Abstract
The specific pair of heat shock protein 70 (Hsp70) and Hsp40 constitutes an essential molecular chaperone system involved in numerous cellular processes, including the proper folding/refolding and transport of proteins. Hsp40 family members are characterized by the presence of a conserved J-domain (JD) that functions as a co-chaperone of Hsp70. Tumorous imaginal disc 1 (Tid1) is a tumor suppressor protein belonging to the DNAJA3 subfamily of Hsp40 and functions as a co-chaperone of the mitochondrial Hsp70, mortalin. In this work, we performed nuclear magnetic resonance spectroscopy to determine the solution structure of JD and its interaction with the glycine/phenylalanine-rich region (GF-motif) of human Tid1. Notably, Tid1-JD, whose conformation was consistent with that of the DNAJB1 JD, appeared to stably interact with its subsequent GF-motif region. Collectively with our sequence analysis, the present results demonstrate that the functional and regulatory mode of Tid1 resembles that of the DNAJB1 subfamily members rather than DNAJA1 or DNAJA2 subfamily proteins. Therefore, it is suggested that an allosteric interaction between mortalin and Tid1 is involved in the mitochondrial Hsp70/Hsp40 chaperone system.
Keywords: DNAJ family, GF-motif, Hsp40, Hsp70, J-domain, Nuclear magnetic resonance (NMR), Tid1
INTRODUCTION
Molecular chaperones, essentially including heat shock proteins (Hsps), are recognized as a protein quality control machinery that operates to ensure cellular proteostasis (i.e., protein homeostasis) (1-3). The specific pair of Hsp70 and Hsp40, including their bacterial orthologs, DnaK and DnaJ, respectively (4), is one of the canonical chaperone systems that is ubiquitously present in all living organisms and interacts with many different substrates, via an adenine nucleotide-dependent cycle (5-7). In this machinery, Hsp70, which contains ATPase and substrate-binding domains, functions as a core chaperone, whereas Hsp40 acts as a co-chaperone of Hsp70. Hsp40 recruits polypeptide substrates to Hsp70, stimulates the hydrolysis of ATP bound to Hsp70, and stabilizes Hsp70 substrate interactions (8-10).
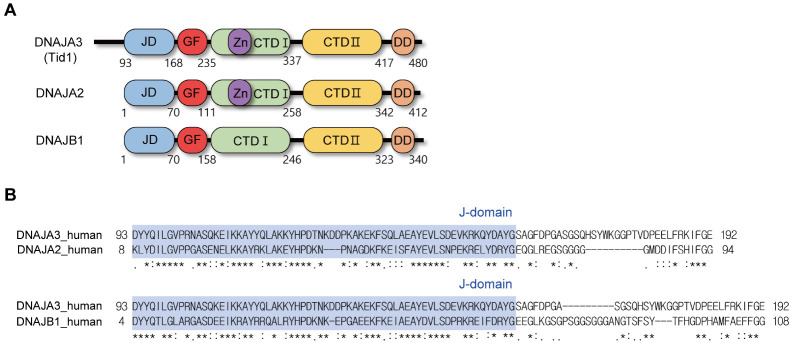
The Hsp40 family proteins generally share several characteristic domains/motifs, including a J-domain (JD), a glycine/phenylalanine-rich region (GF-motif), a cysteine repeat region containing a zinc-finger-like motif, and substrate-binding domains. Particularly, the highly conserved JD is recognized as a principle region of Hsp40 for its interaction with Hsp70, whereas the other regions are presumed to facilitate the targeting of Hsp70 chaperones to specific substrates (11). Both the overall primary sequences and the composition and organization of structural domains vary in individual Hsp40 proteins. For instance, in humans, Hsp40s comprise a variety of subfamilies with more than 40 different members, which vary in their substrate selectivity and individual functions in cells. Based on the structural components, the Hsp40 family is divided into three groups: DNAJA, DNAJB, and DNAJC (8). The former two group members, including DnaJ in bacteria (4) commonly share the JD at their N-terminus (Fig. 1A), whereas the JD is present elsewhere in the DNAJC proteins (12). The N-terminal JD of the DNAJA proteins is followed by a GF-motif, two homologous β-barrel domains containing a zinc-finger-like region in the first sequence, and a C-terminal dimerization domain. The DNAJA members are also subdivided into four subfamilies, from DNAJA1 to DNAJA4. The structural components of the DNAJB group are similar to those of DNAJA, but lack the zinc-finger-like region.
Fig. 1.
Sequence analysis of Tid1. (A) Schematic diagram for comparison of domain organization in DNAJA2, DNAJA3 (Tid1), and DNAJB1 subfamily of Hsp40: JD, J-domain; GF, G/F-rich region; Zn, zinc-finger-like region; CTD-I, C-terminal domain I; CTD-II, C-terminal domain II; DD, dimerization domain. (B) Pairwise sequence alignment of human DNAJA3 (Tid1) with DNAJA2 and DNAJB1 in the region encompassing JD and GF-motif.
Tumorous imaginal disc 1 (Tid1) was identified as a tumor suppressor that is involved in diverse cellular processes, in both the cytosol and mitochondria, that include imaginal discs differentiation, T cell development, and mitochondrial apoptosis signaling (13-16). However, as Tid1 has an N-terminal signal peptide for mitochondrial targeting (Fig. 1A), the protein localizes to the mitochondria and functions as an Hsp40 that participates in the co-chaperone activity of mortalin, the mitochondrial Hsp70 (17, 18). Mortalin and Tid1 constitute the only Hsp70/Hsp40 chaperone system in mitochondria, and in particular, the regulation of apoptosis in mitochondria is initiated by the interaction of Tid1 with mortalin via its JD.
As the domain organization of Tid1 is comparable to that of the DNAJA2-subfamily proteins, except for the presence of the N-terminal mitochondrial targeting sequence, Tid1 is classified as in the DNAJA3 subfamily of Hsp40 (Fig. 1A). However, whether the detailed structure and interdomain interactions, which would provide a molecular basis for substrate selectivity and Hsp70 interaction, are consistent with those of other related proteins remains to be investigated. In particular, it was recently elucidated that the interdomain interaction of JD with the GF-motif differs between the DNAJA2 and DNAJB1 subfamilies (19). Therefore, in the present study, we aimed to determine the three-dimensional structure of Tid1-JD and its interaction with the GF-motif in solution using nuclear magnetic resonance (NMR) spectroscopy.
RESULTS
Sequence analysis of Hsp40 proteins
A recent report by Faust et al. (18) identified that the structural mode of the interaction with Hsp70 is distinct between a DNAJA and a DNAJB Hsp40 family members, i.e., unlike in DNAJA2, the GF-motif in DNAJB1 has a C-terminal α-helix that interferes with the binding of JD to Hsp70. Meanwhile, we have previously identified an α-helix (residues 183-188) at the C-terminus of the GF-motif of Tid1 (20). Therefore, as the DNAJA3 member Tid1, whose domain organization is comparable to that of DNAJA2 (Fig. 1A), might also be close to DNAJB1, we analyzed their sequence homology in the region of JD and GF-motif (Fig. 1B). The sequence comparison of Tid1 in JD revealed 55.6% and 58.7% sequence identity with DNAJA2 and DNAJB1, respectively. Although the sequence variation in the GF-motif was significantly higher than that in the JD, the sequence identity in the GF-motif between Tid1 and DNAJB1 (26.5%) appeared to be higher than that between Tid1 and DNAJA2 (21%). Accordingly, when the sequence homology of Tid1 was analyzed, including both the JD and GF-motif, the sequence identities of Tid1 to DNAJA2 and DNAJB1 were 44% and 48%, respectively. In summary, the amino acid sequence analysis implied that the structure and functional regulation of the Tid1 JD and GF-motif might resemble those of DNAJB1, rather than those of DNAJA2. Therefore, we investigated the solution structure of Tid1 JD and its interaction with the GF-motif by NMR spectroscopy.
Solution structure of the Tid1 JD
The determined NMR structure of the Tid1 JD is shown in Fig. 2, and the structural statistics are shown in Supplementary Table 1. The root mean square deviation (RMSD) of the 20 lowest-energy structures from their average structure was 0.39 ± 0.08 Å for backbone and 0.97 ± 0.10 Å for all heavy atoms, indicating a good convergence of the ensemble structure (Fig. 2A). Overall, the structure showed a typical JD fold composed of four α-helices: α1 (residues 94-99), α2 (105-118), α3 (131-145), and α4 (149-157). The HPD motif, which is known to contain key residues for the interaction of a JD in HSP40 with its cognate Hsp70, adopted a flexible loop conformation between α2 and α3 (Fig. 2B). Although no JD structure was available for DNAJA2, the overall conformation of the Tid1 JD showed a well-matched structural superimposition on the known DNAJB1 JDGF structure (Fig. 2C), as expected from the sequence similarity (Fig. 1B). The RMSD between the JD structures of Tid1 and DNAJB1 for superposition of all Cα atom pairs in the 89-159 region of Tid1 was 2.236. Therefore, we further investigated whether the GF-motif interacts with JD in Tid1, as in DNAJB1.
Fig. 2.
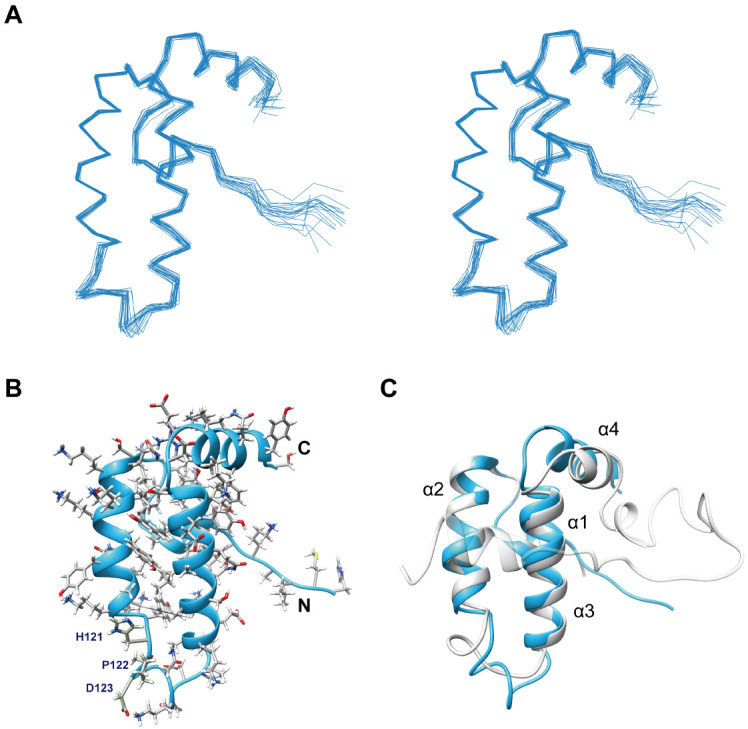
Solution structure of Tid1 JD. (A) Stereo representation of ensemble structure showing superposition of backbone traces for the 20 lowest-energy conformers determined by NMR. (B) Ribbon representation of the lowest-energy structure. Sidechains are presented by stick models and labeled for the HPD motif. (C) Structural comparison between the Tid1 JD (sky blue) and the DNAJB1 JDGF (gray; PDB accession code 6z5n).
Interaction between the JD and GF-motif
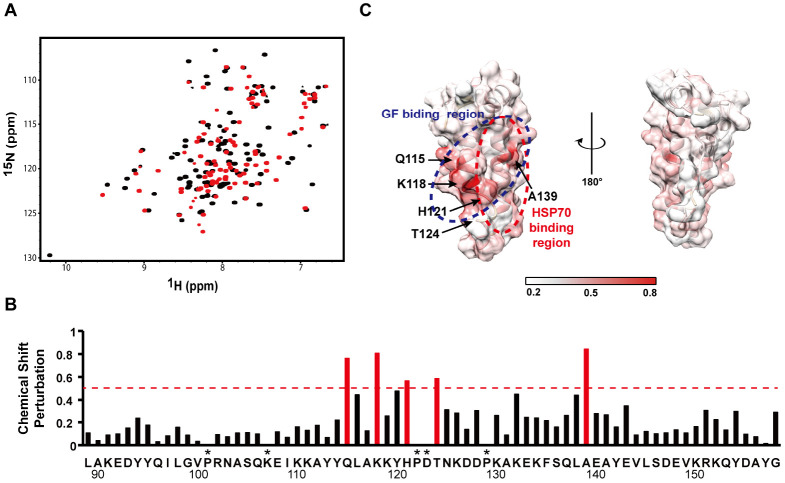
The 2D [1H-15N]HSQC spectrum of JD showed an overall inconsistency with that of JDGF (Fig. 3A), implying alteration of the chemical environment of the JD, probably due to direct interaction with the GF-motif. The most significantly affected residues, which were defined by weighted chemical shift perturbations (CSPs) exceeding 0.45 ppm (Fig. 3B), included Q115, K118, H121, and A139. These amino acids were located mainly on the surface of the α2, α3, and α2-α3 loop regions (Fig. 3C), which mediate the Hsp70 interaction in the JD (19). In particular, the significant CSPs at H121 and T124 suggest that the HPD motif (Fig. 2B) is likely blocked by the GF-motif interaction.
Fig. 3.
Identification of GF-motif-interacting region in JD. (A) Comparison of 2D [1H-15N]HSQC NMR spectrum between JD (red) and JDGF (black). (B) Chemical shift perturbation (CSP) in JD region caused by the GF-motif interaction. The most significantly (> 0.45) perturbed chemical shifts are indicated in red. (C) Mapping of the GF-motif-interaction site on the surface of the JD structure. The GF-motif- and Hsp70-interacting regions are indicated by dotted circles in blue and red, respectively. CSP values are coordinated from white to red colors. Asterisks indicate the residues for which CSPs were not available due to missing assignments.
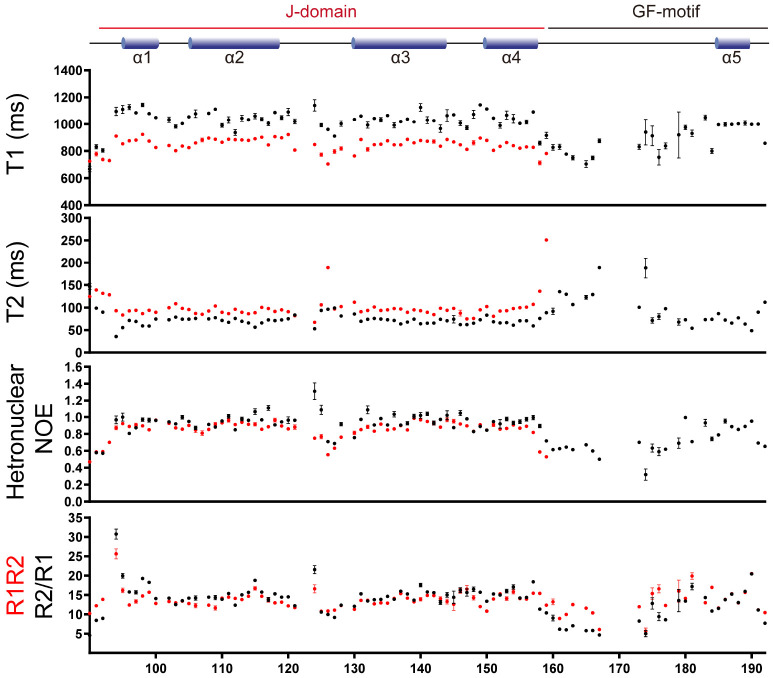
It was confirmed by comparing NMR relaxation dynamics between the JD and JDGF that the overall CSPs are not attributable to a global conformational change of JD (Fig. 4). As expected from the larger size, which increases the rotational correlation time, JDGF showed higher values of longitudinal relaxation time (T1) and lower values of transverse relaxation time (T2) than JD in individual residues. However, the overall patterns of the T1 and T2 traces along the sequence (i.e., relatively high values in helical regions) showed no significant perturbations, except for a few residues in the termini and the flexible loop regions. The hetero-[1H-15N] NOE results were also similar and in good agreement with the determined structure of the JD (i.e., consecutively high values in helical regions). This finding supports the hypothesis that the overall conformation and internal dynamics of the JD, in principle, could be maintained in JDGF. Collectively, the observed CSPs could be attributed to a rigid-body interaction between the JD and the GF-motif. In particular, the region around α5 in the GF-motif could be regarded as the JD-interacting site, as the other part (α4-α5 loop) of the GF-motif in JDGF showed high flexibility, indicated by low T1 and heteronuclear NOE values and high T2 values. The relaxation rates R1 and R2 (i.e., reciprocals of T1 and T2) of JDGF were further analyzed to determine whether α5 could accomplish a stable binding or whether it makes a dynamic or transient contact with the JD. Although the R2/R1 ratio analysis is often used as a simple approach to estimate global correlation time, it cannot distinguish, in principle, between chemical exchange and motional anisotropy (21). In contrast, the product of R1R2, which increases upon the occurrence of chemical exchange, is recognized as a more efficient parameter for estimating the μs-to-ms motional dynamics with attenuating the effect of motional anisotropy (22). As shown in Fig. 4, the average R2/R1 and R1R2 values for the residues showing heteronuclear NOE values higher than 0.7 (22) were 14.8 ± 3.3 and 14.0 ± 2.3, respectively. The smaller deviation of R1R2 (± 2.3) than that of R2/R1 (± 3.3) is likely attributed to the attenuation of motional anisotropy. For example, among the four outlier residues (Y94, T124, D181, and F190) that showed considerable (> 4.6) deviations of R2/R1, R1R2, or both, T124 could be excluded from possible candidates for chemical exchange (i.e., motional dynamics), as it showed highly attenuated R1R2 deviation despite its considerable R2/R1 deviation, indicating motional anisotropy. In contrast, the high R1R2 values of Y94 and F190, which are located at the edges of α1 and α5, respectively, could reflect their motional dynamics, and likely occur due to a general instability at the boundary regions of the helices. Likewise, a localized intrinsic fluctuation at a flexible linker region was probably due to the high R1R2 value with attenuated R2/R1 observed for D181 at the α4-α5 loop. In summary, no significant chemical exchange definable as a dynamic interaction was identified in the present analysis of the relaxation rate. In particular, given that α5 is the JD-interacting site in the GF-motif, the absence of a motional process associated with a chemical exchange in α5 indicates that the GF-motif would stably bind to the JD in Tid1.
Fig. 4.
NMR relaxation dynamics of Tid1 JD and JDGF. Red and black circles in the T1, T2, and heteronuclar NOE analysis indicate the values for JD and JDGF, respectively. The R1R2 (black) and R2/R1 (red) values are for JDGF. Mean values of R1R2 (14.0) and R2/R1 (14.8) are indicated by dashed lines in black and red, respectively.
Structural similarity of Tid1 with DNAJB1
DNAJB proteins constitute a major class of Hsp40 proteins in humans. The aforementioned research by Faust et al. identified that an autoinhibitory mode of DNAJB1—JD binding to Hsp70 is intrinsically blocked by the GF-motif, which can be released upon allosteric interaction (19). The authors also suggested that this regulatory mode, which was not present in DNAJA2, is unique to the DNAJB class, as it is essential for the specific function of DNAJB1-Hsp70 in disaggregating amyloid fibers by controlling substrate targeting. However, our present analysis of the DNAJA3 protein Tid1 indicated that its amino acid sequence is more homologous to DNAJB than DNAJA class proteins (Fig. 1B), despite its similarity to DNAJA2 in domain organization (Fig. 1A). In addition, all structural features and mutual interaction of the JD and GF-motif of Tid1 showed a close resemblance to those of DNAJB2. In particular, as the C-terminal helix of the GF-motif binds to the putative Hsp70-interacting region (i.e., α2-α3 region) of JD, as in DNAJB1, the interaction of Tid1 with Hsp70 via its JD is also expected to be blocked by the GF-motif. It remains to be further investigated whether the regulatory and functional modes of Tid1 would be comparable with those of DNAJB1. However, given that Tid1 is the only member of Hsp40 in mitochondria that interacts with the sole mitochondrial Hsp70, mortalin, we expect that the present results will contribute to catalyzing related research, to provide insight into the functional evolution of the Hsp40/Hsp70 chaperone system in mitochondria.
In the present study, we found that the solution structure of the Tid1 JD was consistent with the known JD structure of DNAJB1. In addition, as the GF-motif stably binds to JD in Tid1, as in DNAJB1, it is postulated that the JD interaction with Hsp70 is intrinsically blocked and require an allosteric interaction with Hsp70 to release that inhibition. The structural resemblance of Tid1 to DNAJB1 suggests that the mitochondrial Hsp40/Hsp70 chaperone system may have evolved to employ the specific functional mode of the major class of Hsp40 and DNAJB proteins.
Data availability statement
NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank under the following accession codes: 36474, 26322 for Tid1-JD and Tid1-JDGF, respectively. The structure of Tid1-JD has been deposited to the Protein Data Bank (PDB) under accession code 7X89.
MATERIALS AND METHODS
Protein preparation
Recombinant proteins corresponding to the Tid1 JD (residues 89-159) and its extended form, JDGF (residues 89-192), which encompasses both the JD and GF-motif, were prepared for NMR, as described previously (20). Briefly, DNA fragments encoding the JD and JDGF were subcloned from the cDNA of human Tid1 into the pCold-I vector plasmids. The Escherichia coli BL21(DE3)pLysS strain transformed with the recombinant plasmids was cultured in M9 minimal media supplemented with [13C]glucose and [15N]NH4Cl, as the sole source of carbon and nitrogen, respectively. The expressed proteins carrying a hexa-histidine (H6) tag were purified via the sequential application of nickel affinity and anion-exchange chromatography. The purified proteins were treated with protease factor-Xa for H6 tag removal, followed by final purification via gel-permeation chromatography.
NMR measurement
The isotope [13C/15N]-enriched protein samples for NMR were dissolved in 20 mM Tris-HCl buffer (pH 8.0) containing 150 mM NaCl, 1 mM dithiothreitol, and 7 % (v/v) D2O. Two-dimensional HSQC spectra and three-dimensional (3D) NMR data were recorded at 25°C on a Bruker 800 or 900 MHz NMR spectrometer equipped with a cryoprobe. The measured NMR spectra were processed using the NMRPipe/NMRDraw software (23) and analyzed with the NMRView program (24). Based on the previously achieved backbone NMR assignments (20), the side-chain assignments of the JD were performed using 3D HBHA(CO)NH, HCCH-COSY, and 15N-HCCH-TOCSY spectra. The combined chemical shift perturbations were represented by [(ΔδNH2 + ΔδN2/25)/2]1/2, where ΔδNH and ΔδN are the chemical shift changes of backbone amide proton (NH) and amide (N), respectively.
Structural determination
Distance restraints for structure calculation were obtained from 3D [15N]- and [13C]-edited NOESY-HSQC NMR spectra, of which NOE cross-peaks were assigned by the auto-assignment function of CYANA 2.1 (25). Dihedral ϕ and φ angle restraints were derived from the prediction of TALOS+ (26) using the previously assigned backbone NMR chemical shifts (20). Initial structures of the Tid1 JD were generated with CYANA 2.1, followed by refinements with the statistical torsion angle potential energy functions (27) using the Charmm program (28). The final 20 structures with the lowest energy were validated by three types of scores:: DOPE (29), normalized DOPE, and dDFIRE (30). Ramachandran plot appearance was measured using MolProbity (31).
Relaxation analysis
The longitudinal relaxation rate (R1) and the transverse relaxation rate (R2) of backbone 15N nuclei, as well as the heteronuclear NOEs between amide 15N and 1H nuclei were assessed using established HSQC-based NMR pulse sequences. Relaxation experiments were performed at 900 MHz (21.2 Tesla). T1 values were measured from 1H-15N correlation spectra recorded with relaxation evolution delays of 50, 100, 200, 300, 500, 700, 900, and 1200 ms. A 3 s delay was used between scans. T2 values were measured from 1H-15N correlation spectra recorded using relaxation evolution delays of 16.96, 33.92, 67.84, 101.76, 135.68, 169.6, 203.52, and 254.4 ms. A delay of 2 s was used between scans. The steady-state 1H-15N heteronuclear NOE values were determined from the peak ratios observed between the two spectra, which were collected with or without pre-saturation of the proton dimension. A delay of 3 s was used between the scans.
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1A5A2015541, 2019R1F1A1057427 and 2019R1A2C1004883) and by ”Regional Inovation Strategy (RIS)“ through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (MOE) (2021RIS-001). The use of NMR was supported by the Korea Basic Science Institute under the R&D program (Project No. C140440), supervised by the Ministry of Science and ICT.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Jo KS, Kim JH, Ryu KS, et al. Unique unfoldase/aggregase activity of a molecular chaperone Hsp33 in its holding-inactive state. J Mol Biol. 2019;431:1468–1480. doi: 10.1016/j.jmb.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 2.Keum M, Ito D, Kim MS, et al. Molecular effects of elongation factor Ts and trigger factor on the unfolding and aggregation of elongation factor Tu induced by the prokaryotic molecular chaperone Hsp33. Biology (Basel) 2021;10:1171. doi: 10.3390/biology10111171.6f6a22656d4b4dc8aa63c07bce5f7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim MJ, Kim J, Im JS, Kang I, Ahn JK. Hepatitis B virus X protein enhances liver cancer cell migration by regulating calmodulin-associated actin polymerization. BMB Rep. 2021;54:614–619. doi: 10.5483/BMBRep.2021.54.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yochem J, Uchida H, Sunshine M, Saito H, Georgopoulos CP, Feiss M. Genetic analysis of two genes, dnaJ and dnaK, necessary for Escherichia coli and bacteriophage lambda DNA replication. Mol Gen Genet. 1978;164:9–14. doi: 10.1007/BF00267593. [DOI] [PubMed] [Google Scholar]
- 5.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B. The Hsp70 chaperone network. Nat Rev Mol Cell Biol. 2019;20:665–680. doi: 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- 8.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell stress & chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:SFAEOD>2.3.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig EA, Marszalek J. How do J-proteins get Hsp70 to do so many different things? Trends Biochem Sci. 2017;42:355–368. doi: 10.1016/j.tibs.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyr DM, Langer T, Douglas MG. DnaJ-like proteins: molecular chaperones and specific regulators of Hsp70. Trends Biochem Sci. 1994;19:176–181. doi: 10.1016/0968-0004(94)90281-X. [DOI] [PubMed] [Google Scholar]
- 11.Kelley WL. The J-domain family and the recruitment of chaperone power. Trends Biochem Sci. 1998;23:222–227. doi: 10.1016/S0968-0004(98)01215-8. [DOI] [PubMed] [Google Scholar]
- 12.Perrody E, Cirinesi AM, Desplats C, et al. A bacteriophage-encoded J-domain protein interacts with the DnaK/Hsp70 chaperone and stabilizes the heat-shock factor sigma32 of Escherichia coli. PLoS Genet. 2012;8:e1003037. doi: 10.1371/journal.pgen.1003037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn BY, Trinh DL, Zajchowski LD, Lee B, Elwi AN, Kim SW. Tid1 is a new regulator of p53 mitochondrial translocation and apoptosis in cancer. Oncogene. 2010;29:1155–1166. doi: 10.1038/onc.2009.413. [DOI] [PubMed] [Google Scholar]
- 14.Proft J, Faraji J, Robbins JC, et al. Identification of bilateral changes in TID1 expression in the 6-OHDA rat model of Parkinson's disease. PloS one. 2011;6:e26045. doi: 10.1371/journal.pone.0026045.81f4a8e5c9a043268d7ae13d6d7df3a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trinh DL, Elwi AN, Kim SW. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget. 2010;1:396–404. doi: 10.18632/oncotarget.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C, Taslima F, Abdelhamid M, et al. Beta-Amyloid increases the expression levels of Tid1 responsible for neuronal cell death and amyloid beta production. Mol Neurobiol. 2020;57:1099–1114. doi: 10.1007/s12035-019-01807-2. [DOI] [PubMed] [Google Scholar]
- 17.Wang SF, Huang KH, Tseng WC, et al. DNAJA3/Tid1 is required for mitochondrial dna maintenance and regulates migration and invasion of human gastric cancer cells. Cancers 12. 2020;(Basel):3463. doi: 10.3390/cancers12113463.eddc325e5326479cbaa2ec6d573e5e74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Syken J, De-Medina T, Munger K. TID1, a human homolog of the Drosophila tumor suppressor l(2)tid, encodes two mitochondrial modulators of apoptosis with opposing functions. Proc Natl Acad Sci. 1999;96:8499–8504. doi: 10.1073/pnas.96.15.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Faust O, Abayev-Avraham M, Wentink AS, et al. HSP40 proteins use class-specific regulation to drive HSP70 functional diversity. Nature. 2020;587:489–494. doi: 10.1038/s41586-020-2906-4. [DOI] [PubMed] [Google Scholar]
- 20.Jo KS, Sim DW, Kim EH, et al. Backbone NMR assignments of a putative p53-binding domain of the mitochondrial Hsp40, Tid1. J Kor Magn Reson Soc. 2018;22:64–70. [Google Scholar]
- 21.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 22.Kneller JM, Lu M, Bracken C. An effective method for the discrimination of motional anisotropy and chemical exchange. J Am Chem Soc. 2002;124:1852–1853. doi: 10.1021/ja017461k. [DOI] [PubMed] [Google Scholar]
- 23.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 24.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 25.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TR, Yang JS, Shin S, Lee J. Statistical torsion angle potential energy functions for protein structure modeling: a bicubic interpolation approach. Proteins. 2013;81:1156–1165. doi: 10.1002/prot.24265. [DOI] [PubMed] [Google Scholar]
- 28.Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H, Skolnick J. Protein structure prediction by pro-Sp3-TASSER. Biophys J. 2009;96:2119–2127. doi: 10.1016/j.bpj.2008.12.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams CJ, Headd JJ, Moriarty NW, et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 2018;27:293–315. doi: 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NMR chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank under the following accession codes: 36474, 26322 for Tid1-JD and Tid1-JDGF, respectively. The structure of Tid1-JD has been deposited to the Protein Data Bank (PDB) under accession code 7X89.