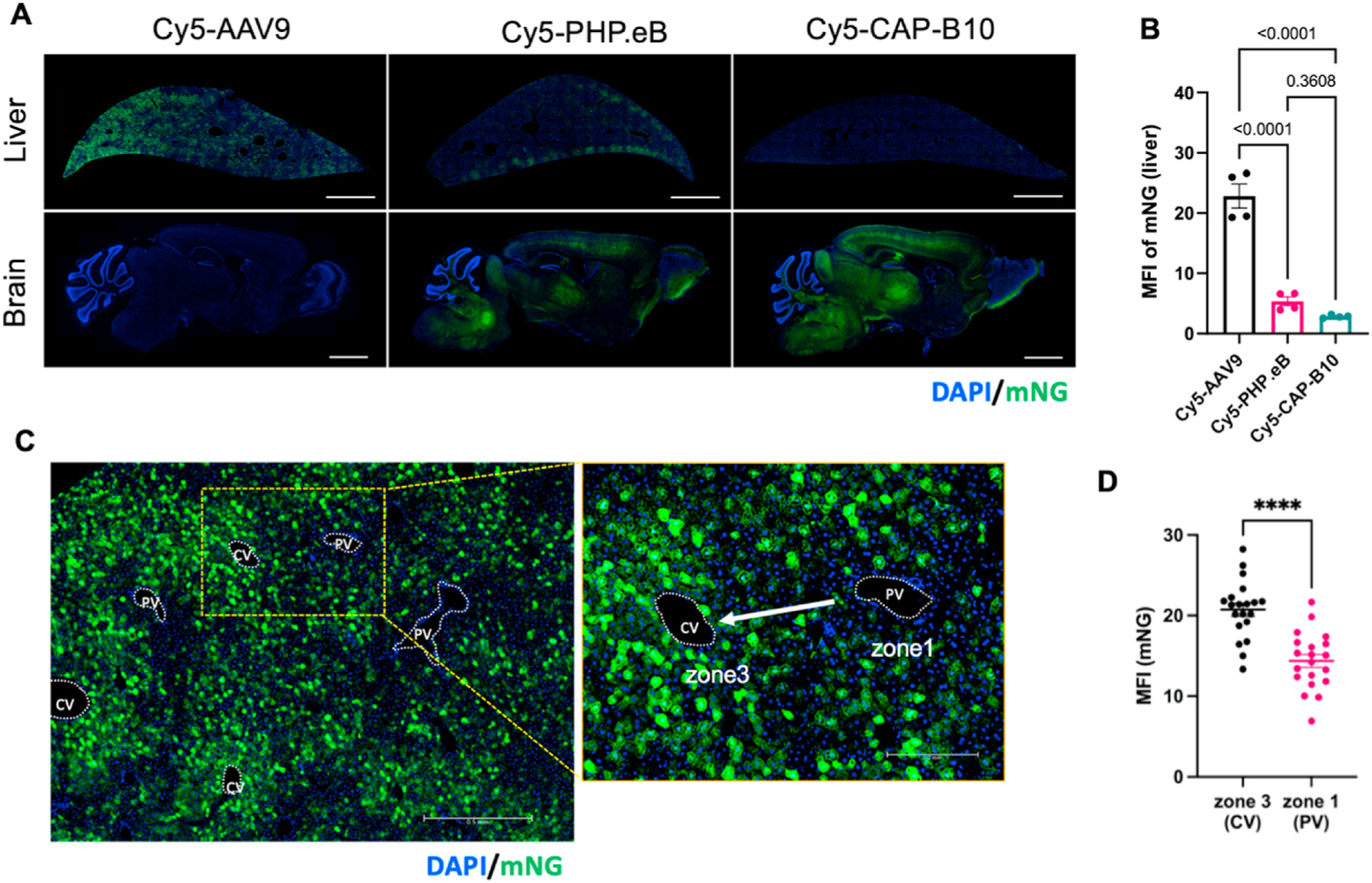
Liver transduction of Cy5-labeled AAV9 variants displays the same zonal patterns as AAV localization. A. Fluorescence microscope images (FMIs) of liver and brain sections from mice collected at 3 weeks systemic p.i. of 7 × 1011 vg Cy5-AAV9, -PHP.eB, and -CAP-B10 packaging CAG-mNeonGreen (mNG). Sliced tissues were stained with DAPI (blue) for morphology. Scale Bar: 2 mm. B. Median fluorecence intensity (MFI) of mNG from whole liver section (n = 4 from 2 mice per group). C. Magnified FMI of mouse liver obtained at 3 weeks p.i. of AAV9:CAG-mNG. In left image, hepatocytes around central veins (CV) express more mNG compared to those around the portal vein (PV). Yellow area is magnified in right image. Sliced tissues were stained with DAPI (blue), allowing the differentiation of portal vein and central vein. Arrow indicates blood flow from PV to CV. D. Comparison of MFI (mNG) of zone 1 (PV) and zone 3 (CV) in livers of mice administered Cy5-AAV9:CAG-mNG. mNG expression in cells around CV was significantly higher than near PV (p < 0.0001). ROI data (n = 20) was pooled from livers of two mice administered Cy5-AAV9. Data and error bars are presented as mean ± SEM. Ordinary one-way ANOVA with Tukey’s multiple comparison test (B) and unpaired two-tailed t-test with Welch’s correction (D) were performed for the statistical analysis. ****P < 0.0001.
Transgene expression in the liver was reduced in previous studies of capsid variants CAP-B10 and PHP.eB (particularly CAP-B10) [20]. Our observation of similar uptake of 64Cu-CAP-B10 and -PHP-eB in the liver as compared to 64Cu-AAV9 (Figs. 3C vs Fig. 4C right) suggests that the reduced transduction in the liver may be related to differences in liver cell uptake of engineered AAVs and AAV9.
We further investigated the spatially-differentiated mNG transduction pattern in the liver of mice injected with Cy5-AAV9. mNG was strongly expressed in hepatocytes around the CV (zone 3) and diminished around the PV (zone 1). Thus, mNG intensity was inverse to the capsid flow direction (Fig. 5C). Quantification of the MFI in zone 3 (CV) vs zone 1 (PV) demonstrated a significantly higher transduction of mNG in zone 3 (CV) than in zone 1 (PV) (Fig. 5D). The data suggest that the zonal distribution of the Cy5-AAV9 capsids observed at 4 h p.i. are related to the zonal transduction of gene.
3. Discussion
Recently, a series of AAV capsids have been developed to reduce transduction in the liver, spleen, kidneys, and lungs when compared with naturally-occurring AAV serotypes. New capsids have shown the potential to cross the BBB in non-human primates [20] and accomplish cell specific transduction, and we evaluated one such capsid here. Novel imaging combinations are needed to answer fundamental questions regarding vector pharmacokinetics and the resulting protein expression in both target and clearance organs. Here, we established an AAV imaging platform by conjugating an optical or PET imaging tag using a tetrazine-AAV biorthogonal reaction. Also, for the first time, we uniquely combine both capsid and reporter gene imaging with PET to noninvasively assess AAV brain localization and transduction over time and space.
For consideration of liver accumulation and transduction, we combined optical reporter gene imaging and capsid tagging with PET tagging. While the immunotoxicity of rAAV is less severe than other viral vectors, recent tragic patient deaths highlight the need for improved vectors [7,37]. The inherent hepatotropism of some current AAVs motivated their development and translation but also limits the dose and associated toxicity. Severe liver dysfunction has been reported and associated with recent deaths in a report by Greer et al. [7]. Therefore, there is an urgent need to enhance efficiency and reduce off-target toxicity. Our non-invasive AAV imaging strategy, which includes capsid and reporter gene imaging, may serve this unmet need.
CAP-B10 shares a modification of the AA588 loop of PHP.eB, which was selected to enhance transgene expression in the brain, and also has a modification of the AA455 loop, which was selected to reduce the transgene expression in peripheral organs. Our capsid imaging differentiated the systemic pharmacokinetics of CAP-B10 and PHP.eB. Through the analysis facilitated by PET imaging of the capsid, we found that the volume of distribution was 30% lower with 64Cu-CAP-B10 than 64Cu-PHP.eB and the early retention of 64Cu-CAP-B10 in the liver was 49% lower than for 64Cu-PHP.eB. Since the volume of distribution is proportional to the receptor density and ligand receptor association constant, we hypothesize that the differences are due to differences in receptor affinity. The additional modification of the AA455 loop may play a role in cell-surface receptor binding: 1) as the furthest protruding loop, 2) due to its proximity to the AA588 loop which is the second furthest protruding, and 3) as an integral part of the three-fold symmetry of the AAV [27]. As a result of the reduced liver uptake, the blood circulation time of CAP-B10 was enhanced. Therefore, ultimately the brain accumulation was similar to 64Cu-PHP.eB (although slowed).
In the liver, we found that the accumulation of AAV9 and the engineered AAV capsids was high (~15% of the injected dose per cc for all capsids considered and not significantly different between capsids at 21 h). However, liver transduction from the engineered capsids was low. We hypothesized that this disparity between the uptake and transduction might result from a different cellular localization of AAVs within the liver. Our optical imaging results from Cy5-labeled AAVs indicate early uptake of the PHP.eB and CAP-B10 primarily by Kupffer cells. CAP-B10 accumulation overlapped with the locations of Kupffer cells to a greater extent than PHP.eB. The enhanced Kupffer cell uptake of the engineered AAVs was in part responsible for the reduction in transfected hepatocytes. In a previous report by Carestia et al., the total accumulation could not be measured but individual AAVs monitored in real time by 30 min were observed on the surface of both Kupffer and endothelial cells and were internalized by Kupffer cells over time. However, Kupffer cell transfection was not observed on day-2 post injection [10]. Taken together, these studies indicate that AAV internalization by Kupffer cells does not appear to result in transfection. Incorporating the information from PET and optical imaging, the data suggest that AA588 modification in engineered AAVs altered the cellular tropism in the liver, and the AA455 loop modification in CAP-B10 reduced the receptor affinity for both the brain and liver. Similarly, in the spleen, the uptake of 64Cu-PHP.eB and -CAP-B10 was higher than 64Cu-AAV9 and the gene transduction of PHP.eB and CAP-B10 was lower as reported by Goertsen et al. [20]. The rapid uptake of engineered AAVs in the reticuloendothelial system (RES) may contribute to the enhanced clearance and reduced transduction.
Our data support zonal variations in the accumulation, uptake and transduction of hepatocytes. In mice, Zone 3 (central venous hepatocytes) is thought to play a role in drug metabolism and detoxification with enhanced uptake of therapeutics. Thus, the local conditions may impact zone-dependent liver transduction. AAV9 capsids were particularly localized in zone 3, suggesting that they traversed through the liver with reduced Kupffer cell uptake before densely concentrating in hepatocytes near the central vein. This dense accumulation coincided with similar transduction for AAV9.
PET reporter gene and probe systems have been exploited to non-invasively assess expression of the transgene in gene therapy [38]. Several reporter genes such as herpes simplex virus-1 thymine kinase (HSV1-tk), sodium iodide symporter (NIS), and somatostatin receptor subtype 2 (SSTr2) are readily available, however, the paired PET probes have poor BBB penetration. Dopamine 2 receptor (D2R) with [18F] Fallypride has been effectively applied but has high background in the pituitary and striatum [12,39]. The PKM2 reporter gene and [18F] DASA-23 probe pair has an advantage of potentially monitoring gene therapy in the central nervous system because [18F]DASA-23 freely crosses the BBB and PKM2 has low expression in brain. The PKM2/[18F] DASA-23 pair is not ideal for imaging the abdomen due to the slow clearance of the probe and expression of the protein in metabolically-active organs. Therefore, we compared the brain kinetics to the entire body kinetics across capsids to establish the unique opportunity for brain reporter gene imaging with this pair.
In the brain, PET reporter gene imaging with PKM2 reporter gene pointed to spatial variations in protein expression which may result from differing cellular tropism, quantified the unexpectedly rapid protein expression after engineered AAV systemic injection, and provided a noninvasive method to monitor AAV-mediated protein expression over time. The enhanced PHP.eB and CAP-B10 capsid brain accumulation was largely correlated with increased brain transduction, as compared with AAV9. In our comparison of PET images from the capsid distribution and reporter protein expression, 64Cu-PHP.eB accumulation and transduction were spatially similar across the cerebral cortex, midbrain, thalamus, and pons. However, cerebellar reporter gene imaging intensity was low with both PET and optical reporter proteins, while capsid accumulation was high. The differences may be associated with cell density or cellular specificity. Indeed, the cerebellum has a lower neuronal cell density as compared to the cortex and olfactory bulb [40], and PHP.eB has been shown to display neurotropism. The confirmation of the spatial distribution of the PET reporter with fluorescent protein images suggests that longitudinal imaging of large animal or human transduction is feasible, including capsid sub-region localization and longitudinal PET reporter protein imaging. In future work, the PKM2 reporter gene (~2000 bp) would be applied to monitor the expression of the therapeutic gene being delivered into the brain either by delivery in a single AAV or multiple AAVs.
When PHP.eB:PKM2 was systemically administered using PHP.eB, we found that the [18F]DASA-23 signal was significantly increased within the brain within 7 days. This rapid PKM2 protein production supports previous research [4] showing a low level of endothelial transduction resulting from engineered viruses as early as 24 h and significant transduction after 7 days. Here, we further tracked the protein production with PET imaging in the same PHP.eB cohort for more than 34 weeks. When administered in the AAV9 capsid, PKM2 mRNA gradually increased from 1 week to 3 weeks and was significantly higher than the saline control group. However, the transcribed PKM2 protein from AAV9 administration was not sufficient to be detected by [18F] DASA-23.
The PKM2 protein also plays a role in inflammatory responses [41], but inflammation has not been detected as a result of the introduction of the PKM2 reporter gene. PKM2 is upregulated in many cancers, but it is not independently oncogenic [42] and is upregulated in healthy tissues outside of the brain. Here, TSPO, a translocator protein extensively exploited as an indicator of neuroinflammation, was not enhanced with the overexpression of the PKM2 gene in the brain [43–45]. In the future, the 18 KDa TSPO protein can also be tracked noninvasively with [11C] DPA-713.
As shown here, molecular imaging can precisely track the location and transduction of capsids. Tracking the clearance or degradation of the nucleic acids is more challenging. For the radiolabeled AAVs tracked with PET, the radioactive molecule can be separated from the chelator within cells and, therefore, clear more slowly from the liver (or brain) than the degraded nucleic acid. In addition, the fluorescent tag can be degraded within intracellular vesicles or the nucleus at the time that the nucleic acids separate from the capsid. Thus, the whole-body clearance of radioactivity from the body can overestimate the duration of the intact capsid, while the unquenched fluorescence can underestimate the AAV stability. Still, early time point imaging tracks tissue and cellular accumulation, and with the addition of reporter gene imaging, the location and kinetics of transduction can be accurately followed over time.
In summary, combining optical and PET imaging technologies allowed us for the first time to visualize AAV uptake and transduction in both target and clearance organs across scale within the same subject. PET pharmacokinetic imaging has the potential to quantify the ligand receptor association constant and monitor the resulting organ-specific and off-target accumulation. Optical pharmacokinetic imaging further characterizes the cellular specificity of viral accumulation. PET reporter gene imaging quantified the rate of protein expression in the brain after systemic AAV injection and provided a noninvasive method to monitor protein expression over time and assess differences in cellular tropism across the brain. These powerful technologies allow for zonal and cell-specific evaluation in a manner that is information-rich and characterizes the complex dynamics of viral accumulation and transduction. Therefore, developing imaging techniques to assess the PK and transduction of such variants in larger animals and human studies is compelling as they can provide for mechanistic insight and understanding of species related differences.
4. Methods
4.1. Materials & reagents
The detailed list of materials and reagents is provided in the Supplementary information.
4.2. AAV production
AAV9, AAV-PHP.eB, and AAV.CAP-B10 were produced as previously described [46]. Briefly, AAV were harvested 5 days after triple transfection in HEK293 cells by PEG precipitation of 3 and 5 days media and osmotic lysis of cell pellets. Crude AAV were then purified by extraction from iodixanol density gradients and buffer exchanged into DPBS. Viral titers were determined by qPCR on a WPRE element present in all packaged AAV genomes as detailed below. AAVs used in all experiments are listed in Table S3.
4.3. Radiolabeling of AAVs
All radiolabeling experiments were conducted under the Controlled Radiation Authorization (CRA) approved by Stanford University (Palo Alto, CA). To radiolabel AAV9 (4 × 1012 vg), PHP.eB (5 × 1012 vg), and CAP-B10 (5 × 1012 vg), the respective AAVs in 1xPBS (0.2 mL) were mixed with 0.1 M Na2CO3 solution (20 μL, pH 9.2) and 2 mM tetrazine-PEG5-NHS (Tz-PEG5-NHS, 2 nmol, 1 μL in DMSO). The AAV mixtures were incubated at 25 ◦C for 30 min, added 0.1 mL 1xPBS, and transferred to a mini-dialysis device (20 kDa molecular weight cut-off (MWCO)). The AAV mixtures were dialyzed in 1xPBS for 4 h (15 mL) and transferred for overnight dialysis (0.5 L). Dialyzed Tz-AAVs were further reacted with [64Cu]Cu-chelated (NOTA)8-TCO for 0.5–1 h, freshly prepared from a reaction of 64CuCl2 (111–148 MBq (3–4 mCi), 4–5 μL) and 10 μM (NOTA)8-TCO (40–100 pmol, 4–10 μL) in ammonium citrate buffer (20 μL, pH 6.5). The incorporation of Cu-64 to (NOTA)8- TCO was monitored by instant thin-layer chromatography (iTLC) and completed in 30 min. Radiolabeled AAVs were purified by a 100 kDa MWCO centrifugal filter unit (Thermo Fisher Scientific) with three cycles of dilution with 1xPBS (15 mL) containing 0.001% Pluronic F-68 (Gibco) and concentrated in ~200 μL volume.
4.4. Animal models
All animal experiments were conducted with a protocol approved by the Administrative Panel for Laboratory Animal Care (APLAC) at Stanford University. AAVs were evaluated in wild-type 8–12 week old C57BL/6 mice (Charles River).
4.5. PET/CT imaging and biodistribution
For the PK and biodistribution of AAVs in Figure 1, 64Cu-AAV9 (112 ± 18 KBq/mouse, n = 3), 64Cu-PHP.eB (370 ± 17 KBq/mouse, n = 4), and 64Cu-CAP-B10 343 ± 21 KBq/mouse, n = 4) were administered via the tail vein of C57BL/6 mice on the Siemens Inveon PET/CT scanner after the start of image acquisition.
For the longitudinal monitoring of PKM2 gene expression with [18F] DASA-23 as in Fig. 2, PHP.eB:EF1A-PKM2 (2 × 1011 vg in 0.12 mL, n = 4), AAV9:EF1A-PKM2 (2 × 1011 vg in 0.12 mL, n = 4), and saline (0.12 mL, n = 4) were systemically injected to C57BL/6 mice. At 1, 3, 8, 16, and 34 weeks p.i. of viral particles, [18F]DASA-23 (63.7 ± 21.8 GBq/ μmol) was administered via tail vein injection. PET scanning was carried out for 30 min on a 3D printed mice bed in the PET/CT scanner. [18F] DASA-23 was produced at the Stanford Cyclotron & Radiochemistry Facility as previously reported [13–16,19]. The list mode data acquired for 30 min was reconstructed and analyzed as described in PET/CT image analysis. Details regarding the PET imaging procedure and analysis are included in the Supplementary Methods.
4.6. Gene expression assay
Brain tissue was collected, cut less than 0.5 cm thick, and added to the sterilized 5 mL cryotube. The brain sample was immediately stabilized in RNAprotect tissue reagent (Qiagen) under ice. After removing the RNA protective reagent, mRNA was extracted using an RNeasy Midi kit (Qiagen) by the manufacturer’s protocol. During this process, genomic DNA was removed by DNase. The concentrations of extracted mRNA from all mouse brains were diluted to be identical. cDNA was prepared with SuperScript IV VILO Master mix (ThermoFisher Scientific) as in the manufacturer’s protocol. TaqMan qPCR (Assay ID for PKM2: Hs00987261_g1, for TSPO: Mm00437828_m1 and for β-actin: Mm02619580_g1) was performed to calculate the ΔΔCt of the target gene (PKM2 and TSPO) over the housekeeping gene (β-actin).
4.7. Fluorescence dye labeling and characterization
AAV9 (8 × 1012 vg), PHP.eB (9 × 1012 vg), and CAP-B10 (9 × 1010 vg) in 1xPBS (0.3 mL) were mixed with 0.1 M Na2CO3 solution (25 μL, pH 9.2) and 2 mM tetrazine-PEG5-NHS (6 nmol, 3 μL in DMSO). The AAV mixtures were incubated at 25 ◦C for 30 min, added 0.1 mL 1xPBS, and transferred to a mini-dialysis device (20 kDa molecular weight cut-off (MWCO)). The AAV mixtures were dialyzed in 1xPBS for 4 h (15 mL) and overnight (0.5 L). Dialyzed Tz-AAVs were further reacted with 1 mM Cy5-TCO (1 μL) for 1 h. Cy5-AAVs were purified by a 100 kDa MWCO centrifugal filter with three cycles of dilution with 1xPBS (15 mL) containing 0.001% Pluronic F-68 and concentrated in ~200 μL volume. The number of labels per particle was calculated by dividing the fluorescent dye number obtained from the standard curve by the vector genome (vg) counted by qPCR. Details regarding the qPCR for counting vector genome is included in the Supplementary Methods.
4.8. Fluorescent immunohistochemistry and confocal microscopy
Cy5-AAV9 (2 × 1011 vg/mouse, n = 4), -PHP.eB (2 × 1011 vg/mouse, n = 4), -CAP-B10 (2 × 1011 vg/mouse, n = 4) in 1xPBS (0.15 mL), and saline (n = 2) were systemically administered to mice via the tail vein. Two mice per group and one mouse from the saline group as a negative control were euthanized by Euthasol under deep isoflurane at 4 h and 3 weeks p.i. Immediately after mice were euthanized, the left ventricle was cannulated, and mice were perfused with 1xPBS (10 mL) and 4% PFA (10 mL). Details regarding the tissue fixation and fluorescence image processing are included in the Supplementary Methods.
4.9. Statistics and reproducibility
All statistical analyses were performed in GraphPad Prism software (Prism 9.0). The statistical tests with confidence intervals, effect sizes, degree of freedom and P values can be found in the source data. Sample size for each experiment and the biological replicates across experiments are shown in the appropriate figure caption.
Supplementary Material
Acknowledgments
We thank Dr. Weiyu Chen and Dr. Sadaf Aghevilan for the technical assistance in biodistribution and titer of AAVs, Christoper Acosta for the technical advice in Western blot, and the CLOVER Center in the Beck-man Institute at Caltech for producing AAVs. PET/CT and microscope imaging were performed at the Stanford Center for Innovation in In Vivo Imaging (SCi3) and Canary Center of Stanford University. This work was supported by NIHCA112356 and NIHEB028646.
Footnotes
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.biomaterials.2022.121701.
Data availability
Additional methods are found in Supplementary Methods section. The authors declare that the image and quantitative data supporting the findings of this study are available within the paper and supplementary data. The raw PET images and associated data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- [1].Bedbrook CN, Deverman BE, Gradinaru V, Viral strategies for targeting the central and peripheral nervous systems, Annu. Rev. Neurosci 41 (2018) 323–348. [DOI] [PubMed] [Google Scholar]
- [2].Fischell JM, Fishman PS, A multifaceted approach to optimizing AAV delivery to the brain for the treatment of neurodegenerative diseases, Front. Neurosci 15 (2021), 747726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chan KY, et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems, Nat. Neurosci 20 (2017) 1172–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Deverman BE, et al. , Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain, Nat. Biotechnol 34 (2016) 204–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu X, Jingwen Y, Yuanguo C, Pharmacokinetic Study of Viral Vectors for Gene Therapy: Progress and Challenges, 2011.
- [6].Seo JW, et al. , Positron emission tomography imaging of novel AAV capsids maps rapid brain accumulation, Nat. Commun 11 (2020), 2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Greer C, Kozyak B, Stedman H, Challenges at the crossroads: Myopathy trials in 2020 hindsight, Mol. Ther 29 (2021) 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kietzmann T, Metabolic zonation of the liver: the oxygen gradient revisited, Redox Biol. 11 (2017) 622–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bell P, et al. , Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates, Mol. Genet. Metabol 104 (2011) 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Carestia A, et al. , Modulation of the liver immune microenvironment by the adeno-associated virus serotype 8 gene therapy vector, Mol. Ther. Methods Clin. Dev 20 (2021) 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Witney TH, et al. , PET imaging of tumor glycolysis downstream of hexokinase through noninvasive measurement of pyruvate kinase M2, Sci. Transl. Med 7 (2015), 310ra169. [DOI] [PubMed] [Google Scholar]
- [12].Yaghoubi SS, Campbell DO, Radu CG, Czernin J, Positron emission tomography reporter genes and reporter probes: gene and cell therapy applications, Theranostics 2 (2012) 374–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Beinat C, Alam IS, James ML, Srinivasan A, Gambhir SS, Development of [18F] DASA-23 for imaging tumor glycolysis through noninvasive measurement of pyruvate kinase M2, Mol. Imag. Biol 19 (2017) 665–672. [DOI] [PubMed] [Google Scholar]
- [14].Beinat C, et al. , The utility of [18F]DASA-23 for molecular imaging of prostate cancer with positron emission tomography, Mol. Imag. Biol 20 (2018) 1015–1024. [DOI] [PubMed] [Google Scholar]
- [15].Beinat C, et al. , A clinical PET imaging tracer ([18F]DASA-23) to monitor pyruvate kinase M2-induced glycolytic reprogramming in glioblastoma, Clin. Cancer Res 27 (2021) 6467–6478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Beinat C, et al. , Human biodistribution and radiation dosimetry of [18F]DASA-23, a PET probe targeting pyruvate kinase M2, Eur. J. Nucl. Med. Mol. Imag 47 (2020) 2123–2130. [DOI] [PubMed] [Google Scholar]
- [17].Beinat C, Patel CB, Xie Y, Gambhir SS, Evaluation of glycolytic response to multiple classes of anti-glioblastoma drugs by noninvasive measurement of pyruvate kinase M2 using [18F]DASA-23, Mol. Imag. Biol 22 (2020) 124–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brown D, et al. , Deep parallel characterization of AAV tropism and AAV-mediated transcriptional changes via single-cell RNA sequencing, Front. Immunol 12 (2021), 730825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Haywood T, et al. , Positron emission tomography reporter gene strategy for use in the central nervous system, Proc. Natl. Acad. Sci. U.S.A 116 (2019) 11402–11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Goertsen D, et al. , AAV capsid variants with brain-wide transgene expression and decreased liver targeting after intravenous delivery in mouse and marmoset, Nat. Neurosci 25 (2021) 106–115. [DOI] [PubMed] [Google Scholar]
- [21].Castle MJ, et al. , Postmortem analysis in a clinical trial of AAV2-NGF gene therapy for alzheimer’s disease identifies a need for improved vector delivery, Hum. Gene Ther 31 (2020) 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Nagahara AH, et al. , MR-guided delivery of AAV2-BDNF into the entorhinal cortex of non-human primates, Gene Ther. 25 (2018) 104–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Girod A, et al. , Genetic capsid modifications allow efficient re-targeting of adeno-associated virus type 2, Nat. Med 5 (1999) 1052–1056. [DOI] [PubMed] [Google Scholar]
- [24].Michelfelder S, et al. , Peptide ligands incorporated into the threefold spike capsid domain to Re-direct gene transduction of AAV8 and AAV9 in vivo, PLoS One 6 (2011), e23101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Michelfelder S, et al. , Successful expansion but not complete restriction of tropism of adeno-associated virus by in vivo biopanning of random virus display peptide libraries, PLoS One 4 (2009), e5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Adachi K, Enoki T, Kawano Y, Veraz M, Nakai H, Drawing a high-resolution functional map of adeno-associated virus capsid by massively parallel sequencing, Nat. Commun 5 (2014) 3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].DiMattia MA, et al. , Structural insight into the unique properties of adeno-associated virus serotype 9, J. Virol 86 (2012) 6947–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bartlett JS, Samulski RJ, Fluorescent viral vectors: a new technique for the pharmacological analysis of gene therapy, Nat. Med 4 (1998) 635–637. [DOI] [PubMed] [Google Scholar]
- [29].Leopold PL, et al. , Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells, Hum. Gene Ther 9 (1998) 367–378. [DOI] [PubMed] [Google Scholar]
- [30].Liu Y, Joo KI, Wang P, Endocytic processing of adeno-associated virus type 8 vectors for transduction of target cells, Gene Ther. 20 (2013) 308–317. [DOI] [PubMed] [Google Scholar]
- [31].Liu Y, et al. , Site-specific modification of adeno-associated viruses via a genetically engineered aldehyde tag, Small 9 (2013) 421–429. [DOI] [PubMed] [Google Scholar]
- [32].Bell CL, et al. , The AAV9 receptor and its modification to improve in vivo lung gene transfer in mice, J. Clin. Investig 121 (2011) 2427–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Oliveira BL, Guo Z, Bernardes GJL, Inverse electron demand Diels–Alder reactions in chemical biology, Chem. Soc. Rev 46 (2017) 4895–4950. [DOI] [PubMed] [Google Scholar]
- [34].Logan J, A review of graphical methods for tracer studies and strategies to reduce bias, Nucl. Med. Biol 30 (2003) 833–844. [DOI] [PubMed] [Google Scholar]
- [35].Gambhir SS, et al. , A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography, Proc. Natl. Acad. Sci. USA 97 (2000) 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Qin JY, et al. , Systematic comparison of constitutive promoters and the doxycycline-inducible promoter, PLoS One 5 (2010), e10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Large EE, Silveria MA, Zane GM, Weerakoon O, Chapman MS, Adeno-associated virus (AAV) gene delivery: dissecting molecular interactions upon cell entry, Viruses 13 (2021), 1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Concilio SC, Russell SJ, Peng KW, A brief review of reporter gene imaging in oncolytic virotherapy and gene therapy, Mol. Ther. Oncolytics 21 (2021) 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Serganova I, Blasberg RG, Molecular imaging with reporter genes: has its promise been delivered? J. Nucl. Med 60 (2019) 1665–1681. [DOI] [PubMed] [Google Scholar]
- [40].Keller D, Ero C, Markram H, Cell densities in the mouse brain: a systematic review, Front. Neuroanat 12 (2018), 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Alves-Filho JC, Pålsson-McDermott EM, Pyruvate kinase M2: a potential target for regulating inflammation, Front. Immunol 7 (2016), 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Iqbal MA, Gupta V, Gopinath P, Mazurek S, Bamezai RN, Pyruvate kinase M2 and cancer: an updated assessment, FEBS Lett. 588 (2014) 2685–2692. [DOI] [PubMed] [Google Scholar]
- [43].Endres CJ, et al. , Initial evaluation of 11C-DPA-713, a novel TSPO PET ligand, in humans, J. Nucl. Med 50 (2009) 1276–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Simmons DA, et al. , TSPO-PET imaging using [18F]PBR06 is a potential translatable biomarker for treatment response in Huntington’s disease: preclinical evidence with the p75NTR ligand LM11A-31, Hum. Mol. Genet 27 (2018) 2893–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].James ML, et al. , [18F]GE-180 PET detects reduced microglia activation after LM11A-31 therapy in a mouse model of alzheimer’s disease, Theranostics 7 (2017) 1422–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Challis RC, et al. , Systemic AAV vectors for widespread and targeted gene delivery in rodents, Nat. Protoc 14 (2019) 379–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Additional methods are found in Supplementary Methods section. The authors declare that the image and quantitative data supporting the findings of this study are available within the paper and supplementary data. The raw PET images and associated data that support the findings of this study are available from the corresponding author upon reasonable request.


