Abstract
Glioblastoma (GBM), a malignant brain tumor, is a world-wide health problem because of its poor prognosis and high rates of recurrence and mortality. Apolipoprotein C1 (APOC1) is the smallest of apolipoproteins, implicated in many diseases. Recent studies have shown that APOC1 promotes tumorigenesis and development of several types of cancer. In this study we investigated the role of APOC1 in GBM tumorigenesis. Using in silico assays we showed that APOC1 was highly expressed in GBM tissues and its expression was closely related to GBM progression. We showed that APOC1 protein expression was markedly increased in four GBM cell lines (U251, U138, A172 and U87) compared to the normal brain glia cell lines (HEB, HA1800). In U251 cells, overexpression of APOC1 promoted cell proliferation, migration, invasion and colony information, which was reversed by APOC1 knockdown. APOC1 knockdown also markedly inhibited the growth of GBM xenografts in the ventricle of nude mice. We further demonstrated that APOC1 reduced ferroptosis by inhibiting KEAP1, promoting nuclear translocation of NRF2 and increasing expression of HO-1 and NQO1 in GBM cells. APOC1 also induced ferroptosis resistance by increasing cystathionine beta-synthase (CBS) expression, which promoted trans-sulfuration and increased GSH synthesis, ultimately leading to an increase in glutathione peroxidase-4 (GPX4). Thus, APOC1 plays a key role in GBM tumorigenesis, conferring resistance to ferroptosis, and may be a promising therapeutic target for GBM.
Keywords: glioblastoma, tumorigenesis, APOC1, ferroptosis, lipid ROS, NRF2, CBS
Introduction
Glioma is a type of tumor that occurs in the brain and spinal cord, the most common primary intracranial tumor, which accounts for 40%–50% of brain tumors [1]. According to the World Health Organization (WHO) classification of CNS tumors, diffuse gliomas can be classified into grades I, II, III, and IV [2]. Grade IV glioma, also known as glioblastoma (GBM), is the most common malignancy among the pathological types of glioma. Up to now, clinical treatment of GBM included surgery, radiation therapy, chemotherapy, targeted therapy and immune therapy [3]. However, the prognosis of GBM is still poor and the recurrence rate after initial treatment is very high [4]. For GBM patients, the median overall survival is only 12–15 months [5] and the five-year survival rate does not exceed 5% [6]. Genome sequence analysis has revealed that mutations in IDH1/2, BRAF, PTEN, 1p-19q co-deletion, and MGMT methylation are some of the molecular pathogenetic characteristics of glioma that can be used for diagnosis and treatment [2]. However, the molecular mechanism of GBM progression is still unclear and there is no target drug for GBM.
APOC1 belongs to the apolipoprotein family and is the smallest apolipoprotein. It participates in composition of triglyceride-rich lipoproteins and high-density lipoproteins, and functions in the metabolism of lipoproteins, such as very low-density lipoprotein (VLDL) and low-density lipoprotein (LDL) [7, 8]. Studies showed that elevated expression of APOC1 was closely related to cancers such as pancreatic cancer and significantly related to poor prognosis [9]. Patients with gastric cancer also had higher expression of APOC1 in tumor tissues than in adjacent normal tissues and prognosis was clearly influenced by APOC1 level [10]. In addition, there are a few studies on APOC1 in other cancers, such as non-small cell lung cancer [11], acute myeloid leukemia [12], nephroblastoma [13] and papillary thyroid carcinoma [14]. Although evidence has revealed connections between some types of cancer and the apolipoproteins including APOA, APOB, APOC, APOE and others, there have been few studies of apolipoproteins in GBM. APOE has been used as a drug carrier in GBM because of its ability to pass through the blood-brain barrier [15]. APOC1 was found to be highly expressed in GBM [16], but no investigation into the mechanism of its function in GBM is currently available.
Ferroptosis of cells is characterized as iron-catalyzed non-apoptotic cell death that differs from apoptosis, autophagy, and necrosis [17]. Cells undergoing ferroptosis have a condensed membrane, a ruptured outer membrane and dysmorphic small mitochondria [18]. Ferroptosis is characterized by iron dependence and accumulation of lipid-reactive oxygen species (lipid ROS). Excessive peroxidation caused by decreased cellular glutathione (GSH), loss of glutathione peroxidase-4 (GPX4), and inactivation of system XC- led to lipid-toxic ROS accumulation, which resulted in ferroptosis of cells [19]. Ferroptosis was associated with cancers and some therapeutic strategies were based on regulating ferroptosis. Though the main treatment for GBM is surgery, researchers are examining new ways to treat GBM by inducing ferroptosis [20]. Stimulation of ferroptosis in GBM may increase the efficacy of radiation and chemotherapy and inhibit tumor growth [21]. A number of recent studies have shown that many genes that are highly expressed in cancers are associated with ferroptosis. Nuclear factor erythroid 2-related factor 2 (NRF2) plays an important role in regulating intracellular redox homeostasis, and inhibition of NRF2 increased sensitivity of cells to ferroptosis [22]. The KEAP1-NRF2 pathway was reported to be involved in GBM by promoting cell proliferation and diminishing ferroptosis [23]. Our data showed that APOC1 influenced the level of ROS and lipid ROS in GBM cells, which means APOC1 may participate in ferroptosis regulation. However, it was unclear whether APOC1 participated in ferroptosis regulation through the KEAP1-NRF2 pathways in GBM.
In this study, we first explored the functional role and underlying mechanism of APOC1 action in GBM. Our results showed that APOC1 was highly expressed in GBM patients and positively correlated with GBM progression, and that overexpression of APOC1 promoted proliferation, migration, invasion and colony formation of GBM cells. Furthermore, our results demonstrated that APOC1 overexpression conferred ferroptosis resistance by inhibiting KEAP1 which promoted nuclear translocation of NRF2 and increased the expression of HO-1 and NQO1 in GBM cells. Overexpression of APOC1 also reduced ferroptosis by increasing expression of cystathionine beta-synthase (CBS), which raised GSH level and GPX4 expression and finally reduced lipid ROS. These results suggest that APOC1 plays an important role in tumorigenesis and development of GBM by increasing ferroptosis resistance, and may be a promising therapeutic target for GBM.
Materials and methods
Cell culture and tumor specimens
The normal human glial cells, HEB and HA1800, were bought from ScienCell, while human GBM cell lines, U251, U138, A172 and U87 were purchased from the American Type Culture Collection (ATCC). U251, U138, A172 and U87 were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco, New Zealand). HEB and HA1800 were cultured in DMEM with 20% FBS at 37 °C in a humidified incubator containing 5% CO2. GBM and adjacent normal tissue samples were collected from GBM patients. The use of human GBM specimens from the Peking Union Medical College Hospital, Beijing, China was approved by Institutional Review Board (IRB) (S-424). The human GBM tissue array was kindly gifted by Dr. Hong-qing Cai (Cancer Hospital Chinese Academy of Medical Sciences).
Chemical reagents
Erastin, Ferrostatin-1, Z-VAD-FMK and Necrosulfonamide were bought from MedChemExpress (Monmouth Junction, NJ, USA). The cell counting kit-8 (CCK-8) was from MeilunBio (Dalian, China), DCFH-DA was from Sigma-Aldrich (St. Louis, MO, USA), and C11 BODIPY was from Thermo-Fisher Scientific (Carlsbad, CA, USA). The glutathione assay kit was from BioVision (Milpitas, CA, USA). FerroOrange was from Dojindo (Kumamoto, Japan).
Transfection
U87 and U251 cells were cultured in 60 mm dishes for 24 h to 70% confluence before transfection. U251 cells were transfected with an APOC1-expressing plasmid (OriGene, Rockville, USA) for 24 h using the transfection reagent, Lipofectamine™ 3000 (Invitrogen, Grand Island, NY, USA) according to instructions. Recombinants were selected by incubation of cells with 600 μg/mL G418 for 3 weeks. U87 and U251 cells were transfected with specific siRNAs (GenePharm, Shanghai GenePharma Co., Ltd.) for 48 h before detection. Expression of APOC1 was verified by Western blotting and RT-qPCR. The sequences of specific interfering RNAs are listed in Supplementary Table 1.
Experimental treatments and cell viability assay
U87 and U251 cells were seeded in 96-well plates at 3 × 103/well and cultured for 24 h at 37 °C. Cells were first incubated with Ferrostatin-1, Z-VAD-FMK and Necrosulfonamide for 1 h before addition of Erastin to a concentration of 10 μM and incubation for 24 h. After treatment, the medium was discarded and cell growth was assessed with the CCK-8 assay according to instructions. Relative growth of U87 and U251 cells treated was determined at 24, 48, and 72 h using the CCK-8 protocol with absorbance readings at 450 nm measured with a SpectraMax M5 plate-reader (Molecular Devices, Sunnyvale, CA, USA). The data were processed by GraphPad Prism 7.
Transwell assays of cell migration and invasiveness
Cell migration and invasion ability were measured using 24-well plates with transwell inserts of 8 μm pore size (Corning Costar, USA). For the invasion assay, the bottom well was coated with 12.5% Matrigel (Corning Biocoat, USA). Treated cells were harvested and suspended in FBS-free medium at 5 × 104 cells/mL. Aliquots of 200 μL of cell suspension were added to the upper chamber and 1 mL of DMEM plus 10% FBS was added to the lower chamber. After incubation for 24 h, the cells that had passed through the membrane were fixed in 4% paraformaldehyde for 20 min at room temperature (RT), stained with 1% crystal violet for 15 min, and counted using a brightfield microscope (Nikon, Japan).
Soft agar colony formation assay
The colony formation assay was carried out in 6-well plates. DMEM with 10% FBS and 0.7% agar was added as the bottom layer for 2 mL/well and 1 mL DMEM containing 3000 treated cells with 10% FBS and 0.35% agar was added as the top layer. The plates were then incubated at 37 °C for 3 weeks and colonies were stained with MTT (3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium bromide, 200 μL/well) and photoed.
Three-dimensional (3D) Matrigel cell culture
50 µL cells (1000/well) were mixed with an equal volume of ice-cold BD Matrigel (BD Bioscience, Franklin Lakes, NJ, USA) and seeded into a 96-well plate. After culturing at 37 °C in 5% CO2 for 2 weeks, the cells were imaged by fluorescence microscopy (Nikon Eclipse Ti-U, Japan).
Reverse transcriptase quantitative real-time PCR (RT-qPCR)
Cells were harvested and homogenized in TRIzol for 5 min at RT. Lysates were mixed with chloroform and centrifuged at 12,000 × g for 15 min at 4 °C, and the aqueous supernatants were transferred into new RNase-free centrifuge tubes. An equal volume of isopropanol was added, gently mixed with the supernatant, and left at RT for 30 min. Samples were centrifuged at 12,000 × g for 15 min, the pellets were dissolved in RNase-free water, and the RNA concentration and quality were determined. One microgram of total RNA was used for cDNA synthesis with MonScript RTIII All-in-One mix (Monad, China) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using a reaction mixture containing 1 × AceQ Universal SYBR qPCR master mix (Vazyme Biotech Co., Ltd., Nanjing, China) and 200 nmol/L gene-specific APOC1 primers (Supplementary Table 2). Assays were carried out three times on a CFX thermocycler (Bio-Rad, Hercules, CA, USA).
Immunohistochemistry (IHC)
Immunohistochemistry (IHC) was carried out on 5 µm paraffin-embedded AT specimens, which were incubated for about 24 h at 37 °C. Sections were deparaffinized, rehydrated and washed in PBS. Antigen retrieval was done by incubating the sections in citrate buffer at 100 °C for 10 min, then in 3% H2O2 at RT to block endogenous peroxidases. The slides were incubated with monoclonal anti-APOC1 rabbit antibody (ab19288, Abcam, St. Louis, MO, USA) at a dilution of 1:200 overnight at 4 °C, washed and incubated 1 h with biotinylated secondary antibody. Stained cells were visualized using Vectastain with diaminobenzidine (DAB) as substrate and cells were counterstained with hematoxylin (Sigma–Aldrich). The negative controls were treated with rabbit serum alone (Santa Cruz Biotechnology). A Nikon Eclipse Ti microscope was used to image the stained sections and staining density was determined by ImageJ software.
Immunofluorescence (IF)
U251 cells were detached with trypsin, centrifuged at 400 × g for 5 min, resuspended in 10% DMEM and seeded into 15 mm glass bottom cell culture dishes (NEST, USA) at 1 × 104 cells per dish. After incubation with or without Erastin for 24 h, the cells were fixed with 4% paraformaldehyde for 20 min, then permeabilized with 0.2% Triton-100 for 10 min, and blocked with 5% bovine serum albumin (BSA) for 30 min at RT. Incubation with primary antibodies against APOC1, KEAP1 and CBS was done at 4 °C overnight. After washing three times with PBS, the cells were stained with fluorescent second antibodies (1:500): green for APOC1 (4412, anti-rabbit IgG, Cell Signaling Technology, Danvers, USA) and red for KEAP1 and CBS (4409, anti-mouse IgG, Cell Signaling Technology, Danvers, USA) at 37 °C for 1 h. After washing three times with PBS, cells were stained with DAPI for 30 min at RT [24]. Images were acquired with a Leica TCS SP8X confocal microscope.
ROS and lipid ROS detection
ROS: Cells were treated with or without Erastin for 24 h, then digested, centrifuged, and incubated with 2 μM DCFH-DA at 37 °C for 30 min. Cells were washed three times with PBS.
Lipid ROS: Cells in 60 mm dishes were treated with or without Erastin for 24 h, then BODIPY C11 was added to a final concentration of 10 μM and incubation at 37 °C was continued for 30 min. Cells were trypsinized, centrifuged and washed three times with PBS.
Both the ROS level and lipid ROS level were detected using an Accuri C6 Plus flow cytometer (BD, San Diego, CA, USA) and data were analyzed by FlowJo ver10 Software (Tristar, CA, USA).
Intracellular Fe2+ assay
U251 cells were seeded into 96-well glass-bottom plates (Sunnyvale, CA, USA) at 1 × 103 per well, and incubated with or without Erastin for 24 h, after which they were washed three times with HBSS and incubated with 1 μM FerroOrange at 37 °C for 30 min. The fluorescence intensity represents the content of Fe2+. Images were acquired with the Leica TCS SP8X confocal microscope.
In silico analysis
The GDAC database (http://gdac.broadinstitute.org/) was used to determine the expression of APOC1 in tumor and normal tissues. The Oncomine database (https://www.oncomine.com/) was used to determine the APOC1 expression in different types of glioma and other cancers. The Betastasis database (https://www.betastasis.com/) was used to compare the survival rates between GBM patients with high and low APOC1 expression. The GEPIA database (http://gepia.cancer-pku.cn/) was used to determine the APOC1 expression in different tissues from GBM and non-GBM subjects. The UALCAN database (http://ualcan.path.uab.edu/analysis.html) was used to analyze the data on APOC1 expression in normal and GBM patients from the TCGA database.
Western blotting
After treatment, cells were lysed in RIPA buffer (Applygen Technologies, Shanghai, China) containing protease and phosphatase inhibitors (Thermo Fisher Scientific, Carlsbad, CA, USA) on ice for 30 min. Total protein was collected by centrifugating the lysates at 12,000 × g for 15 min at 4 °C. Proteins were separated on a 10%–12% SDS polyacrylamide gel, then transferred to a polyvinylidene fluoride (PVDF) membrane and blocked in 5% skimmed milk for 2 h at RT. The membranes were incubated with primary antibodies at 4 °C overnight. The details about the antibodies are listed in Supplementary Table 3. After washing five times with TBST, the blots were incubated with HRP-conjugated secondary antibodies (7074, anti-rabbit IgG and 7076, anti-mouse IgG, Cell Signaling Technology, Danvers, USA). Target signals were detected using Super ECL (Applygen, Beijing, China) on a Tanon 4600 Imaging System (Tanon, Beijing, China).
Co-immunoprecipitation (CO-IP)
Cells were seeded in 100 mm plates and were harvested, suspended in 600 μL lysis buffer with PMSF for 30 min on ice, then centrifuged at 12,000 × g for 10 min at 4 °C. Supernatants were collected and 40 μL samples were mixed with 10 μL of protein A/G PLUS agarose (sc-2003, Santa Cruz Biotech, USA), 10 μL DDK-agarose (Bimake, Shanghai, China), respectively, then, rotated at 4 °C overnight. Afterwards, the beads were washed five times by centrifugation at 1500 r/min, for 5 min. The supernatants were discarded, the beads were mixed with an equal volume of 2× loading buffer, heated at 100 °C for 5 min, and processed for Western blotting analysis [25].
U87 glioma orthotopic assay in nude mice
Animal studies were performed according to the principles of the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Ethics Committee for Laboratory Animal Care and Use of the Institute of Materia Medica, Chinese Academy of Medical Science and Peking Union Medical College (Beijing, China). Athymic nude mice (17–19 g) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). U87-shNC, U87-shAPOC1-1 and U87-shAPOC1-2 cells were harvested and resuspended in phosphate buffered saline (PBS) to a concentration of 2 × 103 cells/μL. An aliquot of 5 μL cells was slowly injected into the right ventricle to a depth of 3.3 mm at a rate of 1 μL/min. After two weeks, MRI was performed to measure the tumor volume and brains were collected for analysis.
Statistical analysis
The results are represented as mean ± SD. Statistically significant differences between groups were determined by Student’s t test or one-way ANOVA using GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA). P < 0.05 was considered to be statistically significant.
Results
APOC1 is highly expressed in GBM patients
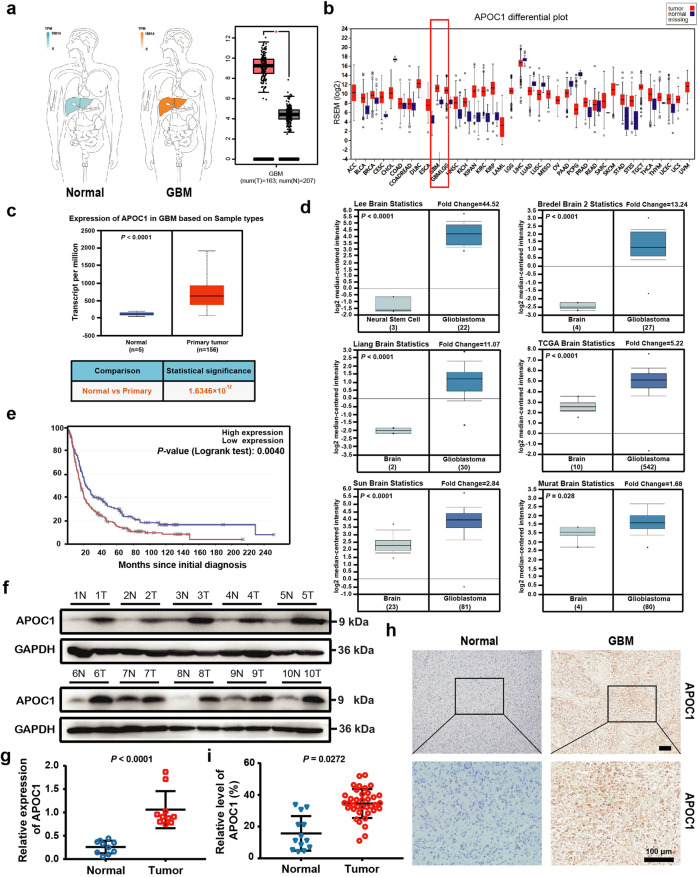
To determine the expression level of APOC1 in GBM patients and how it was associated with GBM progression, an in silico assay was carried out using data from the GDAC (http://gdac.broadinstitute.org/), GEPIA (http://gepia.cancer-pku.cn/) and Oncomine databases (https://www.oncomine.com/). It can be seen in Fig. 1a–d and Supplementary Fig. S1a that APOC1 was highly expressed in GBM, and there was significantly higher APOC1 expression in oligodendroglioma, anaplastic astrocytoma and GBM than in healthy brain tissue (Supplementary Fig. S1b, c). Survival analysis using the Betastasis database (https://www.betastasis.com/) showed that GBM patients with higher APOC1 expression had lower survival rates (Fig. 1e and Supplementary Fig. S1d). Although APOC1 was highly expressed in GBM patients, there was no significant difference in APOC1 expression between patients of different race, gender and age according to UALCAN data (Supplementary Fig. S1e). However, African American GBM patients with high expression of APOC1 did have lower survival rates (Supplementary Fig. S1f). To determine whether elevated expression of APOC1 protein in GBM tissue was associated with glioma progression, expression of APOC1 in ten paired samples of GBM tissues and adjacent normal brain tissues from GBM patients was assessed by Western blotting; APOC1 was highly expressed in GBM tissues compared with adjacent tissues (Fig. 1f, g). Immunohistochemistry (IHC) was carried out to measure expression of APOC1 in an array of 37 GBM patient tissues, and representative photos are shown in Fig. 1h. Results from IHC showed that expression of APOC1 was higher in GBM than adjacent normal brain tissues (Fig. 1h, i), and that there was higher APOC1 expression in high-grade glioma, especially GBM, than in low-grade glioma (Supplementary Fig. S1g, h). Thus, we concluded that APOC1 was highly expressed in patients with GBM and that the expression was closely related to the progression of GBM.
Fig. 1. APOC1 is highly expressed in GBM patients.
a There is higher APOC1 expression in GBM than that in normal tissues. APOC1 expression in tumor and normal tissues was analyzed in website (http://gepia.cancer-pku.cn/). b Expression of APOC1 is higher in GBM than normal tissues. Expression of APOC1 in tumor and normal tissues was analyzed in website (http://gdac.broadinstitute.org/). c APOC1 expression is high in GBM patients. Data of APOC1 expression in normal and GBM patients from TCGA database were analyzed in website (http://ualcan.path.uab.edu/analysis.html). d APOC1 is highly expressed in GBM. APOC1 expression between normal and GBM patients was analyzed in website (https://www.oncomine.org/). e Patients with high APOC1 expression have lower survival rates. Survival rate of GBM patients with high APOC1 expression and low APOC1 expression was analyzed in website (https://www.betastasis.com/). f APOC1 was highly expressed in GBM tissues than adjacent normal tissues. Western blotting analysis of APOC1 expression in GBM patients (n = 10). g Quantitative analysis of APOC1 expression in the GBM tissues and adjacent brain tissues. Bars indicates SD, P values represented the significant difference between GBM tissues and adjacent brain tissues, Student’s t test. h Immunohistochemical analysis of APOC1 expression in 37 glioma patient tissues and 13 adjacent normal tissues. Representative images showing the APOC1 protein in normal tissue and glioma tissue. Scale bar = 100 μm. i Quantitative analysis of APOC1 expression in the 37 glioma tissues and 13 normal tissues. Bars indicates SD, P values represented the significant difference between normal tissues and GBM tissues, Student’s t test.
APOC1 promotes tumorigenicity of GBM cells
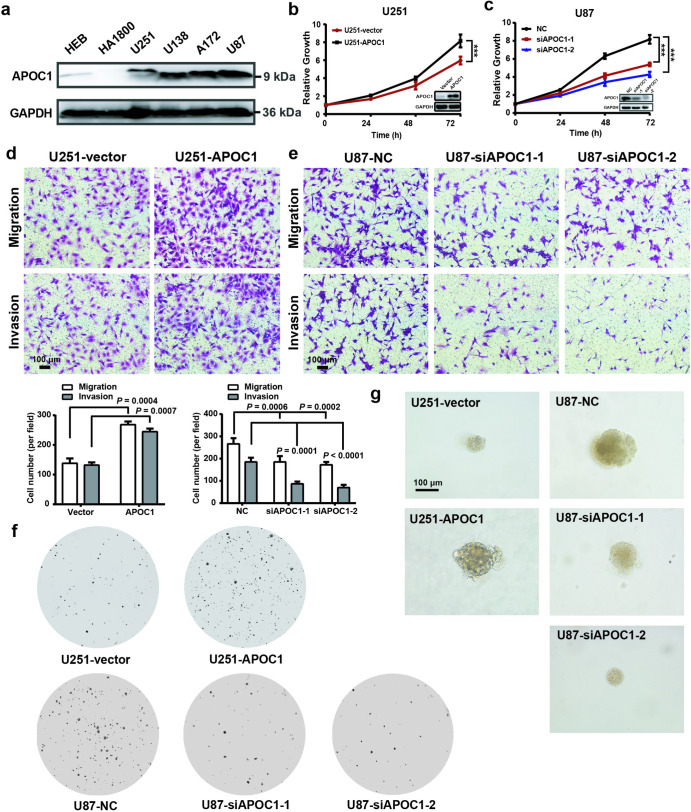
To explore the function of APOC1 in GBM, the expression APOC1 in two normal brain glial cell lines and four GBM cell lines was assessed by Western blotting. APOC1 expression in GBM cell lines was higher than in normal brain cell lines (Fig. 2a). U251 cells were transfected with an APOC1 expression plasmid, stable clones with high APOC1 expression were selected and the effects of APOC1 overexpression were determined (Supplementary Fig. S2a). In contrast, APOC1 expression in U87 cells was efficiently knocked down by APOC1 siRNAs (Supplementary Fig. S2b). Assays of proliferation, migration and invasion, colony formation and growth in 3D Matrigel were carried out to explore the effects of APOC1 on GBM. APOC1 overexpression promoted proliferation of U251 cells while knockdown inhibited U87 cells proliferation (Fig. 2b, c). Overexpression of APOC1 significantly enhanced migration and invasion of U251 cells whereas inhibiting APOC1 expression reduced migration and invasion of U87 cells (Fig. 2d, e). In addition, overexpression of APOC1 promoted U251 cells colony formation while downregulation of APOC1 reduced colony formation of U87 cells (Fig. 2f). As shown in Fig. 2g, APOC1 overexpression enhanced the growth of U251 cells while APOC1 knockdown inhibited growth of U87 cells in 3D Matrigel. Overall, these results suggest that APOC1 plays a key functional role in GBM development.
Fig. 2. APOC1 improved proliferation, migration, invasion, and colony formation of GBM cells.
a APOC1 expression in normal glial cells (HEB and HA1800) and GBM cells (U251, U138, A172 and U87). b Overexpression of APOC1 increased proliferation of U251 cells in 24, 48, and 72 h. c Knockdown of APOC1 inhibited proliferation of U87 cells in 24, 48, and 72 h. d Overexpression of APOC1 improved the migration and invasion of U251-APOC1 cells. Scale bar = 100 μm. e Knockdown of APOC1 inhibited the migration and invasion ability of U87-siAPOC1 cells. Scale bar = 100 μm. f Overexpression of APOC1 improved the colony formation of U251-APOC1 cells whereas knockdown of APOC1 inhibited colony formation of U87-siAPOC1 cells. g 3D matrigel assay showed overexpression of APOC1 promoted growth of U251-APOC1 cells whereas knockdown of APOC1 inhibited growth of U87-siAPOC1 cells in 3D Matrigel. Scale bar = 100 μm. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test or one-way ANOVA, ***P < 0.001 vs. NC.
APOC1 knockdown increases ferroptosis in GBM cells
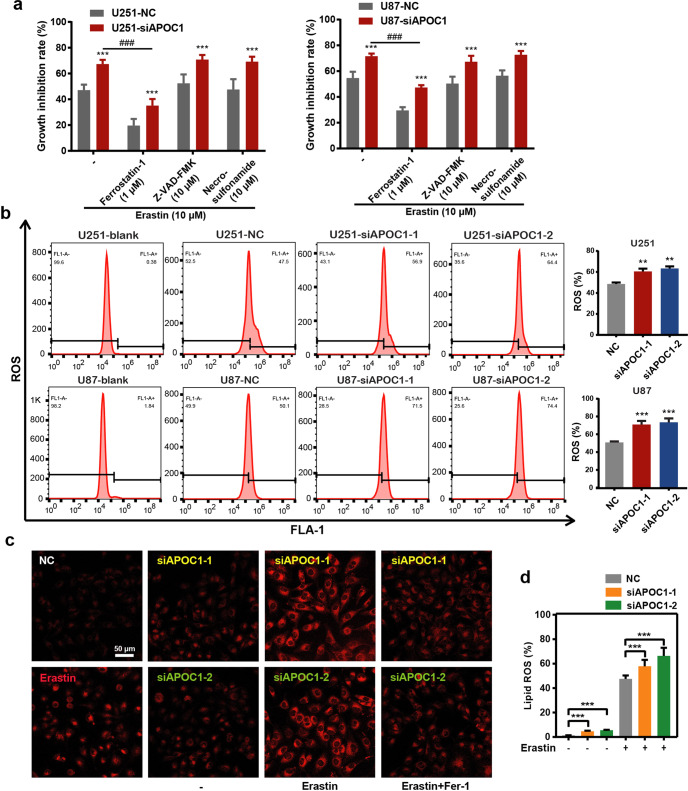
Many recent studies have shown that ferroptosis is associated with cancer progression. To determine whether APOC1 influenced ferroptosis in GBM, Erastin (ferroptosis inducer), Ferrostatin-1 (Fer-1) (ferroptosis inhibitor), Z-VAD-FMK (apoptosis inhibitor), and Necrosulfonamide (Nec) (necrosis inhibitor) were used to treat U87-NC, U87 siAPOC1 cells, U251-NC and U251 siAPOC1 cells. The morphology of U251 cells undergoing ferroptosis (with Erastin for 24 h) is shown in Supplementary Fig. S2c. Results showed that cell growth was inhibited by ferroptosis, which could be reversed by Fer-1 but not Z-VAD-FMK or Necrosulfonamide. APOC1 knockdown significantly increased ferroptosis of cells induced by Erastin (Fig. 3a). These results confirmed that APOC1 downregulation increased ferroptosis in U87 and U251 cells, but did not affect apoptosis or necroptosis. It is well known that oxidative stress from excess ROS and accumulation of lipid ROS is a trigger for ferroptosis. To explore the potential link between APOC1 expression and ROS in cells, ROS levels were determined by flow cytometry. APOC1 knockdown increased ROS accumulation in U87 and U251 cells (Fig. 3b), whereas APOC1 overexpression reduced ROS levels in U251 cells (Supplementary Fig. S2d). The concentration of Fe2+ was increased in Erastin-treated U251-siAPOC1 cells compared to untreated cells, which was blocked by Fer-1 (Fig. 3c). APOC1 downregulation was also found to increase lipid ROS levels (Fig. 3d). Altogether, these results verified that knockdown of APOC1 expression increases both Fe2+ concentration and ROS level.
Fig. 3. Knockdown of APOC1 expression significantly increased ferroptosis of GBM cells induced by Erastin through increasing Fe2+ concentration and ROS level.
a Knockdown of APOC1 promoted ferroptosis of U87 and U251 cells induced by Erastin. Cells were treated with Erastin (10 μM) with or without ferroptosis inhibitor, Ferrostatin-1 (1 μM), apoptosis inhibitor, Z-VAD-FMK (10 μM), necrosis inhibitor, Necrosulfonamide (10 μM) for 24 h. b Knockdown of APOC1 increased ROS production in U87 and U251 cells. Quantitative analysis of ROS level in indicated treatment U251 and U87 cells. c Knockdown of APOC1 increased intracellular Fe2+ in U251 cells. Representative images of Fe2+ in three assays were shown. Scale bar = 50 μm. d Knockdown of APOC1 increased lipid ROS level in U87 cells. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test or one-way ANOVA, **P < 0.01, ***P < 0.001 vs. NC.
APOC1 knockdown increases ROS levels by reducing expression of HO-1 and NQO1
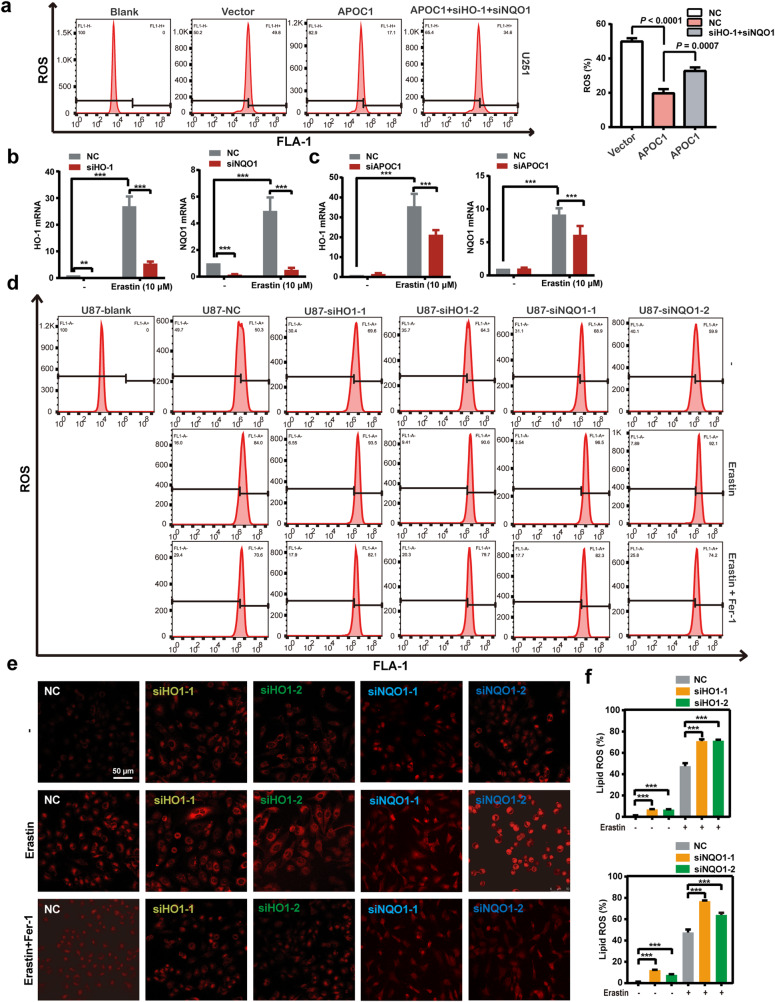
HO-1 and NQO1 are proteins involved with oxidative stress and the regulation of intracellular ROS production. The above data showed that knockdown of APOC1 expression increased ROS levels. To determine if there was a link between APOC1 level and HO-1, NQO1 in the accumulation of cellular ROS, HO-1 and NQO1 were knocked down by siHO-1 and siNQO1 in APOC1-overexpressing U251 cells and the changes in ROS level were measured by flow cytometry. Overexpression of APOC1 decreased ROS level, but this could be blocked by knockdown of HO-1 and NQO1 (Fig. 4a). Ferroptosis induction with Erastin upregulated HO-1 and NQO1 mRNA levels in U87 cells, which could be blocked by specific HO-1 or NQO1 siRNAs (Fig. 4b). Knockdown of APOC1 significantly reduced HO-1 and NQO1 mRNA levels in cells undergoing ferroptosis (Fig. 4c), which suggests that APOC1 may regulate HO-1 and NQO1 expression in ferroptosis. To investigate whether HO-1 and NQO1 expression were related to the negative regulation of ROS in ferroptosis, siHO-1 or siNQO1 were used to inhibit expression of HO-1 and NQO1, respectively, and ROS level was checked by flow cytometry. Knockdown of HO-1 or NQO1 expression increased ROS level in cells and Erastin-mediated induction of ferroptosis further increased ROS accumulation, which could be reversed by Fer-1 in U87 and U251 cells (Fig. 4d and Supplementary Fig. S3a-c). Reduction of HO-1 or NQO1 expression increased Fe2+ concentration (Fig. 4e) and lipid ROS (Fig. 4f) in cells treated with Erastin. Taken together, these results demonstrate that APOC1 knockdown increases ROS production by reducing expression of HO-1 and NQO1.
Fig. 4. Knockdown of APOC1 promoted ferroptosis by reduced expression of HO-1 and NQO1 expression.
a Overexpression of APOC1 attenuated ROS level which can be reversed by knockdown of HO-1 and NQO1 expression in U251 cells. b mRNA expression of HO-1 and NQO1 was increased in U87 cells after treatment of Erastin for 24 h. c Knockdown of APOC1 expression decreased HO-1 and NQO1 mRNA level in U87 cells treated by Erastin for 24 h. d Knockdown of HO-1 or NQO1 expression increased ROS level of U87 cells treated by Erastin for 24 h. e Knockdown of HO-1 or NQO1 increased intracellular Fe2+ of U251 cells in Erastin-induced ferroptosis. Representative images of Fe2+ with identical results in three assays were shown. Scale bar = 50 μm. f Knockdown of HO-1 or NQO1 increased lipid ROS level of U87 cells. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test or one-way ANOVA, **P < 0.01, ***P < 0.001 vs. NC.
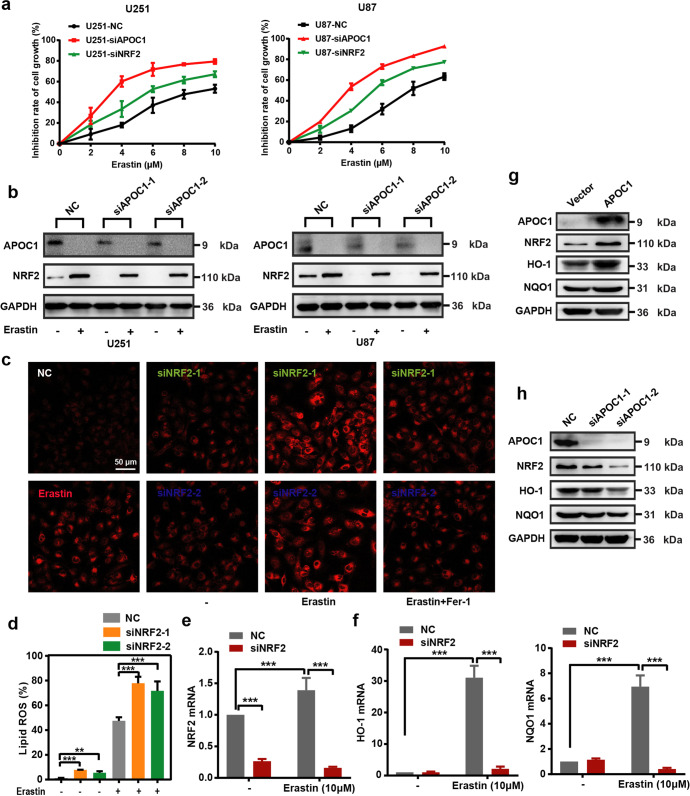
APOC1 knockdown confers sensitivity to Erastin by reducing NRF2-mediated HO-1 and NQO1 expression
NRF2 is a transcription factor that regulates heme oxygenase-1 (HO-1) and quinone oxidoreductase 1 (NQO1) expression and suppresses ferroptosis [26]. To investigate whether APOC1 regulated ferroptosis by controlling NRF2 expression, APOC1 and NRF2 expression were knocked down by APOC1-siRNA and NRF2-siRNA respectively, and sensitivity of U87 cells and U251 cells to Erastin was evaluated. Knockdown of APOC1 or NRF2 increased sensitivity of U87 and U251 cells to Erastin in a dose-dependent manner (Fig. 5a). U87 and U251 cells undergoing ferroptosis had higher NRF2 expression compared with control cells, and this could be attenuated by inhibiting APOC1 expression (Fig. 5b). NRF2 downregulation also increased the Fe2+ level in Erastin-treated U251 cells (Fig. 5c), and significantly increased lipid ROS levels in U87 cells undergoing ferroptosis (Fig. 5d). These results supported the hypothesis that inhibition of NRF2 expression made U87 and U251 cells more sensitive to Erastin. To further explore how APOC1 knockdown increased ferroptosis of cells through its effects on the NRF2 pathway, the downstream targets of NRF2, HO-1 and NQO1, were quantitated by Western blotting. Ferroptosis induction by Erastin significantly increased the expression of HO-1 and NQO1 in cells, while NRF2 knockdown inhibited the Erastin-induced increase of HO-1 and NQO1 mRNA, which is similar to the effect of APOC1 (Fig. 5e, f). APOC1 upregulation improved expression of NRF2, HO-1 and NQO1 (Fig. 5g), whereas knockdown of APOC1 reduced expression of NRF2, HO-1 and NQO1 (Fig. 5h). Collectively, these results are consistent with the idea that APOC1 expression contributes to ferroptosis resistance by stimulating the NRF2/HO-1 and NQO1 pathway.
Fig. 5. Knockdown of APOC1 conferred sensitivity of cells to Erastin by reducing NRF2-mediated HO-1 and NQO1 expression.
a Knockdown of APOC1 or NRF2 in U87 and U251 cells increased cell sensitivity to Erastin. b Knockdown of APOC1 decreased expression of NRF2 protein in cells treated by Erastin. The experiments were performed in triplicate. c Knockdown of NRF2 increased intracellular Fe2+ in U251 cells. Representative images of Fe2+ with identical results in three assays were shown. Scale bar = 50 μm. d Knockdown of NRF2 increased lipid ROS level of U87 cells in Erastin-induced ferroptosis. e, f NRF2 regulated HO-1 and NQO1 expression during ferroptosis in U87 cells. g Overexpression of APOC1 upregulated NRF2, HO-1 and NQO1 expression in U251 cells. h Knockdown of APOC1 reduced protein expression of NRF2, HO-1 and NQO1 in U87 cells. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test or one-way ANOVA, **P < 0.01, ***P < 0.001 vs. NC.
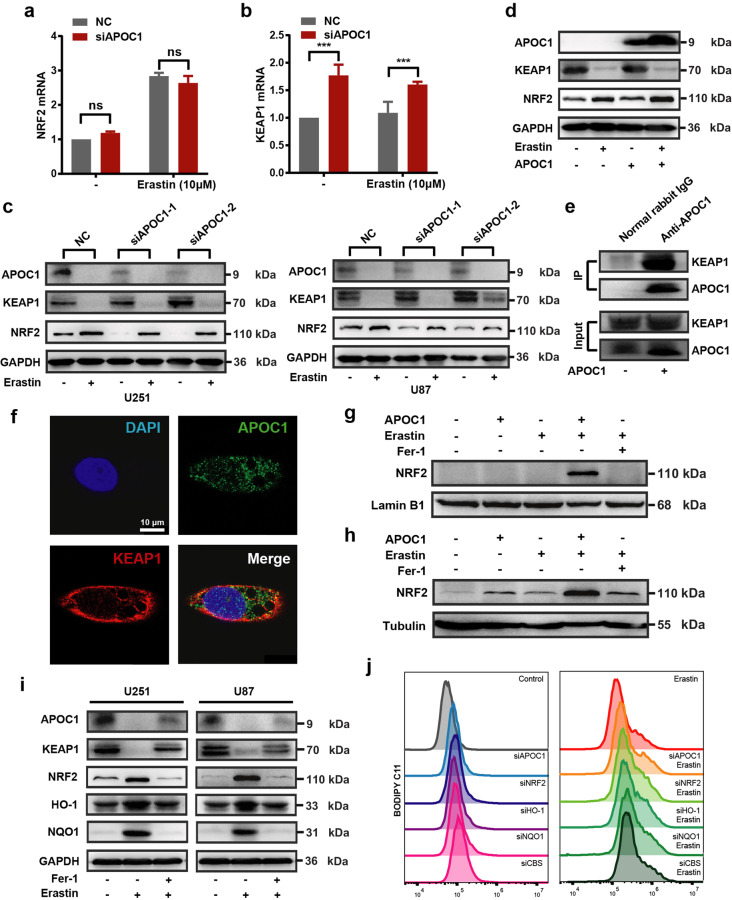
APOC1 promotes translocation of NRF2 via the KEAP1-NRF2 pathway
Fan et al. showed that NRF2 diminished ferroptosis through the KEAP1-NRF2 pathway, thereby promoting cell proliferation in GBM [23]. Overexpression of KEAP1 induced NRF2 degradation and reduced its translocation [22]. To further elucidate the effects of APOC1 knockdown on Erastin-induced ferroptosis through the KEAP1/NRF2/HO-1 and NQO1 pathways, the expression of KEAP1 and NRF2 was measured in U87-siAPOC1 cells. There was no significant difference in NRF2 mRNA expression between U87-NC cells and U87-siAPOC1 cells, whether treated with Erastin or not (Fig. 6a); knockdown of APOC1 significantly increased KEAP1 mRNA expression (Fig. 6b). Expression of NRF2 protein was significantly increased in cells treated with Erastin (Fig. 6c). APOC1 knockdown increased KEAP1 protein levels (Fig. 6c) while APOC1 overexpression reduced expression of KEAP1 and improved expression of NRF2 (Fig. 6d). The co-IP assay showed that APOC1 bound to KEAP1 (Fig. 6e), and immunofluorescence data showed that overlap between APOC1 and KEAP1 was increased in Erastin-treated cells (Fig. 6f and Supplementary Fig. S4), which indicated that APOC1 might interact strongly with KEAP1 in cells undergoing ferroptosis. Thus, APOC1 could increase NRF2 level by regulating KEAP1 expression. Overexpression of APOC1 enhanced translocation of NRF2 from cytoplasm to nucleus (Fig. 6g, h). In cells undergoing Erastin-mediated ferroptosis, the expression of NRF2, HO-1 and NQO1 was increased while KEAP1 expression was reduced (Fig. 6i). The lipid ROS level was significantly increased in Erastin-treated U87 cells subjected to APOC1, NRF2, HO-1 or NQO1 knockdown (Fig. 6j). Taken together, these results showed that knockdown of APOC1 increased KEAP1 expression, which inhibited NRF2 translocation and accelerated the ferroptosis process.
Fig. 6. APOC1 promote translocation of NRF2 by KEAP1-NRF2 pathway.
a, b Knockdown of APOC1 increased mRNA expression of KEAP1 but did not change mRNA expression of NRF2 in U87 cells treated by Erastin for 24 h. c Knockdown of APOC1 increased protein expression of KEAP1 and attenuated NRF2 expression in U87 and U251 cells treated by Erastin for 24 h. d Overexpression of APOC1 enhanced NRF2 expression while decreased KEAP1 protein expression in U251 cells. e Co-Immunoprecipitation assay showed APOC1 can interact with KEAP1. f Immunofluorescence images of APOC1 and KEAP1 in U251 cell. Green for APOC1 and red for KEAP1. Scale bar = 10 μm. Data are representative images of three assays with identical results. g, h APOC1 promotes nuclear translocation of NRF2. i Expression of APOC1, KEAP1, NRF2, HO-1 and NQO1 in U87 and U251 cells treated by Erastin with or without Fer-1 for 24 h. j Representative image of lipid ROS with identical results in three assays was shown. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test, ***P < 0.001, n.s. nonsignificant vs. NC.
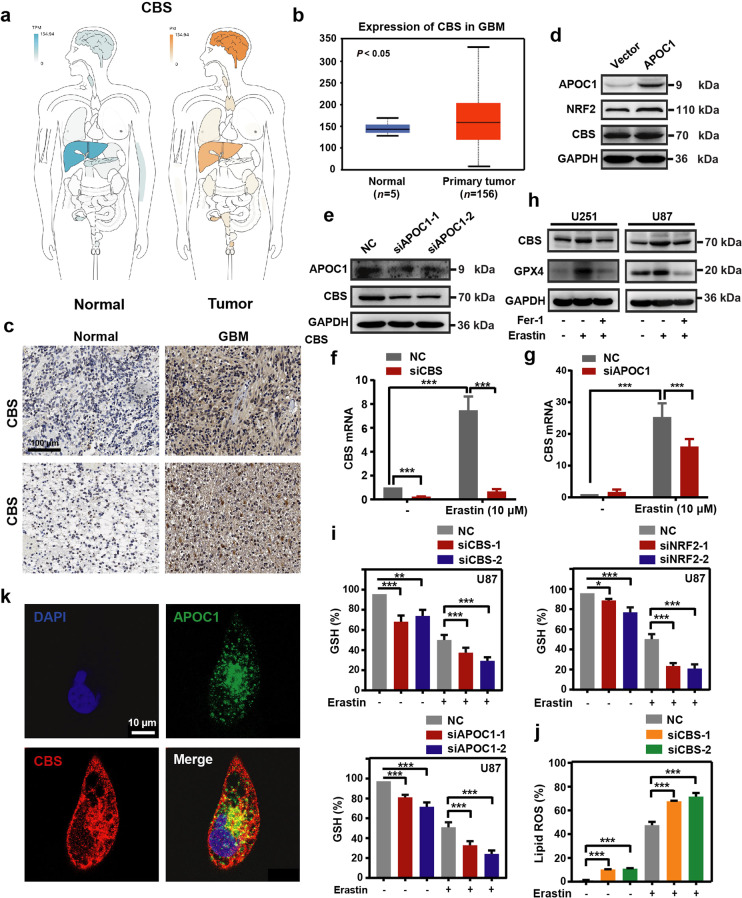
Upregulation of APOC1 enhanced cystathionine beta-synthase expression that protected GBM cells from ferroptosis
Cystathionine beta-synthase (CBS) is a key enzyme in cysteine synthesis, which catalyzes the first step in the trans-sulfuration pathway [27] and plays an important part in the ferroptosis process [28]. In our previous data, comparison of RNA-Seq data from U251- APOC1 with U251 transfected with empty vector showed that expression of CBS was significantly increased when APOC1 was overexpressed. To determine the expression of CBS in GBM and explore the role of CBS in ferroptosis and the relationship between APOC1 and CBS, bioinformatics analysis was carried out using the GEPIA and Oncomine databases. We found higher CBS expression in GBM than in normal tissues (Fig. 7a, b and Supplementary Fig. S5a). Results from IHC also showed that CBS was highly expressed in GBM brain tissues than adjacent normal brain tissues (Fig. 7c). Overexpression of APOC1 increased the mRNA and protein expression of CBS (Fig. 7d and Supplementary Fig. S5b), while knockdown of APOC1 significantly reduced CBS expression (Fig. 7e). Compared with U87-NC, treatment with Erastin significantly increased the CBS mRNA level and Erastin-induced upregulation of CBS was blocked by specific siRNA interference (Fig. 7f). The knockdown efficiency of siCBS was shown in Supplementary Fig. S5c. Knockdown of APOC1 significantly reduced the mRNA level of CBS in Erastin-treated cells (Fig. 7g). It was shown in Fig. 7h that Erastin significantly increased the levels of CBS and GPX4 protein, which could be blocked by Fer-1. Moreover, knockdown of CBS expression significantly reduced GSH level of U87 cells (Fig. 7i and Supplementary Fig. S5d). Knockdown of NRF2 expression reduced CBS expression (Supplementary Fig. S5e) and downregulation of APOC1 and NRF2 mRNA also decreased GSH level (Fig. 7i and Supplementary Fig. S5d), which means that CBS may be regulated by the APOC1/NRF2 pathway. Knockdown of CBS increased Fe2+ concentration (Supplementary Fig. S5f) and lipid ROS (Fig. 7j). Moreover, APOC1 may interact with CBS and the overlap between APOC1 and CBS was increased in cells treated by Erastin (Fig. 7k and Supplementary Fig. S5g). These results together are consistent with the hypothesis that CBS is involved in ferroptosis, and that it may be regulated directly by APOC1 or through activation of APOC1/NRF2.
Fig. 7. CBS participates in ferroptosis resistance which can be regulated by APOC1.
a, b CBS is highly expressed in brain of GBM. CBS expression in GBM was analyzed in GEPIA database (http://gepia.cancer-pku.cn/) (a) and Oncomine database (https://www.oncomine.org/) (b). c Immunohistochemical analysis of CBS expression in GBM patients. Representative images showed expression of CBS protein was higher in GBM than adjacent normal tissues. Scale bar = 100 μm. d Overexpression APOC1 improved CBS expression in U251 cells. e Knockdown of APOC1 decreased CBS expression in U87 cells. f mRNA level of CBS increased in Erastin-induced ferroptosis for 24 h. g Knockdown of APOC1 decreased mRNA expression of CBS in cells treated by Erastin for 24 h. h CBS and GPX4 were highly expressed in U87 and U251 cells treated by Erastin for 24 h. i Knockdown of APOC1, NRF2, or CBS attenuates GSH level in Erastin-induced ferroptosis in U87 cells. j Knockdown of CBS increases lipid ROS level in U87 cells treated by Erastin for 24 h. k Immunofluorescence images of APOC1 and CBS. Green for APOC1 and red for CBS. Scale bar = 10 μm. Data are representative images of three assays with identical results. Experiments were performed in triplicate. Data are presented as mean ± SD (n = 3). Statistical significance was determined by Student’s t test or one-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001 vs. NC.
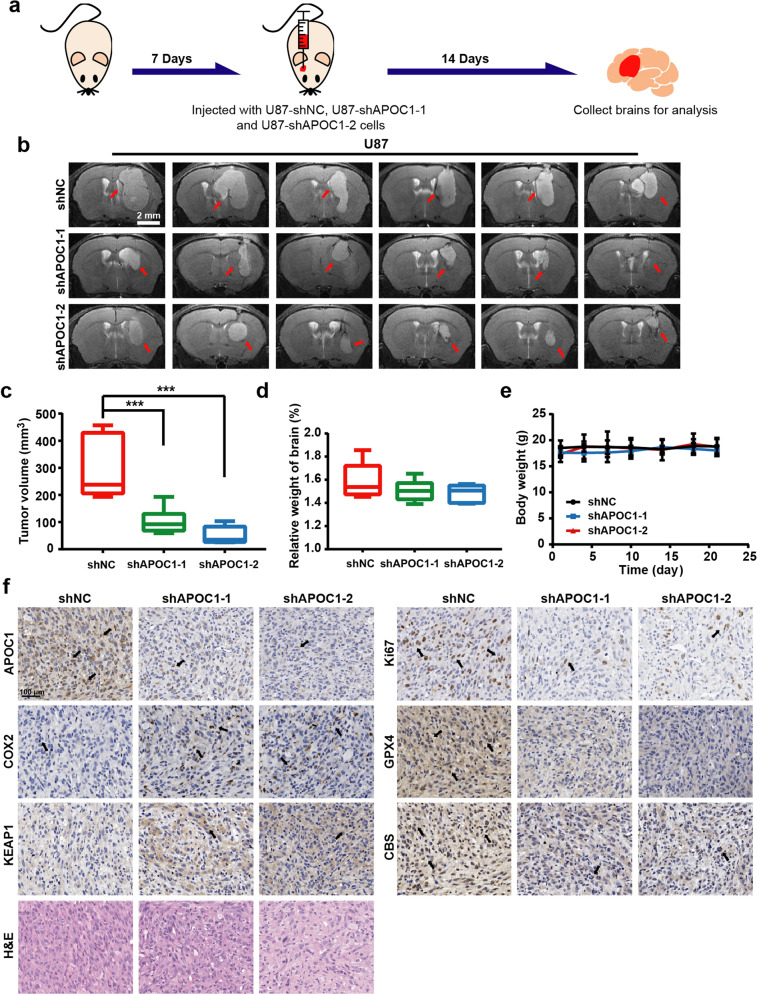
APOC1 knockdown suppresses GBM tumor growth and enhances sensitivity of cells to ferroptosis in vivo
To investigate the effects of APOC1 on the growth of GBM in vivo, U87-shNC cells, U87-shAPOC1-1 cells, and U87-shRNA-2 cells were used to produce orthotopic xenograft tumors in the right brains of athymic nude mice and MRI was performed to monitor the tumor volume throughout the experiment (Fig. 8a). The U87-shNC cells grew more aggressively in the brain than U87-shAPOC1-1 cells and U87- shAPOC1-2 cells, which confirmed the in vitro results, showing that APOC1 knockdown significantly inhibited growth of GBM cells (Fig. 8b). The final tumor volume and relative brain weight of mice in the U87-shAPOC1-1 and U87-shAPOC1-2 groups were significantly lower than those of mice in the U87-shNC group (Fig. 8c, d). No obvious weight loss or abnormal behavior was observed (Fig. 8e). By H&E staining, it was shown that the tumor tissue in the NC group was denser than that in the APOC1 knockdown group (Fig. 8f). COX2 encoded by PTGS2 was highly expressed in ferroptosis tissue [29, 30]. Our IHC results also showed higher COX2 expression in the siAPOC1 group than in the shNC group. Protein level of Ki67, GPX4, and CBS was decreased, while KEAP1 was increased in the shAPOC1 group (Fig. 8f), which was consistent with the above results. These data suggested that knockdown of APOC1 suppressed tumorigenesis and enhanced sensitivity of GBM to ferroptosis by regulating both the KEAP1 and CBS pathways in vivo.
Fig. 8. Down-regulation of APOC1 inhibited GBM growth in vivo.
a Schematic of APOC1 function in orthotopic GBM tumor growth in vivo. b Representative MRI images of intracranial tumors from U87-shNC, U87-shAPOC1-1, and U87-shRNA-2. Scale bar = 2 mm. c Tumor volumes of U87 orthotopic model. The tumor volume was calculated with formula: V tumor volume = L maximum length × W maximum width × T thickness of the tumor slice. d Relative weight of brain in U87-shNC, U87-shAPOC1-1, and U87-shRNA-2 group. e Body weight change of U87-shNC, U87-shAPOC1-1, and U87-shRNA-2 group. f H&E staining and immunohistochemical results of APOC1, Ki67, COX2, KEAP1, CBS, and GPX4 expression in brain tissues of GBM orthotopic nude mice model. Representative images of each protein with identical results were shown in the shNC, shAPOC1-1, and shAPOC1-2 tumor tissues. Scale bar = 100 μm. Data are presented as mean ± SD (n = 6). Statistical significance was determined by one-way ANOVA, ***P < 0.001 vs. shNC.
Discussion
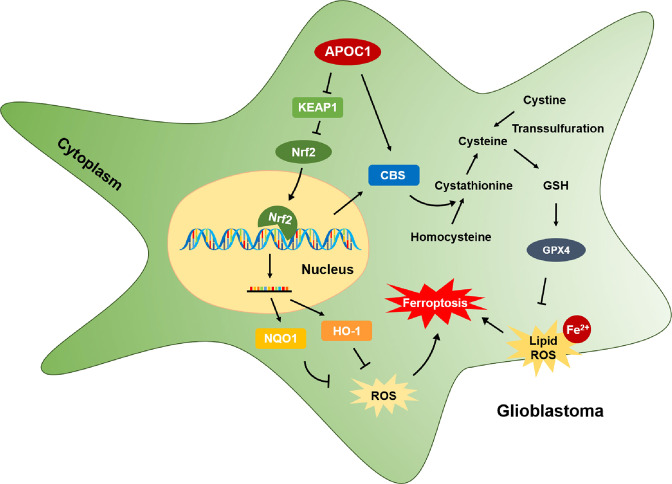
GBM is a severe disease of central nervous system with high mortality and poor prognosis. Since the molecular mechanism of GBM is still unclear and currently there is no good therapeutic target, GBM is difficult to treat successfully owing to its high rate of metastasis and recurrence. Thus, the search for novel, effective therapeutic targets and exploring the molecular mechanisms of the related pathways is of great significance for diagnosis and treatment of patients with GBM. In this study, we found that APOC1 was highly expressed in GBM tissues compared with adjacent normal brain tissues. APOC1 was also highly expressed in GBM cell lines compared with normal brain cell lines. Overexpression of APOC1 promoted tumorigenicity by increasing proliferation, migration, invasion and colony formation of GBM cells; knockdown of APOC1 inhibited growth of GBM cells and xenografted tumors. These results suggest that APOC1 may play an important role in the proliferation and metastasis of GBM. This study is also the first to reveal that knockdown of APOC1 could increase the level of ROS and lipid ROS and promote ferroptosis in cells treated with Erastin through regulation of the KEAP1/NRF2/HO-1 and NQO1 pathways and the CBS/GPX4 axis (Fig. 9). In light of these findings, it seems highly likely that APOC1 could be a potential drug target for GBM therapy.
Fig. 9. Mechanism of APOC1 in ferroptosis regulation of GBM.
APOC1 reduced the sensitivity of GBM cells to ferroptosis by up-regulating the KEAP1/NRF2/HO-1 and NQO1 pathway and the CBS/GPX4 axis, which finally decreasing intracellular ROS and Fe2+ level.
Studies have shown that targeting ferroptosis may become a new strategy for inhibiting the progression of GBM [31, 32]. Ferroptosis is a newly discovered form of regulated cell death different from apoptosis and necrosis. It is defined as an iron- and ROS-dependent form of regulated necrosis characterized by lipid peroxidation [33]. Compared with traditional anticancer therapies, the current body of evidence offers proof that triggering ferroptosis may be an effective new way to eradicate cancers, especially those aggressive malignancies with multiple drug resistance [34, 35]. Researchers have designed and developed several anticancer drugs that cause ferroptosis induction and are testing them on various malignancies including GBM [36]. For example, combining temozolomide with the ferroptosis inducer Erastin can enhance the effect of temozolomide on glioblastoma by increasing sensitivity of GBM cells to ferroptosis through inhibiting the cystine-glutamate anti-transporter system Xc- [37]. Some genes have been found to confer ferroptosis resistance on cancer cells and targeting these genes may become a new therapy for malignancies that are resistant to traditional therapies. Our study is the first to show how APOC1 influenced growth and metastasis of GBM cells through intervention with the ferroptosis process. Knockdown of APOC1 increased the level of Fe2+ and lipid ROS in cells. The NRF2 signaling pathway is a defensive transduction pathway that reduces oxidative stress and the effects of chemical stimulation [38, 39]. NRF2 is an important inhibitor of ferroptosis because it can inhibit cellular uptake of iron and limit ROS production [40]. KEAP1 is part of the E3 ubiquitin ligase which regulates the activity of NRF2 by mediating its ubiquitination and proteasome-dependent degradation. When cells undergo oxidative stress, the cysteine sensor within KEAP1 drives NRF2 to escape from ubiquitination through a complex molecular mechanism [41]. NRF2 will then translocate from the cytoplasm into the nucleus and initiate the transcription of antioxidant proteins. HO-1 and NQO1 are downstream targets of the NRF2 signaling pathway [42]. Sun et al. found that the P62/KEAP1/NRF2 pathway had a vital part in ROS-mediated ferroptosis by upregulating downstream anti-oxidation proteins [22]. In this study, we elucidated the role and mechanism of APOC1 in ferroptosis of GBM cells, and showed that knockdown of APOC1 attenuated the expression of HO-1and NOQ1 which caused an increase of intracellular Fe2+ and lipid ROS. We further clarified the role that the APOC1-mediated KEAP1-NRF2 signaling pathway plays in neutralizing ferroptosis to protect GBM cells from death. Knockdown of APOC1 increased KEAP1 expression by promoting KEAP1 transcription. Interestingly, both the mRNA and protein levels of KEAP1 were increased in U87-siAPOC1 compared with U87-siNC, whereas the protein level of NRF2 was decreased with unchanged mRNA level. This reveals that downregulation of NRF2 protein by knockdown of APOC1 was caused by KEAP1 mediated NRF2 degradation rather than by directly altering the transcription of NRF2. The transcription of KEAP1 did not change in ferroptosis, but the protein level decreased significantly, which suggests that a large amount of KEAP1 protein was degraded during ferroptosis; APOC1 knockdown increased the KEAP1 protein level in ferroptosis. Results of CO-IP and immunofluorescence assays also showed that APOC1 can interact with KEAP1. Therefore, it seems likely that inhibition of APOC1 promotes the expression of KEAP1 by reducing its degradation; but the mechanism of how APOC1 affects the degradation of KEAP1 remains to be determined. The translocation of NRF2 from cytoplasm to nucleus was significantly increased during ferroptosis in cells with high APOC1 expression. The above results indicated that APOC1 can interact with KEAP1 and inhibit NRF2 degradation, thereby increasing the level of NRF2 and promoting the translocation of NRF2 from nucleus to cytoplasm, facilitating the expression of the antioxidant proteins, HO-1 and NQO1. Therefore, we hypothesized that APOC1 conferred ferroptosis resistance on GBM cells though its action on the KEAP1-NRF2-mediated antioxidant pathway.
Besides the aforementioned genes, some systems were also involved in ferroptosis, such as system XC-, the glutathione (GSH) synthesis system [43, 44]. Cysteine is a key compound required for the synthesis of glutathione (GSH), which is the main antioxidant for maintaining redox homoeostasis [45, 46]. GSH metabolism and anti-oxidant capacity regulates sensitivity to ferroptosis. Cellular cysteine for GSH synthesis can be obtained by cells in two ways: cystine imported from extracellular sources by system XC- and converted to cysteine, and synthesis by CBS-mediated trans-sulfurization [47]. System XC- plays an important role in ferroptosis by exerting material exchange between the extracellular and intracellular milieus. SLC3A2 and SLC7A11 are two components of the disulfide-linked heterodimers of system XC- that import extracellular cystine in exchange for intracellular glutamate [48]. Although, SLC7A11 was reported to be a downstream target of NRF2 [49, 50], we did not find any change in SLC7A11 expression in the APOC1-mediated NRF2 pathway in GBM cells. Therefore, we concluded that APOC1 did not affect ferroptosis progression of GBM cells through system XC-. GSH is the substrate of GPX4, which reduces lipid peroxides [51]. CBS is the gene encoding cystathionine-β-synthetase, which catalyzes the first step in the homocysteine trans-sulfurization pathway, and irreversibly converts homocysteine into cysteine [52]. It has been reported that CBS is involved in ferroptosis resistance of cancers, such as ovarian [47], neuroblastoma [53], and breast [54]. Here we reported that both APOC1 and CBS were highly expressed in GBM, and that overexpression of APOC1 significantly increased both mRNA and protein expression of CBS. APOC1 is not a transcription factor, so the upregulation of CBS may be associated with the APOC1-mediated NRF2 pathway. Immunofluorescence data indicated that the overlap between APOC1 and CBS was high, which can be enhanced in Erastin-induced ferroptosis. Knockdown of CBS also reduced intracellular GSH levels and increased lipid ROS. These results suggest that APOC1 may increase CBS expression by NRF2 activation or directly interacting with CBS to promote synthesis of GSH and upregulate GPX4, which reduces lipid peroxidation and leads to ferroptosis resistance.
Conclusions
In summary, we discovered that APOC1 was highly expressed in both GBM cells and tissues of patients with GBM. Overexpression of APOC1 promoted GBM tumorigenesis by increasing cell proliferation, migration, invasion and colony formation. APOC1 reduced ROS levels and conferred ferroptosis resistance by activating NRF2 and its target genes, NQO1 and HO-1, and interacting with KEAP1. APOC1 also contributed to ferroptosis resistance through the APOC1/KEAP1/NRF2 and APOC1/CBS/GPX4 pathways. Together, these results provide a clear indication that APOC1 may be an effective drug target in GBM.
Supplementary information
Acknowledgements
This research was funded by Beijing Natural Science Foundation (7212157), CAMS Innovation Fund for Medical Sciences (2021-I2M-1-029), National Natural Science Foundation of China (81703536, 81803584, 81703565), Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025, 2018ZX09711001-012), Peking Union Medical College Graduate Innovation Fund (2019-1007-07).
Author contributions
JHW, YW and GHD developed the hypothesis, designed the experiments, and revised the manuscript. XJZ and WLC conducted all experiments and wrote the main manuscript. WL, WQF, JYL and LWR collected data. SL, BBG, YHY, YZZ, HY performed the statistical analysis. JY provided the study materials and patients. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this article was revised: In this article fig. 4 has been updated.
These authors contributed equally: Xiang-jin Zheng, Wen-lin Chen.
Change history
8/16/2022
A Correction to this paper has been published: 10.1038/s41401-022-00969-5
Change history
5/13/2024
A Correction to this paper has been published: 10.1038/s41401-024-01271-2
Contributor Information
Yu Wang, Email: ywang@pumch.cn.
Jin-hua Wang, Email: wjh@imm.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41401-022-00917-3.
References
- 1.Chen R, Smith-Cohn M, Cohen A, Colman H. Glioma Subclassifications and Their Clinical Significance. Neurotherapeutics: the journal of the American Society for Experimental. NeuroTherapeutics. 2017;14:284–97. doi: 10.1007/s13311-017-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–20. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma–are we there yet? Neuro Oncol. 2013;15:4–27. doi: 10.1093/neuonc/nos273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar AA, Abraham Koshy A. Regression of recurrent high-grade glioma with temozolomide, dexamethasone, and levetiracetam: case report and review of the literature. World Neurosurg. 2017;108:990.e11–e16. doi: 10.1016/j.wneu.2017.08.136. [DOI] [PubMed] [Google Scholar]
- 5.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 6.McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34:981–98. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Jong MC, van Dijk KW, Dahlmans VE, Van der Boom H, Kobayashi K, Oka K, et al. Reversal of hyperlipidaemia in apolipoprotein C1 transgenic mice by adenovirus-mediated gene delivery of the low-density-lipoprotein receptor, but not by the very-low-density-lipoprotein receptor. Biochem J. 1999;338:281–7. doi: 10.1042/bj3380281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jong MC, Dahlmans VE, van Gorp PJ, van Dijk KW, Breuer ML, Hofker MH, et al. In the absence of the low density lipoprotein receptor, human apolipoprotein C1 overexpression in transgenic mice inhibits the hepatic uptake of very low density lipoproteins via a receptor-associated protein-sensitive pathway. J Clin Invest. 1996;98:2259–67. doi: 10.1172/JCI119036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takano S, Yoshitomi H, Togawa A, Sogawa K, Shida T, Kimura F, et al. Apolipoprotein C-1 maintains cell survival by preventing from apoptosis in pancreatic cancer cells. Oncogene. 2008;27:2810–22. doi: 10.1038/sj.onc.1210951. [DOI] [PubMed] [Google Scholar]
- 10.Yi J, Ren L, Wu J, Li W, Zheng X, Du G, et al. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for gastric cancer. Ann Transl Med. 2019;7:380. doi: 10.21037/atm.2019.07.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin Y, Yang Y, Su Y, Ye X, Liu W, Yang Q, et al. Identification a novel clinical biomarker in early diagnosis of human non-small cell lung cancer. Glycoconj J. 2019;36:57–68. doi: 10.1007/s10719-018-09853-z. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y, Zhang J, Guo F, Zhao W, Zhan Y, Liu C, et al. Identification of Apolipoprotein C-I peptides as a potential biomarker and its biological roles in breast cancer. Med Sci Monit. 2016;22:1152–60. doi: 10.12659/MSM.896531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Wang J, Dong R, Yang S, Zheng S. Identification of novel serum biomarkers in child nephroblastoma using proteomics technology. Mol Biol Rep. 2011;38:631–8. doi: 10.1007/s11033-010-0149-4. [DOI] [PubMed] [Google Scholar]
- 14.Fan Y, Shi L, Liu Q, Dong R, Zhang Q, Yang S, et al. Discovery and identification of potential biomarkers of papillary thyroid carcinoma. Mol Cancer. 2009;8:79. doi: 10.1186/1476-4598-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouyang J, Jiang Y, Deng C, Zhong Z, Lan Q. Doxorubicin Delivered via ApoE-directed reduction-sensitive polymersomes potently inhibit orthotopic human glioblastoma xenografts in nude mice. Int J Nanomed. 2021;16:4105–15. doi: 10.2147/IJN.S314895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evangelou P, Groll M, Oppermann H, Gaunitz F, Eisenlöffel C, Müller W, et al. Assessment of ApoC1, LuzP6, C12orf75 and OCC-1 in cystic glioblastoma using MALDI-TOF mass spectrometry, immunohistochemistry and qRT-PCR. Med Mol Morphol. 2019;52:217–25. doi: 10.1007/s00795-019-00223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, Dixon S. Mechanisms of ferroptosis. Cell Mol life Sci: CMLS. 2016;73:2195–209. doi: 10.1007/s00018-016-2194-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hassannia B, Vandenabeele P, Vanden Berghe T. Targeting ferroptosis to iron out cancer. Cancer Cell. 2019;35:830–49. doi: 10.1016/j.ccell.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Cao F, Yin H, Huang Z, Lin Z, Mao N, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Fu X, Jia J, Wikerholmen T, Xi K, Kong Y, et al. Glioblastoma therapy using codelivery of cisplatin and glutathione peroxidase targeting siRNA from iron oxide nanoparticles. ACS Appl Mater interfaces. 2020;12:43408–21. doi: 10.1021/acsami.0c12042. [DOI] [PubMed] [Google Scholar]
- 21.Chen T, Chuang J, Ko C, Kao T, Yang P, Yu C, et al. AR ubiquitination induced by the curcumin analog suppresses growth of temozolomide-resistant glioblastoma through disrupting GPX4-Mediated redox homeostasis. Redox Biol. 2020;30:101413. doi: 10.1016/j.redox.2019.101413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology (Baltimore. Md). 2016;63:173–84. [DOI] [PMC free article] [PubMed]
- 23.Fan Z, Wirth A, Chen D, Wruck C, Rauh M, Buchfelder M, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6:e371. doi: 10.1038/oncsis.2017.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye Z, Wang D, Lu Y, He Y, Yu J, Wei W, et al. Vacuolin-1 inhibits endosomal trafficking and metastasis via CapZβ. Oncogene. 2021;40:1775–91. doi: 10.1038/s41388-021-01662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang J, Li J, Qi W, Qiu W, Li P, Li B, et al. Inhibition of SREBP by a small molecule, betulin, improves hyperlipidemia and insulin resistance and reduces atherosclerotic plaques. Cell Metab. 2011;13:44–56. doi: 10.1016/j.cmet.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Dodson M, Castro-Portuguez R, Zhang D. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol. 2019;23:101107. doi: 10.1016/j.redox.2019.101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Blake S, Chan K, Pearson R, Kang J. βCystathionine -synthase in physiology and cancer. BioMed Res Int. 2018;2018:3205125. doi: 10.1155/2018/3205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Cai H, Hu Y, Liu F, Huang S, Zhou Y, et al. A pharmacological probe identifies cystathionine β-synthase as a new negative regulator for ferroptosis. Cell Death Dis. 2018;9:1005. doi: 10.1038/s41419-018-1063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun Y, Chen P, Zhai B, Zhang M, Xiang Y, Fang J, et al. The emerging role of ferroptosis in inflammation. Biomed Pharmacother Biomed Pharmacother. 2020;127:110108. doi: 10.1016/j.biopha.2020.110108. [DOI] [PubMed] [Google Scholar]
- 30.Friedmann Angeli J, Krysko D, Conrad M. Ferroptosis at the crossroads of cancer-acquired drug resistance and immune evasion. Nat Rev Cancer. 2019;19:405–14. doi: 10.1038/s41568-019-0149-1. [DOI] [PubMed] [Google Scholar]
- 31.Yee P, Wei Y, Kim S, Lu T, Chih S, Lawson C, et al. Neutrophil-induced ferroptosis promotes tumor necrosis in glioblastoma progression. Nat Commun. 2020;11:5424. doi: 10.1038/s41467-020-19193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong X, Zeng Y, Liu Y, You L, Yin X, Fu J, et al. Aloe-emodin: a review of its pharmacology, toxicity, and pharmacokinetics. Phytother Res: PTR. 2020;34:270–81. doi: 10.1002/ptr.6532. [DOI] [PubMed] [Google Scholar]
- 33.Tang D, Chen X, Kang R, Kroemer G. Ferroptosis: molecular mechanisms and health implications. Cell Res. 2021;31:107–25. doi: 10.1038/s41422-020-00441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun X, Niu X, Chen R, He W, Chen D, Kang R, et al. Metallothionein-1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology (Baltimore. Md). 2016;64:488–500. [DOI] [PMC free article] [PubMed]
- 35.Chaudhary N, Choudhary B, Shah S, Khapare N, Dwivedi N, Gaikwad A, et al. Lipocalin 2 expression promotes tumor progression and therapy resistance by inhibiting ferroptosis in colorectal cancer. Int J Cancer. 2021;149:1495–511. doi: 10.1002/ijc.33711. [DOI] [PubMed] [Google Scholar]
- 36.Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater (Deerfield Beach, Fla) 2019;31:e1904197. doi: 10.1002/adma.201904197. [DOI] [PubMed] [Google Scholar]
- 37.Chen L, Li X, Liu L, Yu B, Xue Y, Liu Y. Erastin sensitizes glioblastoma cells to temozolomide by restraining xCT and cystathionine-γ-lyase function. Oncol Rep. 2015;33:1465–74. doi: 10.3892/or.2015.3712. [DOI] [PubMed] [Google Scholar]
- 38.Bellezza I, Giambanco I, Minelli A, Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim Biophys Acta Mol Cell Res. 2018;1865:721–33. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Jiang T, Harder B, Rojo de la Vega M, Wong P, Chapman E, Zhang D. p62 links autophagy and Nrf2 signaling. Free Radic Biol Med. 2015;88:199–204. doi: 10.1016/j.freeradbiomed.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mou Y, Wang J, Wu J, He D, Zhang C, Duan C, et al. Ferroptosis, a new form of cell death: opportunities and challenges in cancer. J Hematol Oncol. 2019;12:34. doi: 10.1186/s13045-019-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ulasov A, Rosenkranz A, Georgiev G, Sobolev A. Nrf2/Keap1/ARE signaling: Towards specific regulation. Life Sci. 2022;291:120111. doi: 10.1016/j.lfs.2021.120111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jaramillo M, Zhang D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–91. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Liu Y, Du T, Yang H, Lei L, Guo M, et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc. Cell Death Differ. 2020;27:662–75. doi: 10.1038/s41418-019-0380-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ursini F, Maiorino M. Lipid peroxidation and ferroptosis: The role of GSH and GPx4. Free Radic Biol Med. 2020;152:175–85. doi: 10.1016/j.freeradbiomed.2020.02.027. [DOI] [PubMed] [Google Scholar]
- 45.Paul B, Sbodio J, Snyder S. Cysteine metabolism in neuronal redox homeostasis. Trends Pharmacol Sci. 2018;39:513–24. doi: 10.1016/j.tips.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conrad M, Sato H. The oxidative stress-inducible cystine/glutamate antiporter, system x (c) (-): cystine supplier and beyond. Amino acids. 2012;42:231–46. doi: 10.1007/s00726-011-0867-5. [DOI] [PubMed] [Google Scholar]
- 47.Liu N, Lin X, Huang C. Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br J Cancer. 2020;122:279–92. doi: 10.1038/s41416-019-0660-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Xia X, Huang P. xCT: A critical molecule that links cancer metabolism to redox signaling. Mol Ther: J Am Soc Gene Ther. 2020;28:2358–66. doi: 10.1016/j.ymthe.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong H, Qiang Z, Chai D, Peng J, Xia Y, Hu R, et al. Nrf2 inhibits ferroptosis and protects against acute lung injury due to intestinal ischemia reperfusion via regulating SLC7A11 and HO-1. Aging. 2020;12:12943–59. doi: 10.18632/aging.103378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen D, Tavana O, Chu B, Erber L, Chen Y, Baer R, et al. NRF2 is a major target of ARF in p53-independent tumor suppression. Mol Cell. 2017;68:224–32.e4. doi: 10.1016/j.molcel.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hayashima K, Kimura I, Katoh H. Role of ferritinophagy in cystine deprivation-induced cell death in glioblastoma cells. Biochem Biophys Res Commun. 2021;539:56–63. doi: 10.1016/j.bbrc.2020.12.075. [DOI] [PubMed] [Google Scholar]
- 52.Casique L, Kabil O, Banerjee R, Martinez J, De, Lucca M. Characterization of two pathogenic mutations in cystathionine beta-synthase: different intracellular locations for wild-type and mutant proteins. Gene. 2013;531:117–24. doi: 10.1016/j.gene.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Floros K, Chawla A, Johnson-Berro M, Khatri R, Stamatouli A, Boikos S, et al. MYCNMYCN upregulates the transsulfuration pathway to suppress the ferroptotic vulnerability in -amplified neuroblastoma. Cell Stress. 2022;6:21–9. doi: 10.15698/cst2022.02.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Erdélyi K, Ditrói T, Johansson H, Czikora Á, Balog N, Silwal-Pandit L, et al. Reprogrammed transsulfuration promotes basal-like breast tumor progression via realigning cellular cysteine persulfidation. Proc Natl Acad Sci USA 2021;118:e2100050118. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.