Background:
Advances in next-generation sequencing technologies are changing the ways cancer diagnosis and treatment, which leads to a new branch of precision medicine: “Precision Oncology”. This study aims to deliver a structured overview to carry out a bibliometric analysis of precision oncology research over the past 10 years retrospectively.
Methods:
Bibliometric methods including clustering analysis and co-occurrence visualized study were conducted based on publications of academic databases Web of Science Main Collection from 1st January 2012, to 31st December 2021. This study analyzed the information about related research outputs, countries, institutions, authors, cited papers, and hot topics.
Results:
7163 papers related to precision oncology were identified. Since 2014, the number of articles has proliferated, and oncology precision has attracted significant attention from scholars worldwide in recent years. The USA leads the research in this field, and the League of European Research Universities is the primary research institution. Research institutions from Asia paid more attention to this field through high-level international cooperation. Besides, there are still many issues expected to be explored and evaluated correctly. Such as the considerable uncertainty that pharmacogenomic methods have no significant influence on patient outcomes.
Conclusions:
Precision oncology serves as an essential method in clinical treatment, and is closely related to biological study, including biochemistry, molecular and genetics, advanced technology, and pharmacology discovery. The future research prospect would be the broad involvement of social participation and global cooperation in oncology precision research to acquire better results via the balance of technology and public health policy.
Keywords: bibliometric analysis, cancer, mapping knowledge domain, oncology precision, precision medicine
1. Introduction
Precision medicine is defined as targeted treatments toward individuals’ needs based on the genetic, biomarker, phenotypic, and psychosocial characteristics.[1] It was first put forward by American medical scholars in 2011. The former US president Obama Barack declared a proposal for precision medicine study in January 2015.[2] And the prospect of precision medicine’s application has been dramatically improved in the past 10 years by the development of large-scale biologic databases, like the human genome sequence, computational tools for analyzing large sets of data, and robust methods for characterizing patients(including proteomics, metabolomics, genomics, diverse cellular assays, mobile health technology).[2] Moreover, Jones et al[3] found a growing emphasis on recent drug discovery efforts on the epigenome (DNA methylation and histone modifications). Xiong et al[4] developed a machine-learning technique to facilitate precision medicine application and whole-genome annotation. Robinson et al[5] established a multi-institutional clinical sequencing infrastructure for the development of the precision medicine framework for metastatic, which could have impact on individual treatment decisions. Therefore, rare diseases and uncommon cancers could get real-time guided, individualized prevention and therapeutics to improve results by existing technological advancements.[4]
Cancer is a hot research field because it is an increasing cause of death worldwide.[2] And the advancement in next-generation sequencing technologies is changing the ways of cancer diagnosis and treatment.[6] “Precision Oncology” is a branch of precision medicine.[7] Oncology rationally sits as precision medicine application’s pioneer.[6] Cancer is a genomic disease. Research has already revealed that inherited genetic variations have significant effects on the prevalence of cancer, and most cancers attribute to the combined impact of proto-oncogene and tumor suppressors.[6] They work together to control the molecular pathways and genetic expression.[6] Based on this understanding, cancer’s influence risk assessment, diagnostic categories, and therapeutic strategies (attention to drugs and antibodies focus on fighting the impact on specific molecular drivers) have taken advantage of the oncogenic biological mechanisms.[2] Oncology has become a unique verifying field for the genomics-driven framework in medical research. The Cancer Genome Atlas and the International Cancer Genome Consortium have inspected about thirty thousand multiple cancer genomes to contribute to the classification of numerous targeted genomic cancer causes.[6] Moreover, advanced precision diagnostics and targeted anticancer agents have been applied in clinical use. And several have been confirmed to be extraordinary.[8] For example, Mullane et al[9] stated that PARP-1 makes a difference in targetable alterations for castration-resistant prostate cancer. Druker et al[10] proved that imatinib mesylate is highly effective against a blood cancer: chronic myeloid leukemia carrying the BCL-ABL1 chromosomal translocation. And tamoxifen is used to develop a molecularly driven treatment aiming at estrogen receptors in breast cancer.[6]
Bibliometrics analysis takes literature as the unit of measurement, time as the axis sets mathematics, statistics, and philology as a whole. It is a more useful quantitative measurement compared with the “subjective” and “objective” methods (intuitive assessment methods, e.g., peer review). It has been used since scientific papers were first published in the seventeenth century.[11] Nowadays, it is a common trend to combine bibliometrics, quantitative and statistic methods to describe publications’ patterns, scientific development, funding purposes, scholars’ contribution, international cooperation, and citation within a given research field,[12] which can provide an overview of a global dynamics of science in the past few years.[13] And the bibliometric method has already been widely applied to medical-related topics such as cancer,[12] mental health,[14] Dendritic Epidermal T Cell (DETC) research,[15] nonspecific low back pain research,[16] and hepatology.[17] Several researchers have reported a lot of productive work, instrumental in specific policy-making and clinical guidelines development.[18]
Although a large number of systematic reviews and meta-analysis had been conducted to state the research status of precision oncology, a little study provided an overview growing trend in this field. So, the paper’s primary purpose is to develop a summative evaluation of existing research and potential innovative research aspects of precision medicine applied in the cancer research field from 2012 to 2021. It covers quantity, keywords, clustering, geographic feature, co-work, and bibliographic coupling analyses, which are meaningful for understanding the external characteristics and internal focus of the existing literature.[16] They also provided information about the contributions of authors, institutions, and countries in this research field.[19] The paper consists of three chapters: methodology, results, and discussion, conclusion. The first part describes the study design, including data acquisition, exclusion process, and data analysis methods. Part two begins by laying out the evaluated result, then having in-depth discussions. And the design or analysis restrictions of this study were considered. The third part gives summative conclusions following above.
2. Material and methods
This study carried out a retrospective search based on the bibliometrics analysis methods using the Web of Science core collection database, which is considered the largest comprehensive academic information resource in the world and the ideal database for bibliometrics analysis. This database covers over 12,000 international journals that have the highest impact, highly qualified scientific findings, and more than 8700 influential core academic journals in natural science, biomedicine, and other research fields.[20] Several studies have shown that the Web of Science core collection database plays a vital role in discipline development research. The edition of 2021 Journal Citation Reports, issued by the Institute for Scientific Information, has also been chosen as the source of our required data.[21,22]
The search strategy in the Web of Science core collection is as follows: ((“Precision Medicine” [TS] OR “Precise Medicine” [TS]) AND (“Cancer” [TS] OR “Oncology” [TS] OR “Tumor” [TS])).
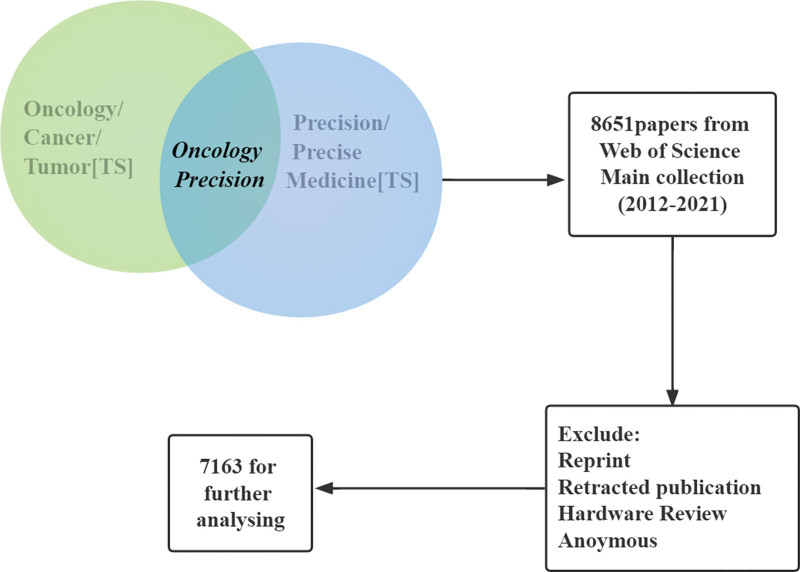
According to the retrieval formula, 8651 papers were obtained. Then, we developed criteria related to the research objectives based on the topic, period, and other characteristics of these papers to guide the other paper selection. The inclusion criteria were that the period of publications is from January 1, 2012 to December 31, 2021, and the search database is limited to the Web of Science core collection. And the search field is precision oncology. The exclusion criteria were that we sifted out date papers, reprints, and hardware reviews. Projects having anonymous authors and repeated research papers were also excluded.
Finally, 7163 of them were included in the analysis according to the developed standards. Figure 1 shows the selection processes. Data were collected from January 17, 2022 to January 22, 2022. (We believe the data-collection date would only have a slight impact on the conclusion as these data arise from global precision medical research on cancer in a given period and are used for comparative reasons.) The selection process is visualized in Figure 1. Finally, related data was saved in TXT format and sorted into different tables according to their categories. Diverse clustering results were presented as a content schema with texts, figures, and tables using Excel, VOSview-1.6.5, and CiteSpace-5.8. R3.
Figure 1.
Selection process.
3. Results
3.1. Increase pattern of the annual publication
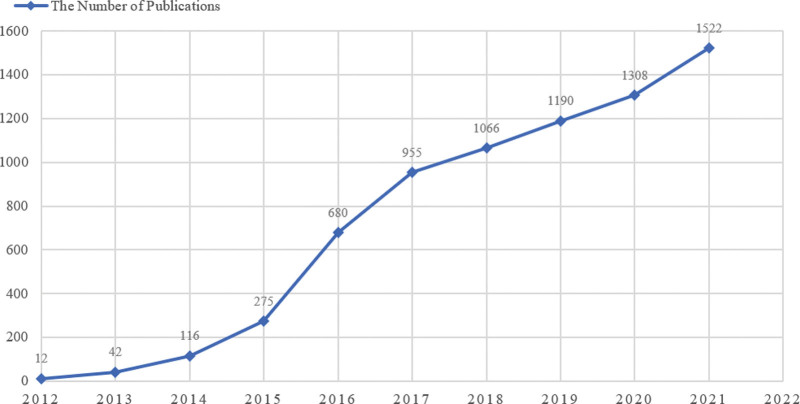
The timely changed papers number can highlight the development tendency of a research field from the bibliometric perspective.[23] From 2012 to 2021, the number of annual publications on oncology precision grew year by year as shown in Figure 2. From the figure, only 12 (0.17%) papers were published in 2012, while it increased 126.75 times by 1522 (21.23%) in 2021. The comparison represents a sharply increasing awareness in Oncological precision medicine research. Furthermore, the changing pattern can be roughly divided into three stages: Initial Stage (2012–2013). At this stage, the progress slowed to a crawl. The annual increment was only 30. Stage of Rapid Development (2014–2017). During this period, the number of papers increased significantly, with an average annual growth of 228. Stage of Steady-state Development (2018–2021). At this stage, the publication number was in large quantities, with an average of 1271 papers being published each year. Over third times more articles were published in the third stage (6483, 75.683%) than that in first two stages. From the above discussion, the conclusion can be reached that Oncology precision medicine has become a considerable issue in medical research. Significantly more work will be done in this area, and it is expected to reach new heights by 2025.
Figure 2.
Annual publication pattern.
3.2. Geographical distribution of research output
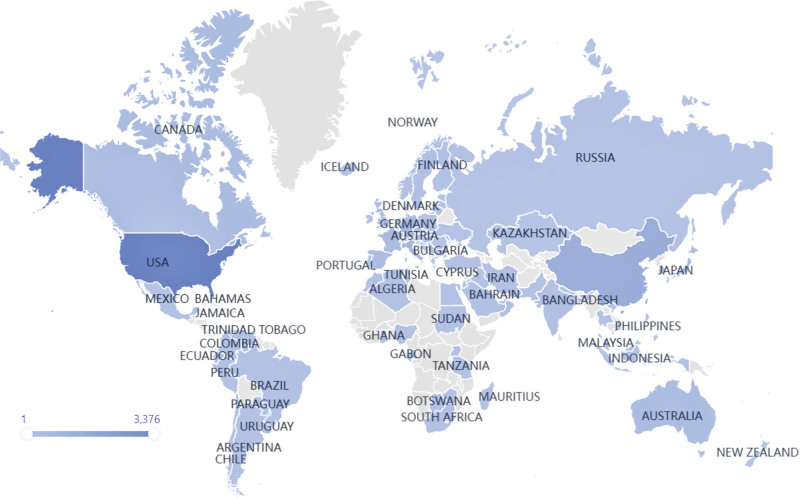
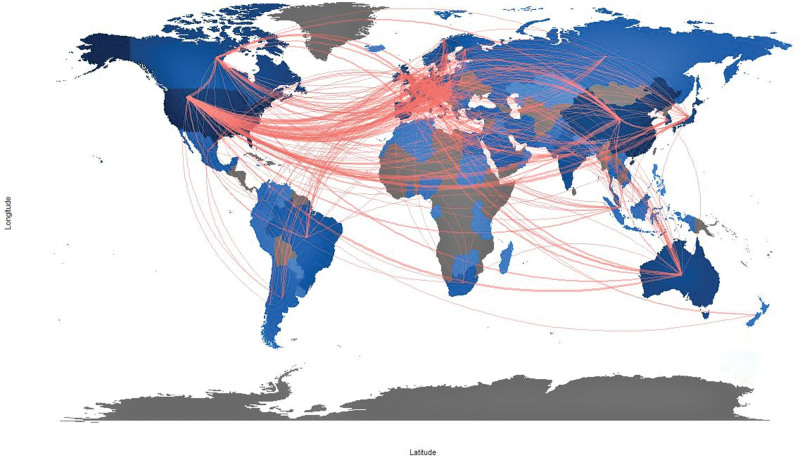
National publications can objectively reflect the research level and strength in an academic field of a country or territory.[24] This study selected papers on a global scale to figure out the actual research activities conditions. The global spatial clusters in cancer precision research were visualized in Figure 3, which were mainly concentrated in the USA, Western Europe, and Asia. Statistics demonstrated that 109 countries or territories had papers in this domain, and 9.7% had papers exceeding 100. A special concern needs to be given to the USA, which had the largest number of papers (3376, 47.13%). These results indicate that the USA has grabbed and controlled the top priority in this research field with the advanced medical levels. And there has been a discernible gap in related research between the USA and other countries. China ranked second with 1175 papers (16.40%), staying one step ahead of other countries and having shown favorable development prospects, followed by Italy (600, 8.38%), Germany (457, 6.38%), and the UK (452, 6.31%). Moreover, nine developed countries (USA, UK, Germany, Italy, Canada, France, Australia, Spain, Netherlands, and Japan) are in the top ten countries/territories. It provides direct evidence for the developed countries’ leading position in academic research. And it is not startling to find this domination as it has appeared in many research fields. This phenomenon may be closely correlated with the top economic and scientific technology level of these countries. In addition, the strength of oncology medicine research reflects the clinical application status of this method to some extent, and there may be significant inequality in the application of precision medicine in cancer treatment among countries. And the main points for broad and fair access to precision cancer medicine rely on the national health system and overall technology level. So, these two parts may be the main focus of promoting the application of oncology medicine in the future.
Figure 3.
Worldwide publication distribution.
3.3. Prolific institutions and their research impact
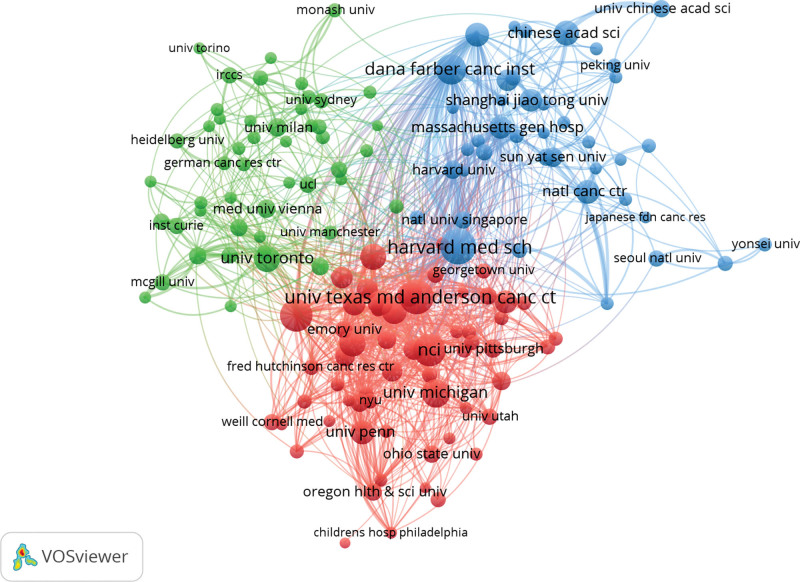
To figure out the main force of this research field, we chose to list the distribution of research institutions. A total of 6059 institutions were involved in the publication of “precision medicine” and “cancer” from 2012 to 2021. League of European Research Universities was the most active institution in this research field with the largest number of 567 papers (7.91%). Table 1 shows the detailed information for the top 10 research institutions (ranked by the number of papers). These institutions listed in Table 1 are considered the academic leader in this field. Their research results and academic papers represent the highest standard in the world. Furthermore, six of them are high education institutions. It was learned that universities plan an essential role in oncology precision research and extend their in-depth learning. Then, this study used VOSviewer to build a co-work institution network (Fig. 4). In this figure: Each institution names’ font size and the circle show its publication number. The thickness of the linking line shows the cooperative intensity between institutions. The different colors represent different clustering statuses. The figure illustrated there had been fixed co-work nets among these research institutions. The co-work net is also stratified by region (Fig. 5), especially within the USA and Europe. Moreover, it is worth mentioning that research institutions from China mainland, Japan, Singapore, and Taiwan paid more attention to this field through high-level international cooperation. The rapid increase in related input and output may be closely linked with the progress of scientific research and the growth of economic strength in Asia. The research has been considered by these Asia advanced economies. They are also willing to work together or with emerging developing countries, which draws the attention of the world.
Table 1.
Top 10 institutions of oncology precision medicine during 2012–2021.
| Rank | Research Institution | Total publications | Country |
|---|---|---|---|
| 1 | League of European Research Universities LERU | 567 | EUR |
| 2 | Harvard University | 444 | USA |
| 3 | University of California System | 377 | USA |
| 4 | University of Texas System | 332 | USA |
| 5 | National Institutes of Health NIH USA | 266 | USA |
| 6 | UDICE French Research Universities | 256 | EUR |
| 7 | UTMD Anderson Cancer Center | 228 | USA |
| 8 | Harvard Medical School | 227 | USA |
| 9 | Institut national de la santé et de la recherche médicale | 223 | EUR |
| 10 | Dana Farber Cancer Institute | 221 | USA |
Figure 4.
Institutions co-work net of research on oncology precision.
Figure 5.
Institutions belonged to countries’ collaboration pattern of research on oncology precision.
3.4. Productivity and co-work across authorship
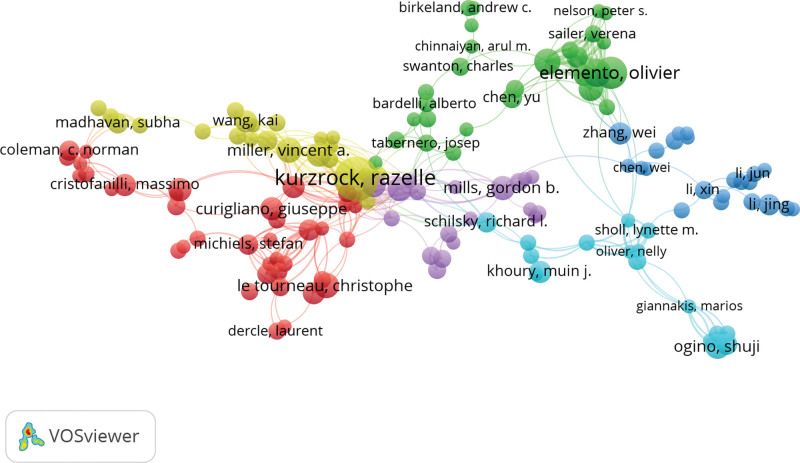
The publication of authors and their co-work nets could help determine whether there are outstanding scholars who lead the research way in this field, and provide information about the status of the collaborations among authors, which is important for future research development.[16] In this study, 35,175 authors from 37 different countries were recorded in the Affiliation field of Web of Science Main Collection related to the precision medicine applied in the cancer research field. The top 10 authors who made the most outstanding contribution have 353 papers. Table 2 provides detailed information on the top 10 authors’ publication amount and research impact. Among the table, Total publications (TP) and h-index were reported. TP can be understood as the output frequency of a researcher in one research field. The h-index is another measurement method. It represents an author who has published at least h papers having at least h citations, which is simple and handy to measure the impact of a researcher.[25] The paper tried to estimate the academic level of researchers related to cancer precision medicine on a global level by employing these indexes and got a surprising output: the author’s rank in the number of papers does not exactly match the h-index’s rank. Wang, Yu (Shanghai Jiao Tong University) had the largest number of publications but ranked seven based on the h-index. Li, Yan ((Shanghai University of Traditional Chinese Medicine) and Zhang, Yan (Ohio State University) met the same situation. They have many papers in this field, while the papers’ quality may not be as good as expected. Then Kurzrock, Razelle (Medical College of Wisconsin), and Wang, Jing (University of Texas M.D.Anderson Cancer Center), whose h-index ranks first and second respectively had satisfying results with a great number of publications. So, we are awarded that there may not exist a most reliable independent parameter to evaluate the quality of research. It is a cue for the further analyzing approach: the way to earn an accurate and completed result should be on the integration of various indicators instead of isolated indicators. In addition, the co-work net for authors was estimated in Figure 6. From the visualization network, there had been 5 main cooperation clusters. But there was no clear co-work pattern among the most influential researchers. For future research, promoting collaborations among high-quality authors’ may be a good way to improve the paper’s quality.
Table 2.
Top 10 productive authors in publications of oncology precision medicine during 2012–2021.
| Author | TP | TP Rank | H-index | Rank (h-index) | Institution |
|---|---|---|---|---|---|
| Wang, Yu | 46 | 1 | 15 | 7 | Shanghai Jiao Tong University |
| Kurzrock, Razelle | 40 | 2 | 133 | 1 | Medical College of Wisconsin |
| Li, Yan | 39 | 3 | 12 | 8 | Shanghai University of Traditional Chinese Medicine |
| Zhang, Yan | 37 | 4 | 5 | 10 | Ohio State University |
| Wang, Jing | 36 | 5 | 77 | 2 | University of Texas M.D. Anderson Cancer Center |
| Jing Li | 34 | 6 | 29 | 5 | Georgia Institute of Technology |
| Chen, Yangshan | 33 | 7 | 10 | 9 | Sun Yat Sen University |
| Liu, Yan | 31 | 8 | 18 | 6 | Capital Medical University |
| Elemento, Olivier | 29 | 9 | 74 | 3 | Cornell University |
| Liu, Joyce F. | 28 | 10 | 39 | 4 | Dana-Farber Cancer Institute |
TP = total publications.
Figure 6.
Authors co-work net of research on oncology precision.
3.5. Most frequently cited papers and citation visualization analysis
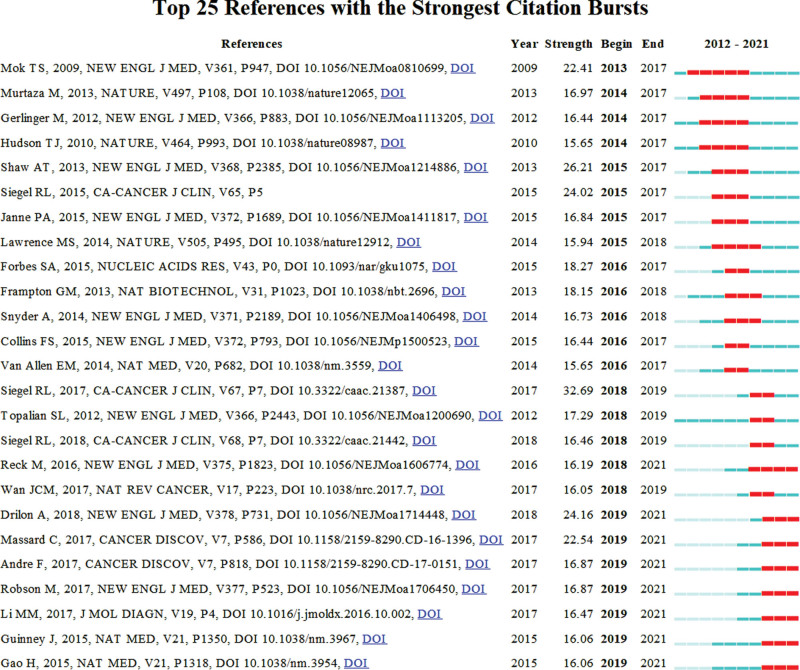
Citation analysis is a formal and scholarly founded method. It can measure the impact of research. This study used CiteSpace to filter out the 25 most cited papers involved in cancer precision medicine and get their burst characteristics to identify the classic papers in the field across time. The table of the Top 25 references with the strongest citation bursts (Fig. 7) could reflect the annual change trend, evolution, and inheritance relation of studies in this field.[26] These most cited papers supported some essential conclusions as follows.
Figure 7.
Top 25 References with the Strongest Citation Bursts.
Mok et al[27] conducted a phase 3 open-label study to get the conclusion that compared to carboplatin-paclitaxel, Gefitinib is the better medicine for pulmonary adenocarcinoma among nonsmokers or ever smokers in East Asia (HR for progression or death: 0.74, 95% CI: 0.65–0.85, P < .001). Especially for patients who have the epidermal growth factor receptor (EGFR) gene mutation. Meanwhile, these two medicines would have side effects. For gefitinib, it would be rash, acne, and diarrhea. For carboplatin-paclitaxel, neurotoxic effects, neutropenia, and alopecia are much more common.[27] This outcome was cited chiefly between 2013 to 2017.
Murtaza et al[28] followed up with 6 patients having advanced breast, ovarian and lung cancers and got plasma samples at regular intervals to report that quantification of allele fractions in plasma pointed out developed representation of mutant alleles in relationship with the appearance of therapy resistance, including an activating mutation in PIK3CA after treatment with paclitaxel. These results support that exome-wide analysis of circulating tumor DNA was another meaningful invasive biopsy method for acquired drug resistance-related mutation identification.[28] This paper was cited 1155 times during 2014 to 2017.
Gerlinger, Marco et al stated that intratumor heterogeneity, which relates to heterogeneous protein function, would have negative impacts on the tumor genomics landscape portrayed using one tumor-biopsy sample.[29] This effect may result in therapeutic failure via Darwinian selection, which hinders the development of precision medicine and biomarkers.[29] This study was cited 5158 times during 2014 to 2017.
Furthermore, most of the papers of the 25 in Figure 7 were published in the New England Journal of Medicine, Nature, and Ca-A Cancer Journal for Clinicians, which have great influence. So, these journals are worth studying carefully, which may have promising beneficial effects in later research on cancer precision medicine. And the papers listed below would be of great significance to be used as references.
3.6. Research hotpots
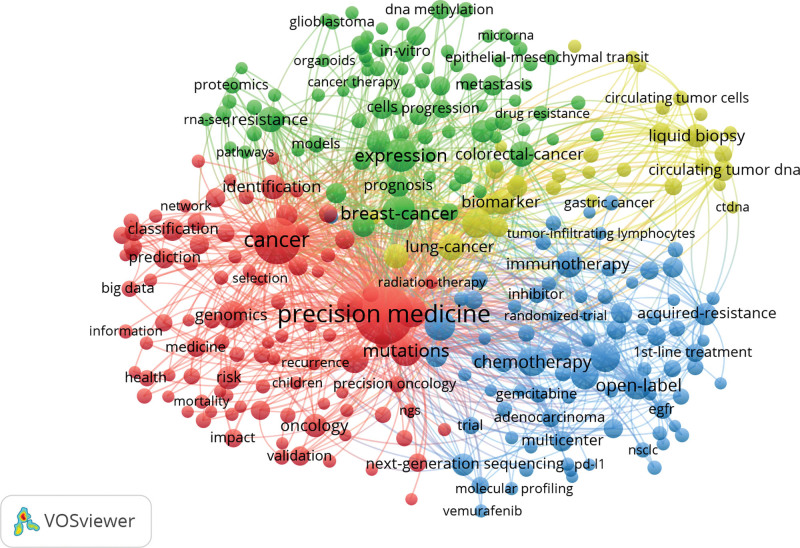
Co-word analysis is a complex network analytical method. It attempts to extract data from a paper-set, then creates networks based on co-occurrences, likenesses, or other links to that data. This method promotes building and visualizing the structure inherent to a set of publications.[30] Co-occurrence analysis and clustering research are important indicators for tracking scientific development, which can be used to show the aroused general interests.[30] In this study, paper records were loaded into the VOSviewer Tool to create a keywords co-occurrence and clustering network to identify the research topics of oncology precision medicine papers.[31] The network was pruned by dropping isolated nodes and edges to increase the map’s clarity. The final network consisted of the top 300 nodes (Fig. 8). Nodes were sized relative to the appearance count of keywords; the thickness of lines was an indication determined by the occurrence frequency of keywords at the same time among these papers, which can show the correlation between keywords. Furthermore, VOSviewer stratified the keywords and applied different colors to keywords. Colored lay in the communities identified by the community detection algorithm. The color represented various clustering status.[32]
Figure 8.
Co-occurrence and clustering network of keywords.
These relationships were divided into specific thematic groups based on their interrelated terminology and the frequency of co-occurrence links drawn in Figure 8. The top 300 most used keywords could be classified into four clusters: “targeted cancer study”, “biological study (include biochemistry, molecular and genetics”, “new technology study”, and “pharmacology study”:
(1) In the “targeted cancer study’’ cluster, the most frequent meaningful values were acute lymphoblastic-leukemia, breast-cancer, lung-cancer, colorectal-cancer, ovarian-cancer, adenocarcinoma, gastric-cancer, and cholangiocarcinoma.
According to the result above, precision cancer treatment was commonly used in specific cancers having confirmed tumor-related biomarkers. For example. Studies approved that: 87% of lung-cancers are identified as non-small cell lung carcinoma, including three types based on molecular subtypes: adenocarcinoma, squamous cell carcinoma, and large cell carcinoma histological pattern.[33] They are all caused by specific genetic alterations that promote and retain lung tumorigenesis.[33] These driver mutations and the related constitutively active mutant signaling proteins play essential roles in tumor cell surviving.[34] So, the novel targeted therapies research on lung cancer has increased dramatically in the past few years.[35] So, more research focus is expected to put in cancer-associated gene loci and molecular receptors, etc., to make attribution to expanding application area of oncology precise medicine.
(2) In the cluster of “biological study”, the most pointed keywords were biobank, bioinformatics, genome-wide association, DNA methylation, messenger-RNA, EGFR mutations, endothelial growth-factor, KRAS, BRAF, microsatellite instability, androgen receptor, estrogen-receptor, and cell-free DNA.
Genomic instability is defined as genetic and genomic abnormalities of the cancer development stage, including gene, molecular, and cell-level mechanisms. For example, mitotic checkpoint signaling, DNA damage checkpoints’ defect, DNA damage repair, DNA replication, and cytokinesis. These changes would lead to mutations, aneuploidy, the change of focal copy number and translocations. And they also provide possible intervening pathways for cancer treatment. For instance, Catanese and Lordick[36] finished a literature review and found that there have already been three predictive biomarkers for gastric cancer therapeutic algorithm recognized: Human Epidermal Growth Factor Receptor 2, Microsatellite Instability, and Programmed Death-Ligand 1 (PD-L1) expression.
(3) In the “new technology study” cluster, the main keywords were artificial intelligence, big data, molecular diagnostics, radiotherapy, tumor xenografts, 1st-line treatment, adjuvant chemotherapy, neoadjuvant chemotherapy, mass-spectrometry, and positron-emission-tomography.
These keywords pointed out some essential technological methods used frequently in the oncology precision. And with the development of modern technology, more accurate, and sophisticated therapies have been applied in precise cancer treatment. Jin et al[37] reported that the hot technology of Artificial intelligence is widely used in biological information analysis, including evaluating lymph node metastasis, drug treatment response, and surgical coaching, assessment, and guidance. Besides, developments in delivery and imaging technologies promote the advances in image-guided and adaptive radiotherapy.[35] Kong et al[38] used data from 42 stage II/III NSCLC patients treated with radiation therapy to get that this method makes a difference in improving 2-year local-regional tumor control rates while the overall tumor control rate was 52%. And the 5-year survival rate was 30%.
(4) In the “pharmacology study” cluster, the frequently used keywords were pharmacogenomics, sorafenib, bevacizumab, cetuximab, cisplatin, crizotinib, erlotinib, gefitinib, gemcitabine, pembrolizumab, and vemurafenib.
These results show that there have already been several specific medicines for targeted oncology treatment. However, the use of targeted drugs poses many problems. One notable drawback is that the wrong choice of patients makes molecularly targeted therapies be useless and even have worse outcomes than standard chemotherapy.[39] For example, gefitinib is a tyrosine kinase inhibitor of EGFR which can considerably extend survival year for non-small-cell lung cancer patients with activating EGFR mutations. But it would have worse efficacy than standard chemotherapy for patients without activating EGFR mutations.[39] Another complex issue is that different supervisory decision lead to diverse access levels among different countries.[40] A typical example is that the European Medicines Agency controls the assessment of new medicines in the European Union. But European Medicines Agency’s approval cannot be applied in every specific Europe country. Besides, there would be diverse regulatory approval processes, access to testing and matched drugs across countries based on their different healthcare-system.[40] Moreover, some scholars have opposite attitudes toward cancer precision medicine. They stated that pharmacogenomic tests have no direct influence on patient outcomes, but rather improve the clinician’s decisions about the treatment.3 The problems listed above need worth further study in the future.
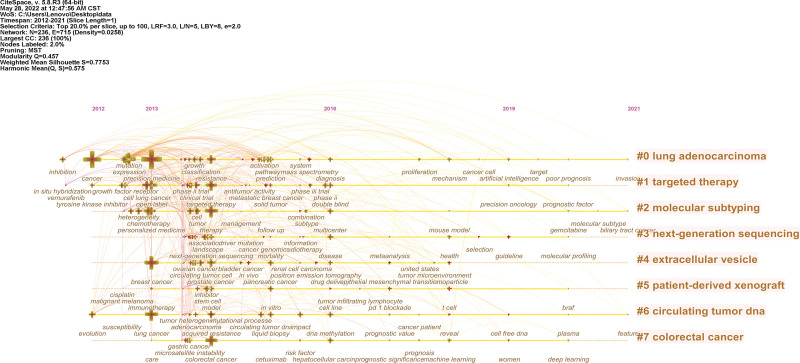
In addition, CiteSpace was used to draw a figure to show the development of hot research topics during 2012 to 2021 (Fig. 9). Seven sought-after topics were found in this figure: “lung adenocarcinoma”, “targeted therapy”, “molecular subtyping”, “next-generation sequencing”, “extracellular vesicle”, “patient-derived xenograft”, “circulating tumor DNA”, “colorectal cancer”. The timeline changes illustrate that lung adenocarcinoma and colorectal cancer are two outstanding research fields using precision medicine. During 2013 to 2016, the investigation of mutation expression, growth classification, and activation pathway leads to the development of lung adenocarcinoma treatment. The research on colorectal cancer pays much attention to microsatellite instability, Cetuximab, machine learning, and deep learning. And the focused biological mechanisms range from liquid biopsy, DNA methylation to cell free DNA and plasma across 2013 to 2021. The keywords of targeted therapy changed from situ hybridization growth in 2012 to artificial intelligence in 2019. The next-generation sequencing has been involved in the gemcitabine and biliary tract cancer recently, which shows specific progress within this topic. The extracellular vesicle-linked study is another hotpot, which has a strong association with circulating tumor cells, renal cell carcinoma, ovarian cancer, and bladder cancer. Moreover, the patient-derived xenograft is a meaningful method for some tumor treatment, especially for breast cancer, prostate cancer, and pancreatic cancer. And the lympho-epithelial mesenchymal transition was introduced to this field in 2017. Finally, the research of circulating tumor DNA transformed from immunotherapy in 2013, stem cell model in 2014, in vitro in 2015 to cell line 2016, pd 1 blockade in 2017, t cell in 2018, and braf in 2020.
Figure 9.
Top 25 Timeline view of main topics.
4. Discussion
This paper focused on the global development of oncology precision during the past ten years by the bibliometric method. The key findings can be summarized as follows. The research on oncology precision developed relatively slowly from 2012 to 2013, but since 2014 the number of papers has grown rapidly. It is shown that oncology precision has attracted great attention from scholars worldwide in recent years. The USA leads the research in this field and the League of European Research Universities is the leading research institution. They represent the highest academic standards in the world. And research institutions from China mainland, Japan, Singapore, and other East Asian countries/regions hold more important places and have significant co-work net. They emerged as developing players in the field. To a certain extent, there is a considerable positive correlation between the progress of scientific research and the growing economic status in Asia. The high-level co-work mode among scholars could lead to shaping research strategies globally, which is expected to consider further in the future. There have been some advanced applications of precision medicine in cancer treatment which have had positive impacts on cancer diagnosis and clinical treatment. However, there are still many issues expected to be explored and evaluated properly. Such as the larger uncertainty that pharmacogenomic methods have no significant influence on patient outcomes. And the key problem which restrains further development of this field is how to raise the actual clinical outcomes effectively and further understanding of the pathogenesis of cancer at the molecular or cell-level. It is predicted that adequate precise clinical practice data from well-conducted trials and advancements in molecular biology will be carried out in the following years. The future research prospect would be the wide involvement of social participation and global cooperation in oncology precision research to acquire better results via the balance of technology and public health policy.
And yet, several limitations of the study should also be considered. First, this paper only got publications from the Web of Science Main Collection, which mainly includes the studies in English. Some essential papers in other languages would be ignored. Besides, search strategy is limited to the keywords: “precise” or “precision” narrowly. It may reduce the breadth and depth of the results. In addition, we found some papers which did not have a complete affiliation and were deleted. Such as, some articles were not portioned out to a fixed country. So, the data for their countries would be underestimated.
5. Conclusion
All in all, the results of the study showed that Oncology Precision plays an important role in the field of medicine year by year. The biological study includes biochemistry, molecular and genetics, advanced technology, and pharmacology discovery linked with precision oncology closely. However, they are still some issues restricting its development. So, more precise and innovative studies in oncology precision research are expected to get a concern in the future.
Author contributions
The authors are very grateful to the referees and anonymous reviewers for their helpful comments and suggestions. The authors also thank the authors of the original studies included in this analysis.
BZ and YH designed the experiments and drafted the manuscript. BZ, YH, XL, PB, and SY mainly performed the experiments and analyzed the data. HW, AB, and RL reviewed the manuscript. All authors read and approved the final manuscript.
Conceptualization: Hongyun Wang, Ruifeng Li.
Data curation: Baoyue Zhang, Youliang Huang, Xinyue Lu, Shuang Yang, Pengfei Bao.
Formal analysis: Baoyue Zhang, Youliang Huang, Xinyue Lu, Shuang Yang, Pengfei Bao, Hongyun Wang.
Software: Shuang Yang, Pengfei Bao.
Supervision: Ruifeng Li.
Visualization: Shuang Yang, Pengfei Bao.
Writing – original draft: Baoyue Zhang, Youliang Huang.
Writing – review & editing: Bo Ao, Ruifeng Li.
Abbreviations:
- EGFR =
- epidermal growth factor receptor
- HR =
- hazard ratio
- TP =
- total publications
This work was supported by the “Social Science Foundation of Beijing” (Grant no. 20GLB019) and the Key project of the Beijing University of Chinese Medicine (Grant no. 2020-JYB-ZDGG-067).
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
How to cite this article: Zhang B, Ao B, Lu X, Yang S, Bao P, Wang H, Li R, Huang Y. Global research trends on precision oncology: A systematic review, bibliometrics, and visualized study. Medicine 2022;101:43(e31380).
Contributor Information
Baoyue Zhang, Email: baoyue.zhang.21@ucl.ac.uk.
Bo Ao, Email: zryhyyab@163.com.
Xinyue Lu, Email: luxinyue22@mails.ucas.ac.cn.
Shuang Yang, Email: shuang0925@yeah.net.
Pengfei Bao, Email: bpfbucm@163.com.
Hongyun Wang, Email: 700600@bucm.edu.cn.
References
- [1].Jameson JL, Longo DL. Precision medicine – personalized, problematic, and promising. N Engl J Med. 2015;372:2229–34. [DOI] [PubMed] [Google Scholar]
- [2].Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jones PA, Issa JPJ, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41. [DOI] [PubMed] [Google Scholar]
- [4].Xiong HY, Alipanahi B, Lee LJ, et al. The human splicing code reveals new insights into the genetic determinants of disease. Science. 2015;347:1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Froeling FEM, Casolino R, Pea A, et al. Molecular subtyping and precision medicine for pancreatic cancer. J Clin Med. 2021;10:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Malod-Dognin N, Petschnigg J, Pržulj N. Precision medicine – a promising, yet challenging road lies ahead. Curr Opin Syst Biol. 2018;7:1–7. [Google Scholar]
- [8].Agusti A, Bel E, Thomas M, et al. Treatable traits: toward precision medicine of chronic airway diseases. Eur Respir J. 2016;47:410–9. [DOI] [PubMed] [Google Scholar]
- [9].Mullane SA, Van Allen EM. Precision medicine for advanced prostate cancer. Curr Opin Urol. 2016;26:231–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Druker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. [DOI] [PubMed] [Google Scholar]
- [11].Perianes-Rodriguez A, Waltman L, van Eck NJ. Constructing bibliometric networks: a comparison between full and fractional counting. J Informetr. 2016;10:1178–95. [Google Scholar]
- [12].Min HS, Yun EH, Park J, et al. Cancer news coverage in Korean newspapers: an analytic study in terms of cancer awareness. J Prev Med Public Health. 2020;53:126–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tatry MV, Fournier D, Jeannequin B, et al. EU27 and USA leadership in fruit and vegetable research: a bibliometric study from 2000 to 2009. Scientometrics. 2014;98:2207–22. [Google Scholar]
- [14].Zeinoun P, Akl EA, Maalouf FT, et al. The Arab Region’s contribution to global mental health research (2009-2018): a bibliometric analysis. Front Psychiatry. 2020;11:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Deng Z, Wang H, Chen Z, et al. Bibliometric Analysis of Dendritic Epidermal T Cell (DETC) research from 1983 to 2019. Front Immunol. 2020;11:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Weng LM, Zheng YL, Peng MS, et al. A bibliometric analysis of nonspecific low back pain research. Pain Res Manag. 2020;2020:5396734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yu Q, Tan S, Ren Y, et al. Bibliometric analysis of the 100 most-cited articles in the field of hepatology. Gastroenterol Hepatol. 2020;43:349–57. [DOI] [PubMed] [Google Scholar]
- [18].Carpenter CR, Cone DC, Sarli CC. Using publication metrics to highlight academic productivity and research impact. Acad Emerg Med. 2014;21:1160–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ma C, Su H, Li H. Global research trends on prostate diseases and erectile dysfunction: a bibliometric and visualized study. Front Oncol. 2020;10:627891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zhou X, Zhao G. Global liposome research in the period of 1995–2014: a bibliometric analysis. Scientometrics. 2015;105:231–48. [Google Scholar]
- [21].Bernabò N, Greco L, Mattioli M, et al. A scientometric analysis of reproductive medicine. Scientometrics. 2016;109:103–20. [Google Scholar]
- [22].Huang Y, Huang Q, Ali S, et al. Rehabilitation using virtual reality technology: a bibliometric analysis, 1996–2015. Scientometrics. 2016;109:1547–59. [Google Scholar]
- [23].Chen CM. CiteSpace II: detecting and visualizing emerging trends and transient patterns in scientific literature. J Am Soc Inf Sci Technol. 2006;57:359–77. [Google Scholar]
- [24].Meho LI, Yang K. Impact of data sources on citation counts and rankings of LIS faculty: web of science versus scopus and google scholar. J Am Soc Inf Sci Technol. 2007;58:2105–25. [Google Scholar]
- [25].Hirsch JE. Does the h index have predictive power? Proc Natl Acad Sci USA. 2007;104:19193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Valencia GE, Roldan J, Duarte JL. Bibliometric and visualization analysis of sliding mode control research. Contemp Eng Sci. 2018;11:3673–80. [Google Scholar]
- [27].Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. [DOI] [PubMed] [Google Scholar]
- [28].Murtaza M, Dawson SJ, Tsui DWY, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–12. [DOI] [PubMed] [Google Scholar]
- [29].Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Michalopoulos A, Falagas ME. A bibliometric analysis of global research production in respiratory medicine. Chest. 2005;128:3993–8. [DOI] [PubMed] [Google Scholar]
- [31].Belter CW. A bibliometric analysis of NOAA’s Office of ocean exploration and research. Scientometrics. 2013;95:629–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Matricardi PM, Kleine-Tebbe J, Hoffmann HJ, et al. EAACI molecular allergology user’s guide. Pediatr Allergy Immunol. 2016;27(Suppl 23):1–250. [DOI] [PubMed] [Google Scholar]
- [33].Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. [DOI] [PubMed] [Google Scholar]
- [34].Travis WD, Brambilla E, Riely GJ. New pathologic classification of lung cancer: relevance for clinical practice and clinical trials. J Clin Oncol. 2013;31:992–1001. [DOI] [PubMed] [Google Scholar]
- [35].Constanzo J, Wei L, Tseng H-H, Naqa IE. Radiomics in precision medicine for lung cancer. Transl Lung Cancer Res. 2017;6:635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Catanese S, Lordick F. Targeted and immunotherapy in the era of personalised gastric cancer treatment. Best Pract Res Clin Gastroenterol. 2021;50–51:101738. [DOI] [PubMed] [Google Scholar]
- [37].Jin P, Ji X, Kang W, et al. Artificial intelligence in gastric cancer: a systematic review. J Cancer Res Clin Oncol. 2020;146:2339–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kong F-M, Ten Haken RK, Schipper M, et al. Effect of midtreatment PET/CT-adapted radiation therapy with concurrent chemotherapy in patients with locally advanced non-small-cell lung cancer: a phase 2 clinical trial. JAMA Oncol. 2017;3:1358–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kou T, Kanai M, Kamada M, et al. A platform for comprehensive genomic profiling in human cancers and pharmacogenomics therapy selection. Brown JB, ed. In: Computational Chemogenomics. New York, NY: Springer, 2018:413–24. [DOI] [PubMed] [Google Scholar]
- [40].Thunnissen E, Weynand B, Udovicic-Gagula D, et al. Lung cancer biomarker testing: perspective from Europe. Transl Lung Cancer Res. 2020;9:887. [DOI] [PMC free article] [PubMed] [Google Scholar]