Abstract
The dissociation-enhanced lanthanide fluorescent immunoassays (DELFIA) were developed for the detection of staphylococcal enterotoxin B, Yersinia pestis-specific F1 antigen, and Venezuelan equine encephalitis virus. These assays were compared to previously developed enzyme-linked immunosorbent assays (ELISAs) by determining the sensitivity or limit of detection (LOD), the dynamic range, and the reproducibility of each assay in a number of different sample matrices. The sensitivity and specificity of each assay were then determined by using a small panel of blinded spiked and nonspiked samples. All three DELFIAs demonstrated at least 1 log greater sensitivity than corresponding ELISAs utilizing the same reagents and showed an increase in dynamic range of at least 2 log10 concentrations. This increased LOD resulted in higher sensitivity rates for the DELFIA. The specificity of all of the assays evaluated was 100%, and no sample matrix effects were observed in either format. However, the reproducibility of the DELFIA was poor due to randomly distributed wells exhibiting excessive background signal (hot wells), which occurred throughout the evaluation. As this technology matures, the reproducibility of these assays should improve, as will the ability to identify hot wells. Despite its sensitivity, the logistical burden associated with the DELFIA and the technical expertise required to complete assays and interpret the data limit the application of this technology to reference or large clinical laboratories.
Rapid diagnosis is essential for the surveillance and control of epidemic diseases. Early diagnosis gives health-care providers the information necessary for effective treatment and control of disease outbreaks. Rapid identification of infectious agents has also taken on greater importance in recent years with the increased threat of biological warfare and terrorism. Microorganisms such as Yersinia pestis and Venezuelan equine encephalitis (VEE) virus, as well as toxins produced by bacteria such as Staphylococcus aureus (staphyloccal enterotoxin B [SEB]), can cause natural disease in humans, as well as be used as biological weapons (5). Many of these agents can also be effectively transmitted through contaminated food and water (3). Because of the diverse symptomology and progression of disease associated with different pathogens, detection of the causative agent in individuals suspected to have been exposed to an agent requires highly sensitive and specific means of detection. Aerosol infectious doses for these agents are low. As little as 30 ng of SEB can incapacitate a person, while as little 1.7 μg/person is lethal. One-hundred Y. pestis organisms are enough to infect a human, while even fewer VEE virus particles are required (8). In addition to this challenge, a wide variety of clinical specimens (nasal swabs, blood, serum, urine, or tissues) and environmental samples (food, water, plant material, or soil) containing even lower concentrations of agent may need to be tested to identify suspected pathogens. SEB may be found in urine as early as 12 h after an aerosol exposure at concentrations of as low as 625 pg/ml by our enzyme-linked immunosorbent assay (ELISA) and as early as 6 h by a newly developed electrochemiluminescent assay (100 pg/ml) (C. A. Rossi, unpublished data). VEE virus was recovered from serum as early as 72 h after aerosol exposure at concentrations detectable only by plaque assay (10 to 100 PFU) (B. Walker, unpublished data).
Several immunological methodologies have been applied to the antigenic detection of microorganisms. The earliest methods utilized radioactive isotopes. Although sensitive, the expense and hazards associated with handling radioactive materials severely restricted the wide spread application of these techniques. ELISAs were developed as an alternative to radioactive assays. Typically, an ELISA can detect as little as 100 to 500 pg of a low-molecular-weight analyte per ml or 106 to 107 organisms per ml and does not require specialized handling procedures. Another alternative to the use of radioactivity is the use of time-resolved fluorometry (TRF).
TRF has been used for a variety of applications, such as the detection of antibodies, microorganisms, drugs, and other therapeutic agents (2, 6, 10, 11, 22). Theoretically, TRF offers an increase in sensitivity and a wider dynamic range than other assays such as the ELISA. The basis of the TRF technology is the use of lanthanide chelate labels that have unique fluorescence properties. The labels have an intense long-lived fluorescence and a large stokes shift (∼10 times greater than that of fluorescein), all of which contribute to an increased signal-to-noise ratio (9). These properties also minimize the effect of any background fluorescence inherent in the sample matrix. Four different chelates are available, each with its unique narrow emission spectra, making multiplexing in a single well possible. One format, dissociation-enhanced lanthanide fluorescent immunoassay (DELFIA; Wallac Oy, Turku, Finland), has been applied to the detection of analytes in various samples (2, 6, 10, 11, 15, 22). This method has the added advantage of increased sensitivity due to efficient dissociation of the bound chelate (within a few minutes) and the formation of long-lasting fluorescent micelles. This dissociation occurs with the addition of a low-pH enhancement solution (Perkin-Elmer-Wallac). Assay formats using DELFIA technology include competitive and noncompetitive designs and are applicable for protein-protein binding assays, ligand receptor binding studies, and assays measuring the inhibition of enzyme activity.
The major emphasis of our laboratory is the development of rapid, sensitive immunoassays which can be conducted in portable field medical laboratories by personnel with limited training. The mobility and degree of operator training dictates that equipment be simple to operate and have a small footprint. Expendable supplies must be minimized. Thus, a single immunoassay platform and assay format are essential. Currently, we utilize ELISAs in our mobile laboratories. These assays are very reproducible, easy to perform and interpret, and use a single assay format to detect a large number of microorganisms. However, due to the lack of sensitivity of these assays for at least some microorganisms, an alternative detection technology is required. Because of the great diversity of potential biological warfare agents and endemic diseases that can be encountered, each potential platform is evaluated for its ability to detect a broad range of microorganisms. To eliminate potential differences between reagents, whenever possible, an assay format using reagents identical to that of our existing ELISA is used.
This study evaluated the ability of the DELFIA technology to detect SEB toxin, Y. pestis-specific F1 antigen, and VEE virus in a number of different sample matrices (phosphate-based buffer, serum, and urine) compared to our previously developed, in-house horseradish peroxidase (HRP)-based ELISA and to determine if this platform is suited for use in portable field medical laboratories. Since this was a preliminary evaluation, no attempt was made to achieve statistical significance or to determine clinical sensitivity and specificity.
MATERIALS AND METHODS
Reagents.
Purified SEB toxin was provided by Toxinology Division, U.S. Medical Research Institute of Infectious Disease (USAMRIID), Fort Detrick, Md., and was stored at −20°C at 2 mg/ml until used. An SEB/SED-specific monoclonal antibody (2B; Igen, Inc., Rockville, Md.) was used as the capture antibody, and polyclonal rabbit serum (Toxin Technology, Inc., Madison, Wis.) was used for the detection of SEB toxin.
Recombinant F1 antigen was provided by the Bacteriology Division, USAMRIID, and was stored at −20°C at 0.7 mg/ml until used (1). Hyperimmune polyclonal rabbit serum was used as the capture antibody and an F1-specific monoclonal antibody (6H3) was used for the detection of Y. pestis F1 antigen (both provided by John Ezzell, USAMRIID).
VEE, Trinidad donkey strain, viral antigen was prepared from infected hamster kidney (BHK) cells, inactivated by cobalt irradiation and stored at −70°C until used (17). The antigen had a titer of 1010 PFU/ml as determined by plaque assay (18). Ascites fluid from an alphavirus-specific monoclonal antibody (SLK-42; Alan Schmaljohn, USAMRIID) was used as the capture antibody, and hyperimmune polyclonal rabbit serum prepared in our laboratory was used for the detection of VEE antigen. Polyclonal rabbit anti-VEE virus hyperimmune serum was prepared by purification of VEE on sucrose gradients and immunization of rabbits with viable virus harvested from these gradients combined with complete, and then incomplete, Freund's adjuvant. All research was conducted in compliance with the Animal Welfare Act and other Federal statutes and regulations relating to animals and experiments involving animals and adheres to principles stated in the Guide for the Care and Use of Laboratory Animals (NIH publication 86-23, 1985 edition).
All antibodies were protein G purified as previously described (14) and then lyophilized before being labeled with europium for use in the DELFIA.
ELISA.
ELISAs previously developed in our laboratory for the detection of these organisms were used in this study. The format was a double-antibody sandwich identical to that previously described (16). This identical format is the basis for more than 29 antigen detection assays previously developed in our laboratory (7, 16, 21). Optimal concentrations of capture and detector antibodies specific for the assays described in our current efforts were determined by serial cross-titrations of reagents. Capture antibody was diluted in 0.01 M phosphate-buffered saline (pH 7.4; PBS), and 100 μl was adsorbed onto Maxisorb U-bottom 16-well microtiter strips (Nunc, Naperville, Ill.). Optimal coating dilutions were 1:1,000 for the SEB, 1:2,500 for the F1, and 1:500 for the VEE assays. Control wells were similarly coated with negative nonspecific antibodies produced in the same species as the specific antibodies. After an overnight incubation at 4°C, the plates were washed three times (300 μl per well) with PBS supplemented with 0.1% Tween (PBS-T; Sigma, St. Louis, Mo.). All subsequent antigen and antibody reagents added to the plates were diluted in PBS-T containing 5% skim milk (Difco, Detroit, Mich.). After the addition of each reagent, the plates were incubated at 37°C for 1 h and then washed three times in PBS-T. Antigen-spiked samples were added to both specific and nonspecific antibody-coated wells. Bound antigen was detected by incubating microtiter plates with detector antibody, followed by a HRP-labeled species-specific antibody: goat anti-rabbit immunoglobulin G (IgG), heavy and light chain specific for the SEB and VEE assays (catalog number 074-1506; Kirkegaard and Perry, Gaithersburg, Md.), and goat anti-mouse IgG, heavy and light chain specific for the plague F1 assay (catalog number 605-250; Boehringer Mannheim, Indianapolis, Ind.). The substrate used was 2,2′-azino-di(3-ethylbenzthiazoline sulfonate) (Kirkegaard and Perry). Plates were read spectrophometrically at 410 nm by using a 490-nm reference filter. Results were expressed as optical density (OD) values.
DELFIA.
All equipment, platform-specific reagents, and protocols were those specified by Perkin-Elmer-Wallac in order to achieve optimal platform system performance. The DELFIA was performed as the ELISA described above with the following exceptions. DELFIA buffer (Perkin-Elmer-Wallac, Gaithersburg, Md.) was used to dilute the antigen-spiked samples and detector antibody. Plates were vigorously washed by using an OEM M12/2R Columbus plus washer (Tecan) between each step. A wash step consisted of four cycles at 700 μl per well programmed on overflow mode. These settings produced a vortex within the well and resulted in thorough washing between steps and lower background signals. After the plates were coated, all subsequent incubations were done for 1 h at room temperature at low speed on a model 1296-004 DELFIA Plateshaker (Perkin-Elmer-Wallac). Detector antibodies were directly labeled with europium (Eu) either by Perkin-Elmer-Wallac or by using a DELFIA Eu-labeling kit (Perkin-Elmer-Wallac). Detector antibodies were reconstituted in carbonate buffer at pH 9 and labeled with a 40-fold excess of europium chelate according to the manufacturer's procedures. Unbound europium was removed by gel filtration using a Sephadex G-25 column (Pharmacia, Uppsala, Sweden). The protein concentrations were determined by using MicroBCA assay (Pierce, Rockford, Ill.) according to the manufacturer's instructions with bovine serum albumin as the standard. Europium labeling efficiency was determined as recommended by Perkin-Elmer-Wallac. Optimal concentrations of Eu-labeled detector antibody were determined by serial checkerboard titrations. DELFIA enhancement solution (100 μl/well) (Perkin-Elmer-Wallac) was used to dissociate Eu3+ from solid-phase bound Eu-labeled detector antibody. Plates were allowed to incubate for an additional 5 min at room temperature on the shaker on low speed. Fluorescence was read using the 1420 Victor2 multilabel counter (Perkin-Elmer-Wallac). Emission peaks were captured as TRF counts at 613 nm from the top of the wells after excitation of the sample at 340 nm. Results were expressed as TRF counts.
Performance parameters.
Performance for each assay was measured by determining the sensitivity or limit of detection (LOD) and the reproducibility of the assay within a plate (intraplate variation) and from day to day (interplate variation) for a number of different sample matrices. The matrices evaluated included human serum (except for SEB assays), human urine, and PBS supplemented with 0.3% Tween 20 (PBS-0.3T). Serum used in these analyses did not contain any agent-specific antibodies as determined by antibody ELISA (data not shown). Sample matrices were spiked with antigen and added at a 1:2 dilution in triplicate to the microtiter plate and diluted fourfold down the microtiter plate to produce matrix-specific standard curves and to determine intraassay variation. The linear range of the assay was defined as the area of the standard curve that exhibited a regression coefficient greater than 0.969. Wells in which unusually high or low TRF counts or OD values occurred were examined for the presence of an outlier value by applying the Dixon test (4). These randomly distributed wells exhibiting excessive background signal are commonly referred to as hot wells. Outliers or hot wells successfully identified by this test were removed from subsequent calculations. The Dixon test was only used to exclude sample replicates from analysis. It was not used to exclude a sample from analysis. The DELFIA was also run on three consecutive days to determine interassay variation. In addition, a small panel of blinded spiked and nonspiked samples representing the various sample matrices were evaluated, in triplicate, to determine the relative assay sensitivity and specificity. Sample sets were prepared by spiking each set with SEB or F1 between 100 and 0.1 ng/ml in 10-fold increments or concentrations of VEE virus of between 1 × 109 and 5 × 106 PFU/ml in half-log increments. Sample concentrations were chosen based on what levels might be expected in biological specimens taken from patients and the relative sensitivity of the ELISA.
RESULTS
Europium labeling of antibodies.
Europium labeling of antibodies resulted in molar incorporation ratios of 6.8 (F1), 9.4 (SEB), and 4.8 (VEE). There was no discernible loss of ELISA reactivity for Eu-labeled antibodies compared to unlabeled antibodies (data not shown). Optimal concentrations of Eu-labeled antibodies were determined by serial checkerboard titrations and found to be 1 μg/ml for the SEB toxin and VEE virus assays and 0.25 μg/ml for the F1 assay. In comparison, optimal concentrations of ELISA detector antibodies were 2 μg/ml for the SEB toxin assay, 10 μg/ml for the F1 assay, and 20 μg/ml for the VEE virus assay.
ELISA.
As is typical for ELISA, adjusted OD values were determined for each sample by subtracting negative capture antibody values from positive capture antibody values. A positive ELISA value was determined by calculating a cutoff value that was equal to three times the standard deviation of the matrix-specific negative controls plus the average OD reading for the same controls, rounded up to the nearest tenth. In the ELISA, cutoff values from samples prepared in all three matrices were similar. No outliers were identified using the Dixon test.
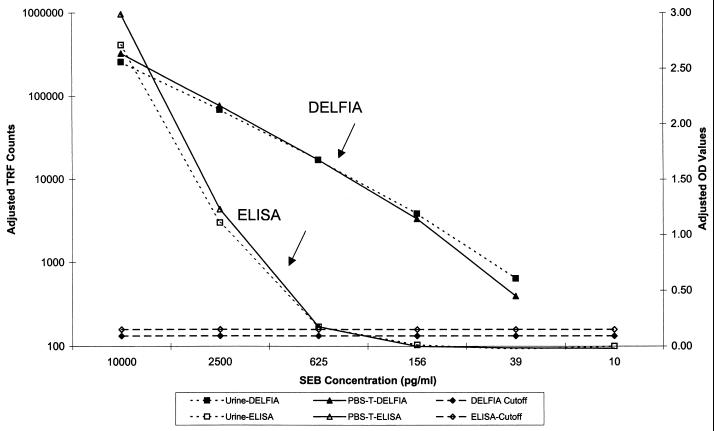
The SEB detection ELISA was linear between 10 and 1 ng/ml with an LOD of 625 pg/ml (Fig. 1 and Table 1). This linear range is identical to that observed in over 150 replicates of this assay conducted over 11 years (data not shown). Intraplate variation was <11% for values within the linear portion of the curve (n = 3). Interplate variation was not measured; however, coefficients of variation for most ELISAs are typically between 15 and 25% (data not shown). Variation between matrices was <20% (n = 6). Sensitivity of the assay was 75% (6 of 8) using blinded samples spiked with SEB; however, it was 100% (6 of 6) for concentrations within the LOD of the assay. The specificity was 100% (9 of 9).
FIG. 1.
SEB DELFIA and ELISA standard curves prepared in different sample matrices. Urine and PBS-0.3T matrices were spiked with 10,000 pg of SEB toxin per ml and diluted fourfold. Three replicates of each concentration were used for each determination. DELFIA results are expressed as TRF counts, whereas ELISA results are expressed as OD values. Assay cutoff values were equal to three times the standard deviation of negative control values (all matrices noted above) plus the average value for the same controls. The ELISA cutoff was then rounded up to the nearest tenth.
TABLE 1.
Performance of ELISA and DELFIA in detecting SEB, Y. pestis F1 antigen, and VEE virus
| Parameter | SEB toxin
|
Y. pestis F1 antigen
|
VEE virus
|
|||
|---|---|---|---|---|---|---|
| ELISA | DELFIA | ELISA | DELFIA | ELISA | DELFIA | |
| Assay LOD | 0.625 ng/ml | 0.039 ng/ml | 25 ng/ml | 0.024 ng/ml | 1.25E+07 PFU/mlc | 3.13E+06 PFU/ml |
| % Intraassay variation (n = 3) | <11 | <37 | <13 | <52 | <20 | <18 |
| % Interassay variation (n = 9) | NDa | <52 | ND | <68 | ND | <30 |
| % Intermatrix variation (n) | <20 (6) | <33 (6) | <9 (9) | <26 (9) | <9 (9) | <25 (9) |
| Upper limit of linear range | 10 ng/ml | >10 ng/ml | 500 ng/mlb | >25 ng/ml | 5E+08 PFU/mlb | >5E+07 PFU/ml |
| Lower limit of linear range | 1 ng/ml | 0.039 ng/ml | 50 ng/mlb | 0.024 ng/ml | 5E+07 PFU/mlb | 3.13E+06 PFU/ml |
| % Sensitivity (no. positive/total no.) | ||||||
| Spiked samples | 75 (6/8) | 100 (8/8) | 25 (3/12) | 91.7 (11/12) | 50 (6/12) | 83.3 (10/12) |
| Within LOD | 100 (6/6) | 100 (8/8) | 100 (3/3) | 91.7 (11/12) | 100 (6/6) | 83.3 (10/12) |
| % Specificity (n = 9) | 100 | 100 | 100 | 100 | 100 | 100 |
ND, not determined.
Based on historical data.
That is, 1.25 × 107.
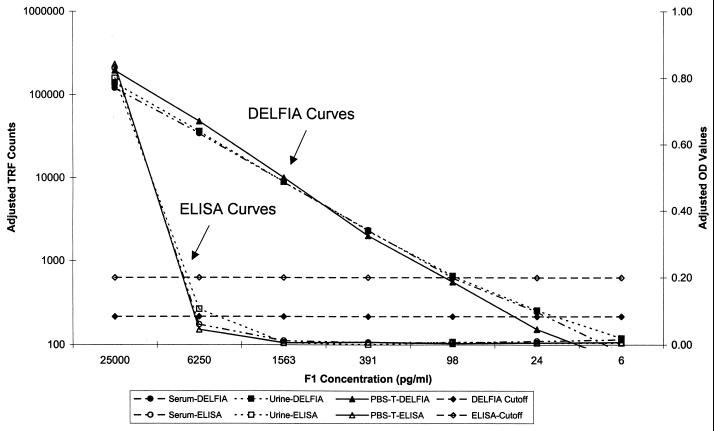
The linearity of the F1 detection ELISA is between 500 and 50 ng/ml and was established in over 38 replicates of this assay conducted over 9 years (data not shown). The LOD of the assay was 25 ng/ml (Fig. 2 and Table 1). Intraplate variation within the linear range was <13% (n = 3) and between matrices was <9% (n = 9). The sensitivity of the assay was 25% (3 of 12) using blinded samples spiked with F1 and 100% (3 of 3) for concentrations within the LOD of the assay. The specificity was 100% (9 of 9).
FIG. 2.
Y. pestis F1 antigen DELFIA and ELISA standard curves prepared in different sample matrices. F1 antibody negative serum, urine, and PBS-0.3T matrices were spiked with 25,000 pg of F1 antigen per ml and diluted fourfold. Three replicates of each concentration were used for each determination. DELFIA results are expressed as counts, whereas ELISA results are expressed as OD values. Assay cutoff values were equal to three times the standard deviation of negative control values (all matrices noted above) plus the average value for the same controls. The ELISA cutoff was then rounded up to the nearest tenth.
The linearity of the VEE virus detection ELISA is between 5 × 108 and 5 × 107 PFU/ml and was established in more than 40 replicates of this assay conducted over 12 years (data not shown). The LOD of the assay was 1.25 × 107 PFU/ml (Fig. 3 and Table 1). Intraplate variation was <20% (n = 3) and between matrices was <9% (n = 9). The sensitivity of the assay was 50% (6 of 12) using blinded samples spiked with VEE virus and 100% (6 of 6) for concentrations within the LOD of the assay. The specificity was 100% (9 of 9).
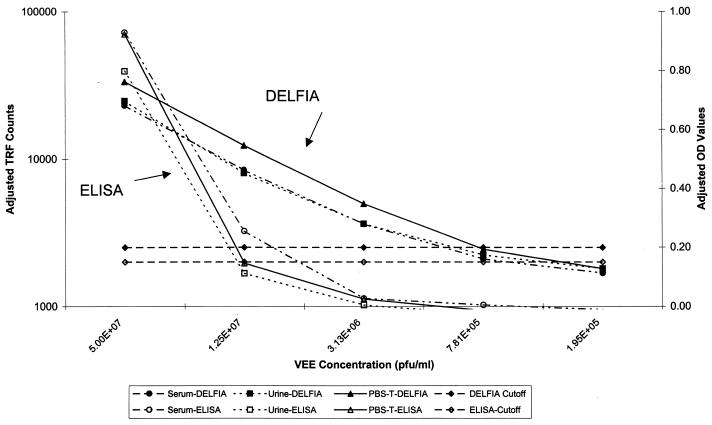
FIG. 3.
VEE virus DELFIA and ELISA standard curves prepared in different sample matrices. VEE antibody-negative serum, urine, and PBS-0.3T matrices were spiked with 5.00E + 07 PFU of VEE virus per ml and diluted fourfold. Three replicates of each concentration were used for each determination. DELFIA results are expressed as TRF counts, whereas ELISA results are expressed as OD values. Assay cutoff values were equal to three times the standard deviation of negative control values (all matrices noted above) plus the average value for the same controls. The ELISA cutoff was then rounded up to the nearest tenth.
DELFIA.
As with the ELISA, adjusted TRF values were determined for each sample, and cutoff values were calculated. In the DELFIA, cutoff values from samples prepared in all three matrices were similar. A total of four wells were identified as outliers by the Dixon test, and these were removed from subsequent calculations (two for F1 analysis and two for SEB analysis).
The SEB detection by DELFIA was linear between 10 and .039 ng/ml, with an LOD of 39 pg/ml (Fig. 1 and Table 1). Assay linearity may have covered a larger range, but higher concentrations of analyte were not tested. Intraplate variation was <37% for values within the linear portion of the curve (n = 3). Interplate variation was <52% (n = 9). Variation between matrices was <33% (n = 6). The sensitivity of the assay was 100% (8 of 8) using blinded samples spiked with SEB. All concentrations tested were within the LOD of the assay. The specificity was 100% (9 of 9).
The F1 detection by DELFIA was linear between 25 and .024 ng/ml, with an LOD of 24 pg/ml (Fig. 2 and Table 1). The assay linearity may have covered a larger range, but higher concentrations of analyte were not tested. Intraplate variation was <52% for values within the linear portion of the curve (n = 3). Interplate variation was <68% (n = 9). Variation between matrices was <26% (n = 9). The sensitivity of the assay was 91.7% (11 of 12) using blinded samples spiked with F1. All concentrations tested were within the LOD of the assay. The specificity was 100% (9 of 9).
The VEE virus detection DELFIA was linear at between 5 × 107 and 3.13 × 106 PFU/ml, with an LOD of 3.13 × 106 PFU/ml (Fig. 3 and Table 1). The assay linearity may have covered a larger range, but higher concentrations of analyte were not tested. Intraplate variation was <18% for values within the linear portion of the curve (n = 3). Interplate variation was <30% (n = 9). Variation between matrices was <25% (n = 9). The sensitivity of the assay was 83.3% (10 of 12) using blinded samples spiked with VEE virus. All concentrations tested were within the LOD of the assay. The specificity was 100% (9 of 9).
DISCUSSION
DELFIA and ELISA assays were developed that can detect SEB toxin, F1 antigen of Y. pestis, and VEE virus. Since a large variety of sample matrices may be presented to the clinical laboratory for agent identification or patient diagnosis during a disease outbreak or possible biowarfare scenario, a number of different sample matrices (phosphate-based buffer, serum, and urine) were examined. Due to the high prevalence of SEB antibodies in serum (≥70% [12, 13, 19, 20]) and the rapid clearance of the toxin from the bloodstream (8), serum is an inappropriate sample from which to identify SEB toxin. Therefore, this matrix was not tested in our SEB experiments.
The ELISA format used was one that is an industry standard and has proven to be simple and easy to perform. For simplicity and ease of transition, we chose to develop a noncompetitive DELFIA format that mimicked the ELISA. This strategy also enabled us to evaluate the DELFIA platform by eliminating differences between reagents. While both formats required an overnight incubation, the DELFIA required 80 min less time to perform due to the use of a directly labeled detector antibody and faster substrate development. However, the transition of our ELISA to the DELFIA format was not straightforward. This assay requires the use of a specialized washer, a plate shaker, special assay buffers, and rigorous decontamination procedures. Experiments conducted with standard ELISA equipment resulted in unacceptably high background readings (data not shown). The assay requires a liter of wash buffer per plate and dedicated pipetters to dispense enhancement solution. The labeled detector antibody must be filtered through a 0.2-μm (pore-size) filter just before addition to the plate. Despite these precautions, a number of hot wells still occurred over the course of this study. The majority of these hot wells were not identified as outliers by the Dixon test. This resulted in the high DELFIA intra- and interplate variation that was observed and directly affects the reproducibility of the assay.
Eu labeling of antibodies was easy and simple using standard chemistries, but unbound Eu tag must be totally removed to minimize assay background. The manufacturer suggests that labeling should be conducted at a site not used for conducting the assay and should use dedicated equipment and columns for this procedure. Labeling was reproducible, as evidenced by the results from the labeling of two different lots of SEB rabbit antibody (data not shown).
ELISA standard curves were linear over 1 log10 concentration, while DELFIA standard curves were linear over at least 3 log10 concentrations. This wide dynamic range eliminates the need to run a large number of serial dilutions, thus allowing a greater number of unknowns to be tested in a single assay.
The DELFIA assays were at least 1 log more sensitive than ELISA utilizing the same reagents. This increased LOD resulted in higher sensitivity rates for the DELFIA. The specificity of the assays were 100% when readings were adjusted to account for the reactivity of the sample with a nonspecific capture antibody. However, in experiments conducted without subtracting TRF counts from nonspecific capture antibody wells and using our standard method for determining assay cutoff values, false-positive results were seen (79%, 19 of 24) (data not shown). This finding indicates that major modifications would be required to the way in which the cutoff is calculated, which in turn would adversely affect the LOD of the DELFIA. Use of these nonadjusted readings also resulted in substantial differences between matrices (i.e., matrix effects [data not shown]).
In conclusion, the DELFIA was a rapid and highly sensitive method of detection, which shows great promise in clinical diagnostic applications. Like the ELISA, it can detect analytes in a number of different sample matrices, though at much lower concentrations. Currently, the many special requirements (equipment and technical) and frequent hot wells make these assays cumbersome to perform and hard to interpret, especially for junior laboratory personnel. These findings are similar to those previously reported (10). Future improvements in this technology should simplify this format and potentially eliminate these problems. A more rigorous evaluation, including large numbers of spiked and clinically relevant samples, would be warranted at that time.
ACKNOWLEDGMENTS
We thank Tamara Lewis and Michael Robich for technical help and Brian Walker and Jeff Teska for reviewing the manuscript.
REFERENCES
- 1.Andrews G P, Heath D G, Anderson G W, Jr, Welkos S L, Friedlander A M. Fraction 1 capsular antigen (F1) purification from Yersinia pestis C092 and an Escherichia coli recombinant strain and efficacy against lethal plague challenge. Infect Immun. 1996;64:2180–2187. doi: 10.1128/iai.64.6.2180-2187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard G, Helmick B, Madden S, Gilbourne C, Patel R. The measurement of prion protein in bovine brain tissue using differential extraction and DELFIA® as a diagnostic test for BSE. Luminescence. 2000;15:357–362. doi: 10.1002/1522-7243(200011/12)15:6<357::AID-BIO621>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 3.Benenson A, editor. Control of communicable diseases manual. 16th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 4.Bliss C I. Statistics in biology. New York, N.Y: McGraw-Hill Book Company; 1967. pp. 153–155. [Google Scholar]
- 5.Christopher G W, Cieslak T J, Pavin J W, Eitzen E M. Biological warfare: a historical perspective. JAMA. 1997;278:412–417. [PubMed] [Google Scholar]
- 6.Crooks S R, Ross P, Thompson C S, Haggan S A, Elliott C T. Detection of unwanted residues of ivermectin in bovine milk by dissociation-enhanced lanthanide fluoroimmunoassay. Luminescence. 2000;15:371–376. doi: 10.1002/1522-7243(200011/12)15:6<371::AID-BIO622>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 7.Dohm D J, Logan T M, Linthicum K J, Rossi C A, Turell M J. Transmission of Crimean-Congo hemorrhagic fever virus by Hyalomma impeltatum (Acari: Ixodidae) after experimental infection. J Med Entomol. 1996;33:848–851. doi: 10.1093/jmedent/33.5.848. [DOI] [PubMed] [Google Scholar]
- 8.Franz D R, Jahrling P B, Friedlander A M, McClain D J, Hoover D L, Bryne W R, Pavin J A, Christopher G W, Eitzen E M. Clinical recognition and management of patients exposed to biological warfare agents. JAMA. 1997;278:399–411. doi: 10.1001/jama.278.5.399. [DOI] [PubMed] [Google Scholar]
- 9.Hemmila I A, Dakubu S, Mukkala V-M, Siitara H, Lovgren T. Europium as a label in time-resolved immunofluorometric assays. Anal Biochem. 1984;137:335–343. doi: 10.1016/0003-2697(84)90095-2. [DOI] [PubMed] [Google Scholar]
- 10.Hierholzer J C, Johanasson K H, Anderson L J, Tsou C J, Halonen P E. Comparison of monoclonal time-resolved fluoroimmunoassay with monoclonal capture-biotinylated detector enzyme immunoassay for adenovirus antigen detection. J Clin Microbiol. 1987;25:1662–1667. doi: 10.1128/jcm.25.9.1662-1667.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hierholzer J C, Bingham P G, Coombs R A, Johansson K H, Anderson L J, Halonen P E. Comparison of monoclonal antibody time-resolved fluoroimmunoassay with monoclonal antibody capture-biotinylated detector enzyme immunoassay for respiratory syncytial virus and parainfluenza virus antigen detection. J Clin Microbiol. 1989;27:1243–1249. doi: 10.1128/jcm.27.6.1243-1249.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jozefczyk Z. Specific human antibodies to enterotoxin A, B, and C1 of Staphylococcus: their increased synthesis in staphylococcal infection. J Infect Dis. 1974;130:1–7. doi: 10.1093/infdis/130.1.1. [DOI] [PubMed] [Google Scholar]
- 13.Jozefczyk Z, Robbins R N, Spitz J M, Bergdoll M S. Antibodies to staphylococcal enterotoxin in laboratory personnel. J Clin Microbiol. 1980;11:438–439. doi: 10.1128/jcm.11.4.438-439.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kijek T M, Rossi C A, Moss D, Parker R W, Henchal E A. Rapid and sensitive immunomagnetic-electrochemiluminescent detection of staphyloccocal enterotoxin B. J Immunol Methods. 2000;236:9–17. doi: 10.1016/s0022-1759(99)00234-3. [DOI] [PubMed] [Google Scholar]
- 15.Knipping G, Gogg-Fassolter B, Frohnwiesser B, Krempler F, Kostner G M, Malle E. Quantification of apolipoprotein D by an immunoassay with time-resolved fluorescence spectroscopy. J Immunol Methods. 1997;202:85–95. doi: 10.1016/s0022-1759(96)00240-2. [DOI] [PubMed] [Google Scholar]
- 16.Ksiazek T G, Rollin P E, Jahrling P B, Johnson E, Dalgard D W, Peters C J. Enzyme immunosorbent assay for Ebola virus antigens in tissues of infected primates. J Clin Microbiol. 1992;30:947–950. doi: 10.1128/jcm.30.4.947-950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meegan J M, Yedlounschnig R J, Peleg B A. Enzyme-linked immunosorbent assay for detection of antibodies to Rift Valley fever virus in ovine and bovine sera. Am J Vet Res. 1981;48:1139–1143. [PubMed] [Google Scholar]
- 18.McClain D J, Pittman P R, Ramsburg H H, Nelson G O, Rossi C A, Mangiafico J A, Schmaljohn A L, Malinoski F J. Immunological interference from sequential administration of live-attenuated alphavirus vaccine. J Infect Dis. 1998;177:634–641. doi: 10.1086/514240. [DOI] [PubMed] [Google Scholar]
- 19.McGann V G, Rollins J B, Mason D W. Evaluation of resistance to staphylococcal enterotoxin B: acquired antibodies of man and monkey. J Infect Dis. 1971;124:206–213. doi: 10.1093/infdis/124.2.206. [DOI] [PubMed] [Google Scholar]
- 20.Notermans S, Van Leeuwin W J, Dufrenne J, Bergdoll M S. Serum antibodies to enterotoxins produced by Staphylococcus aureus with specific reference to enterotoxin F and toxin shock syndrome. J Clin Microbiol. 1983;18:1055–1060. doi: 10.1128/jcm.18.5.1055-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turell M J, Rossi C A, Tammariello R F, Bailey C L. Reduced recovery of Rift Valley Fever virus associated with assay of mosquito (Diptera: Culcidae) larval pools. J Med Entomol. 1986;23:416–422. doi: 10.1093/jmedent/23.4.416. [DOI] [PubMed] [Google Scholar]
- 22.Willson V J, Lockley W J, Mather A, Singh J, Gilbert C M, Bayliss M A, Wilkinson D. A time-resolved fluorescence immunoassay for the determination of a novel respiratory therapeutic agent, AR-C68397XX (Vrozan) in human plasma. J Pharm Biomed Anal. 2000;23:947–954. doi: 10.1016/s0731-7085(00)00374-5. [DOI] [PubMed] [Google Scholar]