Background:
Whether metformin is related to nonalcoholic fatty liver disease (NAFLD) is controversial. Our aim was to investigate the relationship between metformin and NAFLD that may predict the metformin potential of these lesions and new prevention strategies in NAFLD patients.
Methods:
The meta-analysis was analyzed by Revman 5.3 softwares systematically searched for works published through July 29, 2022. Network pharmacology research based on databases, Cytoscape 3.7.1 software and R software respectively.
Results:
The following variables were associated with metformin in NAFLD patients: decreased of alanine aminotransferase (ALT) level (mean difference [MD] = −10.84, 95% confidence interval [CI] = −21.85 to 0.16, P = .05); decreased of aspartate amino transferase (AST) level (MD = −4.82, 95% CI = −9.33 to −0.30, P = .04); decreased of triglyceride (TG) level (MD = −0.17, 95% CI = −0.26 to −0.08, P = .0002); decreased of total cholesterol (TC) level (MD = −0.29, 95% CI = −0.47 to −0.10, P = .003); decreased of insulin resistance (IR) level (MD = −0.42, 95% CI = −0.82 to −0.02, P = .04). In addition, body mass index (BMI) (MD = −0.65, 95% CI = −1.46 to 0.16, P = .12) had no association with metformin in NAFLD patients. 181 metformin targets and 868 NAFLD disease targets were interaction analyzed, 15 core targets of metformin for the treatment of NAFLD were obtained. The effect of metformin on NAFLD mainly related to cytoplasm and protein binding, NAFLD, hepatitis B, pathway in cancer, toll like receptor signaling pathway and type 2 diabetes mellitus (T2DM). The proteins of hypoxia inducible factor-1 (HIF1A), nuclear factor erythroid 2-related factor (NFE2L2), nitric oxide synthase 3 (NOS3), nuclear receptor subfamily 3 group C member 1 (NR3C1), PI3K catalytic subunit alpha (PIK3CA), and silencing information regulator 2 related enzyme 1 (SIRT1) may the core targets of metformin for the treatment of NAFLD.
Conclusion:
Metformin might be a candidate drug for the treatment of NAFLD which exhibits therapeutic effect on NAFLD patients associated with ALT, AST, TG, TC and IR while was not correlated with BMI. HIF1A, NFE2L2, NOS3, NR3C1, PIK3CA, and SIRT1 might be core targets of metformin for the treatment of NAFLD.
Keywords: candidate drug, meta-analysis, metformin, network pharmacology, nonalcoholic fatty liver disease, therapeutic effect
1. Introduction
nonalcoholic fatty liver disease (NAFLD) has become the most common chronic liver disease in the world and the incidence rate of NAFLD has been increasing in recent years.[1] NAFLD is characterized by abnormal accumulation of triglycerides in hepatocytes, not due to secondary hepatic steatosis caused by excessive alcohol consumption or other reasons.[2] NAFLD is usually divided into 2 histological categories including nonalcoholic fatty liver (NAFL) and nonalcoholic steatohepatitis (NASH) which cause of liver disease with prevalence estimates ranging from 25% to 45%.[3] NAFLD includes a series of progressive liver diseases, ranging from simple steatosis to NASH, with fibrosis usually developing into cirrhosis and hepatocellular carcinoma (HCC).[4] Diet and exercise therapy are the cornerstone for NAFLD, but there is still no ideal treatment drug at present.
Previous studies have shown that obesity, insulin resistance (IR), dyslipidemia, abnormal glucose metabolism and hypertension are risk factors of NAFLD.[5] Liver lipid metabolism disorder, IR, the role of cytokines, increased expression of cytochrome P450 (CYP2E1, CYP4A), oxidative stress, lipid peroxidation, immune response, genetic factors and other related factors are all involved in the pathogenesis of NAFLD.[6] Therefore, improving IR and adjusting the balance of glucose and lipid metabolism may be an important measure for the prevention and treatment of NAFLD. In addition, NAFLD is not only related to morbidity and mortality of liver, but also with the increased risk of other diseases such as chronic kidney disease and colorectal carcinoma.[7]
Many researchers have conducted a series of animal experiments and clinical studies on the efficacy of metformin on NAFLD.[8] Metformin is an insulin sensitizer which may improve insulin sensitivity by increasing the binding of peripheral insulin and insulin receptor, increasing the clearance of blood sugar and improving insulin sensitivity.[9] Furthermore, the gastrointestinal discomfort which caused by metformin can reduce the food intake and body weight of obese patients.[10] Previous clinical studies have shown that metformin may improve the liver enzymes involved in lipid metabolism in patients with NAFLD and regulate the expression of liver fat hormones and adipokines.[11] However, it remains controversial including studies on the ineffectiveness of metformin to NAFLD have also been reported.[12] In addition, the effect and mechanism of metformin on NAFL is unclear. Therefore, the aim of the meta-analysis and network pharmacology research was to further assess the effects of metformin on NAFLD and figured out the most likely mechanism of metformin on NAFLD.
2. Methods
We followed the methods of Jingxin Mao et al 2020 and 2022, respectively.[13,14]
2.1. Search strategy
The applicable posted articles including PubMed, Embase databases, and ISI Web of Science databases were used to investigated until July 29, 2022. The following keywords were used in searching: “metformin” OR “met” AND “nonalcoholic fatty liver disease OR NAFLD OR nonalcoholic fatty liver NAFL OR nonalcoholic steatohepatitis OR NASH OR fatty liver” AND clinic trial OR treatment OR relationship. Relevant articles have been used to expand the search scope, and all retrieved studies, evaluations and convention abstracts had been retrieved by using the computer. If more than one posted studies describe the same population, we extract solely the most entire or latest one immediately. Two authors independently accomplished the decision procedure and finally resolved the differences via discussion.
2.2. Selection criteria
The selection/determination strategy used the following criteria: English language studies; Randomized controlled/managed clinical trial or potential or retrospective original research or cohort that explored the impact of metformin on NAFLD; To diagnosed of NAFLD used to be primarily based on standardized ultrasound examination or pathologic examination, The relevant records together with alanine aminotransferase (ALT), aspartate amino transferase (AST), total cholesterol (TC), triglyceride (TG), body mass index (BMI) and homeostasis model assessment of insulin resistance index (HOMA-IR) had been all analyzed statistically.
The exclusion standards had been adapted to exclude studies from meta-analysis as following: Non-English language studies; Reviews, case reports, editorials, letters to editors, congress abstracts, commentaries, exercise guidelines, conferences or convention records; Insufficient data (for instance: less than 30 participants in the study); Single diagnostic criteria for NAFLD without imaging examination (for instance: only using fatty liver index or serum liver enzyme index); Studying length past 20 years, Autoimmune, drug-induced liver disease/disorder or liver damage.
2.3. Data extraction
Two authors abstracted the following data from the included articles: first author, country, publication years, study design, case number, diagnosis method, covariate adjustment, met dose of treatment group and Newcastle-Ottawa quality assessment scale (NOS). Any disagreements were resolved by a third investigator. The NOS was finally been used to assess the quality of the study.
2.4. Explore the anti-NAFLD target of metformin
Validated and predicted targets of metformin were obtained from the SuperPred (https://prediction.charite.de/index.php) and SwissTarget Prediction (http://www.swisstargetprediction.ch/) databases. Using “nonalcoholic fatty liver disease” and “nonalcoholic hepatic disease” as keywords to search for NAFLD-related targets in CTD database (http://ctdbase.org/). The metformin targets retrieved above were mapped with NAFLD disease targets to obtain potential targets of metformin in the treatment of NAFLD, which were imported into the Uniprot database (https://www.uniprot.org/) and converted into corresponding gene names. Eliminate non-human targets in preparation for the subsequent topological analysis of protein protein interaction (PPI) networks.
2.5. PPI network construction, analysis and core gene screening
Import the gene names obtained in 2.4 into the STRING database, the species is limited to homo sapiens, the confidence is set to > 0.17, and the others are the default settings, create a PPI network map and save the TSV format file. The above results were imported into the cytohubba plugin of Cytoscape 3.7.1 software, and the maximal clique centrality (MCC) calculation method was selected to screen out the core target genes.
2.6. Biological function and pathway enrichment analysis of common drug-disease targets
The clusterProfiler R software was used to conduct GO functional enrichment analysis and KEGG pathway enrichment analysis on the potential targets of metformin in the treatment of NAFLD. P < .05 indicated that the difference was statistically significant. The above results were visualized using the R language gg-Plot2 package.
2.7. Component-target molecular docking validation
Using the Zinc database (http://zinc.docking.org/) to search the molecular structure of metformin and download its MOL2 format. Core target genes were screened in section 2.5 through the PDB database (http://www.rcsb.org/) to obtain the suitable protein structure and download it in PDB format. With AutoDockTools software, the molecular structure of metformin was optimized, and the core gene protein structure was dewatered and hydrogenated, and the molecule and protein storage format was converted to “pdbqt” format. Using Vina software for molecular docking, collect binding energy scores, and finally use PyMOL software for visual analysis.
2.8. Ethics statement
The meta-analysis and network pharmacology research were strictly followed the ethical approval that referenced publication articles which related to human care, handling, sampling and administration procedures should be approved by ethics committee.
2.9. Patient and public involvement
Patients and the public were not involved in the design, conduct, reporting, or dissemination plans of this research.
2.10. Statistical analysis
Using Ravman Manager softwares (version 5.3, Cochrane Collaboration) for statistical evaluation and analysis. The magnitude of the impact of each studying was utilized to calculated by the mean difference (MD) and the odds ratio (OR) of 95% confidence interval (CI) respectively. A P value <.05 was once regarded statistically significant. Furthermore, the heterogeneity was quantified the use of the Q test and the I2 statistic. While P > .1 and I2 < 50%, a fixed-effect model was finally used otherwise a random-effect model was finally applied. In addition, Begg funnel plots were used to calculated for viable publication bias.
3. Results
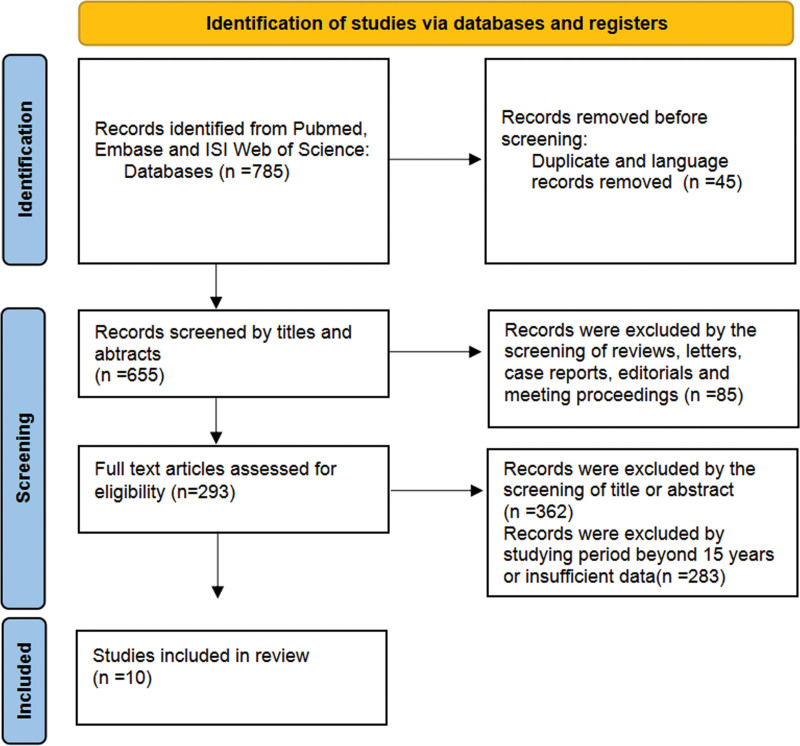
After searching, a total of 10 studies which met our selection criteria were finally included in the present meta-analysis. The selection flowchart of study was presented in Figure 1. The basic characteristics of the studies was included in Table 1. In addition, the network pharmacology of metformin on NAFLD was presented in the manuscript. Through network pharmacology data mining, 181 metformin targets and 868 NAFLD disease targets were obtained, and 38 potential targets of metformin in the treatment of NAFLD were obtained by mapping. 15 core target interaction relationships were obtained after PPI network calculation by MCC method. GO enrichment analysis and KEGG pathway analysis further elucidate the functional information of biological processes, cellular components closely with cytoplasm and protein binding, molecular functions pathways that mainly related to NAFLD, hepatitis B, pathway in cancer, toll like receptor signaling pathway and type 2 diabetes mellitus. Molecular docking results showed that the binding energies of metformin and core targets were all < 0 kJ mol−1, and the binding energies of hypoxia inducible factor-1 (HIF1A), nuclear factor erythroid 2-related factor (NFE2L2), nitric oxide synthase 3 (NOS3), nuclear receptor subfamily 3 group C member 1 (NR3C1), PI3K catalytic subunit alpha (PK3CA), and silencing information regulator 2 related enzyme 1 (SIRT1) and metformin were <−5 kJ mol−1 significantly.
Figure 1.
Flow chart of the study selection process.
Table 1.
Basic characteristics of included studies on the association between hypothyroidism and NAFLD.
| First author | Country | Publication yrs | Study design | Case number | Duration (mo) |
Diagnosis method | Covariate adjustment | Met dose of treatment group |
NOS |
|---|---|---|---|---|---|---|---|---|---|
| Feng[15] | China | 2018 | Prospective, randomized trial | 93 | 24 | Ultrasonography | Age, sex, AST, ALT, insulin resistance, blood glucose, body weight, BMI, TG, TC, HOMA- IR, HDL, LDL | 250 mg first wk, 500 mg second wk, and 1000 mg third wk | 8 |
| Garinis[16] | Italy | 2010 | Prospective study | 50 | 6 | Ultrasonographic | Age, sex, AST, ALT, insulin resistance, glucose, BMI, TG, TC, HOMA- IR, HDL, LDL | 1000 mg/d | 7 |
| Handzlik- Orlik[17] | Poland | 2018 | Open-label, randomized study | 42 | 6 | Ultrasound | AST, ALT, Insulin, BMI, TG, TC, HOMA- IR, GGTP | 500 mg first wk, 2000 mg second wk | 8 |
| Haukeland [18] |
Norway | 2009 | Proof-of-concept study | 44 | 6 | Biopsy | Weight, AST, ALT, insulin, BMI, TG, TC, HOMA-IR, free fatty acid | 500–3000 mg/d | 7 |
| Idilman[19] | Turkey | 2008 | Prospective study | 129 | 6 | Biochemical, radiological and histological criteria |
Age, gender, weight, AST, ALT, BMI, TG, TC, HOMA, body fat content, plasma glucose | 850 mg/d | 9 |
| Loomba[20] | American | 2009 | Open-label study | 28 | 12 | MRI and CT, imaging and liver biopsy | Age, gender, weight, AST, ALT, BMI, TG, TC, total bilirubin, LDL, HDL | 500 mg once daily, first wk, 500 mg twice daily, second wk to 3 wk, 1000 mg twice daily |
8 |
| Nar[21] | Turkey | 2008 | Prospective study | 34 | 6 | Ultrasonographic | Age, gender, AST, ALT, BMI, TG, TC, hypertension, glucose, LDL, HDL, leptin | 850–1700 mg/d | 7 |
| Nobili[22] | Italy | 2008 | Open-label, observational pilot study | 57 | 24 | Biopsy and ultrasonographic | Age, gender, AST, ALT, γ-GGT, BMI, TG, TC, HOMA | 1.5 g/d | 8 |
| Sofer[23] | Israel | 2011 | Single-center study | 63 | 4 | Ultrasonography | Age, gender, BMI, AST, ALT, TG, HOMA-IR, HOMA-β, HDL, LDL | 850–1700 mg/d | 8 |
| Uygun[24] | Turkey | 2004 | Randomized controlled study |
36 | 6 | Ultrasonography | Age, gender, AST, ALT, BMI, TG, TC, glucose, index of insulin resistance |
850 mg/d | 7 |
ALT = alanine aminotransferase, AST = aspartate amino transferase, HDL = high density lipoprotein, HOMA = homeostasis model assessment, HOMA-IR = insulin resistance index, LDL = low density lipoprotein, NAFLD = nonalcoholic fatty liver disease, NOS = Newcastle-Ottawa quality assessment scale, TG = triglyceride, TC = total cholesterol, γ-GGT = γ- glutamyl transferase.
3.1. Effect of metformin on NAFLD patients (Table 2 )
Table 2.
Effect of metformin on NAFLD patients.
| Related indicators | MD | 95% CI | P value |
|---|---|---|---|
| ALT | −10.84 | −21.85 to 0.16 | .05 |
| AST | −4.82 | −9.33 to −0.30 | .04 |
| BMI | −0.65 | −1.46 to 0.16 | .12 |
| TG | −0.17 | −0.26 to −0.08 | .0002 |
| TC | −0.29 | −0.47 to −0.10 | .003 |
| IR | −0.42 | −0.82 to −0.02 | .04 |
CI = confidence interval, MD = mean difference, NAFLD = nonalcoholic fatty liver disease.
3.1.1. Effect of metformin on ALT level
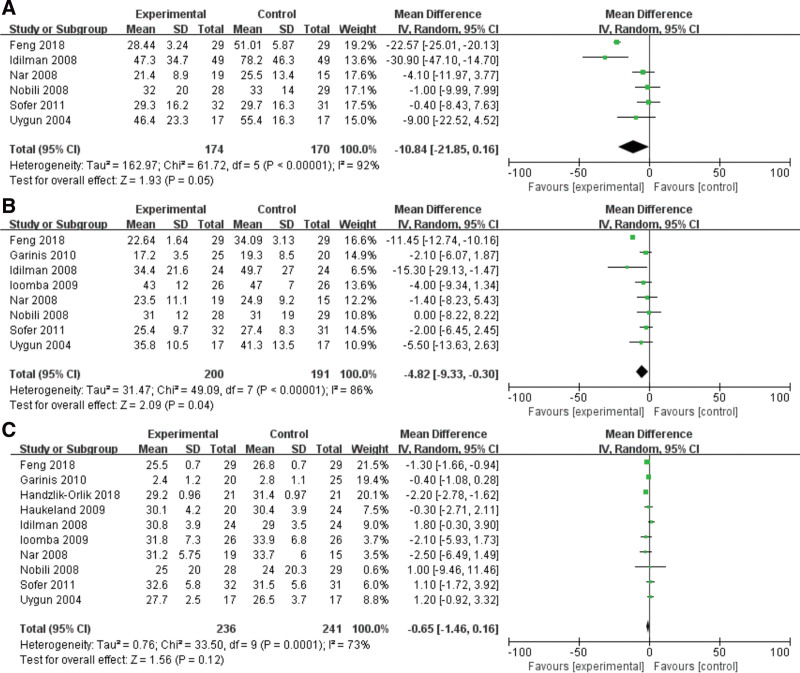
A random-effects model and input continuous data were selected using inverse variance method to calculated (P < .00001, I2 = 92%). The results indicated that a significant association was existed between metformin and ALT, metformin may decrease the level of ALT in NAFLD patients (MD = −10.84, 95% CI = −21.85 to 0.16, P = .05) (Fig. 2A).
Figure 2.
Forest plots of the effect of metformin on (A) ALT, (B) AST and (C) BMI level. ALT = alanine aminotransferase, AST = aspartate amino transferase, BMI = body mass index.
3.1.2. Effect of metformin on AST level
Total of 8 studies were investigated for the relationship between AST and NAFLD patients. A random-effects model and input continuous data were selected using inverse variance method to calculated (P < .00001, I2 = 86%). It was revealed that metformin could decreased the level of AST in NAFLD patients (MD = −4.82, 95% CI = −9.33 to −0.30, P = .04) (Fig. 2B).
3.1.3. Effect of metformin on BMi
Ten included studies were explored for the effect of metformin on BMI in the NAFLD patients. A random-effects model was utilized to analyze the data (P = .0001, I2 = 73%). It was demonstrated that metformin was not related to BMI in patients with NAFLD (MD = −0.65, 95% CI = −1.46 to 0.16, P = .12) (Fig. 2C).
3.1.4. Effect of metformin on TG level
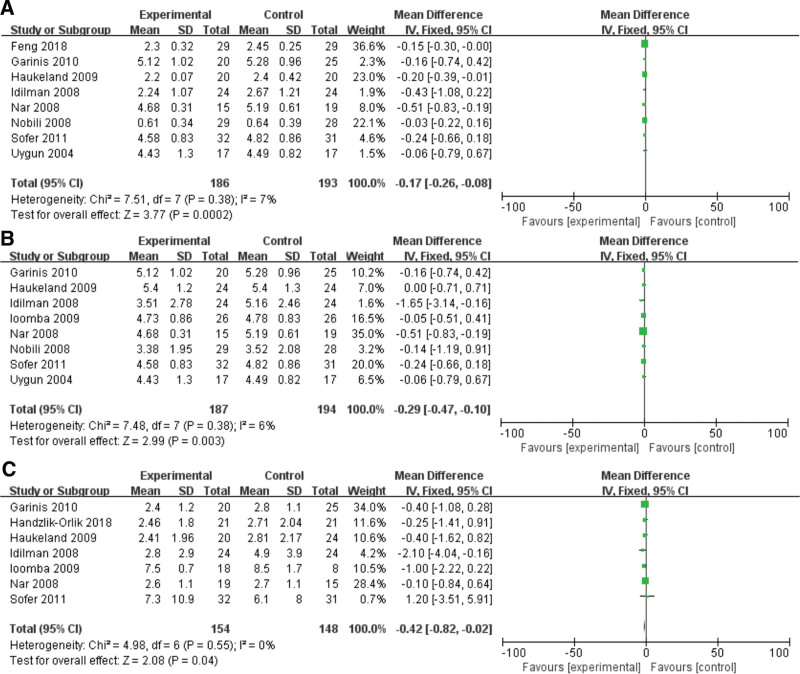
Eight studies were analyzed for the correlation between metformin and the level of TG in NAFLD patients. A fixed-effects model was used to analyze the data (I2 = 7%). In the meta-analysis, the unit of measurement has been unified as mmol/L. (TG: 88.5 mg/dL = 1 mmol/L). It was demonstrated that metformin was significantly relatively related to TG in NAFLD patients (MD = −0.17, 95% CI = −0.26 to −0.08, P = .0002) (Fig. 3A).
Figure 3.
Forest plots of the effect of metformin on (A) TG, (B) TC and (C) IR level. IR = insulin resistance, TG = triglyceride, TC = total cholesterol.
3.1.5. Effect of metformin on TC level
In the meta-analysis, the unit of measurement has been unified as mmol/L (TC: 100 mg/dL = 2.6 mmol/L). A random-effects model was utilized to analyze the data (I2 = 82%). Six included studies were evaluated for the relationship between metformin and TC in NAFLD patients. It was revealed that metformin can significantly improve the high level of TC in NAFLD patients (MD = −0.29, 95% CI = −0.47 to −0.10, P = .003) (Fig. 3B).
3.1.6. Effect of metformin on IR
A fixed-effects model was utilized to analyze the data (I2 = 0%). Seven included studies were investigated for the relationship between metformin and IR in NAFLD patients, and the IR was evaluated by HOMA-IR. It was revealed that metformin was closely associated with HOMA-IR in NAFLD patients which means metformin could significantly reduce the level of IR (MD = −0.42, 95% CI = −0.82 to −0.02, P = .04) (Fig. 3C).
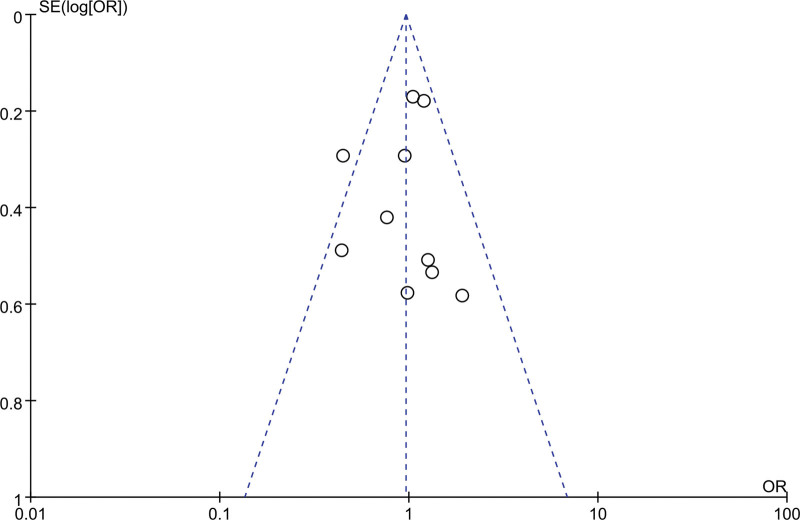
3.1.7. Publication bias and sensitivity analysis
Cochrane funnel plot was used to explore the publication bias. No obvious asymmetric distribution exhibits in Figure 4 which indicating that there was no publication bias.
Figure 4.
Funnel plots for publication bias analysis of the included articles.
3.2. Network pharmacology of metformin on NAFLd
3.2.1. Metformin anti-NAFLD targets
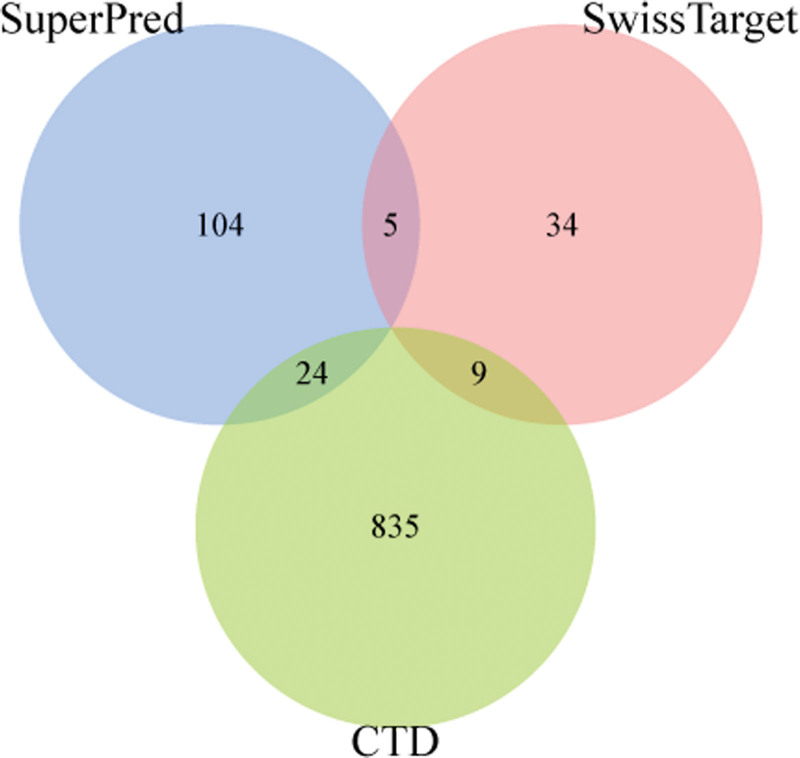
A total of 868 genes were collected as NAFLD disease targets through CTD database. 133 verified targets and 48 predicted targets of metformin were retrieved using SuperPred database and SwissTargetPrediction database, and 38 intersection targets were obtained after mapping with NAFLD targets point (Table 3 and Fig. 5), predicted to be the potential target of metformin for the treatment of NAFLD.
Table 3.
NAFLD related targets from metformin.
| Gene names | Entry | Protein names | Length (bp) |
|---|---|---|---|
| PLAU | P00749 | Urokinase-type plasminogen activator | 431 |
| ADORA1 | P30542 | Adenosine receptor A1 | 326 |
| METAP2 | P50579 | Methionine aminopeptidase 2 | 478 |
| CA1 | P00915 | Carbonic anhydrase 1 | 261 |
| TYMP | P19971 | Thymidine phosphorylase | 482 |
| VDR | P11473 | Vitamin D3 receptor | 427 |
| FGR | P09769 | Tyrosine-protein kinase | 529 |
| RPS6KA5 | O75582 | Ribosomal protein S6 kinase alpha-5 | 802 |
| ACACA | Q13085 | Acetyl-CoA carboxylase 1 | 2346 |
| IKBKB | O14920 | Inhibitor of nuclear factor kappa-B kinase | 756 |
| PIK3CA | P42336 | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 1068 |
| NR4A1 | P22736 | Nuclear receptor subfamily 4 group A member 1 | 598 |
| DPP9 | Q86TI2 | Dipeptidyl peptidase 9 | 863 |
| HIF1A | Q16665 | Hypoxia-inducible factor 1-alpha | 826 |
| NFE2L2 | Q16236 | Nuclear factor erythroid 2-related factor 2 | 605 |
| NR3C1 | P04150 | Glucocorticoid receptor | 777 |
| SIRT3 | Q9NTG7 | NAD-dependent protein deacetylase sirtuin-3, mitochondrial | 399 |
| AURKB | Q96GD4 | Aurora kinase B | 344 |
| GLS | O94925 | Glutaminase kidney isoform, mitochondrial | 669 |
| MAP4K2 | Q12851 | Mitogen-activated protein kinase kinase kinase kinase 2 | 820 |
| CCR2 | P41597 | C-C chemokine receptor type 2 | 374 |
| PIK3R1 | P27986 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 724 |
| NFKB1 | P19838 | Nuclear factor NF-kappa-B p105 subunit | 968 |
| SIRT1 | Q96EB6 | NAD-dependent protein deacetylase sirtuin-1 | 747 |
| TDO2 | P48775 | Tryptophan 2,3-dioxygenase | 406 |
| NR1I2 | O75469 | Nuclear receptor subfamily 1 group I member 2 | 434 |
| APEX1 | P27695 | DNA-(apurinic or apyrimidinic site) endonuclease | 318 |
| CYP3A4 | P08684 | Cytochrome P450 | 503 |
| RPS6KB1 | P23443 | Ribosomal protein S6 kinase beta-1 | 525 |
| NOS2 | P35228 | Nitric oxide synthase, inducible | 1153 |
| NOS1 | P29475 | Nitric oxide synthase, brain | 1434 |
| SLC47A1 | Q96FL8 | Multidrug and toxin extrusion protein 1 | 570 |
| NOS3 | P29474 | Nitric oxide synthase, endothelial | 1203 |
| INMT | O95050 | Indolethylamine N-methyltransferase | 263 |
| SLC22A1 | O15245 | Solute carrier family 22 member 1 | 554 |
| HRH2 | P25021 | Histamine H2 receptor | 359 |
| ESR2 | Q92731 | Estrogen receptor beta | 530 |
| ACHE | P22303 | Acetylcholinesterase | 614 |
NAFLD = nonalcoholic fatty liver disease.
Figure 5.
The venn analysis of metformin on NAFLD. NAFLD = Nonalcoholic fatty liver disease.
3.2.2. Biofunction and pathway enrichment analysis of metformin-potential targets of NAFLD
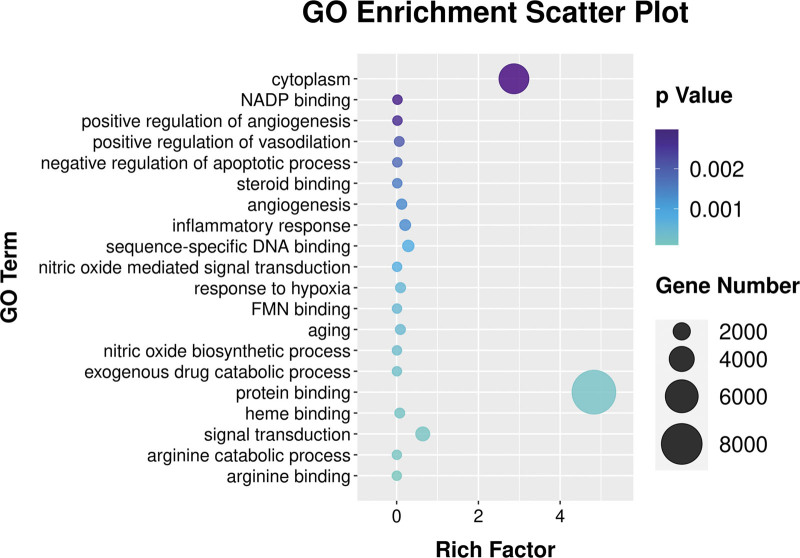
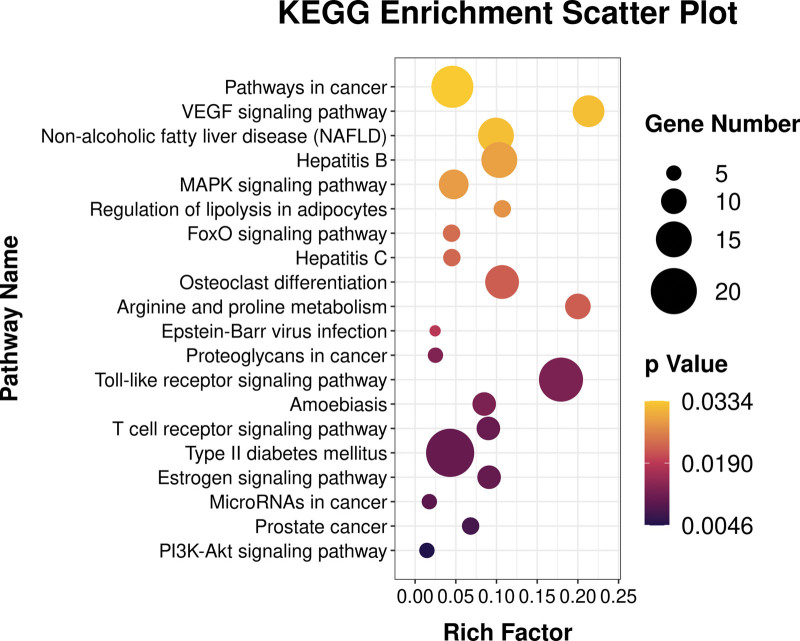
The clusterProfiler R software was used to analyze the biological processes and molecular functions of potential targets involved in NAFLD, and to explored the possible molecular mechanism of metformin in the treatment of NAFLD. The GO function enrichment results were sorted by significance (P < .05), and the top 20 items in various analyses were selected for R software visualization (Fig. 6). The potential targets of metformin in the treatment of NAFLD mainly involve cytoplasm, NADP binding, positive regulation of angiogenesis, positive regulation of vasodilation, negative regulation of apoptotic process, steroid binding, angiogenesis, inflammatory response, sequence-specific DNA binding, nitric oxide mediated signal transduction, response to hypoxia, FMN binding, aging, nitric oxide biosynthetic process, exogenous drug catabolic process, protein binding, heme binding, signal transduction, arginine catabolic process, arginine binding. KEGG pathway enrichment analysis screened the top 20 signaling pathways for visual analysis (P < .05) (Fig. 7). The NAFLD signaling pathways involved mainly including pathways in cancer, VEGF signaling pathway, non-alcoholic fatty liver disease (NAFLD), Hepatitis B, MAPK signaling pathway, regulation of lipolysis in adipocytes, FoxO signaling pathway, Hepatitis C, Osteoclast differentiation, Arginine and proline metabolism, Epstein-Barr virus infection, Proteoglycans in cancer, Toll-like receptor signaling pathway, Amoebiasis, T cell receptor signaling pathway, Type II diabetes mellitus, Estrogen signaling pathway, MicroRNAs in cancer, Prostate cancer, PI3K-Akt signaling pathway.
Figure 6.
GO enrichment analysis of metformin on NAFLD. NAFLD = Nonalcoholic fatty liver disease.
Figure 7.
KEGG pathway enrichment analysis of metformin on NAFLD. NAFLD = Nonalcoholic fatty liver disease.
3.2.3. The results of PPI network construction, analysis and core gene screening
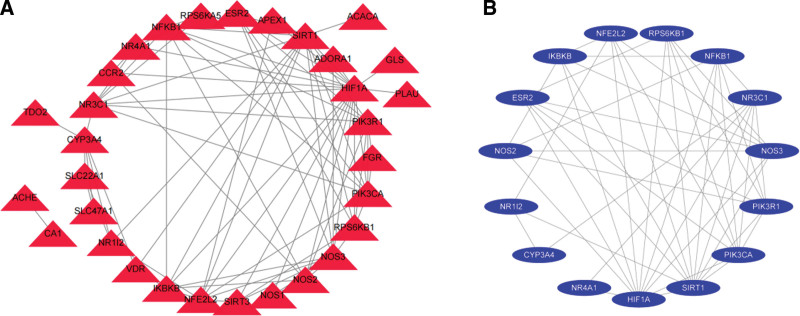
Import the potential targets mined in metformin anti-NAFLD targets into the STRING database, and save the PPI network map and TSV format files. The nodes in the figure represent proteins, and the edges represent the interaction between proteins, which fully reflects the complexity of the molecular mechanism of metformin in the treatment of NAFLD. The nodes in the figure represent proteins, and the edges represent the interaction between proteins, which fully reflects the complexity of the molecular mechanism of metformin in the treatment of NAFLD (Fig. 8A). Import the above files into the cytohubba plug-in, and use the MCC method to calculate and screen out HIF1A, SIRT1, NOS3, PIK3CA, NR3C1, NFE2L2, PIK3R1, NFKB1, IKBKB, NOS2, RPS6KB1, CYP3A4, ESR2, NR1I2, and NR4A1 targets in the core position (Table 4 and Fig. 8B). Its interaction with other proteins is more closely related, and these 15 targets can be predicted to be the key targets of metformin in the treatment of NAFLD.
Figure 8.
PPI network analysis diagram of potential targets of metformin on NAFLD. NAFLD = nonalcoholic fatty liver disease, PPI = protein protein interaction.
Table 4.
Core targets of metformin on NAFLD and their topological characteristics.
| Gene names | Targets names | Betweenness centrality | Degree |
|---|---|---|---|
| HIF1A | Hypoxia-inducible factor 1-alpha | 0.3527774 | 18 |
| SIRT1 | NAD-dependent protein deacetylase sirtuin-1 | 0.20641763 | 14 |
| NOS3 | Nitric oxide synthase, endothelial | 0.04175313 | 10 |
| PIK3CA | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | 0.05492052 | 9 |
| NR3C1 | Glucocorticoid receptor | 0.17371514 | 8 |
| NFE2L2 | Nuclear factor erythroid 2-related factor 2 | 0.05083879 | 8 |
| PIK3R1 | Phosphatidylinositol 3-kinase regulatory subunit alpha | 0.03997753 | 8 |
| NFKB1 | Nuclear factor NF-kappa-B p105 subunit | 0.03621829 | 8 |
| IKBKB | Inhibitor of nuclear factor kappa-B kinase | 0.02620307 | 7 |
| NOS2 | Nitric oxide synthase, inducible | 0.02237358 | 7 |
| RPS6KB1 | Ribosomal protein S6 kinase beta-1 | 0.01082576 | 7 |
| CYP3A4 | Cytochrome P450 | 0.15933485 | 6 |
| ESR2 | Estrogen receptor beta | 0.00339315 | 6 |
| NR1I2 | Nuclear receptor subfamily 1 group I member 2 | 0.07004283 | 4 |
| NR4A1 | Nuclear receptor subfamily 4 group A member 1 | 0.01549404 | 4 |
NAFLD = nonalcoholic fatty liver disease
3.2.4. Component-target molecular docking validation
A total of 6 core targets with degree >8 and metformin among the 10 core target genes screened in PPI network were entered into the PDB and Zinc databases, to obtained the molecular structure of metformin and target protein structure respectively. The interaction between proteins and small molecule receptors was simulated by molecular docking technology, and the binding energy values of docking were collected. The low-energy stable conformation between ligand and receptor indicates that there is a greater possibility of interaction between the 2. The binding energies of metformin and core target proteins are all negative, suggesting that ligands and receptors can bind spontaneously. The binding energies of HIF1A, NFE2L2, NOS3, NR3C1, PK3CA, SIRT1 and metformin are <−5 kJ mol−1 among them. It was proved that the above targets had stronger binding activity to metformin (Table 5 and Fig. 9).
Table 5.
The results of molecular docking.
| Compound | Target | PDB | Energy (kcal/mol) |
|---|---|---|---|
| metformin | HIF1A | 1H2K | −5.0 |
| metformin | SIRT1 | 4IG9 | −5.0 |
| metformin | NOS3 | 1M9J | −6.4 |
| metformin | PIK3CA | 2ENQ | −5.0 |
| metformin | NR3C1 | 1M2Z | −6.4 |
| metformin | NFE2L2 | 2F1U | −6.1 |
PDB = protein data bank.
Figure 9.
Molecular docking results of metformin on NAFLD with core targets. NAFLD = Nonalcoholic fatty liver disease.
4. Discussion
The incidence of NAFLD and NASH is increasing due to the obesity and diabetes epidemics.[25] It has been demonstrated that the benefits of metformin in inhibiting hepatic gluconeogenesis, improving hepatic fatty acid metabolism (including inhibition of adipose tissue lipolysis), increasing fatty acid oxidation, inhibiting lipogenesis and enhancing insulin sensitivity.[26] Metformin as a treatment for NAFLD/NASH has been partly examined by pilot studies and randomized controlled clinical trials in the past. In addition, previous studies have been reported these beneficial effects of metformin on liver histology in NAFLD/NASH patients to to reduce mortality and fibrosis.[27,28] In addition, Metformin was also suggested to reduce risks for HCC and protect against NASH-related HCC.[29] NAFLD is among the most common causes of liver disease worldwide. Increasing exercise and reasonable planning of diet are all conventional treatments for patients with NAFLD which is expected to reduce weight, but this process takes a long time and is prone to sustained liver injury.[30] Previous studies have shown that NAFLD is closely related to metabolic syndrome including obesity, diabetes, hyperlipidemia and hypertension are risk factors of NAFLD.[31] In addition, some researches has demonstrated that pathophysiological mechanism of these risk factors is IR which exists in more than 90% of NAFLD patients.[32] The pathogenesis of NAFLD is complex and closely related to type 2 diabetes mellitus (T2DM).[33] Generally accepted, it is interpreted as the “two-hit” hypothesis, which means the systemic multiple system damage caused by IR and its secondary disorder of glucose and lipid metabolism.[34] Therefore, correcting the disorder of glucose metabolism and improving IR maybe an important measure to treat NAFLD patients.
Metformin is a recognized the first-line drug to treatment for T2DM. It increases the insulin sensitivity of the peripheral and liver, reduces the production of basic liver glucose, and increases the glucose uptake and utilization of insulin-stimulated peripheral tissues.[35] In recent years, new indications for metformin treatment have appeared, including polycystic ovary syndrome and obesity.[36] Due to NAFLD patients are often accompanied by decreased blood glucose, dyslipidemia, and obesity, metformin may have a beneficial effect on liver steatosis.[37] The exact mode of action of metformin in liver steatosis has not been fully explored, but it may be involved in the destruction of the mitochondrial oxidation process.[38]
The present meta-analysis revealed that metformin may has a certain therapeutic effect on NAFLD that short-term (no more that 6 months), low-dose (500 to 3000 mg/d) metformin could ameliorate the levels of ALT, AST, TG and TC in NAFLD patients, and significantly improve HOMA-IR finally. It was showed that metformin can improve the disorder of lipid metabolism, reduce the steatosis and inflammatory response of the liver, and this effect does not depend on the hypoglycemic effect and exists independently.
Transaminase especially ALT and AST considered to be the common indicator for evaluating whether liver cells are damaged.[39] In a variety of liver diseases including viral hepatitis, alcoholic liver disease, drug-induced liver damage, autoimmune hepatitis and other diseases, ALT and AST always exhibit an upward trend.[40] ALT is mainly found in liver cells, while AST is mainly distributed in myocardium.[41] It was reported that ALT and AST can be used as a biomarker to diagnosis of progress of NAFLD even the increased in the normal range.[42] However, in the laboratory examination of early NAFLD patients, the rise of enzyme has no significant specificity, and it is easy to ignore the occurrence of liver injury. In present study, it was found that metformin has closely association with ALT and AST in NAFLD patients. With the increase of metformin dosage and time, both of ALT (MD = −10.84, P = .05) and AST (MD = −4.82, P = .04) showed a downward trend.
BMI = body weight (kg)/[height (m)2], which is a usually research measure used to divide the weight of study participants into broad groups, typically underweight, normal weight, overweight and corpulent classes (obese categories).[43] It was reported that generally the mean or median BMI is about 24 to 27 in Western population-based studies.[44, 45] Therefore, the consequence of adopting the world health organization (WHO) classification is that ~50% or additional of the final adult population may continuously be within the overweight (now preobese) and corpulent classes.
It was also revealed that BMI which related to obese is the most useful predictive factor for the onset of NAFLD which means a poorer prognosis ultimately.[46] In addition, obesity has reduced insulin sensitivity and higher susceptibility to NAFLD, which is also the main reason for the multiple body fatness of NAFLD patients.[47] However, according to our analysis data, metformin was not significantly improve BMI in patients with NAFLD (MD = −0.65, P = .12). These relatively conflicting findings between different studies might be related to different characteristics of the patients studied, including sample sizes and proportions of different types of NAFLD. Furthermore, the negligible effect of metformin on weight has been shown before.[48]
Eight studies were analyzed for the relationship between metformin and the level of TG in NAFLD patients. The present study has demonstrated that metformin was closely association with TG which exhibits a downtrend in NAFLD patients (MD = −0.17, P = .0002). It was revealed that the liver can not only decompose and utilize the stored body fat, but also store the digested and absorbed fat as body fat, which is dynamically balanced in the normal population.[49] However, once this balance is broken, TG is deposited in the liver to form fatty liver in metabolic disorders.[50] In addition, NAFLD is closely related to dyslipidemia and elevated blood sugar. When the liver accumulates fat, its gluconeogenesis strengthens, resulting in increased blood sugar, hyperglycemia can stimulate increased insulin secretion, causing hyperinsulinemia, which further promotes liver synthesis of TG aggravates the lipid deposition of the liver and forms a vicious circle.[51] Hyper TG causes increased release of free fatty acids, which interferes with the binding of insulin to receptors in the surrounding tissues, resulting in insulin resistance.[52]
TC refers to the cholesterol contained in various lipoproteins in serum which including he sum of bound cholesterol and free cholesterol.[53] TC is the main component of the cell membrane, therefore the serum concentration of TC can be used as an indicator of lipid metabolism.[54] A large amount of TG synthesized in NAFLD patients with fatty liver may reducing the high-density lipoprotein cholesterol levels and affecting synthesis of high-density lipoprotein cholesterol.[55] When extensively TG is produced in the NAFLD patient’s body, the level of TC may also significantly increased, which is also a sign of severe damage to the patient’s liver.[56] In present study, the effect of metformin on TC (MD = −0.29, P = .003) and TG (MD = −0.17, P = .002) levels were statistically significant, indicating that metformin can significantly improve the blood lipid levels of patients with NAFLD.
Previous study have proved the closely relationship between NAFLD and IR which indicating NAFLD is consider to be an early marker of IR.[57] Meantime, NAFLD is also a metabolic stress-induced liver injury that related to IR which may causes hepatic steatosis and genetic susceptibility.[58] Furthermore, it was also found that metformin may improves liver function, HOMA-IR in NAFLD patients.[59] In present study, it was revealed that patients with NAFLD have severe IR, which is closely related to the pathogenesis of NAFLD. Furthermore, metformin was effectively on improving IR (MD = −0.42, P = .04).
The network pharmacology of metformin on NAFLD results showed that the core targets, significant GO function enrichment and KEGG pathways enrichment. In present study, a total of 181 metformin targets and 868 NAFLD disease targets were interaction analyzed, and 38 potential and 15 core targets of metformin for the treatment of NAFLD were obtained. The effect of metformin on NAFLD mainly related to cytoplasm and protein binding of GO function enrichment respectively, as well as NAFLD, hepatitis B, pathway in cancer, toll like receptor signaling pathway and type 2 diabetes mellitus respectively of KEGG pathways enrichment. It was demonstrated that NAFLD is related to elevated cytoplasmic calcium signaling in hepatocytes[60] which is similar with our predicted GO function enrichment. In addition, sterol regulatory element binding protein has also been reported to play a critical role in NAFLD lipogenesis and correlates with a poor prognosis in NAFLD[61] which is also consistent with our predicted result. Interestingly, the NAFLD and hepatitis B pathway were also obtained in the KEGG pathways enrichment which further proves the correctness of our conjecture and analysis. It was revealed that the co-existence of NAFLD and chronic hepatitis B is common among patients that may synergistically exacerbate liver fibrosis and hepatocellular carcinoma.[62] It was reported that NAFLD-induced lipid accumulation may accelerate tumorigenesis through activation of PI3K. However, the protein kinase Akt is a key factor in hepatic insulin output to glucose.[63] Therefore, pathway in cancer is related to NAFLD. Previous study suggests that intestinal dysbiosis, intestinal barrier dysfunction, and activated toll-like receptor 4 signaling play key roles in the pathogenesis of NAFLD.[64] It was demonstrated that toll like receptor signaling pathway is closely with NAFLD. It has been found that NAFLD is not an isolated disease, but part of a metabolic disorder caused by high energy intake, obesity, sedentary lifestyle, and key IR and T2DM.[65] The prevalence of NAFLD and T2DM has risen markedly because of both of them may share similar risk factors, epidemiology, and pathophysiology. Furthermore, the presence of T2DM significantly increased the odds of NASH and fibrosis compared with NAFLD without T2DM.[66] The above studies confirmed NAFLD is strongly connection with T2DM which is similar with our predicted result. It was reported that the higher the degree value of the target topology analysis, the better the connectivity of this type of targets and the regulation of the entire network.[67] Based on the principle, the proteins of HIF1A, NFE2L2, NOS3, NR3C1, PK3CA, and SIRT1 were considered to be the core targets of metformin for the treatment of NAFLD.
5. Conclusion
Taken together, this systematic evaluation and network pharmacology research have shown that metformin could improve biochemical and metabolic characteristics in the progression and the development of NAFLD patients. Furthermore, metformin was related to ALT, AST, TG, TC and IR while was not correlated with BMI in NAFLD patients. HIF1A, NFE2L2, NOS3, NR3C1, PIK3CA and SIRT1 may be the core targets of metformin in the treatment of NAFLD. Therefore, metformin might be a promising drug for the treatment of NAFLD due to its metabolic effect and safety curative effect. These studies have extended the knowledge on the complicated mechanism of NAFLD, and provided the prevention options of NAFLD in patients with metabolic diseases.
6. Strengths and limitations of this study
There still exist some limitations in the present meta-analysis and network pharmacology research. Firstly, the heterogeneity is significant in some studies. We attempted to explore the heterogeneity between studies through a random effects model to give a more conservative effect estimate. Secondly, criteria of diagnosis cannot reach uniformity. Due to liver biopsy is not feasible in the general population, some non-standardization methods such as ultrasound, computed tomography, magnetic resonance imaging, and spectroscopy are commonly used to diagnose NAFLD. Thirdly, only 10 suitable studies were included for investigating the effect of metformin on NAFLD patients. Fourth, the manuscript is only based on network pharmacology software and database for prediction without experimental validation. In future studies, large-scale and long-term randomized controlled trials should be conducted in different populations to provide more important evidence. In addition, the experimental validation for predicting key targets of metformin on NAFLD should be carried out at the cellular and animal levels respectively.
Author contributions
JXM conceived and designed the research. YSH, XDW, and JXM conducted statistical analysis and wrote the paper. LZ, CL, TLL and EL abstracted the total data from the included articles in the meta-analysis. YSH, JXM, XDW, LDZ and LZ completed the data analysis of network pharmacology research. All authors contributed to manuscript revision, read and approved the submitted version.
Conceptualization: Jingxin Mao.
Data curation: Yuanshe Huang, Xiaodong Wang, Chen Yan, Lidan Zhang, Lai Zhang, Tianlei Liu.
Formal analysis: Lai Zhang, Tianlei Liu.
Funding acquisition: Yuanshe Huang.
Investigation: Yuanshe Huang, Xiaodong Wang, Chen Yan, Chen Li.
Methodology: Yuanshe Huang, Xiaodong Wang, Chen Yan, Lidan Zhang, E Liang, Jingxin Mao.
Resources: Jingxin Mao.
Software: Xiaodong Wang, Chen Li, Lai Zhang, E Liang, Tianlei Liu.
Supervision: E Liang.
Validation: Chen Yan, Chen Li, Lidan Zhang.
Visualization: Yuanshe Huang, Xiaodong Wang, Jingxin Mao.
Writing – original draft: Yuanshe Huang.
Writing – review & editing: Chen Yan, Jingxin Mao.
Acknowledgments
The authors would like to thank Prof Min Chen of the College of Pharmaceutical Sciences, Southwest university for helpful discussions on topics related to this work.
Abbreviations:
- ALT =
- alanine aminotransferase
- AST =
- aspartate amino transferase
- BMI =
- body mass index
- CI =
- confidence interval
- HCC =
- hepatocellular carcinoma
- HIF1A =
- hypoxia inducible factor-1
- HOMA-IR =
- insulin resistance index
- IR =
- insulin resistance
- MCC =
- maximal clique centrality
- MD =
- mean difference
- NAFLD =
- nonalcoholic fatty liver disease
- NAFL =
- nonalcoholic fatty liver
- NASH =
- nonalcoholic steatohepatitis
- NFE2L2 =
- nuclear factor erythroid 2-related factor
- NOS =
- Newcastle-Ottawa quality assessment scale
- NOS3 =
- nitric oxide synthase 3
- NR3C1 =
- nuclear receptor subfamily 3 group C member 1
- OR =
- odds ratio
- PIK3CA =
- PI3K catalytic subunit alpha
- PPI =
- protein protein interaction
- SIRT1 =
- silencing information regulator 2 related enzyme 1
- T2DM =
- type 2 diabetes mellitus
- TC =
- total cholesterol
- TG =
- triglyceride
- WHO =
- world health organization
The authors have no conflicts of interest to disclose.
This work was supported by 2020 Ministerial Project of China (No. 2020YYCXCQSJ050), Anshun University/Innovation Center for Efficient Agricultural of Guizhou Mountain Characteristics/Branch of learning in Agricultural resources and environment, Guizhou Province Department of Education Project (QJH KY [2020] 063) and Science and Technology Bureau of Anshun [grant no. ASKN (2020) 07] respectively.
YSH, CY, JXM and XDW contributed equally to this work.
All data generated or analyzed during this study are included in this published article [and its supplementary information files]
How to cite this article: Huang Y, Wang X, Yan C, Li C, Zhang L, Zhang L, Liang E, Liu T, Mao J. Effect of metformin on nonalcoholic fatty liver based on meta-analysis and network pharmacology. Medicine 2022;101:43(e31437).
Contributor Information
Yuanshe Huang, Email: 285275167@qq.com.
Chen Yan, Email: chen.li@fu-berlin.ed.
Chen Li, Email: chen.li@fu-berlin.ed.
Lidan Zhang, Email: 975575681@qq.com.
Lai Zhang, Email: 975575681@qq.com.
E Liang, Email: 1013653671@qq.com.
Tianlei Liu, Email: tianlei.liu@vip.163.com.
References
- [1].Smolle E, Kessler SM, Golob N, et al. Non-alcoholic fatty liver disease. Met Syn. 2016;65:641–57. [Google Scholar]
- [2].Kamath S, Chavez AO, Gastaldelli A, et al. Coordinated defects in hepatic long chain fatty acid metabolism and triglyceride accumulation contribute to insulin resistance in non-human primates. PLoS One. 2011;6:e27617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gunn NT, Shiffman ML. The use of liver biopsy in nonalcoholic fatty liver disease: when to biopsy and in whom. Clin Liver Dis. 2018;22:109–19. [DOI] [PubMed] [Google Scholar]
- [4].Su Q, Kumar V, Sud N, et al. MicroRNAs in the pathogenesis and treatment of progressive liver injury in NAFLD and liver fibrosis. Adv Drug Deliv Rev. 2018;129:54–63. [DOI] [PubMed] [Google Scholar]
- [5].Reinehr T, Toschke AM. Onset of puberty and cardiovascular risk factors in untreated obese children and adolescents: a 1-year follow-up study. Arch Pediatr Adolesc Med. 2009;163:709–15. [DOI] [PubMed] [Google Scholar]
- [6].Aubert J, Begriche K, Knockaert L, et al. Increased expression of cytochrome P450 2E1 in nonalcoholic fatty liver disease: mechanisms and pathophysiological role. Clin Res Hepatol Gastroenterol. 2011;35:630–7. [DOI] [PubMed] [Google Scholar]
- [7].Ionică FE, Negreș S, Bejenaru L, et al. Pharmacological approaches for nonalcoholic fatty liver disease. Rom J Diabetes Nutr Metab Dis. 2016;23:313–8. [Google Scholar]
- [8].Li Y, Liu L, Wang B, et al. Metformin in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Bio Rep. 2013;1:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gupta A, Bisht B, Dey CS. Peripheral insulin-sensitizer drug metformin ameliorates neuronal insulin resistance and Alzheimer’s-like changes. Neuropharmacology. 2011;60:910–20. [DOI] [PubMed] [Google Scholar]
- [10].Yasuda N, Inoue T, Nagakura T, et al. Metformin causes reduction of food intake and body weight gain and improvement of glucose intolerance in combination with dipeptidyl peptidase IV inhibitor in Zucker fa/fa rats. J Pharmacol Exp Ther. 2004;310:614–9. [DOI] [PubMed] [Google Scholar]
- [11].Kim Y-H, Lee YJ, Jeong Y-Y, et al. The effect of metformin on liver lipid accumulation in mice fed a high-fat diet. J Korean Soc Appl Biol Chem. 2010;53:198–205. [Google Scholar]
- [12].Shyangdan D, Clar C, Ghouri N, et al. Insulin sensitisers in the treatment of non-alcoholic fatty liver disease: a systematic review. Health Technol Assess. 2011;15:1–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Mao J, Zhang Q, Zhang H, et al. Risk factors for lymph node metastasis in papillary thyroid carcinoma: a systematic review and meta-analysis. Front Endocrinol. 2020;11:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jiang G, Sun C, Wang X, et al. Hepatoprotective mechanism of Silybum marianum on nonalcoholic fatty liver disease based on network pharmacology and experimental verification. Bioengineered. 2022;13:5216–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Feng WH, Bi Y, Li P, et al. Effects of liraglutide, metformin and gliclazide on body composition in patients with both type 2 diabetes and non-alcoholic fatty liver disease: a randomized trial. J Diabetes Investig. 2019;10:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Garinis G, Fruci B, Mazza A, et al. Metformin versus dietary treatment in nonalcoholic hepatic steatosis: a randomized study. Int J Obes (Lond). 2010;34:1255–64. [DOI] [PubMed] [Google Scholar]
- [17].Handzlik G, Holecki M, Kozaczka J, et al. Evaluation of metformin therapy using controlled attenuation parameter and transient elastography in patients with non-alcoholic fatty liver disease. Pharmacol Rep. 2019;71:183–8. [DOI] [PubMed] [Google Scholar]
- [18].Haukeland JW, Konopski Z, Eggesbø HB, et al. Metformin in patients with non-alcoholic fatty liver disease: a randomized, controlled trial. Scand J Gastroenterol. 2009;44:853-–860. [DOI] [PubMed] [Google Scholar]
- [19].Idilman R, Mizrak D, Corapcioglu D, et al. Clinical trial: insulin-sensitizing agents may reduce consequences of insulin resistance in individuals with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2008;28:200–8. [DOI] [PubMed] [Google Scholar]
- [20].Loomba R, Lutchman G, Kleiner D, et al. Clinical trial: pilot study of metformin for the treatment of non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2009;29:172–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nar A, Gedik O. The effect of metformin on leptin in obese patients with type 2 diabetes mellitus and nonalcoholic fatty liver disease. Acta Diabetol. 2009;46:113–8. [DOI] [PubMed] [Google Scholar]
- [22].Nobili V, Manco M, Ciampalini P, et al. Metformin use in children with nonalcoholic fatty liver disease: an open-label, 24-month, observational pilot study. Clin Ther. 2008;30:1168–76. [DOI] [PubMed] [Google Scholar]
- [23].Sofer E, Boaz M, Matas Z, et al. Treatment with insulin sensitizer metformin improves arterial properties, metabolic parameters, and liver function in patients with nonalcoholic fatty liver disease: a randomized, placebo-controlled trial. Metabolism. 2011;60:1278–84. [DOI] [PubMed] [Google Scholar]
- [24].Uygun A, Kadayifci A, Isik AT, et al. Metformin in the treatment of patients with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2004;19:537–44. [DOI] [PubMed] [Google Scholar]
- [25].Zhou J, Massey S, Story D, et al. Metformin: an old drug with new applications. Int J Mol Sci. 2018;19:2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barbero-Becerra V, Santiago-Hernandez J, Villegas-Lopez F, et al. Mechanisms involved in the protective effects of metformin against nonalcoholic fatty liver disease. Curr Med Chem. 2012;19:2918–23. [DOI] [PubMed] [Google Scholar]
- [27].Bugianesi E, Gentilcore E, Manini R, et al. A randomized controlled trial of metformin versus vitamin E or prescriptive diet in nonalcoholic fatty liver disease. Am J Gastroenterol. 2005;100:1082–90. [DOI] [PubMed] [Google Scholar]
- [28].Said A, Akhter A. Meta-analysis of randomized controlled trials of pharmacologic agents in non-alcoholic steatohepatitis. Ann Hepatol. 2017;16:538–47. [DOI] [PubMed] [Google Scholar]
- [29].Zhang Y, Wang H, Xiao H. Metformin actions on the liver: protection mechanisms emerging in hepatocytes and immune cells against NASH-related HCC. Int J Mol Sci. 2021;22:5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Romero-Gómez M, Zelber-Sagi S, Trenell M. Treatment of NAFLD with diet, physical activity and exercise. J Hepatol. 2017;67:829–46. [DOI] [PubMed] [Google Scholar]
- [31].Yeh MM, Brunt EM. Pathology of nonalcoholic fatty liver disease. American J Clin Path. 2007;128:837–47. [DOI] [PubMed] [Google Scholar]
- [32].Salgado ALFA, Carvalho L, Oliveira AC, et al. Insulin resistance index (HOMA-IR) in the differentiation of patients with non-alcoholic fatty liver disease and healthy individuals. Arq Gastroenterol. 2010;47:165–9. [DOI] [PubMed] [Google Scholar]
- [33].Tilg H, Moschen AR, Roden M. NAFLD and diabetes mellitus. Nat Rev Gastroenterol Hepatol. 2017;14:32. [DOI] [PubMed] [Google Scholar]
- [34].Durazzo M, Belci P, Collo A, et al. Focus on therapeutic strategies of nonalcoholic fatty liver disease. Int J Hepatol. 2012;2012:464–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Popovic-Pejicic S, Soldat-Stankovic V. Metformin: new perspectives for an old antidiabetic drug. Cardiovasc Endocrinol. 2015;4:17–21. [Google Scholar]
- [36].Gateva A, Kamenov Z. Cardiovascular risk factors in Bulgarian patients with polycystic ovary syndrome and/or obesity. Obstet Gynecol Int. 2012;2012:306347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Linden MA, Lopez KT, Fletcher JA, et al. Combining metformin therapy with caloric restriction for the management of type 2 diabetes and nonalcoholic fatty liver disease in obese rats. Appl Physiol Nutr Metab. 2015;40:1038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Maslak E, Zabielski P, Kochan K, et al. The liver-selective NO donor, V-PYRRO/NO, protects against liver steatosis and improves postprandial glucose tolerance in mice fed high fat diet. Biochem Pharmacol. 2015;93:389–400. [DOI] [PubMed] [Google Scholar]
- [39].Valizadeh R, Nikbakht M, Davodi M, et al. The effect of eight weeks elected aerobic exercise on the levels of (AST, ALT) enzymes of men patients with have fat liver. Procedia Soc Behav Sci. 2011;15:3362–5. [Google Scholar]
- [40].Cummings D, Ferstenberg R. Recommendations for the evaluation for autoimmune liver disease in non-alcoholic fatty liver disease. Hepatology. 2018;68:1203. [DOI] [PubMed] [Google Scholar]
- [41].Nsiah K, Dzogbefia V, Ansong D, et al. Pattern of AST and ALT changes in relation to hemolysis in sickle cell disease. Clin Med Insights Blood Disord. 2011;4:CMBD. S3969. [Google Scholar]
- [42].Vutukuru SS, Prabhath NA, Raghavender M, et al. Effect of arsenic and chromium on the serum amino-transferases activity in Indian major carp, Labeo rohita. Int J Environ Res Public Health. 2007;4:224–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gosse MA. How accurate is self-reported BMI? Nutr Bull. 2014;39:105–14. [Google Scholar]
- [44].Nuttall FQ. Body mass index: obesity, BMI, and health: a critical review. Nutr Today. 2015;50:117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ogden CL, Fryar CD, Carroll MD, et al. Mean body weight, height, and body mass index, United States 1960–2002. Adv Data. 2004;27:1–17. [PubMed] [Google Scholar]
- [46].Miyake T, Kumagi T, Hirooka M, et al. Body mass index is the most useful predictive factor for the onset of nonalcoholic fatty liver disease: a community-based retrospective longitudinal cohort study. J Gastroenterol. 2013;48:413–22. [DOI] [PubMed] [Google Scholar]
- [47].Coudriet GM, Delmastro-Greenwood MM, Previte DM, et al. Treatment with a catalytic superoxide dismutase (SOD) mimetic improves liver steatosis, insulin sensitivity, and inflammation in obesity-induced type 2 diabetes. Antioxidants (Basel). 2017;6:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Golay A. Metformin and body weight. Int J Obes (Lond). 2008;32:61–72. [DOI] [PubMed] [Google Scholar]
- [49].Pietilainen KH, Rissanen A, Kaprio J, et al. Acquired obesity is associated with increased liver fat, intra-abdominal fat, and insulin resistance in young adult monozygotic twins. Am J Physiol Endocrinol Metab. 2005;288:E768–74. [DOI] [PubMed] [Google Scholar]
- [50].Kallwitz ER, Kumar M, Aggarwal R, et al. Ethnicity and nonalcoholic fatty liver disease in an obesity clinic: the impact of triglycerides. Dig Dis Sci. 2008;53:1358–63. [DOI] [PubMed] [Google Scholar]
- [51].Guiducci L, Lionetti V, Burchielli S, et al. A dose-response elevation in hepatic glucose uptake is paralleled by liver triglyceride synthesis and release. Endocr Res. 2011;36:9–18. [DOI] [PubMed] [Google Scholar]
- [52].Bo S, Seletto M, Choc A, et al. The acute impact of the intake of four types of bread on satiety and blood concentrations of glucose, insulin, free fatty acids, triglyceride and acylated ghrelin. A randomized controlled cross-over trial. Food Res Int. 2017;92:40–7. [DOI] [PubMed] [Google Scholar]
- [53].Lemieux I, Lamarche B, Couillard C, et al. Total cholesterol/HDL cholesterol ratio vs LDL cholesterol/HDL cholesterol ratio as indices of ischemic heart disease risk in men: the Quebec Cardiovascular Study. Arch Intern Med. 2001;161:2685–92. [DOI] [PubMed] [Google Scholar]
- [54].Marseille-Tremblay C, Ethier-Chiasson M, Forest JC, et al. Impact of maternal circulating cholesterol and gestational diabetes mellitus on lipid metabolism in human term placenta. Mol Reprod Dev. 2008;75:1054–62. [DOI] [PubMed] [Google Scholar]
- [55].Corey KE, Lai M, Gelrud LG, et al. Non–high-density lipoprotein cholesterol as a biomarker for nonalcoholic steatohepatitis. Clin Gastroenterol Hepatol. 2012;10:651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Elshazly MB, Quispe R, Michos ED, et al. Patient-level discordance in population percentiles of the total cholesterol to high-density lipoprotein cholesterol ratio in comparison with low-density lipoprotein cholesterol and non–high-density lipoprotein cholesterol: the very large database of lipids study (VLDL-2B). Circulation. 2015;132:667–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Singh GK, Vitola BE, Holland MR, et al. Alterations in ventricular structure and function in obese adolescents with nonalcoholic fatty liver disease. J Pediatr. 2013;162:1160–1168. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Day CP. Genetic and environmental susceptibility to non-alcoholic fatty liver disease. Dig Dis. 2010;28:255–60. [DOI] [PubMed] [Google Scholar]
- [59].Zhang L, Zhang Y, Jiang Y, et al. Upregulated SOCC and IP3R calcium channels and subsequent elevated cytoplasmic calcium signaling promote nonalcoholic fatty liver disease by inhibiting autophagy. Mol Cell Biochem. 2021;476:3163–75. [DOI] [PubMed] [Google Scholar]
- [60].Ahmed MH, Byrne CD. Modulation of sterol regulatory element binding proteins (SREBPs) as potential treatments for non-alcoholic fatty liver disease (NAFLD). Drug Discov Today. 2007;12:740–7. [DOI] [PubMed] [Google Scholar]
- [61].Zhang J, Lin S, Jiang D, et al. Chronic hepatitis B and non-alcoholic fatty liver disease: conspirators or competitors?. Liver Int. 2020;40:496–508. [DOI] [PubMed] [Google Scholar]
- [62].Zhang X. NAFLD related-HCC: the relationship with metabolic disorders. Adv Exp Med Biol. 2018;2018:55–62. [DOI] [PubMed] [Google Scholar]
- [63].Fang J, Sun X, Xue B, et al. Dahuang Zexie decoction protects against high-fat diet-induced NAFLD by modulating gut microbiota-mediated toll-like receptor 4 signaling activation and loss of intestinal barrier. Evid Based Complement Alternat Med. 2017;2017:2945803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Ferguson D, Finck BN. Emerging therapeutic approaches for the treatment of NAFLD and type 2 diabetes mellitus. Nat Rev Endocrinol. 2021;17:484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Marušić M, Paić M, Knobloch M, et al. NAFLD, insulin resistance, and diabetes mellitus type 2. Can J Gastroenterol. 2021;2021:6613827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Peng Q, Schork N. Utility of network integrity methods in therapeutic target identification. Front Genet. 2014;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]