Stressed mitochondria activate the mitochondrial unfolded protein response (UPRmt) to improve organismal health. Li et al. show that increased ATFS-1 translation, mediated by v-ATPase/TORC1 and involving multiple cellular organelles including lysosomes and ribosomes, plays an essential role in UPRmt activation and mild mitochondrial stress-induced longevity.
Abstract
To adapt mitochondrial function to the ever-changing intra- and extracellular environment, multiple mitochondrial stress response (MSR) pathways, including the mitochondrial unfolded protein response (UPRmt), have evolved. However, how the mitochondrial stress signal is sensed and relayed to UPRmt transcription factors, such as ATFS-1 in Caenorhabditis elegans, remains largely unknown. Here, we show that a panel of vacuolar H+-ATPase (v-ATPase) subunits and the target of rapamycin complex 1 (TORC1) activity are essential for the cytosolic relay of mitochondrial stress to ATFS-1 and for the induction of the UPRmt. Mechanistically, mitochondrial stress stimulates v-ATPase/Rheb-dependent TORC1 activation, subsequently promoting ATFS-1 translation. Increased translation of ATFS-1 upon mitochondrial stress furthermore relies on a set of ribosomal components but is independent of GCN-2/PEK-1 signaling. Finally, the v-ATPase and ribosomal subunits are required for mitochondrial surveillance and mitochondrial stress-induced longevity. These results reveal a v-ATPase-TORC1-ATFS-1 signaling pathway that links mitochondrial stress to the UPRmt through intimate crosstalks between multiple organelles.
Introduction
Mitochondria are essential organelles participating in numerous cellular processes, such as energy harvesting, intermediate metabolism, calcium buffering, apoptosis, and immune response (Mishra and Chan, 2014; Nunnari and Suomalainen, 2012; West and Shadel, 2017). Although mitochondria possess their own genome, most mitochondrial proteins are encoded in the nucleus. Therefore, the expression of the mitochondrial proteome requires tight coordination between the two genomes to adapt to changes in the cellular milieu and extracellular environment (Mottis et al., 2019; Zhu et al., 2022). The mitochondrial unfolded protein response (UPRmt), a branch of the mitochondrial stress response (MSR), is an adaptive transcriptional response that aims at resolving protein-folding stress by orchestrating the remodeling of gene expression programs after multiple forms of mitochondrial stress (Mottis et al., 2019; Shpilka and Haynes, 2018). In general, the activation of the UPRmt preserves fitness during aging and delays the onset of age-related-diseases (Higuchi-Sanabria et al., 2018; Lima et al., 2022; Sun et al., 2016; Vafai and Mootha, 2012).
Although first described in mammalian cells (Zhao et al., 2002), the UPRmt has been mainly studied in the nematode Caenorhabditis elegans. In C. elegans, the UPRmt is induced when the stress-activated transcription factor-1 (ATFS-1) translocates to the nucleus in response to mitochondrial perturbations (Nargund et al., 2012). In addition, a number of other transcription factors/co-factors, histone methyltransferases, demethylases, acetyltransferases, and deacetylases work together with ATFS-1 during UPRmt activation (Haynes et al., 2007; Li et al., 2021; Merkwirth et al., 2016; Shao et al., 2020; Tian et al., 2016; Yuan et al., 2020; Zhu et al., 2020). Despite the fact that some evidence implicated the involvement of TORC1 components in UPRmt signaling (Baker et al., 2012; Haynes et al., 2007; Runkel et al., 2013; Shpilka et al., 2021), these studies either did not explore in detail the molecular mechanisms involved (Baker et al., 2012; Haynes et al., 2007; Runkel et al., 2013) or primarily focused on the role of TORC1 activity in development-associated UPRmt, which is for mitochondrial network expansion (Shpilka et al., 2021). Thus, the function of TORC1 during mitochondrial stress and the mechanism of how TORC1 mediates UPRmt activation remains to be revealed. In mammalian cells, mitochondrial dysfunction triggers the integrated stress response (ISR; Costa-Mattioli and Walter, 2020; Pakos-Zebrucka et al., 2016), in which the phosphorylation of the eukaryotic translation initiation factor 2α (EIF2α) results in the translation of the ATF4, ATF5, and CHOP transcription factors that jointly coordinate a gene expression program considered the functional equivalent of the UPRmt (Mottis et al., 2019; Shpilka and Haynes, 2018). However, little is known about how the mitochondrial stress signal is transmitted through the cytosol and sensed by these UPRmt transcription factors/co-factors. Furthermore, whether the communication between mitochondria and other cellular organelles, such as the lysosomes and ribosomes, contribute to the activation of the UPRmt remains poorly understood.
Here, we demonstrate that stressed mitochondria increase TORC1 activity through a v-ATPase- and Rheb-dependent mechanism in C. elegans. Activated TORC1 thereby leads to increased translation of the UPRmt transcription factor, ATFS-1, a process mediated by cytosolic ribosomes. The accumulated ATFS-1 protein, which is excluded from mitochondria (Nargund et al., 2012), then translocates to the nucleus and mediates the induction of a specific panel of UPRmt effector genes. Many of these UPRmt effectors play positive roles in the recovery of mitochondrial function, metabolic reprogramming, and lifespan extension. Collectively, our findings reveal a pivotal role of v-ATPase-TORC1-ATFS-1 signaling in UPRmt activation and mild mitochondrial stress-induced longevity. Furthermore, the current study highlights that cytosolic relay of the mitochondrial stress signal from mitochondria to the nucleus also relies on the tight coordination of multiple cellular organelles, including lysosomes and ribosomes.
Results
V-ATPase mediates UPRmt activation in C. elegans
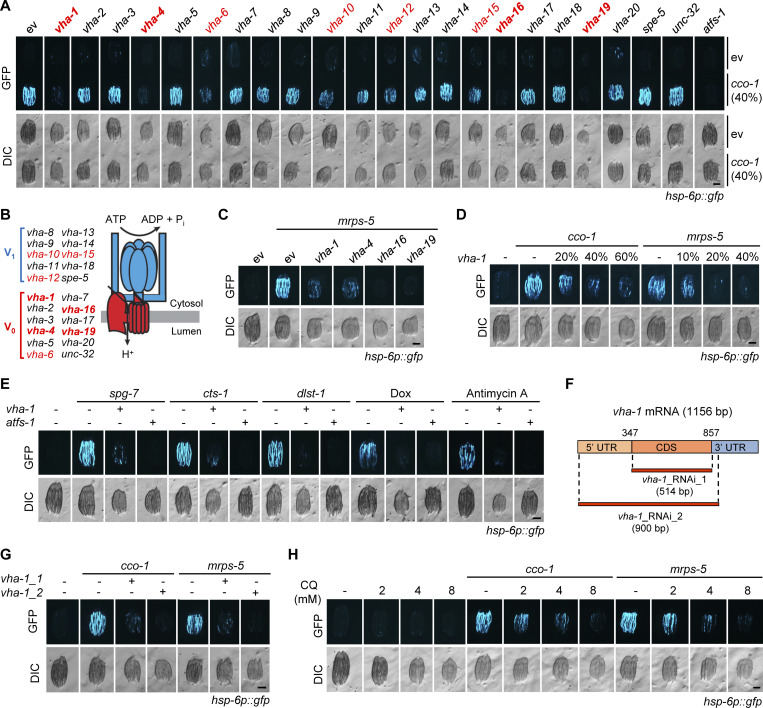
We performed an RNA interference (RNAi) screen to identify genes required for UPRmt activated by cco-1 (cytochrome c oxidase-1) RNAi (Durieux et al., 2011) using the UPRmt reporter hsp-6p::gfp strain of C. elegans (Yoneda et al., 2004). RNAi against multiple subunits of the vacuolar H+-ATPase (v-ATPase; Lee et al., 2010), i.e., vha-1, vha-4, vha-16, and vha-19, attenuated cco-1 (40%) RNAi-induced UPRmt activation to a similar extent as the silencing of atfs-1 (Nargund et al., 2012), while vha-6, vha-10, vha-12, and vha-15 RNAi demonstrated a more moderate impact on UPRmt suppression (Fig. 1, A and B). Similar effects were also found when using another mitochondrial stress inducer, i.e., the RNAi of mrps-5 (mitochondrial ribosomal protein S5; Houtkooper et al., 2013; Fig. 1 C). Moreover, in agreement with previous results reported in drp-1 mutant worms (Wei and Ruvkun, 2020), RNAi of unc-32, vha-7, vha-14, and vha-18 also attenuated the UPRmt to some extent, in response to a lower load of cco-1 (20%) RNAi (Fig. S1 A). The suppressive effect of vha-1 RNAi on UPRmt was furthermore dose dependent (Fig. 1 D). Likewise, genetic or pharmacological activation of the UPRmt by either knockdown of spg-7, cts-1, and dlst-1, or by treatment with antimycin A and doxycycline (Dox; Houtkooper et al., 2013; Liu et al., 2014) was abolished by vha-1 RNAi (Fig. 1 E). Moreover, another RNAi clone (vha-1_RNAi_2) targeting a broader region of the vha-1 mRNA and showing better knockdown efficiency (Fig. S1 B) compared with the one used in the RNAi screen (vha-1_RNAi_1) had an even more pronounced effect in suppressing the UPRmt (Fig. 1, F and G). As an alternative approach to inhibit v-ATPase function, we used two classical small-molecule inhibitors of v-ATPase, Bafilomycin A1 (BafA1) and Concanamycin A (ConA; Bowman et al., 1988; Drose et al., 1993). Both inhibitors abrogated UPRmt activation induced by either cco-1 or mrps-5 RNAi in a concentration-dependent manner (Fig. S1, C and D). Likewise, inhibition of lysosomal acidification by using chloroquine (CQ; Homewood et al., 1972) also dose-dependently blocked the activation of the UPRmt (Fig. 1 H). Notably, RNAi of vha-1, vha-4, vha-16, and vha-19 did not affect the activation of the endoplasmic reticulum (ER) UPR (UPRER) induced by tunicamycin or hsp-3 RNAi or the cytosolic UPR (UPRCYT)/heat shock response in C. elegans (Fig. S1, E–G), suggesting a specific role of v-ATPase in relaying the stress signal from mitochondria.
Figure 1.
Identification of a panel of v-ATPase subunits that are essential for the UPRmt activation in C. elegans. (A) RNAi of multiple v-ATPase subunits attenuated UPRmt activation induced by cco-1 RNAi in hsp-6p::gfp worms. For RNAi treatment, RNAi targeting v-ATPase subunits or atfs-1 occupied 60%, cco-1 RNAi occupied 40%, control RNAi (empty vector [ev]) was used to supply to a final 100% of RNAi for all conditions. DIC, differential interference contrast image. (B) The two functional domains of v-ATPase: the V1 domain, which hydrolyzes ATP to generate the energy required for pumping protons, and the membrane-anchoring V0 domain, which transports H+ across the lipid bilayer. Subunits whose RNAi suppressed the UPRmt were highlighted in red, and the best four of them in red bold. (C) RNAi of vha-1, vha-4, vha-16, and vha-19 (25%) attenuated mrps-5 RNAi-induced UPRmt activation in hsp-6p::gfp worms. (D) vha-1 RNAi inhibited UPRmt activation induced by cco-1 or mrps-5 (40%) RNAi in a dose-dependent manner. (E) vha-1 RNAi attenuated UPRmt activation induced by spg-7, cts-1 or dlst-1 RNAi, and by doxycycline (Dox; 30 μg/ml) or antimycin A (2.5 μM). (F) Schematic diagram showing the regions on mRNA targeted by the two different vha-1 RNAi obtained from either the Vidal (vha-1_RNAi_1) or Ahringer library (vha-1_RNAi_2). (G) Both vha-1 RNAi as indicated in F disrupted the UPRmt induced by either cco-1 or mrps-5 RNAi. Note the more robust effect of vha-1_RNAi_2 in UPRmt suppression as compared to that of vha-1_RNAi_1. (H) The lysosomal acidification inhibitor chloroquine (CQ) suppressed UPRmt activation in a dose-dependent manner. hsp-6p::gfp worms were fed with control or cco-1 (40%) RNAi, in combination with 2–8 mM CQ. Scale bars, 0.3 mm.
Figure S1.
Impact of vha-1, vha-4, vha-16 or vha-19 RNAi in different stress responses and gene expression in C. elegans. (A) RNAi of multiple v-ATPase subunits attenuated UPRmt activation induced by cco-1 RNAi in hsp-6p::gfp worms. For RNAi treatment, RNAi targeting v-ATPase subunits occupied 80%, cco-1 RNAi occupied 20%. DIC, differential interference contrast image. (B) qRT-PCR analysis of vha-1 mRNA (n = 4 biologically independent samples) in worms fed with control (ev), or vha-1 RNAi obtained from either the Vidal (vha-1_RNAi_1) or Ahringer library (vha-1_RNAi_2). (C and D) The v-ATPase inhibitors, Bafilomycin A1 (BafA1; C) and Concanamycin A (ConA; D), dose-dependently suppressed the UPRmt induced by mrps-5 or cco-1 RNAi. (E–G) RNAi of vha-1, vha-4, vha-16 or vha-19 (25%) did not block the endoplasmic reticulum (ER) UPR (UPRER) induced by tunicamycin (5 μg/ml; E) or hsp-3 (40%) RNAi (F) or the cytosolic UPR (UPRCYT)/heat shock response induced by heat shock at 30°C for 8 h (G). (H) Venn diagram of the differentially expressed genes in worms fed with RNAi as indicated. (I) Functional clustering of the 4,563 genes that were commonly affected by vha-1, vha-4, vha-16, and vha-19 RNAi. (J) Heat map of the transcripts of representative mitophagy/autophagy-related genes in worms fed with RNAi as indicated; results are based on the RNA-seq dataset. Scale bars, 0.3 mm. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (**P < 0.01; ***P < 0.001).
Transcriptional induction of UPRmt genes requires v-ATPase subunits
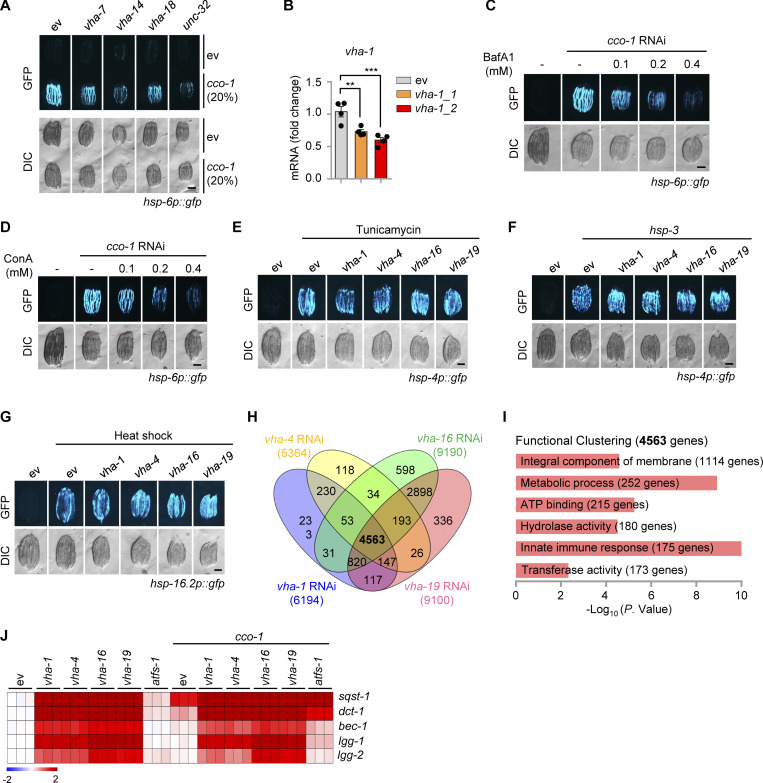
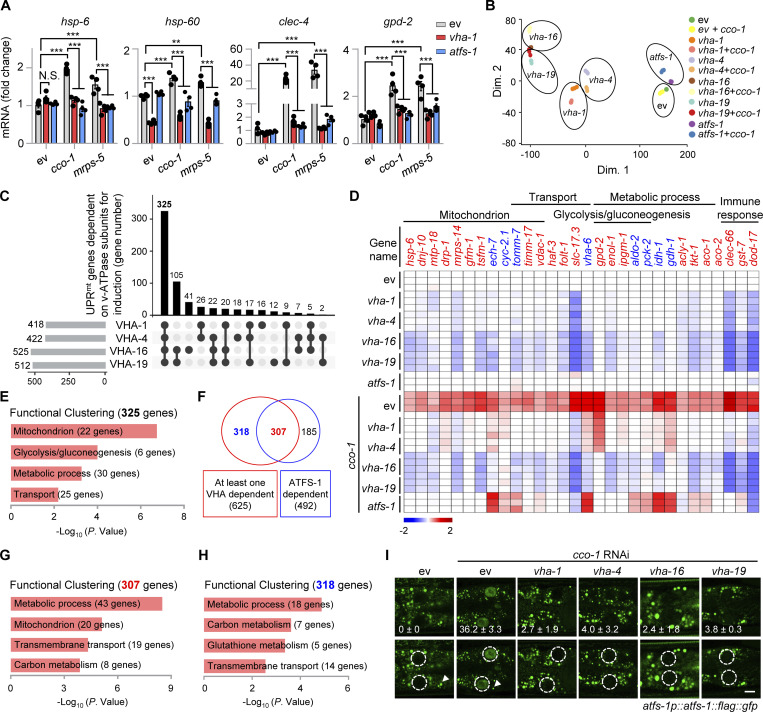
Quantitative real-time PCR (qRT-PCR) revealed that vha-1 RNAi strongly blocked the induction of multiple prototypical UPRmt transcripts (e.g., hsp-6, hsp-60, clec-4, and gpd-2) in response to cco-1 or mrps-5 knockdown (Fig. 2 A), indicating that v-ATPase controls the transcriptional activation of the UPRmt. We thus performed RNA sequencing (RNA-seq) on total RNA isolated from hsp-6p::gfp worms fed with cco-1 RNAi, in combination with vha-1, vha-4, vha-16, vha-19, and atfs-1 RNAi (Fig. 2 B and Table S1). Between 5,364 and 9,190 differentially expressed genes (DEGs; Log2FC > 1 or < −1, adjusted P < 0.05) were identified in response to RNAi targeting each of the four v-ATPase subunits, and 4,563 of them were commonly regulated (Fig. S1 H). Gene ontology (GO) analysis revealed that 1,114 (24.4%) of these DEGs were related to “integral component of membrane” (Fig. S1 I), confirming a key role of v-ATPase in membrane-associated biological processes (Forgac, 2007). RNAi of cco-1 led to the upregulation of 1,382 transcripts, among which 625 (45.2%) were dependent on at least one of the four v-ATPase subunits for induction, and 325 were dependent on all the four subunits of v-ATPase (Fig. 2 C and Table S1). These 325 genes were enriched for multiple mitochondrial pathways including “mitochondrion” (e.g., hsp-6), “glycolysis/gluconeogenesis” (e.g., gpd-2), “metabolic processes” (e.g., idh-1), and “transport” (e.g., folt-1; Fig. 2, D and E). Meanwhile, some innate immune genes, such as C-type lectin clec-66, were also included in this gene set (Fig. 2 D). In line with the observation that vha-16 and vha-19 RNAi had superior effects in suppressing the UPRmt in hsp-6p::gfp worms (Fig. 1, A and C), induction of 105 UPRmt transcripts were exclusively dependent on VHA-16 and VHA-19, but not VHA-1 and VHA-4 (Fig. 2 C). Finally, among the 625 UPRmt transcripts regulated by the v-ATPase, 307 genes were dependent on ATFS-1 as well, covering 62.4% (307/492) of the ATFS-1-dependent program, and 318 genes were solely dependent on v-ATPase (Fig. 2 F). Interestingly, both the two gene clusters were enriched for “metabolic process,” “transmembrane transport,” and “carbon metabolism” (Fig. 2, G and H), confirming a vital role of v-ATPase in the rewiring of global metabolism in response to mitochondrial stress. In contrast, the transcripts of another branch of the MSR, i.e., mitophagy/autophagy (e.g., sqst-1 and dct-1), were conversely increased upon RNAi of v-ATPase subunits (Fig. S1 J). Notably, the nuclear-localized UPRmt transcription factor ATFS-1 during mitochondrial stress (Nargund et al., 2012) was no longer detectable in the atfs-1p::atfs-1::flag::gfp worms fed with RNAi of vha-1, vha-4, vha-16, and vha-19 (Fig. 2 I). Altogether, these results suggest that v-ATPase controls the transcriptional activation of a large set of UPRmt genes, an effect likely achieved through the regulation of ATFS-1.
Figure 2.
Transcriptional induction of a large set of UPRmt genes requires VHA-1, VHA-4, VHA-16, and VHA-19. (A) qRT-PCR analysis of transcripts (n = 4 biologically independent samples) in hsp-6p::gfp worms fed with control (ev), cco-1 or mrps-5 (40%) RNAi in combination with vha-1 (40%) or atfs-1 (60%) RNAi. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (**P < 0.01; ***P < 0.001). (B) Principal-component analysis (PCA) of the RNA-seq profiles of worms fed with the indicated RNAi. (C) Upset plot of cco-1 RNAi-induced UPRmt genes that are dependent on VHA-1, VHA-4, VHA-16, or VHA-19 for induction. (D) Heat-maps of representative UPRmt transcripts that are regulated by VHA-1, VHA-4, VHA-16, or VHA-19. The color-coded heat map represents gene expression differences in log2(fold change, FC) relative to the control RNAi condition. Genes dependent on at least one VHA (−1, −4, −16 or −19) and ATFS-1 are highlighted in red. Genes dependent on at least one VHA, but not ATFS-1, are highlighted in blue. (E) Functional clustering of the 325 UPRmt genes as indicated in C. (F) Venn diagram of the UPRmt genes dependent on at least one VHA (−1, −4, −16 or −19), that are in common with ATFS-1-dependent UPRmt genes, based on the RNA-seq dataset. (G and H) Functional clustering of the 307 (G) and 318 (H) UPRmt genes as indicated in F. (I) Photomicrographs of the most proximal two intestinal cells in atfs-1p::atfs-1::flag::gfp worms fed with control or cco-1 (40%) RNAi, in combination with vha-1, vha-4, vha-16, or vha-19 (25%) RNAi. The nuclei were outlined with white dashed-line circles (bottom panels). The punctae (white arrowheads) are endogenous autofluorescence lysosomes. Mean percentages (±SEM) of worms with nuclear accumulation of ATFS-1::GFP are indicated (n = 3 independent experiments). Scale bar, 10 μm.
Mitochondrial stress induces TORC1 activation and increases ATFS-1 expression
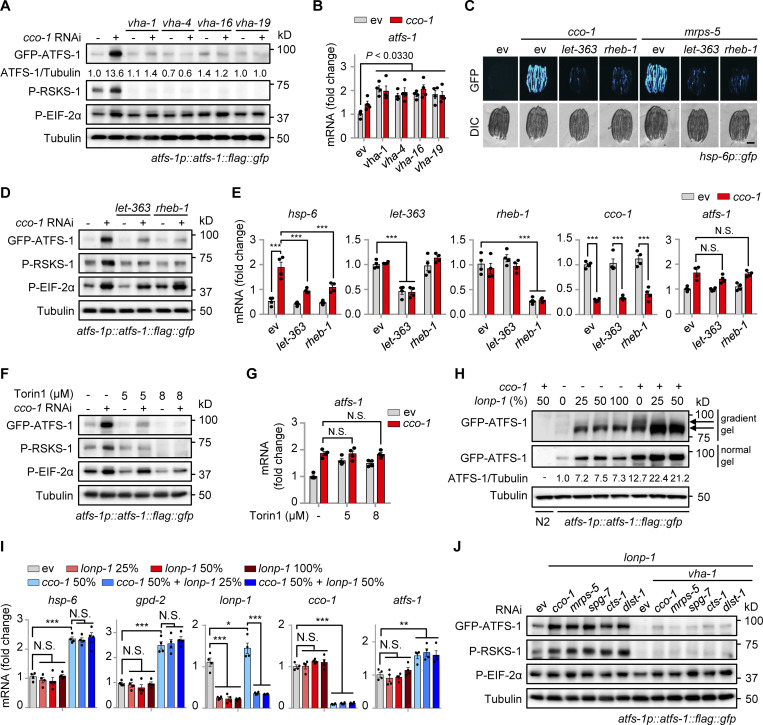
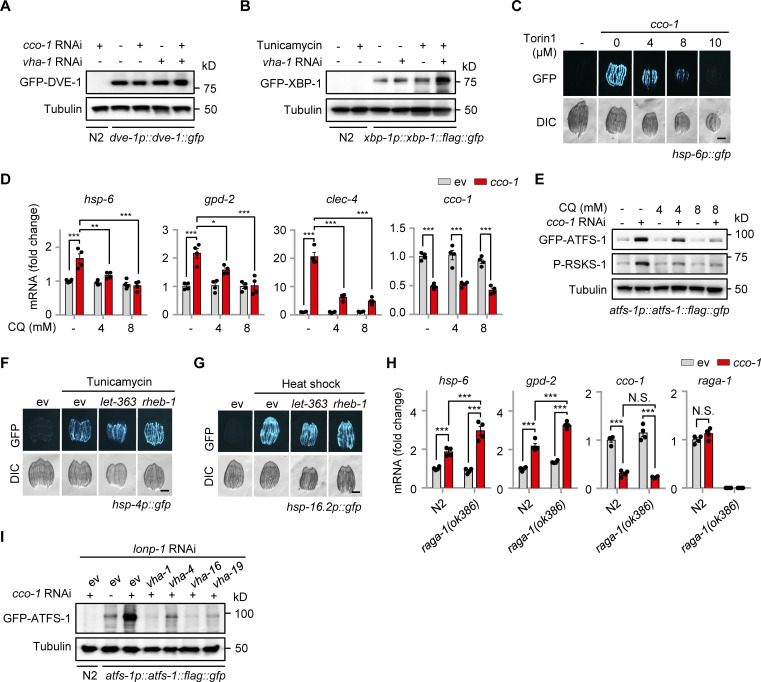
To further explore the molecular mechanism of how v-ATPase regulates the UPRmt, we examined the expression of ATFS-1 (Nargund et al., 2015; Nargund et al., 2012) in atfs-1p::atfs-1::flag::gfp worms fed with vha-1, vha-4, vha-16, and vha-19 RNAi in the absence or presence of mitochondrial stress. In worms fed with cco-1 RNAi, the protein level of ATFS-1 was increased by more than tenfold compared with the unstressed condition, and this response was almost completely blocked by vha-1, vha-4, vha-16, and vha-19 RNAi (Fig. 3 A). The v-ATPase has been shown to not only act as an ATP-driven proton pump to maintain the acidic environment of the lysosomal lumen but also as an essential mediator for the activation of pathways, including mTORC1 signaling (Lawrence and Zoncu, 2019; Shimobayashi and Hall, 2014; Wolfson and Sabatini, 2017; Zoncu et al., 2011). We found that phosphorylation of RSKS-1, the C. elegans ortholog of mammalian S6 kinase (S6K) and a common readout of mTORC1 activity, was robustly increased in a v-ATPase-dependent manner in response to mitochondrial stress (Fig. 3 A). By contrast, mitochondrial stress-induced phosphorylation of EIF-2α (Baker et al., 2012), the worm ortholog of mammalian EIF2α, was only modestly decreased (upon vha-1, vha-4, and vha-19 RNAi) or unaffected (upon vha-16 RNAi; Fig. 3 A). Meanwhile, the atfs-1 mRNA level was even higher in v-ATPase RNAi-fed worms compared with that in control worms (Fig. 3 B). Unlike ATFS-1, the protein level of DVE-1 (Haynes et al., 2007), another key transcription factor of the UPRmt, was not altered by mitochondrial stress or vha-1 RNAi (Fig. S2 A). Additionally, vha-1 silencing conversely lead to more protein expression of XBP-1 (Calfon et al., 2002; Fig. S2 B), an essential transcription factor of the UPRER.
Figure 3.
Mitochondrial stress induces v-ATPase-dependent TORC1 activation and increases ATFS-1 expression. (A and B) Western blots (A) and atfs-1 mRNA levels (n = 4 biologically independent samples; B) of atfs-1p::atfs-1::flag::gfp worms fed with control, vha-1, vha-4, vha-16, or vha-19 (25%) RNAi with or without cco-1 (50%) RNAi. (C) RNAi of let-363 or rheb-1 attenuated UPRmt activation. hsp-6p::gfp worms were fed with control, let-363 (75%), or rheb-1 (75%) RNAi, in combination with cco-1 (25%) or mrps-5 (25%) RNAi. Scale bar, 0.3 mm. (D and E) Western blots (D) and qRT-PCR analysis of transcripts (n = 4 biologically independent samples; E) of atfs-1p::atfs-1::flag::gfp worms fed with control, let-363 (60%) or rheb-1 (60%) RNAi, with or without cco-1 (40%) RNAi. (F and G) Western blots (F) and qRT-PCR analysis of transcripts (n = 4 biologically independent samples; G) of atfs-1p::atfs-1::flag::gfp worms fed with control or cco-1 (40%) RNAi, co-treated with or without 5 or 8 μM Torin1. (H and I) Western blots (H) and qRT-PCR analysis of transcripts (n = 4 biologically independent samples; I) in atfs-1p::atfs-1::flag::gfp worms fed with control, lonp-1 and/or cco-1 RNAi. Wild-type (N2) worms fed with cco-1 RNAi were used as a negative control. Western blots were performed with either normal or 4–12% gradient gels. (J) Western blots of atfs-1p::atfs-1::flag::gfp worms fed with control, cco-1, mrps-5, spg-7, or cts-1 (50%) RNAi in the presence of lonp-1 (25%) RNAi, and/or vha-1 (25%) RNAi. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (*P < 0.05; **P < 0.01; ***P < 0.001). Source data are available for this figure: SourceData F3.
Figure S2.
Impact of mitochondrial stress, ER stress, and TORC1 signaling regulators in gene expression and stress responses. (A) Western blots of dve-1p::dve-1::gfp worms fed with control, cco-1 (50%) and/or vha-1 (25%) RNAi. The wild-type (N2) worms fed with cco-1 (50%) RNAi were used as a negative control. (B) Western blots of N2 or xbp-1p::xbp-1::flag::gfp worms fed with control or vha-1 (25%) RNAi, with or without tunicamycin (2.5 μg/ml). (C) Torin1 suppressed cco-1 RNAi-induced UPRmt activation in a dose-dependent manner. hsp-6p::gfp worms were fed with control or cco-1 (40%) RNAi, in combination with 4–10 μM Torin1. (D and E) qRT-PCR analysis of transcripts (n = 4 biologically independent samples; D) and Western blots (E) of atfs-1p::atfs-1::flag::gfp worms fed with control or cco-1 (40%) RNAi, co-treated with or without 4 or 8 mM CQ. (F and G) RNAi of let-363 and rheb-1 (75%) did not block the UPRER induced by tunicamycin (5 μg/ml; F) or the UPRCYT induced by heat shock at 30°C for 8 h (G). (H) qRT-PCR analysis of transcripts (n = 4 biologically independent samples) of wild-type or raga-1(ok386) worms fed with control or cco-1 (50%) RNAi. (I) Western blots of N2 or atfs-1p::atfs-1::flag::gfp worms fed with control, vha-1, vha-4, vha-16 or vha-19 (25%) RNAi and/or cco-1 (50%) RNAi, in the presence of lonp-1 (25%) RNAi. Scale bars, 0.3 mm. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (*P < 0.05; **P < 0.01; ***P < 0.001). Source data are available for this figure: SourceData FS2.
Similar to the effects of v-ATPase RNAi, suppression of worm TORC1 activity by either RNAi of let-363 (the worm ortholog of human mTOR) and rheb-1 (the worm ortholog of mTORC1 upstream activator, Rheb, Ras homolog enriched in brain; Inoki et al., 2003), or by applying the TORC1 catalytic inhibitor Torin1 (Thoreen et al., 2009) attenuated UPRmt induced by cco-1 or mrps-5 knockdown (Fig. 3 C and Fig. S2 C). Notably, we found that ATFS-1 accumulation and RSKS-1 phosphorylation in response to cco-1 RNAi were abrogated by RNAi of let-363 or rheb-1 and by Torin1 treatment, while EIF-2α phosphorylation and atfs-1 mRNA expression were barely affected (Fig. 3, D–G). Consistent with the importance of lysosomes in TORC1 activation (Lawrence and Zoncu, 2019), the lysosomal acidification inhibitor CQ attenuated the induction of UPRmt genes, TORC1 activity (Fedele and Proud, 2020; Jewell et al., 2015), and the accumulation of ATFS-1 in a dose-dependent manner (Fig. S2, D and E), confirming that the intact lysosomal function/pH is essential for TORC1 and UPRmt activation. Moreover, let-363 or rheb-1 RNAi did not affect the activation of the UPRER or UPRCYT (Fig. S2, F and G). Intriguingly, knockout of raga-1, the sole worm ortholog of the evolutionarily conserved ras-related GTPase RagA and RagB that are essential for the activation of the mTORC1 by exogenous amino acids (Sancak et al., 2008; Schreiber et al., 2010), lead to even more robust induction of UPRmt genes in response to cco-1 RNAi (Fig. S2 H), suggesting that mitochondrial stress probably represents a unique intrinsic signal for TORC1 activation independent of the Rag GTPase (Hesketh et al., 2020).
The protein level of ATFS-1 has been reported to be mainly controlled by the mitochondrial Lon protease homolog (LONP-1; Nargund et al., 2012; Shpilka et al., 2021; Yang et al., 2022), which mediates the degradation of ATFS-1 in the mitochondrial matrix so that little ATFS-1 is available for the transcriptional activation of the UPRmt in unstressed worms. Indeed, lonp-1 RNAi led to the upregulation of ATFS-1 protein without affecting its mRNA level and UPRmt activity (Fig. 3, H and I). However, more accumulation of ATFS-1 protein in response to cco-1 RNAi was detected even in the background of 50% of lonp-1 RNAi (Fig. 3 H), a condition when lonp-1 transcript was firmly knocked down (Fig. 3 I). As expected, lonp-1 RNAi only increased the accumulation of mitochondrial-localized ATFS-1 (the lower band, produced after the cleavage of its mitochondrial targeting sequence; Nargund et al., 2012), while cco-1 RNAi led to the accumulation of both the mitochondrial-localized and unprocessed forms of ATFS-1 (Fig. 3 H). Importantly, RNAi of vha-1, vha-4, vha-16, and vha-19 blocked the accumulation of ATFS-1 induced by mitochondrial stress when lonp-1 was silenced (Fig. S2 I). Likewise, other mitochondrial stress inducers, such as mrps-5, spg-7, cts-1, and dlst-1 RNAi, also increased ATFS-1 expression and RSKS-1 phosphorylation in the background of lonp-1 RNAi, and these responses were furthermore abolished by vha-1 RNAi (Fig. 3 J). These results suggest that v-ATPase regulates ATFS-1 protein expression upstream of LONP-1 and through a previously uncharacterized LONP-1-independent mechanism.
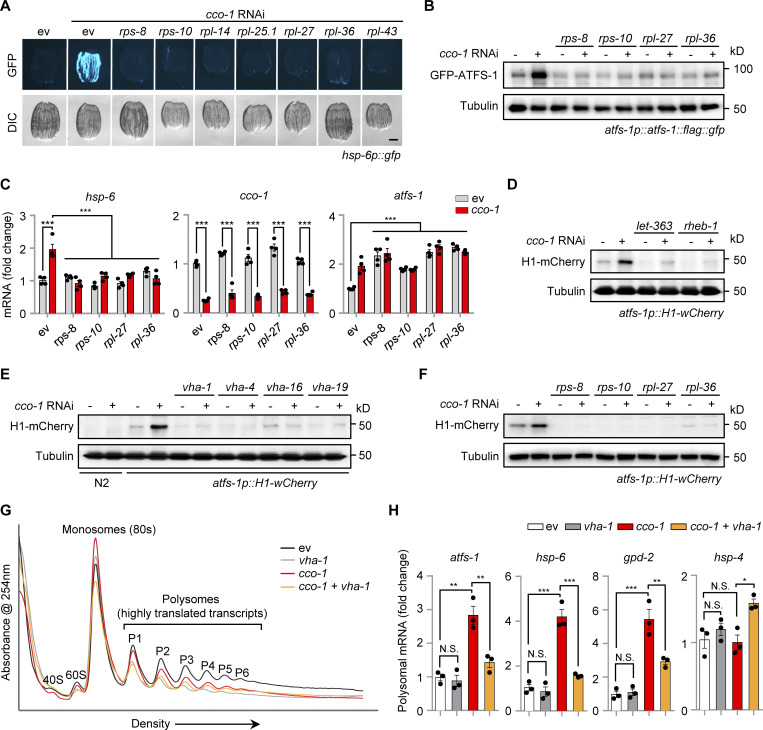
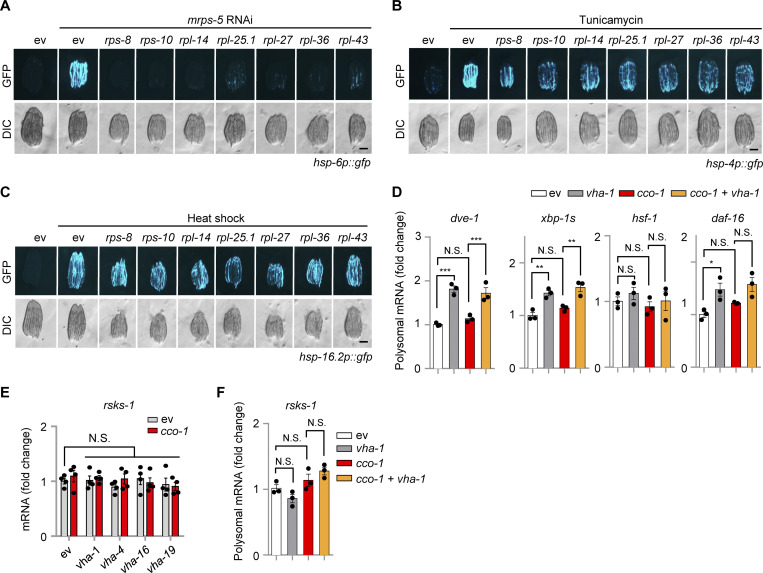
Knockdown of ribosomal subunits blocks the UPRmt and ATFS-1 translation
To test whether increased accumulation of ATFS-1 protein upon mitochondrial stress is caused either by the direct increase of atfs-1 translation or by the suppression of other yet unknown ATFS-1 degradation pathways (e.g., through a ULP-4-mediated SUMOylation mechanism [Gao et al., 2019]), we first knocked down a set of ribosomal subunits, including the small (rps-8 and rps-10) and large ones (rpl-14, rpl-25.1, rpl-27, rpl-36 and rpl-43) in the UPRmt reporter hsp-6p::gfp strain. RNAi of each individual ribosomal subunit remarkably blocked cco-1 or mrps-5 RNAi-induced UPRmt activation, while tunicamycin-induced UPRER was only partially attenuated, and the UPRCYT/heat shock response was not affected (Fig. 4 A and Fig. S3, A–C). In support of these results, paraquat-induced UPRmt was also reported to rely on multiple ribosomal subunits in a previous screen study (Runkel et al., 2013). Importantly, RNAi of rps-8, rps-10, rpl-27, and rpl-36 almost completely blocked ATFS-1 expression in the atfs-1p::atfs-1::flag::gfp strain upon mitochondrial stress (Fig. 4 B), while, in apparent contrast, the mRNA level of atfs-1 was even upregulated in response to their knockdown (Fig. 4 C). Next, we took advantage of the atfs-1 reporter strain atfs-1p::H1-wCherry (Murray et al., 2012), which was constructed such that the expression of the Histone–mCherry reporter protein is under the strict control of the upstream intergenic sequences (including both the promoter and 5′-UTR regions) of atfs-1. Thus, the expression/translation of this Histone–mCherry is controlled in a similar fashion as that of the endogenous ATFS-1 protein. In addition, since the translated Histone–mCherry is not degraded by LONP-1 or other ATFS-1-specific-targeting enzymes, the atfs-1p::Histone-wCherry transgenic strain is therefore an ideal system to study ATFS-1 translation regulation, independent of its degradation. Similar to the results acquired with the atfs-1p::atfs-1::flag::gfp worms (Fig. 3, A and D; and Fig. 4 B), we found that cco-1 RNAi led to a robust upregulation of the Histone–mCherry reporter protein in the atfs-1p::H1-wCherry worms, which was almost completely blocked by RNAi targeting let-363, rheb-1, v-ATPase, or the ribosomal subunits (Fig. 4, D–F). Finally, we applied polysome profiling whereby free ribosomal subunits, monosomes (mRNA with one ribosome associated), and polysomes (mRNA with two or more ribosomes associated, the highly translated ribosome-mRNA fraction), were separated over a sucrose density gradient and quantified by optical density. Similar to the results as shown for mrps-5 RNAi treatment (Molenaars et al., 2020), cco-1 RNAi led to a shift from polysomes to monosomes (Fig. 4 G), confirming an adaptive cytosolic translation reduction in response to mitochondrial stress (D’Amico et al., 2017; Molenaars et al., 2020; Suhm et al., 2018). Importantly, increased polysomal mRNA of atfs-1, as well as that of UPRmt targets hsp-6 and gpd-2, was detected in response to cco-1 RNAi, a phenomenon which is strongly attenuated with vha-1 RNAi co-treatment (Fig. 4 H). Of note, the overall cellular mRNA level of atfs-1 was rather higher on vha-1 RNAi (Fig. 3 E). In contrast, the polysomal mRNA of other transcription factors or typical reporter genes (e.g., hsp-4, dve-1, xbp-1s, hsf-1, and daf-16) involved in different stress responses was not affected by mitochondrial stress and was either unchanged or even upregulated after the co-treatment of vha-1 RNAi (Fig. 4 H and Fig. S3 D). Of note, neither vha-1 RNAi nor mitochondrial stress altered the overall total and polysomal mRNA levels of rsks-1 (Fig. S3, E and F). Together, these results suggest that increased translation of atfs-1, mediated by v-ATPase/TORC1 and cytosolic ribosomes, is a key mechanism that leads to the accumulation of ATFS-1 protein for UPRmt activation in response to mitochondrial stress.
Figure 4.
Knockdown of ribosomal subunits blocks UPRmt activation and ATFS-1 translation upon mitochondrial stress. (A) RNAi of multiple ribosomal subunits attenuated UPRmt activation induced by cco-1 RNAi. RNAi targeting cco-1 occupies 40%, RNAi targeting ribosomal subunits occupies 60%. Scale bar, 0.3 mm. (B and C) Western blots (B) and qRT-PCR analysis of transcripts (n = 4 biologically independent samples; C) in atfs-1p::atfs-1::flag::gfp worms fed with control, rps-8, rps-10, rpl-27 or rpl-36 (60%) RNAi with or without cco-1 (40%) RNAi. (D–F) Western blots of atfs-1p::H1-wCherry worms fed with control or cco-1 (40%) RNAi, in combination with let-363 or rheb-1 (60%) RNAi (D), vha-1, vha-4, vha-16, or vha-19 (25%) RNAi (E), or rps-8, rps-10, rpl-27 or rpl-36 (60%) RNAi (F). (G) Representative polysome profiles of worms fed with control or cco-1 (50%) RNAi, with or without or vha-1 (25%) RNAi. The relative positions of the ribosome subunits (40 and 60 S), monosomes (80 S), and polysomes (P1-P6) are indicated. (H) qRT-PCR analysis of transcripts (n = 3 biologically independent samples) in the polysomal fractions (highly translated) of worms as indicated in G. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (*P < 0.05; **P < 0.01; ***P < 0.001). Source data are available for this figure: SourceData F4.
Figure S3.
Impact of ribosomal subunit RNAi in stress responses in C. elegans. (A) RNAi of ribosomal subunits strongly blocked the UPRmt activation induced by mrps-5 RNAi. RNAi targeting mrps-5 occupies 40%; RNAi targeting ribosomal subunits occupies 60%. (B and C) RNAi of ribosomal subunits only partially attenuated the UPRER induced by tunicamycin (5 μg/ml; B) and did not affect the UPRCYT induced by heat shock at 30°C for 8 h (C). (D) qRT-PCR analysis of transcripts (n = 3 biologically independent samples) in the polysomal fractions of worms fed with control or cco-1 (50%) RNAi, with or without or vha-1 (25%) RNAi. (E and F) Total mRNA (n = 4 biologically independent samples; E) and polysomal mRNA (n = 3 biologically independent samples; F) level of rsks-1 in worms fed with control or cco-1 (50%) RNAi, with or without or vha-1 (25%) RNAi. Scale bars, 0.3 mm. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (*P < 0.05; **P < 0.01; ***P < 0.001).
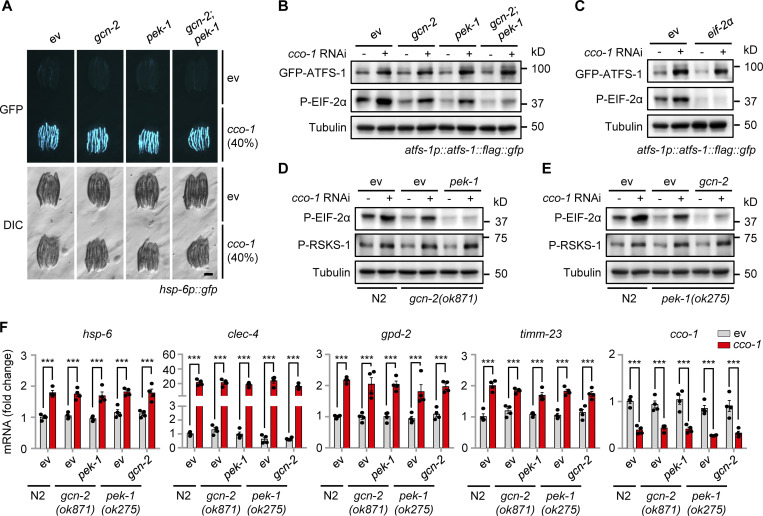
Mitochondrial stress-induced ATFS-1 is independent of the GCN-2/PEK-1 signaling
In mammals, phosphorylation of EIF2α by four dedicated kinases (GCN2, PERK, HRI, and PKR) serves to attenuate the general cytosolic translation in response to a variety of intra- and extracellular stresses (e.g., amino-acid starvation, viral infection, oxidative and unfolded protein stress), and meanwhile stimulates the translation of ATF4, ATF5, and CHOP, the mammalian functional orthologs of ATFS-1, to activate the ISR (Costa-Mattioli and Walter, 2020; Pakos-Zebrucka et al., 2016; Quiros et al., 2017). We thus questioned whether a similar mechanism also exists in C. elegans. Surprisingly, RNAi of gcn-2 and/or pek-1, which encode the only two known corresponding worm EIF-2α kinases, GCN-2 and PEK-1 (Baker et al., 2012; Shen et al., 2001), failed to block cco-1 RNAi-induced UPRmt in hsp-6p::gfp worms (Fig. 5 A). Meanwhile, EIF-2α phosphorylation was attenuated by either gcn-2 or pek-1 RNAi, in both basal and mitochondrial stress conditions (Fig. 5 B). Moreover, although only GCN-2 is required for EIF-2α phosphorylation in the clk-1(qm30) mitochondrial mutant (Baker et al., 2012; Lakowski and Hekimi, 1996), less of EIF-2α phosphorylation was detected with the cosilencing of both gcn-2 and pek-1 upon cco-1 RNAi (Fig. 5 B). In line with the results acquired in hsp-6p::gfp worms (Fig. 5 A), mitochondrial stress-induced upregulation of ATFS-1 was not affected by either gcn-2, pek-1, or eif-2α RNAi (Fig. 5, B and C). Furthermore, comparable levels of RSKS-1 phosphorylation and UPRmt transcript induction upon cco-1 RNAi were found in the gcn-2 or pek-1 knockout worm mutants, even in conditions with full suppression of EIF-2α phosphorylation, as compared with that in WT (N2) worms (Fig. 5, D–F). Likewise, activation of the UPRmt was also not affected in autophagy-defective mutants (Fig. S4). Collectively, these results suggest that increased expression of ATFS-1 in response to mitochondrial stress is independent of the GCN-2/PEK-1 signaling as well as the autophagic process per se.
Figure 5.
Increased expression of ATFS-1 in response to mitochondrial stress is independent of the GCN-2/PEK-1 signaling. (A) RNAi of gcn-2 and/or pek-1 did not affect UPRmt activation. hsp-6p::gfp worms were fed with control, gcn-2, and/or pek-1 RNAi, in combination with cco-1 RNAi. Scale bar, 0.3 mm. (B) Western blots of atfs-1p::atfs-1::flag::gfp worms fed with control, gcn-2, and/or pek-1 RNAi, with or without cco-1 RNAi. (C) Western blots of atfs-1p::atfs-1::flag::gfp worms fed with control, eif-2α and/or cco-1 RNAi. (D) Western blots of WT (N2) or gcn-2(ok871) worms fed with control, pek-1, and/or cco-1 RNAi. (E) Western blots of wild-type or pek-1(ok275) worms fed with control, gcn-2, and/or cco-1 RNAi. (F) qRT-PCR analysis of transcripts (n = 4 biologically independent samples) in wild-type, gcn-2(ok871) or pek-1(ok275) worms fed with control, pek-1, gcn-2 and/or cco-1 RNAi. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (***P < 0.001). Source data are available for this figure: SourceData F5.
Figure S4.
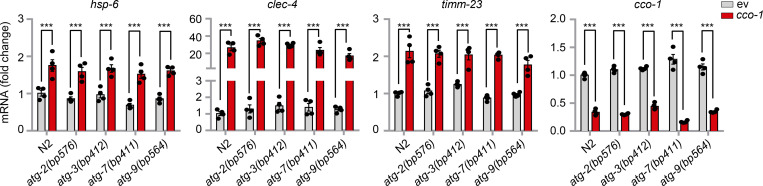
Autophagy defective mutants have normal activation of the UPRmt in response to mitochondrial stress. qRT-PCR analysis of transcripts (n = 4 biologically independent samples) in wild-type (N2) or autophagy defect worm mutants fed with control or cco-1 RNAi. Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (***P < 0.001).
A crucial role of v-ATPase and ribosomal subunits in mitochondrial surveillance
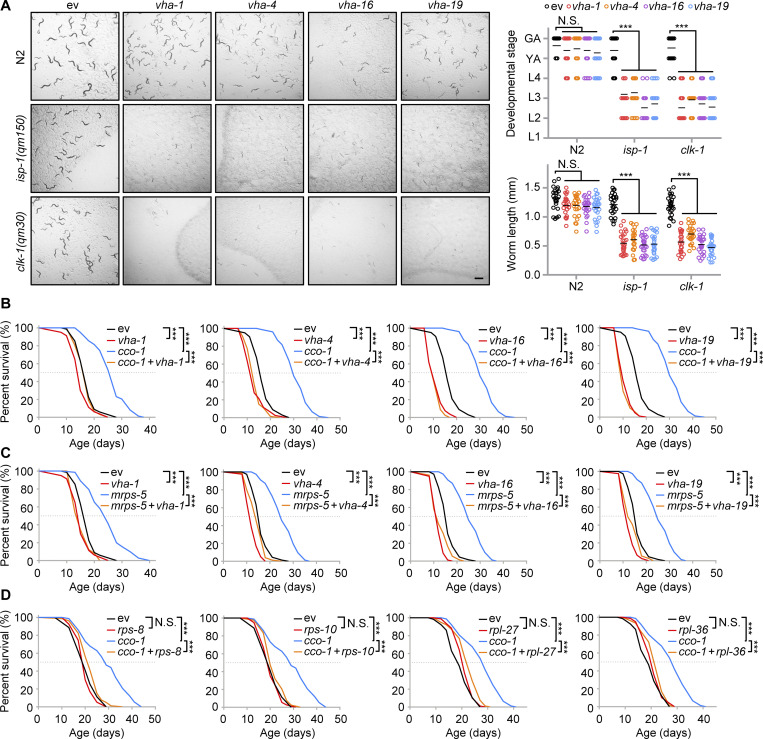
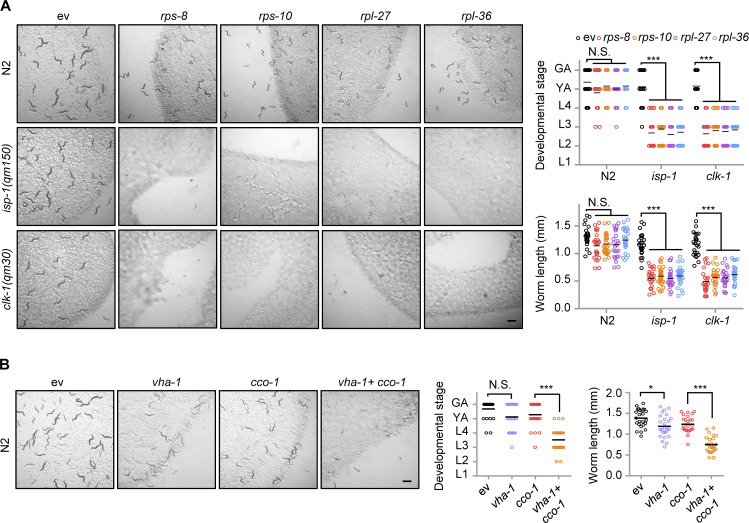
To explore the functions of v-ATPase and ribosomal subunits in mitochondrial homeostasis and adaptations upon stresses, we raised WT and the mitochondrial respiration mutants isp-1(qm150) and clk-1(qm30) (Feng et al., 2001; Lakowski and Hekimi, 1996), and on control, vha-1, vha-4, vha-16, vha-19, rps-8, rps-10, rpl-27, or rpl-36 RNAi bacteria. Compared to C. elegans fed with control RNAi, RNAi targeting v-ATPase or ribosomal subunits led to severe synthetic growth defects of the mitochondrial stressed mutants, whereas the development of WT worms was only slightly delayed (Fig. 6 A and Fig. S5 A). Similar results were also found in worms fed with vha-1 RNAi and/or cco-1 RNAi (Fig. S5 B). Thus, mitochondrial respiration mutants heavily rely on v-ATPase and ribosomal subunits to maintain growth. We then questioned whether v-ATPase and ribosomal subunits also contribute to mild mitochondrial stress-induced lifespan extension in C. elegans (Durieux et al., 2011; Houtkooper et al., 2013). In line with an essential role of the v-ATPase subunits in the UPRmt (Fig. 1, A–C), RNAi of vha-1, vha-4, vha-16, and vha-19 strongly attenuated the lifespan extension induced by cco-1 or mrps-5 RNAi (Fig. 6, B and C). Consistently, the silencing of ribosomal subunits including rps-8, rps-10, rpl-27, and rpl-36 also blunted the lifespan extension induced by cco-1 RNAi (Fig. 6 D). Thus, v-ATPase and ribosomal components play a crucial role in mitochondrial surveillance and regulate the longevity induced by mild mitochondrial stress in C. elegans.
Figure 6.
A crucial role of v-ATPase and ribosomal subunits in mitochondrial surveillance and mitochondrial stress-induced longevity. (A) Representative brightfield pictures of wild-type (N2), isp-1(qm150), or clk-1(qm30) worms fed with control (ev), vha-1 (20%), vha-4 (20%), vha-16 (10%), or vha-19 (20%) RNAi since maternal L4 stage. The developmental stage and body length of the F1 progeny were quantified on Day 4 after hatching (n = 25 worms for each condition). Scale bar, 1 mm. GA, gravid adult; YA, young adult; L1-4, larval stage 1–4. (B and C) RNAi of v-ATPase subunits attenuated mitochondrial stress-induced lifespan extension. Survival of worms fed with control, vha-1 (20%), vha-4 (20%), vha-16 (10%), or vha-19 (20%) RNAi, with or without cco-1 (50%; B) or mrps-5 (50%; C) RNAi. (D) RNAi of ribosomal subunits attenuates mitochondrial stress-induced lifespan extension. Survival of worms fed with control, rps-8 (60%), rps-10 (60%), rpl-27 (60%), or rpl-36 (60%) RNAi, with or without cco-1 (40%) RNAi. Statistical analysis was performed by log-rank test (**P < 0.01; ***P < 0.001).
Figure S5.
A key role of ribosomal subunits and VHA-1 in mitochondrial surveillance. (A) Representative brightfield pictures of wild-type (N2), isp-1(qm150) or clk-1(qm30) worms fed with control, rps-8 (60%), rps-10 (60%), rpl-27 (60%), or rpl-36 (60%) RNAi since the maternal L4 stage. Scale bar, 1 mm. The developmental stage (top right) and body length (bottom right) of the F1 progeny were quantified at day 4 after hatching (n = 25 worms for each condition). GA, gravid adult; L1-4, larval stages 1–4; YA, young adult. (B) Representative brightfield pictures of worms fed with control, vha-1 (20%), and/or cco-1 (50%) RNAi. The developmental stage and body length of the F1 progeny were quantified at day 4 after hatching (n = 25 worms for each condition). Error bars denote SEM. Statistical analysis was performed by ANOVA followed by Tukey post-hoc test (*P < 0.05; ***P < 0.001).
Discussion
The UPRmt was initially defined as a transcriptional response triggered by the presence or accumulation of unfolded proteins/peptides from mitochondria (Shpilka and Haynes, 2018; Zhao et al., 2002). In C. elegans, almost all the well-characterized UPRmt regulators, such as ATFS-1 (Nargund et al., 2012), DVE-1 (Haynes et al., 2007), MET-2/LIN-65 (Tian et al., 2016), JMJD-3.1/JMJD-1.2 (Merkwirth et al., 2016), SET-6/BAZ-2 (Yuan et al., 2020), HDA-1 (Shao et al., 2020), NuRD (Zhu et al., 2020), and CBP-1 (Li et al., 2021), are localized or translocated in the nucleus upon mitochondrial stress. However, how the mitochondrial stress signal is sensed in the cytosol and relayed to these UPRmt regulators is only partially elucidated. Moreover, whether other protein complexes and organelles, such as the lysosomes and ribosomes, also play a role in the UPRmt activation process remains elusive.
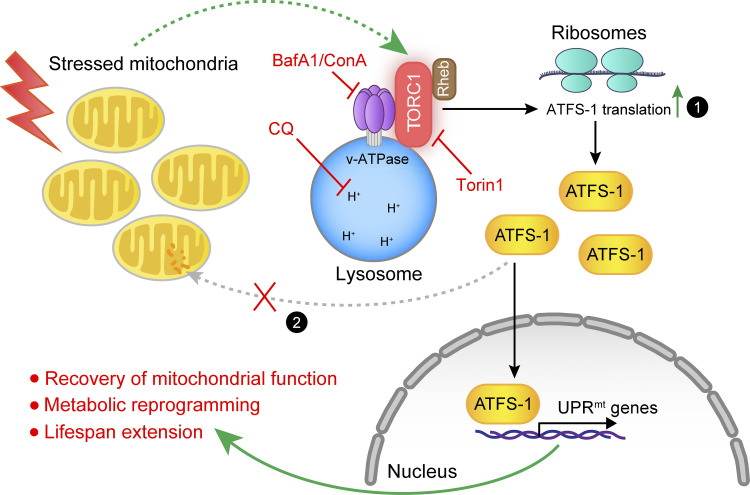
Here, we demonstrated that the mitochondrial stress is transduced through a v-ATPase-TORC1-ATFS-1 signaling pathway in the cytosol involving distinct organelles (Fig. 7). In this signaling network, stressed mitochondria increase TORC1 activity through a v-ATPase- and Rheb-dependent mechanism. Activated TORC1 thereby leads to increased translation of the UPRmt transcription factor, ATFS-1, which is dependent on the cytosolic ribosomes (Fig. 7; indicated by mechanism 1). The accumulated ATFS-1 protein then translocates to the nucleus and mediates the induction of a specific panel of UPRmt effector genes. Many of these UPRmt effectors play positive roles in the recovery of mitochondrial function, metabolic reprogramming, and lifespan extension. Importantly, genetic or pharmacological disruption of any components in this pathway robustly suppressed the UPRmt, but not other similar stress responses, such as UPRER and UPRCYT. Our work thus reveals that in addition to the attenuated mitochondrial import of ATFS-1 in response to mitochondrial stress (Nargund et al., 2012; Fig. 7; indicated by mechanism 2), v-ATPase/TORC1-mediated upregulation of ATFS-1 translation is also essential to ensure that enough ATFS-1 is translocated to the nucleus for UPRmt activation. Of note, v-ATPase/TORC1-mediated translation of ATFS-1 seems to be a prerequisite step for the increased accumulation and the subsequent nuclear localization of ATFS-1 for UPRmt activation during mitochondrial stress, as evidenced by the GFP-ATFS-1 nuclear localization results (Fig. 2 I), supporting that the two mechanisms likely act as a whole in MSR.
Figure 7.
Model for v-ATPase/TORC1-mediated regulation of ATFS-1 translation and UPRmt activation in C. elegans. When mitochondria are stressed in response to various intracellular or extracellular stimuli, the activity of TORC1 is increased through a v-ATPase- and Rheb-dependent mechanism, which could be blocked by both inhibitors of v-ATPase, Bafilomycin A1 (BafA1), and Concanamycin A (ConA); the lysosomal acidification inhibitor, chloroquine (CQ); as well as the TORC1 inhibitor, Torin1. Activated TORC1 thereby stimulates cytosolic ribosomes to translate the UPRmt transcription factor ATFS-1 (mechanism 1). Under the basal nonstressed condition, ATFS-1 is transported and degraded in the mitochondria; during mitochondrial stress, in addition to increased ATFS-1 translation, mitochondrial import efficiency is also decreased (mechanism 2; Nargund et al., 2012). Both mechanisms together result in nuclear accumulation of ATFS-1, where it induces a diverse panel of UPRmt effector genes. Many of these UPRmt effectors play positive roles in mitochondrial function recovery, metabolic reprogramming, and lifespan extension.
Consistent with a central role of TORC1 signaling in the UPRmt, TORC1 and RSKS-1 were reported to be indispensable for the increased UPRmt activity to support mitochondrial network expansion during development, a condition when TORC1 activity is already known to be active (Shpilka et al., 2021). How the TORC1 activity is activated in response to mitochondrial stress remains an important direction for future work. One possibility is that the unfolded proteins/peptides produced upon mitochondrial stress could somehow be transported from the mitochondria to the lysosomes and thereby digested to amino acids within the lysosomes, which could then lead to TORC1 activation at the lysosomal surface (Lawrence and Zoncu, 2019; Shimobayashi and Hall, 2014; Wolfson and Sabatini, 2017; Zoncu et al., 2011). We found that the cco-1 RNAi-induced UPRmt activation is independent of raga-1 (Fig. S2 H), suggesting that mitochondrial stress likely represents a unique intrinsic signal for TORC1 activation by lysosome-derived amino acids (Hesketh et al., 2020), which apparently differs from what is observed during development or upon stimulation with exogenous amino acids (Sancak et al., 2008; Schreiber et al., 2010). In support of this model, a Rab5-mediated mitochondrion-endosome-lysosome pathway functions in mitochondrial quality control and is activated upon mitochondrial dysfunction, independent of the autophagic process (Hammerling et al., 2017; Sugiura et al., 2014). More mitochondrial proteins/peptides were also detected in Rab5-positive endosomes in response to mitochondrial stress (Hammerling et al., 2020). Of note, stressed mitochondria might also directly communicate with lysosomes via mitochondria–lysosome membrane contact sites (Wong et al., 2019). Finally, v-ATPase has been also found to participate in endosomal membrane fusion processes (Peters et al., 2001), and it may thereby facilitate the transportation of mitochondria-derived unfolded proteins/peptides to the lysosomes, together with Rab5 and other cofactors.
In addition to the vital role of v-ATPase/mTOR signaling upon mitochondrial stress, we have noticed that the basal expression of some UPRmt transcripts (e.g., hsp-60) decreased after the RNAi of certain v-ATPase subunits (Fig. 2, A and D; and Table S1), indicating that v-ATPase functions in maintaining basal UPRmt activity as well. Nevertheless, the difference between basal and stress conditions is somehow artificial, especially considering that cells and organisms are constantly exposed to various intra- and extracellular cues, and different wild C. elegans strains differ at the level of UPRmt activity under basal conditions (Yin et al., 2017).
Key components in the v-ATPase-TORC1-ATFS-1 pathway are well conserved in mammals, suggesting that a similar mechanism may also exist in mammalian cells. As a case in point, mTORC1 activity has been shown to be essential for ATF4 activation downstream of growth signals (Ben-Sahra et al., 2016; Torrence et al., 2021). Further studies are therefore required to explore whether genetically or pharmacologically targeting this v-ATPase/TORC1-driven MSR pathway could have therapeutic applications against mitochondria-associated diseases, metabolic disorders, as well as normal aging in other organisms.
Limitations of the study
While our current study reveals an indispensable role of v-ATPase/TORC1-mediated ATFS-1 translation in UPRmt activation and mitochondrial stress-associated lifespan extension in C. elegans, several limitations exist. First, how the lysosomes or the v-ATPase/TORC1 complex senses the mitochondrial stress is not well addressed in the current work. To fill this gap, high-resolution imaging and systematic proteomic analysis of the changes in the content of endosomal and lysosomal vesicles in response to mitochondrial stress would be required in the future. Second, we did not manage to find a way to quantitate and compare the magnitude of the effects of v-ATPase/TORC1-mediated ATFS-1 translation versus the suppression of mitochondrial import as nonmutually exclusive mechanisms for UPRmt activation. Third, despite that the decreased expression of ATFS-1 protein is sufficient to explain the reduced activation of UPRmt upon v-ATPase/TORC1 inhibition in C. elegans, we cannot exclude that TORC1 or other enzymes, such as CBP-1 (Li et al., 2021), could still contribute to the UPRmt through posttranslational modifications (e.g., phosphorylation, acetylation) of ATFS-1. Finally, it has been extensively reported that TORC1 signaling inhibition extends the lifespan in multiple animal models (Saxton and Sabatini, 2017; Shimobayashi and Hall, 2014). However, in our current study as well as in other published work (Shpilka et al., 2021), TORC1 activity seems also to be essential for the mitochondrial stress response, which generally extends the lifespan as well. Thus, how these two intertwined processes coordinate to determine overall organismal health and lifespan still requires further investigation.
Materials and methods
C. elegans strains
The Bristol strain (N2) was used as the wild-type strain. SJ4100 [zcIs13(hsp-6p::GFP)], MQ887 [isp-1(qm150)IV], MQ130 [clk-1(qm30) III], SJ4005 [zcIs4(hsp-4p::GFP)], CL2070 [dvIs70 (hsp-16.2p::GFP)], SJ4197 (zcIs39[dve-1p::dve-1::GFP]), OP506 (wgIs506 [xbp-1::TY1::EGFP::3xFLAG + unc-119(+)]). OP675 (wgIs675 [atfs-1::TY1::EGFP::3xFLAG + unc-119(+)]), VC222 [raga-1(ok386)], HZ1683 [atg-2(bp576)], HZ1684 [atg-3(bp412)], HZ1686 [atg-7(bp411)], HZ1687 [atg-9(bp564)], RW12006 (stIs12006 [atfs-1::H1-wCherry + unc-119(+)]), RB967 [gcn-2(ok871)], and RB545 [pek-1(ok275)] were obtained from the Caenorhabditis Genetics Center (CGC). Worms were routinely maintained at 20°C and fed with Escherichia coli (E. coli) OP50 (RRID:WB-STRAIN:WBStrain00041971) on Nematode Growth Medium (NGM) plates.
RNA interference and drug treatment
E. coli strain HT115(DE3; RRID:WB-STRAIN:WBStrain00041080) was obtained from the CGC, transformed with the empty vector L4440, and used as the RNAi control. RNAi clones were obtained from either the Ahringer or Vidal library (Kamath et al., 2003; Rual et al., 2004) and further verified by sequencing or qRT-PCR. The accession codes for vha-1 RNAi clones were 10008-A6 (vha-1_RNAi_1) from the Vidal library and III-5A20 (vha-1_RNAi_2) from the Ahringer library. Double RNAi experiments were performed by mixing bacterial cultures normalized to their optical densities (OD600) before seeding. For treatment of worms with compounds, stock solutions of Tunicamycin (Cat. T7765; Sigma-Aldrich), Concanamycin A (Cat. C9705; Sigma-Aldrich), Bafilomycin A1 (Cat. S1413; Selleckchem), or chloroquine (CQ, Cat. C6628; Sigma-Aldrich) were added to the NGM with final concentrations, as indicated in the figure legends, just before pouring the plates.
UPRmt induction and imaging in C. elegans
For RNAi-induced UPRmt, RNAi bacteria were cultured in lysogeny broth (LB) medium containing 100 μg/ml ampicillin at 37°C overnight. The bacteria were then seeded onto NGM plates containing 2 mM IPTG and 25 mg/ml carbenicillin. L4/young adult worms were picked onto the RNAi bacteria-seeded plates and cultured at 20°C until their progenies reached the young adult stage. A total of 5–10 progenies were then randomly picked and aligned in 10 mM tetramisole (Cat. T1512; Sigma-Aldrich) droplets on NGM plates. Fluorescent images, with the same exposure time for each condition within each of the experiments, were acquired using a Nikon SMZ1000 microscope. All images are compared relatively only to each negative and positive controls in the same batch of the experiment. For compound-induced UPRmt, antimycin A (Cat. A8674; Sigma-Aldrich) with a final concentration of 2.5 μM or Dox (Cat. D9891; Sigma-Aldrich) with a final concentration of 30 μg/ml was added into the NGM just before pouring the plates. For imaging GFP-tagged ATFS-1, atfs-1p::atfs-1::flag::gfp (OP675) worms were mounted in 10 mM tetramisole (Cat. T1512; Sigma-Aldrich) on 2% agarose pads. The most proximal two intestinal cells in each worm were assessed with a Zeiss LSM 700 confocal microscope. At least 20 nuclei were analyzed for each condition. All images were acquired at room temperature with the software provided with the corresponding microscope.
Lifespan experiments
Lifespan experiments were performed at 20°C as described previously (Houtkooper et al., 2013). Briefly, 80–100 worms were used per condition and scored every other day, and those that disappeared or exploded at the vulva were censored. Worms were transferred to fresh plates every week. All RNAi treatments for lifespan started at the maternal L4 stage.
RNA extraction and RNA-seq analysis
For worm samples, synchronized worm eggs were seeded onto NGM plates and cultured at 20°C until the worms reached L4/young adult stage. Worms were then harvested with M9 buffer and snap-frozen in liquid nitrogen. For RNA extraction, 1 ml of TriPure Isolation Reagent (Cat. 11667165001; Roche) was added to each sample tube. The worms were then frozen with liquid nitrogen and thawed in a water bath quickly eight times to rupture cell membranes. Total RNA was then extracted using a column-based kit (Cat. 740955.250; Macherey-Nagel). All RNA-seq was performed by BGI with the BGISEQ-500 platform.
For RNA-seq results, the raw data were filtered by removing adaptor sequences, contamination, and low-quality (phred quality <20) reads. Qualified reads were then mapped to either the worm “Caenorhabditis_elegans.WBcel235.89” with STAR aligner version 2.6.0a. Reads were counted using htseq-count version 0.10.0 using these flags: -f bam -r pos -s no -m union -t exon -i gene_id. Differential expression of genes was calculated by Limma–Voom. The genes with a Benjamini–Hochberg adjusted P value <0.05 were defined as statistically significant. Genes whose expressions were significantly upregulated with log2FC > 0.393 (the fold change for hsp-6, which encodes the UPRmt maker protein HSP-6) in cco-1 RNAi condition and were then downregulated by more than 25% of the log2FC after vha-1/-4/-16/-19 or atfs-1 RNAi co-treatment, compared with the log2FC of cco-1 RNAi condition, were considered as VHA- or ATFS-1-dependent. Functional clustering was conducted using the DAVID (Database for Annotation, Visualization, and Integrated Discovery) database (Huang et al., 2009). Heat-maps were generated by Morpheus (https://software.broadinstitute.org/morpheus). UpSet plot was generated by Intervene (https://asntech.shinyapps.io/intervene/; Khan and Mathelier, 2017).
Quantitative RT-PCR (qRT-PCR)
Worms were harvested and total RNA was extracted as described above for RNA-seq. cDNA was synthesized using the Reverse Transcription Kit (Cat. 205314; Qiagen). qRT-PCR was conducted with the LightCycler 480 SYBR Green I Master kit (Cat. 04887352001; Roche). Primers used for qRT-PCR are listed in Table S2. Primers for worm pmp-3 were used as the normalization control.
Western blots
Proteins were extracted with Radio-immunoprecipitation Assay (RIPA) buffer supplied with protease and phosphatase inhibitors, as described previously (Houtkooper et al., 2013). For Western blots, the GFP-Flag tagged ATFS-1 in atfs-1p::atfs-1::flag::gfp or XBP-1 in xbp-1p::xbp-1::flag::gfp worms was detected by the anti-FLAG M2 antibody (Cat. F1804, 1:1,000, RRID:AB_262044; Sigma-Aldrich). The GFP tagged DVE-1 in dve-1p::dve-1::gfp worms were detected by the anti-GFP antibody (Cat. 2956, CST, 1:1,000, RRID:AB_1196615). P-RSKS-1 was detected using the phospho-Drosophila p70 S6 kinase (Thr398) antibody (Cat. 9209, 1:500, RRID:AB_2269804; CST), as validated in another study (Heintz et al., 2017). P-EIF-2α was detected using the P-EIF2α antibody (Cat. 3597, 1:500, RRID:AB_390740; CST), as validated in another study (Baker et al., 2012). Other antibodies used were: Tubulin (Cat. T5168, 1:2,000, RRID:AB_477579; Sigma-Aldrich), mCherry (Cat. 43590, 1:1,000, RRID:AB_2799246; CST), and HRP-labeled anti-rabbit (Cat. 7074, 1:5,000, RRID:AB_2099233; CST) and anti-mouse (Cat. 7076, 1:5,000, RRID:AB_330924; CST) secondary antibodies. Western blots were performed with either normal SDS-PAGE or 4–12 % gradient gels (Cat. M00654; GenScript).
Polysome profiling
Polysome profiling was conducted as described previously (Gobet et al., 2020; Molenaars et al., 2020). Briefly, worms were collected and lysed in a polysome lysis buffer (100 mM KCl, 10 mM MgCl2, 0.1% NP-40, 2 mM DTT, 0.5 mM Cycloheximide [Cat. S7418; Selleckchem], and RNasin Ribonuclease Inhibitor [Cat. N2611; Promega]) with a Dounce homogenizer. The samples were then centrifuged at 1,200 g for 10 min to remove debris, and the supernatant was normalized using a DC protein assay (Cat. 5000112; Bio-Rad). Using the Gradient Master 108 programmable gradient pourer (Biocomp), sucrose gradients (17.5–50%) were generated in gradient buffer (20 mM Tris-HCl, 150 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 100 μg/ml Cycloheximide). Homogenized worm lysates containing 800 µg of total protein were then loaded onto the sucrose gradients and centrifuged at 32,000 rpm for 2.5 h in an SW40Ti rotor in a Beckman L7 ultracentrifuge (Beckman Coulter). After centrifugation, the gradients were fractionated and measured for RNA content (absorbance at 254 nm) using a Piston Gradient Fractionator (Biocomp) connected to a UV monitor (Bio-Rad). The polysomal fractions (P1–P6) were then collected and combined for RNA extraction using a column-based kit (Cat. 740955.250; Macherey-Nagel).
Statistical analysis
No statistical methods were used to predetermine the sample size. Investigators were not blinded to allocation during experiments and outcome assessment. All experiments, except for the RNA-seq, were repeated at least twice, and similar results were acquired. All statistical analyses were performed using Graphpad Prism 8 software. Data distribution was assumed to be normal, but this was not formally tested. Differences between the two groups were assessed using two-tailed unpaired Student’s t tests. Analysis of variance (ANOVA) followed by Tukey post-hoc test (one-way ANOVA for comparisons between groups, and two-way ANOVA for comparisons of magnitude of changes between different groups from different treatments or cell lines) was used when comparing more than two groups. Survival analyses were performed using the Kaplan–Meier method, and the significance of differences between survival curves was calculated using the log-rank (Mantel-Cox) method.
Online supplemental material
Fig. S1 shows the impact of vha-1, vha-4, vha-16, or vha-19 RNAi in different stress responses and gene expression in C. elegans. Fig. S2 shows the impact of mitochondrial stress, ER stress, and TORC1 signaling regulators in gene expression and stress responses. Fig. S3 shows the impact of ribosomal subunit RNAi in stress responses in C. elegans. Fig. S4 shows that autophagy-defective mutants have normal activation of the UPRmt in response to mitochondrial stress. Fig. S5 shows the key role of ribosomal subunits and VHA-1 in mitochondrial surveillance. Table S1 shows the RNA-seq results of worms fed with RNAi targeting v-ATPase subunits and/or cco-1 RNAi. Table S2 shows the list of primers used for qRT-PCR in this study.
Data availability
Original reagents are available upon request. The raw and processed sequencing datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database with the accession numbers: GSE179517.
Supplementary Material
show the RNA-seq results of worms fed with RNAi targeting v-ATPase subunits and/or cco-1 RNAi.
show the list of primers used for qRT-PCR in this study.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. S2.
Acknowledgments
We thank the Caenorhabditis Genetics Center for providing the C. elegans strains. We thank all members of J. Auwerx and K. Schoonjans laboratories for helpful discussions.
This work was supported by grants from the Ecole Polytechnique Federale de Lausanne (EPFL), the European Research Council (ERC-AdG-787702), the Swiss National Science Foundation (SNSF 31003A_179435), and the GRL grant of the National Research Foundation of Korea (NRF 2017K1A1A2013124). T.Y. Li was supported by the “Human Frontier Science Program” (LT000731/2018-L). A.W. Gao was supported by the Accelerator prize given by the United Mitochondrial Disease Foundation (PF-19-0232). X. Li was supported by the China Scholarship Council (201906050019).
The authors declare no competing financial interests.
Author contributions: T.Y. Li and J. Auwerx conceived the project. T.Y. Li and A.W. Gao performed most of the experiments. T.Y. Li., A.W. Gao, X. Li, H. Li, and A. Lalou performed data analysis. A.W. Gao, Y.J. Liu, N. Neelagandan, and F. Naef contributed to the polysome profiling experiment. K. Schoonjans and J. Auwerx supervised and financed the study. T.Y. Li, A.W. Gao, and J. Auwerx wrote the manuscript with comments from all authors.
References
- Baker, B.M., Nargund A.M., Sun T., and Haynes C.M.. 2012. Protective coupling of mitochondrial function and protein synthesis via the eIF2alpha kinase GCN-2. PLoS Genet. 8:e1002760. 10.1371/journal.pgen.1002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Sahra, I., Hoxhaj G., Ricoult S.J.H., Asara J.M., and Manning B.D.. 2016. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 351:728–733. 10.1126/science.aad0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, E.J., Siebers A., and Altendorf K.. 1988. Bafilomycins: A class of inhibitors of membrane ATPases from microorganisms, animal cells, and plant cells. Proc. Natl. Acad. Sci. USA. 85:7972–7976. 10.1073/pnas.85.21.7972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon, M., Zeng H., Urano F., Till J.H., Hubbard S.R., Harding H.P., Clark S.G., and Ron D.. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415:92–96. 10.1038/415092a [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli, M., and Walter P.. 2020. The integrated stress response: From mechanism to disease. Science. 368:eaat5314. 10.1126/science.aat5314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amico, D., Sorrentino V., and Auwerx J.. 2017. Cytosolic proteostasis networks of the mitochondrial stress response. Trends Biochem. Sci. 42:712–725. 10.1016/j.tibs.2017.05.002 [DOI] [PubMed] [Google Scholar]
- Drose, S., Bindseil K.U., Bowman E.J., Siebers A., Zeeck A., and Altendorf K.. 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry. 32:3902–3906. 10.1021/bi00066a008 [DOI] [PubMed] [Google Scholar]
- Durieux, J., Wolff S., and Dillin A.. 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 144:79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedele, A.O., and Proud C.G.. 2020. Chloroquine and bafilomycin A mimic lysosomal storage disorders and impair mTORC1 signalling. Biosci. Rep. 40:BSR20200905. 10.1042/BSR20200905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, J., Bussiere F., and Hekimi S.. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 1:633–644. 10.1016/s1534-5807(01)00071-5 [DOI] [PubMed] [Google Scholar]
- Forgac, M. 2007. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8:917–929. 10.1038/nrm2272 [DOI] [PubMed] [Google Scholar]
- Gao, K., Li Y., Hu S., and Liu Y.. 2019. SUMO peptidase ULP-4 regulates mitochondrial UPR-mediated innate immunity and lifespan extension. Elife. 8:e41792. 10.7554/eLife.41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobet, C., Weger B.D., Marquis J., Martin E., Neelagandan N., Gachon F., and Naef F.. 2020. Robust landscapes of ribosome dwell times and aminoacyl-tRNAs in response to nutrient stress in liver. Proc. Natl. Acad. Sci. USA. 117:9630–9641. 10.1073/pnas.1918145117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerling, B.C., Najor R.H., Cortez M.Q., Shires S.E., Leon L.J., Gonzalez E.R., Boassa D., Phan S., Thor A., Jimenez R.E., et al. 2017. A Rab5 endosomal pathway mediates Parkin-dependent mitochondrial clearance. Nat. Commun. 8:14050. 10.1038/ncomms14050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerling, B.C., Shires S.E., Leon L.J., Cortez M.Q., and Gustafsson A.B.. 2020. Isolation of Rab5-positive endosomes reveals a new mitochondrial degradation pathway utilized by BNIP3 and Parkin. Small GTPases. 11:69–76. 10.1080/21541248.2017.1342749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes, C.M., Petrova K., Benedetti C., Yang Y., and Ron D.. 2007. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell. 13:467–480. 10.1016/j.devcel.2007.07.016 [DOI] [PubMed] [Google Scholar]
- Heintz, C., Doktor T.K., Lanjuin A., Escoubas C., Zhang Y., Weir H.J., Dutta S., Silva-Garcia C.G., Bruun G.H., Morantte I., et al. 2017. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature. 541:102–106. 10.1038/nature20789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesketh, G.G., Papazotos F., Pawling J., Rajendran D., Knight J.D.R., Martinez S., Taipale M., Schramek D., Dennis J.W., and Gingras A.C.. 2020. The GATOR-Rag GTPase pathway inhibits mTORC1 activation by lysosome-derived amino acids. Science. 370:351–356. 10.1126/science.aaz0863 [DOI] [PubMed] [Google Scholar]
- Higuchi-Sanabria, R., Frankino P.A., Paul J.W. 3rd, Tronnes S.U., and Dillin A.. 2018. A futile battle? Protein quality control and the stress of aging. Dev. Cell. 44:139–163. 10.1016/j.devcel.2017.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homewood, C.A., Warhurst D.C., Peters W., and Baggaley V.C.. 1972. Lysosomes, pH and the anti-malarial action of chloroquine. Nature. 235:50–52. 10.1038/235050a0 [DOI] [PubMed] [Google Scholar]
- Houtkooper, R.H., Mouchiroud L., Ryu D., Moullan N., Katsyuba E., Knott G., Williams R.W., and Auwerx J.. 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 497:451–457. 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D.W., Sherman B.T., and Lempicki R.A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Inoki, K., Li Y., Xu T., and Guan K.L.. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17:1829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell, J.L., Kim Y.C., Russell R.C., Yu F.X., Park H.W., Plouffe S.W., Tagliabracci V.S., and Guan K.L.. 2015. Metabolism. Differential regulation of mTORC1 by leucine and glutamine. Science. 347:194–198. 10.1126/science.1259472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath, R.S., Fraser A.G., Dong Y., Poulin G., Durbin R., Gotta M., Kanapin A., Le Bot N., Moreno S., Sohrmann M., et al. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 421:231–237. 10.1038/nature01278 [DOI] [PubMed] [Google Scholar]
- Khan, A., and Mathelier A.. 2017. Intervene: A tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinf. 18:287. 10.1186/s12859-017-1708-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski, B., and Hekimi S.. 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science. 272:1010–1013. 10.1126/science.272.5264.1010 [DOI] [PubMed] [Google Scholar]
- Lawrence, R.E., and Zoncu R.. 2019. The lysosome as a cellular centre for signalling, metabolism and quality control. Nat. Cell Biol. 21:133–142. 10.1038/s41556-018-0244-7 [DOI] [PubMed] [Google Scholar]
- Lee, S.K., Li W., Ryu S.E., Rhim T., and Ahnn J.. 2010. Vacuolar (H+)-ATPases in Caenorhabditis elegans: What can we learn about giant H+ pumps from tiny worms? Biochim. Biophys. Acta. 1797:1687–1695. 10.1016/j.bbabio.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Li, T.Y., Sleiman M.B., Li H., Gao A.W., Mottis A., Bachmann A.M., El Alam G., Li X., Goeminne L.J.E., Schoonjans K., and Auwerx J.. 2021. The transcriptional coactivator CBP/p300 is an evolutionarily conserved node that promotes longevity in response to mitochondrial stress. Nat. Aging. 1:165–178. 10.1038/s43587-020-00025-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, T., Li T.Y., Mottis A., and Auwerx J.. 2022. Pleiotropic effects of mitochondria in aging. Nat. Aging. 2:199–213. 10.1038/s43587-022-00191-2 [DOI] [PubMed] [Google Scholar]
- Liu, Y., Samuel B.S., Breen P.C., and Ruvkun G.. 2014. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 508:406–410. 10.1038/nature13204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth, C., Jovaisaite V., Durieux J., Matilainen O., Jordan S.D., Quiros P.M., Steffen K.K., Williams E.G., Mouchiroud L., Tronnes S.U., et al. 2016. Two conserved histone demethylases regulate mitochondrial stress-induced longevity. Cell. 165:1209–1223. 10.1016/j.cell.2016.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra, P., and Chan D.C.. 2014. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat. Rev. Mol. Cell Biol. 15:634–646. 10.1038/nrm3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaars, M., Janssens G.E., Williams E.G., Jongejan A., Lan J., Rabot S., Joly F., Moerland P.D., Schomakers B.V., Lezzerini M., et al. 2020. A conserved mito-cytosolic translational balance links two longevity pathways. Cell Metabol. 31:549–563.e7. 10.1016/j.cmet.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottis, A., Herzig S., and Auwerx J.. 2019. Mitocellular communication: Shaping health and disease. Science. 366:827–832. 10.1126/science.aax3768 [DOI] [PubMed] [Google Scholar]
- Murray, J.I., Boyle T.J., Preston E., Vafeados D., Mericle B., Weisdepp P., Zhao Z., Bao Z., Boeck M., and Waterston R.H.. 2012. Multidimensional regulation of gene expression in the C. elegans embryo. Genome Res. 22:1282–1294. 10.1101/gr.131920.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund, A.M., Fiorese C.J., Pellegrino M.W., Deng P., and Haynes C.M.. 2015. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPRmt. Mol. Cell. 58:123–133. 10.1016/j.molcel.2015.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund, A.M., Pellegrino M.W., Fiorese C.J., Baker B.M., and Haynes C.M.. 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science. 337:587–590. 10.1126/science.1223560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari, J., and Suomalainen A.. 2012. Mitochondria: In sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka, K., Koryga I., Mnich K., Ljujic M., Samali A., and Gorman A.M.. 2016. The integrated stress response. EMBO Rep. 17:1374–1395. 10.15252/embr.201642195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, C., Bayer M.J., Buhler S., Andersen J.S., Mann M., and Mayer A.. 2001. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 409:581–588. 10.1038/35054500 [DOI] [PubMed] [Google Scholar]
- Quiros, P.M., Prado M.A., Zamboni N., D’Amico D., Williams R.W., Finley D., Gygi S.P., and Auwerx J.. 2017. Multi-omics analysis identifies ATF4 as a key regulator of the mitochondrial stress response in mammals. J. Cell Biol. 216:2027–2045. 10.1083/jcb.201702058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual, J.F., Ceron J., Koreth J., Hao T., Nicot A.S., Hirozane-Kishikawa T., Vandenhaute J., Orkin S.H., Hill D.E., van den Heuvel S., and Vidal M.. 2004. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 14:2162–2168. 10.1101/gr.2505604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runkel, E.D., Liu S., Baumeister R., and Schulze E.. 2013. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 9:e1003346. 10.1371/journal.pgen.1003346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak, Y., Peterson T.R., Shaul Y.D., Lindquist R.A., Thoreen C.C., Bar-Peled L., and Sabatini D.M.. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 320:1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton, R.A., and Sabatini D.M.. 2017. mTOR signaling in growth, metabolism, and disease. Cell. 169:361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- Schreiber, M.A., Pierce-Shimomura J.T., Chan S., Parry D., and McIntire S.L.. 2010. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase Raga-1. PLoS Genet. 6:e1000972. 10.1371/journal.pgen.1000972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, L.W., Peng Q., Dong M., Gao K., Li Y., Li Y., Li C.Y., and Liu Y.. 2020. Histone deacetylase HDA-1 modulates mitochondrial stress response and longevity. Nat. Commun. 11:4639. 10.1038/s41467-020-18501-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., Ellis R.E., Lee K., Liu C.Y., Yang K., Solomon A., Yoshida H., Morimoto R., Kurnit D.M., Mori K., and Kaufman R.J.. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 107:893–903. 10.1016/s0092-8674(01)00612-2 [DOI] [PubMed] [Google Scholar]
- Shimobayashi, M., and Hall M.N.. 2014. Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15:155–162. 10.1038/nrm3757 [DOI] [PubMed] [Google Scholar]
- Shpilka, T., Du Y., Yang Q., Melber A., Uma Naresh N., Lavelle J., Kim S., Liu P., Weidberg H., Li R., et al. 2021. UPR(mt) scales mitochondrial network expansion with protein synthesis via mitochondrial import in Caenorhabditis elegans. Nat. Commun. 12:479. 10.1038/s41467-020-20784-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpilka, T., and Haynes C.M.. 2018. The mitochondrial UPR: Mechanisms, physiological functions and implications in ageing. Nat. Rev. Mol. Cell Biol. 19:109–120. 10.1038/nrm.2017.110 [DOI] [PubMed] [Google Scholar]
- Sugiura, A., McLelland G.L., Fon E.A., and McBride H.M.. 2014. A new pathway for mitochondrial quality control: Mitochondrial-derived vesicles. EMBO J. 33:2142–2156. 10.15252/embj.201488104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhm, T., Kaimal J.M., Dawitz H., Peselj C., Masser A.E., Hanzen S., Ambrozic M., Smialowska A., Bjorck M.L., Brzezinski P., et al. 2018. Mitochondrial translation efficiency controls cytoplasmic protein homeostasis. Cell Metabol. 27:1309–1322.e6. 10.1016/j.cmet.2018.04.011 [DOI] [PubMed] [Google Scholar]
- Sun, N., Youle R.J., and Finkel T.. 2016. The mitochondrial basis of aging. Mol. Cell. 61:654–666. 10.1016/j.molcel.2016.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen, C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., and Gray N.S.. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032. 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, Y., Garcia G., Bian Q., Steffen K.K., Joe L., Wolff S., Meyer B.J., and Dillin A.. 2016. Mitochondrial stress induces chromatin reorganization to promote longevity and UPR(mt). Cell. 165:1197–1208. 10.1016/j.cell.2016.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrence, M.E., MacArthur M.R., Hosios A.M., Valvezan A.J., Asara J.M., Mitchell J.R., and Manning B.D.. 2021. The mTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. Elife. 10:e63326. 10.7554/eLife.63326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafai, S.B., and Mootha V.K.. 2012. Mitochondrial disorders as windows into an ancient organelle. Nature. 491:374–383. 10.1038/nature11707 [DOI] [PubMed] [Google Scholar]
- Wei, W., and Ruvkun G.. 2020. Lysosomal activity regulates Caenorhabditis elegans mitochondrial dynamics through vitamin B12 metabolism. Proc. Natl. Acad. Sci. USA. 117:19970–19981. 10.1073/pnas.2008021117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West, A.P., and Shadel G.S.. 2017. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17:363–375. 10.1038/nri.2017.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson, R.L., and Sabatini D.M.. 2017. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metabol. 26:301–309. 10.1016/j.cmet.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.C., Kim S., Peng W., and Krainc D.. 2019. Regulation and function of mitochondria-lysosome membrane contact sites in cellular homeostasis. Trends Cell Biol. 29:500–513. 10.1016/j.tcb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Liu P., Anderson N.S., Shpilka T., Du Y., Naresh N.U., Li R., Zhu L.J., Luk K., Lavelle J., et al. 2022. LONP-1 and ATFS-1 sustain deleterious heteroplasmy by promoting mtDNA replication in dysfunctional mitochondria. Nat. Cell Biol. 24:181–193. 10.1038/s41556-021-00840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J.A., Gao G., Liu X.J., Hao Z.Q., Li K., Kang X.L., Li H., Shan Y.H., Hu W.L., Li H.P., and Cai S.Q.. 2017. Genetic variation in glia-neuron signalling modulates ageing rate. Nature. 551:198–203. 10.1038/nature24463 [DOI] [PubMed] [Google Scholar]
- Yoneda, T., Benedetti C., Urano F., Clark S.G., Harding H.P., and Ron D.. 2004. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117:4055–4066. 10.1242/jcs.01275 [DOI] [PubMed] [Google Scholar]
- Yuan, J., Chang S.Y., Yin S.G., Liu Z.Y., Cheng X., Liu X.J., Jiang Q., Gao G., Lin D.Y., Kang X.L., et al. 2020. Two conserved epigenetic regulators prevent healthy ageing. Nature. 579:118–122. 10.1038/s41586-020-2037-y [DOI] [PubMed] [Google Scholar]
- Zhao, Q., Wang J., Levichkin I.V., Stasinopoulos S., Ryan M.T., and Hoogenraad N.J.. 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21:4411–4419. 10.1093/emboj/cdf445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, D., Li X., and Tian Y.. 2022. Mitochondrial-to-nuclear communication in aging: An epigenetic perspective. Trends Biochem. Sci. 47:645–659. 10.1016/j.tibs.2022.03.008 [DOI] [PubMed] [Google Scholar]
- Zhu, D., Wu X., Zhou J., Li X., Huang X., Li J., Wu J., Bian Q., Wang Y., and Tian Y.. 2020. NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 6:eabb2529. 10.1126/sciadv.abb2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu, R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., and Sabatini D.M.. 2011. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science. 334:678–683. 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
show the RNA-seq results of worms fed with RNAi targeting v-ATPase subunits and/or cco-1 RNAi.
show the list of primers used for qRT-PCR in this study.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. S2.
Data Availability Statement
Original reagents are available upon request. The raw and processed sequencing datasets have been deposited in the NCBI Gene Expression Omnibus (GEO) database with the accession numbers: GSE179517.