Abstract
Purpose:
RB1 mutations and loss of retinoblastoma (Rb) expression represent consistent but not entirely invariable hallmarks of small cell lung cancer (SCLC). The prevalence and characteristics of SCLC retaining wild-type Rb are not well-established. Furthermore, the performance of targeted next-generation sequencing (NGS) versus immunohistochemistry for Rb assessment is not well-defined.
Experimental Design:
A total of 208 clinical SCLC samples were analyzed by comprehensive targeted NGS, covering all exons of RB1, and Rb IHC. On the basis of established coordination of Rb/p16/cyclinD1 expression, p16-high/cyclinD1-low profile was used as a marker of constitutive Rb deficiency.
Results:
Fourteen of 208 (6%) SCLC expressed wild-type Rb, accompanied by a unique p16-low/cyclinD1-high profile supporting Rb proficiency. Rb-proficient SCLC was associated with neuroendocrine-low phenotype, combined SCLC with non-SCLC (NSCLC) histology and aggressive behavior. These tumors exclusively harbored CCND1 amplification (29%), and were markedly enriched in CDKN2A mutations (50%) and NSCLC-type alterations (KEAP1, STK11, FGFR1). The remaining 194 of 208 SCLC were Rb-deficient (p16-high/cyclinD1-low), including 184 cases with Rb loss (of which 29% lacked detectable RB1 alterations by clinical NGS pipeline), and 10 cases with mutated but expressed Rb.
Conclusions:
This is the largest study to date to concurrently analyze Rb by NGS and IHC in SCLC, identifying a 6% rate of Rb proficiency. Pathologic-genomic data implicate NSCLC-related progenitors as a putative source of Rb-proficient SCLC. Consistent upstream Rb inactivation via CDKN2A/p16↓ and CCND1/cyclinD1↑ suggests the potential utility of CDK4/6 inhibitors in this aggressive SCLC subset. The study also clarifies technical aspects of Rb status determination in clinical practice, highlighting the limitations of exon-only sequencing for RB1 interrogation.
Translational Relevance.
This is the largest study to date to systematically examine retinoblastoma (Rb) status in small cell lung cancers (SCLC) by broad targeted next-generation sequencing (NGS) and protein IHC. The study highlights major limitations of targeted NGS for identifying RB1 inactivating alterations, and illustrates the value of IHC for Rb, p16, and cyclin D1 for establishing Rb functional status. Using these methods, we describe previously minimally characterized and controversial SCLC subset with expression of wild-type Rb, and identify several distinctive characteristics of these tumors, including clinical aggressiveness and consistent alterations associated with upstream functional Rb inhibition, suggesting their potential sensitivity to CDK4/6 inhibitors. More broadly, given emerging data on Rb deficiency as an important biomarker of tumor progression and therapy outcomes in a variety of tumors, the insights regarding diagnostic interrogation of Rb status using SCLC as a model may have implications across multiple cancer types.
Introduction
Small cell lung cancer (SCLC) is an exceptionally aggressive malignancy with limited therapeutic options (1, 2). The molecular hallmarks of SCLC include inactivating genomic alterations of the retinoblastoma gene (RB1), leading to the loss of Rb protein expression, and concomitant TP53 alterations. The central role of the dual RB1/TP53 inactivation in SCLC pathogenesis is supported by decades of studies showing their nearly invariable inactivation in clinical samples and cell lines of SCLC, as well as a demonstration that their co-disruption in lung epithelium generates murine models of SCLC (3–6). However, several investigators have reported cases of SCLC that lack identifiable RB1 genomic alterations (7–10) or retain Rb protein expression (11, 12), with studies primarily in SCLC cell lines documenting the retained expression of wild-type Rb (9, 13–15). However, there have been no comprehensive studies concurrently analyzing RB1 genomic alterations and protein expression in a large series of clinical samples of SCLC to clarify the prevalence and characteristics of SCLC with the expression of wild-type Rb protein (Rb proficiency). This question is particularly relevant given the recent evidence that Rb-proficient SCLC cell lines exhibit selective sensitivity to CDK4/6 inhibitors (9, 13).
Also lacking is a comprehensive characterization of the laboratory methods available in clinical practice to assess Rb status. Sequencing of RB1 gene presents special challenges because it is a large gene (∼178 kb) that contains more introns than average (n = 26), with intronic sequences accounting for ∼99% of the total gene length (16). RB1 genomic alterations are enriched in intronic splice-site mutations and structural variants (4, 16); these may not be detected by targeted sequencing methods utilized in clinical practice, which generally cover exclusively or primarily exonic regions. IHC methods to assess Rb expression are well-established in pathology clinical practice. However, benchmarking of targeted sequencing versus IHC for Rb assessment in SCLC at scale has not been performed.
Considering the potential complexity of Rb status assessment, in this study, we also sought to evaluate not only RB1 genomic alterations and expression but also the functional status of Rb through interrogation of key regulators of the G1–S cell-cycle checkpoint in the Rb pathway. Data from cell lines and clinical samples representing a broad spectrum of tumor types has described a tightly coordinated expression pattern between Rb and its upstream regulators—cyclin-dependent kinase (CDK) inhibitor p16INK4A (p16), encoded by CDKN2A, and cyclin D1, encoded by CCND1 (17–19). As such, in tumors with mutationally-inactivated Rb, p16 consistently undergoes compensatory upregulation due to the loss of an Rb-mediated negative feedback loop on p16 expression (20, 21). On the basis of this principle, overexpression of p16 is an established biomarker of HPV-driven tumors in routine pathology practice, where high p16 expression reflects Rb constitutive inactivation by viral oncoproteins (22). In addition, mutationally-inactivated Rb is invariably associated with low or absent expression of cyclin D1 given the mutual exclusivity of Rb loss and cyclin D1 upregulation (11, 17–19). Taken together, Rb-deficient status could be inferred from a concurrent p16-high/cyclin D1-low expression profile. Here, we sought to test application of this approach in clinical samples of SCLC characterized for RB1 genomic alterations and protein expression.
In this study, we performed comprehensive profiling of Rb status in 208 SCLC patient samples by a combination of clinical methods used for pathologic and molecular tumor diagnosis, including a broad targeted next-generation sequencing (NGS) assay covering all exons of RB1 and Rb IHC in conjunction with IHC for p16 and cyclin D1 to further evaluate Rb functional status. Detailed clinicopathologic and genomic analyses were performed to elucidate the characteristics of SCLC showing evidence of Rb proficiency.
Materials and Methods
Sample selection and study design
The study was performed with the approval of the Institutional Review Board (IRB) of Memorial Sloan Kettering Cancer Center (MSKCC), and was conducted in accordance with the Declaration of Helsinki. All patients included in this study signed an informed consent form according to the protocol approved by the MSKCC IRB. The specimens comprised clinical samples of SCLC diagnosed at MSKCC primarily between January 2014 and March 2021 from patients who consented to molecular testing. Cases of SCLC transformation of lung adenocarcinoma on targeted therapy were excluded. Only cases adequately profiled by both Rb IHC and targeted DNA sequencing were included. Criteria for the diagnosis of SCLC was based on the 2021 WHO classification (23), see Supplementary Materials and Methods for details. Detailed annotation of demographic variables, histopathology, IHC profiles, and genomic alterations was performed.
IHC
Rb IHC was performed using a mouse anti-Rb protein mAb (clone 13A10, Leica; Supplementary Table S1). Stromal cells served as the positive internal control. Cases lacking adequate internal control were excluded.
All Rb-proficient SCLC and a set of Rb-deficient cases were analyzed for conventional neuroendocrine markers [synaptophysin [SYN], chromogranin A (CgA), CD56/NCAM, and INSM1], TTF-1, Ki-67, novel markers for SCLC transcriptional subtypes (ASCL1, NEUROD1, POU2F3, YAP1; refs. 1, 24), and markers of Rb functional status (p16 and cyclin D1). Detailed IHC protocols and the total number of cases with evaluable results for each marker are summarized in Supplementary Tables S1 and S2, respectively.
Marker expression was evaluated by a semiquantitative H-scoring method (24). H-scores were derived by multiplying the percentage of positive tumor cells (0–100%) by staining intensity ordinal values (0 = no signal, 1 = weak, 2 = moderate, and 3 = strong) yielding a range of possible H-scores from 0 to 300. For each marker, H-scores were also dichotomized into two-tier categories. Detailed scoring criteria for each marker are summarized in Supplementary Table S1.
For the analysis of p16 and cyclin D1, cases from this cohort were supplemented with a set of 77 SCLC with Rb loss by IHC from a previously-described tissue microarray (TMA; ref. 24).
Comprehensive targeted DNA sequencing by MSK-IMPACT
Genomic profiling of matched tumor/normal samples was performed using the MSK-IMPACT platform as described previously (25). Briefly, the MSK-IMPACT assay is a clinically validated custom hybridization capture-based platform that sequences the entire coding region and selected noncoding regions of 341 (v3), 410 (v4), 468 (v5), or 505 (v6) genes (full list in Supplementary Table S3) for the detection of single-nucleotide variants, indels, copy-number alterations, and selected structural variants. Germline variants were filtered out based on the matched germline DNA using a bioinformatic pipeline. Confirmation of whole-gene level amplifications and deletions as well as intragenic deletions was performed by manual inspection of log-ratio copy-number plots using the integrative genomics viewer and GISTIC V2.0 (Supplementary Materials and Methods). Cases with inadequate tumor purity (<20%) or insufficient coverage (<100×) were excluded. Copy-number alterations, ploidy, purity, and clonal assessment (cancer cell fraction) were assessed using the Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS) pipeline, as described in the Supplementary Materials and Methods.
RB1 genomic profiling
All exons of the RB1 gene (NM_000321) were covered in each version of the MSK-IMPACT assay. In addition, the latest version (V6, used for 29 tumors) also covered RB1 regions of the 5′UTR and introns 6, 8, and 23. Mutation calling was performed for all exons and off-target intronic regions with sufficient coverage (Supplementary Materials and Methods). The length of flanking intronic coverage for RB1 is shown in Supplementary Fig. S1 (100× coverage was achieved for up to 50 bp from exon/intron junction for ∼70% of samples, and up to 100 bp for ∼20% of samples). For splice-site mutations, only canonical mutations (i.e., splice acceptor or donor sites located 2 bp into the intron from the intron/exon junctions) were included in the clinical analysis pipeline. For cases lacking RB1 alterations on routine clinical pipeline, a manual review of RB1 sequencing reads was performed using the integrative genomics viewer and log-ratio copy number plots, as described in Supplementary Materials and Methods. Noncanonical splice-site mutations (i.e., mutations located more than 2 bp into the intron from the exon/intron junction) were identified and their likely impact on splicing was estimated using the SpliceAI deep learning-based tool. FACETS output was reviewed for RB1 gene-specific copy-number alterations including loss of heterozygosity (LOH) and homozygous loss (Supplementary Materials and Methods).
Statistical analysis
Statistical analysis was conducted using IBM SPSS Statistics V27.0 (IBM), cBioPortal platform for cancer genomics (26), and GraphPad Prism V9 (GraphPad Software). Fisher and Freeman–Halton modified exact tests and Mann–Whitney U tests were calculated for categorical and continuous variables, respectively. For multiple comparisons, the Benjamin–Hochberg procedure was applied to adjust the FDR. Overall survival was calculated using the Kaplan–Meier approach from the time of diagnostic specimen collection to the time of death. Patients were otherwise censored at the time of the last clinical follow-up. Survival curves were compared using the log-rank test.
Data availability
Data are available in a repository (cBioPortal for Cancer Genomics) that can be accessed via a DOI link upon request.
Results
Correlation of Rb protein expression and RB1 genomic alterations
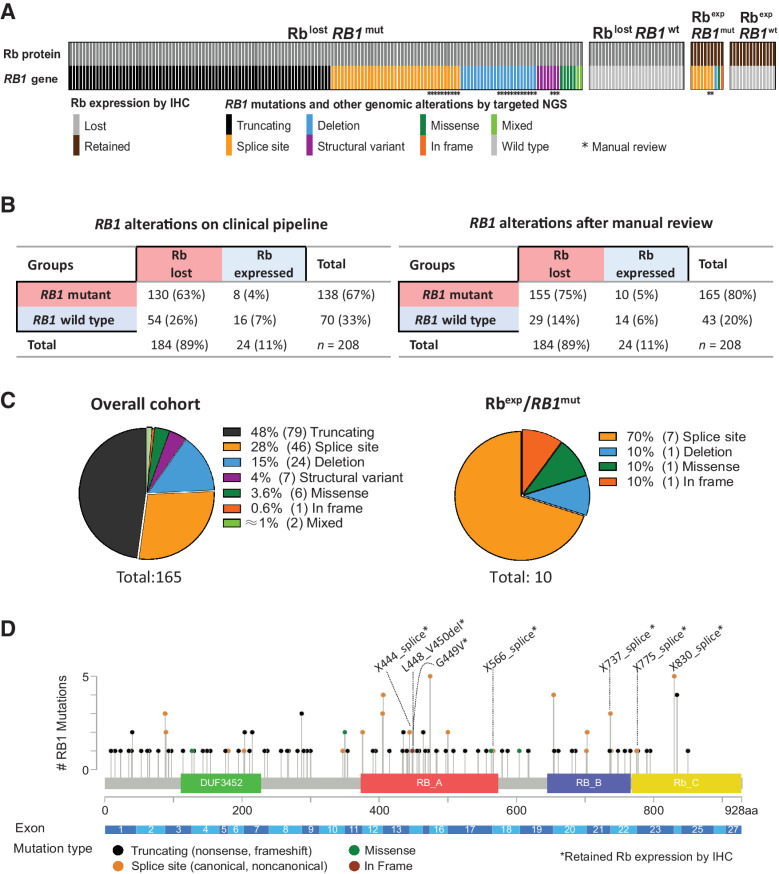
The study cohort comprised 208 de novo SCLC cases with evaluable NGS and Rb IHC. The overall clinicopathologic characteristics of the cohort are summarized in Supplementary Table S4. By IHC, 184 cases (89%) showed complete loss of Rb expression, whereas 24 cases (11%) showed retained Rb expression (Fig. 1A and B; examples in Supplementary Fig. S2). By NGS, only 138 (67%) of SCLC had RB1 alterations identified by clinical pipeline. After manual review of NGS data (see next section), RB1 alterations were identified in 27 additional cases, for a total of 165 RB1-mutated cases (80%). Among cases with Rb loss, RB1 alterations were detected in 71% (130/184) of cases without manual review and 84% (155/184) with manual review. On the basis of IHC and manually reviewed molecular data, the following groups were identified: Rb-lost/RB1-mutated (n = 155; 75%), Rb-lost/RB1-wild type (n = 29; 14%), Rb-expressed/RB1-mutated (n = 10; 5%), and Rb-expressed/RB1-wild type (n = 14; 6%).
Figure 1.
The spectrum of RB1 genomic alterations and Rb protein expression profiles in SCLC. A, Grouping of SCLC (n = 208) according to Rb immunoexpression and the presence of RB1 genomic alterations. *, Detected by manual review only. mut, mutated; wt, wild type; exp, expressed. B, Contingency table summarizing the total rates of subgroups based on RB1 mutations and Rb expression for alterations detected by routine clinical pipeline (left) and full set of alterations after manual review (right). C, Pie charts with the distribution of RB1 mutation types detected in the entire cohort (left) and those associated with Rb expression (right). D, Genomic mapping of selected RB1 mutations (nonsense, frameshift, splicing, missense, and in-frame variants). *, Associated with Rb expression.
The spectrum of RB1 genomic alterations in relation to Rb protein expression
RB1 genomic alterations (n = 165, total) comprised nonsense and frameshift (truncating) mutations (48%), splice-site mutations (28%), deletions (15%), and other uncommon events (Fig. 1C; Supplementary Table S5). RB1 mutations occurred across the entire RB1 gene with only a minority of mutations occurring as recurrent events (Fig. 1D). RB1 alterations that were identified on manual review but not on initial clinical reporting (n = 27) comprised noncanonical splice-site mutations (n = 12), deletions (n = 12), and structural variants (n = 3; details for each case provided in Supplementary Table S6). In silico analysis using SpliceAI algorithm predicted that nearly all noncanonical splice-site mutations had pathogenic or likely pathogenic effects on splicing (Supplementary Table S6).
In 10 cases with expressed/mutated Rb, RB1 alterations were primarily splice-site mutations (70%; Fig. 1C and D). Genomic mapping suggested that these alterations were concentrated around the exons encoding key functional domains of Rb: the E2F transcription factor binding pocket (RB-A and RB-B) and the C-terminal domains (Fig. 1D; Supplementary Fig. S3).
We also performed FACETS to assess the allelic configuration of the RB1 locus (Supplementary Tables S5 and S7). This revealed that SCLC lacking Rb expression exhibited LOH or homozygous deletions at RB1 locus in 178 of 182 evaluable cases (98%), in line with a two-hit model of RB1 inactivation. Similarly, tumors with expressed/mutated Rb exhibited RB1 locus LOH in 9 of 10 cases. In SCLC with expressed/wild-type Rb, RB1 LOH was also seen in 9 of 13 evaluable cases, consistent with previous observations of single copy RB1 loss in a variety of tumors lacking RB1 mutations or protein loss (19).
Rb functional status adjudicated by p16INK4A and cyclin D1 IHC profiles
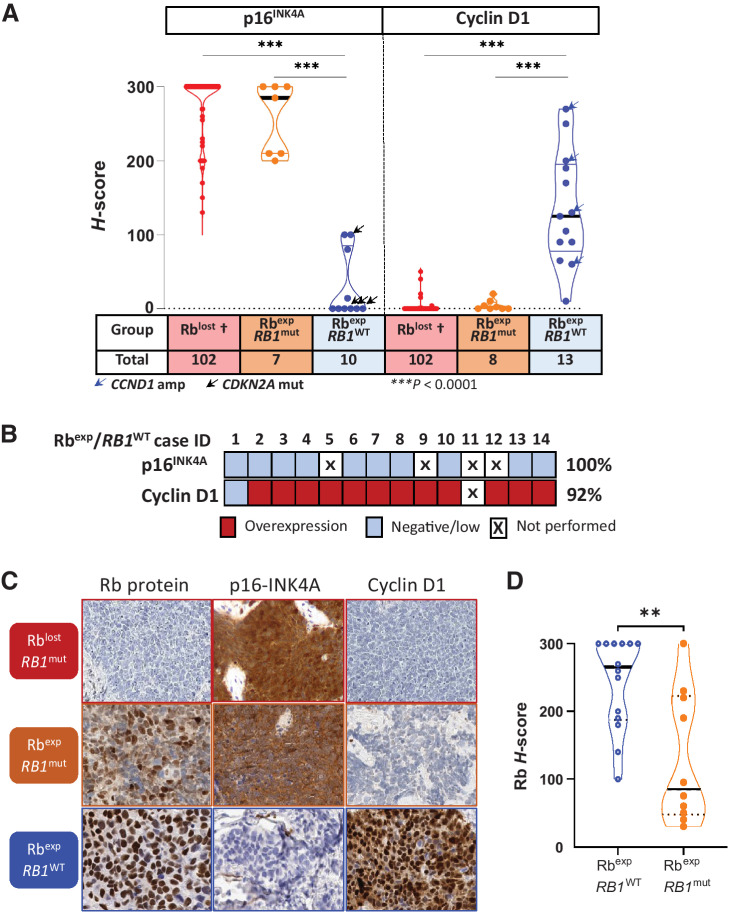
The above findings indicated that targeted NGS, even with manual review, had incomplete sensitivity for RB1 alterations, and that SCLC can express mutated Rb, raising a question of whether expressed Rb in SCLC was functional or nonfunctional. This prompted us to assess markers of functional Rb status in relation to groups defined by RB1 mutations and Rb protein expression. p16 and cyclin D1 expression was thus assessed by IHC in all SCLC with expressed Rb that had sufficient residual tissue, and in a control group of SCLC with lost Rb expression (n = 102).
As anticipated, all 102 SCLC with Rb loss had consistent p16-high expression (p16 H-score >100; median H-score: 300). In contrast, all Rb-expressed/wild-type SCLCs were p16-low (median H-score: 0). Remarkably, all Rb-expressed/mutant cases were p16-high (median H-score: 285), supporting that expressed Rb in this group is nonfunctional (Fig. 2A–C).
Figure 2.
p16 and cyclin D1 as ancillary markers of Rb functional status. A, Violin dot-plot graph depicting p16 and cyclin D1 H-scores in relation to groups defined by Rb expression and RB1 alterations, and demonstrating a consistent p16high/cyclin D1low profile for Rblost and Rbexp/RB1mut tumors, whereas all Rbexp/RB1WT tumors demonstrated the opposite p16low profile, with cyclin D1high in most cases. †, Rb-lost group in this analysis included 25 cases from the current cohort and 77 cases from TMAs (see Materials and Methods). Statistical significance: ***, P < 0.0001. B, Heatmap of Rbexp/RB1WT cases summarizing the incidence of p16low (H-score ≤ 100) and cyclin D1high (H-score ≥ 50), with the thresholds defined using the distribution of scores in Rb-lost SCLC (see Supplementary Table S1). C, Photomicrographic examples of Rb, p16, and cyclin D1 profiles in SCLC groups defined by Rb expression and RB1 alterations. D, Violin dot-plot graph showing H-scores of Rb immunoexpression in RB1 wild-type (RbexpRB1WT) tumors compared with those with RB1 mutations (RbexpRB1MUT). Statistical significance: **, P = 0.0057. Exp, expressed; mut, mutated; WT, wild type.
Similarly, cyclin D1 was negative/low in all SCLC with Rb loss. In line with the p16 data, all Rb-expressed/mutant tumors also exhibited negative/low cyclin D1 profile. Conversely, the Rb-expressed/wild-type group was uniquely associated with cyclin D1-high expression in 92% of cases, although the level of expression had a wide range. Overall, p16 and cyclin D1 data provided robust support for the lack of functional Rb in cases without Rb expression and in cases with expressed/mutated Rb (Rb-deficient SCLC), and for a preserved Rb functional status in tumors with expressed/wild-type Rb (Rb-proficient SCLC).
We also assessed whether there was a difference in the level of Rb expression in tumors with expressed/mutated versus expressed/wild-type Rb. For expressed/wild-type group, Rb expression in most cases was strong and diffuse, although few cases showed lower levels (median H-score: 265, range: 100–300). Conversely, in expressed/mutated group, Rb scores exhibited a broad range, but most cases had H scores below 100 (median H-score: 85; range: 30–300; P = 0.0057; Fig. 2D, examples in Supplementary Fig. S4). Thus, low Rb levels can serve as a clue to the expression of a mutated protein, but high levels do not exclude this possibility.
Clinicopathologic characteristics of Rb-proficient SCLC
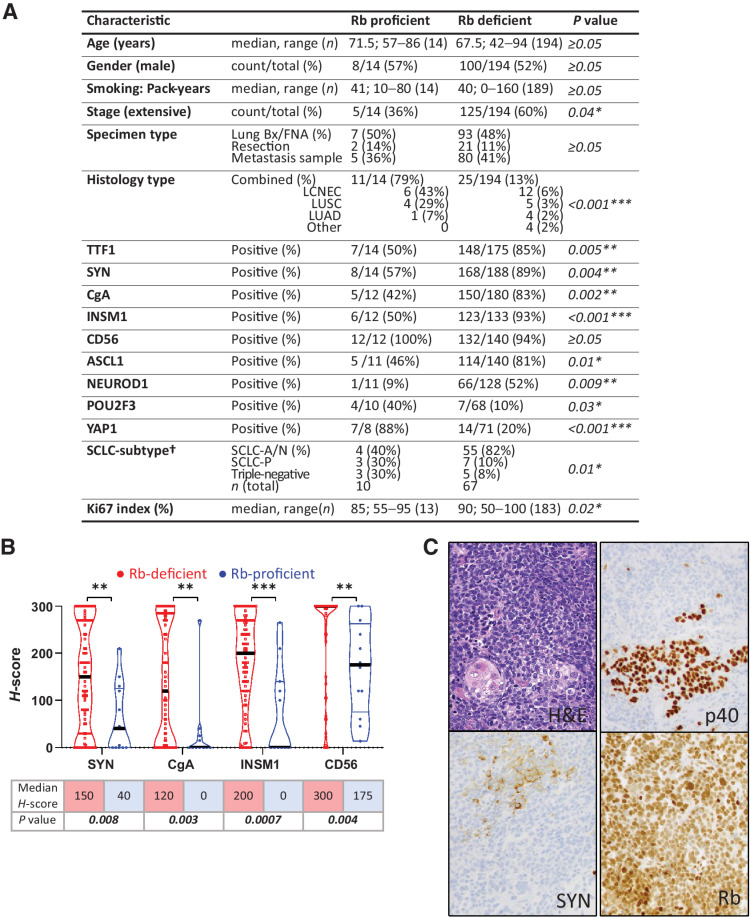
Patient characteristics and pathologic findings for Rb-proficient compared to Rb-deficient SCLC are shown in Fig. 3A. Detailed clinicopathologic and IHC findings for individual Rb-proficient SCLC are summarized in Supplementary Table S8. Although the overall patient and sample characteristics were similar (age, gender, smoking history, primary vs. metastatic sample site, and method of sampling), Rb-proficient tumors were marginally enriched in limited-stage disease. Notably, Rb-proficient SCLC were significantly enriched in combined SCLC containing both non–small cell carcinoma (NSCLC) and SCLC components (79% vs. 13%, respectively; P < 0.001). The histotypes of NSCLC components included large cell neuroendocrine carcinoma (LCNEC; n = 6), lung squamous cell carcinoma (LUSC; n = 4), and lung adenocarcinoma (LUAD; n = 1). In addition, Rb-proficient SCLC had significantly lower expression of all standard markers of neuroendocrine differentiation in SCLC components—SYN, CgA, INSM1, and CD56, as measured by either a proportion of positive cases (Fig. 3A) or the extent of expression (Fig. 3B). TTF1—a marker that generally parallels neuroendocrine expression in SCLC—was also significantly lower in SCLC areas of Rb-proficient SCLC. This “neuroendocrine-low” phenotype was congruent with significantly lower expression of ASCL1 and NEUROD1—the transcriptional subtype markers associated with neuroendocrine-high state, and concomitant enrichment in POU2F3 and YAP1—the marker associated with neuroendocrine-low state (Fig. 3A).
Figure 3.
Clinicopathologic characteristics of Rb-proficient compared with Rb-deficient SCLC. A, Demographic, histopathologic, and IHC characteristics. †, SCLC subtypes were assigned only to cases with data available for ASCL1, NEUROD1, and POU2F3. SCLC-A/N: tumor with expression of ASCL1 and/or NEUROD1. SCLC-P: tumors expressing exclusively POU2F3. Triple-negative: tumors lacking ASCL1, NEUROD1, and POU2F3 expression. B, Violin dot plots showing the quantitative distribution of H-scores for conventional neuroendocrine marker expression in Rb-proficient versus Rb-deficient SCLC. C, Example of a combined SCLC (top component: p40−, SYN+) with LUSC (bottom component: p40+, SYN−; bottom left: keratin pearl) showing retained Rb expression in both components. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001; not significant, P ≥ 0.05.
Despite predominantly neuroendocrine-low phenotype and association with NSCLC components, the pathologic characteristics of SCLC areas in Rb-proficient tumors were those of prototypical SCLC (examples in Fig. 3C; Supplementary Fig. S2), including typical histomorphology and extremely high Ki67 proliferation index (mean 80%, range 55–95%; Supplementary Table S8). In combined carcinomas, SCLC areas accounted for 30% to 90% of tumor cellularity. The clonal nature of SCLC and NSCLC components is such tumors was supported by clonal or near-clonal distribution of TP53 and CDKN2A mutations based on cancer cell fraction analysis (Supplementary Materials and Methods; Supplementary Table S8) and concordant Rb, cyclin D1 and p16 expression by IHC in SCLC and NSCLC components in all cases (Supplementary Table S8).
Genomic characteristics of Rb-proficient SCLC
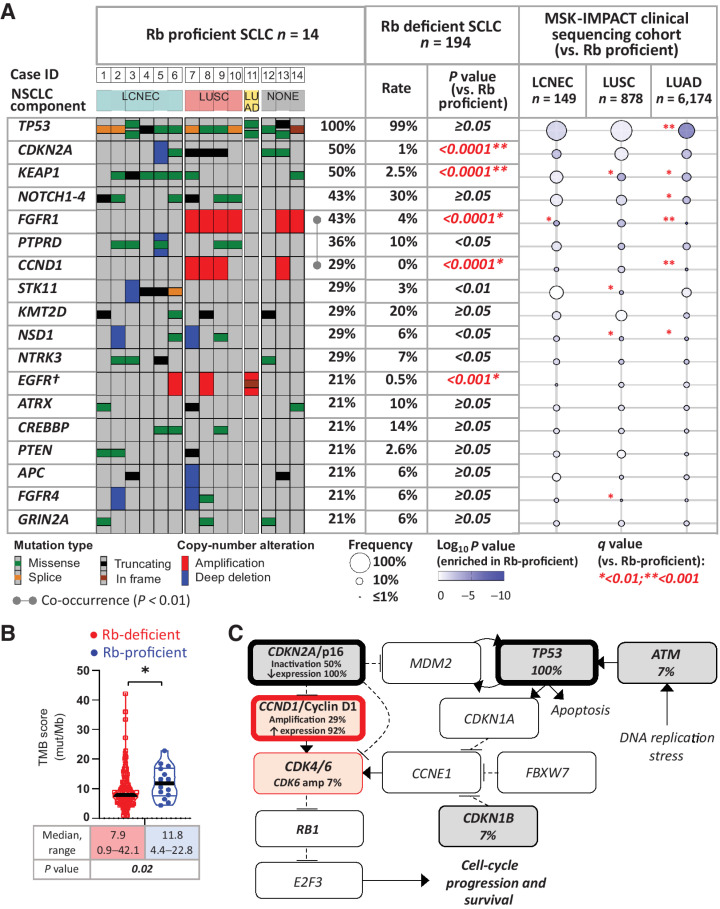
Mutational profiling of the 14 Rb-proficient SCLC in comparison to 194 Rb-deficient SCLC is shown in Fig. 4A. Rb-proficient SCLC harbored a median of 12.5 nonsynonymous mutations per sample (range: 5–26) with a median coverage of 678× (range: 110–861×). Tumor mutation burden (TMB) in Rb-proficient tumors was higher than in Rb-deficient ones (median 11.8 vs. 7.9 mut/Mb; respectively; P = 0.02; Fig. 4B). Tumor purity, sample coverage, and ploidy levels were similar for Rb-proficient and Rb-deficient cases (Supplementary Table S7).
Figure 4.
Genomic profile of Rb-proficient SCLC. A, OncoPrint depicting the most prevalent recurrent genomic alterations in Rb-proficient SCLC. Tumors were grouped by NSCLC components in combined carcinomas. Alteration rates in Rb-proficient SCLC were compared with those in Rb-deficient SCLC. In addition, gene alteration prevalence in other major lung cancer types (LCNEC, LUSC, and LADC) are represented using the bubble plot based on the prospective MSK-IMPACT clinical sequencing cohort as registered in cBioPortal (accessed January 2022). All comparisons versus Rb-proficient SCLC. †, Shown are amplifications and canonical driver alterations. For EGFR, group comparison results are based on amplification events only. B, Violin dot plots showing the TMB scores in Rb-proficient versus Rb-deficient SCLC. C, Cell-cycle pathway by PathwayMapper (cBioPortal) showing a high rate of alterations converging on activation of CDK4/6.
Top recurrently altered genes in Rb-proficient SCLC demonstrated several notable differences relative to those in Rb-deficient SCLC (Fig. 4A; see Supplementary Table S9 for a full list). All Rb-proficient SCLC harbored TP53 mutations, but compared with Rb-deficient tumors, they were uniquely associated with amplification of the CCND1 (29% vs. 0%; P < 0.0001) and exhibited marked enrichment in CDKN2A alterations (50% vs. 1%; P < 0.0001), respectively. Of note, unlike Rb-deficient SCLC, CDKN2A mutations in Rb-deficient SCLC (n = 2) were not accompanied by the loss of p16 expression. Rb-proficient tumors were also markedly enriched in alterations typical of NSCLC, including FGFR1 amplifications (43% vs. 4%, P < 0.0001), EGFR amplifications (21% vs. 0.5%, P < 0.001), KEAP1 mutations (50% vs. 2.5%, P < 0.0001), and STK11 mutations/loss (29% vs. 3%, P < 0.01), respectively (Fig. 4A; Supplementary Fig. S5). Enrichment analysis identified CDKN2A (q < 0.0001), KEAP1 (q < 0.001), CCND1 (q < 0.01), and FGFR1 (q < 0.01) as the most differentially altered genes in Rb-proficient compared with Rb-deficient SCLC (Supplementary Fig. S6). Other recurrent alterations involved NOTCH1–4, PTPRD, KM2TD, NSD1, NTRK3, ATRX, CREBBP, PTEN, APC, FGFR4, and GRIN2A, with rates comparable with those in Rb-deficient tumors.
The frequencies of individual alterations in Rb-proficient SCLC were also compared with those in other major types of lung carcinoma—LCNEC (n = 149), LUSC (n = 878), and LUAD (n = 6,174), which were profiled as part of MSK-IMPACT clinical sequencing cohorts. The distribution of NSCLC-type alterations clearly tracked with the histology of combined NSCLC components in Rb-proficient SCLC: FGFR1 amplifications (characteristic of LUSC) occurred exclusively in combination with LUSC, whereas STK11/KEAP1 co-mutations (characteristic of LCNEC) were seen exclusively in combination with LCNEC histology. A single canonical EGFR mutation occurred in SCLC combined with LUAD. In addition, two histologically-pure SCLC also harbored FGFR1 amplifications.
Correlation gene–gene matrix analysis demonstrated significant co-occurrence of FGFR1 and CCND1 amplifications in Rb-proficient SCLC (P = 0.005; Fig. 4A; Supplementary Fig. S7), which was not detected in other lung tumors, including LUSC (not shown).
Cell-cycle regulatory alterations, incorporating genomic and IHC data, are summarized in Fig. 4C. This highlights the high rate of alterations converging on CDK4/6 activation, including the inactivating mutation and loss of expression of CDKN2A/p16, and amplification and overexpression of CCND1/cyclin D1. Single cases with CDK6 amplification and CDKN1B mutation were also detected.
Survival and treatment of Rb-proficient SCLC
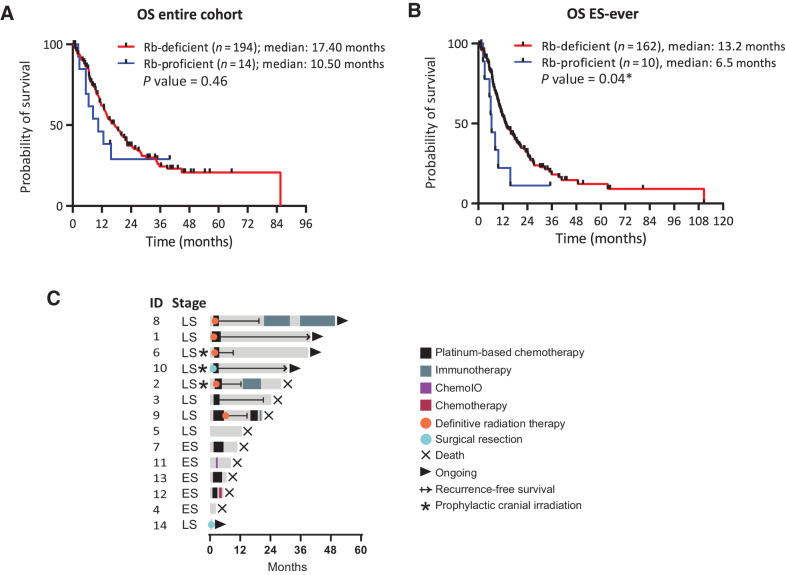
Overall survival analysis for the entire cohort revealed a trend toward worse survival for patients with Rb-proficient compared to Rb-deficient SCLC (median survival 10.5 months vs. 17.4 months, respectively; P = 0.46; Fig. 5A). Given the observed imbalance in stage at diagnosis (Fig. 3A), we also assessed outcome in patients with extensive stage disease at either presentation or progression. For such patients, Rb-proficient SCLC exhibited significantly worse survival (median survival 6.5 months vs. 13.2 months, respectively; P = 0.04; Fig. 5B).
Figure 5.
Survival and treatment analysis of Rb-proficient SCLC. Comparison of overall survival (A) and survival restricted to patients with extensive-stage (ES)-ever (B). C, Swimmer plot depicting treatment modality, time on treatment, and outcome.
A swimmer plot summarizing therapeutic interventions in patients with Rb-proficient SCLC including time on treatment and outcomes is presented in Fig. 5C. To explore chemosensitivity of Rb-proficient SCLC, treatment outcomes were assessed for the patients who received platinum/etoposide-based regimens. Of the 5 patients with extensive-stage disease, four received first-line platinum/etoposide-based regimens. One patient (patient ID4) died prior to initiation of therapy. For the 4 evaluable patients (patient ID 7, 9, 12, 13), the median time to progression from the initiation of therapy was 2.3 months (range: 2.0–3.9 months), consistent with primary chemorefractory disease (27).
SCLC with expression of mutated Rb: clinicopathologic and genomic characteristics
Above, we described that 10 of 208 (5%) SCLC exhibited expression of Rb protein despite the presence of RB1 mutations. Such tumors exhibited p16-high/cyclin D1-low profile, supporting expression of a nonfunctional protein. To determine whether characteristics of these tumors were equivalent to those in other Rb-deficient SCLC, we compared the clinicopathologic and genomic features in these groups and found that all examined parameters (patient demographics, tumor histology, genomic features) were comparable (Supplementary Fig. S8).
Discussion
In this study, we performed analysis of RB1 genomic alterations by broad targeted NGS concurrently with Rb protein IHC in 208 clinical samples of SCLC and further documented Rb functional status by p16/cyclin D1 expression profiles. We provide comprehensive benchmarking of the performance of targeted NGS relative to IHC, highlighting the major limitation of exon-only sequencing for the detection of RB1 genomic events and illustrating a rare subset of SCLC with expression of mutated, nonfunctional Rb. Finally, we provide a detailed clinicopathologic and genomic characterization of a here-to-fore poorly defined but highly distinctive group of SCLC with expression of wild-type Rb (Rb-proficient SCLC), including identification of molecular alterations associated with unique therapeutic vulnerabilities in this subset of tumors.
Rb-proficient SCLC
We identified that Rb proficiency accounts for 6% of SCLC. This subset was defined as SCLC harboring expressed/wild-type Rb, in which preserved Rb functionality was confirmed by an exclusive p16-low/cyclin D1-high phenotype. Most prior studies have assessed Rb in SCLC by either sequencing or IHC alone; however, as illustrated here, each method in isolation has limitations for establishing Rb proficiency. Combined genomic and protein expression approaches have been primarily limited to SCLC cell lines, where expression of wild-type Rb was noted in 10% to 16% of cases (9, 13–15). To our knowledge, this is the first study to identify and comprehensively characterize this subset in SCLC patient samples.
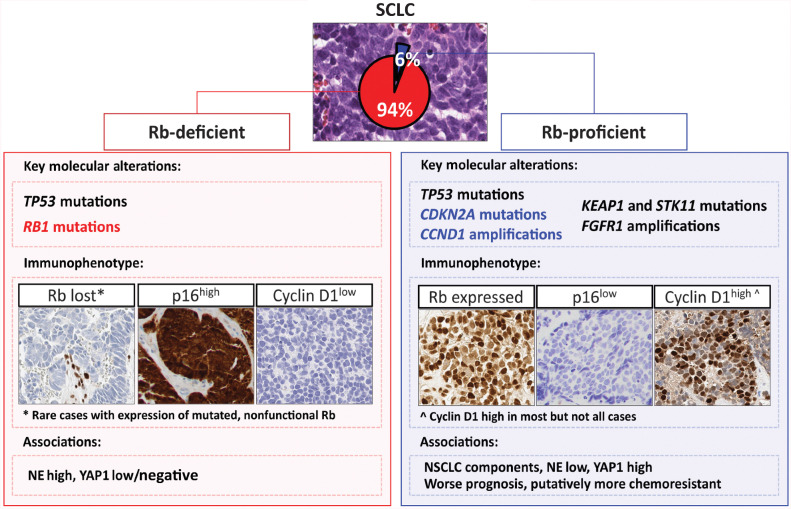
We found that Rb-proficient SCLC were associated with several highly distinctive characteristics (Fig. 6). First, these tumors were strongly associated with the presence of NSCLC histologic components: NSCLC components were detected in 78% of Rb-proficient SCLC compared with 13% of Rb-deficient cases. In addition, Rb-proficient tumors showed marked enrichment in NSCLC-type genomic alterations. For example, FGFR1 amplifications are rarely seen in Rb-deficient SCLC, but they were found in 43% of Rb-proficient tumors, specifically corresponding to SCLC combined with squamous histologic components. Even in cases lacking demonstrable NSCLC components, the presence of NSCLC-type alterations suggests a link with NSCLC obscured by SCLC overgrowth or incomplete sampling. These pathologic and molecular findings suggest a consistent histogenetic origin of Rb-proficient SCLC from NSCLC-type progenitors, which contrasts with the presumed predominant neuroendocrine precursor origin for conventional SCLC (28). Supporting this hypothesis, a non-neuroendocrine origin of SCLC with FGFR1 activation has been documented in mice (29).
Figure 6.
Diagram summarizing the distinct clinicopathologic and genomic characteristics of Rb-proficient SCLC. Enrichment in NSCLC histologic components and NSCLC-type genomic alterations in Rb-proficient SCLC suggests putative origin of these tumors from an overlapping precursor of NSCLC, with greater potential for divergent differentiation.
Despite the putatively distinct derivation and unusual Rb status, SCLC components in Rb-proficient tumors had classic histopathologic features of SCLC. We postulate that the enrichment in NSCLC histologic components reflects the propensity of these tumors for divergent differentiation possibility as a result of their origin from an overlapping precursor of NSCLC. Major genomic alterations in Rb-proficient combined carcinomas (TP53, CDKN2A) were clonal or near-clonal, in line with recent studies that confirmed clonal nature of combined SCLC and implicated transcriptional reprogramming rather than mutational events in plasticity between NSCLC and SCLC states (30). Future studies will be needed to elucidate the specific molecular mechanisms that underlie such plasticity in an Rb-proficient background. FGFR1/CCND1 co-occurrence may be of interest in this regard as it was shown to promote proliferation and phenotypic plasticity in LUSC cell lines (31).
Second, Rb-proficient SCLC were associated with low expression of neuroendocrine markers, predominance of ASCL1/NEUROD1-negative phenotype, and elevated expression of POU2F3 and YAP1. These features correspond to what has been termed as “variant” or “non-neuroendocrine” subtype of SCLC (32, 33). Indeed, in prior studies, primarily in SCLC cell lines, Rb expression in SCLC was noted to be associated with NE-low phenotype (13) and elevated YAP1 expression (9). We speculate that the enrichment in “variant” phenotype parallels the high rate of combined SCLC/NSCLC subtype among Rb-proficient SCLC, because combined SCLC have been previously found to be associated with ASCL1/NEUROD1-low but POU2F3/YAP1-high profile (24). Indeed, for the analysis restricted to tumors with combined histology, Rb-proficient and Rb-deficient tumors showed similar phenotypic characteristics (not shown).
Third, we observed that Rb-proficient SCLC demonstrated features of increased clinical aggressiveness. Although limited by small numbers, it is notable that all evaluable patients exhibited primary chemoresistance to platinum/etoposide therapy, a treatment associated with a response rate of over 60% (1, 34). This is in agreement with prior suggestions that the lack of RB1 alterations in SCLC might be associated with shorter survival and chemo-refractoriness (8, 9). Shorter doubling time and treatment resistance have been long noted as a feature of “variant” SCLC cell lines (32), and ectopic Rb expression in SCLC cell lines can lead to increased drug resistance (15). The latter suggests that Rb status itself may represent a determinant of chemosensitivity in SCLC. The NE-low/YAP1-high phenotype has also been associated with chemoresistance (35).
Finally, a feature exclusive to Rb-proficient tumors was the high rate of CCND1 amplifications (29%) and CDKN2A mutations accompanied by the loss of p16 expression (50%). In prior studies, CCND1 and CDKN2A alterations were also almost entirely absent in Rb-deficient SCLC (4, 7, 9). The strict exclusivity of RB1 inactivation with CCND1 and CDKN2A aberrations reflects a well-established pan-cancer phenomenon, consistent with Rb loss obviating the evolutionary advantage imparted by CDK4/6 activation by CCND1/cyclin D1 and CDKN2A/p16 (19). At the protein level, all evaluable Rb-proficient SCLC were p16-low and most cases were cyclin D1-high, supporting a consistent downstream CDK4/6 activation in these tumors and the resultant functional (reversible) Rb inhibition via CDK4/6-mediated phosphorylation.
Despite clinical aggressiveness and poor response to standard chemotherapy, distinct molecular features of Rb-proficient SCLC suggest several potential therapeutic approaches to these tumors. First, the high rate of alterations converging on CDK4/6 activation suggests that Rb-proficient SCLC may be selectively vulnerable to CDK4/6 inhibitors (36). In fact, cell line data support selective CDK4/6 sensitivity of SCLC with expressed or wild-type Rb (9, 13). Second, the high rate of FGFR1 amplifications may be of interest as a potential target of anti-FGFR agents (37). Furthermore, particularly high TMB in Rb-proficient SCLC may suggest potentially increased sensitivity of this subset to immune checkpoint inhibitors (38). The potential of unique treatment approaches to Rb-proficient SCLC, if confirmed in clinical studies, would provide a strong rationale for a routine assessment of Rb status in SCLC samples.
Technical aspects of clinical Rb interrogation
Our study provides several important insights into technical aspects of Rb assessment in clinical practice. First, here we document that clinical NGS, covering primarily exonic regions, fails to identify RB1 genomic alterations in 29% of cases exhibiting complete loss of Rb protein expression. The limitations of exon-only sequencing in RB1 interrogation in SCLC are not unexpected given the known high prevalence of intronic splice-site mutations and structural variants in RB1 by WGS (31% and 12%, respectively; ref. 4). However, this is the first study to quantify the sensitivity of clinical targeted NGS for RB1 alterations relative to IHC. Our rate of detection of RB1 mutations in SCLC is in line with other exome-only sequencing studies, which have failed to identify RB1 alterations in 25% to 48% of cases (7–10). In contrast, by WGS, the lack of bi-allelic RB1 inactivation was seen in 7% of conventional SCLC (4), closely in line with the prevalence of Rb proficiency in our study.
We found that manual review can improve the detection of RB1 alterations by targeted NGS, yet 16% of cases with complete loss of Rb expression by IHC still remained with no detectable genomic RB1 alterations. Furthermore, functional consequences of noncanonical splice-site mutations, even if predicted as pathogenic in silico, are uncertain. In fact, uncertain functional impact applies more broadly to all types of non-truncating mutations in tumor suppressor genes, and such mutations comprise more than half of RB1 alterations. Overall, our findings indicate that targeted sequencing, even with manual improvement, has limited sensitivity for the detection of RB1 alterations, and incorporation of orthogonal methods of Rb assessment is necessary for the accurate determination of Rb status in clinical practice.
We find that IHC represents a robust method of establishing Rb deficiency, readily documenting complete loss of Rb expression in 89% of SCLC. The methods for Rb IHC have evolved over the years, with the current widely-used Rb monoclonal antibody (clone 13A10) readily distinguishing lost from retained Rb expression.
A critical observation in this study relates to the description of a group of SCLC with expressed but mutated Rb, representing 5% of SCLC. A consistently p16-high/cyclin D1-low profile in these tumors supports their Rb-deficient status (i.e., expression of a nonfunctional Rb protein). Clinicopathologic and genomic characteristics of such tumors were entirely equivalent to SCLC with lost Rb. Interestingly, mutations associated with preserved expression of Rb comprised predominantly splice-site alterations, likely reflecting preserved stability of some mis-spliced proteins (39). The possibility of SCLC expressing mutated Rb has in fact been noted in the classic SCLC cell line studies from 1990s, which identified mutated and aberrantly migrating Rb in gel electrophoresis, which was functionally-inactive based on in vitro protein binding and phosphorylation studies (17, 40, 41). To our knowledge, this is the first demonstration and detailed characterization of this SCLC subset in patient samples.
The existence of SCLC subset with expressed/mutated Rb has major implications for interpretation of Rb IHC in practice. This indicates that detection of expressed Rb, especially at low levels, requires adjudication by additional methods to determine Rb functional status since approximately half of Rb-expressing SCLC represent expression of a mutated protein. Our data support the utility of p16 and cyclin D1 as robust ancillary markers to clarify the functional status of expressed Rb. Given the consistent reciprocity of Rb and p16 expression in this series and prior studies (11, 14, 18, 42), p16 alone would be sufficient for clarification of Rb functional status (p16-high supporting Rb deficiency and p16-low supporting Rb proficiency). However, cyclin D1-high status may serve as additional supporting evidence of Rb proficiency, although the absent or low cyclin D1 expression is of lesser utility given that we observed rare Rb-proficient case without detectable cyclin D1. Overall, our data on the strong coordination in the expression of Rb, p16, and cyclin D1 are in line with prior literature (11, 17–19), but here we describe the utility of profiling for these markers to clarify Rb functional status in SCLC exhibiting Rb expression. We note that some discordances in Rb with p16 and cyclin D1 relationships analyzed by IHC alone in prior studies could be a result of unsuspected mutations in the expressed Rb.
Several other methods have been proposed for assessing Rb functional status, including Rb phosphorylation and protein binding (41), p16/cyclin D1 mRNA ratio (42), and analysis of gene expression signatures (19, 43–45). However, these methods are currently confined to experimental laboratory settings, whereas p16 and cyclin D1 IHC offers distinct advantage of being widely available in diagnostic pathology laboratories, making the results readily applicable in current practice.
Our findings regarding the clinical laboratory methods of Rb assessment may have implications beyond SCLC given that Rb pathway is emerging as a critical marker of tumor aggressiveness and therapeutic outcomes in a variety of tumor types (19, 43–46) and given the growing interest in exploiting Rb pathway for cancer therapy (47). Among lung cancers, RB1 mutations have emerged as markers of treatment failure and predisposition to small cell transformation of EGFR-mutant adenocarcinomas treated with tyrosine kinase inhibitors (48). In addition, RB1 alterations are emerging as biomarkers for treatment selection in large cell neuroendocrine carcinoma (2, 49). Given that the complexity of RB1 genomic interrogation applies to other tumor types, our observations regarding the complementary nature of targeted NGS and IHC may have broad application across tumor types.
From the perspective of pathologic diagnosis of SCLC, the loss of Rb expression by IHC has been proposed as one of ancillary markers to support the diagnosis of SCLC in routine practice (50). Here we clarify that while lost in the majority of SCLC, retained expression is observed in 11% of SCLC and thus does not exclude the possibility of SCLC.
In conclusion, this is the first comprehensive IHC-NGS study in clinical samples of SCLC, in which we characterize a distinctive Rb-proficient subset and assess technical aspects of Rb status determination in clinical practice highlighting the complementary roles of sequencing and IHC. Given the evidence from this and other studies on increased aggressiveness of Rb-proficient SCLC and potential of unique treatment approaches for these tumors, routine testing for Rb in SCLC could become clinically informative.
Supplementary Material
Acknowledgments
This work was supported by the Ning Zhao and Ge Li Family Initiative for Lung Cancer Research and New Therapies; Fiona and Stanley Druckenmiller Center for Lung Cancer Research; NCI grants U24CA213274, R35CA263816, and P30 CA008748; the Molecular Diagnostics Service in the Department of Pathology; and the Marie-Josee and Henry R. Kravis Center for Molecular Oncology.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
H.A. Yu reports personal fees and other support from AstraZeneca, Cullinan, Daiichi, Blueprint Medicine, and Janssen; other support from Novartis, Pfizer, Lilly, and ERASCA; and personal fees from C4 Therapeutics outside the submitted work. M. Offin reports personal fees from Novartis, Jazz, PharmaMar, Targeted Oncology, OncLive, and American Society for Radiation Oncology and grants from Druckenmiller Foundation outside the submitted work. C.M. Rudin reports personal fees from AbbVie, Amgen, AstraZeneca, Bristol Myers Squibb, D2G Oncology, Genentech/Roche, Ipsen, Jazz, Kowa, Merck, Syros, Bridge Medicines, Earli, and Harpoon Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
C.A. Febres Aldana: Conceptualization, data curation, software, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.C. Chang: Data curation, formal analysis, investigation, methodology, writing–review and editing. R. Ptashkin: Data curation, formal analysis, investigation, methodology, writing–review and editing. Y. Wang: Data curation, formal analysis, investigation, methodology, writing–review and editing. E. Gedvilaite: Data curation, formal analysis, investigation, methodology, writing–review and editing. M.K. Baine: Data curation, formal analysis, investigation, methodology, writing–review and editing. W.D. Travis: Investigation, writing–review and editing. K. Ventura: Data curation, writing–review and editing. F. Bodd: Data curation, writing–review and editing. H.A. Yu: Investigation, writing–review and editing. A. Quintanal-Villalonga: Investigation, writing–review and editing. W.V. Lai: Investigation, writing–review and editing. J.V. Egger: Data curation, formal analysis, investigation, visualization, writing–review and editing. M. Offin: Data curation, formal analysis, investigation, visualization, writing–review and editing. M. Ladanyi: Investigation, writing–review and editing. C.M. Rudin: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, writing–review and editing. N. Rekhtman: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Rudin CM, Brambilla E, Faivre-Finn C, Sage J. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rekhtman N. Lung neuroendocrine neoplasms: recent progress and persistent challenges. Modern Pathol 2022;35:36–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer 2008;8:671–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Harbour JW, Lai S-L, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science 1988;241:353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gazdar AF, Hirsch FR, Minna JD. From mice to men and back: An assessment of preclinical model systems for the study of lung cancers. J Thorac Oncol 2016;11:287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dowlati A, Lipka MB, McColl K, Dabir S, Behtaj M, Kresak A, et al. Clinical correlation of extensive-stage small-cell lung cancer genomics. Ann Oncol 2016;27:642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McColl K, Wildey G, Sakre N, Lipka MB, Behtaj M, Kresak A, et al. Reciprocal expression of INSM1 and YAP1 defines subgroups in small cell lung cancer. Oncotarget 2017;8:73745–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beasley MB, Lantuejoul S, Abbondanzo S, Chu W-S, Hasleton PS, Travis WD, et al. The P16/cyclin D1/Rb pathway in neuroendocrine tumors of the lung. Hum Pathol 2003;34:136–42. [DOI] [PubMed] [Google Scholar]
- 12. Igarashi T, Jiang S-X, Kameya T, Asamura H, Sato Y, Nagai K, et al. Divergent cyclin B1 expression and Rb/p16/cyclin D1 pathway aberrations among pulmonary neuroendocrine tumors. Modern Pathol 2004;17:1259–67. [DOI] [PubMed] [Google Scholar]
- 13. Sonkin D, Vural S, Thomas A, Teicher BA. Neuroendocrine negative SCLC is mostly RB1 WT and may be sensitive to CDK4/6 inhibition. bioRxiv 2019;516351. [Google Scholar]
- 14. Otterson GA, Kratzke RA, Coxon A, Kim YW, Kaye FJ. Absence of p16INK4 protein is restricted to the subset of lung cancer lines that retains wildtype RB - PubMed. Oncogene 1994;11:3375–8. [PubMed] [Google Scholar]
- 15. Shimizu E, Coxon A, Otterson GA, Steinberg SM, Kratzke RA, Kim YW, et al. RB protein status and clinical correlation from 171 cell lines representing lung cancer, extrapulmonary small cell carcinoma, and mesothelioma - PubMed. Oncogene 1994;9:2441–8. [PubMed] [Google Scholar]
- 16. Jung H, Lee KS, Choi JK. Comprehensive characterisation of intronic mis-splicing mutations in human cancers. Oncogene 2021;40:1347–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaye FJ. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene 2002;21:6908–14. [DOI] [PubMed] [Google Scholar]
- 18. Yuan J, Knorr J, Altmannsberger M, Goeckenjan G, Ahr A, Scharl A, et al. Expression of p16 and lack of pRB in primary small cell lung cancer. J Pathology 1999;189:358–62. [DOI] [PubMed] [Google Scholar]
- 19. Knudsen ES, Nambiar R, Rosario SR, Smiraglia DJ, Goodrich DW, Witkiewicz AK. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun Biology 2020;3:158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol 1996;16:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parry D, Bates S, Mann DJ, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J 1995;3:503–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mahajan A. Practical issues in the application of p16 immunohistochemistry in diagnostic pathology. Hum Pathol 2016;51:64–74. [DOI] [PubMed] [Google Scholar]
- 23. Beasly M, Brambilla E, MacMahon H, Osamura R, Papotti M, Rekhtman N, et al. Small cell lung carcinoma. In:Board TWC of TE, editor. In:Thoracic tumours WHO classification of tumours. 5th ed.Lyon, France: International Agency for Research on Cancer; 2021. p 139–43. [Google Scholar]
- 24. Baine MK, Hsieh M-S, Lai WV, Egger JV, Jungbluth AA, Daneshbod Y, et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol 2020;15:1823–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ganti AKP, Loo BW, Bassetti M, Blakely C, Chiang A, D'Amico TA, et al. Small cell lung cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19:1441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Gene Dev 2015;29:1447–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ferone G, Song J-Y, Krijgsman O, van der Vliet J, Cozijnsen M, Semenova EA, et al. FGFR1 oncogenic activation reveals an alternative cell of origin of SCLC in Rb1/p53 mice. Cell Rep 2020;30:3837–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quintanal-Villalonga A, Taniguchi H, Zhan YA, Hasan MM, Chavan SS, Meng F, et al. Multi-omic analysis of lung tumors defines pathways activated in neuroendocrine transformation. Cancer Discov 2021;11:3028–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y, Lu T, Li Z, Lu S. FGFR1 regulates proliferation and metastasis by targeting CCND1 in FGFR1 amplified lung cancer. Cell Adhes Migr 2020;14:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;6:2924–30. [PubMed] [Google Scholar]
- 33. Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gong J, Salgia R. Managing patients with relapsed small-cell lung cancer. J Oncol Pract 2018;14:359–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ito T, Matsubara D, Tanaka I, Makiya K, Tanei Z, Kumagai Y, et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci 2016;107:1527–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov 2016;6:353–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krook MA, Reeser JW, Ernst G, Barker H, Wilberding M, Li G, et al. Fibroblast growth factor receptors in cancer: genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Brit J Cancer 2021;124:880–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell 2018;33:853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maquat LE. Defects in RNA splicing and the consequence of shortened translational reading frames. Am J Hum Genet 1996;2:279–86. [PMC free article] [PubMed] [Google Scholar]
- 40. Horowitz JM, Park SH, Bogenmann E, Cheng JC, Yandell DW, Kaye FJ, et al. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc National Acad Sci U S A 1990;87:2775–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaye FJ, Kratzke RA, Gerster JL, Horowitz JM. A single amino acid substitution results in a retinoblastoma protein defective in phosphorylation and oncoprotein binding. Proc National Acad Sci U S A 1990;87:6922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mizuarai S, Machida T, Kobayashi T, Komatani H, Itadani H, Kotani H. Expression ratio of CCND1 to CDKN2A mRNA predicts RB1 status of cultured cancer cell lines and clinical tumor samples. Mol Cancer 2011;10:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen WS, Alshalalfa M, Zhao SG, Liu Y, Mahal BA, Quigley DA, et al. Novel RB1-loss transcriptomic signature is associated with poor clinical outcomes across cancer types. Clin Cancer Res 2019;25:4290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Malorni L, Piazza S, Ciani Y, Guarducci C, Bonechi M, Biagioni C, et al. A gene expression signature of retinoblastoma loss-of-function is a predictive biomarker of resistance to palbociclib in breast cancer cell lines and is prognostic in patients with ER positive early breast cancer. Oncotarget 2016;7:68012–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ertel A, Dean JL, Rui H, Liu C, Witkiewicz A, Knudsen KE, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell Cycle 2010;9:4153–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Witkiewicz AK, Ertel A, McFalls J, Valsecchi ME, Schwartz G, Knudsen ES. RB-pathway disruption is associated with improved response to neoadjuvant chemotherapy in breast cancer. Clin Cancer Res 2012;18:5110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Knudsen ES, Pruitt SC, Hershberger PA, Witkiewicz AK, Goodrich DW. Cell cycle and beyond: exploiting new RB1 controlled mechanisms for cancer therapy. Trends Cancer 2019;5:308–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Offin M, Chan JM, Tenet M, Rizvi HA, Shen R, Riely GJ, et al. Concurrent RB1 and TP53 alterations define a subset of EGFR-mutant lung cancers at risk for histologic transformation and inferior clinical outcomes. J Thorac Oncol 2019;14:1784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rekhtman N, Pietanza MC, Hellmann MD, Naidoo J, Arora A, Won H, et al. Next-generation sequencing of pulmonary large cell neuroendocrine carcinoma reveals small cell carcinoma–like and non–small cell carcinoma–like subsets. Clin Cancer Res 2016;22:3618–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Thunnissen E, Borczuk AC, Flieder DB, Witte B, Beasley MB, Chung J-H, et al. The use of immunohistochemistry improves the diagnosis of small cell lung cancer and its differential diagnosis. An international reproducibility study in a demanding set of cases. J Thorac Oncol 2017;12:334–46. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available in a repository (cBioPortal for Cancer Genomics) that can be accessed via a DOI link upon request.