Background:
In recent years, many studies have found possible links between gene polymorphisms and venous thromboembolism (VTE). By identifying genetic risk factors before facing environmental risk factors such as surgical interventions and COVID-19 vaccination, we could rapidly respond to the risk of VTE. The aim of this study was to perform an umbrella review of genetic variants related to VTE. Integrative gene analysis of VTE was performed to identify critical genetic variations.
Methods:
This study conducted an umbrella review of systematic reviews and meta-analyses. All included studies were selected from the PubMed/MEDLINE database. To select eligible studies, the following variables were extracted: first author name; effect size of each study genetic variant; year of publication; the number of studies included in each article; ethnicity, sample size, P values, and heterogeneity estimates. To assess cumulative evidence in genetic epidemiology about effects of gene polymorphisms on VTE, Human Genome Epidemiology Network’s Venice criteria were used. Methodological quality assessment was conducted with JBI Critical Appraisal Checklist for Systematic Reviews and Research Syntheses.
Results:
Genes provided in the present study with genetic variants associated with VTE were FVL (G1691A), Prothrombin (G20210A), MTHFR (C677T, A1298C), PAI-1 (4G/5G), factor VII activating protease (1601G > A), and endothelial protein C receptor (g.6936A_G, c.4600A_G). Among them, variants in FVL, Prothrombin, MTHFR, and PAI-1 showed high significance. Particularly, variants in Prothrombin (G20210A), MTHFR (C677T), and PAI-1 (4G/5G) had more than 2 types of model significance.
Conclusion:
The present study performed a systematic review of genetic variants associated with VTE. Our results could lead to a more comprehensive understanding of VTE etiology. These results could give a strategy of prediagnosis about evaluating individual risks of VTE who might be exposed to environmental risk factors.
Keywords: femoral head osteonecrosis, genetic variant, polymorphism, steroid, umbrella review
1. Introduction
Venous thromboembolism (VTE) is a serious clinical disease. It includes pulmonary embolism (PE) and deep vein thrombosis as 2 different forms of the same disease.[1] VTE is also known as a silent killer. It has high morbidity and mortality due to abnormal coagulation of blood.[2,3] Coagulation causes thrombosis in superficial leg, arm, cerebral, renal, and portal veins.[4] VTE is the third most frequent vascular disorder.[5] It affects roughly 1 to 2 people per 1000 people each year.[6] The probability of a recurring VTE event after a first VTE event is substantial. Around 30% of people who have an incident VTE event have a recurrence within 10 years.[2]
Recent studies have shown that VTE is a complex multi-factor disease in which polygenetic factors play a principal role.[7,8] Synergistic gene-gene and gene-environment interactions contribute to the increase of VTE. For example, the presence of heterozygosity for factor V Leiden (FVL), a VTE gene factor, plus the use of oral contraceptives, a VTE environment factor, can result in a 34-fold increase in thrombotic risk.[9,10] Checking genetic risk factors before getting environmental risk factors (e.g., surgical interventions, pregnancy, and COVID-19 vaccination) is important.[11,12] Therefore, exploring genetic variants associated with VTE occurrence would be important to discover the direction of prevention and treatment of VTE.
It has become critical to comprehend the significance of genetic variants in coagulation factors that might contribute to the development of VTE. Genetic variants known to be the most prominent factors in VTE include defects in natural anticoagulants, methylenetetrahydrofolate reductase (MTHFR), prothrombin, protein C, plasminogen activator inhibitor-1 (PAI-1), and FVL.[13–15] Studies have shown that these genetic defects could cause thrombotic disorders with a complexity of underlying mechanisms. Indeed, profiling individual genetic risk could be a useful prevention strategy for VTE.[8,16]
Many studies published in recent years have found possible links between gene polymorphisms and VTE.[17–22] By identifying genetic risk factors before facing environmental risk factors such as surgical interventions and COVID-19 vaccination, we could rapidly respond to the risk of VTE. Therefore, the aim of this study was to perform an umbrella review of genetic variants related to VTE. Integrative gene analysis of VTE was performed to identify critical genetic variations.
2. Methods
2.1. Search strategy and selection criteria for eligible studies
This study conducted an umbrella review for systematic reviews and meta-analyses. All included studies were selected from the PubMed/MEDLINE database. The search strategy included keywords “(‘venous thrombosis’[MeSH Terms] OR (‘venous’[All Fields] AND ‘thrombosis’[All Fields]) OR ‘venous thrombosis’[All Fields] OR (‘deep’[All Fields] AND ‘vein’[All Fields] AND ‘thrombosis’[All Fields]) OR ‘deep vein thrombosis’[All Fields]) AND (‘venous thromboembolism’[MeSH Terms] OR (‘venous’[All Fields] AND ‘thromboembolism’[All Fields]) OR ‘venous thromboembolism’[All Fields]) AND (‘genes’[MeSH Terms] OR ‘genes’[All Fields] OR ‘gene’[All Fields]) AND (‘meta’[Journal] OR ‘meta’[All Fields])” applied to the title/abstract/keywords field to find diverse systematic meta-analyses and reviews. Two authors independently screened retrieved publications. In case of disagreements, the final decision was resolved according to consensus. When disagreements could not be resolved, a third investigator was included. Full texts of publications were selected to find eligible studies after title/abstract was screened.
Selection criteria for eligible studies were: studies on genetic variants based on systematic reviews and gene meta-analyses related to VTE; studies that provided definite information about statistical processes and results; and studies written in English. Exclusion criteria were: systematic meta-analyses without a quantitative synthesis of the evidence; non-human studies; studies with huge errors or poor quality; studies without specific statistical results including overall P values; studies without examining risk factors; studies with publication bias; and studies with heterogeneity.
2.2. Data extraction
For each eligible reference, 1 author extracted data and then a second author screened it. To select eligible studies, the following variables were extracted: first author name; effect size of each study genetic variant; year of publication; the number of studies included in each article; ethnicity, sample size, P values, and heterogeneity estimates.
2.3. Statistical analysis
To assess cumulative evidence in genetic epidemiology about effects of gene polymorphisms on VTE, Human Genome Epidemiology Network’s Venice criteria were used.[23] The guideline included the index of the amount of evidence, the extent of replication, and protection from bias. Evidence criteria were categorized into 3 levels (strong, moderate, or weak). According to these criteria, large sample sizes and a large amount of evidence would ensure appropriate power for detecting eligible results. A specific description of these criteria is provided in Table 1. Methodological quality assessment was conducted with JBI Critical Appraisal Checklist for Systematic Reviews and Research Syntheses. This assessment comprised 11 items evaluating the methodological quality of studies. Each question was answered as “yes,” “no,” or “unclear.”
Table 1.
Description of the extension of the human genome epidemiology network’s Venice criteria to assess genetic main effect.
| Criteria of consideration | Category | Proposed operationalization |
|---|---|---|
| Amount of evidence | A | Sample size over 1000 |
| B | Sample size 100-1000 | |
| C | Sample size under 100 | |
| Replication | A | I2 < 50% |
| B | 25% < I2 < 50% | |
| C | I2 > 50% | |
| Protection from bias | A | Consideration biases such as bias in genotyping, population stratification, and Selective reporting biases. |
| B | ||
| Bias in genotype definition = Not reported what was done/No quality control checks/Appropriate quality control checks. | ||
| C | ||
| Population stratification = Not reported what was done/Nothing done/Same descent group/Adjustment for reported descent/Family-based design/Genomic control, PCA or a similar method | ||
| Selective reporting biases = Meta-analysis of published data/Retrospective efforts to include unpublished data/Meta-analysis within consortium. |
2.4. Ethical consideration
Ethical approval was not necessary because of the nature of the present study.
3. Results
3.1. Number of articles and types identified
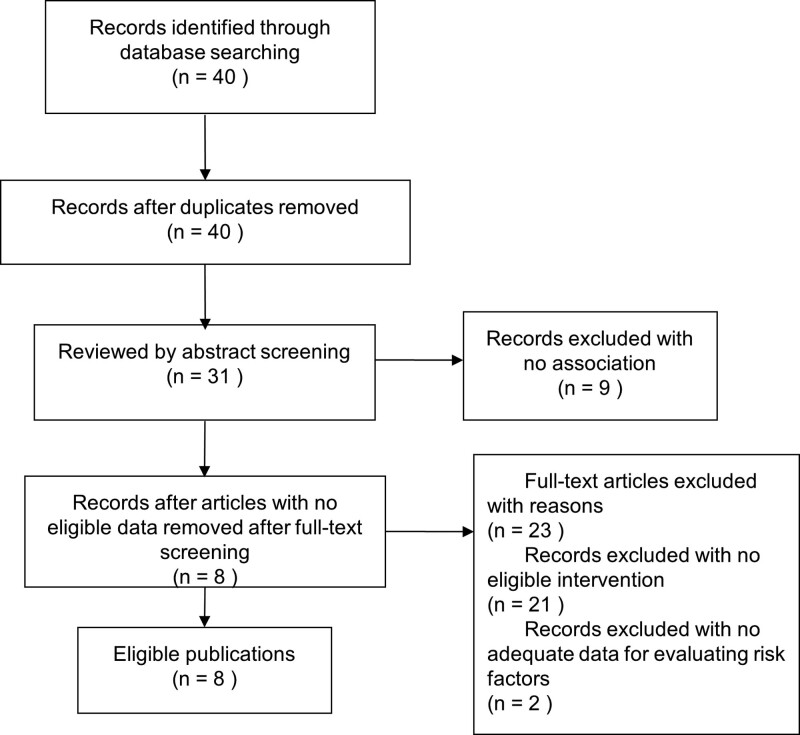
Our search strategy identified 40 unique references from the database. After excluding duplicates and articles unrelated to main topics by abstract screening, 31 publications remained. Of these 31 publications, 23 full-text articles without eligible intervention (n = 21) or without adequate data for evaluating risk factors (n = 2) were excluded. Finally, 8 eligible publications met the criteria of inclusion and exclusion with significant results, proper qualitative evaluation, and convincing data (Fig. 1).[17–22,24,25] These 8 eligible articles included 6 genes and 7 genetic variants.
Figure 1.
Flowchart showing the study selection process using PRISMA.
3.2. Main findings of meta-analyses of SNP-based studies
In the study, 4 genetic variants of 4 genes were significantly associated with VTE. FVL mutation in the G1691A gene was investigated in 3 articles, including the study of Antonio Marchiori et al (including 10 studies), the study of Xindie Zhou et al (including 14 studies), and the study of Wai Khoon Ho et al (including 10 studies).[21,22,24] The pooled risk ratio (RR) for G1691A gene mutation in the study of Antonio Marchiori et al showed significant results in a heterozygous model (fixed-effects model: RR = 1.39, 95% CI: 1.15-1.67, P = .0005; random-effects model: RR = 1.45, 95% CI: 1.13-1.85, P = .0003). The study of Wai Khoon Ho et al also showed significant results for G1691A gene mutation in a heterozygous model (fixed-effects model: RR = 1.41, 95% CI: 1.14-1.75, P = .002). However, the study of Xindie Zhou et al did not show any significance for G1691A gene mutation.
Prothrombin mutation in the G20210A gene was investigated in 3 articles, including they study of Antonio Marchiori et al (including 10 studies), the study of Xindie Zhou et al (including 7 studies), and the study of Wai Khoon Ho et al (including 9 studies).[21,22,24] Pooled odds ratio (OR) for prothrombin G20210A gene mutation in their study of Xindie Zhou et al showed significant results in a dominant model (fixed-effects model: OR = 2.16, 95% CI: 1.27-3.69, P = .005). The study of Wai Khoon Ho et al also showed significant results for prothrombin G20210A gene mutation in a heterozygous model (fixed-effects model: OR = 1.72, 95% CI: 1.27-2.31, P = .001). However, the study of Antonio Marchiori et al did not show any significance for prothrombin G20210A gene mutation.
MTHFR mutation in C677T gene was investigated in 3 articles, including the study of Xindie Zhou et al (including 2 studies), the study of Peijin Zhang et al (including 24 studies), and the study of Miao Gao et al (including 31 studies).[19,20,22] Pooled OR for MTHFR C677T gene mutation in the study of Peijin Zhang et al showed significant results in an allele model (random-effects model: OR = 1.48, 95% CI: 1.32-1.67, P = .005), a heterozygous model (random-effects model: OR = 1.33, 95% CI: 1.11-1.60, P = .008), and a dominant model (random-effects model: OR = 1.53, 95% CI: 1.28-1.84, P = .002). The study of Miao Gao et al also showed significant results for MTHFR C677T gene mutation in a reverse allele model (random-effects model: OR = 0.80, 95% CI: 0.71-0.90, P = .00001), a reverse recessive model (random-effects model: OR = 0.68, 95% CI: 0.56-0.83, P = .0002), a reverse dominant model (random-effects model: OR = 0.82, 95% CI: 0.72-0.94, P = .005), a reverse heterozygous model (random-effects model: OR = 0.65, 95% CI: 0.52-0.81, P = .0001), and a reverse homozygous model (random-effects model: OR = 0.73, 95% CI: 0.60-0.89, P = .002). However, the study of Xindie Zhou et al did not show any significance for MTHFR C677T gene mutation.
PAI-1 4G/5G gene mutation was investigated in 1 article by Qiang Zhang et al (including 27 studies).[25] The pooled OR for PAI-1 4G/5G gene mutation in the study of Qiang Zhang et al showed significant results in a recessive model (random-effects model: OR = 1.34, 95% CI: 1.10-1.63, P = .004) and homozygous model (random-effects model: OR = 1.59, 95% CI: 1.17-2.15, P = .003).
Factor VII activating protease 1601G > A gene mutation and endothelial protein C receptor g.6936A_G, c.4600A_G mutations have also been investigated by Da Li et al and Jessica Dennis et al However, there were no significant results. All genetic variants data are provided in Table 2. Main findings of eligible studies are shown in Table 3.
Table 2.
Genetic variants derived from eligible meta-analyses of venous thromboembolism (VTE).
| Genetic variant | Polymorphism | Reference | Ethnicity | Sample size (case/control) | Included studies | Minor allele & Reference allele | Genetic model | Type of model | Reported OR (or RR) (95% CI) | P value for genetic main effect | Heterogeneity | Venice criteria | Evidence class |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Factor V Leiden (G1691A) | rs6025 | Antonio Marchiori et al[24] | mixed | 496/2707 | 10 | A/G | heterozygous | Fixed-effects model | 1.39 [1.15-1.67] (RR) | .0005 | 37.20% | ABB | Moderate |
| Factor V Leiden (G1691A) | rs6025 | Antonio Marchiori et al | mixed | 496/2707 | 10 | A/G | heterozygous | Random-effects model | 1.45 [1.13-1.85] (RR) | .0003 | 37.20% | ABB | Moderate |
| Factor V Leiden (G1691A) | rs6025 | Xindie Zhou et al[22] | Caucasian | 1032/2903 | 14 | A/G | dominant | Fixed-effects model | 1.41 [1.03-1.94] | .03 | 8.00% | AAA | Strong |
| Factor V Leiden (G1691A) | rs6025 | Wai Khoon Ho et al[21] | mixed | 535/3104 | 10 | A/G | heterozygous | Fixed-effects model | 1.41 [1.14-1.75] | .002 | 41.40% | ABB | Moderate |
| Prothrombin (G20210A) | rs1799963 | Xindie Zhou et al | Caucasian | 558/1214 | 7 | A/G | dominant | Fixed-effects model | 2.16 [1.27-3.69] | .005 | 44.00% | ABA(equivalent to AAA) | Strong |
| Prothrombin (G20210A) | rs1799963 | Antonio Marchiori et al | mixed | 466/2742 | 10 | A/G | heterozygous | Fixed-effects model | 1.20 [0.89-1.61] | .23 | 0% | AAB(equivalent to AAA) | Strong |
| Prothrombin (G20210A) | rs1799963 | Antonio Marchiori et al | mixed | 466/2742 | 10 | A/G | heterozygous | Random-effects model | 1.36 [1.02-1.82] | .03 | 0% | AAB(equivalent to AAA) | Strong |
| Prothrombin (G20210A) | rs1799963 | Wai Khoon Ho et al | mixed | 469/2903 | 9 | A/G | heterozygous | Fixed-effects model | 1.72 [1.27-2.31] | .001 | 28.20% | ABB | Moderate |
| MTHFR (C677T) | rs1801133 | Xindie Zhou et al | Caucasian | 119/239 | 2 | T/C | recessive | Fixed-effects model | 2.36 [1.03-5.42] | .04 | 0.00% | BAA(equivalent to AAA) | Strong |
| MTHFR (C677T) | rs1801133 | Peijin Zhang et al[20] | Asian | 2339/4048 | 24 | T/C | allele | Random-effects model | 1.48 [1.32-1.67] | .005 | 47.90% | ABA(equivalent to AAA) | Strong |
| MTHFR (C677T) | rs1801133 | Peijin Zhang et al | Asian | 2339/4048 | 24 | T/C | heterozygous | Random-effects model | 1.33 [1.11-1.60] | .008 | 45.60% | ABA(equivalent to AAA) | Strong |
| MTHFR (C677T) | rs1801133 | Peijin Zhang et al | Asian | 2339/4048 | 24 | T/C | homozygous | Fixed-effects model | 2.11 [1.79-2.48] | .112 | 26.80% | ABA(equivalent to AAA) | Strong |
| MTHFR (C677T) | rs1801133 | Peijin Zhang et al | Asian | 2339/4048 | 24 | T/C | dominant | Random-effects model | 1.53 [1.28-1.84] | .002 | 51.60% | ABA(equivalent to AAA) | Strong |
| MTHFR (C677T) | rs1801133 | Peijin Zhang et al | Asian | 2339/4048 | 24 | T/C | recessive | Fixed-effects model | 1.83 [1.59-2.11] | .693 | 0% | AAA | Strong |
| MTHFR (C677T) | rs1801133 | Miao Gao et al[19] | mixed | 7960/10567 | 31 | C/T | reverse recessive | Random-effects model | 0.68 [0.56-0.83] | .0002 | 69% | ACB | Weak |
| MTHFR (C677T) | rs1801133 | Miao Gao et al | mixed | 7960/10567 | 31 | C/T | reverse dominant | Random-effects model | 0.82 [0.72-0.94] | .005 | 63% | ACB | Weak |
| MTHFR (C677T) | rs1801133 | Miao Gao et al | mixed | 7960/10567 | 31 | C/T | reverse heterozygous | Random-effects model | 0.65 [0.52-0.81] | .0001 | 69% | ACB | Weak |
| MTHFR (C677T) | rs1801133 | Miao Gao et al | mixed | 7960/10567 | 31 | C/T | reverse homozygous | Random-effects model | 0.73 [0.60-0.89] | .002 | 60% | ACB | Weak |
| MTHFR (C677T) | rs1801133 | Miao Gao et al | mixed | 7960/10567 | 31 | C/T | reverseallele | Random-effects model | 0.80 [0.71-0.90] | .00001 | 76% | ACB | Weak |
| MTHFR (A1298C) | rs1801131 | Miao Gao et al | mixed | 917/1735 | 6 | A/C | dominant | Random-effects model | 0.97 [0.71-1.32] | .84 | 4% | AAC | Weak |
| MTHFR (A1298C) | rs1801131 | Miao Gao et al | mixed | 917/1735 | 6 | A/C | overdominant | Random-effects model | 0.91 [0.77-1.08] | .29 | 0% | AAC | Weak |
| MTHFR (A1298C) | rs1801131 | Miao Gao et al | mixed | 530/1000 | 6 | A/C | homozygous | Random-effects model | 0.90 [0.66-1.23] | .52 | 0% | AAC | Weak |
| MTHFR (A1298C) | rs1801131 | Miao Gao et al | mixed | 462/885 | 6 | A/C | heterozygous | Random-effects model | 1.01 [0.67-1.52] | .96 | 35% | ABC | Weak |
| MTHFR (A1298C) | rs1801131 | Miao Gao et al | mixed | 917/1735 | 6 | A/C | allele | Random-effects model | 0.95 [0.83-1.07] | .39 | 0% | AAB(equivalent to AAA) | Strong |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al[25] | Mixed | 2908/4860 | 27 | 4G/5G | allele | Random-effects model | 1.25 [1.05-1.49] | .014 | 84.30% | ACB | Weak |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al | Mixed | 2908/4860 | 27 | 4G/5G | dominant | Random-effects model | 1.38 [1.06-1.81] | .019 | 78.70% | ACB | Weak |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al | Mixed | 2908/4860 | 27 | 4G/5G | recessive | Random-effects model | 1.34 [1.10-1.63] | .004 | 65.80% | ACB | Weak |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al | Mixed | 2908/4860 | 27 | 4G/5G | overdominant | Random-effects model | 0.98 [0.83-1.17] | .856 | 64.80% | ACC | Weak |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al | Mixed | 2908/4860 | 27 | 4G/5G | homozygous | Random-effects model | 1.59 [1.17-2.15] | .003 | 74.20% | ACB | Weak |
| PAI-1 (4G/5G) | rs1799889 | Qiang Zhang et al | Mixed | 2908/4860 | 27 | 4G/5G | heterozygous | Random-effects model | 1.27 [1.00-1.65] | .049 | 73.50% | ACB | Weak |
| FSAP (1601G > A) | rs7080536 | Da Li et al[17] | Caucasian | 2411/2850 | 7 | G/A | allele | Fixed-effects model | 1.33 [1.07-1.66] | .01 | 25% | ABA(equivalent to AAA) | Strong |
| FSAP (1601G > A) | rs7080536 | Da Li et al | Caucasian | 2411/2850 | 7 | G/A | Heterozygous | Fixed-effects model | 1.34 [1.06-1.68] | .01 | 11% | AAA | Strong |
| FSAP (1601G > A) | rs7080536 | Da Li et al | Caucasian | 2411/2850 | 7 | G/A | Dominant | Fixed-effects model | 1.33 [1.06-1.66] | .01 | 20% | AAA | Strong |
| FSAP (1601G > A) | rs7080536 | Da Li et al | Caucasian | 2411/2850 | 7 | G/A | Homozygous | Fixed-effects model | 1.48 [0.43-5.12] | .54 | 0% | AAA | Strong |
| FSAP (1601G > A) | rs7080536 | Da Li et al | Caucasian | 2411/2850 | 7 | G/A | recessive | Fixed-effects model | 1.46 [0.42-5.06] | .55 | 0% | AAA | Strong |
| PROCR (g.6936A_G, c.4600A_G) | rs867186 | Jessica Dennis et al[18] | mixed | 4821/6070 | 11 | G/A | allele | Random-effects model | 1.22 [1.11-1.33] | .197 | 20% | AAB(equivalent to AAA) | Strong |
| PROCR (g.6936A_G, c.4600A_G) | rs867186 | Jessica Dennis et al | mixed | 4736/6010 | 11 | G/A | heterozygous | Random-effects model | 1.21 [1.05-1.40] | .063 | 43% | ABB | Moderate |
| PROCR (g.6936A_G, c.4600A_G) | rs867186 | Jessica Dennis et al | mixed | 3825/4979 | 11 | G/A | homozygous | Random-effects model | 1.81 [1.29-2.56] | .694 | 0% | AAB(equivalent to AAA) | Strong |
| PROCR (g.6936A_G, c.4600A_G) | rs867186 | Jessica Dennis et al | mixed | 4821/6070 | 11 | G/A | dominant | Random-effects model | 1.25 [1.08-1.44] | .041 | 47% | ABB | Moderate |
| PROCR (g.6936A_G, c.4600A_G) | rs867186 | Jessica Dennis et al | mixed | 4821/6070 | 11 | G/A | recessive | Random-effects model | 1.76 [1.24-2.48] | .739 | 0% | AAB(equivalent to AAA) | Strong |
Evidence class is classified as strong, moderate, and weak. When scored A in every criterion, evidence is categorized as strong. When scored no C in every criterion but no AAA, evidence is categorized as moderate. Weak evidence is recorded with C in one out of 3 criteria.
Table 3.
Evidence across the systematic reviews of risk factors.
| Author | Title | Main findings |
|---|---|---|
| Antonio Marchiori et al[26] | The risk of recurrent venous thromboembolism among heterozygous carriers of factor V Leiden or prothrombin G20210A mutation. A systematic review of prospective studies | Specified disease name: DVT (Deep Vein Thrombosis), PE (Pulmonary embolism) |
| 10 eligible studies included | ||
| Finding: Significant association between the FVL polymorphism and VTE was found under the heterozygous model (RR: 1.45 95% CI: 1.13-1.18). Heterozygous carriage of FVL is clearly associated with an increased risk of recurrent thromboembolism. | ||
| Da Li et al[15] | Association between FSAP 1601G > A polymorphism and venous thromboembolism risk: A meta-analysis | Specified disease name: VTE |
| 7 eligible studies included | ||
| Finding: Significant association between the FSAP 1601G > A polymorphism and VTE was found under allele (OR: 1.33, 95% CI: 1.07– | ||
| 1.66), the heterozygous model (OR: 1.34, 95% CI: 1.06–1.68), and the dominant model (GG,OR: 1.33, 95% CI: 1.06–1.66). FSAP 1601 > A polymorphism of 3 genetic comparison models may be associated with VTI susceptibility. | ||
| Jessica Dennis et al[18] | The endothelial protein C receptor (PROCR) Ser219Gly variant and risk of common thrombotic disorders: a HuGE review and meta-analysis of evidence from observational studies | Specified disease name: VTE |
| 11 eligible studies included | ||
| Finding: Significant association between the PROCR (g.6936A_G, c.4600A_G) variant and VTE was found under allele model (OR:1.22, 95% CI: 1.11-1.33). PROCR variant of allele model may increase risk of common thrombotic disorders. | ||
| Miao Gao et al[22] | Meta-analysis of the relationship between methylenetetrahydrofolate reductase C677T and A1298C polymorphism and venous thromboembolism in the Caucasian and Asian | Specified disease name: VTE |
| 31 eligible studies included | ||
| Finding: Significant association between the MTHFR C677T mutation and VTE was found under reverse recessive model (OR: 0.68, 95% CI: 0.56-0.83), reverse dominant model (OR: 0.82, 95% CI: 0.72-0.94), heterozygote model (OR: 0.65, 95% CI: 0.52-0.81), homozygote model (OR: 0.73, 95% CI: 0.60-0.89), and allele model (OR: 0.80, 95% CI: 0.71-0.90). MTHFR A1298C polymorphism did not show significance with VTE in all genetic models. MTHFR C677T of 5 genetic comparison models may be associated with VTI susceptibility. | ||
| Peijin Zhang et al[20] | Association Between MTHFR C677T Polymorphism and Venous Thromboembolism Risk in the Chinese Population: A Meta-Analysis of 24 Case-Controlled Studies | Specified disease name: VTE |
| 24 eligible studies included | ||
| Finding: Significant association between the MTHFR C677T polymorphism and VTE was found under allele model (OR: 1.48, 95% CI: 1.32-1.67), heterozygous model (OR: 1.33, 95% CI: 1.11-1.60), homozygote model (OR: 2.11, 95% CI: 1.79-2.48), dominant model (OR: 1.53, 95% CI: 1.28-1.84) and recessive model (OR: 1.83, 95% CI: 1.59-2.11). The findings support the associations of MTHFR C677T polymorphism with VTE risk in the Chinese population | ||
| Qiang Zhang et al[25] | Plasminogen activator inhibitor-1 (PAI-1) 4G/5G promoter polymorphisms and risk of venous thromboembolism—a meta-analysis and systematic review | Specified disease name: VTE |
| 27 eligible studies included | ||
| Finding: Significant association between the PAI-1 4G/5G polymorphism and VTE was found under allele model (OR: 1.25, 95% CI: 1.05-1.49), dominant model (OR: 1.38, 95% CI: 1.06-1.81), recessive model (OR: 1.34, 95% CI: 1.10-1.63), heterozygote model (OR: 1.27, 95% CI: 1.00-1.65), and homozygote model (OR: 1.59, 95% CI: 1.17-2.15). The findings support the associations of PAI-1 4G/5G polymorphism with VTE risk in the Asian population. | ||
| Wai Khoon Ho et al[21] | Risk of recurrent venous thromboembolism in patients with common thrombophilia | Specified disease name: VTE |
| 10 eligible studies included (FVL), 9 eligible studies included (prothrombin G20210A) | ||
| Finding: Significant association between the FVL and VTE was found under the heterozygous model (OR: 1.41, 95% CI: 1.14-1.75). The significant association between the prothrombin G20210A and VTE was found under the heterozygous model (OR: 1.72, 95% CI: 1.27-2.31). FVL and prothrombin G20210A of the heterozygous model are each associated with an increased risk of VTE. | ||
| Xindie Zhou et al[22] | Who are at risk for thromboembolism after arthroplasty? A systematic review and meta-analysis | Specified disease name: VTE |
| 14 eligible studies included (FVL), 7 eligible studies included (prothrombin), 2 eligible studies included (MTHFR) | ||
| Finding: Significant association between the FVL and VTE was found under the dominant model (OR: 1.41, 95% CI: 1.03-1.94). The significant association between the prothrombin G20210A and VTE was found under the dominant model (OR: 2.16, 95% CI: 1.27-3.69). The significant association between the MTHFR and VTE was found under the recessive model (OR: 2.36, 95% CI: 1.03-5.42). FVL, prothrombin G20210A, and MTHFR may increase the risk of VTE in Caucasian populations who are scheduled for arthroplasty. |
3.3. Qualitative methodological appraisal of eligible meta-analyses
Results of qualitative methodological appraisal of included studies using JBI Critical Appraisal Checklist for Systematic Reviews and Research Syntheses are shown in Table 4. Studies of Antonio Marchiori et al, Jessica Dennis et al, and Wai Khoon Ho et al showed the lowest quality score at 9. On the other hand, studies of Miao Gao et al, Peijin Zhang et al, Qiang Zhang et al, and Xindie Zhou et al showed the highest score of 11. Only the study of Da Li et al showed a score of 10.
Table 4.
The critical appraisal results of the included studies using the JBI- meta-analyses using critical appraisal checklist for systematic reviews and research syntheses.
| References | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | Total | |
| Antonio Marchiori et al[24] | Y | Y | Y | Y | N | Y | Y | Y | Y | N | Y | 9 |
| Da Li et al[17] | Y | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | 10 |
| Jessica Dennis et al[18] | Y | N | Y | Y | Y | Y | Y | Y | Y | N | N | 9 |
| Miao Gao et al[19] | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 11 |
| Peijin Zhang et al[20] | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 11 |
| Qiang Zhang et al[25] | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 11 |
| Wai Khoon Ho et al[21] | Y | Y | Y | N | Y | Y | Y | Y | Y | Y | N | 9 |
| Xindie Zhou et al[22] | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | Y | 11 |
0 = No, 1 = Yes, JBI = Joanna Briggs Institute.
4. Discussion
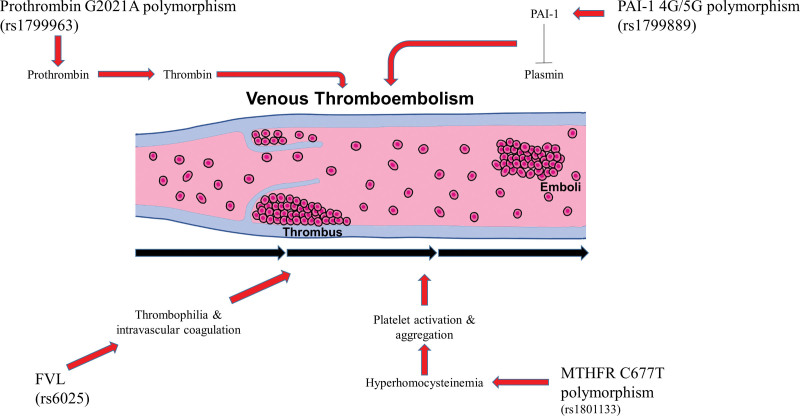
Genes reviewed in the present study with genetic variants associated with VTE included FVL (G1691A), prothrombin (G20210A), MTHFR (C677T, A1298C), PAI-1 (4G/5G), factor VII activating protease (1601G > A), and endothelial protein C receptor (g.6936A_G, c.4600A_G). Among them, variants of FVL, prothrombin, MTHFR, and PAI-1 showed high significance. Particularly, variants of prothrombin (G20210A), MTHFR (C677T), and PAI-1 (4G/5G) had more than 2 types of model significance.
Prothrombin gene mutation can alter the transcription of prothrombin gene, consequently generating thrombin, a key factor in blood clotting.[22] In Caucasians, prothrombin G20210A is one of the most common inherited thrombophilic disorders with a prevalence of 2%-3%.[28] In the presence of prothrombin G20210A mutation, the risk of the first episode of VTE is increased by 2- to 3-fold.[31] Gohil et al have found that patients with prothrombin G20210A polymorphism have a higher risk of deep vein thrombosis or PE.[32] Wahlandr K et al have also reported that prothrombin gene G20210A mutations could be potential risk factors for VTE after environmental factors such as a hip or knee replacement surgery.[33] Therefore, prothrombin G20210A could increase the risk of VTE by excessive generation of blood clotting factors. It could be accelerated by environmental factors.
Factor V plays an essential role as a regulator of thrombin formation.[34] FVL is a typical polymorphism of Factor V causing a coagulation disorder.[35] Several studies have reported that total hip replacement or joint replacement surgery could cause VTE in patients who have FVL genotype.[26,33,36–38] Especially, numerous studies have provided convincing evidence that heterozygosity for either mutation of FVL and prothrombin G20210A can significantly increase the risk of VTE.[39–41] The present study also showed that a heterozygous genetic model of FVL and prothrombin G20210A mutation had a high significance in VTE with a moderate or strong evidence class. These results support the relationship of the risk of VTE with genetic variants of FVL and prothrombin G20210A gene.
In the present study, MTHFR C677T gene mutation showed high significance results in various genetic models. MTHFR is an important enzyme regulating folate metabolism and DNA methylation.[20] MTHFR activity deficiency can lead to hyperhomocysteinemia which can alter platelet function causing blood coagulation, ultimately contributing to the etiology of VTE.[19] Recently, the association between MTHFR gene polymorphism and VTE has been studied, showing controversial results. Several studies have demonstrated that MTHFR C677T mutation is significantly associated with VTE.[42,43] In contrast, the MEGA study of Irene D. Bezemer et al has concluded that mildly increased homocysteine levels as a result of MTHFR 677T mutation have no association with VTE.[44] Some studies have suggested that MTHFR might affect the risk of VTE in cooperation with other genetic variants or environmental risk factors.[45,46]
PAI-1 plays an important role in fibrinolysis as an inhibitor of plasminogen activators.[47] The study of Baglin T et al has discovered that the 4G allele of PAI-1 may bind to the transcription region and increase mRNA transcription level of PAI-1.[48] In the present study, PAI-1 4G/5G polymorphism also showed a significant relationship with the risk of VTE in a recessive and homozygous genetic model. Particularly, the 4G/4G homozygous genetic model of PAI-1 with an increasing level of PAI-1 is known to cause a state of low fibrinolysis associated with the risk of VTE.[25] However, some studies did not show any significant results.[49]
Recently, many studies have reported that COVID-19 has a close association with VTE. In the case report of Jose Ramon Fiore et al, a young COVID-19 patient who had a homozygous prothrombin G20210A mutation suffered severe systemic thrombosis.[29] Similarly, numerous studies have reported the relationship between COVID-19 and several polymorphisms associated with VTE.[30,50] Vaccines against COVID-19 might increase the risk of VTE.[27] Some environmental risk factors such as cytokines that are increased after vaccination against COVID-19 might have an interrelationship with genetic variants associated with VTE. Further studies are needed to investigate prognostic genetic markers for VTE in COVID-19 patients for preventing vaccination side effect. The overall gene mechanisms of VET in the present study was shown in Figure 2.
Figure 2.
Overall gene mechanisms related to venous thromboembolism (VTE).
The present study has several limitations. First, to collect vast volumes of data, we eliminated gender characteristics from our study. Second, each data set did not have a specific ethnic classification. Third, data were limited by studies available at the time.
5. Conclusion
The present study performed a systematic review of genetic variants associated with VTE. Our results could lead to a more comprehensive understanding of the mechanism of VTE etiology. These results could give a strategy of prediagnosis to evaluate individual risks of VTE who might be exposed to environmental risk factors.
Author’s contributions
Conceptualization: Sangyeob Lee, Jun-Il Yoo.
Data curation: Sangyeob Lee, Jun-Il Yoo.
Formal analysis: Sangyeob Lee, Jun-Il Yoo.
Investigation: Sangyeob Lee, Chang Han Lee, Min Seok Seo.
Methodology: Sangyeob Lee, Jun-Il Yoo.
Project administration: Jun-Il Yoo.
Resources: Jun-Il Yoo.
Supervision: Sangyeob Lee, Jun-Il Yoo.
Validation: Chang Han Lee.
Visualization: Sangyeob Lee.
Writing – original draft: Sangyeob Lee, Jun-Il Yoo.
Writing – review & editing: Jun-Il Yoo.
Abbreviations:
- FVL =
- factor V Leiden
- MTHFR =
- methylenetetrahydrofolate reductase
- PAI-1 =
- plasminogen activator inhibitor-1
- PE =
- pulmonary embolism
- RR =
- risk ratio
- VTE =
- venous thromboembolism
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
The authors have no conflicts of interest to disclose.
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI22C0494).
Level of Evidence: Level I
How to cite this article: Lee S, Lee CH, Seo MS, Yoo JI. Integrative analyses of genes about venous thromboembolism: An umbrella review of systematic reviews and meta-analyses. Medicine 2022;101:43(e31162).
Contributor Information
Sangyeob Lee, Email: ychkhk1101@naver.com.
Chang Han Lee, Email: ychkhk1101@naver.com.
Min Seok Seo, Email: majestyno1@naver.com.
References
- [1].Schulman S, Ageno W, Konstantinides SV. Venous thromboembolism: past, present and future. Thromb Haemost. 2017;117:1219–29. [DOI] [PubMed] [Google Scholar]
- [2].Heit JA, Spencer FA, White RH. The epidemiology of venous thromboembolism. J Thromb Thrombolysis. 2016;41:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hunter R, Lewis S, Noble S, et al. “Post-thrombotic panic syndrome”: a thematic analysis of the experience of venous thromboembolism. Br J Health Psychol. 2017;22:8–25. [DOI] [PubMed] [Google Scholar]
- [4].Ariëns RA, de Lange M, Snieder H, et al. Activation markers of coagulation and fibrinolysis in twins: heritability of the prethrombotic state. Lancet. 2002;359:667–71. [DOI] [PubMed] [Google Scholar]
- [5].van Schouwenburg IM, Gansevoort RT, Mahmoodi BK, et al. Increased risk of arterial thromboembolism after a prior episode of venous thromboembolism: results from the prevention of REnal and vascular ENd stage disease (PREVEND) study. Br J Haematol. 2012;159:216–22. [DOI] [PubMed] [Google Scholar]
- [6].Ho WK, Hankey GJ, Eikelboom JW. The incidence of venous thromboembolism: a prospective, community-based study in Perth, Western Australia. Med J Aust. 2008;189:144–7. [DOI] [PubMed] [Google Scholar]
- [7].Wassel CL, Rasmussen-Torvik LJ, Callas PW, et al. A genetic risk score comprising known venous thromboembolism loci is associated with chronic venous disease in a multi-ethnic cohort. Thromb Res. 2015;136:966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hotoleanu C. Genetic risk factors in venous thromboembolism. Adv Exp Med Biol. 2017;906:253–72. [DOI] [PubMed] [Google Scholar]
- [9].Cushman M. Inherited risk factors for venous thrombosis. Hematology. 2005;2005:452–7. [DOI] [PubMed] [Google Scholar]
- [10].Vandenbroucke JP, Koster T, Briët E, et al. Increased risk of venous thrombosis in oral-contraceptive users who are carriers of factor V leiden mutation. Lancet. 1994;344:1453–7. [DOI] [PubMed] [Google Scholar]
- [11].Prandoni P. Acquired risk factors for venous thromboembolism in medical patients. Hematology Am Soc Hematol Educ Program. 2005;et al. :458–61. [DOI] [PubMed] [Google Scholar]
- [12].Andraska EA, Kulkarni R, Chaudhary M, et al. Three cases of acute venous thromboembolism in females following vaccination for COVID-19. J Vasc Surg Venous Lymphat Disord. 2021;et al. :14–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Coppens M, Reijnders JH, Middeldorp S, et al. Testing for inherited thrombophilia does not reduce the recurrence of venous thrombosis. J Thromb Haemost. 2008;6:1474–7. [DOI] [PubMed] [Google Scholar]
- [14].Chai W, Zhang Z, Ni M, et al. Genetic association between methylenetetrahydrofolate reductase gene polymorphism and risk of osteonecrosis of the femoral head. Biomed Res Int. 2015;2015:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Souto JC, Almasy L, Borrell M, et al. Genetic susceptibility to thrombosis and its relationship to physiological risk factors: the GAIT study. Am J Hum Genet. 2000;67:1452–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].van Hylckama Vlieg A, Baglin CA, Bare LA, et al. Proof of principle of potential clinical utility of multiple SNP analysis for prediction of recurrent venous thrombosis. J Thromb Haemost. 2008;6:751–4. [DOI] [PubMed] [Google Scholar]
- [17].Da L, Jiahui Z, Xiaoqiang L. Association between FSAP 1601G > a polymorphism and venous thromboembolism risk: a meta-analysis. Phlebology. 2020;35:345–53. [DOI] [PubMed] [Google Scholar]
- [18].Dennis J, Johnson CY, Adediran AS, et al. The endothelial protein C receptor (PROCR) Ser219Gly variant and risk of common thrombotic disorders: a HuGE review and meta-analysis of evidence from observational studies. Blood. 2012;119:2392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao M, Feng N, Zhang M, et al. Meta-analysis of the relationship between methylenetetrahydrofolate reductase C677T and A1298C polymorphism and venous thromboembolism in the Caucasian and Asian. Biosci Rep. 2020;40:BSR20200860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhang P, Gao X, Zhang Y, et al. Association between MTHFR C677T polymorphism and venous thromboembolism risk in the Chinese population: a meta-analysis of 24 case-controlled studies. Angiology. 2015;66:422–32. [DOI] [PubMed] [Google Scholar]
- [21].Ho WK, Hankey GJ, Quinlan DJ, et al. Risk of recurrent venous thromboembolism in patients with common thrombophilia: a systematic review. Arch Intern Med. 2006;166:729–36. [DOI] [PubMed] [Google Scholar]
- [22].Zhou X, Qian W, Li J, et al. Who are at risk for thromboembolism after arthroplasty? A systematic review and meta-analysis. Thromb Res. 2013;132:531–6. [DOI] [PubMed] [Google Scholar]
- [23].Ioannidis JPA, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. [DOI] [PubMed] [Google Scholar]
- [24].Marchiori A, Mosena L, Prins MH, et al. The risk of recurrent venous thromboembolism among heterozygous carriers of factor V leiden or prothrombin G20210A mutation. a systematic review of prospective studies. Haematologica. 2007;92:1107–14. [DOI] [PubMed] [Google Scholar]
- [25].Zhang Q, Jin Y, Li X, et al. Plasminogen activator inhibitor-1 (PAI-1) 4G/5G promoter polymorphisms and risk of venous thromboembolism – a meta-analysis and systematic review. Vasa. 2020;49:141–6. [DOI] [PubMed] [Google Scholar]
- [26].Ryan DH, Crowther MA, Ginsberg JS, et al. Relation of factor V leiden genotype to risk for acute deep venous thrombosis after joint replacement surgery. Ann Intern Med. 1998;128:270–6. [DOI] [PubMed] [Google Scholar]
- [27].Chih-Cheng L, I-Tzu C, Chien-Ming C, et al. COVID-19 vaccines: concerns beyond protective efficacy and safety. Expert Rev Vaccines. 2021;20:1013–25. [DOI] [PubMed] [Google Scholar]
- [28].Rosendaal FR, Doggen CJ, Zivelin A, et al. Geographic distribution of the 20210 G to A prothrombin variant. Thromb Haemost. 1998;79:706–8. [PubMed] [Google Scholar]
- [29].Fiore JR, Ciarallo M, Di Stefano M, et al. Severe systemic thrombosis in a young COVID-19 patient with a rare homozygous prothrombin G20210A mutation. Infez Med. 2021;29:259–62. [PubMed] [Google Scholar]
- [30].Fallerini C, Daga S, Benetti E, et al. SELP Asp603Asn and severe thrombosis in COVID-19 males. J Hematol Oncol. 2021;14:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].De Stefano V, Rossi E, Paciaroni K, et al. Screening for inherited thrombophilia: indications and therapeutic implications. Haematologica. 2002;87:1095–108. [PubMed] [Google Scholar]
- [32].Gohil R, Peck G, Sharma P. The genetics of venous thromboembolism. A meta-analysis involving approximately 120,000 cases and 180,000 controls. Thromb Haemost. 2009;102:360–70. [DOI] [PubMed] [Google Scholar]
- [33].Wåhlander K, Larson G, Lindahl TL, et al. Factor V leiden (G1691A) and Prothrombin Gene G20210A mutations as potential risk factors for venous thromboembolism after total hip or total knee replacement surgery. Thromb Haemost. 2002;87:580–5. [PubMed] [Google Scholar]
- [34].Rosing J, Tans G. Coagulation factor V: an old star shines again. Thromb Haemost. 1997;78:427–33. [PubMed] [Google Scholar]
- [35].Bertina RM, Koeleman BP, Koster T, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–7. [DOI] [PubMed] [Google Scholar]
- [36].Woolson ST, Zehnder JL, Maloney WJ. Factor V leiden and the risk of proximal venous thrombosis after total hip arthroplasty. J Arthroplasty. 1998;13:207–10. [DOI] [PubMed] [Google Scholar]
- [37].Della Valle CJ, Issack PS, Baitner A, et al. The relationship of the factor V leiden mutation or the deletion-deletion polymorphism of the angiotensin converting enzyme to postoperative thromboembolic events following total joint arthroplasty. BMC Musculoskelet Disord. 2001;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Bowler DJM, Bale E, O’Byrne J. Factor V leiden: prevalence and thromboembolic complications after total hip replacement in Ireland. Ir J Med Sci. 2007;176:273–7. [DOI] [PubMed] [Google Scholar]
- [39].Simioni P, Tormene D, Spiezia L, et al. Inherited thrombophilia and venous thromboembolism. Semin Thromb Hemost. 2006;32:700–8. [DOI] [PubMed] [Google Scholar]
- [40].Crowther MA, Kelton JG. Congenital thrombophilic states associated with venous thrombosis: a qualitative overview and proposed classification system. Ann Intern Med. 2003;138:128–34. [DOI] [PubMed] [Google Scholar]
- [41].Bauer KA. The thrombophilias: well-defined risk factors with uncertain therapeutic implications. Ann Intern Med. 2001;135:367–73. [DOI] [PubMed] [Google Scholar]
- [42].Abudureheman K, Mahemuti A, Xia Y, et al. [Association between gene polymorphisms of methylenetetrahydrofolate reductase and plasma homocysteine in Uygur patients with venous thromboembolism]. Zhonghua Xin Xue Guan Bing Za Zhi. 2012;40:1030–6. [PubMed] [Google Scholar]
- [43].Jang MJ, Jeon YJ, Choi W-I, et al. The 677C>T mutation of the MTHFR gene increases the risk of venous thromboembolism in Koreans and a meta-analysis from Asian population. Clin Appl Thromb Hemost. 2013;19:309–14. [DOI] [PubMed] [Google Scholar]
- [44].Bezemer ID, Doggen CJM, Vos HL, et al. No association between the common MTHFR 677C→T polymorphism and venous thrombosis: results from the MEGA study. Arch Intern Med. 2007;167:497–501. [DOI] [PubMed] [Google Scholar]
- [45].Fujimura H, Kawasaki T, Sakata T, et al. Common C677T polymorphism in the methylenetetrahydrofolate reductase gene increases the risk for deep vein thrombosis in patients with predisposition of thrombophilia. Thromb Res. 2000;98:1–8. [DOI] [PubMed] [Google Scholar]
- [46].Cattaneo M, Tsai MY, Bucciarelli P, et al. A common mutation in the methylenetetrahydrofolate reductase gene (C677T) Increases the risk for deep-vein thrombosis in patients with mutant factor V (Factor V:Q506). Arterioscler Thromb Vasc Biol. 1997;17:1662–6. [DOI] [PubMed] [Google Scholar]
- [47].Zhao L, Huang P. Plasminogen activator inhibitor-1 4G/5G polymorphism is associated with type 2 diabetes risk. Int J Clin Exp Med. 2013;6:632–40. [PMC free article] [PubMed] [Google Scholar]
- [48].Baglin T. Inherited and acquired risk factors for venous thromboembolism. Semin Respir Crit Care Med. 2012;33:127–37. [DOI] [PubMed] [Google Scholar]
- [49].Yilmaz E, Akar E, Akar N. Effect of plasminogen activator inhibitor-1 4G/5G polymorphism in Turkish deep vein thromboembolic patients with and without prothrombin 20210 G-A. Turk J Haematol. 2004;21:83–6. [PubMed] [Google Scholar]
- [50].Abu-Farha M, Al-Sabah S, Hammad MM, et al. Prognostic genetic markers for thrombosis in COVID-19 Patients: a focused analysis on D-Dimer, homocysteine and thromboembolism. Front Pharmacol. 2020;11:587451. [DOI] [PMC free article] [PubMed] [Google Scholar]