Abstract
Purpose:
We hypothesized that inhibition and trapping of PARP1 alone would be sufficient to achieve antitumor activity. In particular, we aimed to achieve selectivity over PARP2, which has been shown to play a role in the survival of hematopoietic/stem progenitor cells in animal models. We developed AZD5305 with the aim of achieving improved clinical efficacy and wider therapeutic window. This next-generation PARP inhibitor (PARPi) could provide a paradigm shift in clinical outcomes achieved by first-generation PARPi, particularly in combination.
Experimental Design:
AZD5305 was tested in vitro for PARylation inhibition, PARP-DNA trapping, and antiproliferative abilities. In vivo efficacy was determined in mouse xenograft and PDX models. The potential for hematologic toxicity was evaluated in rat models, as monotherapy and combination.
Results:
AZD5305 is a highly potent and selective inhibitor of PARP1 with 500-fold selectivity for PARP1 over PARP2. AZD5305 inhibits growth in cells with deficiencies in DNA repair, with minimal/no effects in other cells. Unlike first-generation PARPi, AZD5305 has minimal effects on hematologic parameters in a rat pre-clinical model at predicted clinically efficacious exposures. Animal models treated with AZD5305 at doses ≥0.1 mg/kg once daily achieved greater depth of tumor regression compared to olaparib 100 mg/kg once daily, and longer duration of response.
Conclusions:
AZD5305 potently and selectively inhibits PARP1 resulting in excellent antiproliferative activity and unprecedented selectivity for DNA repair deficient versus proficient cells. These data confirm the hypothesis that targeting only PARP1 can retain the therapeutic benefit of nonselective PARPi, while reducing potential for hematotoxicity. AZD5305 is currently in phase I trials (NCT04644068).
Translational Relevance.
Here we describe the primary pharmacology of a next-generation PARP inhibitor (PARPi) and trapper, AZD5305, designed to address the limitations of current clinical PARPi, particularly in combination with first-line chemotherapy. In biochemical and cellular assays, AZD5305 shows high selectivity for PARP1 versus other PARP family enzymes. In multiple in vivo models, AZD5305 shows robust anticancer efficacy both as monotherapy and in combination. In rat preclinical toxicology models, AZD5305 causes minimal hematologic toxicity as monotherapy and is differentiated from first-generation PARPi in combination. Overall, AZD5305 demonstrates that potent and selective inhibition and trapping of PARP1 represents a promising avenue to achieve improved outcomes for patients treated with PARPi.
Introduction
The clinically approved PARP inhibitors (PARPi) olaparib, rucaparib, niraparib, and talazoparib, have demonstrated efficacy in ovarian, breast, prostate, and pancreatic cancer, particularly in homologous recombination repair (HRR)-deficient tumors, including BRCA-mutant (BRCAm) tumors (1). Despite providing significant benefit and improved tolerability compared with standard-of-care chemotherapy, PARPi still display some undesirable side effects (2, 3). In particular, clinical cytopenias of varying severity are common to this class of compounds, suggesting that these may be driven by broad PARPi pharmacology. Moreover, combination of these drugs with already poorly tolerated chemotherapies has been limited by overlapping hematologic toxicities (2).
All approved PARPi have limited selectivity for PARP1 over PARP2 (as well as other members of the PARP family; ref. 4). In terms of enzymatic function, both PARP1 and PARP2 bind to sites of DNA damage and build chains of poly-ADP-ribose (PAR) on themselves and other proteins in a process called PARylation (5). PARP1 has the most prominent role in cellular PARylation accounting for 80%–95% of overall activity with PARP2 responsible for the remaining 5%–20% (6). As a consequence of catalytic inhibition, PARPs can remain tightly bound to the chromatin through a process called “PARP-DNA trapping” (7). This ternary complex (inhibitor-PARP-DNA) is stabilized by the presence of the drug and causes a physical block to the replisome machinery during DNA replication, leading to replication fork collapse and DNA double-strand breaks (DSB; ref. 8). Normal cells effectively repair DSB through the HRR pathway. In cancers with DNA repair defects, particularly those with HRR-deficiency, accumulation of unrepaired DSB results in unsupportable genomic instability that ultimately leads to cell death, a process known as synthetic lethality (1).
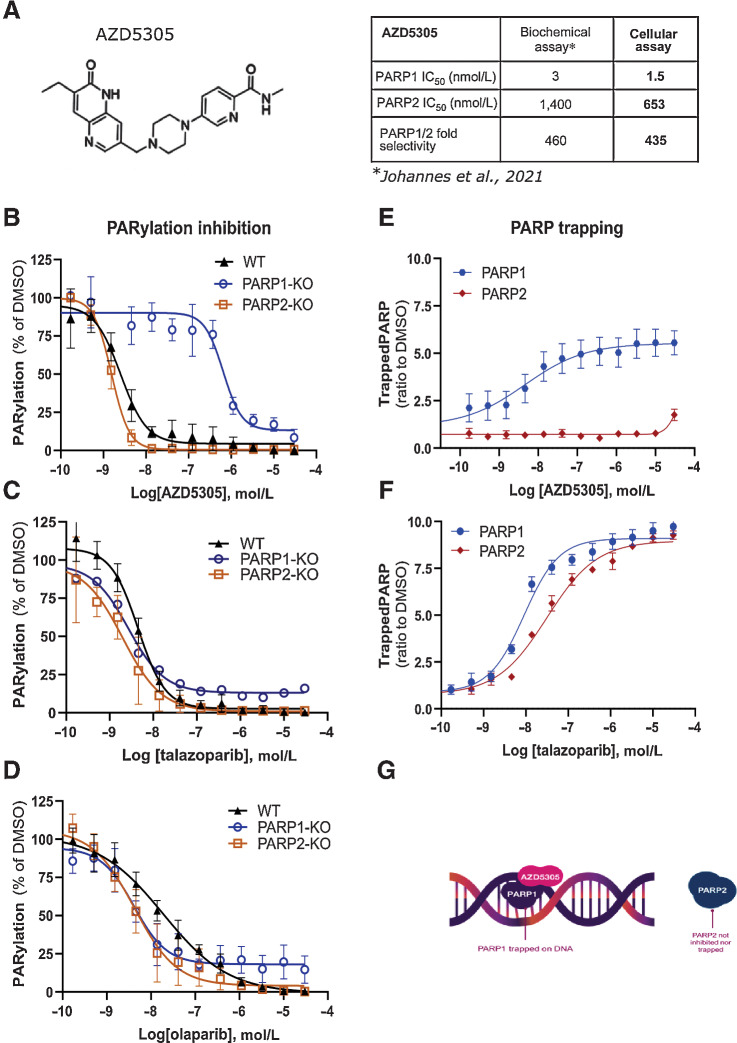
It has previously been demonstrated that synthetic lethality with BRCA mutations was solely caused by PARP1 inhibition and trapping, and that trapping of PARP2 may not be required for anticancer activity (6, 7). Furthermore, in animal models, PARP2 has been shown to play a key role in the survival of hematopoietic/stem progenitor cells (9, 10) suggesting that inhibition and trapping of PARP2, a feature shared by all the currently approved PARPi, may disproportionately drive the hematological toxicity observed in the clinic. These observations led us to hypothesise that a highly selective PARP1 inhibitor and trapper would retain anticancer efficacy, while displaying reduced hematological toxicity (4), potentially expanding clinical options, including combinations with standard-of-care agents. Therefore, we initiated a drug discovery programme that led to the development of AZD5305 (4), which selectively inhibits and traps PARP1 but not PARP2 (Fig. 1A).
Figure 1.
AZD5305 inhibits and traps selectively PARP1 in cells. A, Left: chemical structure of AZD5305. Right: table summarizing AZD5305 IC50 in binding and inhibition of PARP1 and PARP2 in biochemical and cellular assays. B–D, PARylation inhibition in A549 isogenic cell lines: WT (solid triangle), PARP1-KO (empty circle), PARP2-KO (empty square) upon dose–response of AZD5305 (B), talazoparib (C), or olaparib (D). E and F, Trapping profiles of PARP1 (blue circle) and PARP2 (red diamond) in A549 cells, measured upon dose–response treatments of AZD5305 (E) or talazoparib (F). Details of all experiments are described in Materials and Methods section. Each dose–response curve is the mean of four independent experiments; the error bars indicate ±SEM. G, Scheme representing the selective targeting of AZD5305.
Here we describe the pharmacologic characterization of AZD5305 in preclinical in vitro and in vivo models. Overall, AZD5305 shows potent antiproliferative activities in vitro, as well as robust and durable anticancer efficacy in vivo. AZD5305 also shows a favorable toxicity profile, with minimal effects on bone marrow in a rat in vivo model.
Materials and Methods
PARP inhibitors, cell lines, and cell culture
All PARPi compounds were synthetized by AstraZeneca as described by Johannes and colleagues (4), dissolved in DMSO at 10 mmol/L stock concentration, and stored protected from light.
A full list of the cell lines and media used in this study can be found in Supplementary Table S1. All cell lines were obtained from ATCC, Horizon Discovery, or BioIVT; authenticated by the AstraZeneca cell bank using short tandem repeat analysis using CellCheck (IDEXX Bioanalytics); and validated free of Mycoplasma contamination using the STAT-Myco assay (IDEXX Bioanalytics).
Colony formation assay
Cells were seeded in 24-well plates (500–1,500 cells/well, depending on the doubling times) and incubated overnight. PARPi were dispensed by an automated digital D300 HP dispenser (Tecan) in titration dilutions, covering 0.1 nmol/L–40 μmol/L; each concentration was tested in triplicate and DMSO was used as untreated control. Plates were incubated at 37°C, 5% CO2 for 8–13 days, to allow colony formation. Cells were then fixed and stained with Blue-G-250 brilliant blue [#B8522–1EA, Sigma, reconstituted in 25% (v/v) methanol and 5% (v/v) acetic acid] for 15 min then thoroughly washed with dH2O. Plates were scanned with GelCount (Oxford OPTRONIX). Colonies were analyzed by total optical density measured with ImageJ software, using a 24-well plate ROI mask. Data analysis was performed by normalization on the vehicle treated of the respective plate set as 1. Data were plotted and IC50s were calculated with GraphPad Prism software.
PARylation inhibition assay
Cells were seeded into 384-well plates (Greiner, Austria; 781090) using a Multidrop Combi (Thermo Fisher Scientific) and incubated overnight at 37°C and 5% CO2 in a rotating incubator. PARPi compounds were added with Echo555 (LabCyte), with final compound concentration range between 1.6 pmol/L and 30 μmol/L. After 1 hour incubation with PARPi, H2O2 was added at final concentration of 10 mmol/L and incubated for 5 min in culture conditions. Cells were then fixed in ice-cold methanol for 15 minutes at 4°C and processed for immunostaining with primary antibody Poly/Mono-ADP Ribose (Cell Signaling Technology, #83732) followed by secondary goat anti-rabbit AlexaFluor-488 antibody (Invitrogen, #A-11008); nuclei were counterstained with DAPI. The nuclear PAR fluorescence intensity (F.I.) was acquired and analyzed using CellInsight CX5 HCS Platform (Thermo Fisher Scientific); dose–response curves were generated with GraphPad Prism software.
PARP trapping assay
Cells were plated at a density of 6,000 cells per well into 384-well plates (Greiner, Austria; 781090) using a Multidrop Combi. PARPi were added with Echo555 (LabCyte) at final concentration range between 1.6 pmol/L and 30 μmol/L and incubated for 4 hours; Methyl methanesulfonate (MMS; Sigma Aldrich, 129925) was added at final 0.01%. Cell media was removed and a pre-extraction step was performed for 10 min at 4°C with cold cytoskeleton (CSK) buffer (10 mmol/L PIPES pH = 6.8, 300 mmol/L sucrose, 200 mmol/L NaCl, 3 mmol/L MgCl2) supplemented with 0.6% Triton X-100. Cells were then fixed with ice-cold methanol for 15 minutes at −20°C; blocking solution (PBS + 0.1% Tween20 + 3% BSA) was added and incubated for 1 hour at room temperature. Immunostaining of PARP1 or PARP2 was performed by adding the primary antibody (PARP1 antibody, ProteinTech, 13371–1-AP; PARP2 antibody, Active Motif, 39743) diluted 1:1,000 in PBS + 1% BSA and incubated overnight at 4°C, followed by incubation with secondary goat anti-rabbit AlexaFluor 488 antibody for 1 hour at room temperature. DNA was counterstained with Hoechst 33342 (Invitrogen, H3570) diluted 1:5,000 in PBS + 1% BSA. The nuclear F.I. was acquired and analyzed using CellInsight CX5 HCS Platform (Thermo Fisher); dose–response curves were generated with GraphPad Prism Software.
DNA damage and cell-cycle analysis by immunofluorescence and flow cytometry
Cells were seeded in 96-well plates for immunofluorescence and in 12-well plates for flow cytometry analysis. Compounds were added using a D300 HP dispenser (Tecan) from compound stocks dissolved in DMSO, in duplicate for each condition. Plates were incubated for 24, 48, or 72 hours. During the last 30 minutes of incubation, EdU was added to cells, at final concentration of 10 μmol/L and then processed for immunofluorescence or flow cytometry analysis. For immunofluorescence analysis, cells were washed and fixed in 4% paraformaldehyde (PFA) for 15 minutes at room temperature and then permeabilized in PBS + 0.1% Triton X-100 for 10 minutes at room temperature. Blocking was performed using 0.5% BSA + 0.2% gelatin from cold water fish skin (Sigma, catalog no. G7765) in PBS for 1 hour at room temperature. EdU Click-IT reaction was performed following the manufacturer's instructions (Thermo Fisher Scientific). Cells were then washed three times with PBG; primary antibodies were incubated overnight at 4°C (γH2AX, Merck Millipore, 05–636, diluted 1:5,000), followed by secondary antibody (anti-mouse, Alexa Fluor 594 (Thermo, A21099, 1:2,000) and DAPI (Thermo Fisher Scientific, D1306, diluted 1 μg/mL) for 1 hour at room temperature.
For flow cytometry analysis, media and cells were collected and fixed in PFA 4% for 15 minutes at room temperature followed by EdU Click-IT reaction. After washes, DNA was counterstained with DAPI. Samples where then acquired using FACSCelesta (BD Biosciences) and analyzed with FlowJo software.
Xenograft studies
The cells were grown as listed in Supplementary Table S1, and were then implanted subcutaneously into the mice [MDA-MB-436 2.5×106 cells with 50% Matrigel (Corning) to SCID female mice (Envigo); DLD-1 and DLD-1 BRCA2−/− 5 × 106 cells to Nude female mice (Envigo)]. HBCx-9 and HBCx-17 PDX studies were performed at Xentech. Tumor fragments were implanted subcutaneously into female athymic Nude-Foxn1nu mice (Envigo). Animal studies were conducted in accordance with UK Home Office legislation, the Animal Scientific Procedures Act 1986, and the AstraZeneca Global Bioethics policy or Institutional Animal Care and Use Committee guidelines. Experimental work is outlined in project license 70/8894 and P0EC1FFDF, which has gone through the AstraZeneca Ethical Review Process. Mice were maintained in a controlled, specific pathogen-free environment at 20–25°C, 40%–70% humidity, and 12 hours light-to-dark cycle. Tumor length and width were measured by caliper, and tumor volume calculated using the formula volume = (length × width2)*π/6. Animal bodyweight and tumor condition were monitored throughout the study. Mice were randomized into treatment groups when mean tumor volume reached approximately 0.1–0.3 cm3 (efficacy studies) or 0.5–0.6 cm3 [pharmacokinetic/pharmacodynamic (PK/PD) studies]. Animals were treated from the day after the randomization. AZD5305 and olaparib (AZD2281) were administered by oral gavage once daily (QD) at 10 mL/kg final dose volume. Olaparib was formulated in 10% DMSO (Sigma), 30% Kleptose (Roquette). AZD5305 was formulated in water/HCl pH 3.5–4. Carboplatin was prepared fresh on the day of dosing and administered intraperitoneally at 10 mL/kg final dose volume. Carboplatin (Sigma) was formulated in 0.85% physiologic saline. For Xentech studies carboplatin stock (Teva or Sandoz) was diluted to the final concentration in 0.9% NaCl.
At the end of experiments mice were euthanized via cervical dislocation followed by severing of major artery/permanent cessation of circulation. In the PK/PD experiments, tumors were collected at various timepoints relative to the final dose.
PD assay
Tumor tissue samples were homogenized in Fast Prep tubes in 1 mL of lysis buffer (20 mmol/L Tris pH 7.5, 137 mmol/L NaCl, 10% glycerol, 50 mmol/L NaF, 1 mmol/L Na3VO4, 1% SDS, 1% NP40) with phosphatase inhibitor cocktails (Sigma P0044 and P5726) and protease inhibitor (Roche 1836145), using 3 cycles at 6.5 m/s setting for 30 seconds in Fast Prep machine (5 min on ice between the cycles). Samples were sonicated for 15 sec 50% amplitude and returned to ice for 30 minutes. After the incubation samples were spun down at 13,000 RPM for 15 minutes at 4°C and the supernatants were transferred to fresh tubes. Protein concentrations were estimated using a BCA Protein assay kit (Pierce Thermo 23227). For PAR analysis protein lysates were incubated at 100°C for 5 minutes, rested on ice for 1 minute, and then spun down at 10,000 RPM for 2 minutes at 4°C. Supernatants were collected and protein concentrations were adjusted to 1 mg/mL in lysis buffer. PAR levels were analyzed using PARP In Vivo Pharmacodynamic Assay II ELISA kit (Trevigen 4520–096-K). Briefly, samples were diluted to 40 ng/μL using kit diluent buffer and then incubated (50 μL) for 16 hours at 4°C on precoated ELISA plates. After washing in PBS Tween (0.1%), plates were incubated with anti-PAR antibody for 2 hours at room temperature, and then with secondary HRP-conjugated antibody for 1 hour at room temperature. Detection was performed using kit peroxyglow reagents. Luminescence was quantified using a Tecan Safire and PAR levels (pg/mL) estimated from standard curve comparisons.
PK measurements
Samples for plasma pharmacokinetics were obtained by collecting live bleeds using a microsampling method. Animals were placed in the hot box for 5–10 minutes to warm up. A 25 Gauge needle was used to prick one of the tail side veins. Twenty microliters of blood was collected with a micro-capillary pipette and placed into an Eppendorf tube containing 80 μL of PBS. The samples were mixed by inversion and centrifuged at 13,000 RPM for 3 minutes at 4°C. Separated plasma was collected and stored at −80°C. To determine compound levels in plasma samples, each plasma sample (25 μL) was prepared using an appropriate dilution factor and compared against an 11-point standard calibration curve (1–10,000 nmol/L) prepared in DMSO and spiked into blank plasma. Acetonitrile (100 μL) was added with the internal standard, followed by centrifugation at 3,000 RPM for 10 minutes. Supernatant (50 μL) was then diluted in 300 μL water and analyzed via UPLC-MS/MS.
Repeat dose studies in rats
All procedures involving animals were conducted in accordance with UK Home Office legislation [Animals (Scientific Procedures) Act 1986] and AstraZeneca's institutional policies. Female Han Wistar rats (Charles River; age 12–13 weeks) were acclimatized and randomized into treatment groups of n = 8 (monotherapy and 14-day combination studies) or n = 4 (42-day combination study) based on cage mean blood parameters obtained 1–2 weeks prior to study initiation. Four animals were held per cage and 1–2 cages per treatment group.
During monotherapy studies, rats received either 1 mg/kg AZD5305 or vehicle [0.5% w/v hydroxypropyl methylcellulose (HPMC)/0.1% Tween80 in water, adjusted for pH 3–3.2], 57 mg/kg niraparib or vehicle (0.5% w/v HPMC) or 100 mg/kg olaparib (co-administered with 0.5% w/v HPMC, 0.1% Tween 80 because this was a monotherapy arm from a combination study), or vehicle (0.5% w/v HPMC/0.1% Tween 80 in water) orally, daily for 14 days.
In combination studies, rats were dosed with 1 mg/kg AZD5305, 100 mg/kg olaparib, or vehicle daily. In the 14-day study, rats were dosed in combination with a single administration of 30 mg/kg carboplatin or vehicle (0.9% w/v physiologic saline) on day 1. In the 42-day two-cycle study, rats were dosed in combination with 40 mg/kg carboplatin or vehicle on day 1 and day 22. Carboplatin or vehicle control were delivered via a single slow (over 1 min) intravenous bolus injection into the lateral tail vein followed immediately with either vehicle control or test article.
For toxicokinetic analysis of AZD5305, niraparib, and olaparib, and hematology analyses, blood samples were collected via tail vein prick at various time points. All animals were monitored throughout the study and sacrificed on day 15 or day 42, approximately 24 hours after administration of the final dose. Bone marrow (left femurs) was fixed in 10% neutral buffered formalin prior to processing, then sectioned and stained with hematoxylin and eosin (H&E) for histopathologic evaluation. Right femurs were processed for bone marrow progenitor cell population analysis by flow cytometry.
Bone marrow differential analysis
Femur flush processing
Rat femurs were collected and processed within 30 minutes to avoid clotting and bone marrow cell suspensions collected according to published methodology (11). Bone marrow suspensions were kept at 2–8°C for cell counting and flow cytometry analysis.
Nucleated cell counting
Trucount absolute counting tubes (Becton Dickinson) were used to determine nucleated cell/white cell count in the bone marrow suspension. For each sample, 100 μL of bone marrow suspension was pipetted into a labelled Trucount tube. LDS-751 (Thermo Fisher Scientific) working solution was prepared by diluting 20 μL of stock LDS-751 (1 mg LDS-751 in 1 mL methanol) with 3 mL of PBS. Twenty μL of LDS-751 working solution was added to each test sample along with 500 μL of PBS containing 0.5% BSA. Samples were mixed and incubated, protected from light, at room temperature for 20 minutes. Stained test samples were analyzed using FACSDiva software on a FACSCanto II flow cytometer with three lasers (BD Biosciences). Doublets and debris were gated out using forward scatter width (FSC-W) versus FSC height (FSC-H), with the remaining single-cell population labeled as single cells. The total event population positive for both fluorescein isothiocyanate (FITC) and phycoerythrin (PE) were gated as beads. The population classified as single cells but not beads was analyzed for LDS-751 positivity, with cells positive for LDS-751 considered to be nucleated and labeled as total nucleated cells (TNC; data not shown). The stopping gate for acquisition was set to 10,000 TNC or 1 minute. Bead events collected within acquisition were recorded into a worksheet using the lot-specific bead count of the Trucount tubes and the dilution factor of the bone marrow suspension to calculate the absolute nucleated cell/white cell count per mL. An aliquot of bone marrow suspension was adjusted to a cell concentration of 1 × 107 cells/mL using PBS containing 0.5% BSA. In the case of very low cell counts (<3 × 106 cells/mL), cells were concentrated to as close to 1 × 107 cells/mL as possible and adjusted test samples stored at 2–8°C until use in flow cytometry.
CD marker selection
A panel of CD markers was chosen based on published methods (11) to label major lineages in the bone marrow. CD45 (common leukocyte antigen) labeled white blood cells, CD71 (transferrin receptor antigen) erythroid cells, and CD61 (integrin beta 3) platelet lineage cells. Differentiation of myeloid and lymphoid cells was based on the distinct light scatter properties of each cell lineage. Their morphologic differences discretely separate each cell type using the SSC signal for internal complexity and granularity (11). LDS-751 was also utilized as a nuclear stain to gate out mature erythrocytes. For a detailed description of CD marker staining and flow cytometry analysis procedures please refer to Supplementary Methods.
Data availability statement
The data generated in this study are available within the article and its supplementary data files. Additional data requests may be directed to the corresponding author.
Results
AZD5305 inhibits PARylation in cells by selectively blocking PARP1 (but not PARP2) enzymatic activity
To assess the ability of AZD5305 to inhibit the catalytic activity of PARP, a PARylation assay was used to measure the dose response in cells. AZD5305 potently inhibited PARylation with an IC50 value of 2.3 nmol/L (Supplementary Fig. S1A) in A549 WT cells. In comparison, the current clinical PARPi showed potencies ranging between 5.1 and 33 nmol/L, for talazoparib and veliparib, respectively (Supplementary Fig. S1A; Supplementary Table S2).
The selectivity of AZD5305 binding for PARP1 over PARP2 was measured by biochemical in vitro assays as reported in Johannes and colleagues (4) and is summarized in Fig. 1A. To confirm the ability of AZD5305 to selectively target PARP1 in cells, we generated isogenic A549 cell lines depleted for PARP1 (PARP1-KO) or PARP2 (PARP2-KO), using CRISPR-Cas9 technology (Supplementary Fig. S1B and S1C) and compared PARylation inhibition profiles. Upon treatment with AZD5305, a dramatic shift in PARylation inhibition was observed in cells deficient for PARP1 (proficient for PARP2) with a mean IC50 value of 653 nmol/L, compared with WT (where both PARPs are present) or compared to the PARP2-KO cells (proficient for PARP1), for which the IC50 was 1.55 nmol/L (Fig. 1B). These data confirm that AZD5305 is more than 500-fold selective for inhibiting PARP1 over PARP2 in cells. When the same experiments were performed with first-generation PARPi, an overlap of dose–response curves in the three isogenic cell lines was observed, confirming the dual PARP1/PARP2 targeting (Fig. 1C and D).
AZD5305 is a potent and selective trapper of PARP1 in cells
The ability of AZD5305 to trap PARP1 or PARP2 onto the chromatin of cells was evaluated using a recently developed assay based on in situ cell preextraction and immunofluorescence (12, 13). In A549 cells, PARP1 trapping is induced in a dose-dependent manner by single digit nanomolar concentrations of AZD5305 (Fig. 1E). In contrast, no PARP2 trapping was detected up to 30 μmol/L AZD5305 (Fig. 1E). These data demonstrate that AZD5305 is a potent PARP1 selective trapper. In contrast, talazoparib, olaparib, and niraparib all induced trapping of both PARP1 and PARP2 at similar concentrations (Fig. 1F and Supplementary Fig. S1D), whereas for veliparib, no trapping of either PARP1 or PARP2 was observed. These results were consistent with previous reports where trapping was monitored using other methods (7). In summary, AZD5305 is a PARP1 selective inhibitor and trapper, with no activity on PARP2 (Fig. 1G).
AZD5305 is a potent and selective antiproliferative agent
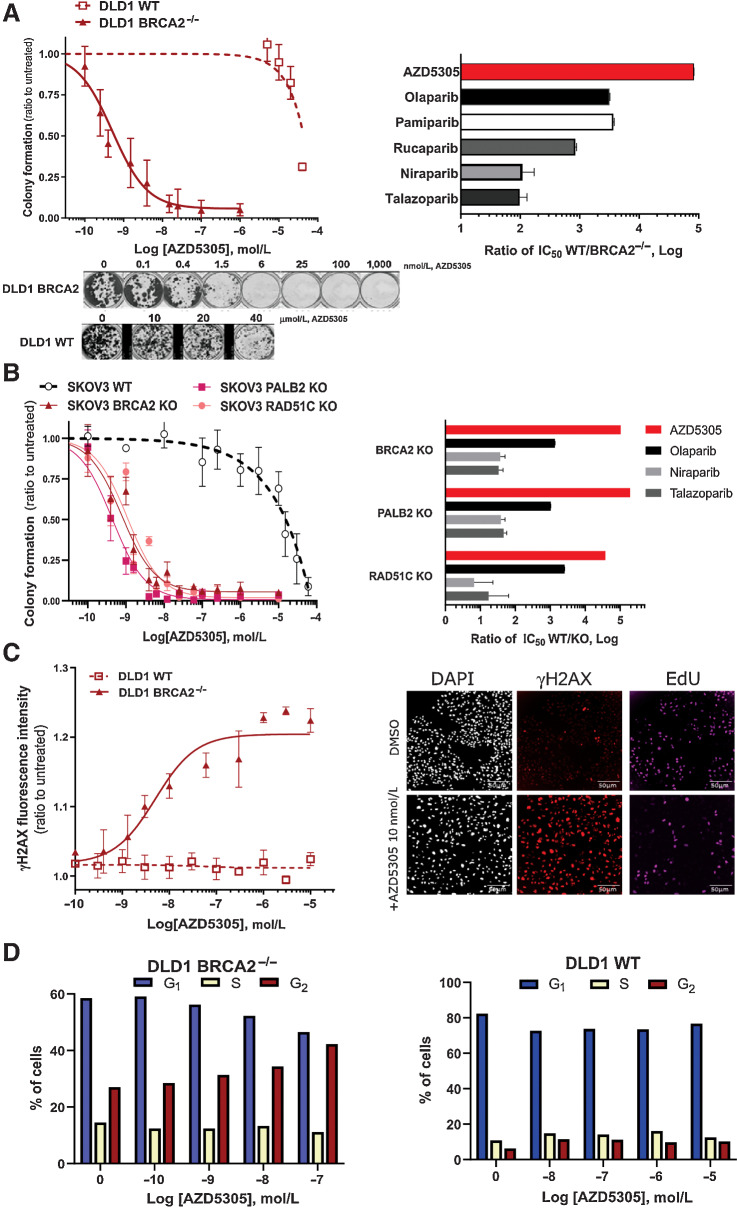
Having confirmed the cell pharmacology properties of AZD5305, we evaluated whether selective inhibition and trapping of PARP1 would retain activity in relevant tumor cell backgrounds. In colony formation (clonogenic) assays, the antiproliferative effects of AZD5305 in different isogenic cell line pairs for BRCA1, BRCA2, PALB2, and RAD51C (Fig. 2; Supplementary Fig. S2) were tested.
Figure 2.
AZD5305 selectively targets cancer cells with HRR-deficiency, inducing DNA damage accumulation and cell-cycle arrest. A, Left, Colony formation assays of DLD-1 isogenic cell line pair (WT in dotted line and BRCA2−/− in solid line) treated with AZD5305 in dose–response. Curves are the mean of six independent experiments; error bars indicate ±SEM. Bottom, Representative images of clonogenic assays. Right, Bar chart representing the differential activity of each PARPi on the BRCA2−/− isogenic cell line pair. Bars are the IC50 ratio of WT over the BRCA2−/− cells for each indicated PARPi. B, Left, Colony formation assays performed in SKOV-3 isogenic pairs: WT (dotted line, empty circle), BRCA2-KO, PALB2-KO, and RAD51C-KO (solid lines and full symbols, as indicated) treated with a dose–response of AZD5305. Curves are the mean of three independent experiments; error bars indicate ±SEM. Right, Bar chart representing the differential activity of each compound on each isogenic cell line pair. Bars are the IC50 ratio of WT over the indicated KO isogenic pair, measured for each indicated PARPi. C, Left, γH2AX measured by immunofluorescence after 72-hour treatments with a dose–response of AZD5305 in DLD-1 BRCA2−/− (solid line and full symbol) or WT cells (dotted line and empty symbol). Curves are the mean of three independent experiments; error bars indicate ±SEM. Right, representative immunofluorescence images of DLD-1 BRCA2−/− cells treated with 10 nmol/L of AZD5305 (bottom) or untreated (top). D, Cell-cycle analysis by flow cytometry of DLD-1 BRCA2−/− (left) and WT cells (right) treated with AZD5305 for 48 hours at indicated concentrations. Bar charts represent the distribution of cells in the different cell-cycle phases, G1, S, and G2; below are representative cell-cycle scatter plots, with DNA and EdU intensity signal on x- and y-axes, respectively.
AZD5305 potently affected the viability of BRCA2−/− cells with an IC50 of 0.4 nmol/L, whereas in WT cells the IC50 was approximately 30 μmol/L (Fig. 2A, left panel; solid and dashed lines respectively). Hence the effect of AZD5305 was over 5 Log (∼100,000 times) more potent in the BRCA2−/− than in the WT cells (Fig. 2A, right panel, top bar). In the same assay, first-generation PARPi also showed preferential activity in the BRCA2−/− cells (Supplementary Fig. S2A). However, for these agents, the ratio between the responses in the isogenic pairs was markedly smaller (Fig. 2A, right) with olaparib, pamiparib, and rucaparib demonstrating an approximately 3 Log difference and niraparib and talazoparib a 2 Log difference. Similar results were obtained also in the UWB1.289 isogenic cell line pair (Supplementary Fig. S2B and S2D), confirming the superior selectivity of AZD5305 over first-generation PARPi in BRCA1-mutant (BRCA1m) cells versus the complemented BRCA1wt cell line.
Next we sought to test the activity of AZD5305 in clinically relevant genetic backgrounds beyond BRCAm (14). For this purpose, we generated ovarian isogenic cell lines depleted of either PALB2, RAD51C, or BRCA2 genes (15). AZD5305 treatment inhibited proliferation in all three isogenic cell lines, shown in Fig. 2B left plot, with IC50s in the sub- or single-digit nanomolar range. In contrast, minimal effects were observed in the WT cells (IC50 13 μmol/L) demonstrating selective antiproliferative activity in HRR-deficient backgrounds. When first-generation PARPi were tested in the same assay (olaparib, niraparib, and talazoparib dose–response curves in Supplementary Fig. S2C), a narrower window between the HRR-proficient and the HRR-deficient cell lines was obtained (Fig. 2B, right bar chart; Supplementary Table S3).
Overall AZD5305 displays greater differential antiproliferative effects between HRR-deficient and -proficient cells compared with first-generation PARPi.
AZD5305 induces accumulation of DNA damage and G2–M arrest selectively in HRR-deficient cells
To gain further insights into the mechanism underlying the antiproliferative effect by AZD5305, we monitored DNA damage (γH2AX), replication (by EdU incorporation) and cell-cycle progression in an isogenic cell pair (Fig. 2C and D).
Induction of γH2AX was observed at low nanomolar concentrations of AZD5305 in the BRCA2−/−, but not in the WT cells. (Fig. 2C, left). In Fig. 2C, the right panels show representative immunofluorescence images of induced increase in DNA damage in BRCA2−/− cells by 10 nmol/L AZD5305. Cell-cycle analysis by flow cytometry confirmed that AZD5305 treatment induced reduction in G1 and replicating cells and accumulation in G2–M phase, specifically in the BRCA2−/− cells (Fig. 2D, left). In contrast, no change was observed in WT cells (Fig. 2D, right).
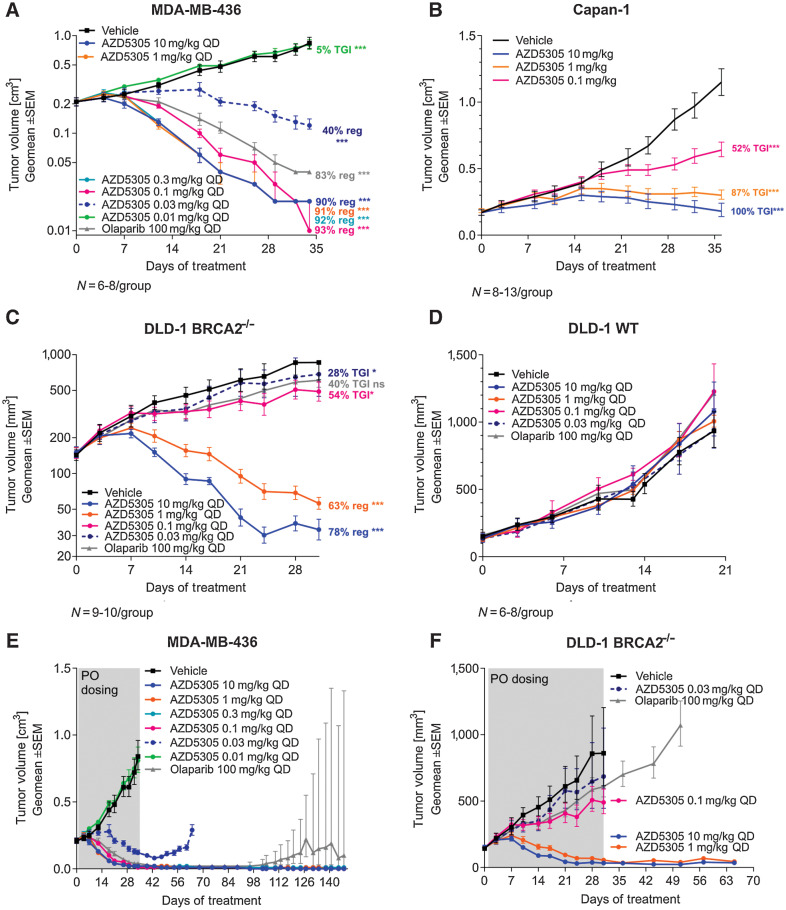
AZD5305 demonstrates sustained antitumor activity in BRCAm xenograft and PDX models in vivo
The antitumor efficacy of AZD5305 monotherapy was evaluated in vivo in BRCAm xenografts and PDX models. In the MDA-MB-436 BRCA1m TNBC model, daily treatment with ≥ 0.1 mg/kg of AZD5305 resulted in profound regressions (≥90%). Further reductions of AZD5305 dose levels to 0.03 mg/kg and 0.01 mg/kg resulted in diminished antitumor efficacy (40% regression and not efficacious, respectively; Fig. 3A). In Capan-1 xenografts, a BRCA2m pancreatic model, daily treatment with 1 or 10 mg/kg of AZD5305 resulted in tumor stasis, while a 0.1 mg/kg oral daily dose led to 52% tumor growth inhibition (TGI; Fig. 3B).
Figure 3.
Efficacy of AZD5305 in BRCAm xenograft tumor models. Antitumor efficacy of AZD5305 dose–responses in MDA-MB-436 BRCA1m TNBC xenograft (A), in Capan-1 BRCA2m pancreatic cancer xenograft (B), and in isogenic xenograft tumor models DLD-1 BRCA2−/− (C) and DLD-1 WT (D). Mice were dosed with indicated doses of AZD5305 or 100 mg/kg olaparib once daily orally for 35 (A, B), 31 (C), or 20 (D) days. E and F, In experiments from A and C, treatment was withdrawn as indicated and tumors were monitored for the regrowth. Graphs depict geomean tumor volume ±SEM and percent tumor growth inhibition (TGI) or regression (reg). Statistical significance was evaluated compared with the vehicle group using a one-tailed t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Furthermore, in a DLD-1 BRCA2−/− xenograft model, daily oral treatment with 10 mg/kg or 1 mg/kg of AZD5305 showed 78% and 63% tumor regression, respectively, with lower doses of 0.1 and 0.03 mg/kg resulting in a modest antitumor effect (54% and 28% TGI, respectively; Fig. 3C). However, as expected, in the isogenic DLD-1 BRCA WT model AZD5305 did not show antitumor efficacy compared with vehicle group at any dose tested (Fig. 3D).
Importantly, the tumor regressions caused by AZD5305 were durable and continued for at least 15 weeks after treatment withdrawal in MDA-MB-436 (Fig. 3E) and for 5 weeks after cessation of treatment in DLD-1 BRCA2−/− tumors (Fig. 3F). HBCx-17, a breast cancer BRCA2m PDX model that showed a similar sensitivity to AZD5305 as MDA-MB-436 (4), also demonstrated sustained responses after cessation of dosing (Supplementary Fig. S3).
All in vivo tumor models were also tested for sensitivity to the dual PARP1/2 inhibitor olaparib. A 100 mg/kg (clinically relevant) daily dose resulted in tumor regressions in MDA-MB-436 and HBCx-17 models; however, some tumor regrowth was observed about 8 weeks after treatment withdrawal (Fig. 3A and E; Supplementary Fig. S3). In DLD-1 BRCA2−/− xenografts, olaparib delivered 40% TGI (Fig. 3C) and, as expected, it was not efficacious in the DLD-1 WT model (Fig. 3D).
In summary, these data show that AZD5305 delivers robust and durable efficacy in the BRCA1/2m models, but not in the BRCA WT/HRR-proficient preclinical models in vivo, confirming that AZD5305 has the potential to be an efficacious targeted therapy for cancers with BRCA genetic alterations.
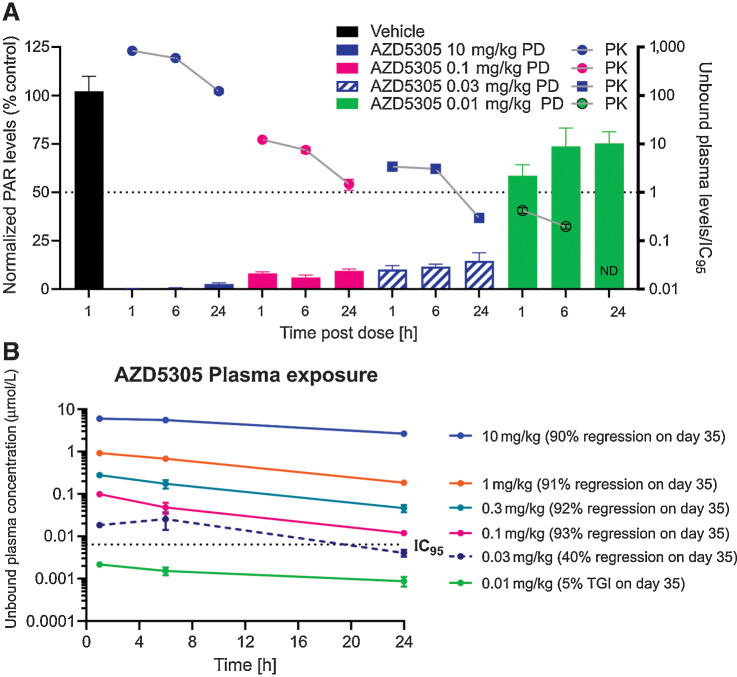
Antitumor efficacy of AZD5305 correlates with PK and PD in BRCA1m tumor models
To understand the relationship between target engagement and efficacy, a measure of total PARylation in tumor lysates was used as PD biomarker. Analysis of MDA-MB-436 tumors at steady-state after 5-day dosing showed that maximally efficacious doses of AZD5305 in this model (≥0.1 mg/kg) inhibited PARylation by >90% compared with the control vehicle group and the effect was durable for up to 24 hours postdose (Fig. 4A). In contrast, doses delivering submaximal efficacy were associated with lower PARylation inhibition. Treatment with AZD5305 at 0.03 mg/kg led to 89%–85% PARylation inhibition for the 24-hour period while the nonefficacious 0.01 mg/kg dose inhibited PARylation by 41% with much shorter duration of effect (1 hour). Likewise, the first-generation inhibitor olaparib, achieved more than 99% inhibition of PARylation 1 hour after dosing, but levels recovered to 24% by 24 hours (Supplementary Fig. S4A). Taken together, these data suggest that maximal efficacy in the MDA-MB-436 is associated with very high and sustained target engagement of ≥90% at trough exposures. These findings were consistent with data obtained in the HBCx-17 PDX model (Supplementary Fig. S4B).
Figure 4.
PK/PD/efficacy relationship of AZD5305 in a BRCA1m TNBC xenograft tumor model. A, PK/PD relationship for AZD5305 in MDA-MB-436 tumor xenograft model. Mice were dosed with indicated dose levels of AZD5305 once daily orally for 5 days. Plasma and tumors were collected at indicated time after the last dose. Bars depict pharmacodynamic (PD) effects (total PARylation in the tumor lysates) and symbols with lines depict pharmacokinetic (PK) unbound plasma levels of AZD5305 coverage of target level (IC95). ND, compound not detected. B, Unbound plasma levels of AZD5305. Mice from experiments of Fig. 3A were bled at indicated time points after the oral dose of AZD5305 on day 7.
To correlate the antitumor efficacy of AZD5305 with PK of the compound at steady state in the MDA-MB-436 model (Fig. 3A), plasma samples were collected at various time points post the day 7 dose. In the maximally efficacious dose groups (≥0.1 mg/kg, achieving >90% tumor regression; Fig. 3A), the unbound plasma levels of AZD5305 were above the DLD-1 BRCA2−/− cells in vitro clonogenic assay IC95 (0.0064 μmol/L) for the duration of the dosing interval of 24 hours (Fig. 4B). In contrast, in the 0.03 mg/kg group, which achieved 40% regression, the unbound plasma levels of AZD5305 were above IC95 for only approximately 17 hours, while in the nonefficacious group (0.01 mg/kg), the unbound plasma concentration of AZD5305 did not reach the IC95 value. These data were consistent with the requirement to maintain high and sustained target engagement in order to achieve maximal efficacy.
In summary, our studies have shown that maintaining free plasma concentrations above the DLD-1 BRCA2−/− cell growth inhibition assay IC95 achieves maximal preclinical efficacy in vivo in the MDA-MB-436 xenograft model. It should be highlighted that these data were consistent with our findings for previous PARP1-selective compounds, AZ’4554 (compound 16; ref. 4) and AZ’5508 (compound 17; ref. 4), which gave us additional confidence in our understanding of the PK/PD/efficacy relationship of PARP1-selective compounds (Supplementary Fig. S4C and S4D).
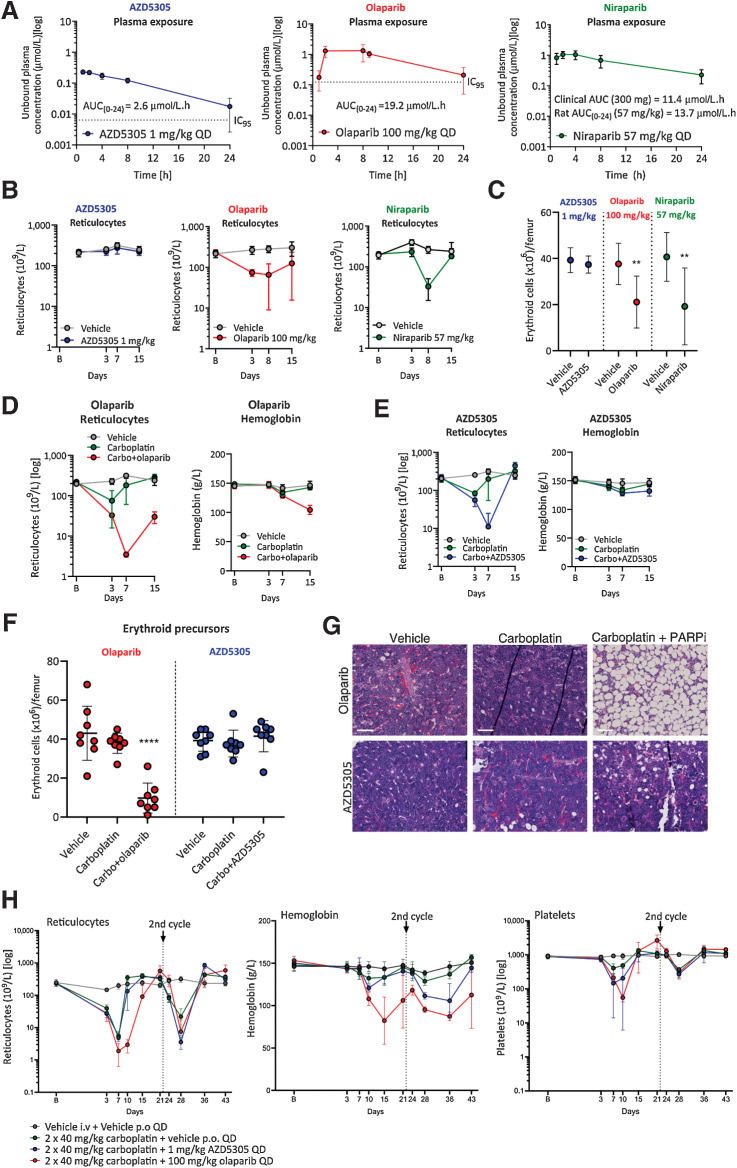
AZD5305 demonstrates reduced hematologic toxicity when compared with dual PARP1/2 inhibitors in rat preclinical models
To determine whether the PARP1 selectivity of AZD5305 would mitigate the hematologic toxicity observed for dual PARP1/2 inhibitors in rat preclinical models, we carried out comparative rat studies (16, 17). AZD5305 and clinical non–PARP1-selective PARPi were dosed once daily for 14 days: AZD5305 1 mg/kg once daily, olaparib 100 mg/kg once daily, or niraparib at 57 mg/kg once daily. AZD5305 and olaparib exposures were targeted to cover their respective cell IC95 for approximately 24 hours (Fig. 5A; dotted lines indicating the IC95). The 57 mg/kg niraparib dose resulted in an unbound AUC(0–24) of 13.7 μmol/L/hour, approximately matching the reported clinical free AUC of 11.4 μmol/L/hour from a 300 mg clinical dose (18). Sequential peripheral blood hematology analysis was performed (Fig. 5B; Supplementary Fig. S5A–S5C) and flow cytometry used to directly assess bone marrow lineage precursors at termination (Fig. 5C; Supplementary Fig. S5D). Consistent with clinical anemia reported for olaparib (14, 19, 20), we observed a sustained reduction in reticulocytes (immature red blood cells) in peripheral blood compared with vehicle controls, although overall red cell mass and hemoglobin were only beginning to show effects within this 14-day study (Fig. 5B; Supplementary Fig. S5A). There were no other significant effects noted in other blood lineages (Supplementary Fig. S5A). Consistent with the effects observed on the peripheral blood, flow cytometry measurement of bone marrow lineage precursor cells revealed reduced erythroid precursor numbers, while myeloid and platelet precursors were unaffected (Fig. 5C; Supplementary Fig. S5D).
Figure 5.
AZD5305 has reduced hematological toxicity in monotherapy and combination with carboplatin in rat pre-clinical models, when compared to dual PARP1/2 inhibitors. Monotherapy (A–C): Rats were dosed once daily (QD) orally for 14 days with the compounds and doses indicated. A, Unbound plasma concentration of AZD5305 (blue; 1 mg/kg), olaparib (red; 100 mg/kg), and niraparib (green; 57 mg/kg) at steady-state on day 14 with resulting unbound AUCs as indicated. The dotted lines represent the efficacious IC95 (derived from the DLD-1 BRCA2−/− clonogenic assays) for AZD5305 and olaparib. B, Reticulocyte counts at baseline (B on x-axis) and at time points indicated, in animal groups treated with vehicle (gray), AZD5305 (blue), olaparib (red), or niraparib (green). C, Terminal erythroid precursor cell counts in AZD5305 (blue), olaparib (red), and niraparib (green) treatment groups as indicated by the legend. Statistical significance was tested relative to vehicle controls using a one-way ANOVA and Dunnett multiple comparison test, where **, P ≤ 0.01. Dots and error bars represent the mean of eight replicates ±SD. Combination (D-H): Rats were dosed with vehicle or once with intravenous (i.v.) carboplatin alone or in combination with PARPi QD for 14 days. D and E, Reticulocyte counts and hemoglobin levels are shown at baseline (B on x-axis) and over time (days; x-axis) for carboplatin+olaparib (red; D) or carboplatin+AZD5305 (blue; E) in comparison with vehicle (gray) and carboplatin controls (green). F, Terminal erythroid precursor cell counts on day (d) 15 for the groups indicated, with olaparib study groups in red and AZD5305 study groups in blue. Statistical significance was tested relative to vehicle controls using a one-way ANOVA and Dunnett multiple comparison test, where ****, P ≤ 0.0001. G, Representative hematoxylin and eosin (H&E)-stained sections of bone marrow from vehicle controls, olaparib and AZD5305 monotherapy, and carboplatin+PARPi treatment groups as indicated. H, Reticulocyte, hemoglobin, and platelet levels over time (days; x-axis) from rats treated with vehicle (gray) or two doses of i.v. carboplatin alone (green) or in combination with continuous daily (QD) AZD5305 (blue) or olaparib (red) as indicated. All error bars represent the standard deviation of the mean of eight (A–C) or four (E) replicates.
Similarly, treatment with 57 mg/kg niraparib resulted in a reduction in reticulocyte counts compared with vehicle controls, translating to a mild reduction on red blood cells and hemoglobin by the end of the study (Fig. 5B; Supplementary Fig. S5C). Terminal erythroid precursor cell counts were also suppressed in the bone marrow, as was observed for olaparib (Fig. 5C). Interestingly, bone marrow myeloid precursor cells were also suppressed, perhaps eliciting a slight reduction in neutrophil counts observed in the hematology on day 15 (Supplementary Fig. S5C and S5D). These data are consistent with both anemia and neutropenia reported for niraparib clinically (2, 14).
In contrast to olaparib and niraparib, daily dosing of AZD5305 did not cause notable responses in any hematologic parameters or in lineage precursor cells in the bone marrow (Fig. 5B and C; Supplementary Fig. S5B and S5D). Even in 1-month rat GLP (good laboratory practice) toxicology studies, where AZD5305 was dosed up to 50 mg/kg (and where the unbound AUC was in excess of 150× the predicted human minimally efficacious exposure), hematologic changes were limited to minimal decreases in white blood cells, neutrophils, lymphocytes, and monocytes (Supplementary Table S4). These results indicate, for the first time, that PARP1 selectivity mitigates the hematotoxicity usually observed with dual PARP1/2i in rat preclinical models.
Next, we investigated whether AZD5305 would also be better tolerated in a chemotherapy combination when compared with dual PARP1/PARP2 inhibitors. We assessed the effects of daily AZD5305 (1 mg/kg) in combination with one cycle of intravenous carboplatin (30 mg/kg) in rat. Concurrent vehicle and carboplatin monotherapy controls were also included. Resulting data were compared to a similarly designed study where daily olaparib (100 mg/kg) was dosed concurrently with carboplatin (30 mg/kg). Resulting unbound exposures from AZD5305 (1 mg/kg) and olaparib (100 mg/kg) gave approximately 24-hour cover of the respective DLD-1 BRCA2−/− IC95 allowing for direct comparison of effects (Supplementary Fig. S6A). The carboplatin 30 mg/kg dose was selected on the basis of the toxicokinetic analysis performed as part of a separate study, where this dose resulted in an AUC(0–24) of 132 μmol/h/L (Supplementary Fig. S5A; or AUC of 1.5 mg/min/mL; see Supplementary Methods for bioanalysis of carboplatin exposure; ref. 21).
The 30 mg/kg carboplatin monotherapy elicited a slight reduction in reticulocyte numbers on day 3 with other hematologic changes remaining minimal, allowing for additional toxicity to be measured in combination with a PARPi (Fig. 5D and E; Supplementary Fig. S6B and S6C). In combination with either PARPi, platelet and neutrophil counts, as well as myeloid and platelet precursor counts in the bone marrow, demonstrated some additional effects compared to carboplatin alone (Supplementary Fig. S6B–S6D). However, due to the variability of these data we investigated these endpoints further in a follow-up study and focused our comparison on the red cell compartment.
Both olaparib and AZD5305 caused strong exacerbation of carboplatin-induced reticulocyte effects, which were particularly notable on day 7 (Fig. 5D and E). However, whilst reticulocytes did not fully recover by the end of the study with continuous olaparib dosing, they were able to fully recover with continuous AZD5305. As a consequence, hemoglobin levels and red blood cell counts were reduced in the carboplatin+olaparib treatment group, but not in the carboplatin+AZD5305 group (Fig. 5D and E; Supplementary Fig. S6B and S6C). This finding was consistent with erythroid precursor cell counts remaining strongly suppressed following combination with olaparib, but not with AZD5305 (Fig. 5F).
Finally, histopathologic analysis of the bone marrow confirmed that animals treated with carboplatin+olaparib showed an increased incidence and severity of both decreased bone marrow cellularity (Fig. 5G; Supplementary Table S5) and increased myeloid:erythroid cell ratio. In contrast, animals in the AZD5305+carboplatin group were histologically unremarkable with exception of 1/8 animals, which had minimal decreased cellularity of the bone marrow (Fig. 5G; Supplementary Table S6).
Next we performed a two-cycle rat in vivo study with a higher dose of carboplatin (40 mg/kg; 228 μmol/L.h) to more closely mimic clinical protocol and better capture clinical thrombocytopenia (Supplementary Fig. S7A). Daily administration of AZD5305 (1 mg/kg) or olaparib (100 mg/kg) was combined with carboplatin (40 mg/kg) every three weeks for two cycles. Resulting unbound exposures from 1 mg/kg AZD5305 administered in combination with carboplatin were in line with previous studies, whereas the unbound exposure for olaparib was approximately 2-fold lower (AUC(0–24) 10.7 μmol/L/hour; Supplementary Fig. S7A vs. Fig. 5A; Supplementary Fig. S6A).
Carboplatin monotherapy (40 mg/kg) caused a marked reduction in reticulocytes in cycle 1 and moderate reduction in cycle 2, with minimal effects on hemoglobin and red blood cells that largely recovered by the end of each cycle (Fig. 5H; Supplementary Fig. S7B). A moderate reduction in platelets was also observed in both cycles 7 days post dose and these fully recovered over time (Fig. 5H). Interestingly, both AZD5305 and olaparib exacerbated carboplatin-induced platelet effects with equal potency in the first cycle, with little additional effect to carboplatin monotherapy in the second cycle (Fig. 5H).
Evaluation of the red cell compartment demonstrated that combination with AZD5305 showed no or little difference in reticulocyte counts or hemoglobin compared with carboplatin monotherapy during the first cycle (Fig. 5H; Supplementary Fig. S7B). However, in the second cycle a stronger suppression of reticulocytes and a marked further decrease in red cell counts and hemoglobin levels was noted for the combination versus carboplatin monotherapy. Critically, however, hemoglobin almost completely recovered by the end of the study, as did the red cells. This was likely due to reticulocytes having rebounded and demonstrating a regenerative response. In contrast, the combination with olaparib caused a stronger and sustained reduction in reticulocytes versus carboplatin monotherapy (Fig. 5H). This sustained reduction resulted in a marked decrease in red blood cells and hemoglobin levels, with little recovery before the second carboplatin dose (Fig. 5H; Supplementary Fig. S7B).
These data suggest that despite AZD5305 and olaparib eliciting similar additional platelet reductions in combination with carboplatin in rat models, at the level of the red cell compartment AZD5305 is differentiated from olaparib by allowing for a more rapid recovery of reticulocytes despite continuous dosing.
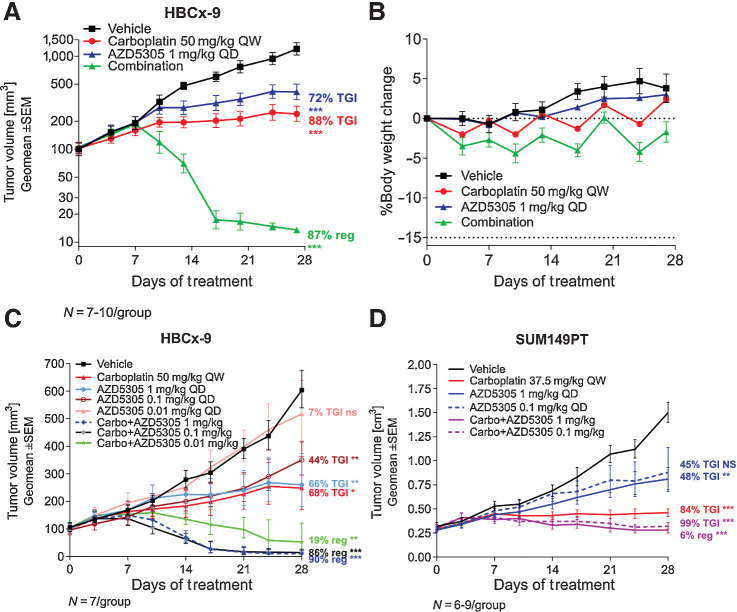
AZD5305 and carboplatin show combination benefit in HBCx-9 PDX and SUM149PT xenograft model
The potential for increased efficacy of AZD5305 in combination with carboplatin was investigated in in vivo efficacy studies in HBCx-9 and SUM149PT models. HBCx-9 is a TNBC PDX model, reported as BRCA1/2 WT with BRCA1 methylation and “BRCAness features” (22). It has also been shown to have a moderate sensitivity to olaparib monotherapy (23). In this model, AZD5305 at 1 mg/kg once daily in combination with 50 mg/kg carboplatin once weekly demonstrated dramatic improvement of the antitumor effect compared with each agent as monotherapy (87% regression vs. 72%–88% TGI; Fig. 6A). Treatments were well tolerated throughout the duration of the study as depicted by minor changes in animal body weight (Fig. 6B). Reducing the dose of AZD5305 from 1 mg/kg to 0.1 mg/kg in combination with carboplatin maintained the antitumor efficacy in HBCx-9 PDX model (Fig. 6C). Further reduction of the AZD5305 combination dose to 0.01 mg/kg was less efficacious in comparison with the other combination groups, however, it still delivered 19% regression and significantly increased benefit when compared with each monotherapy group (Fig. 6C). Similar results were observed in the SUM149PT BRCA1m TNBC xenograft model (Fig. 6D), although in this model the highest efficacy obtained was 6% regression or approximately 99% TGI.
Figure 6.
Antitumor efficacy of AZD5305 in combination with carboplatin in vivo. A, Antitumor efficacy of AZD5305 in combination with carboplatin in the TNBC HBCx-9 PDX. B, Tolerability of the treatments was assessed by monitoring body weight changes throughout the treatment duration. C and D, Anti-tumor efficacy of dose response of AZD5305 in combination with carboplatin in HBCx-9 (C) and SUM149PT (D) tumor models. Mice were dosed with indicated dose levels of AZD5305 once daily orally (PO) and/or with carboplatin once weekly intraperitoneally (IP) for 4 weeks. Graphs depict geomean tumor volume ±SEM and percent tumor growth inhibition (TGI) or regression (reg). Statistical significance was evaluated compared to the vehicle group using a one-tailed t test (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Discussion
Here we present a comprehensive pre-clinical characterization of AZD5305, a potent and highly selective next-generation PARP1 inhibitor and trapper developed to deliver greater efficacy and improved tolerability versus the first-generation PARPi (4). In cells, AZD5305 is exquisitely selective (500 fold) for PARP1 over PARP2 and selectively traps PARP1 onto DNA. These features lead to potent antiproliferative effects in HRR-deficient cells (with mutations in BRCA1, BRCA2, PALB2, RAD51C) and minimal/no effects in HRR-proficient cells. The ability of AZD5305 to specifically target cancer cell models with HRR-deficiency, while sparing HRR-proficient cells is unprecedented (14) and could potentially lead to an improved clinical therapeutic index.
In BRCAm preclinical models, AZD5305 delivers complete and durable in vivo efficacy. Antitumor effects and tumor regression were noted at doses of AZD5305 as low as 0.03 mg/kg daily and following cessation of treatment; mice treated with AZD5305 continued to benefit from the antitumor effects of treatment with long duration of tumor response, exceeding 100 days after treatment was withdrawn. This robust antitumor effect and absence of regrowth in long-term experiments suggests AZD5305 may have the potential to delay or reduce the appearance of resistance, frequently observed with first-generation PARPi (24).
When combined with carboplatin, AZD5305 is well tolerated in mice and shows dose-dependent target engagement and potent anticancer efficacy. AZD5305 potently inhibited tumor PARylation in xenograft and PDX models giving a clear unbound exposure versus response relationship. However, while providing a compelling case for high target engagement as a requirement for efficacy, there were two potential issues with the use of tumor PARylation as a biomarker in isolation. Firstly, the absolute contribution of PARP1 versus PARP2 is difficult to define and therefore a PARP1-selective compound like AZD5305 may show a reduced Emax in cases where PARP2 makes a substantial contribution to total PARylation. Secondly, while target engagement is a prerequisite of trapping pharmacology, it does not directly assess it. Therefore, a clinical exposure target was set based on achieving unbound plasma concentrations ≥ the DLD-1 BRCA2−/− IC95 at Ctrough.
In comparison with first-generation inhibitors, AZD5305 causes minimal hematological toxicity as a monotherapy in rat pre-clinical models at predicted clinically efficacious exposures. Also, in combination with carboplatin, although AZD5305 and olaparib caused comparable decreases in platelets, AZD5305 was notably differentiated from olaparib by allowing for a more rapid recovery of red cell parameters, further demonstrating potential for improved therapeutic index.
In summary, the data presented here demonstrate AZD5305 is a potent and highly selective PARP1 inhibitor and trapper, with improved target engagement and efficacy alongside reduced hematologic toxicity versus first-generation dual PARP1/2 inhibitors. Because of its improved properties, AZD5305 could open up additional therapeutic opportunities in the clinic, beyond those possible with the current PARPi.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Kaoru Sakabe for critical review of the manuscript. We are also grateful to AST and the In vivo team (TDE Bioscience AstraZeneca UK) for supporting the in vivo studies. All authors were employees and shareholders of AstraZeneca at the time of this research. This study was funded by AstraZeneca.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
G. Illuzzi is an employee and shareholder of AstraZeneca. S.J. Gill is an employee and shareholder of AstraZeneca. A. Pike is an employee and shareholder of AstraZeneca. S.E. Critchlow is an employee and shareholder of AstraZeneca. A. Cronin is an employee and shareholder of AstraZeneca. S. Fawell is an employee and shareholder of AstraZeneca. G. Hawthorne is an employee and shareholder of AstraZeneca. K. Jamal is an AstraZeneca employee. J. Johannes reports a patent for WO2021013735 pending. E. Leonard is an employee and shareholder of AstraZeneca. R. Macdonald is an employee and shareholder of AstraZeneca. G. Maglennon is an employee and shareholder of AstraZeneca. M.J. O'Connor is an employee and shareholder of AstraZeneca. H. Southgate is an AstraZeneca employee. E. Leo is an employee and shareholder of AstraZeneca. No disclosures were reported by the other authors.
Authors' Contributions
G. Illuzzi: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. A.D. Staniszewska: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. S.J. Gill: Data curation, formal analysis, investigation, writing–original draft, writing–review and editing. A. Pike: Data curation, formal analysis, investigation, writing–original draft. L. McWilliams: Data curation, formal analysis, validation. S.E. Critchlow: Resources, supervision. A. Cronin: Validation. S. Fawell: Resources, supervision. G. Hawthorne: Formal analysis, validation. K. Jamal: Validation, investigation, methodology. J. Johannes: Supervision. E. Leonard: Formal analysis, validation. R. Macdonald: Validation, investigation. G. Maglennon: Supervision, investigation. J. Nikkilä: Validation, methodology. M.J. O'Connor: Conceptualization, supervision. A. Smith: Data curation, investigation. H. Southgate: Investigation. J. Wilson: Investigation. J. Yates: Data curation, validation, investigation. S. Cosulich: Resources, supervision, writing–original draft, writing–review and editing. E. Leo: Conceptualization, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science 2017;355:1152–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. LaFargue CJ, Dal Molin GZ, Sood AK, Coleman RL. Exploring and comparing adverse events between PARP inhibitors. Lancet Oncol 2019;20:e15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Madariaga A, Bowering V, Ahrari S, Oza AM, Lheureux S. Manage wisely: poly (ADP-ribose) polymerase inhibitor (PARPi) treatment and adverse events. Int J Gynecol Cancer 2020;30:903–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johannes JW, Balazs A, Barratt D, Bista M, Chuba MD, Cosulich S, et al. Discovery of 5-{4-[(7-Ethyl-6-oxo-5,6-dihydro-1,5-naphthyridin-3-yl)methyl]piperazin-1-yl}-N-m ethylpyridine-2-carboxamide (AZD5305): a PARP1-DNA trapper with high selectivity for PARP1 over PARP2 and other PARPs. J Med Chem 2021;64:14498–512. [DOI] [PubMed] [Google Scholar]
- 5. D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 1999;342:249–68. [PMC free article] [PubMed] [Google Scholar]
- 6. Ronson GE, Piberger AL, Higgs MR, Olsen AL, Stewart GS, McHugh PJ, et al. PARP1 and PARP2 stabilise replication forks at base excision repair intermediates through Fbh1-dependent Rad51 regulation. Nat Commun 2018;9:746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murai J, Huang S-YN, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res 2012;72:5588–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murai J, Zhang Y, Morris J, Ji J, Takeda S, Doroshow JH, et al. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J Pharmacol Exp Ther 2014;349: 408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Farrés J, Martín-Caballero J, Martínez C, Lozano JJ, Llacuna L, Ampurdanés C, et al. Parp-2 is required to maintain hematopoiesis following sublethal gamma-irradiation in mice. Blood 2013;122:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Farrés J, Llacuna L, Martín-Caballero J, Martínez C, Lozano JJ, Ampurdanés C, et al. PARP-2 sustains erythropoiesis in mice by limiting replicative stress in erythroid progenitors. Cell Death Differ 2015;22:1144–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saad A, Palm M, Widell S, Reiland S. Differential analysis of rat bone marrow by flow cytometry. Comp Haematol Int 2000;10:97–101. [Google Scholar]
- 12. Illuzzi C, O'Connor MJ, Leo E. A novel assay for PARP-DNA trapping provides insights into the mechanism of action (MoA) of clinical PARP inhibitors (PARPi) [abstract]. Cancer Res 2019;79(13 Suppl):Abstract nr 2077. [Google Scholar]
- 13. Michelena J, Lezaja A, Teloni F, Schmid T, Imhof R, Altmeyer M. Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance. Nat Commun 2018;9:2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pilié PG, Gay CM, Byers LA, O'Connor MJ, Yap TA. PARP inhibitors: extending benefit beyond BRCA-mutant cancers. Clin Cancer Res 2019;25:3759–71. [DOI] [PubMed] [Google Scholar]
- 15. Jamal K, Galbiati A, Armenia J, Illuzzi G, Hall J, Bentouati S, et al. Drug-gene interaction screens coupled to tumour data analyses identify the most clinically-relevant cancer vulnerabilities driving sensitivity to PARP inhibition. bioRxiv 2022;2022.07.29.501846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. U.S. Food and Drug Administration. Application Number: 206162Orig1s000. Pharmacology Review. 2014. [Accessed November 25, 2021]. Available from:https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/206162Orig1s000PharmR.pdf.
- 17. U.S. Food and Drug Administration. Application Number: 208447Orig1s000. Multi-Discipline Review. 2016. [Accessed November 25, 2021]. Available from: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2017/208447orig1s000multidiscipliner.pdf.
- 18. Moore K, Zhang Z-Y, Agarwal S, Burris H, Patel MR, Kansra V. The effect of food on the pharmacokinetics of niraparib, a poly(ADP-ribose) polymerase (PARP) inhibitor, in patients with recurrent ovarian cancer. Cancer Chemother Pharmacol 2018;81:497–503. [DOI] [PubMed] [Google Scholar]
- 19. Hennes ER, Dow-Hillgartner EN, Bergsbaken JJ, Piccolo JK. PARP-inhibitor potpourri: a comparative review of class safety, efficacy, and cost. J Oncol Pharm Pract 2020;26:718–29. [DOI] [PubMed] [Google Scholar]
- 20. Brown JS, Kaye SB, Yap TA. PARP inhibitors: the race is on. Br J Cancer 2016;114:713–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Calvert AH, Newell DR, Gumbrell LA, O'Reilly S, Burnell M, Boxall FE, et al. Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol 1989;7:1748–56. [DOI] [PubMed] [Google Scholar]
- 22. Coussy F, El-Botty R, Château-Joubert S, Dahmani A, Montaudon E, Leboucher S, et al. BRCAness, SLFN11, and RB1 loss predict response to topoisomerase I inhibitors in triple-negative breast cancers. Sci Transl Med 2020;12:eaax2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riches LC, Trinidad AG, Hughes G, Jones GN, Hughes AM, Thomason AG, et al. Pharmacology of the ATM inhibitor AZD0156: potentiation of irradiation and olaparib responses preclinically. Mol Cancer Ther 2020;19:13–25. [DOI] [PubMed] [Google Scholar]
- 24. Mateo J, Lord CJ, Serra V, Tutt A, Balmaña J, Castroviejo-Bermejo M, et al. A decade of clinical development of PARP inhibitors in perspective. Ann Oncol 2019;30:1437–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its supplementary data files. Additional data requests may be directed to the corresponding author.