Abstract
Ischemic stroke (IS) is a severe disease with a high disability, recurrence, and mortality rates. Autophagy, a highly conserved process that degrades damaged or aging organelles and excess cellular components to maintain homeostasis, is activated during IS. It influences the blood–brain barrier integrity and regulates apoptosis. Circular RNAs (circRNAs) are novel non-coding RNAs involved in IS-induced autophagy and participate in various pathological processes following IS. In addition, they play a role in autophagy regulation. This review summarizes current evidence on the roles of autophagy and circRNA in IS and the potential mechanisms by which circRNAs regulate autophagy to influence IS injury. This review serves as a basis for the clinical application of circRNAs as novel biomarkers and therapeutic targets in the future.
Keywords: ischemic stroke, autophagy, circRNA, biomarkers, therapeutic targets
Introduction
Stroke is a leading cause of death and disability worldwide (1) and can be classified as ischemic stroke (IS) or hemorrhagic stroke (2). The major type is IS, accounting for 71% of cases (2). During IS, ischemia and hypoxia cause neuronal and glial anoxic depolarization (3), which increases extracellular levels of glutamate, leading to excess calcium influx and release of calcium from intracellular stores (4). Increased intracellular calcium contributes to neuronal nitric oxide synthase activation with consequent free radical production and the initiation of cell death processes, including apoptosis, necrosis, necroptosis, and autophagy. Current effective treatments for IS include restoration of blood flow through intravenous thrombolysis and neuroscientific intravascular recanalization, both of which reduce disability (2); however, these treatment methods are still limited owing to the limited time window, numerous contraindications (5), and high risk of hemorrhagic complications (6).
Autophagy is activated to varying degrees after IS to restore neuronal homeostasis (2, 7). Autophagy functions in IS by sequestering damaged or aged organelles, superfluous proteins, and cellular components into double membrane-bound vesicles, delivering cytoplasmic cargo to the lysosome, to which it subsequently fuses to form an autolysosome, finally leading to digestion and recycling (8). Autophagy presents a dual effect following ischemic insult. Mild to moderate induction of autophagy can be protective in IS (9), whereas an excessive increase in autophagic activity might be harmful owing to the cytosolic accumulation of autophagosomes and enhanced degradation of essential cellular components (10). Autophagy is divided into two groups according to the role it plays in IS: maladaptive and adaptive autophagy (11).
Circular RNAs are a novel type of non-coding RNAs (12) with a stable and evolutionally conserved covalent loop structure (13). Previous studies have demonstrated that circRNAs are often specifically expressed in tissue and developmental stages (14) and are highly expressed in the mammalian brain (15). CircRNAs are upregulated during neuronal differentiation and are highly enriched in synapses (16). The role of circRNAs has been identified in several human diseases, including neurological disorders, cardiovascular diseases, diabetes mellitus, chronic inflammatory diseases, and cancer (17–21). Interestingly, circRNAs function in ischemic brain injury (22–25); therefore, they are potential biomarkers for IS and may serve as new therapeutic targets.
CircRNAs
The biogenesis and functions of circRNAs
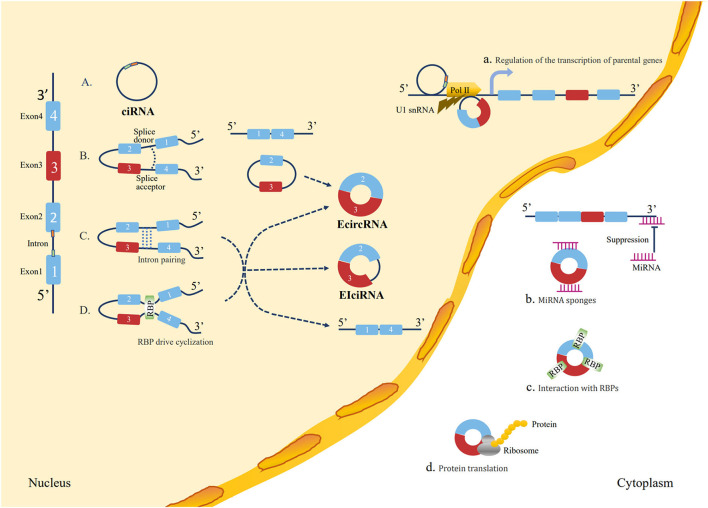
Circular RNAs are a typical class of non-coding RNAs with a covalent loop structure (13), which were first discovered in RNA viruses in 1979 (57) and were originally hypothesized to be a by-product of mis-splicing (58). Due to different splicing processes, circRNAs were divided into three main types, including exonic circRNAs (ecircRNAs), exon-intron circRNAs (EIciRNAs), and circular intronic RNAs (ciRNAs) (59). However, with advances in experimental technology, emerging evidence has indicated that circRNAs are implicated in the occurrence and development of many diseases, such as cancer (60), cardiovascular disorders (61), neurological disorders (62), metabolic disorders (63), and immunological disorders (64). The functions of circRNAs are as follows: acting as a sponge of miRNA or competitive endogenous RNAs (ceRNAs), interacting with RNA-binding proteins (RBPs) and mRNAs, regulating transcription or alternative splicing, and protein translation (65). The biogenesis and functions of circRNAs are illustrated in Figure 1.
Figure 1.
Loop formation mechanisms and biological functions of circRNAs. Loop formation mechanisms: (A) ciRNA formation: the elements near the splice site escape debranching stably so that the intron lariat is formed from the splicing reaction. (B) Lariat-driven circularization: the 5′ splice donor of exon 1 and the 3′ splice acceptor of exon 4 link up end-to-end by exon skipping and form an exon-containing lariat structure. Finally, the ecircRNA forms after introns are removed. (C) Intron pairing-driven circularization: direct base pairing of introns forms a circulation structure, thereby forming ecircRNA or EIciRNA after intron removal. (D) RBP-driven cyclization: RBPs bridge two flanking introns close together and then remove introns to form circRNAs. Biological functions: (a) Regulation of the transcription of parental genes: circRNAs play a regulatory role in the transcription of their parent coding genes. (b) Function as miRNA “sponge”: circRNAs contain a common miRNA response element (MRE) that can bind to miRNA and prevent them from interacting with mRNA. (c) Interaction with RBPs: circRNAs can bind to RBPs to regulate mRNA expression by altering the splicing pattern or mRNA stability. (d) Protein translation: circRNAs have coding potential and can be translated into proteins with ribosomes.
CircRNAs participating in pathological processes in ischemic stroke: Apoptosis, inflammation, oxidative stress, angiogenesis, and autophagy
Ischemic stroke is a complex multifactorial disease in which the cascade response to ischemia is not a single linear process but often involves parallel or cross-interacting mechanisms and events (66, 67). CircRNAs are involved in various mechanisms and events in the ischemic cascade through both the regulations of host genes at the transcriptional level and the ceRNA mechanisms (68), such as apoptosis, inflammation, oxidative stress, angiogenesis, and autophagy. We summarize the evidences of circRNA regulatory roles in these pathophysiological changes of ischemic stroke (Table 1). Yang et al. found that circTTC3 levels were increased in oxygen-glucose deprivation (OGD)-treated astrocytes and middle cerebral artery occlusion/reperfusion (MCAOR) mice (50). The depletion of circTTC3 can decrease cerebral infarction, brain edema, and apoptosis, and attenuates cerebral infarction via the miR-372-3p/TLR4 axis (50). Zhang et al. found that circ_USP36 levels were increased in the serum of atherosclerosis patients and in oxidized low-density lipoprotein (ox-LDL)-stimulated human umbilical vein endothelial cells (HUVECs) (32). Circ_USP36 overexpression can trigger apoptosis and inflammation via the miR-197-3p/ROBO1 axis to promote atherosclerosis, which may finally result in IS (32). Yang et al. found that circPHKA2 levels were decreased in the venous blood of acute IS patients and OGD-induced human brain microvascular endothelial cells (HBMECs) (37). Overexpression of circPHKA2 can inhibit apoptosis, endoplasmic reticulum (ER) stress, and oxidative stress in HBMECs after OGD via the miR-574-5p/SOD2 axis (37). Bai et al. found that circFUNDC1 levels were increased in OGD-treated HBMECs (44). CircFUNDC1 knockdown can promote OGD-blocked cell viability, migration, and angiogenesis of HBMECs by inhibiting phosphatase and tensin homolog (PTEN) by enriching miR-375 (44). Yang et al. found that circ-FoxO3 levels were increased in mouse models with MCAO/R (23). Circ-FoxO3 protected against OGD/R-stimulated endothelial barrier damage and tMCAO-induced BBB collapse in mice by upregulating autophagy (23). Yang et al. found that circSCMH1 levels were significantly decreased in the plasma of patients with acute ischemic stroke, and in the plasma and the peri-infarct cortex of photothrombotic stroke mice (55). CircSCMH1 overexpression contributes to functional recovery post-stroke by enhancing neuronal plasticity, while inhibiting glial activation and peripheral immune cell infiltration (55). Interestingly, a single circRNA can influence more than one pathological process. Circ_0003204 is increased in ox-LDL-induced HUVECs. Circ_0003204 silencing weakens ox-LDL-induced cell viability inhibition, apoptosis, inflammatory response, and oxidative stress, but facilitates proliferation, migration, and angiogenesis (29, 33).
Table 1.
The evidence of CircRNA regulatory roles in the pathophysiological changes of ischemic stroke.
| Pathological process in IS | Cell line/treatment | Animal/disease model | DE CircRNA and expression | Regulating axis | Overexpression role | References | |
|---|---|---|---|---|---|---|---|
| Evidences from basic research | |||||||
| Apoptosis | Arterial SMCs, HEK-293, iPSC | / | Circ_ANRIL | Up | PES1 | Induce apoptosis | (28) |
| Transfection of siRNA | Detrimental | ||||||
| HCtAECs and THP-1 cells | / | Circ_0003204 | Up | MiR-188-3p/TRPC6 | Induce apoptosis | (29) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-942-5p/HDAC9 | Induce apoptosis | (30) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-491-5p/ICAM1 | Induce apoptosis | (31) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-197-3p/ROBO1 | Induce apoptosis | (32) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | Unknown | Induce apoptosis | (33) | |
| Ox-LDL | Detrimental | ||||||
| HBMEC-IM cells | / | Circ_0003423 | Down | MiR-589-5p/TET2 | Inhibit apoptosis | (34) | |
| Ox-LDL | Protective | ||||||
| HUVECs | / | Circ_0003423 | Down | MiR-142-3p/Sirtuin 3, SOD2 | Inhibit apoptosis | (35) | |
| Ox-LDL | Protective | ||||||
| mAS | Mouse | CircCTNNB1 | Down | MiR-96-5p/SRB1 | Inhibit apoptosis | (36) | |
| OGD/R | MCAO | Protective | |||||
| HBMECs | / | CircPHKA2 | Down | MiR-574-5p/SOD2 | Inhibit apoptosis | (37) | |
| OGD | Protective | ||||||
| SK-N-SH | / | Circ_0000647 | Up | MiR-126-5p/TRAF3 | Induce apoptosis | (38) | |
| OGD/R | Detrimental | ||||||
| HBMVECs | / | Circ-Memo1 | Up | MiR-17-5p/SOS1 | Induce apoptosis | (39) | |
| H/R | Detrimental | ||||||
| Apoptosis | HCN-2 | / | CircDlgap4 | Down | MiR-503-3p/NEGR1 | Inhibit apoptosis | (40) |
| OGD | Protective | ||||||
| HUVECs | Mouse | CircDlgap4 | Down | MiR-143/HECTD1 | Inhibit apoptosis | (41) | |
| I/R | I/R | Protective | |||||
| Astrocytes | Rats | Circ 0025984 | Down | MiR-143-3p/TET1/ORP150 | Inhibit apoptosis | (22) | |
| OGD | MCAO | Protective | |||||
| Brain-derived endothelial cells | Mouse | Circ_0072309 | Down | MiR-100/mTOR | Inhibit apoptosis | (42) | |
| OGD | MCAO | Protective | |||||
| HT22 cells | Mouse | CircCDC14A | Up | MiR-23a-3p/CXCL12 | Induce apoptosis | (43) | |
| OGD/R | MCAO | Detrimental | |||||
| HBMECs | / | CircFUNDC1 | Up | MiR-375/PTEN | Induce apoptosis | (44) | |
| OGD | Detrimental | ||||||
| HBMECs | / | CircPHC3 | Up | MiR-455-5p/TRAF3 | Induce apoptosis | (45) | |
| OGD | Detrimental | ||||||
| RGCs | Mouse | CGLIS3 | Up | MiR-203/unknown | Induce apoptosis | (46) | |
| OGD/R | tMCAO | Detrimental | |||||
| HT22 cells | Mouse | Circ-HECTD1 | Up | MicroRNA-133b/TRAF3 | Inhibit apoptosis | (47) | |
| OGD | MCAO | Protective | |||||
| HT22 cells | Mouse | Circ-HECTD1 | Up | MiR-27a-3p/FSTL1 | Inhibit apoptosis | (48) | |
| OGD/R | MCAO | Protective | |||||
| / | Mouse | Circ_008018 | Up | MiR-99a/Akt, GSK3β | Induce apoptosis | (49) | |
| MCAO | Detrimental | ||||||
| Astrocytes | Mouse | CircTTC3 | Up | MiR-372-3p/TLR4 | Induce apoptosis | (50) | |
| OGD | MCAO/R | Detrimental | |||||
| Oxidative Stress | mAS | Mouse | CircCTNNB1 | Down | MiR-96-5p/SRB1 | Inhibit oxidative stress | (36) |
| OGD/R | MCAO | Protective | |||||
| HBMECs | / | CircPHKA2 | Down | MiR-574-5p/SOD2 | Inhibit oxidative stress | (37) | |
| OGD | Protective | ||||||
| SK-N-SH | / | Circ_0000647 | Up | MiR-126-5p/TRAF3 | Induce oxidative stress | (38) | |
| OGD/R | Detrimental | ||||||
| Oxidative Stress | HBMVECs | / | Circ-Memo1 | Up | MiR-17-5p/SOS1 | Induce oxidative stress | (39) |
| H/R | Detrimental | ||||||
| HCtAECs and THP-1 cells | / | Circ_0003204 | Up | MiR-188-3p/TRPC6 | Induce oxidative stress | (29) | |
| Ox-LDL | Detrimental | ||||||
| N2a cells, neurons | Mouse | CircDlgap4 | Down | AUF1/NRF2 mRNA | Inhibit oxidative stress | (51) | |
| OGD | tMCAO | Protective | |||||
| Inflammation | HBMECs | / | Circ_ANRIL | Up | MiR-622 | Induce inflammation | (52) |
| OGD/R | Detrimental | ||||||
| HCtAECs and THP-1 cells | / | Circ_0003204 | Up | MiR-188-3p/TRPC6 | Induce inflammation | (29) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-942-5p/HDAC9 | Induce inflammation | (30) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-491-5p/ICAM1 | Induce inflammation | (31) | |
| Ox-LDL | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | MiR-197-3p/ROBO1 | Induce inflammation | (32) | |
| Ox-LDL | Detrimental | ||||||
| mAS | Mouse | CircCTNNB1 | Down | MiR-96-5p/SRB1 | Inhibit inflammation | (36) | |
| OGD/R | MCAO | Protective | |||||
| SK-N-SH | / | Circ_0000647 | Up | MiR-126-5p/TRAF3 | Induce inflammation | (38) | |
| OGD/R | Detrimental | ||||||
| HBMVECs | / | Circ-Memo1 | Up | MiR-17-5p/SOS1 | Induce inflammation | (39) | |
| H/R | Detrimental | ||||||
| HCN-2 | / | CircDlgap4 | Down | MiR-503-3p/NEGR1 | Inhibit inflammation | (40) | |
| OGD | Protective | ||||||
| HBMECs | / | Circ_0006768 | Down | MiR-222-3p/VEZF1 | Inhibit inflammation | (53) | |
| OGD/R | Protective | ||||||
| Angiogenesis | HBMEC-IMs | / | Circ_0003423 | Down | MiR-589-5p/TET2 | Promote angiogenesis | (34) |
| Ox-LDL | Protective | ||||||
| HUVECs | / | Circ_0003423 | Down | MiR-142-3p/Sirtuin 3, SOD2 | Promote angiogenesis | (35) | |
| OGD | Protective | ||||||
| Angiogenesis | HBMECs | / | Circ_0006768 | Down | MiR-222-3p/VEZF1 | Promote angiogenesis | (53) |
| OGD/R | Protective | ||||||
| HBMECs | / | CircFUNDC1 | Up | MiR-375/PTEN | Inhibit angiogenesis | (44) | |
| OGD | Detrimental | ||||||
| HUVECs | / | Circ_0003204 | Up | Unknown | Inhibit angiogenesis Detrimental | (33) | |
| Ox-LDL | |||||||
| HBMEC | / | CircPHKA2 | Down | MiR-574-5p/SOD2 | Promote angiogenesis | (37) | |
| OGD | Protective | ||||||
| Autophagy | Astrocytes, A172cells | Mouse | CircHECTD1 | Up | MiR142/TIPARP | Promote autophagy Detrimental | (25) |
| OGD-R | tMCAO | ||||||
| Astrocytes | Mouse | CircSHOC2 | Up | MiR-7670-3p/SIRT1 | Inhibit autophagy | (24) | |
| OGD | MCAO | Protective | |||||
| A172 and SK-N-AS | Mouse | Circ_0025984 | Down | MiR-143-3p/TET1 | Inhibit autophagy | (22) | |
| OGD | MCAO | Protective | |||||
| BEnd.3 or HBMECs | Mouse | Circ-FoxO3 | Up | MTOR and E2F1 | Promote autophagy | (23) | |
| OGD/R | MCAO/R | Protective | |||||
| HT22 cells | Mouse | Circ_016719 | Up | MiR-29c/Map2k6 | Promote autophagy | (26) | |
| OGD/R | tMCAO | Detrimental | |||||
| Astrocytes | Mouse | CircAkap7 | Down | MiR-155-5p/ATG12, NRF2 | Promote autophagy | (27) | |
| OGD/R | tMCAO | Protective | |||||
| Other way | HASMCs, MASMCs | / | Circ_ACTA2 | Up | MiR-548f-5p/α-SMA | Facilitate stress fiber formation and cell contraction | (54) |
| Detrimental | |||||||
| Astrocytes, microglia | / | CircSCMH1 | Down | MeCP2 | Enhance neuronal plasticity, inhibit glial activation and peripheral immune cell infiltration | (55) | |
| OGD | Protective | ||||||
| NSCs | Mouse | CircHIPK2 | Up | Smox | Reduce neuronal differentiation | (56) | |
| OGD/R | tMCAO | Detrimental | |||||
PES1, Pescadillo homolog 1; TRPC6, transient receptor potential canonical channel 6; HDAC9, histone deacetylase 9; HUVECs, human umbilical vein endothelial cells; ICAM1, intercellular adhesion molecule 1; ROBO1, roundabout guidance receptor 1; ox-LDL, oxidized low-density lipoprotein; TET2, ten-eleven translocation 2; HBMECs, human brain microvascular endothelial cells; SOD2, superoxide dismutase 2; mAS, mouse astrocytes; OGD/R, oxygen and glucose deprivation/reoxygenation; MCAO, middle cerebral artery occlusion; SRB1, scavenger receptor class B type 1; OGD, oxygen and glucose deprivation; TRAF3, tumor necrosis factor receptor-associated factor 3; HBMVECs, human brain microvascular endothelial cells; H/R, hypoxia/reoxygenation; SOS1, Son of Sevenless 1; NEGR1, neuronal growth regulator 1; I/R, ischemia/reperfusion; TET1, ten-eleven translocation methylcytosine dioxygenase 1; ORP150, oxygen-regulated protein 150; mTOR, mechanistic target of rapamycin; CXCL12, C-X-C motif chemokine 12; PTEN, phosphatase and tensin homolog; FSTL1, follistatin-like 1; Akt, protein kinase B; GSK3β, glycogen synthase kinase 3β; TLR4, Toll-like receptor 4; TRPC6, transient receptor potential canonical channel 6; tMCAO, transient middle cerebral artery occlusion; NRF2, nuclear factor erythroid 2-related factor 2; HDAC9, histone deacetylase 9; ICAM1, intercellular adhesion molecule 1; NEGR1, neuronal growth regulator 1; VEZF1, vascular endothelial zinc finger 1; SIRT3, sirtuin 3; TIPARP, TCDD inducible poly(ADP-ribose) polymerase; SIRT1, sirtuin 1; E2F1, E2F transcription factor 1; Map2k6, Rac-MAPK kinase 6; ATG12, autophagy-related genes 12; HASMCs, human aortic smooth muscle cells; MASMCs, mouse aortic vascular smooth muscle cells; α-SMA, smooth muscle α-actin; MeCP2, methyl-CpG binding protein 2; Smox, spermine oxidase; SK-N-SH, Human neuroblastoma cell line; H/R, hypoxia/reoxygenation; HCN-2, human cortical neuronal cells-2; RGCs, retinal ganglion cells; HCtAECs, human carotid artery endothelial cells; NSCs, neural stem cells.
Autophagy
Overview of autophagy
Autophagy, a degradative and recycling process, is divided into three categories; namely, macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA) (8). The process of autophagy involves the following four steps: initiation, nucleation, elongation, and fusion and degradation (7). With technological advances, the mechanisms and pathways of autophagy have been well-documented, such as the mammalian target of rapamycin (mTOR) pathway, the mitogen-activated protein kinase (MAPK) signaling pathway, and the hypoxia-inducible factor-1 (HIF-1) pathway, among others (69). A disturbance in cellular homeostasis leads to changes in autophagic processes.
The role of autophagy in ischemic stroke
The obstruction of blood flow during a stroke can lead to complex pathophysiological processes (70) including oxidative stress, inflammation, BBB breakdown, calcium overload, excitatory toxicity, and (71) autophagy dysfunctions, leading to neurological disasters. Subsequent recanalization of blood flow (72) results in a secondary injury known as ischemia-reperfusion (I/R) injury.
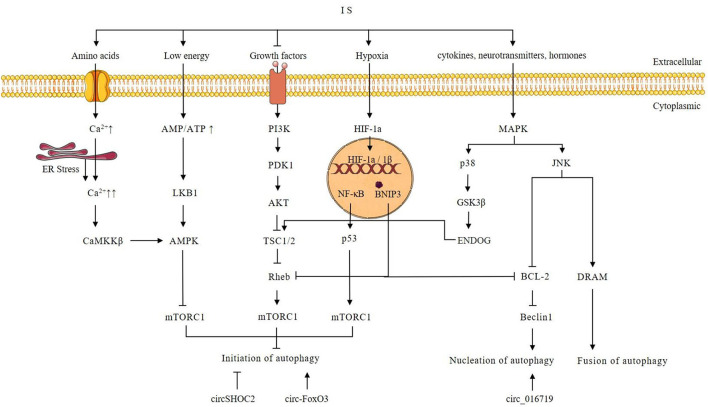
Autophagy is activated during IS and has a dual effect. Autophagy imbalance can aggravate brain damage (maladaptive autophagy), whereas proper autophagy (adaptive autophagy) helps to maintain brain metabolism (73, 74). Maladaptive autophagy is an excessive autophagic activity that accelerates the cytoplasmic accumulation of autophagosomes and the degradation of basic cellular components, triggering apoptosis and necrosis, and eventually causing neuronal cell death and aggravating neurological damage (11). In contrast, adaptive autophagy is a mild to moderate induction of autophagy that promotes neural cell survival (11). During IS, different levels of autophagy are induced in different cell types and time points. Tian et al. found that autophagosome formation in mice heads of the ischemic hemisphere peaked at 1 d after tMCAO, and became gradually weaker from 3 d after tMCAO (75). Moreover, the number of autophagy-induced neurons in the mice head of the ischemic hemisphere was about two times higher compared to the number of autophagic astrocytes at 1 d after tMCAO (75). Liu et al. found that autophagy flux in neurons remained at low levels and was activated immediately upon exposure to OGD, while astrocytes appeared the opposite (76). A series of molecular mechanisms are involved in the regulation of autophagy, including phosphatidylinositol-3-kinase/Akt-mammalian target of rapamycin (PI3k/Akt-mTOR) (28–30), Ca2+/5′-AMP-activated protein kinase/mTOR (Ca2+/AMPK/mTOR) (5, 7, 77, 78), nuclear factor kappa B/p53/mTOR (NF-κB/p53/mTOR) (79, 80), mitogen-activated protein kinase (MAPK) (81–86), and hypoxia-inducible factor 1α/BCL2 interacting protein 3 (HIF-1α/BNIP3) (7, 83, 84, 87–89) signaling pathways. These pathways are illustrated in Figure 2.
Figure 2.
Possible autophagy signaling pathway after ischemic stroke and possible autophagy-regulating mechanism of circRNAs. Intracellular Ca2+ is increased during IS. Increased Ca2+ triggers ER stress and activates CaMKK, which in turn phosphorylates and activates AMPK. AMPK mediates the initiation of autophagy through the inhibition of mTORC1. Growth factors can activate PI3K. Then PI3K activates Akt through phosphorylation, and subsequently, the activated Akt directly phosphorylates and thereby inhibits TSC1/2. The Akt-dependent phosphorylation results in the dissociation of TSC1/2 from the lysosome, where Rheb is localized, promoting Rheb activation. Since GTP-bound Rheb is a potent mTORC1 activator, inhibition of TSC1/2 by AKT-dependent phosphorylation results in mTORC1 activation. Autophagy is inhibited by activated mTORC1. However, under conditions of growth factors or amino acid insufficiency during IS, mTORC1 activity is reduced and induces autophagy. Hypoxia caused by IS activates HIF-1α and induces autophagy through BNIP3 and p53. During IS, an increased AMP/ATP ratio activates LKB1 kinase, which in turn phosphorylates and activates AMPK. AMPK mediates the induction of autophagy through the inhibition of mTORC1. Activation of the p38 MAPK signaling pathway in IS phosphorylates GSK3β, which can subsequently activate ENDOG and TSC1/2. Activated TSC1/2 promotes the initiation of autophagy by inhibiting mTORC1. JNK activation modulates autophagy by promoting Bcl-2/Bcl-xL phosphorylation and upregulating DRAM. Beclin-1 interacts with Bcl2 through its unique BH3 pattern to form the Bcl2-Beclin-1 complex via promoting Beclin-vps34-p50 complex dissociation to inhibit autophagy. DRAM can promote autophagy by stimulating autophagosome-lysosome fusion. Overexpression of circSHOC2 inhibits the initiation of autophagy. Overexpression of circ-FoxO3 can promote the initiation of autophagy through inhibition of mTORC1. Overexpression of circ_016719 promotes nucleation of autophagy.
Maladaptive and adaptive autophagy
Maladaptive autophagy
Blood–brain barrier (BBB) disruption
The BBB is a highly organized multicellular structure composed of capillaries formed by self-fusion of brain microvascular endothelial cells (BMECs) through intact tight junctions, peripheral pericytes, and astrocytes surrounding the capillaries (90). BBB disruption occurs due to central nervous system diseases (91) and may cause secondary brain injuries, including hemorrhage and brain edema (91–94). During IS, blood flow failure causes shrinkage of BMECs, resulting in the translocation of tight junction proteins from the membrane into the cytosol. The increase in BBB permeability elevates the risk for secondary brain damage caused by I/R injury. Various mechanisms regarding the effect of autophagy on BBB during IS have been reported. Zhang et al. found that rapamycin-enhanced autophagy promoted Zonula occludens-1 (ZO-1) reduction in vivo, resulting in increased BBB permeability (95). In 2020, Kim et al. found that ischemia-induced maladaptive autophagy in brain endothelial cells and rat brain capillaries caused ZO-1 degradation, ultimately disrupting BBB integrity and exacerbating neurological damage. Moreover, 3-methyladenine (3-MA) inhibition of autophagy was found to reverse this adverse outcome (96). Liu et al. found that Claudin 5 (CLDN5) is delivered to the autophagosome for autophagy-lysosome-dependent degradation mediated by caveolin-1 (97). CLDN5 is a key molecule in the formation of tight junction chains in the BBB, which seals the intercellular space and maintains the paracellular barrier, and its degradation leads to BBB disruption (98). However, Li et al. recently reported the protective effects of brain endothelial cell autophagy on BBB dysfunction during cerebral I/R injury (99).
Cell death through apoptosis activation
Autophagy causes neural damage by triggering apoptosis. Cao et al. found that the long non-coding RNA (lncRNA) small nucleolar RNA host gene 3 (SNHG3) was overexpressed in the trimethylamine N-oxide (tMAO) mouse model and hypoxia-hypoglycemia/reperfusion cell model, and miR-485 was underexpressed, upregulating autophagy-related 7 (ATG7) to promote autophagy and induce neuronal apoptosis (100). Xie et al. found that neuronal ATG7 deficiency prevented basal autophagy and induced autophagy in the immature brain after hypoxia-ischemia, inhibiting neuronal apoptosis and decreasing neuronal death (101). Autophagy inhibition by 3-MA or lentivirally delivered short hairpin RNAs (shRNAs) against ATG5 and ATG7 significantly reduced the staurosporine (STS)-induced activation of caspase-3 and nuclear translocation of apoptosis-inducing factor (AIF) and provided partial protection against neuronal death (70).
Adaptive autophagy
Adaptive autophagy mitigates IS-related brain damage by the timely removal of old, redundant proteins and damaged organelles (7). Wang et al. found that nicotinamide phosphoribosyltransferase induces autophagy in hypoxic neuron models under OGD in a heterodimeric complex consisting of tuberin Ser1387-TOR-S6 kinase 1 (TSC2Ser1387-TOR-S6K1) signaling pathway in a silent mating type information regulation 2 homolog 1 (SIRT1)-dependent manner, promoting neuronal survival and attenuating cerebral ischemic injury (71). Carloni et al. found that rapamycin-induced autophagy exerted neuroprotective effects through PI3K/Akt-mTOR/Cyclic AMP-responsive element-binding protein (PI3K/Akt-mTOR/CREB) signaling in a young mouse model of cerebral hypoxia-ischemia (9). Ren et al. found that, in comparison with control PC12 cells, OGD/reperfusion (OGD/R) caused an increase in the miR-187-3p level and a decrease in seipin protein levels, reducing autophagic flux and enhancing apoptosis, ultimately increasing ischemia-induced cerebral damage (72). Zhang et al. found that the induction of chloride channel-3 contributed to the formation of the Beclin1 and Vps34 complex, activated autophagy, and attenuated brain ischemic injury (102). Overall, adaptive autophagy attenuates IS-induced brain damage.
Autophagy pathways in IS
MTOR signaling mediated autophagy
The mTOR is a serine/threonine protein kinase comprising rapamycin-sensitive mTOR complex 1 (mTORC1) and rapamycin-less sensitive mTOR complex 2 (mTORC2), of which mTORC1 is the primary regulatory target. mTOR kinase is a key molecule involved in autophagocytosis induction. The activation of mTOR pathways (such as the Akt and MAPK signaling pathways) inhibits autophagocytosis, whereas the negative regulation of mTOR pathways (such as the AMPK and p53 signaling pathways) promotes autophagocytosis. Hei et al. (103) found that IS induces autophagy through mTOR inhibition and alleviates the degree of cerebral ischemia in rats with ischemic injury caused by acute hyperglycemia, which may confirm the view that “moderate autophagy” may have a protective effect on “slow and mild” brain ischemic injury. Moreover, Huang et al. found that curcumin can be activated through the PI3K/Akt-MTOR pathway, alleviating the autophagic activity of nerve cells and improving cerebral IS injury in adult rats (104).
MAPK signaling pathway
The MAPK signaling pathway is composed of p38 extracellular regulated protein kinases (p38-ERK), ERK, and C-Jun N-terminal kinase (JNK) (105). In the early stage of stroke, activation of the p38 signaling pathway promotes Elk1, C/EBP homologous protein (CHOP10), leukocyte cell-derived chemotaxin-2 (LEF2C), and protein kinases MAPKK2/3 to maintain neuronal survival and play roles in anti-inflammatory and anti-apoptotic processes. In contrast, in the late stage of stroke, p38 MAPK is overactivated and promotes the expression of target genes activating transcription factors and caspases (106–108), promoting neuronal apoptosis; thus, differences in IS p38 MAPK signal molecules should be given different interventions. This viewpoint has been supported by relevant studies showing that rhizoma coptidis jiedu soup causes overactivation of ERK and inhibition of JNK, as well as that p38 MAPK signal induction protection autophagy is beneficial for treating IS. The Akt/Smad signaling pathway inhibits JNK and p38 MAPK molecules and negatively regulates OGD-induced autophagy in PC12 cells (73). p38 inhibitors promote cell survival signaling pathways (e.g., ERK) and attenuate ischemic mitochondrial fragmentation or autophagy, reducing ischemic cerebral infarction dead volume and protecting nerve function (86). This evidence suggests that ERK, JNK, and p38 MAPK mediate autophagy in IS. The processes by which ERK activation inhibits autophagy are contrasting to those of JNK and p38 MAPK.
HIF-1α/BNIP3 signaling pathway
The HIF-1α activity is correlated with ischemia-induced neuronal death. In the early stage of acute stroke, HIF-1α/HIF-2α double-knockout mice showed low expression levels of anti-survival factors, such as BNIP3, BNIP3L, and phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1), thus protecting against early acute neuronal cell death and nervous system damage (109). HIF-1α overexpression triggers mitophagy, often accompanied by inhibition of the mTOR pathway and thereby increasing neuronal survival (110). mTOR inactivation by high expression levels of HIF-1α activates AMPK and may affect the survival of bone marrow mesenchymal stem cells (BMSCs) via HIF-1α after transplantation (111). Transplantation of HIF-1α overexpressed BMSCs into MCAO rats reduces the cerebral infarction dead volume and improves neurobehavioral outcomes. In summation, HIF-1α may regulate AMPK and mTOR activation, leading to autophagy which may contribute to the survival of BMSCs.
How circRNAs influence disease progression through the regulation of autophagy
Autophagy is a complex process. Emerging studies have shown that circRNAs can influence disease progression via autophagy regulation, by relying on the transcriptional and post-transcriptional modifications of autophagy-related genes (112). CircRNAs can regulate autophagy by influencing transcription factors at the RNA level. CircST3GAL6 promotes autophagy through FOXP2 transcriptional inhibition of the MET proto-oncogene (MET) to inhibit the PI3K-Akt-mTOR pathway (113). CircRNAs can regulate autophagy directly by influencing the expression of autophagy-related proteins. CircPABPN1 blocks HuR binding to Atg16l1 mRNA, thereby inhibiting autophagy by lowering ATG16L1 expression (114). CircRNAs also can regulate autophagy via RNA methylation. Autophagy-related circRNA (ACR) can inhibit cardiomyocyte autophagy by activating phosphatase and tensin homolog-induced putative kinase 1 (Pink1) expression via its blocking of the Dnmt3B-mediated DNA methylation of the Pink1 promoter. Pink1 inhibits autophagy through the phosphorylating family with sequence similarity 65 member B (FAM65B) (115). This review discusses the circRNA pathways that regulate autophagy in the following section.
MTOR pathway
Akt-MTOR pathway
Xu et al. found that circST3GAL6 was downregulated in gastric cancer (GC) and was associated with poor prognosis of GC patients (113). CircST3GAL6 can upregulate forkhead box P2 (FOXP2) by sponging miR-300. FOXP2, a transcription factor, can suppress the transcriptional activity of MET to promote autophagy via the PI3K–Akt-mTOR pathway (113). Liu et al. found that ACR was decreased in RSC96 cells that were subjected to high glucose (HG) irritation (116). Overexpression of ACR can relieve HG-aroused RSC96 cell autophagy by promoting PI3K/AKT/mTOR pathway activation through a decrease in miR-145-3p expression (116). Wu et al. found that circ_0009910 was highly expressed in acute myeloid leukemia (AML) tissues and cells (117). Circ_0009910 can suppress autophagy by activating the PI3K/AKT signaling pathway via miR-491-5p/β-1, 4-galactosyltransferase 5 in AML cells (117).
AMPK-MTOR pathway
The AMPK pathway can induce autophagy by suppressing mTORC1. Jin et al. found that circRNA_002581 knockdown could induce autophagy to relieve injury in the methionine- and choline-deficient (MCD) diet-induced non-alcoholic steatohepatitis (NASH) mouse model (118). CircRNA_002581 can sponge miR-122 to increase cytoplasmic polyadenylation element-binding protein 1 (CPEB1) expression, which can downregulate PTEN, and then inhibit the AMPK-mTOR pathway to downregulate autophagy in the NASH-like cell model (118). Shang et al. found that circPAN3 was upregulated in bone marrow (BM) samples of AML patients and in doxorubicin (ADM)—resistant cell lines, contributing to AML drug resistance by increasing autophagy (119). CircPAN3 overexpression can upregulate autophagy through the activation of the AMPK/mTOR pathway (119). Chakraborty et al. found abrogation of circHIPK3-induced autophagy in a subset of lung cancer cell lines (120). CircHIPK3 can sponge miR124-3p to increase STAT3 expression, which can downregulate autophagy via the PRKAA/AMPKα/mTOR pathway in A549 cells (120).
MAPK pathway
Yao et al. found that circEIF3K levels were decreased in inflammatory tubal epithelial cells (121). CircEIF3K can bind with MIR139-5p to increase EIF3K and BCL2 expression and MAPK/ERK phosphorylation. This activates the autophagy in inflammatory cells and finally inhibits cell vitality (121). Zhang et al. found that the expression of circ101237 gradually increased with time and promoted autophagy in the cardiomyocytes of 2-day-old mouse models of anoxia/reoxygenation (122). CircRNA_101237 can sponge let-7a-5p to promote IGF2BP3 expression (122). In turn, IGF2BP3 increases the expression of IGF2 (123), which promotes autophagy through the phosphorylation and activation of the MAPK/ERK signaling pathway (124). Jiang et al. found that circ0032821 is upregulated in patients with GC, thereby contributing to GC tumorigenesis through the inhibition of autophagy, which may result from the activation of the MAPK/ERK signaling pathway (125).
Beclin-1 pathway
Gan et al. found that circMUC16 levels were increased in epithelial ovarian cancer tissues (126). CircMUC16 overexpression can promote cancer cell proliferation and migration by activating autophagy through the adsorption of miR199A-5p to mitigate the inhibition of BECN1 (126). Chen et al. found that circMTO1 was upregulated in cervical cancer cell lines and tumors (127). CircMTO1 can promote autophagy by adsorbing miR6893 and promoting the expression of BECN1 to enhance the invasion and migration of cervical cancer cells (127). Guo et al. found that hsa_circ_0023404 overexpression could promote autophagy by upregulating BECN1 to inhibit apoptosis and impart resistance to cisplatin (128).
Additional pathways
Du et al. found that circDNMT1 was increased in eight different breast cancer cell lines and in patients with breast carcinoma (129). CircDNMT1 can promote the nuclear translocation of TP53, which in turn promotes the expression of autophagy-related genes to induce autophagy, thereby enhancing breast cancer progression (129). Cao et al. found that circ0009910 was upregulated in the sera of imatinib-resistant chronic myeloid leukemia patients (130). Circ0009910 can increase the expression of ULK1 by sponging miR-34a-5p. Circ0009910 also can promote the phosphorylation of ULK1 and the expression of BECN1 and LC3-II, thereby promoting autophagy to accelerate imatinib resistance (130). Ren et al. found that circZnf292 was upregulated in ischemic heart disease (131). CircZnf292 can inhibit autophagy by activating the WNT-CTNNB1 and the MTOR signaling pathways through BNIP3 inhibition, thereby reducing damage in the OGD-induced H9c2 cell line (131).
The role of circRNAs as biomarkers and therapeutic targets in IS
Circular RNAs involved in the regulation of nerve cell development, differentiation, material transport, and axonal plasticity are highly expressed in the nervous system (15, 16). Various circRNAs participate in the pathological processes following IS. CircRNAs regulate gene expression post-transcription by competitively binding to miRNAs or RNA-binding proteins (12).
Circulating RNAs, such as Circ_0072309 (42), circular SHOC2 Leucine-Rich Repeat Scaffold Protein (circSHOC2) (24), circUCK2 (132), circCCDC9 (133), and circPHKA2 (37), have been shown to alleviate apoptotic rates and attenuate neuronal injury through miR-100/mTOR, miR-7670-3p/SIRT1, and miR-125b-5p/growth differentiation factor 11 (GDF11), suppressing the Notch receptor 1 (Notch1) signaling pathway and the miR-574-5p/superoxide dismutase 2 (SOD2) axis. Circ-forkhead box O3 (FoxO3) (23) and circRNA 0025984 (22) attenuate neuronal injury through autophagy inhibition. Circ-FoxO3 promotes brain microvessel endothelial cell (BMEC) autophagy via mTORC1 inhibition to clear cytotoxic aggregates. CircRNA 0025984 protects astrocytes from autophagy through the miR-143-3p/Tet methylcytosine dioxygenase 1/150-kDa oxygen-regulated protein (miR-143-3p/TET1/ORP150 pathway). CircDLGAP4 (134) overexpression decreases BBB damage and infarcts areas after stroke by inhibiting endothelial–mesenchymal transition via the miR-143/HECT domain E3 ubiquitin-protein ligase 1 (HECTD1) axis. CircSCMH1 (55) contributes to functional recovery after stroke by enhancing neuronal plasticity and inhibiting glial activation. Circ_0006768 (53) upregulation attenuates HBMEC injury by upregulating vascular endothelial zinc finger 1 (VEZF1) via miR-222-3p inhibition. Circ-Rps5 (135) decreases neuronal damage via the miR-124-3p/SIRT7 axis. The upregulation of circ-FoxO3, circDLGAP4, and circCCDC9 promotes the maintenance of BBB integrity.
However, some significantly upregulated circRNAs showed contrasting effects. Circ-HECTD1 (25) aggregates neuronal deficits by promoting astrocyte autophagy via the miR142-TIPARP axis. Circ cGLIS3 (46), circRNA TLK1 (136), circPHC3 (45), circFUNDC1 (44), and circCDC14A (43) contribute to cell death by activating apoptosis. Overexpression of circ HIPK2 (56) and circSKA3 (137) aggravates neuronal injuries.
CircRNAs affect IS progression by regulating autophagy
CircHECTD1
More and more studies proved that circRNAs can affect IS progression by regulating autophagy, we summarize the evidences from basic research of these related circRNAs in Table 2 and illustrate the pathways in Figure 3. The host gene of circHECTD1, HECTD1, is an E3 ubiquitin ligase that regulates cell migration machinery (138). CircHECTD1 is derived from exons 23 and 24 of HECTD1 and is highly expressed in the brain and lungs (25). In 2018, Han et al. found the upregulation of circHECTD1 in ischemic brain tissues from a transient MCAO (tMCAO) mouse model and in plasma samples from patients with acute IS (25). Functional studies have shown reduced infarct areas, milder neuronal deficits, and less activation of astrocytes resulting from lower circHECTD1 expression in the tMCAO stroke mouse model. A mechanistic study demonstrated that circHECTD1 functions as a sponge to inhibit miR142 activity, thus upregulating TCDD-inducible poly (ADP-ribose) polymerase (TIPARP) and ultimately increasing astrocyte activation via macro-autophagy/autophagy (25). Therefore, downregulation of circHECTD1 expression is associated with reduced cerebral infarction.
Table 2.
The evidence of CircRNA roles in ischemic stroke via autophagy regulation.
| Cell line/treatment | Animal/Disease model | Sample | DE CircRNA and expression | Regulating axis | Autophagy flux |
Apoptosis activation |
DE CircRNA's Roles in IS | References | |
|---|---|---|---|---|---|---|---|---|---|
| Evidences from basic research | |||||||||
| Primary mouse astrocyte, | Male C57BL/6J mouse | Astrocytes | circHECTD1 | Up | circHECTD1/MIR142/ | Upregulated | Undefined | Detrimental | (25) |
| A172 cell | TIPARP/Beclin1 | The nucleation of autophagy | |||||||
| OGD-R | tMCAO | ||||||||
| Primary astrocyte | Male C57BL/6J mice | Astrocytes | circSHOC2 | Up | circSHOC2/miR-7670-3p/SIRT1/Beclin1 | Downregulated | Decreased | Protective | (24) |
| The nucleation of autophagy | |||||||||
| OGD | MCAO | ||||||||
| A172 and SK-N-AS | Male Sprague Dawley rats | Astrocytes | circ_0025984 | Down | circ_0025984/miR-143-3p/TET1//ORP150/GRP78/ATG7 | Upregulated | Increased | Protective | (22) |
| OGD | MCAO | The elongation of autophagy | |||||||
| BEnd.3 or HBMEC | C57BL/6J mice | BMECs | circ-FoxO3 | Up | circ-FoxO3/mTOR/mTORC1 | Upregulated | Undefined | Protective | (23) |
| circ-FoxO3/E2F1/PEKFB3/ | The initiation of autophagy | ||||||||
| OGD/R | MCAO/R | ||||||||
| HT22 cells | Male C57BL/6J mice | Neurons | circ_016719 | Up | circ_016719/miR-29c/Map2k6/ | Upregulated | Increased | Detrimental | (26) |
| Beclin1 | The nucleation of autophagy | ||||||||
| OGD/R | tMCAO | ||||||||
| Primary astrocyte | C57BL/6 mice | Astrocytes | circAkap7 | Down | circAkap7/miR-155-5p/ATG12, NRF2 | Downregulated | Increased | Protective | (27) |
| OGD/R | tMCAO | The elongation of autophagy | |||||||
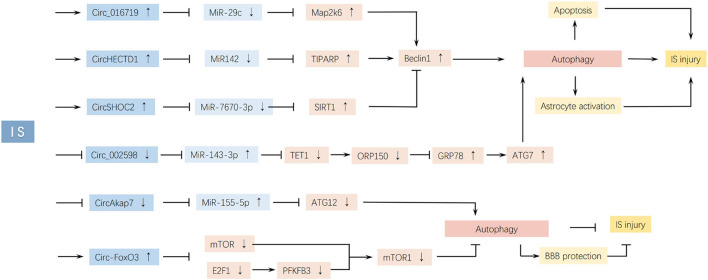
Figure 3.
CircRNAs affect IS injury by regulating autophagy. Changing the expression of circ_016719, circHECTD1, circSHOC2, and circ_002598 can induce IS injury by upregulating autophagy. However, changing the expression of circAkap7 and circ-FoxO3 can inhibit IS injury by upregulating autophagy.
Furthermore, a recent study by He et al. found that circHECTD1 overexpression accelerated endothelial–mesenchymal transition (EndoMT) in human cerebral microvascular endothelial cells (hCMECs) treated with OGD/R, which mimics IS in vitro (139). As EndoMT plays a critical role in BBB dysfunction, this study indicated that increased circHECTD1 expression might damage BBB integrity (140). In addition to IS, circHECTD1 participates in the regulation of various pathological processes and diseases, including acute lung injury (141), hypertrophic scar fibrosis (142), ulcerative colitis (143), gastric cancer (144), hepatocellular carcinoma (145), and glioma (146). CircHECTD1 overexpression reduced colonic injury and inflammation by promoting autophagy in lipopolysaccharide (LPS)-induced Caco-2 cells in dextran sodium sulfate (DSS)-treated mice (143). CircHECTD1 knockdown upregulates HECTD1 protein expression in normal human pulmonary fibroblasts (HPF-a) cells and those exposed to silicon dioxide (SiO2). HECTD1 promotes fibroblast activation and migration via autophagy, promoting pulmonary fibrosis progression (147).
CircSHOC2
CircSHOC2 (circ_0092670) is transcribed from the SHOC2 gene and shows significantly increased expression in ischemic preconditioning astrocyte exosomes (IPAS-EXOs). SHOC2 is a prototypical leucine-rich repeat protein that promotes downstream receptor tyrosine kinase (RTK)/RAS signaling and plays important roles in several cellular and developmental processes (148). In 2020, Chen et al. found that exosomes from ischemia-preconditioned astrocytes inhibited ischemia-induced neuronal apoptosis by downregulating autophagy. The authors further validated that circSHOC2 in these exosomes reduced OGD/R-induced neuronal death. CircSHOC2 suppresses neuronal apoptosis and ameliorates neuronal damage by inhibiting autophagy and acting on the miR-7670-3p/SIRT1 axis (24). CircSHOC2 is highly expressed in IPAS-EXOs but not in neurons. It acts as a sponge for miR-7670-3p without regulating miR-7670-3p expression levels, thereby promoting SIRT1 expression to reduce astrocyte autophagy, decrease apoptosis, and protect neurons from ischemia-induced damage (24). CircSHOC2 overexpression inhibits Gag-LC3 accumulation, LC3-I-to-LC3-II conversion, and translocase of inner mitochondrial membrane 23 (TIMM23) degradation in neurons. The protective effects of circSHOC2 are abolished by an autophagic inhibitor (24). Exosomes function locally or can be stably transferred to recipient cells (149, 150). With a diameter of 30–100 nm, exosomes can cross the BBB (151). Therefore, exosomes with circRNAs are new potential therapeutic agents for tissue recovery after IS.
Circ_0025984
Zhou et al. found that circ_0025984 (hsa_circ_0025984, parent gene SLC38A2) and ten-eleven translocation methylcytosine dioxygenase 1 (TET1) were significantly downregulated, and miR-143-3p was notably upregulated in the tMCAO rat model, which was further demonstrated in astrocytes undergoing OGD/R. The abnormally low expression of circ_0025984 increased astrocyte apoptosis and autophagy through the miR-143-3p/TET1/ORP150 pathway, ultimately causing ischemic cerebral injury. Circ_0025984 overexpression functions as a sponge for miR-143-3p and upregulates TET1, inducing 150-kDa oxygen-regulated protein (ORP150) expression, which decreases glucose-regulated protein 78 kDa (GRP78) levels. GRP78 also induces ATG7 expression (22), promoting autophagy activation. TET1, a member of the Tet family, can convert 5-methylcytosine to 5-hydroxymethylcytosine in a 2-oxoglutarate- and Fe(II)-dependent manner and is involved in DNA demethylation (152). Another study showed that TET1 could also activate the transcriptional expression of glucose metabolism-related genes in hepatocytes (153). TET1 plays a protective role in cells undergoing OGD and induces ORP150 (an ER-associated chapel) expression by binding to its promoter DNA and consequently decreasing its methylation (22). The ORP150, a member of the heat shock protein family, is located in the endoplasmic reticulum (ER), functions as a molecular chaperone in the transport and folding of newly synthesized proteins (154), and prevents ischemia-induced cell death. Its upregulation modulates ATP/ADP exchange in GRP78 and decreases GRP78 expression (a calcium-dependent protein induced to maintain ER and cell homeostasis during calcium dysregulation, oxygen and glucose deprivation, or inflammation), ultimately maintaining calcium homeostasis and inhibiting autophagy and apoptosis (155). GRP78 induces ATG7 expression after OGD (22), thus enabling the transition of LC3-I to LC3-II, subsequently activating autophagy (156), inhibiting aggregation, and promoting the degradation of misfolded proteins (157). Therefore, circ_0025984 overexpression ultimately inhibits elongation and phagosome formation of autophagy (7), decreases apoptosis of astrocytes, and alleviates cerebral injury in MCAO rats.
By regulating autophagy, MiR-143 is involved in many diseases, such as myocardial I/R injury (158–160), Crohn's disease (161), pancreatic cancer (162, 163), acute myeloid leukemia (164), renal cell cancer (165), small-cell lung cancer (166) endometriosis (167), and prostate cancer (168).
Circ-FoxO3
In 2022, Yang et al. found that circ-FoxO3 (hsa_circ_0006404, parent gene FOXO3) alleviated BBB damage by activating autophagy in BMECs and in MCAO/R mice models. They found that circ-FoxO3 was upregulated in brain tissues after I/R injury, especially in BMECs and astrocytes. Furthermore, circ-FoxO3 protected against OGD/R-stimulated endothelial barrier damage and tMCAO-induced BBB collapse in mice by upregulating autophagy (23). Mechanistically, circ-FoxO3 plays a role in the sequestration of mTOR and E2F Transcription factor 1 (E2F1) to inhibit mTORC1 activity, thereby activating autophagy (23). This interaction was reported by Du et al. in 2017, who found that circ-FoxO3 could competitively sequester mTOR and inhibit mTORC1 activity, interact with E2F1 in the cytosol, block E2F1 entrance into the nucleus, and prevent its transcriptional regulation (169). The translocation of mTORC1 to lysosomes is blocked, and mTORC1 is inactivated. Finally, circ-FoxO3 promotes initiation autophagy (23).
Circ-FoxO3 was first reported to have a significant effect on modulating cell proliferation, migration, invasion, and apoptosis through different signaling pathways (170). Ectopic expression of circ-Foxo3 promoted cellular senescence, whereas silencing of circ-Foxo3 decreased cell senescence and apoptosis (169). Circ-Foxo3 is involved in the development and tumorigenesis of many cancers. Circ-Foxo3 is significantly downregulated in esophageal squamous cell cancer, bladder cancer, colorectal cancer, and acute lymphocytic leukemia (171–175). Mechanistically, circ-FoxO3 decreases the interaction between FoxO3 and MDM2 Proto-Oncogene (MDM2), and represses the poly-ubiquitination of FoxO3 modulated by MDM2, which increases FoxO3 activity, promoting p53 upregulated modulator of apoptosis (PUMA) expression and cell apoptosis (176). Circ-FoxO3 retards cell cycle progression by binding to p21 and cyclin-dependent kinase 2 (CDK2). CDK2 initiates the G1-S phase transition by binding to cyclin E (177), which can be stimulated by binding between p21 and CDK2 (178). Circ-FoxO3 overexpression promotes the formation of the circ-FoxO3–p21–CDK2 ternary complex, which hijacks CDK2 together with p21 to avoid the formation of the cyclin E/CDK2 complex, thus blocking the G1-S phase transition and the progression of the cell cycle in the S phase (179).
Circ_016719
In 2019, Tang et al. found that circ_016719 could activate autophagy to partly inhibit cell viability in HT22 cells and mouse models with MCAO/R (26). Circ_016719 is upregulated in brain tissues after I/R injury. Results of mechanistic studies demonstrated that circ_016719 directly sponges miR-29c to increase the expression of Rac-MAPK kinase 6 (Map2k6) (26). Map2k6, also known as MKK6, is a critical upstream regulator of the MAPK pathway, with key roles in cell survival, differentiation, and inflammation (180). Circ_016719 may activate autophagy to promote nucleation via the upregulation of beclin-1 and p53.
CircAkap7
In 2017, Mehta et al. found that circAkap7 is downregulated in tMCAO mice compared to sham mice (181). CircAkap7 (mmu_circ_0000154, mm9_circ_010383) is derived from exon 2 of the Akap7 gene, located on chromosome 10 (1024987113-25009536), whose spliced mature sequence length is 579 bp (27). In 2020, Xu et al. found that exo-circAkap7 treatment decreased tMCAO-induced cerebral inflammation, apoptosis, and oxidative stress, and promoted autophagy. Further mechanistic research demonstrated that circAkap7 directly sponges miR-155-5p to increase the expression of NRF2 and ATG12 in OGD-treated astrocytes to promote autophagy and ameliorate oxidative stress, suggesting that the use of exo-circAkap7 is a potential treatment strategy for cerebral ischemic injury (27). ATG12 is indispensable for the formation of ATG5–ATG12–ATG16L1 complex, which supports the elongation of autophagosomes (182).
Hsa_Circ_0001599
In 2021, Li et al. found that hsa_circ_0001599 was significantly upregulated in patients with large-artery atherosclerotic stroke, and its level positively correlated with stroke neurological severity and infarct volume (183). The parental gene of hsa_circ_0001599, FK506-binding protein 5 (FKBP5), is a member of the immunophilin protein family and plays a role in stress response and inflammation (184). Yu et al. found that FKBP5 was upregulated in patients with acute IS, and it regulated autophagy through the downstream AKT/FoxO3 signaling pathway in the OGD/R cell model (185). Therefore, hsa_circ_0001599 may be a pivotal molecule affecting autophagy after IS.
Conclusion
Stroke is widely known for its high mortality, disability, and recurrence rates, and IS accounts for the majority of the cases. Ischemia and hypoxia activate many pathological processes, such as apoptosis, inflammation, oxidative stress, angiogenesis, and autophagy, among others. CircRNAs have been shown to be differentially expressed during IS and to be participating in all the above processes. One circRNA can influence more than one process in IS, including autophagy, which has become the focus of recent research. However, previous studies have not yielded consistent results. Activated autophagy aggravates neurological dysfunction by exacerbating neuronal death through apoptosis activation and BBB disruption or decreases brain damage by reducing apoptosis. Astrocytes are excellent candidates for regulation in autophagy activation to protect the brain against IS-induced injury owing to their numerical advantages and importance in cerebral parenchymal homeostasis and normal brain function. CircRNAs are differentially expressed during IS and are involved in the regulation of IS injury. CircRNAs influence IS progression, mostly by sponging miRNAs. Four circRNAs have been found to be differentially expressed in astrocytes and influence IS progression by regulating autophagy. Moreover, one circRNA can regulate autophagy through different pathways. These studies provide a new potential treatment for IS through autophagy regulation by altering circRNA expression. However, unresolved questions remain. First, autophagy activation is either a neuroprotective process or a cause of cell death and remains controversial. Second, one circRNA can sponge more than one miRNA, influence many other processed cells, and may contribute to other diseases. Many circRNAs are differentially expressed in IS, and one circRNA can regulate autophagy through different pathways; therefore, determining whether one or more circRNAs could be selected as treatment targets for IS still requires further research. Furthermore, the transportation of the target circRNAs or autophagy regulators into the brain is yet to be fully elucidated. Furthermore, if all circRNAs are altered during IS, the protective and/or detrimental role may be enhanced. Accordingly, more research is required in this direction. Finally, because of the narrow therapeutic time window, blocking the mechanism of neuronal death in the ischemic cascade could be difficult.
Author contributions
XL, LL, and LJ contributed to the review design and revised the manuscript. XL drafted the manuscript. LJ and LL revised the manuscript. All the authors have read and approved the final version of the manuscript.
Funding
This work was funded by Zhejiang Basic Public Welfare Research Program (LGF20H090008 and GC22H095676) and Hangzhou Health Science and Technology Project (ZD20200056, ZD20210007, and A20200341).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
Abbreviations
- IS
Ischemic stroke
- circRNAs
circular RNAs
- RBPs
RNA-binding proteins
- miRNAs
microRNAs
- I/R
ischemia/reperfusion
- HIF-1α
hypoxia-inducible factor 1α
- mTORC1
mTOR complex 1
- mTOR
mammalian target of rapamycin
- AMPK
5′-AMP-activated protein kinase
- LKB1
liver kinase B1
- CAMKKβ
calcium/calmodulin-dependent protein kinase kinase 2
- BNIP3
Bcl-2/adenovirus E1B 19 kDa protein-interacting protein 3
- JNK
c-Jun NH2-terminal kinase
- tMCAO
transient middle cerebral artery occlusion
- pMCAO
permanent middle cerebral artery occlusion
- OGD/R
oxygen-glucose deprivation/reoxygenation
- 3-MA
3-methyladenine
- PTH
Pien Tze Huang
- BMECs
brain microvascular endothelial cells
- CLDN5
claudin 5
- BBB
blood–brain barrier
- SIRT1
silent mating type information regulation 2 homolog 1
- TIPARP
TCDD-inducible poly [ADP-ribose] polymerase
- TET1
Ten-eleven translocation methylcytosine dioxygenase 1
- ER
endoplasmic reticulum
- ORP150
150-kDa oxygen-regulated protein
- LAA
large-artery atherosclerotic
- FKBP5
FK506-binding protein 5.
References
- 1.Feigin VL, Stark BA, Johnson CO, Roth GA, Bisignano C, Abady GG, et al. Global, regional, and national burden of stroke and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol. (2021) 20:795–820. 10.1016/S1474-4422(21)00252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell BC, De Silva DA, Macleod MR, Coutts SB, Schwamm LH, Davis SM, et al. Ischaemic stroke. Nat Rev Dis Primers. (2019) 5:70. 10.1038/s41572-019-0118-8 [DOI] [PubMed] [Google Scholar]
- 3.Obrenovitch TP, Urenjak J, Richards DA, Ueda Y, Curzon G, Symon L, et al. Extracellular neuroactive amino acids in the rat striatum during ischaemia: comparison between penumbral conditions and ischaemia with sustained anoxic depolarisation. J Neurochem. (1993) 61:178–86. 10.1111/j.1471-4159.1993.tb03553.x [DOI] [PubMed] [Google Scholar]
- 4.Wu QJ, Tymianski M. Targeting NMDA receptors in stroke: new hope in neuroprotection. Mol Brain. (2018) 11:15. 10.1186/s13041-018-0357-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tuo QZ, Zhang ST, Lei P. Mechanisms of neuronal cell death in ischemic stroke and their therapeutic implications. Med Res Rev. (2022) 42:259–305. 10.1002/med.21817 [DOI] [PubMed] [Google Scholar]
- 6.Hafez S, Hoda MN, Guo X, Johnson MH, Fagan SC, Ergul A, et al. Comparative analysis of different methods of ischemia/reperfusion in hyperglycemic stroke outcomes: interaction with tPA. Transl Stroke Res. (2015) 6:171–80. 10.1007/s12975-015-0391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galluzzi L, Bravo-San Pedro JM, Blomgren K, Kroemer G. Autophagy in acute brain injury. Nat Rev Neurosci. (2016) 17:467–84. 10.1038/nrn.2016.51 [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. (2020) 383:1564–76. 10.1056/NEJMra2022774 [DOI] [PubMed] [Google Scholar]
- 9.Carloni S, Girelli S, Scopa C, Buonocore G, Longini M, Balduini W, et al. Activation of autophagy and Akt/CREB signaling play an equivalent role in the neuroprotective effect of rapamycin in neonatal hypoxia-ischemia. Autophagy. (2010) 6:366–77. 10.4161/auto.6.3.11261 [DOI] [PubMed] [Google Scholar]
- 10.Shi R, Weng J, Zhao L, Li XM, Gao TM, Kong J, et al. Excessive autophagy contributes to neuron death in cerebral ischemia. CNS Neurosci Ther. (2012) 18:250–60. 10.1111/j.1755-5949.2012.00295.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ajoolabady A, Wang S, Kroemer G, Penninger JM, Uversky VN, Pratico D, et al. Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol Ther. (2021) 225:107848. 10.1016/j.pharmthera.2021.107848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. (2019) 20:675–91. 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 13.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. (2013) 19:141–57. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. (2013) 495:333–8. 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 15.Piwecka M, GlaŽar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. (2017) 357:eaam8526. 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- 16.Rybak-Wolf A, Stottmeister C, GlaŽar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. (2015) 58:870–85. 10.1016/j.molcel.2015.03.027 [DOI] [PubMed] [Google Scholar]
- 17.Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. (2017) 14:1028–34. 10.1080/15476286.2016.1255398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aufiero S, Reckman YJ, Pinto YM, Creemers EE. Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol. (2019) 16:503–14. 10.1038/s41569-019-0185-2 [DOI] [PubMed] [Google Scholar]
- 19.Fang Y, Wang X, Li W, Han J, Jin J, Su F, et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med. (2018) 42:1865–74. 10.3892/ijmm.2018.3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Li K, Lai W, Li X, Wang H, Yang J, et al. Comprehensive circular RNA profiles in plasma reveals that circular RNAs can be used as novel biomarkers for systemic lupus erythematosus. Clin Chim Acta. (2018) 480:17–25. 10.1016/j.cca.2018.01.026 [DOI] [PubMed] [Google Scholar]
- 21.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. (2019) 176:869–81.e13. 10.1016/j.cell.2018.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou D, Huang Z, Zhu X, Hong T, Zhao Y. Circular RNA 0025984 ameliorates ischemic stroke injury and protects astrocytes through miR-143-3p/TET1/ORP150 pathway. Mol Neurobiol. (2021) 58:5937–53. 10.1007/s12035-021-02486-8 [DOI] [PubMed] [Google Scholar]
- 23.Yang Z, Huang C, Wen X, Liu W, Huang X, Li Y, et al. Circular RNA circ-FoxO3 attenuates blood-brain barrier damage by inducing autophagy during ischemia/reperfusion. Mol Ther. (2022) 30:1275–87. 10.1016/j.ymthe.2021.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, Wang H, Zhu Z, Feng J, Chen L. Exosome-shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 Axis. Mol Ther Nucleic Acids. (2020) 22:657–72. 10.1016/j.omtn.2020.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han B, Zhang Y, Zhang Y, Bai Y, Chen X, Huang R, et al. Novel insight into circular RNA HECTD1 in astrocyte activation via autophagy by targeting MIR142-TIPARP: implications for cerebral ischemic stroke. Autophagy. (2018) 14:1164–84. 10.1080/15548627.2018.1458173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang C, Ou J, Kou L, Deng J, Luo S. Circ_016719 plays a critical role in neuron cell apoptosis induced by I/R via targeting miR-29c/Map2k6. Mol Cell Probes. (2020) 49:101478. 10.1016/j.mcp.2019.101478 [DOI] [PubMed] [Google Scholar]
- 27.Xu L, Ji H, Jiang Y, Cai L, Lai X, Wu F, et al. Exosomes derived from CircAkap7-modified adipose-derived mesenchymal stem cells protect against cerebral ischemic injury. Front Cell Dev Biol. (2020) 8:569977. 10.3389/fcell.2020.569977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. (2016) 7:12429. 10.1038/ncomms12429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng W, Li S, Chen S, Yang J, Sun Z. Hsa_circ_0003204 knockdown weakens Ox-LDL-induced cell injury by regulating miR-188-3p/TRPC6 axis in human carotid artery endothelial cells and THP-1 cells. Front Cardiovasc Med. (2021) 8:731890. 10.3389/fcvm.2021.731890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan H, You T, Luo W. circ_0003204 regulates cell growth, oxidative stress, and inflammation in ox-LDL-induced vascular endothelial cells via regulating miR-942-5p/HDAC9 axis. Front Cardiovasc Med. (2021) 8:646832. 10.3389/fcvm.2021.646832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D, Zhang G, Yu K, Zhang X, Jiang A. Circ_0003204 knockdown protects endothelial cells against oxidized low-density lipoprotein-induced injuries by targeting the miR-491-5p-ICAM1 pathway. J Thromb Thrombolysis. (2022) 53:302–12. 10.1007/s11239-021-02606-0 [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Li W, Li H, Zhou M, Zhang J, Fu Y, et al. Circ_USP36 silencing attenuates oxidized low-density lipoprotein-induced dysfunction in endothelial cells in atherosclerosis through mediating miR-197-3p/ROBO1 axis. J Cardiovasc Pharmacol. (2021) 78:e761–72. 10.1097/FJC.0000000000001124 [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Ma X, Mao Z, Shen M, Zhu J, Chen F, et al. Circular RNA has_circ_0003204 inhibits oxLDL-induced vascular endothelial cell proliferation and angiogenesis. Cell Signal. (2020) 70:109595. 10.1016/j.cellsig.2020.109595 [DOI] [PubMed] [Google Scholar]
- 34.Yu H, Pan Y, Dai M, Xu H, Li J. Circ_0003423 alleviates ox-LDL-induced human brain microvascular endothelial cell injury via the miR-589-5p/TET2 network. Neurochem Res. (2021) 46:2885–96. 10.1007/s11064-021-03387-x [DOI] [PubMed] [Google Scholar]
- 35.Wang P, Zhang H, Wang Y. Circ_0003423 alleviates oxidized low-density lipoprotein-induced endothelial cell injury by sponging miR-142-3p and activating Sirtuin 3/superoxide dismutase 2 pathway. J Surg Res. (2022) 277:384–97. 10.1016/j.jss.2022.04.006 [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Chang X, Zhang S, Zhao Q, Lei C. CircRNA CTNNB1 (circCTNNB1) ameliorates cerebral ischemia/reperfusion injury by sponging miR-96-5p to up-regulate scavenger receptor class B type 1 (SRB1) expression. Bioengineered. (2022) 13:10258–73. 10.1080/21655979.2022.2061304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Li X, Zhong C, Peng J, Pang J, Peng T, et al. RNA circPHKA2 relieves OGD-induced human brain microvascular endothelial cell injuries through competitively binding miR-574-5p to modulate SOD2. Oxid Med Cell Longev. (2021) 2021:3823122. 10.1155/2021/3823122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dai Y, Sheng Y, Deng Y, Wang H, Zhao Z, Yu X, et al. Circ_0000647 promotes cell injury by modulating miR-126-5p/TRAF3 axis in oxygen-glucose deprivation and reperfusion-induced SK-N-SH cell model. Int Immunopharmacol. (2022) 104:108464. 10.1016/j.intimp.2021.108464 [DOI] [PubMed] [Google Scholar]
- 39.Ren X, Jing YX, Zhou ZW, Yang JW. Knockdown of circRNA-Memo1 reduces hypoxia/reoxygenation injury in human brain endothelial cells through miRNA-17-5p/SOS1 axis. Mol Neurobiol. (2022) 59:2085–97. 10.1007/s12035-022-02743-4 [DOI] [PubMed] [Google Scholar]
- 40.Qiu L, He J, Chen H, Xu X, Tao Y. CircDLGAP4 overexpression relieves oxygen-glucose deprivation-induced neuronal injury by elevating NEGR1 through sponging miR-503-3p. J Mol Histol. (2022) 53:321–32. 10.1007/s10735-021-10036-8 [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Luo W, Zhang W, Chu H, Wang J, Dai X, et al. circDLPAG4/HECTD1 mediates ischaemia/reperfusion injury in endothelial cells via ER stress. RNA Biol. (2020) 17:240–53. 10.1080/15476286.2019.1676114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Y, Li J, Li J, Xu L, Lian W. The decreased circular RNA hsa_circ_0072309 promotes cell apoptosis of ischemic stroke by sponging miR-100. Eur Rev Med Pharmacol Sci. (2020) 24:4420–9. 10.26355/eurrev_202004_21024 [DOI] [PubMed] [Google Scholar]
- 43.Huo H, Hu C, Lu Y, Zhou J, Mai Z. Silencing of circCDC14A prevents cerebral ischemia-reperfusion injury via miR-23a-3p/CXCL12 axis. J Biochem Mol Toxicol. (2022) 36:e22982. 10.1002/jbt.22982 [DOI] [PubMed] [Google Scholar]
- 44.Bai X, Liu X, Wu H, Feng J, Chen H, Zhou D, et al. CircFUNDC1 knockdown alleviates oxygen-glucose deprivation-induced human brain microvascular endothelial cell injuries by inhibiting PTEN via miR-375. Neurosci Lett. (2022) 770:136381. 10.1016/j.neulet.2021.136381 [DOI] [PubMed] [Google Scholar]
- 45.Xu X, Wu Z, Qiu H, Wu J. Circular RNA circPHC3 promotes cell death and apoptosis in human BMECs after oxygen glucose deprivation via miR-455-5p/TRAF3 axis in vitro. Neuropsychiatr Dis Treat. (2021) 17:147–56. 10.2147/NDT.S288669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang Q, Su DY, Wang ZZ, Liu C, Sun YN, Cheng H, et al. Retina as a window to cerebral dysfunction following studies with circRNA signature during neurodegeneration. Theranostics. (2021) 11:1814–27. 10.7150/thno.51550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dai Q, Ma Y, Xu Z, Zhang L, Yang H, Liu Q, et al. Downregulation of circular RNA HECTD1 induces neuroprotection against ischemic stroke through the microRNA-133b/TRAF3 pathway. Life Sci. (2021) 264:118626. 10.1016/j.lfs.2020.118626 [DOI] [PubMed] [Google Scholar]
- 48.Zhang Z, He J, Wang B. Circular RNA circ_HECTD1 regulates cell injury after cerebral infarction by miR-27a-3p/FSTL1 axis. Cell Cycle. (2021) 20:914–26. 10.1080/15384101.2021.1909885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Ji H, Yao Y, Lai X, Jiang Y, Wu D, et al. Downregulation of circ_008018 protects against cerebral ischemia-reperfusion injury by targeting miR-99a. Biochem Biophys Res Commun. (2018) 499:758–64. 10.1016/j.bbrc.2018.03.218 [DOI] [PubMed] [Google Scholar]
- 50.Yang B, Zang L, Cui J, Wei L. Circular RNA TTC3 regulates cerebral ischemia-reperfusion injury and neural stem cells by miR-372-3p/TLR4 axis in cerebral infarction. Stem Cell Res Ther. (2021) 12:125. 10.1186/s13287-021-02187-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu H, Zheng W, Song Z. circDlgap4 alleviates cerebral ischaemic injury by binding to AUF1 to suppress oxidative stress and neuroinflammation. Mol Neurobiol. (2022) 59:3218–32. 10.1007/s12035-022-02796-5 [DOI] [PubMed] [Google Scholar]
- 52.Jiang S, Zhao G, Lu J, Jiang M, Wu Z, Huang Y, et al. Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol Res. (2020) 53:27. 10.1186/s40659-020-00295-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J, Wang J, Wang Z. Circ_0006768 upregulation attenuates oxygen-glucose deprivation/reoxygenation-induced human brain microvascular endothelial cell injuries by upregulating VEZF1 via miR-222-3p inhibition. Metab Brain Dis. (2021) 36:2521–34. 10.1007/s11011-021-00775-8 [DOI] [PubMed] [Google Scholar]
- 54.Sun Y, Yang Z, Zheng B, Zhang XH, Zhang ML, Zhao XS, et al. A Novel regulatory mechanism of smooth muscle α-actin expression by NRG-1/circACTA2/miR-548f-5p axis. Circ Res. (2017) 121:628–35. 10.1161/CIRCRESAHA.117.311441 [DOI] [PubMed] [Google Scholar]
- 55.Yang L, Han B, Zhang Z, Wang S, Bai Y, Zhang Y, et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation. (2020) 142:556–74. 10.1161/CIRCULATIONAHA.120.045765 [DOI] [PubMed] [Google Scholar]
- 56.Wang G, Han B, Shen L, Wu S, Yang L, Liao J, et al. Silencing of circular RNA HIPK2 in neural stem cells enhances functional recovery following ischaemic stroke. EBioMedicine. (2020) 52:102660. 10.1016/j.ebiom.2020.102751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. (1976) 73:3852–6. 10.1073/pnas.73.11.3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cocquerelle C, Mascrez B, Hétuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J. (1993) 7:155–60. 10.1096/fasebj.7.1.7678559 [DOI] [PubMed] [Google Scholar]
- 59.Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W, et al. CircRNA: a rising star in gastric cancer. Cell Mol Life Sci. (2020) 77:1661–80. 10.1007/s00018-019-03345-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin H, Wang Y, Wang P, Long F, Wang T. Mutual regulation between N6-methyladenosine (m6A) modification and circular RNAs in cancer: impacts on therapeutic resistance. Mol Cancer. (2022) 21:148. 10.1186/s12943-022-01620-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. (2016) 37:2602–11. 10.1093/eurheartj/ehv713 [DOI] [PubMed] [Google Scholar]
- 62.Zhang L, Li Z, Mao L, Wang H. Circular RNA in acute central nervous system injuries: a new target for therapeutic intervention. Front Mol Neurosci. (2022) 15:816182. 10.3389/fnmol.2022.816182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu K, Hu X, Chen H, Li F, Yin N, Liu AL, et al. Downregulation of circRNA DMNT3B contributes to diabetic retinal vascular dysfunction through targeting miR-20b-5p and BAMBI. EBioMedicine. (2019) 49:341–53. 10.1016/j.ebiom.2019.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang C, Wang X, Chen Y, Wu Z, Zhang C, Shi W, et al. The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4(+) T cells of systemic lupus erythematous. Clin Sci (Lond). (2018) 132:2285–98. 10.1042/CS20180403 [DOI] [PubMed] [Google Scholar]
- 65.Li L, Ni Z, Si X, Jiang L, Sang H, Xia W, et al. Emerging clues of regulatory roles of circular RNAs through modulating oxidative stress: focus on neurological and vascular diseases. Oxid Med Cell Longev. (2021) 2021:6659908. 10.1155/2021/6659908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xing C, Arai K, Lo EH, Hommel M. Pathophysiologic cascades in ischemic stroke. Int J Stroke. (2012) 7:378–85. 10.1111/j.1747-4949.2012.00839.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Markus HS. Stroke genetics. Hum Mol Genet. (2011) 20:R124–31. 10.1093/hmg/ddr345 [DOI] [PubMed] [Google Scholar]
- 68.Wang C, Yang Y, Xu M, Mao F, Yang P, Yuan S, et al. Deep sequencing of the rat MCAO cortexes reveals crucial circrnas involved in early stroke events and their regulatory networks. Neural Plast. (2021) 2021:9942537. 10.1155/2021/9942537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang X, Fang Y, Huang Q, Xu P, Lenahan C, Lu J, et al. An updated review of autophagy in ischemic stroke: From mechanisms to therapies. Exp Neurol. (2021) 340:113684. 10.1016/j.expneurol.2021.113684 [DOI] [PubMed] [Google Scholar]
- 70.Grishchuk Y, Ginet V, Truttmann AC, Clarke PG, Puyal J. Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy. (2011) 7:1115–31. 10.4161/auto.7.10.16608 [DOI] [PubMed] [Google Scholar]
- 71.Wang P, Guan YF, Du H, Zhai QW, Su DF, Miao CY, et al. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. (2012) 8:77–87. 10.4161/auto.8.1.18274 [DOI] [PubMed] [Google Scholar]
- 72.Ren Z, Xie P, Lv J, Hu Y, Guan Z, Chen L, et al. miR-187-3p inhibitor attenuates cerebral ischemia/reperfusion injury by regulating Seipin-mediated autophagic flux. Int J Mol Med. (2020) 46:1051–62. 10.3892/ijmm.2020.4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. (2011) 27:107–32. 10.1146/annurev-cellbio-092910-154005 [DOI] [PubMed] [Google Scholar]
- 74.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. (2013) 368:651–62. 10.1056/NEJMra1205406 [DOI] [PubMed] [Google Scholar]
- 75.Tian F, Deguchi K, Yamashita T, Ohta Y, Morimoto N, Shang J, et al. In vivo imaging of autophagy in a mouse stroke model. Autophagy. (2010) 6:1107–14. 10.4161/auto.6.8.13427 [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Tian F, Wang S, Wang F, Xiong L. Astrocyte autophagy flux protects neurons against oxygen-glucose deprivation and ischemic/reperfusion injury. Rejuvenation Res. (2018) 21:405–15. 10.1089/rej.2017.1999 [DOI] [PubMed] [Google Scholar]
- 77.Li L, Li L, Zhou X, Yu Y, Li Z, Zuo D, et al. Silver nanoparticles induce protective autophagy via Ca(2+)/CaMKKβ/AMPK/mTOR pathway in SH-SY5Y cells and rat brains. Nanotoxicology. (2019) 13:369–91. 10.1080/17435390.2018.1550226 [DOI] [PubMed] [Google Scholar]
- 78.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. (2011) 331:456–61. 10.1126/science.1196371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cui DR, Wang L, Jiang W, Qi AH, Zhou QH, Zhang XL, et al. Propofol prevents cerebral ischemia-triggered autophagy activation and cell death in the rat hippocampus through the NF-κB/p53 signaling pathway. Neuroscience. (2013) 246:117–32. 10.1016/j.neuroscience.2013.04.054 [DOI] [PubMed] [Google Scholar]
- 80.Li WL, Yu SP, Chen D, Yu SS, Jiang YJ, Genetta T, et al. The regulatory role of NF-κB in autophagy-like cell death after focal cerebral ischemia in mice. Neuroscience. (2013) 244:16–30. 10.1016/j.neuroscience.2013.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wei Y, Sinha S, Levine B. Dual role of JNK1-mediated phosphorylation of Bcl-2 in autophagy and apoptosis regulation. Autophagy. (2008) 4:949–51. 10.4161/auto.6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shimizu S, Konishi A, Nishida Y, Mizuta T, Nishina H, Yamamoto A, et al. Involvement of JNK in the regulation of autophagic cell death. Oncogene. (2010) 29:2070–82. 10.1038/onc.2009.487 [DOI] [PubMed] [Google Scholar]
- 83.Lorin S, Pierron G, Ryan KM, Codogno P, Djavaheri-Mergny M. Evidence for the interplay between JNK and p53-DRAM signalling pathways in the regulation of autophagy. Autophagy. (2010) 6:153–4. 10.4161/auto.6.1.10537 [DOI] [PubMed] [Google Scholar]
- 84.Wang W, Li J, Tan J, Wang M, Yang J, Zhang ZM, et al. Endonuclease G promotes autophagy by suppressing mTOR signaling and activating the DNA damage response. Nat Commun. (2021) 12:476. 10.1038/s41467-020-20780-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stambolic V, Woodgett JR. Mitogen inactivation of glycogen synthase kinase-3 beta in intact cells via serine 9 phosphorylation. Biochem J. (1994) 303 (Pt 3):701–4. 10.1042/bj3030701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang XM, Zhang L, Wang G, Niu W, He Z, Ding L, et al. Suppression of mitochondrial fission in experimental cerebral ischemia: the potential neuroprotective target of p38 MAPK inhibition. Neurochem Int. (2015) 90:1–8. 10.1016/j.neuint.2015.06.010 [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. (2007) 282:35803–13. 10.1074/jbc.M705231200 [DOI] [PubMed] [Google Scholar]
- 88.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. (2009) 43:67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Choi CH, Lee BH, Ahn SG, Oh SH. Proteasome inhibition-induced p38 MAPK/ERK signaling regulates autophagy and apoptosis through the dual phosphorylation of glycogen synthase kinase 3β. Biochem Biophys Res Commun. (2012) 418:759–64. 10.1016/j.bbrc.2012.01.095 [DOI] [PubMed] [Google Scholar]
- 90.Obermeier B, Daneman R, Ransohoff RM. Development, maintenance and disruption of the blood-brain barrier. Nat Med. (2013) 19:1584–96. 10.1038/nm.3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Profaci CP, Munji RN, Pulido RS, Daneman R. The blood-brain barrier in health and disease: Important unanswered questions. J Exp Med 217. (2020). 10.1084/jem.20190062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stokum JA, Gerzanich V, Simard JM. Molecular pathophysiology of cerebral edema. J Cereb Blood Flow Metab. (2016) 36:513–38. 10.1177/0271678X15617172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jickling GC, Liu D, Stamova B, Ander BP, Zhan X, Lu A, et al. Hemorrhagic transformation after ischemic stroke in animals and humans. J Cereb Blood Flow Metab. (2014) 34:185–99. 10.1038/jcbfm.2013.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Latour LL, Kang DW, Ezzeddine MA, Chalela JA, Warach S. Early blood-brain barrier disruption in human focal brain ischemia. Ann Neurol. (2004) 56:468–77. 10.1002/ana.20199 [DOI] [PubMed] [Google Scholar]
- 95.Zhang S, An Q, Wang T, Gao S, Zhou G. Autophagy- and MMP-2/9-mediated reduction and redistribution of ZO-1 contribute to hyperglycemia-increased blood-brain barrier permeability during early reperfusion in stroke. Neuroscience. (2018) 377:126–37. 10.1016/j.neuroscience.2018.02.035 [DOI] [PubMed] [Google Scholar]
- 96.Kim KA, Kim D, Kim JH, Shin YJ, Kim ES, Akram M, et al. Autophagy-mediated occludin degradation contributes to blood-brain barrier disruption during ischemia in bEnd.3 brain endothelial cells and rat ischemic stroke models. Fluids Barriers CNS. (2020) 17:21. 10.1186/s12987-020-00182-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu J, Weaver J, Jin X, Zhang Y, Xu J, Liu KJ, et al. Nitric oxide interacts with caveolin-1 to facilitate autophagy-lysosome-mediated claudin-5 degradation in oxygen-glucose deprivation-treated endothelial cells. Mol Neurobiol. (2016) 53:5935–47. 10.1007/s12035-015-9504-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, et al. Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice. J Cell Biol. (2003) 161:653–60. 10.1083/jcb.200302070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li H, Gao A, Feng D, Wang Y, Zhang L, Cui Y, et al. Evaluation of the protective potential of brain microvascular endothelial cell autophagy on blood-brain barrier integrity during experimental cerebral ischemia-reperfusion injury. Transl Stroke Res. (2014) 5:618–26. 10.1007/s12975-014-0354-x [DOI] [PubMed] [Google Scholar]
- 100.Cao Y, Pan L, Zhang X, Guo W, Huang D. LncRNA SNHG3 promotes autophagy-induced neuronal cell apoptosis by acting as a ceRNA for miR-485 to up-regulate ATG7 expression. Metab Brain Dis. (2020) 35:1361–9. 10.1007/s11011-020-00607-1 [DOI] [PubMed] [Google Scholar]
- 101.Xie C, Ginet V, Sun Y, Koike M, Zhou K, Li T, et al. Neuroprotection by selective neuronal deletion of Atg7 in neonatal brain injury. Autophagy. (2016) 12:410–23. 10.1080/15548627.2015.1132134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang B, Deng F, Zhou C, Fang S. ClC-3 induction protects against cerebral ischemia/reperfusion injury through promoting Beclin1/Vps34-mediated autophagy. Hum Cell. (2020) 33:1046–55. 10.1007/s13577-020-00406-x [DOI] [PubMed] [Google Scholar]
- 103.Hei C, Liu P, Yang X, Niu J. Inhibition PALi of mTOR signaling confers protection against cerebral ischemic injury in acute hyperglycemic rats. Int J Biol Sci. (2017) 13:878–87. 10.7150/ijbs.18976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huang L, Chen C, Zhang X, Li X, Chen Z, Yang C, et al. Neuroprotective effect of curcumin against cerebral ischemia-reperfusion via mediating autophagy and inflammation. J Mol Neurosci. (2018) 64:129–39. 10.1007/s12031-017-1006-x [DOI] [PubMed] [Google Scholar]
- 105.Kim EK, Choi EJ. Compromised MAPK signaling in human diseases: an update. Arch Toxicol. (2015) 89:867–82. 10.1007/s00204-015-1472-2 [DOI] [PubMed] [Google Scholar]
- 106.Song YQ, Zou HL, Zhao YJ, Yu LQ, Tan ZX, Kong R, et al. Activation of p38-mitogen-activated protein kinase contributes to ischemia reperfusion in rat brain. Genet Mol Res. (2016) 15:1–13. 10.4238/gmr.15038492 [DOI] [PubMed] [Google Scholar]
- 107.Li H, Zhou S, Wu L, Liu K, Zhang Y, Ma G, et al. The role of p38MAPK signal pathway in the neuroprotective mechanism of limb postconditioning against rat cerebral ischemia/reperfusion injury. J Neurol Sci. (2015) 357:270–5. 10.1016/j.jns.2015.08.004 [DOI] [PubMed] [Google Scholar]
- 108.Ferrer I, Friguls B, Dalfó E, Planas AM. Early modifications in the expression of mitogen-activated protein kinase (MAPK/ERK), stress-activated kinases SAPK/JNK and p38, and their phosphorylated substrates following focal cerebral ischemia. Acta Neuropathol. 105:425–37. 10.1007/s00401-002-0661-2 [DOI] [PubMed] [Google Scholar]
- 109.Barteczek P, Li L, Ernst AS, Böhler LI, Marti HH, Kunze R, et al. Neuronal HIF-1α and HIF-2α deficiency improves neuronal survival and sensorimotor function in the early acute phase after ischemic stroke. J Cereb Blood Flow Metab. (2017) 37:291–306. 10.1177/0271678X15624933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gong G, Hu L, Liu Y, Bai S, Dai X, Yin L, et al. Upregulation of HIF-1α protein induces mitochondrial autophagy in primary cortical cell cultures through the inhibition of the mTOR pathway. Int J Mol Med. (2014) 34:1133–40. 10.3892/ijmm.2014.1850 [DOI] [PubMed] [Google Scholar]
- 111.Lv B, Li F, Han J, Fang J, Xu L, Sun C, et al. Hif-1α overexpression improves transplanted bone mesenchymal stem cells survival in rat MCAO stroke model. Front Mol Neurosci. (2017) 10:80. 10.3389/fnmol.2017.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ma Q, Long S, Gan Z, Tettamanti G, Li K, Tian L, et al. Transcriptional and post-transcriptional regulation of autophagy. Cells. (2022) 11:441. 10.3390/cells11030441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu P, Zhang X, Cao J, Yang J, Chen Z, Wang W, et al. The novel role of circular RNA ST3GAL6 on blocking gastric cancer malignant behaviours through autophagy regulated by the FOXP2/MET/mTOR axis. Clin Transl Med. (2022) 12:e707. 10.1002/ctm2.707 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 114.Li XX, Xiao L, Chung HK, Ma XX, Liu X, Song JL, et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. (2020) 40:e00492-19. 10.1128/MCB.00492-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhou LY, Zhai M, Huang Y, Xu S, An T, Wang YH, et al. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. (2019) 26:1299–315. 10.1038/s41418-018-0206-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu Y, Chen X, Yao J, Kang J. Circular RNA ACR relieves high glucose-aroused RSC96 cell apoptosis and autophagy via declining microRNA-145-3p. J Cell Biochem. (2019). 10.1002/jcb.29568 [DOI] [PubMed] [Google Scholar]
- 117.Wu Y, Zhao B, Chen X, Geng X, Zhang Z. Circ_0009910 sponges miR-491-5p to promote acute myeloid leukemia progression through modulating B4GALT5 expression and PI3K/AKT signaling pathway. Int J Lab Hematol. (2022) 44:320–32. 10.1111/ijlh.13742 [DOI] [PubMed] [Google Scholar]
- 118.Jin X, Gao J, Zheng R, Yu M, Ren Y, Yan T, et al. circRNA_002581-miR-122-CPEB1 axis alleviates NASH through restoring PTEN-AMPK-mTOR pathway regulated autophagy. Cell Death Dis. (2020) 11:123. 10.1038/s41419-020-2293-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shang J, Chen WM, Liu S, Wang ZH, Wei TN, Chen ZZ, et al. CircPAN3 contributes to drug resistance in acute myeloid leukemia through regulation of autophagy. Leuk Res. (2019) 85:106198. 10.1016/j.leukres.2019.106198 [DOI] [PubMed] [Google Scholar]
- 120.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. (2020) 16:659–71. 10.1080/15548627.2019.1634945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yao L, Xu B, Li X. Neisseria gonorrhoeae-induced salpingitis is targeted by circular RNA EIF3K via miR-139-5p and regulating MAPK/NF-κB signaling pathway to promotes apoptosis and autophagy bacterial cells. Microb Pathog. (2020) 142:104051. 10.1016/j.micpath.2020.104051 [DOI] [PubMed] [Google Scholar]
- 122.Gan J, Yuan J, Liu Y, Lu Z, Xue Y, Shi L, et al. Circular RNA_101237 mediates anoxia/reoxygenation injury by targeting let-7a-5p/IGF2BP3 in cardiomyocytes. Int J Mol Med. (2020) 45:451–60. 10.3892/ijmm.2019.4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lederer M, Bley N, Schleifer C, Hüttelmaier S. The role of the oncofetal IGF2 mRNA-binding protein 3 (IGF2BP3) in cancer. Semin Cancer Biol. (2014) 29:3–12. 10.1016/j.semcancer.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 124.Panebianco F, Kelly LM, Liu P, Zhong S, Dacic S, Wang X, et al. THADA fusion is a mechanism of IGF2BP3 activation and IGF1R signaling in thyroid cancer. Proc Natl Acad Sci USA. (2017) 114:2307–12. 10.1073/pnas.1614265114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. (2020) 20:74. 10.1186/s12935-020-1151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong M, Long X, et al. CircMUC16 promotes autophagy of epithelial ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer. (2020) 19:45. 10.1186/s12943-020-01163-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chen M, Ai G, Zhou J, Mao W, Li H, Guo J, et al. circMTO1 promotes tumorigenesis and chemoresistance of cervical cancer via regulating miR-6893. Biomed Pharmacother. (2019) 117:109064. 10.1016/j.biopha.2019.109064 [DOI] [PubMed] [Google Scholar]
- 128.Guo J, Chen M, Ai G, Mao W, Li H, Zhou J, et al. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother. (2019) 115:108957. 10.1016/j.biopha.2019.108957 [DOI] [PubMed] [Google Scholar]
- 129.Du WW, Yang W, Li X, Awan FM, Yang Z, Fang L, et al. A circular RNA circ-DNMT1 enhances breast cancer progression by activating autophagy. Oncogene. (2018) 37:5829–42. 10.1038/s41388-018-0369-y [DOI] [PubMed] [Google Scholar]
- 130.Cao HX, Miao CF, Sang LN, Huang YM, Zhang R, Sun L, et al. Circ_0009910 promotes imatinib resistance through ULK1-induced autophagy by sponging miR-34a-5p in chronic myeloid leukemia. Life Sci. (2020) 243:117255. 10.1016/j.lfs.2020.117255 [DOI] [PubMed] [Google Scholar]
- 131.Ren Q, Li H, Wang X. The circular RNA ZNF292 alleviates OGD-induced injury in H9c2 cells via targeting BNIP3. Cell Cycle. (2019) 18:3365–77. 10.1080/15384101.2019.1676585 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Chen W, Wang H, Feng J, Chen L. Overexpression of circRNA circUCK2 attenuates cell apoptosis in cerebral ischemia-reperfusion injury via miR-125b-5p/GDF11 signaling. Mol Ther Nucleic Acids. (2020) 22:673–83. 10.1016/j.omtn.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wu L, Xu H, Zhang W, Chen Z, Li W, Ke W, et al. Circular RNA circCCDC9 alleviates ischaemic stroke ischaemia/reperfusion injury via the Notch pathway. J Cell Mol Med. (2020) 24:14152–9. 10.1111/jcmm.16025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bai Y, Zhang Y, Han B, Yang L, Chen X, Huang R, et al. Circular RNA DLGAP4 ameliorates ischemic stroke outcomes by targeting miR-143 to regulate endothelial-mesenchymal transition associated with blood-brain barrier integrity. J Neurosci. (2018) 38:32–50. 10.1523/JNEUROSCI.1348-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang H, Tu Z, Yang D, Hu M, Zhou L, Li Q, et al. Exosomes from hypoxic pre-treated ADSCs attenuate acute ischemic stroke-induced brain injury via delivery of circ-Rps5 and promote M2 microglia/macrophage polarization. Neurosci Lett. (2022) 769:136389. 10.1016/j.neulet.2021.136389 [DOI] [PubMed] [Google Scholar]
- 136.Wu F, Han B, Wu S, Yang L, Leng S, Li M, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. (2019) 39:7369–93. 10.1523/JNEUROSCI.0299-19.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xu T, Li Y, Zhu N, Su Y, Li J, Ke K, et al. circSKA3 acts as a sponge of miR-6796-5p to be associated with outcomes of ischemic stroke by regulating matrix metalloproteinase 9 expression. Eur J Neurol. (2022) 29:486–95. 10.1111/ene.15164 [DOI] [PubMed] [Google Scholar]
- 138.Deng S, Huang C. E3 ubiquitin ligases in regulating stress fiber, lamellipodium, and focal adhesion dynamics. Cell Adh Migr. (2014) 8:49–54. 10.4161/cam.27480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.He GH, Wang Z, Xu W, Song KP, Xiao H. Knockdown of circHECTD1 inhibits oxygen-glucose deprivation and reperfusion induced endothelial-mesenchymal transition. Metab Brain Dis. (2022) 37:427–37. 10.1007/s11011-021-00891-5 [DOI] [PubMed] [Google Scholar]
- 140.Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu G, et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy. (2018) 14:404–18. 10.1080/15548627.2017.1414755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li H, Niu X, Shi H, Feng M, Du Y, Sun R, et al. circHECTD1 attenuates apoptosis of alveolar epithelial cells in acute lung injury. Lab Invest. (2022) 102:945–956. 10.1038/s41374-022-00781-z [DOI] [PubMed] [Google Scholar]
- 142.Ge X, Sun Y, Tang Y, Lin J, Zhou F, Yao G, et al. Circular RNA HECTD1 knockdown inhibits transforming growth factor-beta/small mothers against decapentaplegic (TGF-β/Smad) signaling to reduce hypertrophic scar fibrosis. Bioengineered. (2022) 13:7303–15. 10.1080/21655979.2022.2048771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xu Y, Tian Y, Li F, Wang Y, Yang J, Gong H, et al. Circular RNA HECTD1 mitigates ulcerative colitis by promoting enterocyte autophagy via miR-182-5p/HuR axis. Inflamm Bowel Dis. (2022) 28:273–88. 10.1093/ibd/izab188 [DOI] [PubMed] [Google Scholar]
- 144.Cai J, Chen Z, Wang J, Wang J, Chen X, Liang L, et al. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis. (2019) 10:576. 10.1038/s41419-019-1814-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jiang QL, Feng SJ, Yang ZY, Xu Q, Wang SZ. CircHECTD1 up-regulates mucin 1 expression to accelerate hepatocellular carcinoma development by targeting microRNA-485-5p via a competing endogenous RNA mechanism. Chin Med J (Engl). (2020) 133:1774–85. 10.1097/CM9.0000000000000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Li W, Wang S, Shan B, Cheng X, He H, Qin J, et al. CircHECTD1 regulates cell proliferation and migration by the miR-320-5p/SLC2A1 axis in glioblastoma multiform. Front Oncol. (2021) 11:666391. 10.3389/fonc.2021.666391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chu H, Wang W, Luo W, Zhang W, Cheng Y, Huang J, et al. CircHECTD1 mediates pulmonary fibroblast activation via HECTD1. Ther Adv Chronic Dis. (2019) 10:2040622319891558. 10.1177/2040622319891558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kwon JJ, Hahn WC, A. Leucine-rich repeat protein provides a SHOC2 the RAS circuit: a structure-function perspective. Mol Cell Biol 41. (2021). 10.1128/MCB.00627-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng T, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. (2019) 144:2501–15. 10.1002/ijc.31977 [DOI] [PubMed] [Google Scholar]
- 150.Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. (2020) 121:2118–26. 10.1002/jcb.28239 [DOI] [PubMed] [Google Scholar]
- 151.Yang T, Martin P, Fogarty B, Brown A, Schurman K, Phipps R, et al. Exosome delivered anticancer drugs across the blood-brain barrier for brain cancer therapy in Danio rerio. Pharm Res. (2015) 32:2003–14. 10.1007/s11095-014-1593-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Koivunen P, Laukka T. The TET enzymes. Cell Mol Life Sci. (2018) 75:1339–48. 10.1007/s00018-017-2721-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang C, Zhong T, Li Y, Li X, Yuan X, Liu L, et al. The hepatic AMPK-TET1-SIRT1 axis regulates glucose homeostasis. Elife. (2021) 10:e70672. 10.7554/eLife.70672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsukamoto Y, Kuwabara K, Hirota S, Ikeda J, Stern D, Yanagi H, et al. 150-kD oxygen-regulated protein is expressed in human atherosclerotic plaques and allows mononuclear phagocytes to withstand cellular stress on exposure to hypoxia and modified low density lipoprotein. J Clin Invest. (1996) 98:1930–41. 10.1172/JCI118994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sanson M, Ingueneau C, Vindis C, Thiers JC, Glock Y, Rousseau H, et al. Oxygen-regulated protein-150 prevents calcium homeostasis deregulation and apoptosis induced by oxidized LDL in vascular cells. Cell Death Differ. (2008) 15:1255–65. 10.1038/cdd.2008.36 [DOI] [PubMed] [Google Scholar]
- 156.Tanida I, Yamasaki M, Komatsu M, Ueno T. The FAP motif within human ATG7, an autophagy-related E1-like enzyme, is essential for the E2-substrate reaction of LC3 lipidation. Autophagy. (2012) 8:88–97. 10.4161/auto.8.1.18339 [DOI] [PubMed] [Google Scholar]
- 157.Xin Q, Ji B, Cheng B, Wang C, Liu H, Chen X, et al. Endoplasmic reticulum stress in cerebral ischemia. Neurochem Int. (2014) 68:18–27. 10.1016/j.neuint.2014.02.001 [DOI] [PubMed] [Google Scholar]
- 158.Ma W, Ding F, Wang X, Huang Q, Zhang L, Bi C. By targeting Atg7 MicroRNA-143 mediates oxidative stress-induced autophagy of c-Kit(+) mouse cardiac progenitor cells. EBioMedicine. (2018) 32:182–91. 10.1016/j.ebiom.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Lv XW, Wang MJ, Qin QY, Lu P, Qin GW. 6-Gingerol relieves myocardial ischaemia/reperfusion injury by regulating lncRNA H19/miR-143/ATG7 signaling axis-mediated autophagy. Lab Invest. (2021) 101:865–77. 10.1038/s41374-021-00575-9 [DOI] [PubMed] [Google Scholar]
- 160.Chen G, Wang M, Ruan Z, Zhu L, Tang C. Mesenchymal stem cell-derived exosomal miR-143-3p suppresses myocardial ischemia-reperfusion injury by regulating autophagy. Life Sci. (2021) 280:119742. 10.1016/j.lfs.2021.119742 [DOI] [PubMed] [Google Scholar]
- 161.Lin XT, Zheng XB, Fan DJ, Yao QQ, Hu JC, Lian L, et al. MicroRNA-143 Targets ATG2B to inhibit autophagy and increase inflammatory responses in crohn's disease. Inflamm Bowel Dis. (2018) 24: 781–91. 10.1093/ibd/izx075 [DOI] [PubMed] [Google Scholar]
- 162.Liu C, Wang JO, Zhou WY, Chang XY, Zhang MM, Zhang Y, Yang XH. Long non-coding RNA LINC01207 silencing suppresses AGR2 expression to facilitate autophagy and apoptosis of pancreatic cancer cells by sponging miR-143-5p. Mol Cell Endocrinol. (2019) 493:110424. 10.1016/j.mce.2019.04.004 [DOI] [PubMed] [Google Scholar]
- 163.Liu YF, Luo D, Li X, Li ZQ, Yu X, Zhu HW. PVT1 knockdown inhibits autophagy and improves gemcitabine sensitivity by regulating the MiR-143/HIF-1?/VMP1 axis in pancreatic cancer. Pancreas. (2021) 50:227–34. 10.1097/MPA.0000000000001747 [DOI] [PubMed] [Google Scholar]
- 164.Zhang H, Kang J, Liu L, Chen L, Ren S, Tao Y. MicroRNA-143 sensitizes acute myeloid leukemia cells to cytarabine via targeting ATG7- and ATG2B-dependent autophagy. Aging. (2020) 12:20111–26. 10.18632/aging.103614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Takai T, Tsujino T, Yoshikawa Y, Inamoto T, Sugito N, Kuranaga Y, et al. Synthetic miR-143 exhibited an anti-cancer effect via the downregulation of K-RAS Networks of Renal Cell Cancer Cells In Vitro and In Vivo. Mol Ther. (2019) 27:1017–27. 10.1016/j.ymthe.2019.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Li Y, Zhang H, Guo J, Li W, Wang X, Zhang C, Sun Q, Ma Z. Downregulation of LINC01296 suppresses non-small-cell lung cancer via targeting miR-143-3p/ATG2B. Acta Biochim Biophys Sin (Shanghai). (2021) 53: 1681–90. 10.1093/abbs/gmab149 [DOI] [PubMed] [Google Scholar]
- 167.Yang H, Hu T, Hu P, Qi C, Qian L. miR 143 3p inhibits endometriotic stromal cell proliferation and invasion by inactivating autophagy in endometriosis. Mol Med Rep. (2021) 23:356. 10.3892/mmr.2021.11995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu J, Li M, Wang Y, Luo J. Curcumin sensitizes prostate cancer cells to radiation partly via epigenetic activation of miR-143 and miR-143 mediated autophagy inhibition. J Drug Target. (2017) 25:645–52. 10.1080/1061186X.2017.1315686 [DOI] [PubMed] [Google Scholar]
- 169.Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. (2017) 38:1402–12. 10.1093/eurheartj/ehw001 [DOI] [PubMed] [Google Scholar]
- 170.Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. (2016) 35:3919–31. 10.1038/onc.2015.460 [DOI] [PubMed] [Google Scholar]
- 171.Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T, et al. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. (2020) 121:2595–605. 10.1002/jcb.29481 [DOI] [PubMed] [Google Scholar]
- 172.Wang C, Tao W, Ni S, Chen Q. Circular RNA circ-Foxo3 induced cell apoptosis in urothelial carcinoma via interaction with miR-191-5p. Onco Targets Ther. (2019) 12:8085–94. 10.2147/OTT.S215823 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 173.Li Y, Qiao L, Zang Y, Ni W, Xu Z. Circular RNA FOXO3 Suppresses Bladder Cancer Progression and Metastasis by Regulating MiR-9-5p/TGFBR2. Cancer Manag Res. (2020) 12:5049–56. 10.2147/CMAR.S253412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Deng YY, Min YJ, Zhou K, Yang QS, Peng M, Cui ZR, et al. Identification of the tumor-suppressive role of circular RNA-FOXO3 in colorectal cancer via regulation of miR-543/LATS1 axis. Oncol Rep 46. (2021). 10.3892/or.2021.8190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhou J, Zhou LY, Tang X, Zhang J, Zhai LL, Yi YY, et al. Circ-Foxo3 is positively associated with the Foxo3 gene and leads to better prognosis of acute myeloid leukemia patients. BMC Cancer. (2019) 19:930. 10.1186/s12885-019-5967-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. (2017) 24:357–70. 10.1038/cdd.2016.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA, et al. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. (1992) 70:993–1006. 10.1016/0092-8674(92)90249-C [DOI] [PubMed] [Google Scholar]
- 178.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. (1993) 75:805–16. 10.1016/0092-8674(93)90499-G [DOI] [PubMed] [Google Scholar]
- 179.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. (2016) 44:2846–58. 10.1093/nar/gkw027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin S, Liu K, Zhang Y, Jiang M, Lu R, Folts CJ, et al. Pharmacological targeting of p38 MAP-Kinase 6 (MAP2K6) inhibits the growth of esophageal adenocarcinoma. Cell Signal. (2018) 51:222–32. 10.1016/j.cellsig.2018.08.008 [DOI] [PubMed] [Google Scholar]
- 181.Mehta SL, Pandi G, Vemuganti R. Circular RNA expression profiles alter significantly in mouse brain after transient focal ischemia. Stroke. (2017) 48:2541–8. 10.1161/STROKEAHA.117.017469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Noda NN, Inagaki F. Mechanisms of autophagy. Annu Rev Biophys. (2015) 44:101–22. 10.1146/annurev-biophys-060414-034248 [DOI] [PubMed] [Google Scholar]
- 183.Li S, Hu W, Deng F, Chen S, Zhu P, Wang M, et al. of Circular RNA hsa_circ_0001599 as a novel biomarker for large-artery atherosclerotic stroke. DNA Cell Biol. (2021) 40:457–68. 10.1089/dna.2020.5662 [DOI] [PubMed] [Google Scholar]
- 184.Maiarù M, Tochiki KK, Cox MB, Annan LV, Bell CG, Feng X, et al. The stress regulator FKBP51 drives chronic pain by modulating spinal glucocorticoid signaling. Sci Transl Med. (2016) 8:325ra.19. 10.1126/scitranslmed.aab3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yu S, Yu M, Bu Z, He P, Feng J. FKBP5 exacerbates impairments in cerebral ischemic stroke by inducing autophagy via the AKT/FOXO3 pathway. Front Cell Neurosci. (2020) 14:193. 10.3389/fncel.2020.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]