Abstract
The identification of mechanisms to promote memory T cells (TMEM) has important implications for vaccination and anti-cancer immunotherapy1–4. Using a CRISPR-based screen for negative regulators of TMEM generation in vivo5, we discovered multiple components of the mammalian canonical Brg1/Brg-associated factor (cBAF)6,7. Several components of the cBAF complex are essential for the differentiation of activated CD8+ T cells into T effector (TEFF) cells, and their loss promotes TMEM formation in vivo. During the first division of activated CD8+ T cells, cBAF and c-Myc8 frequently co-assort asymmetrically to the two daughter cells. Daughter cells with high c-Myc and high cBAF display a cell fate trajectory towards TEFF cells, while those with low c-Myc and low cBAF preferentially differentiate towards TMEM cells. The cBAF complex and c-Myc physically interact to establish the chromatin landscape in activated CD8+ T cells. Treatment of naïve CD8+ T cells with a putative cBAF inhibitor during the first 48 hours of activation, prior to the generation of chimeric antigen receptor T (CAR-T) cells, markedly improves efficacy in a solid tumor model. Our results establish cBAF as a negative determinant of TMEM cell fate and suggest that manipulation of cBAF early in T cell differentiation can improve cancer immunotherapy.
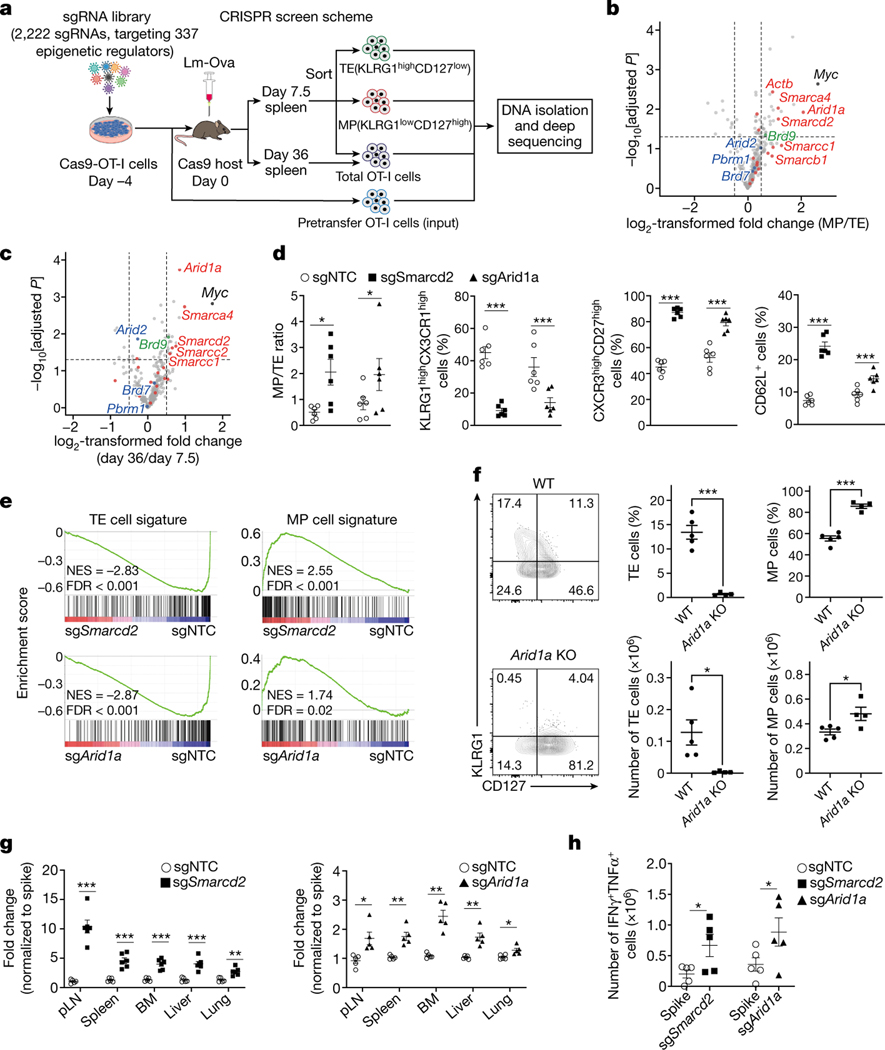
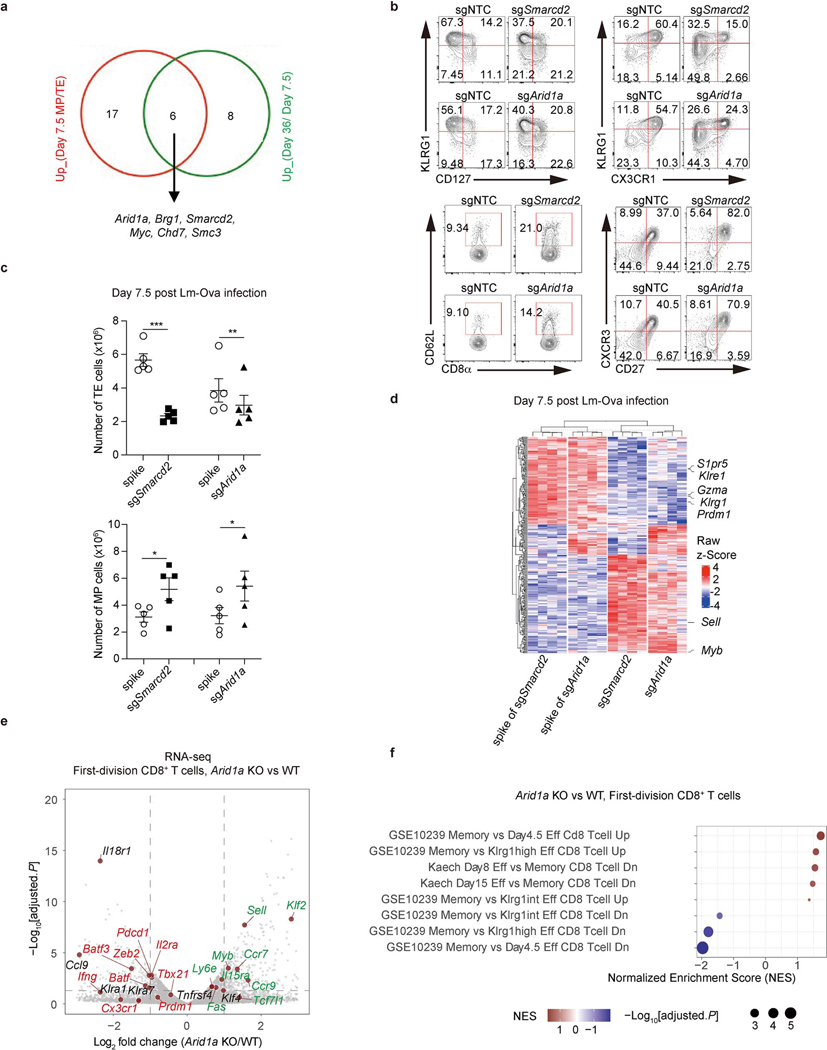
We employed a CRISPR screen for inhibitors of antigen-specific TMEM cell generation in vivo5, using a guide RNA (sgRNA) library designed to target proteins involved in epigenetic modification (see Methods and Fig. 1a). After adoptive transfer of sgRNA-expressing ovalbumin (Ova)-specific CD8+ T (OT-I) cells and challenge with Listeria monocytogenes expressing Ova (Lm-Ova), we performed comparative analysis of sgRNAs in memory precursor (MP; KLRG1loCD127hi) versus terminal effector (TE; KLRG1hiCD127lo)9,10 CD8+ T cells at day 7.5 (Fig.1b), and CD8+ T cell accumulation at 36 days (memory phase) versus 7.5 days (effector phase) (Fig.1c). These analyses revealed a striking enrichment for sgRNAs targeting several components of the cBAF complex, the deletion of which promoted the generation of MP versus TE cells at day 7.5 and memory cell development (day 36 versus day 7.5) (Fig. 1b, c, Extended Data Fig. 1a). Mammalian BAF is a multi-subunit chromatin remodeling complex that contains one of two mutually exclusive ATPase subunits, either Brg1 or Brm. The BAF complexes can be approximately grouped into three distinct assemblies, called canonical BAF (cBAF), polybromo-associated BAF (PBAF), and noncanonical BAF (ncBAF)6,7,11,12. This is an oversimplification, as these complexes likely represent composites produced through combinatorial assembly of the subunits6,12,13. Nevertheless, these complexes differentially localize on chromatin and exhibit distinct functions, including regulation of early T cell development in the thymus14–16. To validate our findings, we ablated the BAF component Smarcd2, or the cBAF-specific component Arid1a, via CRISPR editing in OT-I cells. After adoptive transfer, we challenged the recipient mice with Lm-Ova. Deletion of either component resulted in decreased TE cells and increased MP cells on day 7.5, based on cell numbers of the respective populations, increased MP/TE ratios, reduced expression of TE-associated molecules (e.g. CX3CR1, KLRG1)10,17 and increased memory-associated molecules (e.g. CXCR3, CD27, and CD62L)10,17 (Fig. 1d, Extended Data Fig. 1b, c).
Figure 1. An in vivo CRISPR-Cas9 screen reveals a key role of the cBAF complex in T cell fate decisions.
a, Diagram of the screening system. MP, memory precursor cells (KLRG1loCD127hi); TE, terminal effector cells (KLRG1hiCD127lo). b,c, Volcano plot of the enrichment of sgRNAs in MP relative to TE cells [log2 fold change (MP/TE)] at day 7.5 post-infection (b) or in total cells at day 36 relative to total cells at day 7.5 post-infection (c). BAF components (red), PBAF components (blue) and an ncBAF component (green) are highlighted. b. n=3 replicates (each pooled from 2 mice); c. n=3 replicates (each pooled from 5 mice). d, Quantification of splenic MP/TE ratio and the proportions of TE (KLRG1hiCX3CR1hi) or memory (CXCR3hiCD27hi and CD62L+) populations in the indicated sgRNA-transduced OT-I cells at day 7.5 (n=6 mice). e, Gene set enrichment analysis (GSEA) of the TE or MP cell signatures in sgSmarcd2 or sgArid1a compared to sgNTC-transduced cells at day 7.5 post Lm-Ova infection (n=4 mice). f, Flow cytometry plot (left) and quantification (right) of splenic MP and TE cells at day 9 after IAV-Ova infection. Rosa26CreERT2Arid1afl/fl OT-I and and WT OT-I from littermates treated with oral tamoxifen, and then transferred into C57BL/6 hosts followed by IAV-Ova infection (WT, n=5 mice; Arid1a KO, n=4 mice). g, Quantification of the relative fold change (normalized to spike) of OT-I cells expressing indicated sgRNA in multiple tissues at day >30 post-infection (n=5 mice). h, Quantification of absolute number of IFN-γ+TNF-α+ cells at day 6 after Lm-Ova re-challenge (n=5 mice). Data are representative of one (b, c, e) or two (f) independent experiment; or compiled from two (d, h) or three (g) independent experiments. Data are shown as mean± s.e.m. *p<0.05, **p<0.01, ***p<0.001; two-tailed paired Student’s t-test (d, h) or two-tailed unpaired Student’s t-test (b, c, f, g).
Transcriptional profiling of Smarcd2 or Arid1a-deficient OT-I cells at day 7.5 post-infection revealed enrichment of an MP gene expression signature and reduction of a TE signature9 in Smarcd2- or Arid1a-deficient OT-I cells (Fig. 1e, Extended Data Fig. 1d). We generated Rosa26Cre-ERT2, Arid1afl/fl mice and deleted Arid1a by oral h tamoxifen, followed by activation of CD8+ OT-I cells with anti-CD3ε and anti-CD28 (anti-CD3/CD28) plus ICAM1 in vitro. Acute deletion of Arid1a enhanced TMEM and suppressed TEFF gene signatures following the first cell division in vitro, as revealed by transcriptional profiling (Extended Data Fig. 1e, f). Upon transfer of wild-type (WT) or Arid1a-deficient OT-I cells into mice and IAV-Ova infection, we observed increased numbers and percentages of MP cells in the absence of Arid1a (Fig. 1f), further supporting the pro-memory fate decision upon deletion of cBAF components.
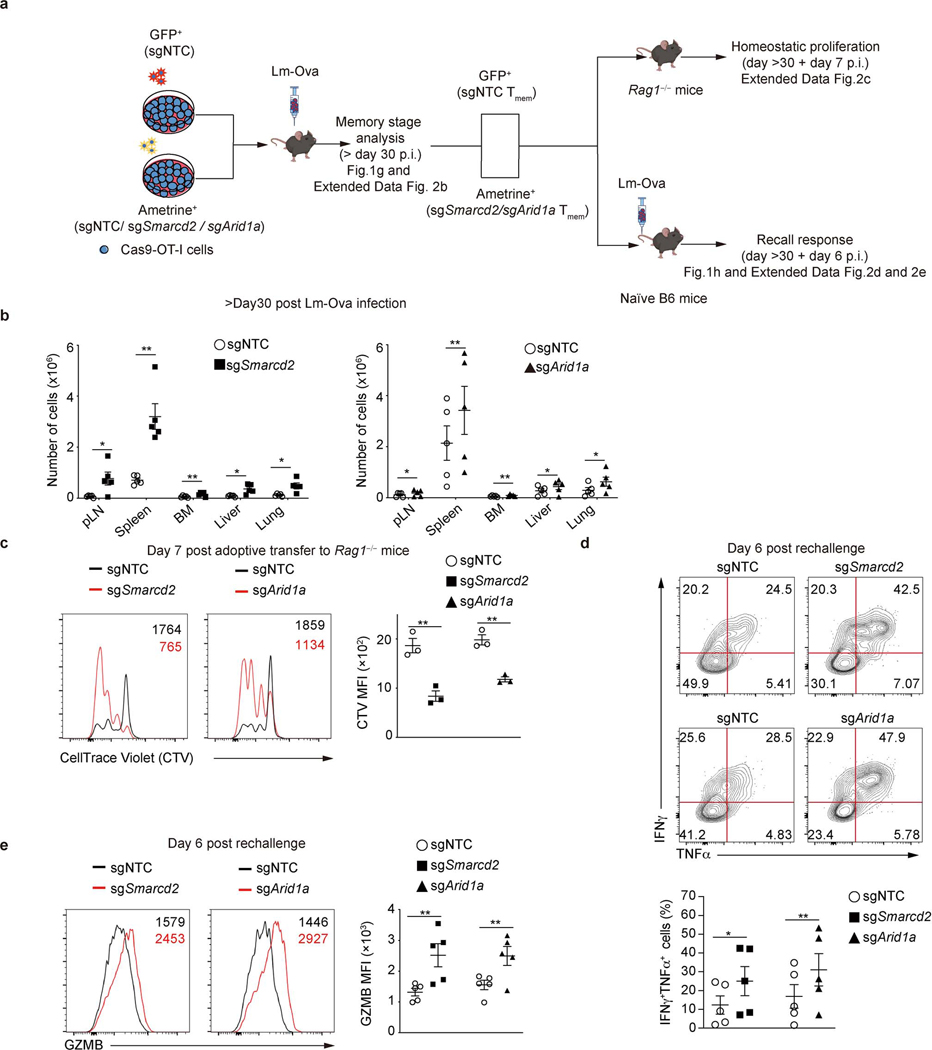
We next analyzed memory responses at day >30 post-infection (Extended Data Fig. 2a). We transduced cells with sgRNA targeting Smarcd2 or Arid1a, and following adoptive transfer and Lm-Ova infection, we observed an increased ratio and absolute number of TMEM in diverse tissues on day >30 post-infection (Fig. 1g, Extended Data Fig. 2b). Thus, TMEM generation was enhanced upon deletion of Smarcd2 or Arid1a. To determine if these TMEM displayed bona fide memory features, we sorted TMEM cells from the donor mice that had been infected with Lm-Ova and transferred them into Rag1–/– animals to assess homeostatic proliferation18, or into naïve hosts for secondary rechallenge with Lm-Ova (Extended Data Fig. 2a). Compared to WT T cells transduced with non-targeting control (NTC), Smarcd2- or Arid1a-deficient T cells displayed increased homeostatic proliferation (Extended Data Fig. 2c), and upon rechallenge, an increased frequency and number of cells expressing both interferon-γ (IFN-γ) and TNF-α (Fig. 1h, Extended Data Fig. 2d) and increased expression of granzyme B (GzmB) (Extended Data Fig. 2e). Therefore, deletion of cBAF components promotes TMEM generation and function.
PBAF and ncBAF share common subunits with cBAF. Ablation of the PBAF-specific component Pbrm16,7,12 (Extended Data Fig. 3a) in CD8+ T cells from Pbrm1fl/flCd4-Cre mice had no effect on total CD8+ T cells, TE or MP generation at 7.5 (Extended Data Fig. 3b, c), or 32 days following Lm-Ova infection (Extended Data Fig. 3d, e). Pbrm1 also did not affect Ova-specific CD8+ T cell numbers in different tissues (Extended Data Fig. 3d) or expression of the marker of central memory T (TCM), CD62L10,17 (Extended Data Fig. 3e). Further, we ablated the ncBAF components Brd9 and Bicra (also called GLTSCR) and found no effect on the generation of MP or TE (Extended Data Fig. 3f). In contrast, deletion Brg1 or Smarcc1 (present in cBAF, PBAF, and ncBAF complexes) promoted MP generation and suppressed TE differentiation, as reflected by increased proportions of CD62L+ and KLRG1loCD127hi cells and decreased proportion of KLRG1hiCD127lo cells (Extended Data Fig. 3g). In contrast to Brg1, ablation of Brm had no effect on TE or MP differentiation (Extended Data Fig. 3g). Altogether, we conclude that the Brg1-containing cBAF complex promotes TE and suppress TMEM generation in CD8+ T cells.
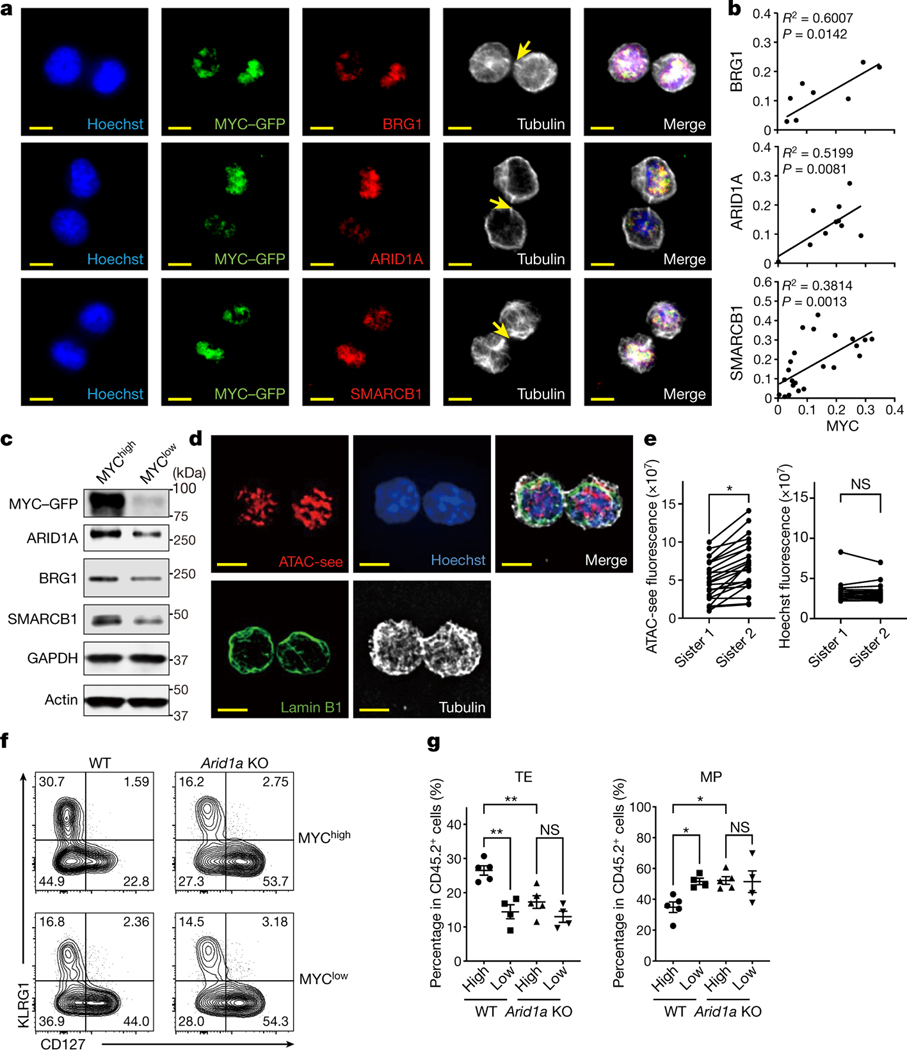
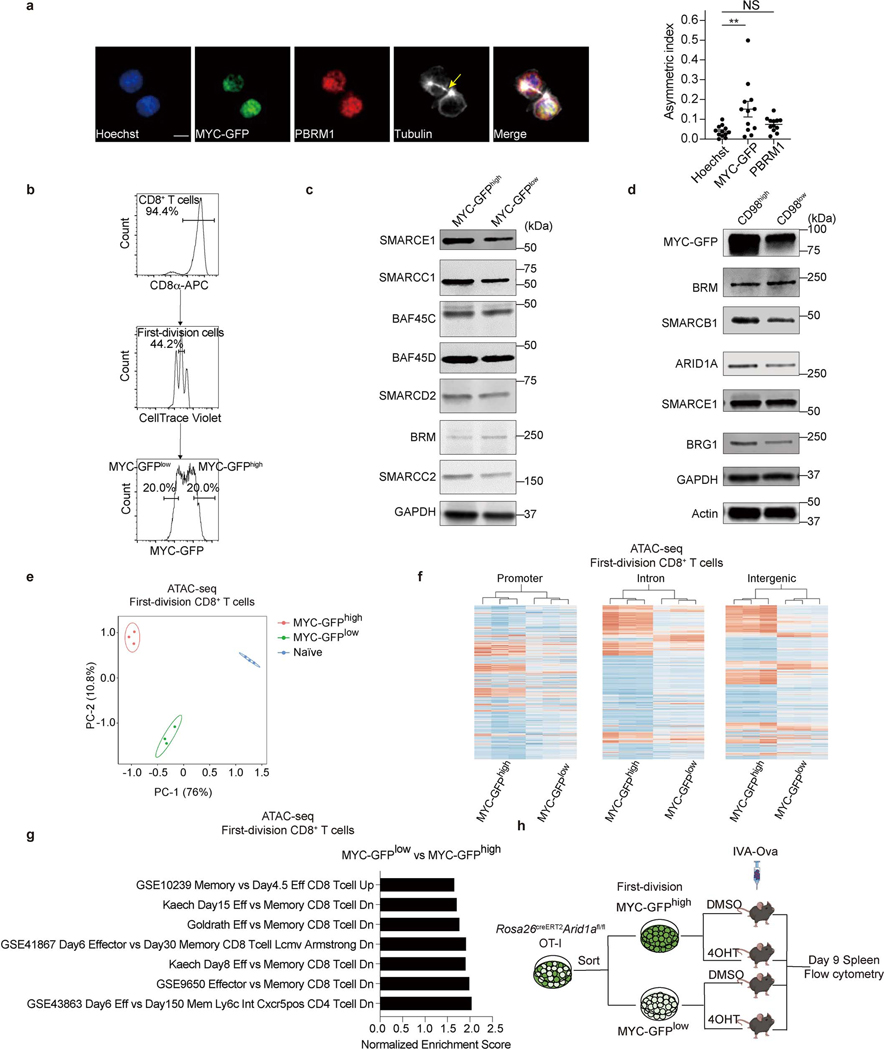
In our screen (Fig. 1b, c), we noted enrichment for sgRNAs targeting the Myc protooncogene in MP versus TE and day 36 TMEM versus day 7.5 TEFF cells. We previously found that ablation of one allele of c-Myc can promote the differentiation of CD8+ T cells to a TMEM phenotype, and that c-Myc protein is sorted asymmetrically into the two daughter cells during the first cell division8. We therefore examined the relationships between c-Myc protein expression and expression of components of the cBAF complex at early time points following TCR stimulation. We activated OT-I containing GFP-tagged c-Myc (GFP inserted onto the C-terminus of c-Myc)8,19 with anti-CD3/CD28 plus ICAM1, and observed telophase cells completing the first division. In dividing T cells with asymmetric distribution of c-Myc in the telophase nuclei, we observed a corresponding asymmetric distribution of cBAF components, Smarcb1, Arid1a, and Brg1 (Fig. 2a, b), while the PBAF component, Pbrm1, was symmetrically distributed (Extended Data Fig. 4a). Sorting of first-division CD8+ T cells based on expression of c-Myc-GFP for immunoblot analysis (Extended Data Fig. 4b) similarly revealed increased components of the cBAF complex in c-Mychi versus c-Myclo cells (Fig. 2c, Extended Data Fig. 4c). Asymmetric expression of CD98 correlates with c-Myc expression in first-division CD8+ T cells8, and we observed increased cBAF components in CD98hi versus CD98lo first-division CD8+ T cells (Extended Data Fig.4d). Therefore, like c-Myc, cBAF often displays asymmetry at the first division of CD8+ T cells.
Figure 2. cBAF asymmetrically segregates to daughter cells in activated CD8+ T cells during the first division.
a, Representative images of conjoined daughter c-Myc-GFP-expressing CD8+ T cells that were fixed and stained for Hoechst (blue), cBAF components (red) and Tubulin (white). Naïve CD8+ T cells were activated by anti-CD3ε (2 μg/ml) and anti-CD28 (1 μg/ml) plus ICAM1 (0.5 μg/ml) for 28 hours. Arrows mark tubulin bridges. Scale bar: 5 μm. b, Quantification of difference in fluorescence intensity/total of BAF components (y-axis) and c-Myc-GFP (x-axis). Each point represents one pair of conjoined daughter cells (Brg1, R2=0.6007, p=0.0142 linear regression, n=9; Arid1a, R2=0.5199, p=0.0081 linear regression, n=12; Smarcb1, R2=0.3814, p=0.0013 linear regression, n=24). c, Immunoblot of BAF components in sorted c-Myc-GFPhi and c-Myc-GFPlo first-division CD8+ T cells activated as in (a) for 36 hours. The gating strategy is in Extended Data Fig. 4b. d, Representative images of conjoined daughter CD8+ T cells activated as in (a) for 28 hours and stained for ATAC-see (red), Hoechst (blue), Lamin B1 (green) and Tubulin (white). Scale bar: 5 μm. e, Quantification of fluorescence intensity of ATAC-see and Hoechst in conjoined sister CD8+ T cells. Each dot represents one cell; data points of sister cells are connected by a line (n=22 cells). f, g, Strategy is shown in Extended data Fig. 4h. Flow cytometry plot (f) and quantification (g) of donor-derived splenic TE (KLRG1hiCD127lo) and MP (KLRG1loCD127hi) cells at day 9 after IAV-Ova challenge (c-Mychi, n=5 mice; c-Myclo, n=4 mice). Data are representative of at least two independent experiments (a−g). Data are shown as mean±s.e.m. *p<0.05, **p<0.01; two-tailed paired Student’s t-test (e) or one-way ANOVA (g).
The BAF complexes control chromatin architecture through nucleosome mobilization, ejection, and histone dimer exchange7,20,21. We therefore visualized the accessible genome during the first division of CD8+ T cells using transposase-accessible chromatin with visualization (ATAC-see), which directly images chromatin accessibility in situ22. We observed many first-division sister cells that differed in the intensity of ATAC-see (Fig. 2d, e). We then performed assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) using sorted, first-division c-Mychi and c-Myclo CD8+ T cells and observed extensive differences in chromatin accessibility between these populations (Extended Data Fig. 4e), with more accessibility detected in the c-Mychi cells in the promoter, intronic, and intergenic regions (Extended Data Fig. 4f). Gene set enrichment analysis of the genes closest to differentially accessible regions revealed that c-Myclo cells and c-Mychi cells exhibited memory-like and effector-like signatures, respectively (Extended Data Fig. 4g). Thus, chromatin accessibility and cBAF expression are coordinately regulated, with higher activities observed in c-Mychi cells.
First-division c-Mychi CD8+ T cells display TE function in vivo, while c-Myclo cells display TMEM function8. We sorted first-division c-Mychi and c-Myclo cells from Arid1afl/fl; Rosa26Cre-ERT2, OT-I mice and then ablated Arid1a by treatment with 4-Hydroxytamoxifen (4OHT) before adoptive transfer into recipient mice and infection with IAV-Ova. At 9 days post-infection, we examined OT-I cells for markers of TE and MP (Extended Data Fig. 4h). Cells that had been c-Mychi at sorting generated more cells with markers of TE cells than those that had been c-Myclo, and the latter generated more cells with MP markers (Fig. 2f, g). Ablation of Arid1a following the sort eliminated this effect; all cells (originally c-Mychi or lo) showed a tendency to differentiate into MP (Fig. 2f, g), indicating the functional requirement of cBAF for T cell fate decisions associated with c-Myc expression levels.
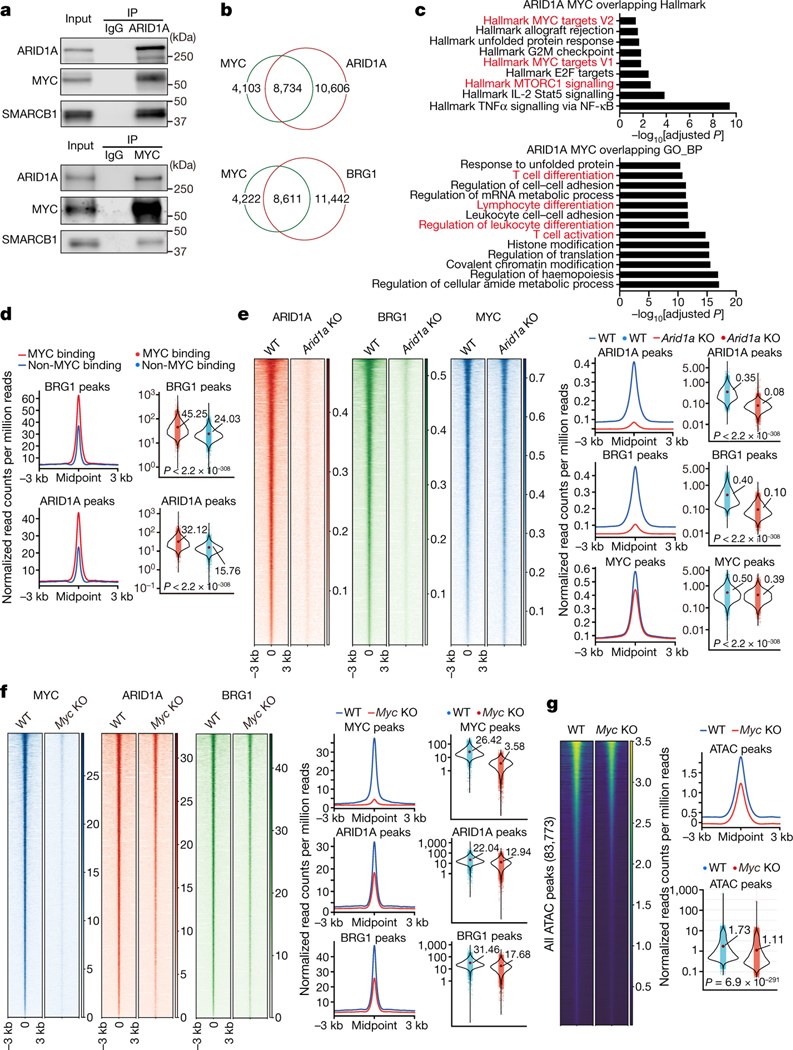
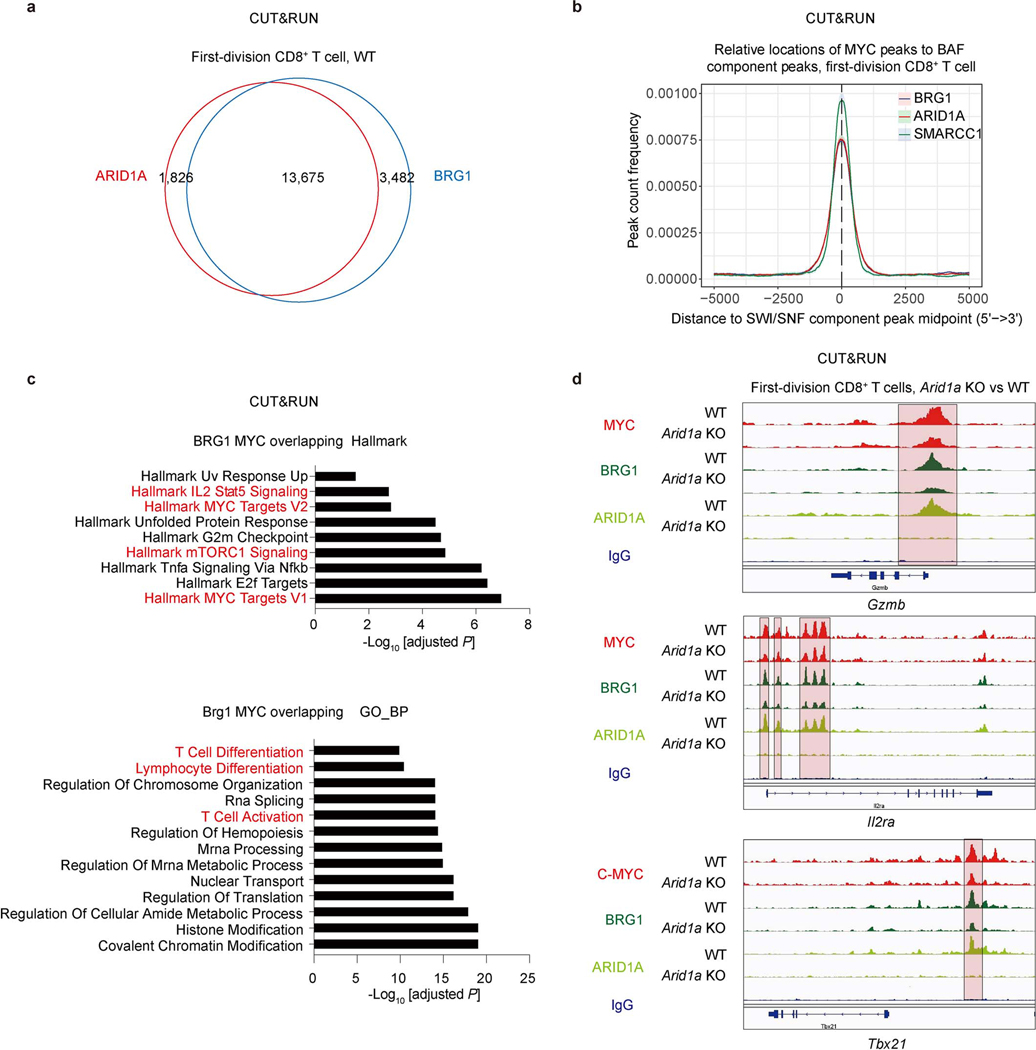
To examine the functional interaction between cBAF and c-Myc, we found that Arid1a and Smarcb1 co-precipitated with c-Myc, and reciprocally, c-Myc and Smarcb1 co-precipitated with Arid1a in activated CD8+ T cells (Fig. 3a), as described for other cells23–25. We then examined chromatin binding of c-Myc, Arid1a, and Brg1 in first-division CD8+ T cells using the CUT&RUN assay26. We observed more than 80% of peaks overlapped between Arid1a and Brg1 (Extended Data Fig. 5a), as expected. Importantly, a substantial proportion of Arid1a (45%) and Brg1 (42%) binding sites were occupied by c-Myc (Fig. 3b, Extended Data Fig. 5b). Gene set enrichment analysis of binding sites occupied by both c-Myc and Arid1a (Fig. 3c), or c-Myc and Brg1 (Extended Data Fig. 5c) revealed Myc target gene sets, gene sets involved in T cell activation and differentiation, and many effector-associated molecules (Extended Data Fig. 5d). Moreover, the binding of Arid1a or Brg1 at c-Myc binding sites, measured by read counts, was more robust than in non-overlapping regions (Fig. 3d).
Figure 3. c-Myc interacts with the cBAF complex and enhances cBAF chromatin binding and function.
a, Endogenous interactions of Arid1a/Smarcb1/c-Myc were analyzed by co-immunoprecipitation (Co-IP). OT-1 were activated as in Fig. 2a for 36 hours. b–e, WT and Arid1a−/− naïve CD8+ T cells were labeled with CellTrace Violet and activated as in Fig. 2a for 36 hours; first-division CD8+ T cells were sorted for CUT&RUN assay (n=2 biological replicates). b, Venn diagram of the overlap between CUT&RUN binding peaks in WT first-division CD8+ T cells for c-Myc, Arid1a and Brg1. c, Over-representation analysis of Arid1a and c-Myc overlapping binding sites. d, Histogram (left) and quantification (right) of Brg1 (upper) and Arid1a (lower) binding signals (read count per million reads normalized to spike-in) in c-Myc binding sites or non-c-Myc binding sites. e, Heatmap (left), histogram (middle) and quantification (right) of Arid1a, Brg1 and c-Myc CUT&RUN signals in WT and Arid1a−/− activated first-division CD8+ cells. Mid, middle. f, g, Naïve CD8+ T cells from Rosa26Cre-ERT2Mycfl/fl mice and WT littermates were treated with 4OHT overnight in IL-2-containing media prior to activation for 36 hours. f, Heatmap (left), histogram (middle) and quantification (right) of c-Myc, Arid1a, Brg1 CUT&RUN signals (read count per million reads normalized to spike-in) in WT and c-Myc−/− activated CD8+ T cells (n=2 biological replicates). g, Heatmap and quantification of all consensus ATAC-seq peaks in WT and c-Myc−/− activated CD8+ T cells (n=3 biological replicates). Data are representative of three (a) or one experiment (b–g). Significance was calculated by one-sided hypergeometric distribution and adjusted with Benjamini-Hochberg procedure (c), or two-tailed unpaired Student’s t-test (d–g).
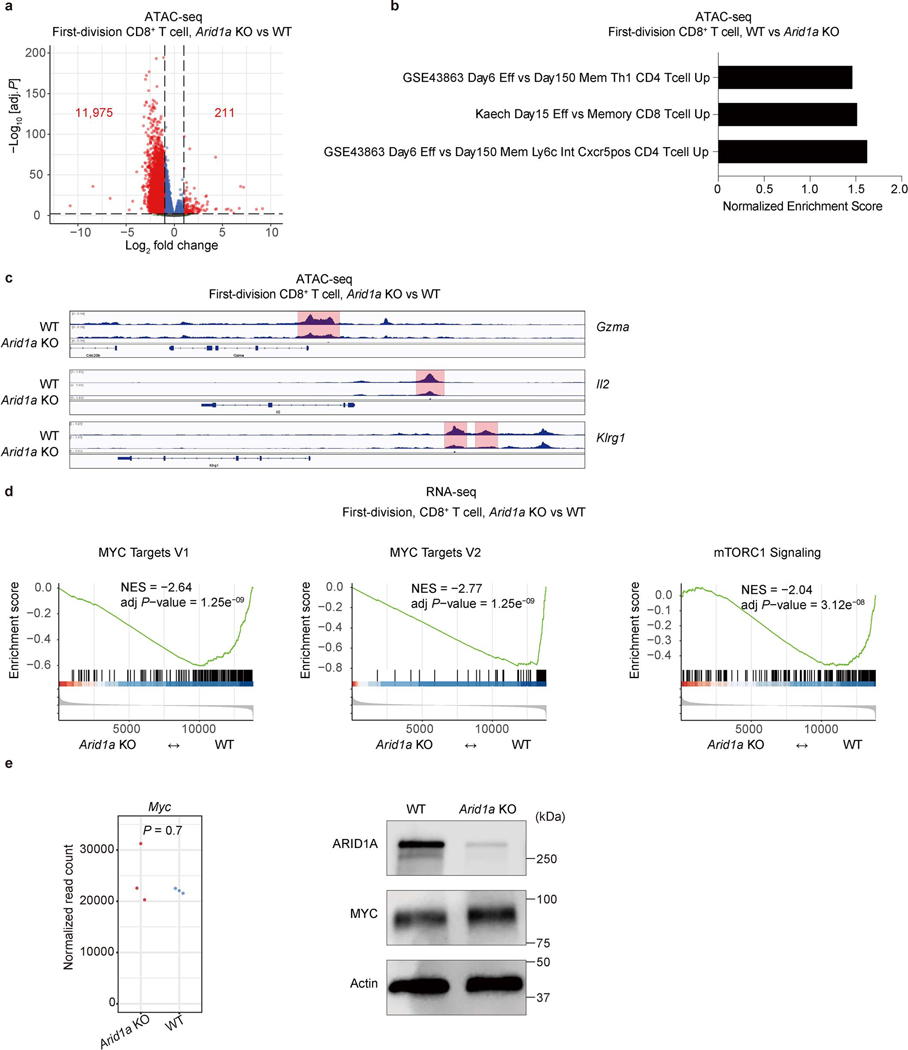
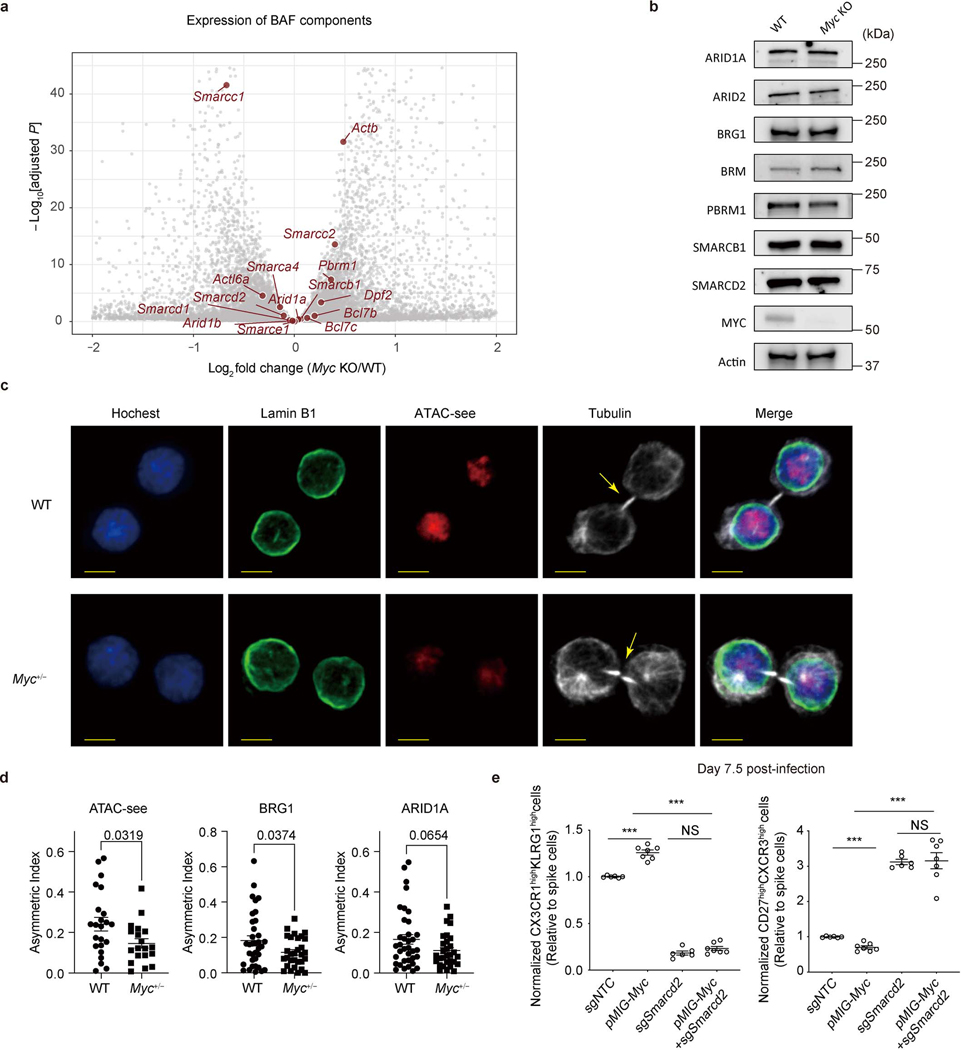
We next examined the functional effects of the interaction between cBAF and c-Myc. In activated CD8+ T cells deficient in Arid1a, accessible chromatin was reduced in ATAC-seq analysis (Extended Data Fig. 6a), including accessible chromatin in gene elements associated with TEFF function (Extended Data Fig. 6b, c). In these Arid1a-deficient cells, chromatin binding of Arid1a and Brg1 was reduced, as expected. While the overall c-Myc binding in Arid1a-deficient activated CD8+ T cells was only modestly reduced (Fig. 3e), the reduced binding sites included gene elements critically involved in TEFF differentiation and function, including those regulating the expression of Granzyme B, IL2Rα, and T-bet, to which c-Myc, Brg1, and Arid1a co-bind (Extended Data Fig. 5d). Consistent with these observations, RNA-seq and gene set enrichment analyses of Arid1a-deficient, first-division CD8+ T cells revealed a reduction of c-Myc as well as TORC1 target genes (Extended Data Fig. 6d). However, ablation of Arid1a did not decrease the expression of c-Myc mRNA or protein itself (Extended Data Fig. 6e). Conversely, in activated CD8+ T cells in which c-Myc had been acutely ablated, the binding of Arid1a and Brg1 to chromatin was reduced (Fig. 3f), and this was associated with reduced chromatin accessibility assessed by ATAC-seq (Fig. 3g). Transcriptome and immunoblot analyses revealed largely comparable expression of various cBAF components in WT and c-Myc-deficient CD8+ T cells (Extended Data Fig. 7a, b).
Acute deletion of c-Myc in CD8+ T cells completely prevents cell division27, while partial reduction of c-Myc levels in Myc+/− CD8+ T cells enhances recall responses CD8hi first-division T cells8. We therefore examined asymmetry of cBAF components and accessible chromatin in the first division of WT and Myc+/– OT-I cells. We found that heterozygous loss of c-Myc significantly reduced the asymmetric distribution of accessible chromatin (Extended Data Fig. 7c). Moreover, quantification of the asymmetric index (difference in fluorescence intensity/total) 8, also showed that heterozygous loss of c-Myc reduced the asymmetric distribution of accessible chromatin and Brg1, with a similar trend for Arid1a (Extended Data Fig. 7d). Together, these results suggest that c-Myc initiates, stabilizes, and/or supports the function of cBAF in activated CD8+ T cells, with such cooperation important for the ability of c-Myc to promote TE and suppress MP cell fate trajectories. To test this hypothesis, we enforced the expression of c-Myc in activated CD8+ T cells. We observed that ectopic c-Myc expression promoted expression of TE-associated markers (CX3CR1 and KLRG1) and reduced expression of memory-associated markers (CD27 and CXCR3), an effect that was eliminated when Smarcd2 was ablated (Extended Data Fig. 7e). These results collectively indicate that cBAF and c-Myc function cooperatively to promote the TE cell fate.
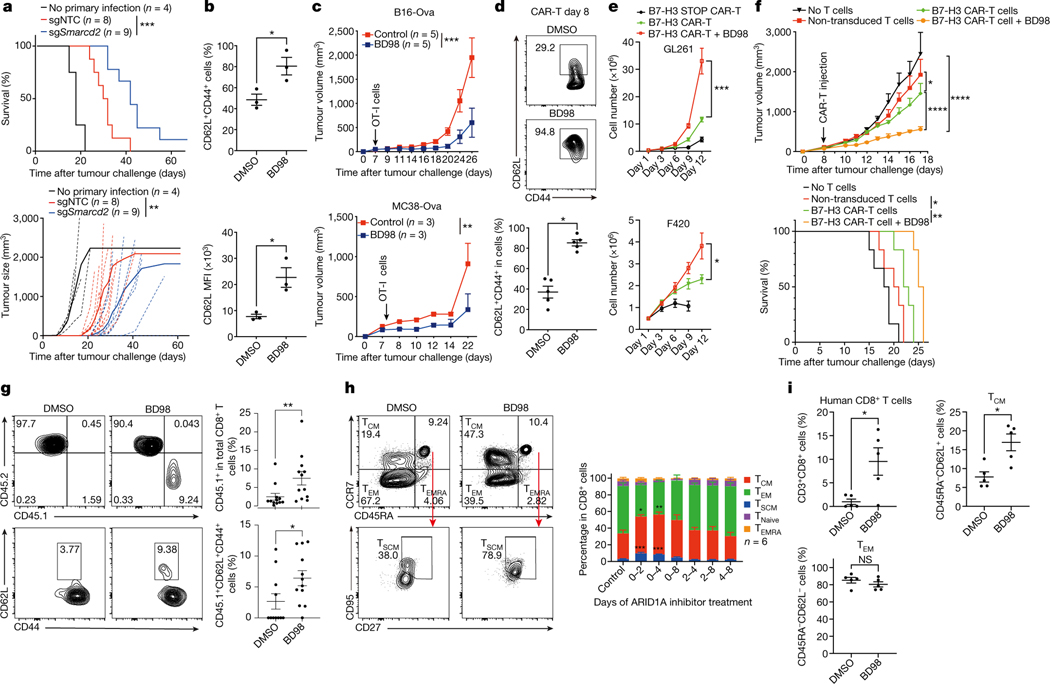
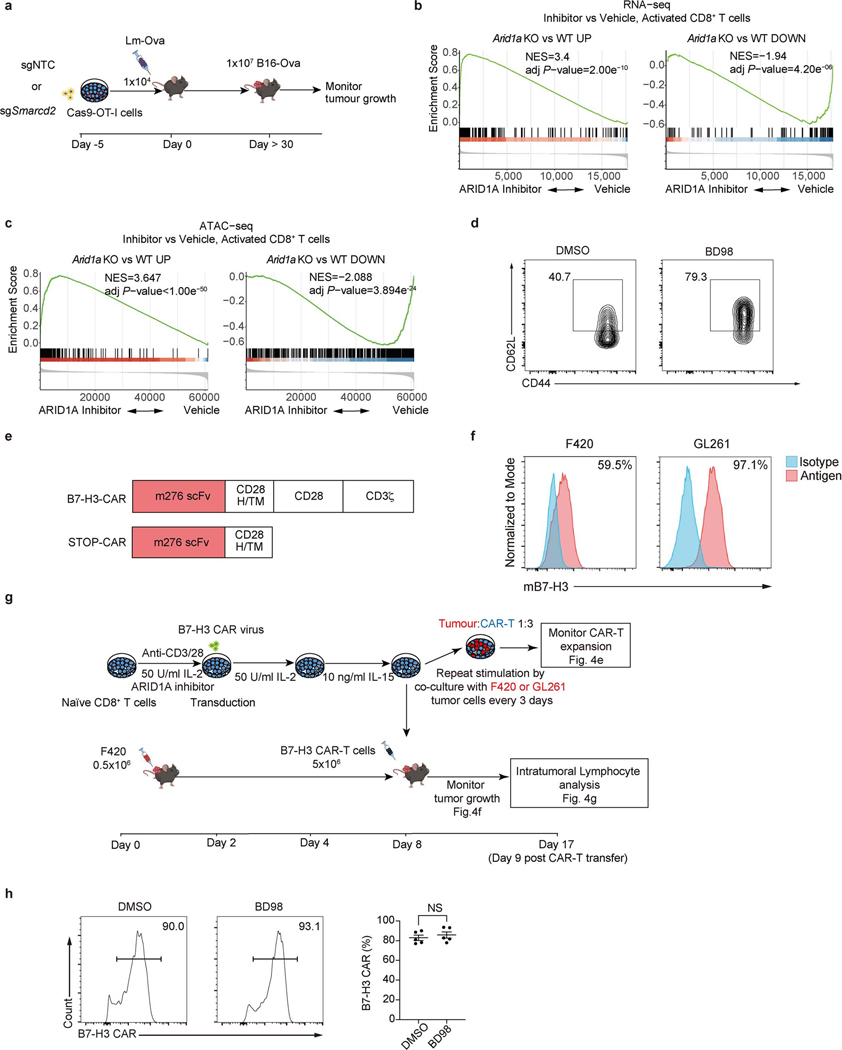
Effective adoptive T cell therapy for cancer is dependent upon T cells with long-term memory potential3,28,29. We therefore asked if manipulation of cBAF can improve anti-tumor therapy. We transduced OT-I cells with sgRNA for Smarcd2 or control sgRNA, transferred them into animals that were subsequently infected with Lm-Ova, and 30 days later inoculated them with B16F10 murine melanoma cells expressing Ova (B16-Ova) (Extended Data Fig. 8a). We observed improved control of tumor growth and enhanced survival by the Smarcd2-ablated CD8+ T cells (Fig. 4a).
Figure 4. Transient inhibition of cBAF during T cell activation improves adoptive T cell therapy.
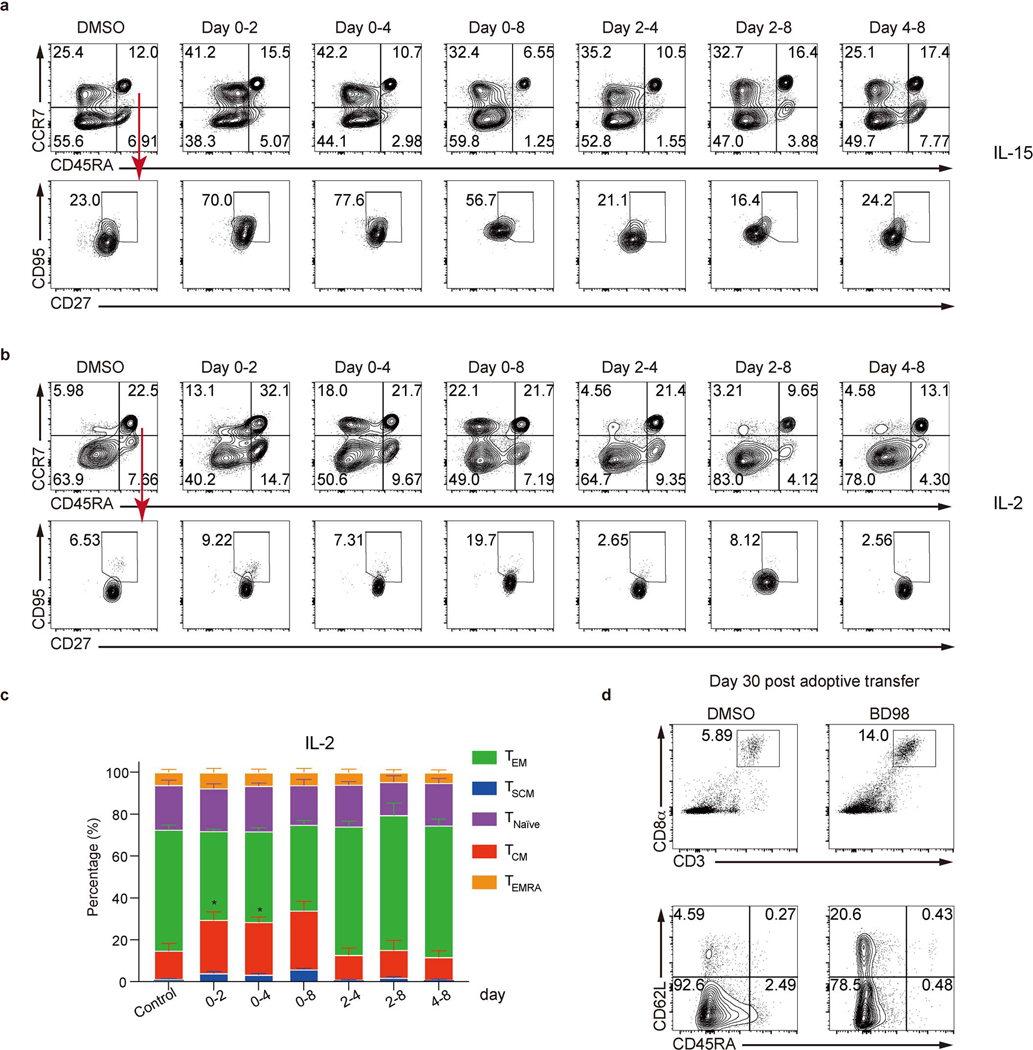
a, Tumor growth and survival of tumor-bearing mice (see Extended Data Fig. 8a). b, Quantification of the percentage of CD62L+CD44+ and the mean fluorescence intensity (MFI) of CD62L in OT-I cells treated with DMSO or BD98 during activation for 48 hours followed additional 2 days culture (n=3 biological replicate). c, Tumor growth of B16-Ova (n=5 mice) and MC38-Ova tumors (n=3 mice) after transfer of OT-I cells with or without BD98 treatment. d–g, B7-H3 CAR-T cells were generated with or without BD98 (see Extended Data Fig. 8g). d, Flow cytometry plot and quantification of CD62L+CD44+ CAR-T cells in vitro (n=5 biological replicates). e, Expansion of CAR-T cells during repeat stimulation with B7-H3-expressing tumor cells (F420 and GL261) in vitro (n=3 biological replates). STOP CAR, a control CAR containing only the single-chain variable fragment and transmembrane domain. f, Tumor growth and survival of F420 tumor-bearing mice treated with indicated CAR-T (n=6 mice). g, Flow cytometry plot and quantification of intratumoral CD45.1+ (upper) and CD62L+ (lower) CAR-T cells (n=12 mice). h, Flow cytometry plot and quantification of human TCM (CD45RA−CCR7+), TEM (CD45RA−CCR7−), and TSCM (CD45RA+CCR7+CD27+CD95+) cells in vitro. BD98 was added for the indicated times (n= 6 biological replicates). i, Quantification of total, TCM-like (CD45RA−CD62L+) and TEM -like (CD45RA−CD62L−) human CD8+ T cells in Nod-Scid-common gamma chain-deficient (NSG) mice at day 30 after transfer of human CD8+ T cells with or without BD98 treatment. Data are representative of two (a, c, f, i) or compiled at least two (b, d, e, g, h) independent experiments. Data are shown as mean±s.e.m. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; two-tailed unpaired Student’s t-test (b, d, g, i), one-way ANOVA (h), two-way ANOVA (a, c, e, f); and log-rank (Mantel-Cox) test (a, f).
Our studies on c-Myc function in T cells suggested that c-Myc acts early in T cell activation to promote cell fate trajectories8, and therefore, if cooperation between cBAF and c-Myc similarly acts early, transient inhibition of cBAF may be sufficient to promote subsequent anti-cancer immunity. To test this hypothesis, we employed BRD-K98645985 (BD98)30,31, originally identified in a screen to detect de-repression of BMI1 and subsequently deemed a likely inhibitor of Arid1a-containing BAF complex30,32. Treatment of CD8+ T cells with BD98 during initial activation resulted in similar transcriptional and chromatin accessibility changes as observed in Arid1a-deficient CD8+ T cells (Extended Data Fig. 8b, c), supporting the inhibitory effect of BD98 on Arid1a function. Next, we activated OT-I cells for 48 hours in the presence or absence of BD98, removed the drug, and cultured the cells for 2 days with interleukin-2 (IL-2). Transient treatment of activated OT-I cells with BD98 promoted the generation of cells with the TCM marker CD62L (Fig. 4b and Extended Data Fig. 8d). We then transferred the OT-I T cells to animals bearing palpable B16-Ova or MC38-Ova tumors. Transient treatment with BD98 improved the ability of OT-I cells to control tumor growth in vivo (Fig. 4c). Therefore, transient inhibition of cBAF function during the activation of CD8+ T cells is effective in promoting their anti-tumor function.
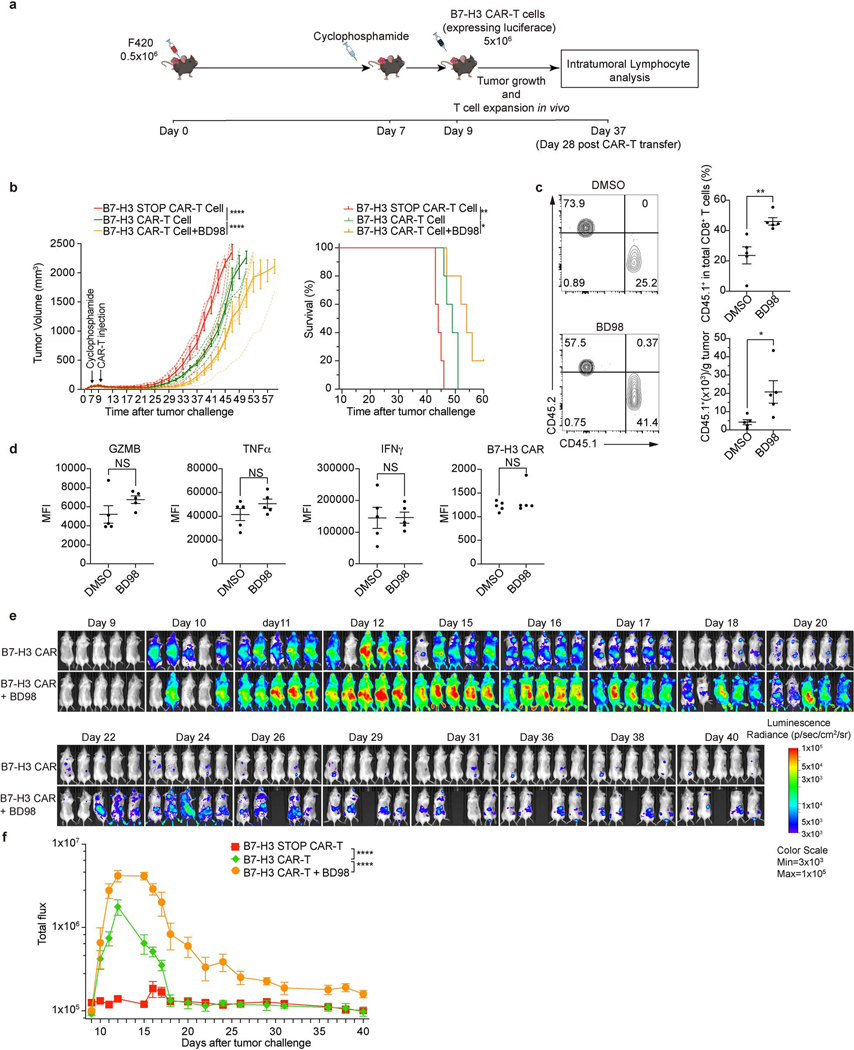
Although the most widely used adoptive T cell therapy employs the use of chimeric antigen receptors (CAR), the use of CAR-T cells for the treatment of solid tumors remains challenging33. We used a CAR targeting murine B7-H334 (Extended Data Fig. 8e) and two B7-H3-positive cell lines, murine osteosarcoma F42035,36 and glioma GL26134 (Extended Data Fig. 8f). We activated murine T cells for 2 days with BD98, followed by transduction to generate CAR-T cells (Extended Data Fig. 8g). After culturing for 6 days, the T cells transiently treated with BD98 generated CAR-T cells with high CD62L expression (Fig. 4d), with no difference in CAR expression versus vehicle-treated controls (Extended Data Fig. 8h). Repeated stimulation with either of two B7-H3-expressing tumor lines resulted in a greater expansion of the CAR-T cells derived from T cells transiently treated with BD98 (Fig. 4e). We then assessed the ability of B7-H3 CAR-T cells to control the growth of F420 cells in vivo. While the CAR-T cells generated from activated T cells that had been transiently treated with BD98 displayed markedly enhanced tumor control (Fig. 4f). CAR-T cells generated from BD98-treated cells were present at a higher frequency 9 days post-transfer than those produced from vehicle-treated CAR-T cells and comprised a higher proportion of CD62L+CD44+ cells (Fig. 4g). As cyclophosphamide lymphodepleting chemotherapy is often employed in the clinical use of CAR-T cells, we repeated this experiment with such conditioning (Extended Data Fig. 9a). Again, we observed better tumor control with CAR-T cells derived from T cells transiently treated with BD98 during initial activation (Extended Data Fig. 9b). We also assessed CAR-T cells in the tumors at 28 days after transfer (37 days after tumor challenge). CAR-T cells generated from transient BD98-treated cells were found at higher numbers than those produced from vehicle-treated CAR-T cells, although the levels of several effector molecules were comparable (Extended Data Fig. 9c, d). Using CAR-T cells generated from luciferase-expressing naïve CD8+ T cells, we followed CAR-T cell persistence longitudinally in tumor-bearing mice in vivo. Transient treatment with BD98 during initial activation prior to CAR-T generation imbued the cells with greater persistence following transfer over the course of several weeks (Extended Data Fig. 9e, f). These results indicate that transient BD98 treatment enables better anti-tumor activity of CAR-T cells, likely by transient inhibition of cBAF function.
We then examined the effect of BD98 on the differentiation of activated, human CD8+ T cells. Human naïve CD8+ T cells from healthy donors were enriched and stimulated with anti-CD3/CD28, then cultured with IL-15 (Fig. 4h, Extended Data Fig. 10a) or IL-2 (Extended Data Fig. 10b, c), two cytokines that respectively promote in vitro TMEM-like and TEFF-like cells37. We found that treatment with BD98 during the first 48 hours was necessary and sufficient to promote the generation of cells with markers of TCM (CD45RA−CCR7+) and T memory stem cells (TSCM, CD45RA+CCR7+CD27+CD95+)4 and reduced numbers of cells with markers of effector memory T cells (TEM, CD45RA−CCR7−) (Fig. 4h, Extended Data Fig. 10a). We then activated human CD8+ T cells with anti-CD3/CD28, with or without transient treatment with BD98 and transferred them into Nod-Scid-common gamma chain-deficient (NSG) mice. We found that transient treatment with BD98 endowed the activated human T cells with enhanced persistence and a greater propensity to express TCM cell markers (Fig. 4i, Extended Data Fig. 10d). Our data indicate that transient inhibition of cBAF function during T cell activation may improve CAR-T therapy for human cancers.
Components of the chromatin remodeling BAF complex are frequently mutated in human cancers6,7,38. In some settings, re-establishment of BAF function has been found to suppress expression of c-Myc39,40. In contrast, in developing T cells, BAF components bind to c-Myc promoters to promote c-Myc expression16. Therefore, the regulation of c-Myc expression and activity by cBAF is likely context-specific.
The fate of activated T cells is determined by many factors, including the strength of T cell receptor engagement, cytokines produced by antigen-presenting cells or in the local environment, metabolite and nutrient availability, among others41–44. Emerging evidence also reveals the importance of epigenetic programs in establishing lineage fate and stability45–47. Here, we found that extensive reprograming of epigenetics and chromatin state occur during the first cell division. Our findings provide insights into observations that gene expression signatures of TE and MP can be detected as early as the first division of activated CD8+ T cells48,49.
We propose that the mechanisms of epigenetic asymmetry in cell fate control can be harnessed to enable better protective immunity in many diseases, including adoptive cancer therapy. It should be noted, however, that in one study the deletion of Arid1a in tumor cells attenuated the response to immunotherapy50. Therefore, these findings do not support the use of such inhibitors in vivo.
Methods
Mice
Mice, including both sexes, were used for the study. We crossed Rosa26-Cas9 knock-in mice7 with OT-I42 transgenic mice to express Cas9 in antigen-specific CD8+ T cells (called Cas9-OT-I mice). The Cas9 mice were fully backcrossed to the C57BL/6 background. Mice bearing floxed Arid1a allele (Arid1afl/fl), in which exon 9 of the Arid1a gene is flanked by loxP sites, were described previously51,52, and crossed with Rosa26Cre-ERT2 knock-in mice52,53. Deletion of exon 9 was validated by RNAseq analysis of arid1a−/−, CD8+ T cells. c-Myc-GFP fusion knock-in mice were provided by B. Sleckman19. T cell-specific deletion of Pbrm154 was achieved by breeding Cd4Cre mice with Pbrm1fl/fl mice. c-Mycfl/fl mice were obtained from F. Alt55 and were backcrossed five generations onto the C57BL/6 background before breeding to Rosa26Cre-ERT2 mice53. C57BL/6, B6-albino56, CAG-luc-eGFP L2G85 transgenic mice57 and Rag1–/– mice were purchased from the Jackson Laboratory. All mice were kept in a specific pathogen-free facility in the Animal Resource Center at St. Jude Children’s Research Hospital. Mice were kept with 12-h light–dark cycles that coincide with daylight in Memphis, TN, USA. The St. Jude Children’s Research Hospital Animal Resource Center housing facility was maintained at 30–70 % humidity and 20–25 °C. All mice were used at 8–16 weeks old. Mouse studies were conducted in accordance with protocols approved by the St. Jude Children’s Research Hospital Committee on Care and Use of Animals and in compliance with all relevant ethical guidelines. All animal experiments were performed with age-matched littermate controls.
Cell line
B16-Ova and MC38-Ova cell line were kindly provided by Dr. Dario Vignali. F420 cell line was provided by Dr. Jason T Yustein. GL261 glioma cell line was purchased from DSMZ German Collection Laboratory. HEK293T and GP+E-86 cell line were purchased from ATCC.
Human naïve CD8+ T cell isolation and stimulation
Human naïve CD8+ T cells were isolated from apheresis rings obtained from the St. Jude Blood Donor Center. Peripheral blood mononuclear cells (PBMC) were isolated using Lymphoprep (07801, StemCell Inc.) following the manufacturer’s protocol. Briefly, blood from the apheresis ring was mixed with PBS containing 2% FBS in 1:1 ratio. The mixture was then layered carefully on lymphoprep solution. Samples were centrifuged at 4,000 rpm for 30 min at room temperature (RT) without braking. The PBMC layer was aspirated, washed twice with 2% FBS/PBS before isolation of naïve CD8+ T cell using the MojoSort Human CD8+ Naïve T Cell Isolation Kit (480046, Biolegend). Naïve CD8+ T cells were then stimulated with ImmunoCult Human CD3/CD28 T Cell Activator (10991, StemCell Inc.) plus 50 U/ml rhIL-2 or 10 ng/ml rhIL-15 for 2 days and expanded with 50 U/ml rhIL-2 or 10 ng/ml rhIL-15. BRD-K98645985 (1 μM; AOB36606, AOBIOUS or HY-114268, MedChemExpress) was added at the indicated time points. All human studies were in compliance with the Declaration of Helsinki. Blood was collected by the Blood Donor Center at St. Jude Children’s Research Hospital. Blood donors provided written consent for research use of their blood products not used in transfusions, which has been reviewed and approved by the Institutional Review Board at St. Jude Children’s Research Hospital.
Viral transduction and genome editing efficiency measurement
Naïve Cas9-expressing OT-I cells were isolated from the spleen and peripheral lymph nodes (pLN) of Cas9-OT-I mice by magnetic bead purification using a naïve CD8α+ T cell isolation kit according to the manufacturer’s instructions (Miltenyi Biotech). Purified naïve OT-I cells were activated in vitro for 18 hours with plate-bound anti-CD3ε (10 μg/ml; 2C11, BE0001, Bio-X-Cell) and anti-CD28 (5 μg/ml; 37.51, BE0015, Bio-X-Cell) antibodies. Viral transduction was performed by spin-infection at 900 g at 25 °C for 3 hours with 10 μg/ml polybrene (Sigma-Aldrich) followed by resting for 3 hours at 37 °C and 5% CO2. Cells were washed and cultured in media supplemented with murine IL-7 (12.5 ng/ml; Peprotech) and IL-15 (25 ng/ml; PeproTech) for 4 days. Cells were sorted based on the expression of fluorescent proteins using a Reflection cell sorter (iCyt) or MoFlo XDP cell sorter before adoptive transfer into recipients. At least two independent guides were designed using the platform from Broad Institute https://portals.broadinstitute.org/gpp/public/analysis-tools/sgrna-design for phenotypic analyses. We verified that at least two guides induced similar phenotypes. The sgRNAs editing efficiency was measured by insertion and deletion (indel) mutation analysis using CRIS.py58 (Supplementary Table 1).
Adoptive transfer, in vivo infection and recall assays
For influenza A virus infection, 5×105 naïve or first-division c-Myc-GFPhi and c-Myc-GFPlo OT-I CD8+ T cells were transferred into CD45.1+ recipient animals intravenously (i.v.), which were infected with 4×103 median embryo infectious dose (EID50) influenza A virus A/X-31 (H3N2) expressing ovalbumin peptide (SIINFEKL) (P. Thomas, St. Jude Children’s Research Hospital). Spleens were collected and analyzed by flow cytometry at nine days after the infection. For Listeria monocytogenes infection, naïve OT-I cells (5×103) or retrovirus-transduced (1−2×104) OT-I cells were adoptively transferred i.v. into naïve C57BL/6 mice (for analysis at effector phase) or into Cas9-expressing hosts (for analysis at memory phase). To examine cell-intrinsic effects, we applied the dual-color transfer systems. Specifically, cells transduced with the indicated sgRNAs (marked by the expression of Ametrine) were mixed at a 1:1 ratio with those transduced with sgNTC (labelled with GFP), followed by adoptive transfer into the same host. For pathogen-induced infections, 3×104 clone-forming units (CFU) of Listeria monocytogenes expressing ovalbumin (Lm-Ova) were injected i.v. To evaluate TMEM recall responses, 5×103 splenic TMEM cells were sorted and transferred to naïve C57BL/6 hosts and re-challenged with 5×104 CFU of Lm-Ova one day after TMEM transfer. The recall responses were analyzed at day 6 after re-challenge. To examine the homeostatic proliferation, a total of 5×105 TMEM cells were labeled with CellTrace Violet (C34557, ThermoFisher Scientific) and transferred into Rag1–/– mice and then analyzed at day 7 post-transfer.
For human CD8+ T cell experiments, naïve CD8+ T cells were activated by human CD3/CD28 T cell activator (10991, Stemcell) with or without an Arid1a inhibitor (BRD-K98645985; 1 μM; AOB36606, AOBIOUS or HY-114268, MedChemExpress) for two days and cultured in rhIL-15 (10 ng/ml, Peprotech) containing medium. 3×106 activated CD8+ T cells were then transferred to NSG mice. After 30 days, splenic cells were analyzed by flow cytometry.
CRISPR-Cas9 mutagenesis screening using the retroviral epigenetic library
Retroviral sgRNA epigenetic library construction
The retroviral sgRNA vector used for library construction was previously described5. A custom mouse epigenetic library targeting 337 genes were selected and guide RNA sequences were designed according to previously published criteria59. The library contains a total of 2,222 gRNAs with six gRNAs targeting one gene and 200 non-targeting controls. The synthesis, purification and quality control of the library were previously described60.
In vivo screening
The in vitro mutagenesis and in vivo screening approaches were modified based on our previous study5. Briefly, retrovirus was produced by co-transfecting the retroviral epigenetic library plasmid with packaging vector (pCL-Eco) in Plat-E cells. At 48 hours after transfection, the supernatant was harvested and frozen at −80 ºC. Naïve Cas9-expressing OT-I cells were isolated from two Cas9-OT-I mice and activated overnight with plate-bound anti-CD3ε (10 μg/ml; 2C11, BE0001, Bio-X-Cell) and anti-CD28 (5 μg/ml; 37.51, BE0015, Bio-X-Cell) antibodies. After activation, T cells were transduced with the retrovirus library at low multiplicity of infection to achieve ~20% transduction efficiency. Cells were washed and cultured in media supplemented with murine IL-7 (12.5 ng/ml; Peprotech) and IL-15 (25 ng/ml; PeproTech) for 4 days to expand and allow gene editing to occur. Transduced cells were sorted based on the expression of Ametrine and an aliquot of 0.5×106 transduced OT-I cells was saved as “day 0 input” (> 200× cell coverage per sgRNA). Transduced OT-I cells (0.2×106) were then transferred i.v. into twenty-one Cas9 expressing hosts followed by Lm-Ova infection (3×104 CFU) 2 hours later. At day 7.5 or 36 after infection, total donor-derived cells, terminal effector (TE, KLRG1hiCD127lo) and memory precursor (MP, KLRG1loCD127hi) cells were sorted and frozen at −80 ºC until genomic DNA extraction. A minimum of 0.5×106 OT-I cells per sample (> 200× cell coverage per sgRNA) was recovered for further analysis.
Library preparation
Genomic DNA was extracted by using the DNeasy Blood & Tissue Kits (69504, Qiagen) according to the manufacturer’s instruction. Two rounds of PCR were performed by using the KOD Hot Start DNA Polymerase (Sigma-Aldrich) with primary PCR to amplify the sgRNAs and second PCR to attach Illumina Nextera adapters to each sample. PCR products were purified using AMPure XP beads (A63881, Beckman Coulter) after each PCR reaction. Primers sequences to amplify sgRNAs for the first PCR were:
Nextera NGS-F: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTTGTGGAAAGGACGAAACACCG; Nextera NGS-R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGCCACTTTTTCAAGTTGATAACGGThe library sequencing was performed using the Hi-seq (Illumina) platform.
Data processing of in vivo pooled CRISPR screening
For data analysis, raw FASTQ files obtained after sequencing were demultiplexed using the HiSeq Analysis software (Illumina). Single-end reads were trimmed and quality-filtered using the CLC Genomics Workbench v.11 (Qiagen) and matched against sgRNA sequences from the sgRNA epigenetic library. Read counts for sgRNAs were normalized against total read counts across all samples. For each sgRNA, the fold change (log2-transformed ratio) for enrichment was calculated between each of the biological replicates and the input experiment. The gene-level false-discovery-rate (FDR)-adjusted p value was calculated among multiple sgRNAs (n=6) of each gene, using a two-tailed paired Student’s t-test between log2-transformed average normalized read counts of MP samples and those of TE samples, between counts of total day 7.5 samples and those of total day 36 samples.
Tissue dissociation of non-lymphoid organs
Lung and liver were collected and minced into small pieces using razor blades. The organs were digested in dissociation buffer containing 1 mg/ml of collagenase IV (LS004188, Worthington Biochemicals) and 0.5 mg/ml of DNase I (DN25-1G, Sigma-Aldrich) at 37 °C for 30 min on a 3D orbital mixer. The cell suspensions were then passed through 70-μm filters to remove undigested tissues and followed by density-gradient centrifugation over Percoll (17089101, GE Healthcare).
Flow cytometry
Antibodies from Biolegend included APC/Cy7 anti-human CD8α (RPA-T8, 301016), BV785 anti-human CD45RA (HI100, 304139), PE anti-human CD62L (DREG-56, 304806), APC anti-human CD27 (M-T271, 356410), PE/Cy7 anti-human CCR7 (G043H7, 353226), BV421 anti-human CD95 (DX2, 305623), BV785 anti-mouse/human CD44 (IM7, 103059), BV510 anti-mouse KLRG1 (2F1/KLRG1, 138421), KIRAVIA Blue 520 anti-mouse CD62L (MEL-14, 104464), BV605 anti-mouse CD127 (A7R34, 135025), BV650 anti-mouse CX3CR1 (SA011F11, 149033), APC anti-mouse CD8α (53–6.7, 100712), Alexa Fluor 700 anti-mouse CD45.2 (104, 109822), APC anti-mouse CD98 (RL388, 128212); PE anti-mouse CXCR3 (CXCR3-173, 12-1831-82, eBioscience); and BUV395 anti-human CD3 (UCHT1, 563548), BUV805 anti-human CD45RO (UCHL1, 748367); BUV496 anti-mouse CD45.1 (A20, 741093), BUV805 anti-mouse CD8α (53–6.7, 612898) were acquired from BD Biosciences. For surface markers, cells were stained for 30 min at 4°C in PBS + 2% (w/v) BSA. To examine intracellular cytokine production, splenocytes were stimulated with 1 μg/ml ovalbumin peptide (SIINFEKL) in the presence of monensin (554724, BD Biosciences) for 5 hours and stained with anti-IFN-γ (XMG1.2, 505826, BioLegend), anti-TNF-α (MP6-XT22, 17-7321-82, Thermo Fisher Scientific) and anti-Granzyme B (GB11, 515405, BioLegend) using a fixation/permeabilization kit (554714, BD Biosciences). Fixable viability dye (Thermo Fisher Scientific) was used for dead-cell exclusion. Samples were acquired on Cytek Aurora (SpectroFlo) or BD LSRII (FACSDiva) flow cytometer and analyzed with FlowJo software.
In vivo tumor transplant experiment
For B16-Ova and MC38-Ova tumors, 0.5×106 tumor cells were injected subcutaneously into naïve mice (7–10 weeks of age). 2×106 OT-I cells were transferred i.v. 7 days after tumor injection. Tumors were measured with callipers and every two days and tumor volumes were calculated by the formula: (width2×length)/2.
For tumor re-challenge assays, sgNTC- or indicated sgRNA-transduced OT-I cells (1×104) were transferred into Cas9-expressing mice (7–10 weeks of age) followed by 3×104 CFU of Lm-Ova infection. At >30 days post infection, B16-Ova cells (1×107) were injected subcutaneously. Tumor size was measured every three days.
For CAR-T tumor models, the murine osteosarcoma cell line F420 was derived from singly floxed p53+/F-Col2.3 transgenic mice35; 0.5×106 F420 cells were injected subcutaneously into naïve mice. 7 days after tumor transplant, 5×106 B7-H3 CAR-T cells were transferred i.v. without lymphodepleting chemotherapy and tumor size was measured every two days.
For CAR-T tumor models with lymphodepleting chemotherapy, 0.5×106 F420 cells were injected subcutaneously into naïve mice. At 7 days after tumor transplant, cyclophosphamide (200mg/kg; Northstar Rx LLC) was injected (i.p.). CAR-T cells were transferred at day 9, and tumor size was measured every two days.
Bioluminescence imaging
After adoptive transfer of CAR-T cells, tumor-bearing mice were injected with D-luciferin (150 mg/kg) 5 minutes before imaging with Xenogen IVIS-200 imaging system (Xenogen). Bioluminescence imaging data were analyzed by Living Image software (v4.7.3, Caliper Life Sciences). Mice were imaged by the St. Jude Center for In Vivo Imaging and Therapeutics (CIVIC).
Generation of murine CAR-T cells
Generation of murine (m) B7-H3-CAR constructs have been previously described34. Briefly, a codon-optimized DNA was generated encoding mB7-H3-specific scFv derived from the m276 monoclonal antibody61 followed by a portion of the murine CD28 molecule (including CD28 extracellular, transmembrane, and cytoplasmic domains) and a CD3ζ domain (with mutated 1st and 3rd ITAMs) as described before62. The sequence was synthesized by GeneArt (Thermo Fisher) and ligated using cut-and-paste cloning into the mouse stem cell virus–based splice-gag vector (MSGV) retroviral backbone between HindIII and SacII cutting sites replacing the ID3–28Z CAR sequence62 with mB7-H3.CD28.CD3ζ sequence. The sequence of final mB7-H3.CD28.ζ CAR construct was verified by sequencing. As a control, we generated a mB7-H3 STOP-CAR (Extended Data Fig. 8c) where CD28.CD3ζ endodomain has been deleted from mB7-H3-CAR using In-Fusion cloning. The sequence of the STOP-CAR construct was verified by sequencing.
The retrovirus for murine CAR-T cell generation was produced as described previously34. Briefly, retroviral particles were generated by transient transfection of 293T cells with the CAR-encoding plasmid, Peg-Pam plasmid encoding MoMLV gag-pol, and plasmid encoding the VSVG envelope. Virus was harvested at 48 hours and filtered with 0.45 mm filter. The VSVG-pseudo typed virus was then used to transduce the GPE86 producer cell line. B7-H3-CAR-expressing GPE86 cells were stained with Alexa Fluor-647 anti-human IgG, F(ab’)2 fragment antibody (109-606-006, Jackson ImmunoResearch) and sorted by using BD FACSAria III.
Murine CAR-T cells were generated as described previously34 with modification. Naïve CD8+ T cells from 6–8 weeks old CD45.1+ mice or from luciferase transgenic mice were activated with plate bound anti-CD3ε (1 μg/ml, 145–2C11, BE0001, BioXcell), anti-CD28 (2 μg/ml, 37.51, BE0015, BioXcell) and 50 U/ml rhIL-2 (200–02, Peprotech). On day 2 post-activation, CD8+ T cells were transduced with retrovirus expressing B7-H3-CAR or control CAR on retronectin (T100B, Takara)-coated non-tissue culture treated plate in complete RPMI medium supplemented with 50 U/ml rhIL-2. At 48 hours after transduction, CAR-T cells were harvested and expanded in the presence of 10 ng/ml rhIL-15 (200–15, Peprotech) for another 3 days and then used for in vivo and in vitro experiments.
Repeat stimulation assay
For repeat stimulation assays, 3×105 tumor cells (GL261, F420) were plated in a 24 well plate and allowed to adhere for 4–6 hours at 37 °C, CAR-T cells (1×106) were then added to the wells containing tumor cells without any exogenous cytokines (tumor : CAR-T cell ratio=1:3). After 3 days of co-culture, CAR-T cells were lifted by gentle pipetting to avoid disrupting adherent tumor cells, and the number of T cells was determined using a hemocytometer. Stimulations and cell counts were repeated every 3 days with the same tumor cells.
Co-immunoprecipitation
For endogenous co-immunoprecipitation, naïve CD8+ T cells were stimulated on plate-bound anti-CD3ε (2 μg/ml, 145-2C11, BE0001, BioXcell), anti-CD28 (1 μg/ml, 37.51, BE0015, BioXcell) and ICAM1 (0.5 μg/ml, Peprotech) for 36 hours. Nuclear extracts were prepared as described63 with some modifications. Briefly, cells were harvested and resuspended in hypotonic buffer (50 mM Tris, pH7.5, 0.1% NP-40, 1mM MgCl2 supplemented with complete, EDTA-free Protease Inhibitor Cocktail (11873580001, Roche)). Nuclei were pelleted at 5000 rpm for 10 min at 4 °C and resuspended in CelLytic M buffer (C2978, Sigma). Lysates were incubated at 4 °C for 30 min and pelleted at 20,000 g for 10 min at 4 °C. Supernatants were collected for co-immunoprecipitation. Nuclear extracts were incubated at 4 °C overnight with 2 μg of the following antibodies: anti-Arid1a (12354, Cell Signaling Technology), anti-c-Myc (5605, Cell Signaling Technology) and rabbit mAb IgG XP Isotype Control (3900, Cell Signaling Technology). Samples were then incubated with Dynabeads protein A (10001D, Thermo Fisher Scientific) for 1 hour at 4 °C. Beads were washed 3 times in RIPA buffer and eluted with 1× sample buffer (1610791, Bio-rad).
Immunoblot
Cells were lysed in CelLytic M buffer (C2978, Sigma). Cell lysates and immunoprecipitated complexes were separated by SDS-PAGE and blotted with the following antibodies: anti-Arid1a (1:1000; D2A8U, 12354S, Cell Signaling Technology), anti-Brg1 (1:1000; EPNCIR111A, ab110641, Abcam), anti-Brg1 (1:1000; G-7, sc-17796, Santa Cruz), anti-c-Myc (1:1000; D84C12, 5605S, Cell Signaling Technology), anti-smarcb1 (1:1000; EPR20189, ab222519, Abcam), anti-Pbrm1 (1:1000; ABE70, Millipore), anti-Brm (1:1000; ab15597, Abcam), anti-Smarcd1 (1:1000; A301-594, Bethyl), anti-Smarcd2 (1:1000; A301-596A, Bethyl), anti-Baf57 (1:1000; EPR8849, ab137081, Abcam), anti-Baf155 (1:1000; D7F8S, 11956, Cell Signaling Technology), anti-Baf170 (1:1000; 8829, Cell Signaling Technology), anti-Baf45c (1:1000; PA5-38011, ThermoFisher Scientific), anti-Baf45d (1:1000; A303-595A, Bethyl), anti-Brd7 (1:1000; B-8, sc376180, Santa Cruz), anti-Brd9 (1:1000; 61537, Active Motif), anti-Actin (1:1000; C-4, sc-47778, Santa Cruz), anti-Gapdh (1:1000; 6C5, MAB374, Millipore).
Immunofluorescence staining and imaging
Naïve CD8+ T cells were stimulated on plate-bound anti-CD3ε (2 μg/ml, 145-2C11, BE0001, BioXcell), anti-CD28 (1 μg/ml, 37.51, BE0015, BioXcell) and ICAM1 (0.5 μg/ml, Peprotech) for 28 hours followed by fixation for 10 min with 4% paraformaldehyde (PFA) (15710, Electron Microscopy Science) at RT. Cells were rinsed with TBS (50 mM Tris pH8.0, 100 mM NaCl) and permeabilized with permeabilization buffer (Tris pH8.0, 100 mM NaCl, 0.3% (v/v) Triton X-100) for 5 min at RT, and then blocked with TBS + 2% BSA (A7030, Sigma) for 30 min at RT. Cells were stained at 4 °C overnight with the following primary antibodies: anti-c-Myc (1:500, D84C12, 5605, Cell Signaling Technology), anti-Arid1a (1:500, EPR13501, ab182560, Abcam), anti-Brg1 (1:500, EPNCIR111A, ab110641, Abcam), anti-Smarcb1 (1:250, EPR20189, 222519, Abcam), anti-tubulin (1:1000, DM1A, T9026, Sigma), anti-tubulin (1:1000, YOL1/34, MA1-80189, Invitrogen), anti-Lamin B1 (1:500, ab16048, Abcam), anti-BAF180 (1:500, ABE70, EMD Millipore), anti-PBRM1 (1:200, BL-39-2C3, A700-019, Bethyl Laboratories). Samples were washed twice with TBS and incubated with for 1 hour at RT with the following secondary anti-bodies: anti-rabbit Alexa Fluor plus 555 (1:2000, A32732, Thermo Fisher Scientific), anti-mouse Alexa Fluor plus 647 (1:2000, A32728, Thermo Fisher Scientific), anti-rat Alexa Flour 647 (1:1000, 712-605-153, Jackson ImmunoResearch). Samples were imaged by Marianas spin disk confocal (Intelligent Imaging Innovations), Prime95B sCMOS camera, and 405, 488, 561 and 640 nm laser lines were used. Dividing sister cells were identified by the presence of an intercellular cytokinetic bridge based on tubulin staining. Sum fluorescent intensities were analyzed by Slide book 6 (Intelligent Imaging Innovations) and Imaris (Oxford Instruments).
ATAC-see
ATCC-see was performed as described22. Briefly, naïve CD8+ T cells were stimulated with anti-CD3/28 and ICAM1 for 28 hours followed by fixation for 10 min at RT with 4% PFA. Cells were permeabilized with lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.01% NP-40) for 10 min at RT. Samples were washed twice with PBS in a humid chamber box at 37 °C. The transposase solution (150 ml 2×TD buffer, 100 nM Tn5-ATTO550N, add H2O to 300 ml) was added to the chambered cover glass and incubated for 30 min at 37 °C. Then, samples were washed three times with stop buffer (0.01% SDS, 50 mM EDTA in PBS) for 15 min at 55 °C. Immunofluorescence staining were performed after ATAC-see staining.
Microarray analysis
A total of 1×105 donor-derived OT-I cells were sorted, and RNA was isolated using the RNeasy Micro Kit (Qiagen) following the manufacturer’s instructions. The RNA Integrity Number (RIN) and concentration of RNA were measured by Agilent 2100 bioanalyzer. RNA (1 ng) was used for subsequent microarray analysis with Clariom S mouse array platform (Thermo Fisher Scientific). To perform microarray analyses, the gene expression signals were summarized with the robust multi-array average algorithm (Affymetrix Expression Console v1.1). The differentially expressed gene analysis was performed using lmFit method implemented in R package limma v.3.34.9 64. FDR was calculated by Benjamini–Hochberg method. Plots were generated using the R package ggplot2 v.2.2.1. Gene set enrichment analysis (GSEA) and functional gene set enrichment using C7 immunological collections from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/) was performed as previously described65. Several gene sets that were frequently used in this study were renamed as follows: MP and TE signatures, which are respectively “IL-7R low vs high eff CD8 T cell DN” and “IL-7R low vs high eff CD8 T cell UP” signatures9.
ATAC-seq
Naïve CD8+ T cells were activated with anti-CD3/CD28 plus ICAM1 for 36 hours and first-division wild-type (WT) CD8+ T cells or CD8+ T cells with acute c-Myc deletion (for c-Myc acute knockout, naïve CD8+ T cells purified from Mycfl/flRosa26Cre-ERT2 mice were treated with 4OHT (4-Hydroxytamoxifen) (H6278, Sigma-Aldrich) overnight with 50 U/ml rhIL-2 before activation) were sorted into medium and processed to generate ATAC-seq libraries immediately as previously described66 with slight modifications. Briefly, 50,000 cells were pelleted at 500 g for 5 min at 4 °C. Samples were washed in 1 ml cold PBS and resuspended in 50 ml cold cell lysis buffer (10 mM Tris pH7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40). Nuclei were pelleted at 1000 g for 10 min and resuspended in 50 μl transposition reaction mixture (25 μl TD buffer, 2.5 μl TDE1, 22.5 μl ddH2O) (20034197, Illumina). Samples were incubated at 42 °C for 45 min. DNA was then purified using MiniElute PCR Purification Kit (28004, Qiagen). Libraries were barcoded and amplified with Q5 High-Fidelity 2×Master Mix (M0492S, NEB). Libraries were quantified using Qubit and size distribution determined by Agilent 4200 TapeStation before sequencing.
Analysis for ATAC-seq data
Paired-end sequencing reads were trimmed with Trim Galore (version 0.5.0) with default parameters. The reads were then aligned to the reference mouse mm10 assembly using Bowtie 2 (version 2.3.5.1) with settings --end-to-end --very-sensitive -X 2000. The resulting alignments, recorded in BAM file, were sorted, indexed, and marked for duplicates with Picard MarkDuplicates function (version 2.19.0). Afterward, the BAM file was filtered with SAMtools (version 1.9), BamTools (version 2.5.1), and scripts of nf-core/chipseq67 to discard reads, mates that were unmapped, PCR/optical duplicates, not primary alignments, mapped to multiple locations, or mapped to ENCODE blacklisted regions68; only reads mapped in proper pair were kept (-F 1804 -f 2 -q 30). MACS (version 2.1.2) was used to call peaks from the BAM file with narrowPeak setting, --extsize 200, and recommended mappable genome size (default value for other parameters). Chromatin accessibility signal was normalized by scaling to 1 million mapped reads using BEDTools (version 2.27.1) and bedGraphToBigWig (version 377), and visualized as heatmaps using deepTools plotHeatmap (version 3.2.1).
In ATAC-seq experiments with spike-in Drosophila S2 cells, two modifications were made from the above analysis: i) a hybrid reference of mouse mm10 and Drosophila melanogaster Ensembl r97 was used, ii) signals were normalized by scaling to per million reads mapped to Drosophila.
Differentially accessibility analysis was performed using DiffBind (version 2.16.0) (summit=250, DESeq2 and other default parameters). Peaks were annotated to the nearest genes with annotatePeak function in R package ChIPseeker (version 1.26.2) at gene level with default parameters. Genes were ranked by Wald statistic from DESeq2 (if a gene was associated with multiple peaks, the mean was used). Gene set enrichment analysis (GSEA) was then performed against MSigDB, GO, Reactome, KEGG with R package clusterProfiler (version 3.18.1).
CUT&RUN
CUT&RUN experiments were performed as previously described26 with slight modifications. For first-division CD8+ T cell samples, naïve CD8+ T cells were stimulated on plate-bound anti-CD3/CD28 plus ICAM1 for 36 hours, and first-division CD8+ T cells were sorted. For c-Myc acute knockout samples, naïve CD8+ T cells purified from Mycfl/flRosa26Cre-ERT2 mice were treated with 4OHT overnight before stimulation; dead cells were removed by using Dead Cell Removal Kit (130-090-101, Miltenyi Biotec). 3×105 cells were washed with wash buffer (20 mM HEPES (H3375, Sigma-Aldrich), 150 mM NaCl (AM9760G, Invitrogen), 0.5 mM spermidine (S0266, Sigma-Aldrich), and protease inhibitor cocktail (5056489001, Sigma-Aldrich)) twice. Cells were then resuspended and bound to concanavalin A-coated magnetic beads (BP531, Bang Laboratories). Samples were placed on the magnetic stand, residual liquid was removed from beads, and beads were resuspended in 100 μl antibody buffer (20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.01% digitonin (300410, Milipore), 2 mM EDTA (AM9260G, Invitrogen), and Protease inhibitor cocktail). Primary antibodies were added to the samples at a final concentration of 1:100 and incubated overnight at 4 °C. The next day, samples were washed with cold Dig-wash buffer (20 mM HEPES, 150 mM NaCl, 0.5 mM spermidine, 0.01% digitonin and protease inhibitor) twice. After washing, pAG-MNase (123461, Addgene) was added to tubes and rotated at 4 °C for 1 h. After two washes with Dig-wash buffer, samples were resuspended in 50 μl Dig-wash buffer. 2 μl of 100 mM CaCl2 (2115, Sigma-Aldrich) was added per sample, briefly vortexed, and immediately placed on ice for 30 min. 50 μl 2×STOP buffer [340 mM NaCl, 20 mM EDTA, 4 mM EGTA (E3889, Sigma-Aldrich), 100 μg/mL RNase A (EN0531, Thermo-Fisher), and 50 μg/mL GlycoBlue (AM9515, Invitrogen)] were added and mixed by gentle vortexing. Then, samples were incubated for 30 min at 37 °C to release CUT&RUN fragments. Fragmented DNA was purified with the NEB Monarch PCR&DNA Cleanup Kit (T1030S, NEB). DNA libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit (E7645S, NEB) and purified with AMPure SPRI beads (B23318, Beckman-Coulter). Libraries were quantified using Qubit and size distribution was determined by Agilent 4200 TapeStation analysis before 15 million paired-end sequencing was performed.
Peaks were annotated to the nearest genes with annotatePeak function in R package ChIPseeker (version 1.26.2) at gene level with default parameters. Enrichment analysis was performed against MSigDB, GO, Reactome, KEGG with the over-representation approach implemented in R package clusterProfiler (version 3.18.1).
CUT&RUN data processing
Paired-end sequencing reads were trimmed with Trim Galore (version 0.5.0) with default parameters. Then, reads were aligned to the reference mouse mm10 assembly using Bowtie 2 (version 2.3.5.1) with settings --end-to-end --very-sensitive --no-mixed --no-discordant -q --phred33 -I 10 -X 700. The resulting alignments, recorded in BAM file, were sorted, indexed, and marked for duplicates with Picard MarkDuplicates function (version 2.19.0). Afterward, the BAM file was filtered with SAMtools (version 1.9), BamTools (version 2.5.1), and scripts of nf-core/chipseq67 to discard reads, mates that were unmapped, PCR/optical duplicates, not primary alignments, mapped to multiple locations, mapped to ENCODE blacklisted regions68, or have more than 4 mismatches (-F 0×004 -F 0×008 -F 0×0100 -F 0×0400 -f 0×001 -q 1). MACS (version 2.1.2) was used to call peaks from the BAM file with IgG control and recommended mappable genome size (default value for other parameters). NarrowPeak mode was used for c-Myc, while broadPeak mode was used for Arid1a and Brg1. Binding signal was normalized by scaling to 1 million mapped reads using BEDTools (version 2.27.1) and bedGraphToBigWig (version 377), and visualized as heatmaps using deepTools plotHeatmap (version 3.2.1).
In CUT&RUN experiments with spike-in E. coli, two modifications were made: i) a hybrid reference of mouse mm10 and E. coli ASM584v2 was used, ii) signals were normalized by scaling to per million reads mapped to E. coli.
RNA-seq
For first-division CD8+ T cell samples, naïve CD8+ T cells were stimulated on plate-bound anti-CD3/CD28 plus ICAM1 for 36 hours, and first-division CD8+ T cells were sorted. For c-Myc acute knockout samples, naïve CD8+ T cells from Mycfl/flRosa26CreERT2 mice were treated with 4OHT overnight before stimulation; dead cells were removed by using Dead Cell Removal Kit (130-090-101, Miltenyi Biotec). Total RNA was isolated using Direct-zol RNA Microprep Kits (R2061, Zymo Research) following the manufacturer’s instruction and quantified using Agilent 4200 TapeStation. Libraries were prepared using KAPA RNA HyperPrep Kit with RiboErase (HMR) (08098131702, Roche) and purified by AMPure SPRI beads (B23318, Beckman-Coulter). Libraries were quantified and size distribution was determined by Agilent 4200 Tapestation before paired-end sequencing was performed.
RNA-seq data processing
Paired-end sequencing reads were mapped by the pipeline of the St Jude Center for Applied Bioinformatics. Briefly, the reads were trimmed with Trim Galore (version 0.5.0) with default parameters. Then, reads were aligned to the reference mouse mm10 assembly plus ERCC spike-in sequences using STAR (version 2.7.5a). The resulting alignments, recorded in BAM file, were sorted, indexed, and marked for duplicates with Picard MarkDuplicates function (version 2.19.0). Transcript quantification was calculated using RSEM69. Differential gene expression analysis was carried out with DESeq2 with Wald test (default parameters)70. Genes were pre-filtered to have at least 5 reads in at least 2 samples. Gene set enrichment analysis (GSEA) was performed against MSigDB, GO, Reactome, KEGG with R package clusterProfiler (version 3.18.1). Wald statistic calculated from DESeq2 was used for gene ranking.
For the RNA-seq experiment with ERCC spike-in. RUVg function in RUVSeq package was used for normalization followed by DESeq2 according to RUVSeq manual71.
Peak set enrichment analysis
Peak set enrichment analysis (PSEA) was developed to examine if the differentially accessible regions up- or down-regulated in Arid1a KO were enriched along the ranked list of accessible regions by the change between Arid1a inhibitor and vehicle. The analysis was based on gene set enrichment analysis and was performed at peak level instead of gene level. It was implemented with GSEA function of R package clusterProfiler. In this study, the geneList parameter was set as the ranked list of accessible regions by the fold change of inhibitor versus vehicle; the TERM2GENE parameter was set using the differentially accessible regions between Arid1a KO and wild type identified as described in the ATAC-seq data analysis section (false positive rate < 0.05 and fold change > 2). The accessible regions were present at least in two replicates of each condition or genotype.
Quantification and statistical analysis
Data are plotted and analyzed by GraphPad Prism (GraphPad Software, version 9.2.0). Statistical significance was calculated with unpaired or paired two-tailed Student’s t-test. Two-way ANOVA was performed to compare tumor growth curves. The log-rank (Mantel-Cox) test was performed for comparing mouse survival curves. P value less than 0.05 was considered significant. Data are presented as mean±s.e.m.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information. All RNA-seq, ATAC-seq, CUT&RUN and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE183619. Publicly available database used in this study are from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/). Source data are provided with this paper.
Extended Data
Extended Data Figure 1. Deletion of Smarcd2 or Arid1a promotes TMEM generation.
a, Venn diagram of the overlapping candidates negatively regulating TMEM generation from enriched genes in MP relative to TE cells at day 7.5 post-infection and in total cells at day 36 relative to total cells at day 7.5. b, Flow cytometry plot of TE (KLRG1hiCD127lo), MP (KLRG1loCD127hi), KLRG1hiCX3CR1hi, CD62L+, and CXCR3hiCD27hi CD8+ T cells in sgNTC- and the indicated sgRNA-transduced OT-I cells at day 7.5 post Lm-Ova infection (n=6 mice). c, Quantification of splenic TE and MP cell numbers at day 7.5 post Lm-Ova infection (n=5 mice). d, Transcriptome profiling by microarray of sgNTC and the indicated sgRNA-transduced OT-I cells at day 7.5 post Lm-ova infection in vivo (n=4 mice). e, f, CellTrace Violet labeled WT and Arid1a−/− naïve CD8+ T cells were activated by anti-CD3/CD28 plus ICAM1 for 36 hours, and first-division cells were sorted for RNA-seq (n=5 mice). e, Volcano plot of differentially expressed genes assessed by RNAseq in WT and Arid1a−/− first-division CD8+ T cells. TEFF and TMEM associated genes are highlighted in red and green respectively. f, Gene set enrichment analysis (GSEA) of RNAseq results from WT and Arid1a−/− first-division CD8+ T cells using the C7 immunological collections. Shown are all enriched TEFF-associated or TMEM-associated gene sets. Data are shown as mean±s.e.m. *p<0.05, **p < 0.01, ***p<0.001; two-tailed paired Student’s t-test (c); two-sided Wald test and adjusted with Benjamini-Hochberg procedure (e); two-sided permutation test and adjusted with Benjamini-Hochberg procedure (f).
Extended Data Figure 2. Deficiency in SWI/SNF promotes bona fide TMEM formation.
a, Diagram of experimental approach to evaluate TMEM function. p.i., post-infection. b, Quantification of numbers of OT-I cells expressing the indicated sgRNA in multiple tissues at day >30 post-infection (n=5 mice per group). c, Flow cytometry analysis (left) and quantification (right) of the mean fluorescence intensity (MFI) of CellTrace Violet (CTV) dilution in donor-derived OT-I cells. Splenic TMEM OT-I cells transduced with either sgNTC (GFP+ spike cells) or the indicated sgRNA (Ametrine+) were isolated from mice that were challenged with Lm-Ova at day >30 prior, labeled with CTV, mixed at a 1:1 ratio, and transferred to Rag1-deficient recipients. Cells were analyzed at day 7 post-transfer (n=3 mice per group). d, e, Splenic TMEM OT-I cells transduced with sgNTC (GFP+ spike cells) or the indicated sgRNA (Ametrine+) were isolated from mice that were challenged with Lm-Ova at day >30 prior, mixed at a 1:1 ratio, and transferred to naïve C57BL/6 recipients followed by Lm-Ova re-challenge. The cells were analyzed at day 6 after re-challenge. d, Flow cytometry plot and quantification of cells expressing both interferon-γ (IFN-γ) and TNF-α after ovalbumin peptide stimulation for 5 hours in the presence of monensin (n=5 per group). e, Quantification of Granzyme B (GzmB) MFI (n=5 per group). Data are compiled from at least two independent experiments (b–e). Data are shown as mean±s.e.m. *p < 0.05, **p < 0.01; two-tailed paired Student’s t-test (b, c, d, e).
Extended Data Figure 3. The cBAF complex but not the PBAF complex affects CD8+ T cell differentiation.
a, Immunoblot of Pbrm1 in naïve CD8+ T cells from Pbrm1fl/fl and Cd4CrePbrm1fl/fl mice. b−e Pbrm1fl/fl and Cd4CrePbrm1fl/fl mice were challenged with Lm-Ova and analyzed at day 7.5 (b, c) or at day >30 post-infection (d, e). b, Flow cytometry analysis (left) and quantification (right) of the frequencies and numbers of total CD8+ T cells (upper) and Ova-specific CD8+ T cells (lower) (Pbrm1fl/fl, n=8 mice; Cd4CrePbrm1fl/fl, n=6 mice). c, Flow cytometry analysis (left) and quantification (right) of Ova-specific TE (KLRG1hiCD127lo), MP (KLRG1loCD127hi), CXCR1hiKLRG1hi and CD62L+ cells in spleen (n=6 per group). d, Flow cytometry analysis (left) and quantification (right) of Ova-specific TMEM cells in the spleen and indicated organs (Pbrm1fl/fl, n=5 mice; Cd4CrePbrm1fl/fl, n=8 mice). e, Percentage of splenic CD62L+ TCM population (Pbrm1fl/fl, n=5 mice; Cd4CrePbrm1fl/fl, n=8 mice). f, Flow cytometry analysis of splenic TE (KLRG1hiCD127lo), MP (KLRG1loCD127hi) and CD62L+ OT-I cells transduced with sgNTC or the indicated sgRNA targeting ncBAF complex components at day 7.5 post Lm-Ova infection (Pbrm1fl/fl, n=5 mice; Cd4CrePbrm1fl/fl, n=8 mice). g, Flow cytometry analysis of splenic TE (KLRG1hiCD127lo), MP (KLRG1loCD127hi) and CD62L+ OT-I cells transduced with sgNTC or the indicated sgRNA at day 7.5 post Lm-Ova infection. Data are compiled from two (b−e) or representative of two (a, f, g) independent experiments. Data are shown as mean±s.e.m. **p<0.01, ns, not significant; two-tailed unpaired Student’s t-test (b−g).
Extended Data Figure 4. cBAF components asymmetrically segregate to daughter cells during the first division after activation.
a, Representative images (left) of conjoined first-division daughter c-Myc-GFP-expressing CD8+ T cells, stained for Hoechst (blue), Pbrm1 (red) and Tubulin (white); and quantification (right) of asymmetric index (difference in fluorescence intensity/total) (n=12 cells). Arrows mark tubulin bridges. Scale bar: 5 μm. b, Sorting strategy for first-division c-Myc-GFPhi and c-Myc-GFPlo CD8+ T cells. Naïve CD8+ T cells were labeled with CellTrace Violet (CTV) and activated with anti-CD3ε (2 μg/ml), anti-CD28 (1 μg/ml) plus ICAM1 (0.5 μg/ml) for 36 hours prior to sorting. c, d, Immunoblot of BAF components in c-Myc-GFPhi and c-Myc-GFPlo (c) or CD98hi and CD98lo (d) first-division CD8+ T cells. e, Principal component analysis of the ATAC-seq data in sorted c-Myc-GFPhi and c-Myc-GFPlo daughter cells from first-division stage (n=3 biological replicates). f, Heatmap of open chromatin regions in sorted c-Myc-GFPhi and c-Myc-GFPlo CD8+ T cells at the promoter, intron and intergenic regions (n=3 biological replicates). g, TEFF and TMEM cell-associated gene sets identified by enrichment analysis of differentially accessible chromatin peaks between c-Myc-GFPhi and Myc-GFPlo CD8+ T cells (n=3 biological replicates). h, Diagram of deletion of Arid1a in sorted c-Myc-GFPhi and c-Myc-GFPlo OT-I cells prior to IAV-Ova infection (related to Fig. 2f, g). Data are representative of one (e−g) or at least two independent experiments (a, c, d). Data are shown as mean±s.e.m. **p < 0.01, ns, not significant; one-way ANOVA (a).
Extended Data Figure 5. The cBAF complex shares binding sites with c-Myc in T cell differentiation-associated chromatin loci.
a–d, WT and Arid1a−/− naïve CD8+ T cells were activated by anti-CD3/CD28 plus ICAM1 for 36 hours, and first-division CD8+ T cells were sorted for CUT&RUN assay. a, Venn diagram of the overlap between CUT&RUN peaks in WT first-division CD8+ T cells for Arid1a and Brg1 (n=2 biological replicates). b, Relative locations of c-Myc peaks to BAF components peaks in chromatin. c, Enrichment analysis of Brg1 and c-Myc shared binding sites in first-division WT CD8+ T cells using Hallmark gene sets (upper) and Gene ontology gene sets (lower). The co-bound sites were annotated to the nearest genes and enrichment was calculated with over-representation approach. d, Genome browser tracks of c-Myc and cBAF shared binding sites at Gzmb, Il2ra and Tbx21 loci. The overlapping peaks are highlighted. Data are representative of one experiment (a−d). The p-value was calculated by hypergeometric distribution, one-sided and adjusted with Benjamini-Hochberg procedure (c).
Extended Data Figure 6. Arid1a deletion reduces chromatin accessibility to sites associated with the TEFF gene signature.
a, Volcano plot of ATAC-seq peaks from WT and Arid1a−/− first-division CD8+ T cells (n=3 biological replicates). b, TEFF and TMEM cell-associated gene sets as analyzed by enrichment analysis of differentially accessible chromatin peaks between first-division WT and Arid1a−/− CD8+ T cells (n=3 biological replicates). c, Genome browser tracks of chromatin accessibility at Gzma, Il2 and Klrg1 loci (n=3 biological replicates). d, Gene set enrichment analysis of Myc targets and mTORC1 signaling gene sets in WT and Arid1a−/− first-division CD8+ T cells from RNA-seq (n=3 biological replicates). e, Normalized read count of Myc mRNA from RNA-seq (left), and immunoblot for c-Myc (right) in WT and Arid1a−/− activated CD8+ T cells. (n=3 biological replicates) p-value was calculated with two-sided Wilcoxon test. Data are representative of one (a−e, RNA-seq) or three (e, immunoblot) experiments. The p-value was calculated with Wald test, two-sided and adjusted with Benjamini-Hochberg procedure (a) or permutation test, two-sided and adjusted with Benjamini-Hochberg procedure (d).
Extended Data Figure 7. cBAF cooperates with c-Myc to restrict CD8+ TMEM differentiation.
a, b, Naïve CD8+ T cells from Rosa26Cre-ERT2Mycfl/fl and Mycfl/wt littermates were treated with 4OHT overnight in IL-2-containing medium prior to activation with anti-CD3/CD28 plus ICAM1 for 36 hours. a, Volcano plot of gene profiles in Myc−/− versus WT activated CD8+ T cells as determined by RNA-seq analysis. BAF components are highlighted (n=5 biological replicates). b, Immunoblot of BAF components in WT and Myc−/− CD8+ T cells. c, d, Naïve CD8+ T cells from WT and Myc+/− mice were activated by anti-CD3/CD28 plus ICAM1 for 28 hours before immunofluorescent staining. Representative image (c) and quantification (d) of conjoined WT and Myc+/− daughter CD8+ T cells that were fixed and stained for Hoechst (blue), ATAC-see (red), Lamin B1 (green) and Tubulin (white). Scale bar: 5 μm. Asymmetric index= (fluorescence intensity in daughter cell 1−fluorescence intensity in daughter cell 2)/(fluorescent intensity in daughter cell 1+ fluorescence intensity in daughter cell 2). e, Quantification of the normalized CX3CR1hiKLRG1hi (left) or CD27hiCXCR3hi (right) ratio in OT-I cells at day 7.5 post Lm-Ova infection. sgNTC (GFP+ spike)- and the indicated constructs (GFP+Ametrine+)-transduced OT-I cells were mixed at a 1:1 ratio and transferred to C57BL/6 hosts followed by Lm-Ova infection. The normalized cell ratio was calculated by dividing the proportions of CX3CR1hiKLRG1hi cells or CD27hiCXCR3hi cells in GFP+Ametrine+ cells by their proportions in GFP+ spike cells. Data are representative of one (a) or two (b, c) or compiled from two independent experiments (d, e). Data are shown as mean±s.e.m. *p<0.05, **p<0.01, ***p<0.001; ns, not significant; two-tailed unpaired Student’s t-test (d, e), two-sided Wald test and adjusted with Benjamini-Hochberg procedure (a).
Extended Data Figure 8. Pharmacological or genetic inhibition of cBAF promotes CD8+ TMEM generation and tumor control.
a, Diagram of tumor rechallenge assay used in Fig. 4a. b, Gene set enrichment analysis of Arid1a knockout (KO) upregulated gene sets and Arid1a KO downregulated gene sets in CD8+ T cells activated for 48 hours in the presence of BD98 versus vehicle. Arid1a KO upregulated and downregulated genes sets were inferred as differentially expressed genes from RNA-seq data of Arid1a KO versus WT activated CD8+ T cells, with thresholds FDR<0.05, fold change>2. c, Peak set enrichment analysis of Arid1a KO upregulated and downregulated chromatin accessibility signatures in CD8+ T cells activated for 48 hours in the presence of BD98 versus vehicle. Arid1a KO upregulated and downregulated ATAC-seq peaks sets were inferred as differentially accessible regions from ATAC-seq of Arid1a KO versus WT activated CD8+ T cells with thresholds FDR<0.05, fold change>2, the top 500 down-regulated peaks (ranked by fold change) were used to avoid inaccurate normalization. d, Representative flow cytometry plot of the frequency of CD44+CD62L+ cells among OT-I CD8+ T cells treated with DMSO or BD98 during anti-CD3/CD28 stimulation with IL-2 for 48 h, followed by culturing in IL-2 containing medium for another 2 days. e, Scheme of murine B7-H3-CAR and STOP-CAR constructs. H/TM: hinge/transmembrane domain. f, Flow cytometry analysis of B7-H3 expression in F420 and GL261 cell lines. The percentage of B7-H3-expressing cells is indicated. g, Diagram of CAR-T cell experiment. h, Flow plot (left) and quantification (right) of B7-H3 CAR expression at day 8 with or without BD98 pretreatment (n=5 biological replicates). Data are representative of at least three independent experiments (d, f, h). Data are shown as mean±s.e.m. ns, not significant; two-tailed unpaired Student’s t-test (h). The p-value was calculated from permutation test, two-sided and adjusted with Benjamini-Hochberg procedure (b,c).
Extended Data Figure 9. Pharmacological inhibition of cBAF promotes CD8+ TMEM generation and tumor control with lymphodepleting chemotherapy.
a, Diagram of CAR-T cell experiments with lymphodepleting chemotherapy. Murine osteosarcoma cell line F420 was injected subcutaneously into naïve B6 (b–d) or B6 albino mice (e, f). At 7 days after tumor inoculation, cyclophosphamide was injected i.p. (200 mg/kg), and CAR-T cells were transferred 2 days after cyclophosphamide treatment. b, Tumor growth and survival curves of F420 tumor-bearing mice treated with B7-H3 CAR-T cells pre-treated with or without BD98 during the first 48 hours of T cell activation (n=5 mice per group). STOP CAR-T: a control CAR containing only the scFv fragment, as a negative control for CAR-T. c, Flow cytometry plots (left) and quantification (right) of intratumoral CD45.1+CD45.2- donor-derived CAR-T cells in F420 tumor bearing mice at day 37 after tumor challenge (day 28 after adoptive transfer of CAR-T cells) (n=5 mice). d, Quantification of Gzmb, TNF-α, IFN-γ and B7-H3 CAR expression levels in intratumoral CAR-T cells (n=5 mice). e, f, Representative bioluminescence images (e) and quantification (f) for CAR-T cells expansion in vivo. The CAR-T cells expressing luciferase were generated from naïve CD8+ T cells isolated from the spleens of CAG-luc-eGFP L2G85 transgenic mice and transferred 2 days after cyclophosphamide injection. Data are representative of one (e, f) or at least two independent experiments (b–d). Data are shown as mean±s.e.m. *p<0.05, **p<0.01, ***p<0.001, ****p<0.001; two-tailed unpaired Student’s t-test (c, d), two-way ANOVA (b, f); and log-rank (Mantel-Cox) test was performed to compare survival curves (b).
Extended Data Figure 10. An putative Arid1a inhibitor promotes human CD8+ TCM and TSCM formation following activation.
a, b, Flow cytometry plot of human TCM (CD45RA−CCR7+), TEM (CD45RA−CCR7−) and TSCM (CD45RA+CCR7+CD27+CD95+) populations. Human naïve CD8+ T cells from healthy donors were enriched and stimulated with anti-CD3/CD28 for 2 days and culturing in IL-15 (a) or IL-2 (b) containing medium. BD98 was added at indicated time points, and the cells were assessed at day 8. c, Quantification of the percentages of TCM, TEM and TSCM cells in (b) (n=6 biological replicates). d, Representative flow cytometry plot of total human CD8+ T cells (upper) in the spleen of NSG mice at day 30 after adoptive transfer, and percentage of TCM-like (CD45RA-CD62L+) cells (lower) in total human CD8+ T cells (n=12 mice per group). Data are representative of two (a, b, d) or compiled from two (c) independent experiments. Data are shown as mean±s.e.m. *p<0.05, **p<0.01, ***p<0.001; one-way ANOVA(c).
Supplementary Material
Acknowledgements:
The authors thank Patrick Fizgerald, Katherine Verbist, Shyam Sirasanagandla, Lacie Harris, Peipei Zhou, and Shaofeng Liu for technical assistance; Cliff Guy for microscope imaging assistance; Nicole Chapman for critical reading and editing of the manuscript; St. Jude Immunology flow cytometry core facility for cell sorting; The Hartwell Center for genome sequencing; Center for Advanced Genome Engineering for targeted sequencing. We also thank Barry Sleckman (UAB) for providing GFP-c-Myc mice. Illustrations were created with BioRender.com. This work was supported by National Institutes of Health grants AI123322 and CA231620 to D.R.G, CA253188 to H.C., National Cancer Institute grant K99CA256262 to D.H, and NCI Cancer Center Support Grant P30 CA021765.
Footnotes
Competing interests: D.R.G consults for Ventus Therapeutics and Inzen Therapeutics. H.C. consults for Kumquat Biosciences, Inc. J.T.Y. consults for Twister Biotech, Inc.
References
- 1.Kaech SM, Wherry EJ & Ahmed R. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2, 251–262, doi: 10.1038/nri778 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Seder RA, Darrah PA & Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol 8, 247–258, doi: 10.1038/nri2274 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Lugli E, Galletti G, Boi SK & Youngblood BA. Stem, Effector, and Hybrid States of Memory CD8(+) T Cells. Trends Immunol 41, 17–28, doi: 10.1016/j.it.2019.11.004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okla K, Farber DL & Zou W. Tissue-resident memory T cells in tumor immunity and immunotherapy. J Exp Med 218, doi: 10.1084/jem.20201605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H et al. In vivo CRISPR screening reveals nutrient signaling processes underpinning CD8(+) T cell fate decisions. Cell 184, 1245–1261 e1221, doi: 10.1016/j.cell.2021.02.021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centore RC, Sandoval GJ, Soares LMM, Kadoch C & Chan HM. Mammalian SWI/SNF Chromatin Remodeling Complexes: Emerging Mechanisms and Therapeutic Strategies. Trends Genet 36, 936–950, doi: 10.1016/j.tig.2020.07.011 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Mittal P & Roberts CWM. The SWI/SNF complex in cancer - biology, biomarkers and therapy. Nat Rev Clin Oncol 17, 435–448, doi: 10.1038/s41571-020-0357-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Verbist KC et al. Metabolic maintenance of cell asymmetry following division in activated T lymphocytes. Nature 532, 389–393, doi: 10.1038/nature17442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joshi NS et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity 27, 281–295, doi: 10.1016/j.immuni.2007.07.010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omilusik KD & Goldrath AW. Remembering to remember: T cell memory maintenance and plasticity. Curr Opin Immunol 58, 89–97, doi: 10.1016/j.coi.2019.04.009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hodges C, Kirkland JG & Crabtree GR. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb Perspect Med 6, doi: 10.1101/cshperspect.a026930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mashtalir N et al. Modular Organization and Assembly of SWI/SNF Family Chromatin Remodeling Complexes. Cell 175, 1272–1288 e1220, doi: 10.1016/j.cell.2018.09.032 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mashtalir N et al. Chromatin landscape signals differentially dictate the activities of mSWI/SNF family complexes. Science 373, 306–315, doi: 10.1126/science.abf8705 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chi TH et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195–199, doi: 10.1038/nature00876 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Astori A et al. ARID1a Associates with Lymphoid-Restricted Transcription Factors and Has an Essential Role in T Cell Development. J Immunol 205, 1419–1432, doi: 10.4049/jimmunol.1900959 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Chi TH et al. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19, 169–182, doi: 10.1016/s1074-7613(03)00199-7 (2003). [DOI] [PubMed] [Google Scholar]
- 17.Kaech SM & Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761, doi: 10.1038/nri3307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wherry EJ et al. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 4, 225–234, doi: 10.1038/ni889 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Huang CY, Bredemeyer AL, Walker LM, Bassing CH & Sleckman BP. Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol 38, 342–349, doi: 10.1002/eji.200737972 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Kassabov SR, Zhang B, Persinger J & Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell 11, 391–403, doi: 10.1016/s1097-2765(03)00039-x (2003). [DOI] [PubMed] [Google Scholar]
- 21.Hargreaves DC & Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21, 396–420, doi: 10.1038/cr.2011.32 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat Methods 13, 1013–1020, doi: 10.1038/nmeth.4031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sammak S, Allen MD, Hamdani N, Bycroft M & Zinzalla G. The structure of INI1/hSNF5 RPT1 and its interactions with the c-MYC:MAX heterodimer provide insights into the interplay between MYC and the SWI/SNF chromatin remodeling complex. FEBS J 285, 4165–4180, doi: 10.1111/febs.14660 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng SW et al. c-MYC interacts with INI1/hSNF5 and requires the SWI/SNF complex for transactivation function. Nat Genet 22, 102–105, doi: 10.1038/8811 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Stojanova A et al. MYC interaction with the tumor suppressive SWI/SNF complex member INI1 regulates transcription and cellular transformation. Cell Cycle 15, 1693–1705, doi: 10.1080/15384101.2016.1146836 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meers MP, Bryson TD, Henikoff JG & Henikoff S. Improved CUT&RUN chromatin profiling tools. Elife 8, doi: 10.7554/eLife.46314 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 35, 871–882, doi: 10.1016/j.immuni.2011.09.021 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gattinoni L, Klebanoff CA & Restifo NP. Paths to stemness: building the ultimate antitumour T cell. Nat Rev Cancer 12, 671–684, doi: 10.1038/nrc3322 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klebanoff CA et al. Central memory self/tumor-reactive CD8+ T cells confer superior antitumor immunity compared with effector memory T cells. Proc Natl Acad Sci U S A 102, 9571–9576, doi: 10.1073/pnas.0503726102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marian CA et al. Small Molecule Targeting of Specific BAF (mSWI/SNF) Complexes for HIV Latency Reversal. Cell Chem Biol 25, 1443–1455 e1414, doi: 10.1016/j.chembiol.2018.08.004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chory EJ et al. Chemical Inhibitors of a Selective SWI/SNF Function Synergize with ATR Inhibition in Cancer Cell Killing. ACS Chem Biol 15, 1685–1696, doi: 10.1021/acschembio.0c00312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dykhuizen EC, Carmody LC, Tolliday N, Crabtree GR & Palmer MA. Screening for inhibitors of an essential chromatin remodeler in mouse embryonic stem cells by monitoring transcriptional regulation. J Biomol Screen 17, 1221–1230, doi: 10.1177/1087057112455060 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim WA & June CH. The Principles of Engineering Immune Cells to Treat Cancer. Cell 168, 724–740, doi: 10.1016/j.cell.2017.01.016 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haydar D et al. Cell-surface antigen profiling of pediatric brain tumors: B7-H3 is consistently expressed and can be targeted via local or systemic CAR T-cell delivery. Neuro Oncol 23, 999–1011, doi: 10.1093/neuonc/noaa278 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S et al. NKD2, a negative regulator of Wnt signaling, suppresses tumor growth and metastasis in osteosarcoma. Oncogene 34, 5069–5079, doi: 10.1038/onc.2014.429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kenneth J Caldwell PN, Alexandra Beckett, Dalia Haydar, Zhongzhen Yi, Giedre Krenciute, Jason Yustein, Stephen Gottschalk, Christopher DeRenzo. Immune Competent Pediatric Sarcoma Models for the Preclinical Evaluation of B7-H3-CAR T-Cell Therapy. MOLECULAR THERAPY 29, 326–326 (2021). [Google Scholar]
- 37.Mitchell DM, Ravkov EV & Williams MA. Distinct roles for IL-2 and IL-15 in the differentiation and survival of CD8+ effector and memory T cells. J Immunol 184, 6719–6730, doi: 10.4049/jimmunol.0904089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C & Allis CD. SWI/SNF complex in cancer. Nat Genet 49, 178–179, doi: 10.1038/ng.3779 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissmiller AM et al. Inhibition of MYC by the SMARCB1 tumor suppressor. Nat Commun 10, 2014, doi: 10.1038/s41467-019-10022-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang SC et al. SWI/SNF component ARID1A restrains pancreatic neoplasia formation. Gut 68, 1259–1270, doi: 10.1136/gutjnl-2017-315490 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapman NM, Boothby MR & Chi H. Metabolic coordination of T cell quiescence and activation. Nat Rev Immunol 20, 55–70, doi: 10.1038/s41577-019-0203-y (2020). [DOI] [PubMed] [Google Scholar]
- 42.Hogquist KA et al. T cell receptor antagonist peptides induce positive selection. Cell 76, 17–27, doi: 10.1016/0092-8674(94)90169-4 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Chang JT, Wherry EJ & Goldrath AW. Molecular regulation of effector and memory T cell differentiation. Nat Immunol 15, 1104–1115, doi: 10.1038/ni.3031 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madden MZ & Rathmell JC. The Complex Integration of T-cell Metabolism and Immunotherapy. Cancer Discov 11, 1636–1643, doi: 10.1158/2159-8290.CD-20-0569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henning AN, Roychoudhuri R & Restifo NP. Epigenetic control of CD8(+) T cell differentiation. Nat Rev Immunol 18, 340–356, doi: 10.1038/nri.2017.146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagert JD et al. Oncohistone mutations enhance chromatin remodeling and alter cell fates. Nat Chem Biol 17, 403–411, doi: 10.1038/s41589-021-00738-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan L et al. Impaired cell fate through gain-of-function mutations in a chromatin reader. Nature 577, 121–126, doi: 10.1038/s41586-019-1842-7 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kakaradov B et al. Early transcriptional and epigenetic regulation of CD8(+) T cell differentiation revealed by single-cell RNA sequencing. Nat Immunol 18, 422–432, doi: 10.1038/ni.3688 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang JT et al. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315, 1687–1691, doi: 10.1126/science.1139393 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Li J et al. Epigenetic driver mutations in ARID1A shape cancer immune phenotype and immunotherapy. J Clin Invest 130, 2712–2726, doi: 10.1172/JCI134402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathur R et al. ARID1A loss impairs enhancer-mediated gene regulation and drives colon cancer in mice. Nat Genet 49, 296–302, doi: 10.1038/ng.3744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao X et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A 105, 6656–6661, doi: 10.1073/pnas.0801802105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Badea TC, Wang Y & Nathans J. A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci 23, 2314–2322 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wurster AL et al. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol 13, 9, doi: 10.1186/1471-2172-13-9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Alboran IM et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55, doi: 10.1016/s1074-7613(01)00088-7 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Hughes ED et al. Genetic variation in C57BL/6 ES cell lines and genetic instability in the Bruce4 C57BL/6 ES cell line. Mamm Genome 18, 549–558, doi: 10.1007/s00335-007-9054-0 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Cao YA et al. Shifting foci of hematopoiesis during reconstitution from single stem cells. Proc Natl Acad Sci U S A 101, 221–226, doi: 10.1073/pnas.2637010100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Connelly JP & Pruett-Miller SM. CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing. Sci Rep 9, 4194, doi: 10.1038/s41598-019-40896-w (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanson KR et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat Commun 9, 5416, doi: 10.1038/s41467-018-07901-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wei J et al. Targeting REGNASE-1 programs long-lived effector T cells for cancer therapy. Nature 576, 471–476, doi: 10.1038/s41586-019-1821-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seaman S et al. Eradication of Tumors through Simultaneous Ablation of CD276/B7-H3-Positive Tumor Cells and Tumor Vasculature. Cancer Cell 31, 501–515 e508, doi: 10.1016/j.ccell.2017.03.005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kochenderfer JN, Yu Z, Frasheri D, Restifo NP & Rosenberg SA. Adoptive transfer of syngeneic T cells transduced with a chimeric antigen receptor that recognizes murine CD19 can eradicate lymphoma and normal B cells. Blood 116, 3875–3886, doi: 10.1182/blood-2010-01-265041 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valencia AM et al. Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling. Cell 179, 1342–1356 e1323, doi: 10.1016/j.cell.2019.10.044 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buenrostro JD, Wu B, Chang HY & Greenleaf WJ ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–21 29 29, doi: 10.1002/0471142727.mb2129s109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ewels PA et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol 38, 276–278, doi: 10.1038/s41587-020-0439-x (2020). [DOI] [PubMed] [Google Scholar]
- 68.Amemiya HM, Kundaje A & Boyle AP. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci Rep 9, 9354, doi: 10.1038/s41598-019-45839-z (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B & Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323, doi: 10.1186/1471-2105-12-323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love MI, Huber W & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, doi: 10.1186/s13059-014-0550-8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Risso D, Ngai J, Speed TP & Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat Biotechnol 32, 896–902, doi: 10.1038/nbt.2931 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its Supplementary Information. All RNA-seq, ATAC-seq, CUT&RUN and microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE183619. Publicly available database used in this study are from the Molecular Signatures Database (http://www.broadinstitute.org/gsea/msigdb/). Source data are provided with this paper.