Abstract
Objective:
Testicular germ cell tumors (TGCT) are the most common solid malignancy in adolescent and young men, with a rising incidence over the past 20 years. Overall, TGCTs are second in terms of the average life years lost per person dying of cancer, and clinical therapeutics without adverse long-term side effects are lacking. Platinum-based regimens for TGCTs have heterogeneous outcomes even within the same histotype that frequently leads to under- and over-treatment. Understanding of molecular differences that lead to diverse outcomes of TGCT patients may improve current treatment approaches. Seminoma is the most common subtype of TGCTs, which can either be pure or present in combination with other histotypes.
Methods:
Here we conducted a computational study of 64 pure seminoma samples from The Cancer Genome Atlas, applied consensus clustering approach to their transcriptomic data and revealed 2 clinically relevant seminoma subtypes: seminoma subtype 1 and 2.
Results:
Our analysis identified significant differences in pluripotency stage, activity of double stranded DNA breaks repair mechanisms, rates of loss of heterozygosity, and expression of lncRNA responsible for cisplatin resistance between the subtypes. Seminoma subtype 1 is characterized by higher pluripotency state, while subtype 2 showed attributes of reprograming into non-seminomatous TGCT. The seminoma subtypes we identified may provide a molecular underpinning for variable responses to chemotherapy and radiation.
Conclusion:
Translating our findings into clinical care may help improve risk stratification of seminoma, decrease overtreatment rates, and increase long-term quality of life for TGCT survivors.
Keywords: Testicular cancer, seminoma, germ cell tumors, subtypes, urological neoplasms
Introduction
Testicular germ cell tumor (TGCT) is the most common solid cancer among men aged 15 to 44 years .1 2 major types of TGCTs are seminomatous (SE) and non-seminomatous (NSE).2 NSE TGCTs include embryonal carcinoma (EC), teratoma (TE), yolk sac tumor (YST), choriocarcinoma and mixed GCTs, which include combinations of any NSE or SE subhistologies. Mixed type of TGCTs in its own turn represents the most common type of NSE, since pure EC, TE, and YST are rare.3 However, seminoma is the most common histological subtype of TGCT among young men 15 to 44 years of age.1,4,5 In 2021 the American Cancer Society estimated around 9470 TGCT cases in the US6 and around 39 000 cases worldwide.7 TGCT is the second cancer type (first being all pediatric cancers combined) based on calculation of life years lost per person dying of cancer.8 NSE tumors are more aggressive compared to SE, requiring more intensive treatment approaches.8-11
Management of patients with seminoma starts with orchiectomy followed by observation, platinum-based chemotherapy (cisplatin) or radiation therapy.12-14 Despite a high patient survival rate, current treatments significantly decrease patients’ quality of life and can cause combinations of around 40 severe adverse long-term side effects like infertility, neurotoxicity, hypercholesterolemia, secondary cancers, and death.15-17 Following chemotherapy, TGCT patients demonstrated a 3.6-dB decline in hearing for every 100 mg/m2 increase in cumulative cisplatin dose.18,19 BEP-treated patients received more than 400 mg/m2 of cisplatin had impaired renal function at the end of treatment and 20% decrease of glomerular filtration rate after 5 years of follow-up.20,21 Moreover, about 20% of seminoma patients will experience relapse, and the reason for this phenomenon is unclear.22,23 Relapsed patients will be treated again using conventional and high-dose chemotherapy with stem cell transplant that aggravate side effects drastically.
Multiple clinical studies demonstrate heterogeneous patient outcomes in the treatment of seminoma patients.24-26 Despite limited understanding of seminoma intratumoral heterogeneity,27,28 the existence and relevance of seminoma subtypes remains unclear and has never been studied in detail.
Recent progress in understanding the molecular heterogeneity of cancer types and intertumoral heterogeneity has suggested improvements for therapeutic strategies.29,30 A large variety of cancer types such as meningioma,31 pancreatic neuroendocrine tumors,32 and squamous cell carcinoma of the head and neck33 reveal the existence of clinically relevant subtypes with distinctive molecular characteristics. Moreover, clinically relevant subtypes for breast34,35 and lung cancer36 have led to different treatment strategies in the clinic, which highlights the importance of subtype-specific therapy. A better understanding of cancer heterogeneity and the identification of subtypes with distinctive clinical characteristics should lay the basis for future applications of personalized cancer therapy aimed to increase the efficiency of patient treatment with reduced toxicity and side effects.37 Various methods for precision cancer medicine are used in clinics for management of cancer patients. Amongst them are diagnostic methods based on genome profiling,38 analysis of tumor proteomic39 and transcriptomic40 data, which have been successfully applied to the most abundant cancer types including lung,41 breast,42 and prostate43 cancers.
Here we conducted a computational study of 64 pure seminoma samples from the TCGA data portal. We applied consensus clustering approach to transcriptomic data that revealed 2 distinct seminoma subtypes. Analysis of transcriptomic, genomic and epigenomic data showed similarity of seminoma subtype 2 with non-seminomatous GCTs. Therefore, we propose that consideration of identified subtypes might help to improve seminoma clinical management by administrating subtype-specific treatment.
Materials and Methods
Data collection
The RNA-sequencing data (HTSeq-count), histopathological slides, DNA methylation data, copy number variation data, single-nucleotide variants, level of lymphocytes infiltration and corresponding patient clinical information (race, ethnicity, clinical stage) were collected for 64 pure seminoma cases (TCGA-TGCT project) from The Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov). Samples acquisition, library preparation, and sequencing details for pure seminoma samples from TCGA were described previously.2 One hundred sixty-two available histopathological slides were evaluated by a pathologist subspecialized in surgical oncology and genitourinary pathology (L.J.). Two cases (TCGA-2G-AAG9 and TCGA-2G-AAH0) were removed from our dataset due to additional types of TGCT (teratoma and embryonal carcinoma) identified on the slides. Two other cases (TCGA-2G-AAFG and TCGA-2G-AAHP) include primary and secondary tumors that were considered as different cases with additional number after case ID, “1” for primary and “2” for secondary tumor respectively (eg, TCGA-2G-AAFG1 and TCGA-2G-AAFG2). Telomer lengths data for pure seminoma TCGA cases was retrieved from previous study.44 List of long non-coding RNAs was retrieved from LNCipedia version 5.2.45
Data processing and consensus clustering
Pure seminomas are commonly infiltrated by lymphocytes.46 To focus on tumor transcriptome, we incorporated an additional filtration step that removes immune cell transcripts (filtered gene set). Immune cell transcripts were defined using the Database of Immune Cell Expression47. ConsensusClusterPlus R-package48 was used to identify transcriptional clusters on the filtered gene set. We used 1000 iterations, 80% sample resampling from 2 to 6 clusters (k2–k6) using hierarchical clustering with average innerLinkage and finalLinkage, as well as Spearman correlation as the similarity metric. Clustering significance was checked in pairwise comparisons using SigClust.49 Gene expression data was median centered and log2 transformed. The R function hclust was used for unsupervised hierarchical clustering of pure seminoma samples with Ward’s method, and the resulting heatmap was cut with the R function cutree. Boxplots were generated using R function ggplot. Comparison P-values for boxplots were calculated using Wilcoxon test.
Identification of differentially expressed genes in seminoma subtypes
Differentially expressed genes for identified seminoma subtypes were obtained using DESeq2 package50 and raw counts from TCGA as the input data. DESeq2 parameters were set by default. At the first step we selected genes with baseMean > 10 (non-filtered gene set) and genes with baseMean > 5 (filtered gene set). Then, we used Log2 Fold Change > 2 and adjusted P-value < .005 to identify genes with significant differential expression. Signature genes differentially expressed in different types of TGCTs were retrieved from previous study.51
Gene set enrichment analysis (GSEA)
GSEA52 was employed to identify characteristically molecular pathways enriched or depleted in the seminoma subtypes. We created a ranked list of genes with distinct level of expression between 2 seminoma subtypes. For that we processed the full list of 53 644 transcripts and removed those that do not have gene names. Then, we removed genes for which all samples have zero counts or extreme count outlier or low mean normalized count (automatic DESeq2 filtering). As the output we generated a list of 14 477 genes. For each of the genes we calculated a rank using formula: (sign of Log2FC) * (−Log10(adjusted P-value)). Ranked gene list was uploaded to GSEA program. The reference gene set “h.all.v7.4.symbols.gmt [Hallmarks]” was obtained from the Molecular Signatures Database (MSigDB). The number of gene set permutations were 1000 times for each analysis. Groups with minimum 5 and maximum 500 genes were selected. The FDR q-value < 0.05 and normalized enrichment score (NES) > 1.5 were considered significant. Top 1000 DEGs were selected based on the gene rank.
Loss of heterozygosity
Previously somatic DNA copy number was determined from Affymetrix SNP 6.0 arrays for 10 522 TCGA samples.53 Ploidy, absolute total copy number and presence or absence of loss of heterozygosity (LOH) for each segment in the genome were determined using the ABSOLUTE algorithm.54 Experimental and data processing details were described previously.53 We downloaded LOH indication for each genome segment of 64 pure seminoma samples from TCGA PanCanAtlas study mentioned above.53 Coordinates of chromosome arms were determined using the UCSC Genome Browser.55 We calculated the fraction of seminoma samples for each subtype that contain LOH in particular chromosome arm at least in one genome segment. Difference between subtypes LOH was evaluated using Chi-squared test. P-value < .05 was considered significant.
Copy number variations analysis
Segmented copy number data for all seminoma samples from TCGA were analyzed using GISTIC2.0 algorithm56 to identify amplified and deleterious regions. GISTIC2.0 q-value cutoff was set at 0.25. Genes located within amplified/deleterious loci between seminoma subtypes were checked for differential expression. Significantly overexpressed genes within amplified loci of seminoma subtypes were analyzed and known anticancer drug targets were selected using DrugBank.57
Methylation analysis
Raw IDAT files for all seminoma samples form TCGA data portal were used for analysis of methylation data. Data from IDAT files were processed and normalized using Minfi package.58 Quantile and functional normalization were applied. For identification of differentially methylated probes and regions we used packages limma59 and DMRcate60 correspondently.
Biomarkers identification
Potential biomarkers were defined among genes differentially expressed between 2 identified seminoma subtypes (adjusted P-value < .005, |Log2FC| > 1). Biomarkers were rated by the specificity (true negative rate). The specificity was calculated based on the level of genes expression using the minimal value for one of subtypes as a threshold. Pseudogenes and novel transcripts were removed from consideration.
Results
Transcriptomic data analysis reveals 2 distinct molecular subtypes of pure seminomas
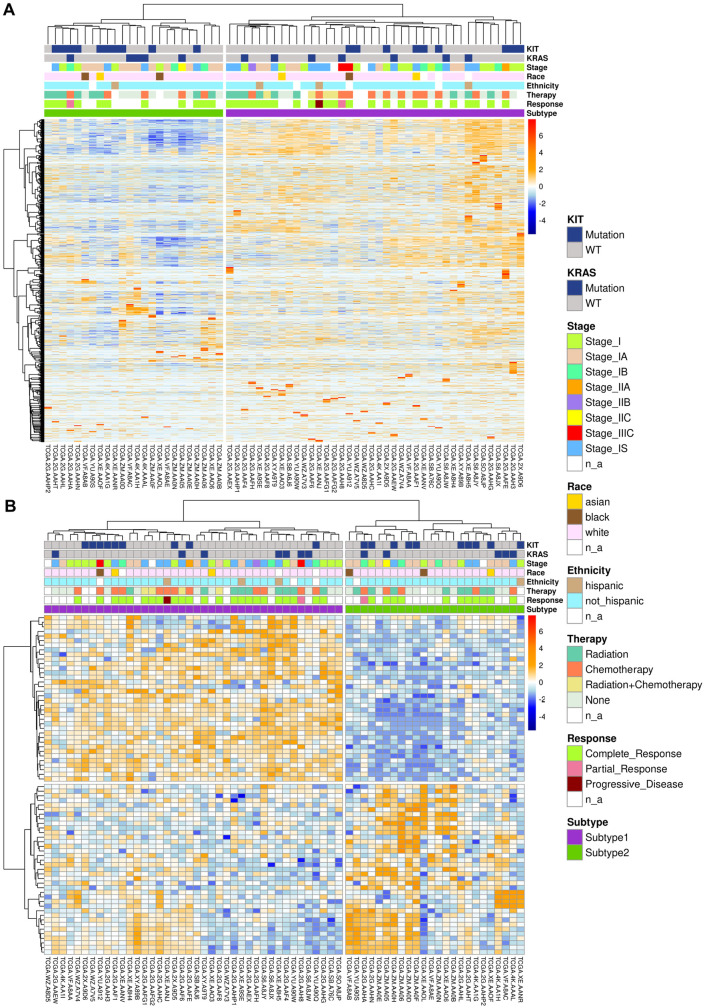
Overall, 64 pure seminoma cases from TCGA database were used for this study. Consensus clustering of 64 samples identified 2 distinct transcriptomic seminoma subtypes (Figure 1). Consensus matrix heatmap represents the results of consensus clustering approach (Supplemental Figure S1) and indicates the frequency of samples occurrence in the same cluster, over repeated sub-samplings of the cohort. The higher the intensity of blue color, the higher the co-clustering. The detailed clinical and pathological information for the seminoma subtypes (Table 1 and Supplemental Table S1) shows significant overrepresentation of seminoma patients of Non-Hispanic ethnicity and White race in the TCGA cohort. Recent studies showed that ethnicity and race could have significant relevance for TGCTs therapeutic response, revealing segment 2q11.1 amplification signature for Latin-origin population61 and 18 novel mutations among Asian patients.62 Therefore, understanding of seminoma subtypes distribution amongst patients from different demographic groups requires additional research using expanded sample cohorts. Study power for our dataset was estimated as 0.75 using RnaSeqSampleSize R package.63
Figure 1.
Clustering of pure seminoma samples based on transcriptomic data: (A) clustering of seminoma samples based on filtered gene set and (B) differentially expressed genes between 2 seminoma subtypes. Columns correspond to samples, rows to genes.
Table 1.
Subtype-based clinical data of seminoma patients.
| Subtype 1 (total = 40) | Subtype 2 (total = 24) | |
|---|---|---|
| Average age | 33.03 | 35.75 |
| KIT mutations | 9 | 10 |
| KRAS mutations | 7 | 5 |
| Stage | ||
| I | 12 | 4 |
| IA | 7 | 11 |
| IB | 3 | 3 |
| IS | 12 | 4 |
| IIA | 1 | 0 |
| IIB | 1 | 0 |
| IIC | 0 | 1 |
| IIIC | 2 | 0 |
| n/a | 2 | 0 |
| Race | ||
| White | 34 | 21 |
| Black | 1 | 2 |
| Asian | 2 | 1 |
| n/a | 3 | 1 |
| Ethnicity | ||
| Hispanic | 3 | 1 |
| Non-Hispanic | 34 | 22 |
| n/a | 3 | 1 |
Two revealed seminoma subtypes differ in pluripotency state and utilized mechanisms of double stranded DNA breaks (DSB) repair
To explore key molecular features of the identified seminoma subtypes, we conducted analysis of differentially expressed genes (DEGs) using the filtered gene set (without B- and T-immune cell transcripts). We found 73 DEGs up- and down-regulated between 2 seminoma subtypes (Figure 1B and Supplemental Table S2). Removing immune cell transcripts is rather stringent criterion helping to avoid potential biases of hierarchical clustering related to immune infiltration of seminomas. However, some of the T- and B- immune cell transcripts are also expressed in tumor cells, so we used the non-filtered set of genes for the remaining analysis. We also conducted DEGs analysis on the whole (non-filtered) set of genes, and generated a list of 229 genes with significant differential expression (Supplemental Table S3).
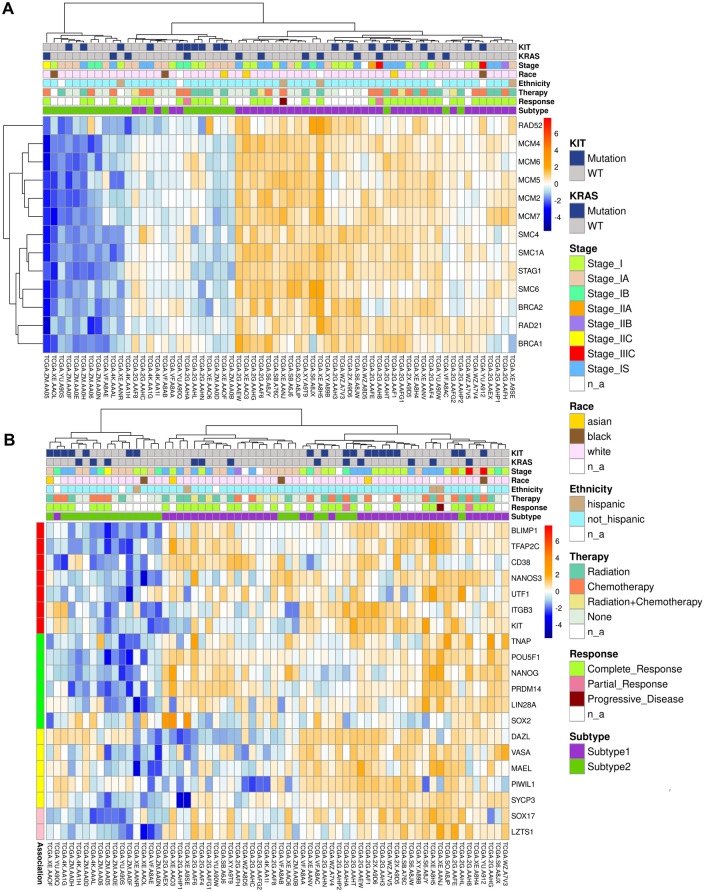
To identify active molecular pathways characterizing either of 2 seminoma subtypes, we analyzed expression of hallmark gene sets.64 We applied GSEA to a pre-ranked list of DEGs (ranking based on the adjusted p-value and a sign of Log2FC). GSEA revealed 3 gene sets for subtype 1 and 21 gene sets for subtype 2 with normalized enrichment score > 1.50 and FDR q-value < 0.05 (Figure 2A). All 3 gene sets detected for subtype 1 play an important role in cell cycle progression: mitotic spindle assembly, G2/M cell cycle transition, G1/S phase progression controlled by E2F transcription factors. We added additional steps to GSEA analysis of subtype 2 allowing us to focus on gene sets with the most prominent activation. We calculated a ratio of top 1000 DEGs per each of the revealed gene sets and selected top 7 gene sets with the ratio >10% (Supplemental Table S4, tab “GSEA”). Those 7 gene sets fell in 3 major categories: metabolism, immune response and DNA damage response. The metabolism group was represented by genes activated by reactive oxygen species (ROS), genes encoding proteins involved in oxidative phosphorylation (OXPHOS), and genes involved in processing of drugs and other xenobiotics. Immune response gene sets included genes up-regulated during allograft rejection and genes regulated by NF-kB in response to tumor necrosis factor (TNF). DNA damage response group comprised of gene sets associated with DNA reparation and p53 signaling. Next, for selected 10 gene sets (3 for subtype 1 and 7 for subtype 2), we conducted a functional analysis of genes represented in top 1000 DEGs (Figure 2B). We have found that for subtype 1, genes BRCA2 and SMC1A were related to all 3 detected gene sets. Both, BRCA2 and SMC1A, are key players of homologous recombination (HR) DNA repair. Moreover, more than a half of top 1000 DEGs related to E2F gene set (Figure 2B, red text) participate in HR repair. We revised a full list of subtype 1 DEGs associated with E2F gene set and found additional well-known players of HR repair (Supplemental Table S4, tab “E2F,” green). We compiled a list of detected HR repair genes in subtype 1 (BRCA2, STAG1, SMC1A, SMC3, SMC4, SMC6, MCM2, MCM4, MCM7, RAD21) and populated it with RAD51 and BRCA1—key mediators of HR repair. The list was used to generate a heatmap of HR repair gene expression in analyzed seminoma samples (Figure 3A). It is notable that the majority of subtype 1 seminomas were characterized by higher activity of HR repair genes in comparison to subtype 2 seminomas. At the same time subtype 2 samples characterized by increased expression level of genes associated with p53 signaling, and p53 is implicated in multiple repair pathways including DSB repair via c-NHEJ.65,66
Figure 2.
Gene sets enriched in seminoma subtype 1 and 2: (A) top gene sets enriched in seminoma subtype 1 (purple) and 2 (green) based on the gene set enrichment analysis, NES > 1.5, FDR p-value < .05 and (B) genes from top 1000 DEGs list related to key gene sets enriched in seminoma subtypes. HR-repair genes highlighted in red.
Figure 3.
Expression of genes related to HR-mediated DNA repair and pluripotency state in revealed 2 seminoma subtypes: (A) heatmap of expression for HR-mediated DNA repair genes in seminoma subtypes and (B) pluripotency stage of revealed seminoma subtypes. The heatmap shows expression of biomarkers of pluripotency (green), early PGC (red), late PGC (yellow), and mediators of PGC specification (pink, SOX17, and LZTS1).
HR repair as well as classical non-homologous end joining (c-NHEJ) repair are 2 key mechanisms utilized by cells to defend against double stranded DNA breaks (DSBs).67 In general, HR repair is more active during S- and G2-phases as it requires a homologous DNA sequence of the sister chromatid. Classical NHEJ is a faster, but error prone process that is active throughout the cell cycle, and is dominant in G0- or G1-phases. The balance between HR and c-NHEJ repair activation in the response to DSBs depends on the type and location of the cell.68 Differentiation of inducible pluripotent or embryonic derived stem cells leads to impairment of DNA damage repair via HR repair and has no effect on c-NHEJ.69 Therefore, high activity of HR repair may be a reflection of a cell stemness status. Group of testicular germ cell cancers unites variety of tumor histological types at the different stages of differentiation, from the most pluripotent embryonal carcinoma to highly differentiated teratoma.70 Primordial germ cell (PGC) considered as a cell of origin for testicular germ cell tumors (TGCTs).71 However, there are several evidences that significant difference in histology and pluripotency state of TGCT subtypes is related to the stem cell hierarchy stage of the initiating cell.72,73 We hypothesized that revealed differences in activity of DSB repair mechanisms maybe related to the differentiation state of the seminoma subtypes. To test that, we built a heatmap of expression for key genes associated with early primordial germ cells (PGCs) (BLIMP1, TFAP2C, DND1, CD38, NANOS3, UTF1, ITGB3, KIT), late PGCs (DAZL, VASA, MAEL, PIWIL, SYCP3), as well as pluripotency (TNAP, POU5F1, NANOG, PRDM14, LIN28A, SOX2)74 (Figure 3B). The heatmap showed that the majority of subtype 1 tumors had elevated level of pluripotency markers in comparison to subtype 2. This finding supports our theory of seminoma subclassification based on the pluripotency state.
On the contrary, subtype 2 of seminoma lacked both, expression of pluripotency and early PGC markers, and had minimal expression of late PGC markers. This shows that subtype 2 seminomas may be at an advanced differentiation stage. Another piece of evidence supporting this is noticeably higher expression level of ROS- and OXPHOS-related genes in the subtype 2 (Figure 2A and B). Various stem cell populations are known for preferential utilization of glycolysis over mitochondrial oxidative metabolism as it allows them to be independent of oxygen level, and also to preserve the genomic integrity by reducing ROS.75 Aerobics metabolism and ROS regulation play a significant role in the stem cell fate change and differentiation76 and is known to be the primary mechanism of ATP production in differentiated (somatic) cells.77
Subtype 2 of seminoma demonstrate molecular features specific for non-seminoma germ cell tumors
Seminoma cells (TCam-2 cell line) can be reprogramed into an EC-like cell fate and further differentiate into mixed non-seminoma TGCT containing different TGCT histological components (EC, YST, seminoma, teratoma).78 To test whether the revealed subtype 2 of seminoma is in a transition stage toward more differentiated TGCTs, we analyzed the expression of signature genes known for other TGCT histological types.51,79 For this analysis we did not use any cutoff for the Log2 FC value, we picked signature genes which expression level was significantly different between subtypes base on adjusted P-value (P-value < .05) (Supplemental Table S5). We found that subtype 2 tumor samples had elevated level of non-seminoma signature genes when subtype 1 demonstrated seminoma features (Supplemental Table S5). The role of the defined genes in GCT cells is not clear, however some of them are important for tumorigenesis in other types of cancer.80-83
A seminoma signature gene LZTS1 (Leucine Zipper Tumor Suppressor 1), has high level of expression in identified seminomas of subtype 1. This gene plays role in cell cycle control moderating the transition from late S to G(2)/M stage.84 It was also shown that expression of LZTS1 correlates with SOX17 and both are important regulators of human pluripotent stem cell (hPSC) endoderm specification. Overexpression of LZTS1 in hPSCs leads to increased expression of SOX17.85 Though there are no information on LZTS1 role in seminomas, or other TGCTs, we know that activation of SOX17 in human embryonic stem cells determines their specification into primordial germ cells and is used as a key marker of the earliest PGCs.74 Therefore, we suggest that LZTS1 may play a role in maintenance of early PGC pluripotency state in subtype 1 seminomas through a positive feedback loop with SOX17, and that can be related to an early PGC cell ancestry.
TGCT signature genes overexpressed in the subtype 2 of seminoma were characteristic for 3 major histotypes of non-seminomatous TGCTs, embryonal carcinoma (EC), yolk sac tumor (YST) and teratoma (Ter) (Supplemental Table S5). GAL and GPC4 gene signatures specific to EC and play an important role in embryonic development and pluripotency. Surface protein glypican 4 (Gpc4) is a component of the signaling machinery regulating embryonic stem cell (ESC) maintenance. In ESC, Gpc4 modulates the response to Wnt ligands and regulates activation of b-catenin signaling. GPC4 is important for teratoma lineage specification as loss of GPC4 in ESC makes ESC incompetent from developing teratomas.86 Elevated expression of GPC4 may be a sign of EC-like stemness of seminoma subtype 2 with an intrinsic tendency to transform into multiple cell lineages including teratoma. Moreover, our data show that subtype 2 seminomas overexpress signature genes of more differentiated TGCTs, as YSTs (APOA2, BMP2, FAM89A, FOXA2, RAGE, VTN) and teratomas (MFAP4, NFKBIZ, and TSPAN8) supporting the hypothesis stated above. Importantly, we found that subtype 2 seminomas have increased expression of FOXA2 (Forkhead Box A2), a transcriptional factor that induces differentiation and microenvironment-triggered reprograming of seminoma cells (TCam-2) into embryonal carcinoma.78 FOXA2 is only upregulated for a limited amount of time and associated with a period of seminoma differentiation into non-seminoma lineages (EC). Once adaptation to the newly acquired cell fate is completed, FOXA2 expression is downregulated and it is not detectable in any TGCT subtypes.78 The fact that we see elevated level of FOXA2 in subtype 2 seminoma supports the idea that this subtype is at an early transitional stage into EC fate. Therefore, histological examination has not revealed significant morphology differences between both seminoma subtypes. However, transcriptomic analysis detects significant changes in multiple molecular process including DNA repair and stemness maintenance. Several recent studies demonstrated that seminomas harbor certain molecular patterns which bring them closer to non-seminomas.87-90 Moreover, a process of reprograming of pure seminoma cells might take place, which results in the progression from seminoma to totipotent embryonal carcinoma cells that have the capacity of originating tumor components of all types of NSE GCTs.87,90 This process should be considered during the design of clinical therapeutic strategies and it increases the risk of poor prognosis.91,92 We hypothesize that revealed subtype 2 can be a precursor of mixed TGCT seminomas.
Genomic and epigenomic features of seminoma subtype 2 revealed similarity with non-seminomatous GCTs
To further understand the biology of identified 2 subtypes of seminomas, we compared their genomic and epigenomic features. TGCTs have very low mutation burden, and only 3 genes were shown to contain recurrent somatic mutations: KIT, KRAS, and NRAS.2 We did not observe any association of a particular mutation pattern of these genes with the revealed seminoma subtypes. Nearly all TGCTs contain significant arm level gain of chromosome 12p (conventional marker of TGCT type II), and moderate arm level gain of chromosomes 7 and 8 .2,93 Both identified seminoma subtypes revealed arm-level gain of all chromosomes mentioned above, as well as arm level loss of chromosomes 9, 11, and 13 (Supplemental Figure S2). We identified 3 genes that are: (1) located in amplified regions, (2) upregulated in subtype 1 and (3) known as targets for approved anticancer drugs (Supplemental Table S6). The first one, Prohibitin 2 (PHB2) is encoded in 12p13.31 chromosome region. PHB2 expression is frequently altered in testicular seminomas. Also, PHB2 protein was detected in plasma membrane fraction of human embryonic stem cells and human embryonal carcinoma cells.94 One of the major known PHB2 function is stabilization of mitochondrial OXPHOS complex and increment of mitochondrial respiration.95 This function is especially important for pluripotent cells. Knock-out of PHB2 in mouse embryonic cells leads to the apoptosis.96 In Drosophila ovaries, PHB2 plays an important role in germ cell maintenance and survival.97 Therefore, inhibition of PHB2 with Rocaglamide98 and its derivative Didesmethylrocaglamide98 can be considered as a treatment for seminoma subtype 1 patients improving their sensitization to oxidative stress caused by chemo- or radiotherapy.99 The second gene, KRAS encoded by 12p12.1 locus is the most frequently mutated gene in human cancers. Approximately 30% of human cancers have mutations in the KRAS gene.100 Although TCGTs revealed low mutation burden, KRAS is among 3 genes that are commonly mutated in this type of cancer.2 KRAS is an important part of RAS/MAPK signaling pathway100 and plays crucial role in cell proliferation.101 Two drugs are known as inhibitors of mutated KRAS: Sotorasib (AMG-510)102 and Adagrasib (MRTX849).103 The third gene encodes interleukin-6 receptor subunit alpha (IL6R) that is located in 1q21.3 chromosome locus. IL6R is a cytokine that is expressed by immune and cancer cells.104 It plays a crucial role in the acute phase response of the immune system and inflammation process.104 Blockade of the IL6 signaling pathway is potential target for immunotherapy of various cancers. Antibodies against IL6 (Siltuximab) and its receptor IL6R (Tocilizumab) have emerged as potential drugs for immunotherapy.105
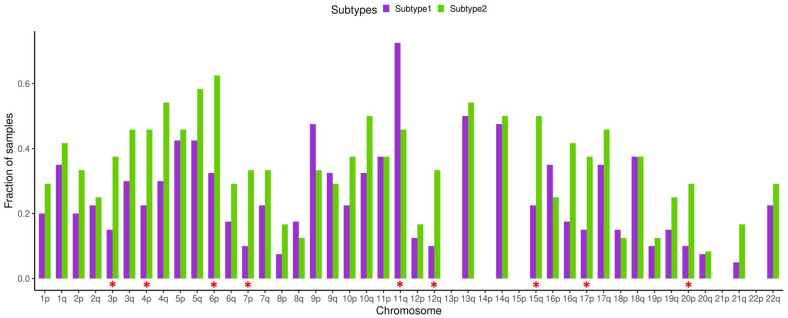
TGCTs have a unique feature that distinguish them from other cancer types, this is highly recurrent chromosome arm level amplifications and loss of heterozygosity (LOH). It was found that TGCTs possess significant enrichment in number of chromosome arms with more than one allele amplified compared to 20 other cancer types.106,107 Comparison of pure seminoma and seminoma originated from mixed TGCT demonstrated significantly higher LOH rate for the mixed seminoma.88 We compared LOH data taken from TCGA PanCanAtlas study53 for the identified seminoma subtypes. Our analysis revealed that for all chromosome arms except 9p and 11q LOH rate is higher for seminoma subtype 2 (Figure 4). Significant difference was detected for arms 3p, 4p, 6p, 7p, 11q, 12q, 15q, 17p, and 20p. For the loci 13p, 14p, 15p, 21p, and 22p no LOH was observed. Increased LOH rate of seminoma subtype 2 may be associated with impaired HR repair system, as we have noticed that in comparison to subtype 1, subtype 2 has decreased expression of genes associated with HR repair due to more advanced differentiation status. There are multiple studies on ovarian cancer that demonstrate that one of the reasons for increased LOH rate is deficiency of homologous recombination related to BRCA1 and BRCA2 mutation status.108-110
Figure 4.
Loss of heterozygosity (LOH) of seminoma subtypes. Fraction of samples with observed LOH. Red asterisks denote significant difference between seminoma subtypes (Chi-squared test P < .05).
Another molecular characteristic that differentiates SE from NSE is dominant telomere elongation in non-seminoma samples.111 Our analysis revealed that telomere elongation is higher for subtype 2, however the difference is not significant (Supplemental Figure S3). Moreover, among genes that have positive correlation between telomere elongation and gene expression in non-seminomas,111 we identified 3 genes which are significantly overexpressed in subtype 2: MT2A (Log2FC = 1.15, P = 3.9E-06), SLC16A3 (Log2FC = 1.1, P = 2.9E-04), and PDHA1 (Log2FC = 0.7, P = 6.2E-05).
Another genomic trait of seminomas versus non-seminomas is the low level of DNA methylation.112,113 If we trace the methylation status of TGCT precursor cells (PGCs), we will find that at the earliest stage when an ESC transforms into a gonocyte (early PGC), it loses its DNA methylation pattern. In the transition from early to late PGC and during further specialization of PGC into spermatogonia, cells will start de novo DNA methylation to re-establish the parental imprinting pattern.73,114 Thus, the DNA methylation level of TGCT cells has a strong association with their stemness and can reflect the transition of TGCT tumor cells into a more differentiated tumor subtype. Previous studies showed that seminoma cells lack DNA methylation.2 We analyzed beta value data to look for a difference between methylated and unmethylated alleles. The average beta value of seminoma subtype 1 was lower than for seminoma subtype 2. However, no significantly differentially methylated probes and regions were identified. This result might be related to unmethylated nature of seminoma.2 Graph of beta value distribution shows 2 major peaks (Supplemental Figure S4). It was shown that the second peak that we observe around beta value .5 to .6 corresponds to lymphocytes that infiltrate seminoma tissue in a large number.2 Therefore, bulk methylation sequencing is not informative for the analysis of differentially methylated regions between 2 seminoma subtypes.
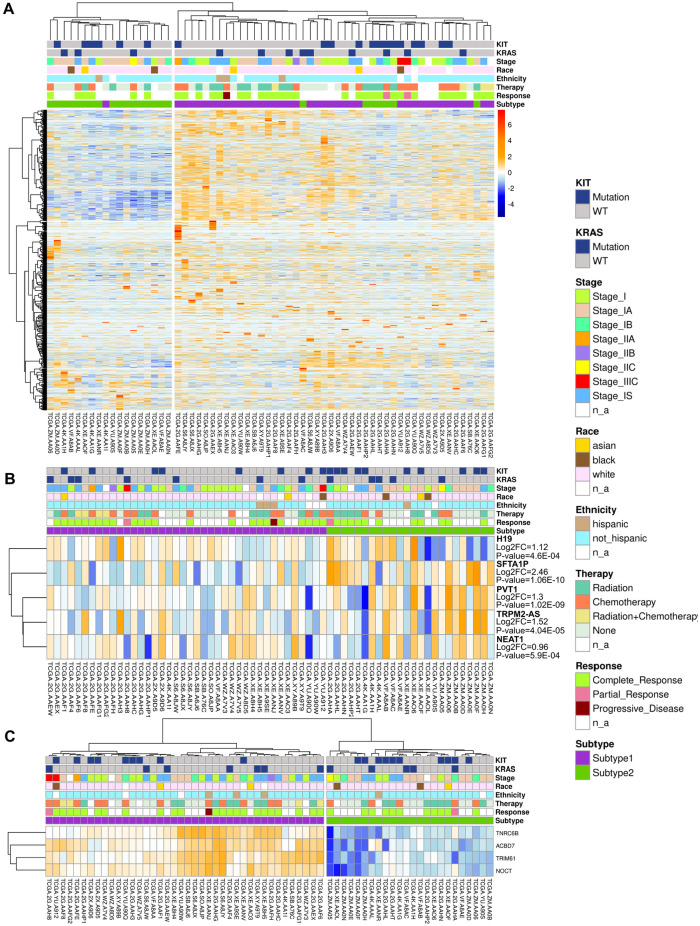
Long non-coding RNA expression pattern of seminoma subtype 2 suggests increased resistance to chemotherapy
Long non-coding RNAs (lncRNAs) were defined relatively recently as non-coding RNAs longer than 200 nucleotides. These molecules have been found to have crucial role in cancer utilizing large variety of functions including oncogenesis and tumor suppression and their function list is rapidly emerging.115 TGCTs are not the exception to this rule, expression levels of several oncogenic lncRNAs have been associated with germ cell tumors.116 Unsupervised clustering of lncRNAs genes based on their expression level showed 2 large clusters which are very similar to identified subtypes based on the whole transcriptome (Figure 5A). LncRNAs play crucial role in the development of cisplatin resistance.117 TGCTs are highly sensitive to chemotherapy and radiotherapy. Interestingly that seminomas are more sensitive than non-seminomas, that might be related to their differentiation status (mature teratomas are the most chemotherapy resistant TGCTs) and active DNA repair mechanisms.118,119 Similar pattern of drug resistance increment was noticed during the development of sperm from PGCs, and therefore it was proposed that seminomas and non-seminomas are derived from cells at different stages of germ-cell differentiation.118 We identified that 5 lncRNAs responsible for cisplatin resistance in different cancer types are overexpressed in seminoma subtype 2 (Figure 5B). H19 was shown to utilize pro-tumorigenic function and promote cisplatin resistance in TGCTs.117,120 Other 4 lncRNAs (NEAT1, PVT1, SFTA1P, TRPM2-AS) are responsible for cisplatin resistance in lung, gastric, and ovarian cancers.117 Advanced differentiation status, HR repair deficiency and overexpression of lncRNA associated with cisplatin resistance allow us to hypothesize that revealed subtype 2 of seminoma is more resistant to genotoxic drugs. Therefore, patients with subtype 2 seminoma may require adjustments of a treatment protocol or development of alternative treatment approaches.
Figure 5.
LncRNA expression in seminoma subtypes and key biomarkers that can be used for their differentiation: (A) unsupervised hierarchical clustering of seminoma samples based on transcriptomic data of lncRNAs, (B) 5 lncRNAs that promote cisplatin resistance and are overexpressed in subtype 2, and (C) potential biomarkers for seminoma subtypes identification (overexpressed in subtype 1).
Potential biomarkers for histological differentiation of seminoma subtypes
We identified 4 potential biomarkers which are capable to distinguish 2 seminoma subtypes (Figure 5C and Supplemental Table S7). All these genes are overexpressed in subtype 1 and show specificity no less than 79%, which is in the specificity range of clinically used biomarkers for various cancer types.121 Overexpressed genes in subtype 2 do not show high enough specificity, so we do not consider them. All of the identified biomarkers were assessed in other cancer types, but were not previously evaluated for TGCTs. Nocturnin (NOCT) shows the highest specificity level of 92%. NOCT is overexpressed in squamous cell lung cancer and has potential as biomarker for this type of cancer.122 TNRC6B shows specificity of 87.5%. Alterations in expression level of this gene was shown to contribute to carcinogenesis.123 Finally, TRIM61 and ACBD7 show specificity of 79.2%. TRIM61 was previously suggested as prognostic biomarker for lung squamous cell cancer,124 while ACBD7 is overexpressed in Hürthle cell carcinoma.125 Combination of potential biomarkers NOCT and TNRC6B results in the highest specificity of 96%. Discussed potential biomarkers were defined using computational analysis and require further experimental validation.
Summing up, our analysis showed that pure seminoma cases can be further subdivided into 2 main subtypes. The subtypes differ in (1) pluripotency stage, (2) activity of DSB DNA repair mechanisms, (3) rate of LOH, (4) expression of lncRNA associated with cisplatin resistance. In comparison to more pluripotent seminoma subtype 1, seminoma subtype 2 shows signs of differentiation into non-seminoma TGCT and may have higher resistance to platinum-based chemotherapy.
Discussion
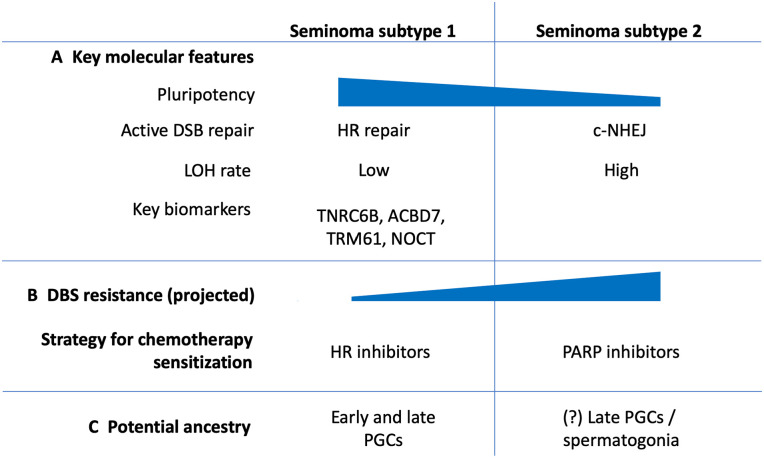
TGCTs represent highly heterogeneous group of cancers, starting with broad separation of seminoma and NSE GCTs.2 Moreover, each TGCT histological type has demonstrated existing internal heterogeneity leading to diverse patient outcomes.118 Our analysis of the dominant TGCT subtype seminoma, demonstrated that on the transcriptomic level seminoma samples can be further classified into 2 subtypes. Subtype 2 revealed several molecular features that can be associated with undergoing differentiation of subtype 2 into non-seminomatous lineages through the stage of embryonal carcinoma (Figure 6). It is important to consider that seminoma subtype 2 showed impaired HR repair in comparison to subtype 1. Subtype 2 depends on more efficient c-NHEJ repair mechanism that allows these tumors to repair DSBs in a timely manner avoiding apoptotic cell death, which may potentially explain greater resistance to chemotherapy. Clinical and therapy response information available for TCGA seminoma cohort doesn’t have complete data on dosages of used drugs, the duration of the therapy and long-term patient outcomes. Out of 38 patients that received any kind of therapy, 17 received radiotherapy, 17 received chemotherapy and 4 patients received both types of therapy (Supplemental Table S1). This data is not enough for identification of correlation between chemotherapy response and identified subtypes. Therefore, an experimental validation considering parameters mentioned above should be conducted on an independent group of patients. Hypothetically, subtype 2 seminoma cells may be responsible for seminoma recurrence after chemotherapy through mechanisms of undertreatment or cisplatin-resistance. Relapsed patients have a 50% chance of disease-specifically mortality; and salvage treatment drastically worsen side effect profiles. However, some patients suffered progressive cancer disease despite high-dose chemotherapy.126 In addition, platinum-base chemotherapy, which is used for TGCTs treatment, significantly decreases patients’ quality of life and can cause complex of around 40 severe long-term side effects including secondary cancers and death.17 Circulating platinum concentration can remain up to 1000 times above the normal level for 20 years after the chemotherapy completion and is associated with many delayed side-effects.127 Therefore, it is important to develop fewer toxic solutions for TGCT therapy. Molecular differences identified between 2 seminoma subtypes will potentially lead to important practical applications. There are multiple studies showing that PARP inhibitors (FDA approved for various ovarian and breast cancers) can be efficiently used against HR-deficient tumors as it induces DSB formation and in addition inhibits alternative repair mechanisms such as microhomology-mediated end joining (MMEJ).128,129 Our analysis showed that subtype 2 seminoma has deficiency in HR repair that makes it a suitable candidate for PARP inhibitor therapy or combination therapy of PARP inhibitors with platinum compounds. However, in some tumors PARP inhibitors may elicit a tumoricidal effect by enhancing c-NHEJ,128,129 therefore preliminary in vitro studies are required. On the contrary to subtype 2, subtype 1 has increased activity of HR repair that also can be used for a new therapy development. For example, tyrosine kinase inhibitor erlotinib disrupts nuclear function of BRCA1 and attenuates HR activity. As the result, it causes sensitization of cancer cells to radiation therapy.130,131 There is also evidence that proteasome inhibitors targeting HR proteins can cause cisplatin sensitization.132
Figure 6.
Summary of molecular features identified for revealed 2 seminoma subtypes.
Conclusions
Computational analysis of omics data of pure seminoma samples from TCGA revealed 2 distinct seminoma subtypes. Based on our computational analysis, seminoma subtype 1 has higher pluripotency rate and demonstrated signs of elevated HR repair activity. On the contrary, seminoma subtype 2 showed features of more differentiated cell type and resembles non-seminoma TGCTs (overexpression of signature genes, high rate of LOH). We also detected that subtype 2 samples had increased expression level of lncRNAs responsible for cisplatin resistance in other cancer types including TGCTs. We hypothesize that drugs targeting HR repair (subtype 1) and other DSB repair mechanisms as c-NHEJ and MMEJ (subtype 2) can increase sensitivity of revealed seminoma subtypes to chemotherapy and irradiation, though in vitro and in vivo studies are required to support the hypothesis. Development of seminoma subtype-specific therapy can help to overcome chemotherapy overtreatment in TGCT patients and improve quality of life for TGCT survivors. Current study has following limitations: (1) experimental validation of biomolecular characteristics of seminoma subtypes is required as the next step; (2) therapy response is not available for all patients from TCGA seminoma cohort that does not allow to define correlation between seminoma subtypes and chemotherapy response, therefore new cohort with known therapy responses is required for validation of main results; (3) patients of White race and Non-Hispanic ethnicity are over represented in TCGA seminoma cohort that creates bias, therefore more ethnicity balanced cohort is required for validation of the results; (4) number of samples with subtype 1 is twice the number with subtype 2 and the unequal sets may bias the accuracy statistics we computed.
Supplemental Material
Supplemental material, sj-docx-1-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-2-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-3-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-4-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-5-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-6-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Acknowledgments
Authors are grateful to TCGA data portal for providing access to TGCT datasets.
Footnotes
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The study is supported by the grants from the National Institutes of Health GM127390 (to N.V.G.), the Welch Foundation I-1505 (to N.V.G.), the Dedman Foundation Scholarship (to A.B.), and from the National Cancer Institute 1K08CA207849 (to K.S.C.).
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author contributions: Kirill E Medvedev: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data Curation, Visualization, Writing—Original Draft, Project administration. Anna V Savelyeva: Conceptualization, Validation, Formal analysis, Investigation, Writing—Original Draft. Kenneth S Chen: Investigation, Methodology, Formal analysis, Writing—Review & Editing. Aditya Bagrodia: Resources, Funding acquisition, Writing - Review & Editing. Liwei Jia: Investigation, Writing - Review & Editing. Nick V Grishin: Conceptualization, Resources, Funding acquisition, Writing—Review & Editing.
Data availability statement: The datasets analyzed during the current study are available from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/). All generated data are included in this manuscript.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Ghazarian AA, Trabert B, Devesa SS, McGlynn KA. Recent trends in the incidence of testicular germ cell tumors in the United States. Andrology. 2015;3:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shen H, Shih J, Hollern DP, et al. Integrated molecular characterization of testicular germ cell tumors. 2018;23:3392-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng L, Lyu B, Roth LM. Perspectives on testicular germ cell neoplasms. Hum Pathol. 2017;59:10-25. [DOI] [PubMed] [Google Scholar]
- 4. McGlynn KA, Cook MB. Etiologic factors in testicular germ-cell tumors. Future Oncol. 2009;5:1389-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghazarian AA, Kelly SP, Altekruse SF, Rosenberg PS, McGlynn KA. Future of testicular germ cell tumor incidence in the United States: forecast through 2026. Cancer. 2017;123:2320-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71:7-33. [DOI] [PubMed] [Google Scholar]
- 7. World Health Organization (WHO). Cancer statistics globcan 2020. Accessed February 1, 2022. https://gco.iarc.fr/today/data/factsheets/populations/900-world-fact-sheets.pdf
- 8. Howlader N, Noone AM, Krapcho M, et al. (eds). SEER Cancer Statistics Review. National Cancer Institute; 1975-2017. https://seer.cancer.gov/csr/1975_2017/. [Google Scholar]
- 9. Gilligan T, Lin DW, Aggarwal R, et al. Testicular cancer, version 2.2020, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2019;17:1529-1554. [DOI] [PubMed] [Google Scholar]
- 10. Boujelbene N, Cosinschi A, Boujelbene N, et al. Pure seminoma: a review and update. Radiat Oncol. 2011;6:90-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shin YS, Kim HJ. Current management of testicular cancer. Korean J Urol. 2013;54:2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bokemeyer C, Kollmannsberger C, Stenning S, et al. Metastatic seminoma treated with either single agent carboplatin or cisplatin-based combination chemotherapy: a pooled analysis of two randomised trials. Br J Cancer. 2004;91:683-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gholam D, Fizazi K, Terrier-Lacombe MJ, Jan P, Culine S, Theodore C. Advanced seminoma—treatment results and prognostic factors for survival after first-line, cisplatin-based chemotherapy and for patients with recurrent disease: a single-institution experience in 145 patients. Cancer. 2003;98:745-752. [DOI] [PubMed] [Google Scholar]
- 14. Soper MS, Hastings JR, Cosmatos HA, Slezak JM, Wang R, Lodin K. Observation versus adjuvant radiation or chemotherapy in the management of stage I seminoma: clinical outcomes and prognostic factors for relapse in a large US cohort. Am J Clin Oncol. 2014;37:356-359. [DOI] [PubMed] [Google Scholar]
- 15. Boer H, Proost JH, Nuver J, et al. Long-term exposure to circulating platinum is associated with late effects of treatment in testicular cancer survivors. Ann Oncol. 2015;26:2305-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Oun R, Moussa YE, Wheate NJ. The side effects of platinum-based chemotherapy drugs: a review for chemists. 2018;47:6645-6653. [DOI] [PubMed] [Google Scholar]
- 17. Chovanec M, Lauritsen J, Bandak M, et al. Late adverse effects and quality of life in survivors of testicular germ cell tumour. Nat Rev Urol. 2021;18:227-245. [DOI] [PubMed] [Google Scholar]
- 18. Haugnes HS, Stenklev NC, Brydøy M, et al. Hearing loss before and after cisplatin-based chemotherapy in testicular cancer survivors: a longitudinal study. Acta Oncol. 2018;57:1075-1083. [DOI] [PubMed] [Google Scholar]
- 19. Frisina RD, Wheeler HE, Fossa SD, et al. Comprehensive audiometric analysis of hearing impairment and tinnitus after cisplatin-based chemotherapy in survivors of adult-onset cancer. J Clin Oncol. 2016;34:2712-2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lauritsen J, Mortensen MS, Kier MGG, et al. Renal impairment and late toxicity in germ-cell cancer survivors. Ann Oncol. 2015;26:173-178. [DOI] [PubMed] [Google Scholar]
- 21. Inai H, Kawai K, Ikeda A, et al. Risk factors for chronic kidney disease after chemotherapy for testicular cancer. Int J Urol. 2013;20:716-722. [DOI] [PubMed] [Google Scholar]
- 22. Warde P, Specht L, Horwich A, et al. Prognostic factors for relapse in stage I seminoma managed by surveillance: a pooled analysis. J Clin Oncol. 2002;20:4448-4452. [DOI] [PubMed] [Google Scholar]
- 23. Escudero-ávila R, Rodríguez-Castaño JD, Osman I, et al. Active surveillance as a successful management strategy for patients with clinical stage I germ cell testicular cancer. Clin Transl Oncol. 2019;21:796-804. [DOI] [PubMed] [Google Scholar]
- 24. Dieckmann KP, Dralle-Filiz I, Matthies C, et al. Testicular seminoma clinical stage 1: treatment outcome on a routine care level. J Cancer Res Clin Oncol. 2016;142:1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kamba T, Kamoto T, Okubo K, et al. Outcome of different post-orchiectomy management for stage I seminoma: Japanese multi-institutional study including 425 patients. Int J Urol. 2010;17:980-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vuky J, Tickoo SK, Sheinfeld J, et al. Salvage chemotherapy for patients with advanced pure seminoma. J Clin Oncol. 2002;20:297-301. [DOI] [PubMed] [Google Scholar]
- 27. Tu S, Bilen MA, Hess KR, et al. Intratumoral heterogeneity: role of differentiation in a potentially lethal phenotype of testicular cancer. Cancer. 2016;122:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brait M, Maldonado L, Begum S, et al. DNA methylation profiles delineate epigenetic heterogeneity in seminoma and non-seminoma. Br J Cancer. 2012;106:414-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsimberidou AM, Fountzilas E, Nikanjam M, Kurzrock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat Rev. 2020;86:102019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsimberidou AM. Initiative for Molecular Profiling and advanced cancer therapy and challenges in the implementation of precision medicine. Curr Probl Cancer. 2017;41:176-181. [DOI] [PubMed] [Google Scholar]
- 31. Liu F, Qian J, Ma C. MPscore: a novel predictive and prognostic scoring for progressive meningioma. Cancers. 2021;13:1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sadanandam A, Wullschleger S, Lyssiotis CA, et al. A cross-species analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discov. 2015;5:1296-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jung AC, Job S, Ledrappier S, et al. A poor prognosis subtype of HNSCC is consistently observed across methylome, transcriptome, and miRNome analysis. Clin Cancer Res. 2013;19:4174-4184. [DOI] [PubMed] [Google Scholar]
- 34. Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288-300. [DOI] [PubMed] [Google Scholar]
- 35. Yin L, Duan JJ, Bian XW, Yu SC. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu H, Piotrowska Z, Hare PJ, et al. Three subtypes of lung cancer fibroblasts define distinct therapeutic paradigms. Cancer Cell. 2021;39:1531-1547.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meric-Bernstam F, Mills GB. Overcoming implementation challenges of personalized cancer therapy. Nat Rev Clin Oncol. 2012;9:542-548. [DOI] [PubMed] [Google Scholar]
- 38. Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schwaederle M, Zhao M, Lee JJ, et al. Association of biomarker-based treatment strategies with response rates and progression-free survival in refractory malignant neoplasms: a meta-analysis. JAMA Oncol. 2016;2:1452-1459. [DOI] [PubMed] [Google Scholar]
- 40. Weidenbusch B, Richter GHS, Kesper MS, et al. Transcriptome based individualized therapy of refractory pediatric sarcomas: feasibility, tolerability and efficacy. Oncotarget. 2018;9:20747-20760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shen H, Zhu M, Wang C. Precision oncology of lung cancer: genetic and genomic differences in Chinese population. NPJ Precis Oncol. 2019;3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Abreu FB, Schwartz GN, Wells WA, Tsongalis GJ. Personalized therapy for breast cancer. Clin Genet. 2014;86:62-67. [DOI] [PubMed] [Google Scholar]
- 43. Ku SY, Gleave ME, Beltran H. Towards precision oncology in advanced prostate cancer. Nat Rev Urol. 2019;16:645-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Barthel FP, Wei W, Tang M, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Volders PJ, Anckaert J, Verheggen K, et al. LNCipedia 5: towards a reference set of human long non-coding RNAs. Nucleic Acids Res. 2019;47:D135-D139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hadrup SR, Braendstrup O, Jacobsen GK, et al. Tumor infiltrating lymphocytes in seminoma lesions comprise clonally expanded cytotoxic T cells. Int J Cancer. 2006;119:831-838. [DOI] [PubMed] [Google Scholar]
- 47. Schmiedel BJ, Singh D, Madrigal A, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701-1715.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y, Hayes DN, Nobel A, Marron JS. Statistical significance of clustering for high-dimension, low–sample size data. J Am Stat Assoc. 2008;103:1281-1293. [Google Scholar]
- 50. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with deseq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sheikine Y, Genega E, Melamed J, Lee P, Reuter VE, Ye H. Molecular genetics of testicular germ cell tumors. Am J Cancer Res. 2012;2:153-167. [PMC free article] [PubMed] [Google Scholar]
- 52. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Taylor AM, Shih J, Ha G, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676-689.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Karolchik D, Hinrichs AS, Kent WJ. The UCSC genome browser. Curr Protoc Bioinformatics. 2009;28:1.4.1-1.4.26. [DOI] [PubMed] [Google Scholar]
- 56. Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wishart DS, Feunang YD, Guo AC, et al.DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074-D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Peters TJ, Buckley MJ, Chen Y, Smyth GK, Goodnow CC, Clark SJ. Calling differentially methylated regions from whole genome bisulphite sequencing with DMRcate. Nucleic Acids Res. 2021;49:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. González-Barrios R, Alcaraz N, Montalvo-Casimiro M, et al. Genomic profile in a non-seminoma testicular germ-cell tumor cohort reveals a potential biomarker of sensitivity to platinum-based therapy. Cancers. 2022;14:2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matsumoto T, Shiota M, Uchiumi T, et al. Genomic characteristics revealed by targeted exon sequencing of testicular germ cell tumors in Japanese men. Int J Urol. 2021;28:40-46. [DOI] [PubMed] [Google Scholar]
- 63. Zhao S, Li CI, Guo Y, Sheng Q, Shyr Y. RnaSeqSampleSize: real data based sample size estimation for RNA sequencing. BMC Bioinformatics. 2018;19:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jiang Y, Dong Y, Luo Y, et al. AMPK-mediated phosphorylation on 53BP1 promotes c-NHEJ. Cell Rep. 2021;34:108713. [DOI] [PubMed] [Google Scholar]
- 66. Moureau S, Luessing J, Harte EC, Voisin M, Lowndes NF. A role for the p53 tumour suppressor in regulating the balance between homologous recombination and non-homologous end joining. Open Biol. 2016;6:160225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ranjha L, Howard SM, Cejka P. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018;127:187-214. [DOI] [PubMed] [Google Scholar]
- 68. Willers H, Dahm-Daphi J, Powell SN. Repair of radiation damage to DNA. Br J Cancer. 2004;90:1297-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mujoo K, Pandita RK, Tiwari A, et al. Differentiation of human induced pluripotent or embryonic stem cells decreases the DNA damage repair by homologous recombination. Stem Cell Reports. 2017;9:1660-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Looijenga LH. Testicular germ cell tumors. Pediatr Endocrinol Rev. 2014;11 Suppl 2:251-262. [PubMed] [Google Scholar]
- 71. Singh SR, Burnicka-Turek O, Chauhan C, Hou SX. Spermatogonial stem cells, infertility and testicular cancer. J Cell Mol Med. 2011;15:468-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Som A, Wen S, Tu SM. Stem cell origin of testicular seminoma. Clin Genitourin Cancer. 2013;11:489-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Baroni T, Arato I, Mancuso F, Calafiore R, Luca G. On the origin of testicular germ cell tumors: from gonocytes to testicular cancer. Front Endocrinol. 2019;10:343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Irie N, Weinberger L, Tang WW, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 2015;160:253-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tsogtbaatar E, Landin C, Minter-Dykhouse K, Folmes CDL. Energy metabolism regulates stem cell pluripotency. Front Cell Dev Biol. 2020;8:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Chaudhari P, Ye Z, Jang YY. Roles of reactive oxygen species in the fate of stem cells. Antioxid Redox Signal. 2014;20:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gatie MI, Kelly GM. Metabolic profile and differentiation potential of extraembryonic endoderm-like cells. Cell Death Discov. 2018;4:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Nettersheim D, Vadder S, Jostes S, Heimsoeth A, Schorle H. TCam-2 cells deficient for SOX2 and FOXA2 are blocked in differentiation and maintain a seminoma-like cell fate in vivo. Cancers. 2019;11:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Alagaratnam S, Lind GE, Kraggerud SM, Lothe RA, Skotheim RI. The testicular germ cell tumour transcriptome. Int J Androl. 2011;34:e133-e150; discussion e150-1. [DOI] [PubMed] [Google Scholar]
- 80. Vecchione A, Baldassarre G, Ishii H, et al. Fez1/lzts1 absence impairs Cdk1/cdc25c interaction during mitosis and predisposes mice to cancer development. Cancer Cell. 2007;11:275-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Meza-Zepeda LA, Kresse SH, Barragan-Polania AH, et al. Array comparative genomic hybridization reveals distinct DNA copy number differences between gastrointestinal stromal tumors and leiomyosarcomas. Cancer Res. 2006;66:8984-8993. [DOI] [PubMed] [Google Scholar]
- 82. Göransson M, Andersson MK, Forni C, et al. The myxoid liposarcoma FUS-DDIT3 fusion oncoprotein deregulates NF-kappab target genes by interaction with NFKBIZ. Oncogene. 2009;28:270-278. [DOI] [PubMed] [Google Scholar]
- 83. Riehl A, Németh J, Angel P, Hess J. The receptor RAGE: bridging inflammation and cancer. Cell Commun Signal. 2009;7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ishii H, Vecchione A, Murakumo Y, et al. FEZ1/LZTS1 gene at 8p22 suppresses cancer cell growth and regulates mitosis. Proc Natl Acad Sci U S A. 2001;98:10374-10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Madrigal P, Pauklin S, Goh KJ, et al. Epigenetic regulations follow cell cycle progression during differentiation of human pluripotent stem cells. BioRxiv. 2020; DOI: 10.1101/2020.06.26.173211. [Google Scholar]
- 86. Fico A, De Chevigny A, Egea J, et al. Modulating Glypican4 suppresses tumorigenicity of embryonic stem cells while preserving self-renewal and pluripotency. Stem Cells. 2012;30:1863-1874. [DOI] [PubMed] [Google Scholar]
- 87. Lobo J, Gillis AJM, Jerónimo C, Henrique R, Looijenga LHJ. Human germ cell tumors are developmental cancers: impact of epigenetics on Pathobiology and clinic. Int J Mol Sci. 2019;20:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Miyai K, Ito K, Nakanishi K, Tsuda H. Seminoma component of mixed testicular germ cell tumor shows a higher incidence of loss of heterozygosity than pure-type seminoma. Hum Pathol. 2019;84:71-80. [DOI] [PubMed] [Google Scholar]
- 89. Miyai K, Ito K, Nakanishi K, Tsuda H. Cell-to-cell variation of chromosomal number in the adult testicular germ cell tumors: a comparison of chromosomal instability among histological components and its putative role in tumor progression. Virchows Arch. 2019;474:711-720. [DOI] [PubMed] [Google Scholar]
- 90. Miyai K, Yonekura Y, Ito K, Matsukuma S, Tsuda H. Gene expression microarray analysis of adult testicular germ cell tumor: a comparison between pure-type seminomas and seminoma components in mixed tumors. Virchows Arch. 2021;479:1177-1186. [DOI] [PubMed] [Google Scholar]
- 91. Ulbright TM. The most common, clinically significant misdiagnoses in testicular tumor pathology, and how to avoid them. Adv Anat Pathol. 2008;15:18-27. [DOI] [PubMed] [Google Scholar]
- 92. Looijenga LH, Gillis AJ, Stoop H, Biermann K, Oosterhuis JW. Dissecting the molecular pathways of (testicular) germ cell tumour pathogenesis; from initiation to treatment-resistance. Int J Androl. 2011;34:e234-e251. [DOI] [PubMed] [Google Scholar]
- 93. Litchfield K, Summersgill B, Yost S, et al. Whole-exome sequencing reveals the mutational spectrum of testicular germ cell tumours. Nat Commun. 2015;6:5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Dormeyer W, van Hoof D, Braam SR, Heck AJ, Mummery CL, Krijgsveld J. Plasma membrane proteomics of human embryonic stem cells and human embryonal carcinoma cells. J Proteome Res. 2008;7:2936-2951. [DOI] [PubMed] [Google Scholar]
- 95. Signorile A, Sgaramella G, Bellomo F, De Rasmo D. Prohibitins: A critical role in mitochondrial functions and implication in diseases. Cells. 2019;8:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kowno M, Watanabe-Susaki K, Ishimine H, et al. Prohibitin 2 regulates the proliferation and lineage-specific differentiation of mouse embryonic stem cells in mitochondria. PLoS One. 2014;9:e81552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Gao Y, Mao Y, Xu RG, et al. Defining gene networks controlling the maintenance and function of the differentiation niche by an in vivo systematic RNAi screen. J Genet Genomics. 2019;46:19-30. [DOI] [PubMed] [Google Scholar]
- 98. Polier G, Neumann J, Thuaud F, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem Biol. 2012;19:1093-1104. [DOI] [PubMed] [Google Scholar]
- 99. Perillo B, Di Donato M, Pezone A, et al. ROS in cancer therapy: the bright side of the moon. Exp Mol Med. 2020;52:192-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Uprety D, Adjei AA. KRAS: from undruggable to a druggable cancer target. Cancer Treat Rev. 2020;89:102070. [DOI] [PubMed] [Google Scholar]
- 101. Zimmermann G, Papke B, Ismail S, et al. Small molecule inhibition of the KRAS-PDEδ interaction impairs oncogenic KRAS signalling. Nature. 2013;497:638-642. [DOI] [PubMed] [Google Scholar]
- 102. Canon J, Rex K, Saiki AY, et al. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature. 2019;575:217-223. [DOI] [PubMed] [Google Scholar]
- 103. Hallin J, Engstrom LD, Hargis L, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov. 2020;10:54-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Weber R, Groth C, Lasser S, et al. IL-6 as a major regulator of MDSC activity and possible target for cancer immunotherapy. Cell Immunol. 2021;359:104254. [DOI] [PubMed] [Google Scholar]
- 105. Kampan NC, Xiang SD, McNally OM, Stephens AN, Quinn MA, Plebanski M. Immunotherapeutic interleukin-6 or interleukin-6 receptor blockade in cancer: challenges and opportunities. Curr Med Chem. 2018;25:4785-4806. [DOI] [PubMed] [Google Scholar]
- 106. Taylor-Weiner A, Zack T, O’Donnell E, et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature. 2016;540:114-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Loveday C, Litchfield K, Proszek PZ, et al. Genomic landscape of platinum resistant and sensitive testicular cancers. Nat Commun. 2020;11:2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep. 2020;10:2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Swisher EM, Lin KK, Oza AM, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75-87. [DOI] [PubMed] [Google Scholar]
- 110. Vanderstichele A, Busschaert P, Olbrecht S, Lambrechts D, Vergote I. Genomic signatures as predictive biomarkers of homologous recombination deficiency in ovarian cancer. Eur J Cancer. 2017;86:5-14. [DOI] [PubMed] [Google Scholar]
- 111. Sun H, Kim P, Jia P, Park AK, Liang H, Zhao Z. Distinct telomere length and molecular signatures in seminoma and non-seminoma of testicular germ cell tumor. Brief Bioinform. 2019;20:1502-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Koul S, McKiernan JM, Narayan G, et al. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol Cancer. 2004;3:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Wermann H, Stoop H, Gillis AJ, et al. Global DNA methylation in fetal human germ cells and germ cell tumours: association with differentiation and cisplatin resistance. J Pathol. 2010;221:433-442. [DOI] [PubMed] [Google Scholar]
- 114. Messerschmidt DM, Knowles BB, Solter D. DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Genes Dev. 2014;28:812-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22-36. [DOI] [PubMed] [Google Scholar]
- 116. Bresesti C, Vezzoli V, Cangiano B, Bonomi M. Long non-coding RNAs: role in testicular cancers. Front Oncol. 2021;11:605606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hu Y, Zhu QN, Deng JL, Li ZX, Wang G, Zhu YS. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther. 2018;11:3185-3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Masters JR, Köberle B. Curing metastatic cancer: lessons from testicular germ-cell tumours. Nat Rev Cancer. 2003;3:517-525. [DOI] [PubMed] [Google Scholar]
- 119. Lobo J, Jerónimo C, Henrique R. Cisplatin resistance in testicular germ cell tumors: current challenges from various perspectives. Cancers. 2020;12:1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Wei J, Gan Y, Peng D, et al. Long non-coding RNA H19 promotes TDRG1 expression and cisplatin resistance by sequestering miRNA-106b-5p in seminoma. Cancer Med. 2018;7:6247-6257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Tang Y, Cui Y, Zhang S, Zhang L. The sensitivity and specificity of serum glycan-based biomarkers for cancer detection. Prog Mol Biol Transl Sci. 2019;162:121-140. [DOI] [PubMed] [Google Scholar]
- 122. Maragozidis P, Papanastasi E, Scutelnic D, et al. Poly(A)-specific ribonuclease and nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Mol Cancer. 2015;14:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Murakami Y, Tamori A, Itami S, et al. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer. 2013;13:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Han P, Liu Q, Xiang J. Monitoring methylation-driven genes as prognostic biomarkers in patients with lung squamous cell cancer. Oncol Lett. 2020;19:707-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Li W, Xia S, Aronova A, et al. CHL1 expression differentiates hürthle cell carcinoma from benign hürthle cell nodules. J Surg Oncol. 2018;118:1042-1049. [DOI] [PubMed] [Google Scholar]
- 126. Rick O, Siegert W, Schwella N, Dubiel M, Krusch A, Beyer J. High-dose chemotherapy as salvage treatment for seminoma. Bone Marrow Transplant. 2002;30:157-160. [DOI] [PubMed] [Google Scholar]
- 127. Gerl A, Schierl R. Urinary excretion of platinum in chemotherapy-treated long-term survivors of testicular cancer. Acta Oncol. 2000;39:519-522. [DOI] [PubMed] [Google Scholar]
- 128. D’Andrea AD. Mechanisms of PARP inhibitor sensitivity and resistance. DNA Repair. 2018;71:172-176. [DOI] [PubMed] [Google Scholar]
- 129. O’Sullivan Coyne G, Karlovich C, Wilsker D, et al. PARP inhibitor applicability: detailed assays for homologous recombination repair pathway components. Onco Targets Ther. 2022;15:165-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Li L, Wang H, Yang ES, Arteaga CL, Xia F. Erlotinib attenuates homologous recombinational repair of chromosomal breaks in human breast cancer cells. Cancer Res. 2008;68:9141-9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Chinnaiyan P, Huang S, Vallabhaneni G, et al. Mechanisms of enhanced radiation response following epidermal growth factor receptor signaling inhibition by erlotinib (tarceva). Cancer Res. 2005;65:3328-3335. [DOI] [PubMed] [Google Scholar]
- 132. Willers H, Azzoli CG, Santivasi WL, Xia F. Basic mechanisms of therapeutic resistance to radiation and chemotherapy in lung cancer. Cancer J. 2013;19:200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-2-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-3-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-4-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-5-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics
Supplemental material, sj-xlsx-6-cix-10.1177_11769351221132634 for Integrated Molecular Analysis Reveals 2 Distinct Subtypes of Pure Seminoma of the Testis by Kirill E Medvedev, Anna V Savelyeva, Kenneth S Chen, Aditya Bagrodia, Liwei Jia and Nick V Grishin in Cancer Informatics