Abstract
MetaMorpheus is a free and open-source software program dedicated to the comprehensive analysis of proteomic data. In bottom-up proteomics, protein samples are digested into peptides prior to chromatographic separation and tandem mass spectrometric analysis. The resulting fragmentation spectra are subsequently analyzed with search software programs to obtain peptide identifications and infer the presence of proteins in the samples. MetaMorpheus seeks to maximize the information gleaned from proteomic data through the use of (a) mass calibration, (b) post-translational modification discovery, (c) multiple search algorithms, which aid in the analysis of data from traditional, crosslinking, and glycoproteomic experiments, (d) isotope-based or label-free quantification, (e) multi-protease protein inference, and f) spectral annotation and data visualization capabilities. This protocol provides detailed descriptions of how use MetaMorpheus and how to customize data analysis workflows using MetaMorpheus tasks to meet the specific needs of the user.
Keywords: Proteomics, Tandem mass spectrometry, Bottom-up, Database search, Open-source, Post-translational modification discovery, Crosslink, Glycopeptides
1. Introduction
Bottom-up proteomics is the foremost method for in-depth identification and characterization of proteins from biological systems. In bottom-up proteomics, proteins are digested generating peptides which are separated, typically by reverse phase high-performance liquid chromatography (RP-HPLC), before mass spectrometric analysis [1]. As peptides elute from the HPLC, they are emitted into the mass spectrometer (MS) using electrospray ionization. Inside the MS, a MS1 spectrum is acquired to determine the mass-to-charge ratio of the eluting intact peptides. These peptides are then isolated and fragmented to produce MS2 spectra, which serve as “bar-codes” to identify each peptide’s amino acid sequence. MS data acquisition is a repetitive process, wherein the acquisition of an MS1 spectrum is followed by acquisition of several MS2 spectra. These experiments generate too much data for reliable and efficient manual analysis, thus database search software programs are commonly employed [2]. These programs compare the observed fragmentation data (MS2 spectra) with the predicted fragment ions of theoretical peptides derived from a reference database.
The principal goal of database search software programs is to correctly identify as many peptide sequences as possible from the acquired fragmentation spectra [3-6]. As the number of quality peptide identifications increases, the ability to accurately and comprehensively characterize the proteins present in the sample also improves. This is important for both discovery-based studies seeking to identify the proteins present, and quantitative studies seeking to determine protein abundance change.
MetaMorpheus is a free and open-source database search software program designed to be user-friendly and maximize the information extracted from bottom-up proteomic experiments. MetaMorpheus contains several features to facilitate extensive data analysis, including (a) mass calibration, (b) post-translational modification (PTM) discovery [7], (c) specialized search algorithms to aid in the analysis of data from various proteomic experiments (traditional, crosslink [8], and glycoproteomic [9]), (d) isotope-based or label-free quantification [10], (e) multi-protease protein inference [11], and (f) spectral annotation and data visualization capabilities. This protocol is devoted to presenting how to construct MetaMorpheus data analysis workflows, enabling users to customize their experience based on their specific experiment. MetaMorpheus version 0.0.310 was used in the development of this protocol (see Note 1).
2. Material
2.1. Mass Spectra Requirements
MetaMorpheus accepts spectra in .mzML (centroided), .mgf, or Thermo .raw formats. Other formats can be converted to .mzML with MSConvert (see Note 2). Regardless of the format, all mass spectra must contain MS2 scans. MetaMorpheus was originally designed for the analysis of high-resolution MS2 data but has since been adapted for the analysis of low-resolution MS2 data (see Note 3).
2.2. Protein Database Requirements
Protein databases for analysis can be supplied in UniProt .XML (see Note 4) or .FASTA formats in either their compressed (.gz) or uncompressed states.
2.3. System Requirements
MetaMorpheus can be installed and operated on any desktop computer with a Windows, MacOS, or Linux 64-bit operating system via the command-line interface (CLI). However, the graphical user interface (GUI) version is currently Windows-only.
There is no formal RAM requirement for MetaMorpheus, but a minimum of 8 GB of RAM is recommended (see Note 5).
MetaMorpheus requires the installation of.NET Core 3.1 (see Note 6).
2.4. Download and Installation
MetaMorpheus can be installed and operated as a graphical user interface (GUI) program in the Microsoft Windows environment or as a command-line interface (CLI) in Microsoft Windows, Apple MacOS, or Linux environments. The latest release of MetaMorpheus can be retrieved from GitHub (https://github.com/smith-chem-wisc/MetaMorpheus/releases). To download the GUI from GitHub, click on the MetaMorpheusInstaller.msi for the latest, or desired release. Follow the directions provided by the installer to complete the installation process. The command-line version of MetaMorpheus can be downloaded by selecting MetaMorpheus_CommandLine.zip for the latest or desired release. Once the zip file has downloaded, contents must be extracted to use the program. A Docker image of the command-line version of MetaMorpheus can be retrieved from Docker Hub by using the following Docker Pull Command:
docker pull smithehemwisc/metamorpheus
3. Methods
MetaMorpheus, similar to many other search software tools, takes in user-supplied spectra files and protein databases. However, a distinguishing feature of MetaMorpheus is the construction of custom data analysis workflows from individual analysis modules called tasks. Tasks, once added to a workflow, are run sequentially, and when appropriate, use information from previous tasks to improve the overall results. The use of individual tasks enables the user to mix and match different analyses to best meet their specific needs. The following protocol will explain the basic set up of MetaMorpheus as well as how to set up each individual task for the creation of custom data analysis workflows.
3.1. Starting MetaMorpheus
Open MetaMorpheus from the start menu or double click the MetaMorpheus desktop icon to open the GUI (see Fig. 1).
Fig. 1.
MetaMorpheus application opens to the About window
3.2. Loading Protein Databases
Select the Databases tab in the menu on the left side of the GUI to open the Protein Databases loading page (see Fig. 2).
Database files can be added by two different approaches: (a) dragging and dropping database file(s) into the GUI (see Note 7), or (b) selecting the +ADD DATABASE button to open a file browsing window enabling navigation to the desired database file(s) for selection.
MetaMorpheus accommodates the use of more than one protein database. This enables the use of a contaminant database as well as the ability to analyze multi-species samples.
To add a database of common contaminant protein sequences, click the ADD DEFAULT CONTAMINANTS button (see Note 8). Users can also supply their own contaminant databases. To have MetaMorpheus recognize a provided database as a contaminant database, right click the file and select Set as contaminant database.
Fig. 2.
Protein Databases window where users specify .FASTA or .XML databases for analysis
3.3. Loading Spectra Files
Select the Spectra tab in the menu on the left side of the GUI to open the Spectra loading page (see Fig. 3).
Mass spectra files can be added in a similar manner to databases either by (a) dragging and dropping the spectra file into the GUI (see Note 7) or by (b) selecting the +ADD SPECTRA button (see Note 9).
Fig. 3.
Spectra window where users specify spectra files for analysis
3.4. Set File-Specific Settings
The ability to specify file-specific search settings is an optional feature of MetaMorpheus that facilitates analysis of complex datasets containing samples generated by differing preparation methods; such as multi-protease experiments.
Select file(s) in the Spectra window requiring a particular file-specific setting.
Click the SET FILE-SPECIFIC SETTINGS button to open a window displaying a list of parameters, which can be set on a file-specific basis. All parameters are disabled by default (see Note 10) (see Fig. 4).
Click the check box next to each parameter to enable editing (see Note 11).
Fig. 4.
File-specific parameters window
3.5. Mass Calibration
The mass accuracy of MS1 and MS2 spectra can vary within a sample, or over multiple samples. Numerous factors can contribute to this variance such as systematic drift, random noise, changes in power supply voltage, vacuum system stability, and varying temperature or humidity. Spectral mass calibration corrects for this undesirable variance and almost always improves the number of confident peptide identifications made by MetaMorpheus [7]. Although it is not strictly necessary, the addition of a Calibration Task to any analysis workflow is recommended.
Select the Tasks tab in the menu on the left side of the GUI to open the Tasks page (see Fig. 5).
Select the +ADD CALIBRATION button above the task panel to open the Calibrate Task window where all parameters for the Calibration Task can be adjusted as needed.
The Calibrate Task window has two parameter sections: (a) Search Parameters and (b) Modifications.
Expansion of the “Search Parameters” section exposes parameters affecting the search component of the mass calibration algorithm. A description of each of these parameters can be found below (see Subheading 3.15, Steps 10, 18, 24, 25, 26, 27, 28, 29, 31, 32, 34, 49, 50 and51).
The selection of the drop-down arrow beneath the “Modifications” header displays lists of PTMs that can be selected as fixed (see Subheading 3.15, Step 12) or variable modifications (see Subheading 3.15, Step 64).
After parameters have been adjusted, select the Add the Calibrate Task button at the bottom of the window to add the Calibration Task to the MetaMorpheus analysis workflow (see Notes 12, 13 and 14).
Fig. 5.
Task window where users can add MetaMorpheus tasks to data analysis workflow
3.6. Global Post-Translational Modification Discovery
Global Post-Translational Modification Discovery (GPTMD) is a tool within MetaMorpheus that enables the identification of peptides containing PTMs, which are not annotated in the supplied database or provided as variable modifications [7]. GPTMD constructs a new reference database in .XML format by annotating the existing database with PTMs discovered by a first-pass search of the provided spectra. It utilizes a mass-tolerant search approach to identify peptide spectral matches (PSMs) with mass shifts (notches) indicative of PTMs selected by the user. GPTMD is superior to many other open-mass search approaches because the discovered PTMs are annotated, instead of being provided as a delta mass value. The GPTMD approach enables variable post-translational modification at targeted positions in the protein to improve search results without incurring the inflated search times or false discovery rates (FDR) typically associated with traditional variable modification searching. GPTMD illuminates larger, and potentially biologically interesting, portions of the proteome which are not identified by other approaches.
Select the Tasks tab in the menu on the left side of the GUI to open the Tasks page (see Fig. 5).
Select the +ADD PTM DISCOVERY button above the task panel to open the GPTMD Task window where all parameters required for post-translational modification discovery can be adjusted.
The GPTMD Task window contains four parameter sections: (a) File Loading Parameters, (b) Search Parameters, (c) Fixed/Variable Modifications and (d) GPTMD Modifications.
Parameters for spectral processing can be found beneath the “File Loading Parameters” header. A detailed description of the involved parameters can be found below (see Subheading 3.15, Steps 8, 9, 33, 58, 60, 61 and63).
Parameters for the first-pass search used for the discovery of candidate post-translationally modified peptides are located under the “Search Parameters” header. A detailed description of the utilized parameters can be found below (see Subheading 3.15, Steps 10, 13, 18, 24, 25, 26, 27, 28, 29, 31, 32, 34, 49, 50 and51).
Selection of the drop-down arrow under the “Fixed/Variable Modifications” header displays lists of PTMs that can be selected as fixed (see Subheading 3.15, Step 12) or variable modifications (see Subheading 3.15, Step 64).
Expansion of the “GPTMD Modifications” section displays the same list of PTMs found in the Fixed/Variable Modifications panel. Selection of a PTM adds its mass shift to the list of notches investigated to identify previously unannotated PTMs (see Notes 15 and 16). Any modifications, except variable oxidation of methionine and fixed carbamidomethylation of cysteine, should be added here. Custom PTMs can be added to MetaMorpheus (see Note 17).
After parameters for the task have been finalized, select the Add the GPTMD Task button at the bottom of the window to add the GPTMD Task to the MetaMorpheus analysis workflow (see Notes 12, 13 and 14).
3.7. Search
Select the Tasks tab in the menu on the left side of the GUI to open the Tasks page (see Fig. 5).
Select the +ADD SEARCH button above the task panel to open the Search Task window where all parameters for the final search of the spectra can be adjusted to fit the user’s needs.
The Search Task window will open to display five expanded sections of parameters: (a) Search Parameters, (b) Modifications, (c) Protein Parsimony, (d) Quantification, and (e) Output Options. Additionally, there is a drop-down menu containing Advanced Options.
The parameters provided in the “Search Parameters” section are the most commonly adjusted. Detailed definitions of each of the parameters in this section can be found below (see Subheading 3.15, Steps 10, 18, 25, 27, 28, 31, 49, 50, 51 and56.
Lists containing all PTM options for fixed (see Subheading 3.15, Step 12) and variable modifications (see Subheading 3.15, Step 64) are displayed beneath the “Modifications” header.
The parameters provided in the “Protein Parsimony” section dictate how peptide identifications undergo protein inference, the process by which peptides are mapped back to their potential protein(s) of origin. Detailed descriptions for each parameter can be found below (see Subheading 3.15, Steps 1, 54 and59).
Quantification parameters inform peptide and protein quantification. Parameters dealing with SILAC and label-free quantification are present within this section and are defined below (see Subheading 3.15, Steps 20, 22, 39, 42, 48 and57).
The “Output Options” section contains parameters dictating what results are reported and how these results are exported. Detailed definitions of all the “Output Options” parameters can be found below (see Subheading 3.15, Steps 3, 11, 65, 67, 68 and69).
Selection of the Advanced Options menu reveals three additional parameter sections: (a) File Loading Parameters, (b) Search Parameters and (c) Post-Search Analysis. Each of these sections contains parameters only adjusted, typically, by more advanced users of MetaMorpheus.
The advanced “File Loading Parameters” section contains parameters regarding spectra file loading and pre-processing. A detailed description of all the involved parameters can be found below (see Subheading 3.15, Steps 8, 9, 33, 40, 41, 45, 58, 60 and61).
The options provided in the advanced “Search Parameters” section are not commonly changed. They provide the user the ability to alter miscellaneous details such as the search algorithm used by MetaMorpheus or how decoy proteins are generated. Detailed definitions of these advanced parameters can be found below (see Subheading 3.15, Steps 7, 13, 14, 15, 17, 21, 23, 24, 26, 29, 32, 34, 43, 44, 53, 55 and62).
The final set of advanced parameters, the “Post-Search Analysis” section, dictates post-analysis processing. A detailed description of these parameters, and their uses can be found below (see Subheading 3.15, Steps 4 and70).
After parameters for the task have been finalized, select the Add the Search Task button at the bottom of the window to add the Search Task to the MetaMorpheus analysis workflow (see Notes 12, 13 and 14).
3.8. Multi-Protease Protein Inference
To utilize MetaMorpheus’ multi-protease protein inference capabilities, mass spectra files from more than one proteolytic digest must be loaded into the Spectra tab of the GUI.
Select a subset of files from the same proteolytic digest and use the Set File-Specific Parameters to set the appropriate protease (see Note 11). Repeat this process until all files have a file-specific protease assigned.
It is recommended to perform Calibration (see Subheading 3.5) and GPTMD (see Subheading 3.6) tasks prior to searching.
Add a Search Task to the MetaMorpheus workflow, ensuring the protein parsimony parameter (see Subheading 3.15, Step 1) is enabled (see Note 18).
3.9. Crosslink Search
Select the Tasks tab in the menu on the left side of the GUI to open the Task page (see Fig. 5).
Select the +ADD XL SEARCH button above the task panel to open the XL Search Task window where all parameters for the search of crosslinked peptides can be adjusted [8].
The XL Search Task window contains four sections of parameters: (a) Crosslink Search, (b) Search Parameters, (c) Modifications, and (d) Output Options.
Parameters within the “Crosslink Search” section are specific to the crosslink experiment performed, such as the chemical crosslinker used and how it was quenched. If the crosslinker used is not one of the provided options, custom crosslinkers can be added to MetaMorpheus (see Note 19). Definitions for each of these crosslink specific parameters can be found below (see Subheading 3.15, Steps 5, 6, 19, 35, 36 and37).
Expansion of the “Search Parameters” sections reveals the parameters used to inform spectra pre-processing and the underlying search algorithm. Detailed definitions for each one of these parameters can be found below (see Subheading 3.15, Steps 8, 14, 18, 25, 26, 28, 29, 31, 33, 34, 49, 50, 51, 56, 58, 60, 61 and63).
The “Modifications” section contains lists of PTMs for selection as fixed (see Subheading 3.15, Step 12) or variable modifications (see Subheading 3.15, Step 64).
MetaMorpheus exports its PSM results using a .psmtsv format. The final parameter section, “Output Options”, provides the option to generate a results file in .pep.XML format in addition to the standard output. The results of the crosslink search task, in the .pep.XML format, can be visualized using ProXL (see Note 20). A more detailed description of this parameter can be found in Subheading 3.15 Step 66.
After parameters for the task have been finalized, select the Add the XL Search Task button at the bottom of the window to add the XL Search Task to the MetaMorpheus analysis workflow (see Notes 12 and 13).
3.10. Glycopeptide Search
Select the Tasks tab in the menu on the left side of the GUI to open the Task page (see Fig. 5).
Select the +ADD GLYCO SEARCH button above the task panel to open the Glyco Search Task window containing all necessary parameters for the identification of N or O-glycosylated peptides [9].
The Glyco Search Task window contains three parameter sections (a) Glyco Search, (b) Search Parameters, and (c) Modifications.
The “Glyco Search” section consists of parameters specific to the glycan searching capabilities of MetaMorpheus. Specific definitions for each of these parameters can be found below (see Subheading 3.15, Steps 2, 10, 16, 19, 30, 38, 46 and47). A unique feature of MetaMorpheus’ Glyco Search Task is the ability to add a custom glycan database for searching (see Note 21).
The parameters within the “Search Parameters” section inform spectra pre-processing and the underlying search algorithm. Detailed definitions for each one of these parameters can be found below (see Subheading 3.15, Steps 8, 14, 18, 25, 26, 27, 28, 29, 31, 33, 34, 49, 50, 51, 58, 60, 61 and63).
The “Modifications” section contains lists of PTMs that can be selected as fixed (see Subheading 3.15, Step 12) or variable modifications (see Subheading 3.15, Step 64).
After parameters for the task have been finalized, select the Add the GlycoSearch Task button at the bottom of the window to add the Glyco Search Task to the analysis workflow (see Notes 12 and 13).
3.11. Starting Analysis in MetaMorpheus
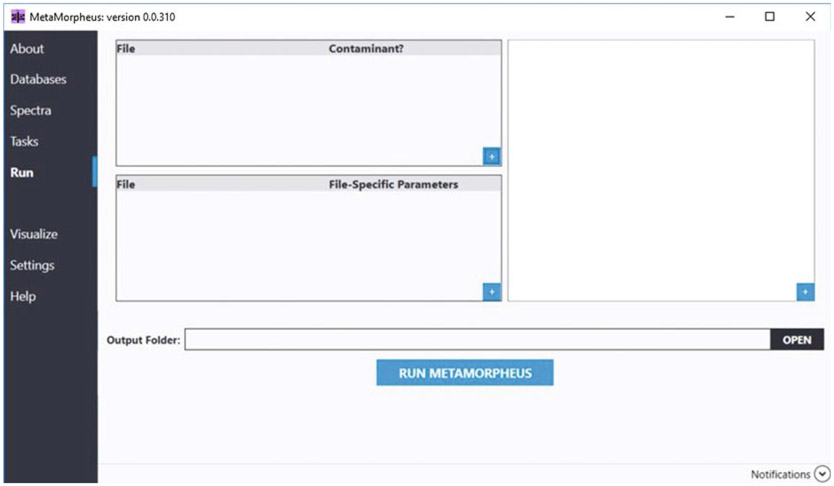
After all desired tasks have been added to the workflow, select the Run tab in the menu on the left side of the GUI (see Fig. 6).
The Run window serves as a summary displaying all databases, spectra files and tasks for analysis (see Note 22). Any missing files or tasks can be added at this time using the small + button at the bottom right of the appropriate panel.
All the results from MetaMorpheus tasks will be placed in an output folder at the location noted within the output folder field. This is automatically set as a time-stamped directory within the directory of the first specified spectra file.
After all parameters and tasks are finalized, select the RUN M ETAMORPHEUS button to run the established workflow.
Analysis can be aborted at any time by clicking the CANCEL RUN button, which appears after the data analysis process has begun. If a run is canceled, the RESET TASKS button can be used to regenerate all tasks in the workflow for future analysis. Once tasks are reset, the RUN METAMORPHEUS button will re-appear, indicating the workflow can once again be run.
Fig. 6.
Run summary page containing all spectra files, databases, and tasks necessary for analysis with MetaMorpheus
3.12. Spectrum Annotation with MetaDraw
Select the Visualize tab in the menu on the left side of the GUI to open MetaDraw, MetaMorpheus’ spectral annotator and data viewer.
A window will open to show the PSM Annotation tab where the spectra files can be uploaded for annotation (see Fig. 7). Files can be added either by dragging and dropping or by clicking the Select button to open a file explorer window.
The PSM or peptide results files (in .psmtsv format) must be also be added using either the drag and drop method or the Select button.
Once all the necessary files have been added, click the Load Files button to populate the Peptide Spectral Matches panel with all MS2 scans from the spectra file corresponding to PSMs in the MetaMorpheus results file.
To display an annotated spectrum, select the row of the desired PSM. The annotated spectra will be displayed in the PSM Annotation window and information about the PSM will be displayed in the Properties panel.
The annotated spectra can be exported as a PDF document using the Export to PDF button.
Fig. 7.
PSM annotation window of MetaDraw within MetaMorpheus. A sample spectra annotation is displayed
3.13. Data Visualization with MetaDraw
From the homepage, select the Visualize tab in the menu on the left side of the GUI to open MetaDraw.
Select the Data Visualization tab next to the PSM Annotation tab in the top left corner (see Fig. 8).
Provide the PSM or peptide results file of interest. The file can be added by dragging and dropping or by using the Select button to browse in file explorer.
Once the results file has been provided, click the Import Files From PSMTSV button to populate the Source file(s) panel with the spectra files containing peptide identifications.
Users can select one or more spectra files or can use the Select all button to generate plots containing peptide identifications above a 1% FDR from the selected spectra files.
Select the desired plot from the Plot Type panel in the bottom left-hand corner. Generated plots are displayed in the Plot panel on the right. A description for each plot type can be found in Table 1.
All plots can be exported as a PDF document using the Export to PDF button.
Fig. 8.
Data Visualization window of MetaDraw within MetaMorpheus. A sample histogram of Precursor PPM errors is displayed
Table 1.
MetaDraw data visualization plots
| Plot type | Description |
|---|---|
| Histogram of Precursor PPM error (around 0 Da mass-notch difference only) | Distribution of precursor mass errors for unmodified peptide identifications (1% FDR). The y-coordinate is the number of identifications in the specific precursor mass error bin (x-coordinate). |
| Histogram of Precursor charges | Distribution of precursor ion charge states for all PSM or peptide identifications (1% FDR) |
| Histogram of fragment charges | Distribution of fragment ion charge states for PSM or peptide identifications (1% FDR) |
| Precursor PPM error vs. RT | Scatter plot where the y-coordinate is the precursor error of each PSM or peptide identification (1% FDR) and the x-coordinate is the experimentally obtained retention time |
| Histogram of PTM spectral counts | This graph displays the number of spectra identified (1% FDR) to contain a specific PTM type. The y-coordinate is the count of PSMs containing a given PTM and the x-axis is the PTM |
| Predicted RT vs. observed RT | Scatter plot comparing the predicted hydrophobicity of a peptide based on its amino acid sequence (y-coordinate) to the observed experimental retention time of the species (x-coordinate) |
3.14. Command-Line Operation of MetaMorpheus
MetaMorpheus can be operated via the command-line interface in Windows, Linux, or MacOS operating systems. To run MetaMorpheus in Windows command line, run ”CMD.exe” with the following arguments:
CMD.exe -d "C:\MyFolder\myDatabase.fasta" -t " C:\MyFolder\myTask1.toml" "C:\MyFolder\myTask2.toml" -s "C:\MyFolder\mySpectraFile.mzML "
To run the .NET Core version of MetaMorpheus in Linux or in MacOS, run "dotnet CMD.dll" with the following arguments:
dotnet CMD.dll -d "\home\myfolder\mydatabase.fasta" -t "\home\myfolder\myTask1.toml" "\horae\myfolder\myTask2.toml" -s "\home\mySpectraFile.mzML"
The command-line arguments for all environments are as follows:
--help: This argument prints a list of all MetaMorpheus command-line arguments and their definitions.
-d: This argument denotes what protein databases will be analyzed (.XML or .FASTA) and is required. Following the argument, provide the file paths for all databases being used, with a space delimiter between each file (see Note 23).
-s: This argument precedes all spectra files to be analyzed and is required. Provide the file paths for all spectra files to be analyzed after this argument with a space delimiter between each file (see Note 23).
-t: This argument indicates which tasks will be performed during the analysis and is required. Tasks are provided as . toml files, supplying all necessary parameters for each task (see Subheading 3.15 for descriptions of all parameters). There are different .toml file formats for each task type (Calibration, GPTMD, Search, XL Search, and Glyco Search). Provide the file paths for all .toml files in the order the tasks are to be performed following the argument, with a space delimiter between each file (see Note 23).
-o: This argument sets the output folder for all results of the MetaMorpheus workflow. This argument is usually optional. If no output folder is explicitly set, then a time-stamped folder will be automatically generated in the directory of the first provided spectra file.
-g: This argument generates .toml files for all tasks containing the default parameter settings. These .toml files can be modified so the parameters fit the experimental data being analyzed. When this argument is used, only the ”--o” argument is required. The ”-o” argument specifies the output where the default .toml files will be written.
-v: This optional argument deals with verbosity, determining the extent to which output and error messages are displayed. The default value for this argument is “normal” but can be set to “none” or “minimal.”
--test: This argument runs a small test search using a yeast database and spectra file included with MetaMorpheus during installation. This command ensures proper installation of MetaMorpheus. When this argument is called, no other command arguments are required.
--version: This argument displays the version information for MetaMorpheus.
3.15. Parameters
This section provides definitions for all MetaMorpheus task parameters. Parameters are organized in alphabetical order, by their name as displayed in the GUI. Following the GUI name, the parameter name, as displayed in .toml setting files, is provided in parenthesis for command-line users. The default values provided for all parameters in MetaMorpheus are designed to facilitate the analysis of most high-resolution MS2 data without requiring alteration.
Apply Protein Parsimony and Construct Protein Groups (DoParsimony): This Search Task parameter indicates if protein parsimony will be performed on the identified peptides (1% PSM-level FDR). Selection of protein parsimony is required for match between runs, protein quantification and multi-protease protein inference.
Child Scan Dissociation (MS2ChildScanDissociationType): This Glyco Search Task parameter specifies the dissociation type used for generating MS3 scans or second dissociation MS2 scans.
Compress Individual File Results (CompressIndividualFiles): This Search Task parameter determines if MetaMorpheus’ individual results files are compressed in order to minimize memory requirements.
Construct Mass Difference Histogram (DoHistogramAnalysis): This post-search analysis parameter within the Search Task allows for the creation of a histogram displaying the observed mass shifts for all peptide identifications (1% FDR). The mass shifts observed for PSMs are clustered into bins and analyzed for peaks corresponding to the molecular weight of a PTM or amino acid substitution. This analysis is primarily useful for interpreting open-mass search results.
Crosslink at Cleavage Sites (CrosslinkAtCleavageSite): This XL Search Task parameter dictates whether or not a crosslink can be identified at a proteolytic cleavage site.
Crosslinker Type (all parameters under the XlSearchParameters.Crosslinker header): This XL Search Task parameter specifies the crosslinker used in the experiment. The crosslinker type can be selected from a list of crosslinkers or a custom crosslinker can be added (see Note 19).
C-Terminal Ions (FragmentationTerminus): This Search Task parameter specifies the generation of fragment ions from the C-terminus (e.g., x-, y-, and z-ions) of all theoretical peptides.
Deconvolute Precursor (DoPrecursorDeconvolution): Present in the GPTMD, Search, XL Search, and Glyco Search Tasks, this parameter enables the identification of multiple peptides from a single MS2 scan. For each MS2 scan, the MS1 isolation window is investigated for precursors that could have been co-fragmented to yield the observed fragmentation pattern.
Deconvolution Max Assumed Charge State (DeconvolutionMaxAssumedChargeState): Present in the GPTMD and Search Tasks, this parameter dictates the maximum expected charge state for a peptide. Any isotopic envelopes with charge states larger than this value are discarded or are incorrectly identified as harmonics.
Dissociation Type (DissociationType): Present in all task types (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter specifies the dissociation type used for the acquisition of MS2 spectra. MetaMorpheus was originally designed for analysis of high-resolution MS2 data, because of this all dissociation types are assumed to be high-resolution, with the exception of the LowCID option (see Note 3).
Filter Results to q-Value (QValueOutputFilter): This Search Task parameter dictates the maximum q-value of peptide identifications in the output files. The filtering of identifications makes the exported result files more manageable for large datasets.
Fixed Modifications (ListOfModsFixed): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter dictates which PTMS are “fixed” and should be applied to every possible location in the database specified. Typically, the only fixed modification necessary is carbamidomethylation of cysteine, which results when reduced samples have been alkylated with iodoacetamide. Other fixed modifications can be selected when appropriate such as TandemMassTag (TMT) labels.
Generate Complementary Ions (AddCompIons): Present in GPTMD and Search Tasks, this parameter adds artificial complementary masses to the experimental MS2 spectrum. Artificial fragment masses are inferred by subtracting the deconvoluted mass of each observed MS2 fragment ion from the observed precursor mass and adding a dissociation type-specific mass shift. This strategy can be helpful in identifying peptides with modifications near a terminus (e.g., C-terminal modifications of tryptic peptides) [12].
Generate Decoy Proteins (DecoyType): Present in all search tasks (Search, XL Search, and Glyco Search), this parameter indicates if MetaMorpheus automatically generates decoy protein sequences from the provided protein database(s). Decoy proteins provide known false-positive sequences which can be used to determine q-values. In the Search and Glyco Search Tasks, decoy proteins can be generated by using either the reversed or slide methods. Reversed decoys are generated by reversing the protein sequence provided in the target database. Slide decoys are generated by non-random shuffling of the amino acids within each provided protein sequence. If the protein database supplied already contains decoy protein sequences, uncheck this feature.
Generate Target Proteins (SearchTarget): This Search Task parameter indicates MetaMorpheus will search for target peptides generated by the in silico digestion of provided database (s). This parameter can be disabled for decoy-only searches, which are useful in analyses where target and decoy databases are searched separately.
Glyco Search (GlycanSearchType): This Glyco Search Task parameter determines whether O-glycopeptides or N-glycopeptides are discovered by the Glyco Search algorithm. Only one class of glycans can be investigated at a time using the Glyco Search Task.
Handle Overlap Between Target and Contaminant Databases (TCAmbiguity): This Search Task parameter specifies the classification of protein entries that are shared between target and contaminant databases as a contaminant entry, target entry or both.
Initiator Methionine (InitiatorMethionineBehavior): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter specifies how MetaMorpheus addresses the potential cleavage of initiator methionine residues in the protein database. The initiator methionine for protein entries can always be cleaved, always be retained, or variable (both cleaved and retained versions are created) in the generation of theoretical peptides. It is recommended to treat the initiator methionine as variable.
Keep Top N Candidates (CrosslinkSearchTopNum or GlycoSearchTopNum): This parameter in the XL Search and Glyco Search Tasks specifies the maximum number of candidate peptides considered per MS2 scan to reduce computational complexity.
LFQ: Quantify peptides/proteins with FlashLFQ (DoQuantification): Selection of this quantification option within the Search Task establishes that FlashLFQ will be used to perform label-free peptide and protein level quantification. An experimental plan in MetaMorpheus is required for label- free quantification (see Note 24). Additional information on FlashLFQ can be found in Chapter 13.
Mass Difference Acceptor Criterion (MassDiffAcceptorType): This Search Task parameter determines the acceptable the mass notch(es) for the difference between a peptide’s observed and theoretical precursor mass. Selections can be made from the provided options (“Exact”, ”1 Missed Monoisotopic Peak”, “1 or 2 Missed Monoisotopic Peaks”, “1, 2 or 3 Missed Monoisotopic Peaks”, “+−3 Missed Monoisotopic peaks”, “-187 and Up”, and “Accept all”). Additionally, MetaMorpheus supports the addition of a custom mass difference acceptor (see Note 25).
Match Between Runs (MatchBetweenRuns): This Search Task parameter indicates match between runs will be utilized as part of the quantification process. Match between runs allows peptides that were fragmented in at least one spectra file to be quantified across all other spectra files. Any peptide identified in one spectra file is searched for in all other files within a small mass-to-charge and retention time window. To learn more about match between runs, refer to the FlashLFQ protocol (see Chapter 13).
Max Fragment Mass (MaxFragmentSize): This Search Task parameter imposes an upper limit for the mass of theoretical fragment ions.
Max Heterozygous variants for Combinatorics (MaxHeterozygousVariants): Present in the Calibration, GPTMD, and Search Tasks, this parameter is only relevant when one of the databases provided is generated via Spritz [13] and contains annotated sequence variants. It dictates the maximum number of variants that can be applied to a single protein sequence, thus determining the number of theoretical variant-containing proteins generated.
Max Missed Cleavages (MaxMissedCleavages): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter specifies the maximum number of missed cleavages allowed during in silico digestion of the protein database(s). The protease utilized affects this parameter because certain proteases, such as Chymotrypsin, are more prone to missed cleavages [14].
Max Modification Isoforms (MaxModficationIsoforms): This parameter is present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search) and specifies the maximum number of different peptide forms (peptidoforms) possible for a single theoretical peptide sequence. A large number variable and/or annotated modifications for a peptide can drastically increase the number of peptidoforms present in the database, making this parameter crucial for controlling database size.
Max Mods Per Peptide (MaxModsForPeptide): This parameter, present in the Calibration, GPTMD, Search, and Glyco Search Tasks, defines the maximum number of PTMs allowed on an individual peptide. As this value increases, so does the number of PTM combinations, search space and computational time.
Max Peptide Length (MaxPeptideLength): Present in all task types (Calibration, GPTMD, Search, XL Search, and Glyco Search) this parameter establishes the maximum length of theoretical peptides generated by in silico database digestion. Any peptides present in the sample longer than the specified value will not be correctly identified.
Max Threads (MaxThreadsToUsePerFile): This parameter specifies the maximum number of threads MetaMorpheus can utilize. The default value is determined based on the CPU running MetaMorpheus and is set to one less than the total number of threads.
Maximum OGlycans Allowed (MaximumOGlycanAllowed): This Glyco Search parameter specifies the maximum number ofO-glycosylation sites possible for a single theoretical peptide. This parameter should be adjusted depending on prior knowledge of the sample being analyzed. For example, mucins are a class of proteins known for heavy O-glycosylation [15]. A sample of primarily mucin proteins should have a higher value set for this parameter than non-mucin samples.
Min Peptide Length (MinPeptideLength): Present in all task types (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter establishes the minimum length of theoretical peptides generated by in silico database digestion. The default value for this parameter is seven, because peptides shorter than this length are difficult to confidently identify.
Min Read Depth for Variants (MinVariantDepth): Found in the Calibration, GPTMD, and Search Tasks, this parameter is only relevant when one or more of the databases provided are generated by Spritz [13] and contain annotated sequence variants. This parameter specifies the read depth, or coverage, that a specific variant must have in the RNA sequencing data in order to be included into theoretical protein sequences. This prevents variants without sufficient transcriptomic support from expanding the search space.
Minimum Intensity Ratio (MinimumAllowedIntensityRatioToBasePeak): This parameter, present in the GPTMD, Search, XL Search, and Glyco Search tasks, establishes the minimum intensity ratio required for each experimental fragment ion. The intensity ratio for each fragment ion is calculated by dividing its intensity by that of the highest intensity peak in the scan. If the minimum intensity threshold is not met, that fragment ion cannot be compared to the theoretical peptide spectra.
Minimum Score Allowed (ScoreCutoff): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter defines the minimum score required to report a PSM. The score, for high-resolution MS2 data, is determined by summing the number of matched fragment ions with the fraction of the total ion current (TIC) accounted for by these matched ions.
MS2 Child Scan Dissociation (MS2ChildScanDissociationType): This XL Search Task parameter specifies the dissociation type used to generate MS3 scans or second dissociation MS2 scans. If this level of fragmentation is not relevant for the spectra being analyzed, the parameter can be set to Null.
MS2 Scan Dissociation Type (DissociationType2): This XL Search Task parameter specifies the dissociation type used to generate MS2 fragmentation spectra.
MS3 Child Scan Dissociation (MS3ChildScanDissociationType): This XL Search Task parameter specifies the dissociation type used to generate MS4 scans or second dissociation MS3 scans. If this level of fragmentation is not relevant for the spectra being analyzed, the parameter can be set to Null.
N-Glycan Database (NGlycanDatabasefile): This Glyco Search Task parameter determine which N-glycan database will be utilized for the identification of N-linked glycopeptides. Custom N-glycan databases can be added if necessary (see Note 21).
No Quantification (DoQuantification): Selection of this quantification option within the Search Task dictates that neither label-free or SILAC quantification will be performed.
Nominal Window Width Thomsons (WindowWidthThomsons): This Search Task parameter specifies the width of the MS1 and MS2 filtering windows in Thomsons (m/z units). Dividing MS1 and MS2 scans into windows helps prevent filtering bias that may result from prevalence of high intensity peaks in the center of the spectrum and lower intensity peaks at low and high m/z ranges.
Normalize Peaks in Each Window (NormalizePeaksAcrossAllWindows): This Search Task parameter enables the normalization of peak intensity values to the most intense peak within the defined window.
Normalize Quantification Results (Normalize): When label-free quantification with FlashLFQ is enabled, this Search Task parameter dictates the normalization of peptide intensity values. This normalization is based on the assumption that the majority of peptides do not change in abundance between conditions (see Chapter 13). The normalization algorithm requires the information provided in the experimental design (see Note 24).
N-Terminal Ions (FragmentationTerminus): This Search Task parameter specifies the generation of fragment ions from the N-terminus (e.g., a-, b-, and c-ions) of all theoretical peptides.
Number of Database partitions (TotalPartitions): The modern, semi-specific, and non-specific search algorithms generate an index of theoretical peptide spectra from the supplied database(s). This index can become prohibitively large and exceed the RAM capacity of the computer. This parameter allows for the search space to be divided into partitions, or sections, before the search is performed to avoid such complications. The theoretical peptides in each partition are searched separately and then aggregated to provide the same results as if the partitioning method was not applied.
Number of Windows (NumberOfWindows): This Search Task parameter defines the number of windows, or sections, the MS1 and MS2 scans are to be divided into for peak filtering. Often, peaks are most intense in the center of the spectrum and less intense on the edges. When filtering is applied to the entire spectrum there is a risk of removing quality peaks in the low and high m/z regions of the spectra and retaining noise peaks in the center. Division of the scans into filtering windows prevents this bias.
O-Glycan Database (OGlycanDatabasefile): This Glyco Search Task parameter determines which O-glycan database will be utilized for the identification of O-linked glycopeptides. Custom O-glycan databases can be added if necessary (see Note 21).
OxoniumIonFilt (OxoniumIonFilt): This Glyco Search Task parameter specifies that only MS2 scans containing an oxonium ion at 204 m/z will be investigated as potential glycopeptides.
Peak-Finding Tolerance (QuantifyPpmTol): This Search Task parameter defines the parent mass tolerance (in ppm) used for label-free quantification.
Precursor Mass Tolerance (PrecursorMassTolerance): This parameter, found in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), establishes the maximum mass difference between the observed and theoretical precursor masses permitted for a PSM. This value is typically specified in ppm but can also be represented in daltons.
Product Mass Tolerance (ProductMassTolerance): Found in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter establishes the maximum mass difference between theoretical and experimental fragment ion permitted for it to be considered a match. This value can also be set in either ppm or daltons (see Note 26).
Protease (Protease): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter establishes the protease used for in silico database digestion. This protease should be the same as was used experimentally to digest the sample. Selection can be made from a provided list of common proteases, or a custom protease can be specified (see Note 27).
Quench Method (XLQuench): This XL Search Task parameter specifies the method(s) utilized to quench the crosslinker.
Report PSM Ambiguity (ReportAllAmbiguity): This Search Task parameter defines how ambiguous peptide spectral matches are reported. When multiple theoretical peptide sequences match the same MS2 spectra, and these PSMs all have the same score, the identification is ambiguous. If this box is unchecked, a random peptide from the multiple ambiguous matches will be reported. Otherwise, all possible sequences are reported.
Require at least Two Peptides to Identify Protein (NoOneHitWonders): This Search Task parameter requires the identification of two unique peptides for the establishment of a protein group in protein parsimony. Historically, this parameter was developed to eliminate the presence of one-hit wonders but has since been considered to be overly stringent and be detrimental to protein parsimony overall [16].
Search Mode (SearchType): This Search Task parameter defines which search algorithm will be used for the task. MetaMorpheus includes 4 different search algorithms (or modes): (a) Classic Search (see Note 28), (b) Modern Search (see Note 29), (c) Semi-Specific Search (see Note 30) and (d) Non-Specific Search (see Note 31).
Separation Type (SeparationType): This parameter in the Search and XL Search Tasks specifies the online separation method utilized prior to mass spectrometric analysis. This determines whether predicted hydrophobicity or electrophoretic mobility values are calculated for the peptides.
SILAC/SILAM: Quantify peptides/proteins with stable isotope labels (DoQuantification and GenerateUnlabeledProteinsForSilac): Selection of this quantification option within the Search Task indicates a portion of the peptides and proteins within the sample have been isotopically labeled enabling relative quantification. Upon selection, additional parameters for SILAC-based quantification appear including a checkbox to quantify unlabeled peptides and a table in which to specify the amino acid labels being used (see Note 32).
Top N Peaks per m/z window (NumberOfPeaksToKeepPerWindow): Present in the GPTMD, Search, XL Search, and Glyco Search Tasks, this parameter indicates the maximum number of peaks allowed in a window with a specified m/z width. This parameter applies to the peak filtering process of MS1 and MS2 scans. The peaks within the window are ordered by intensity prior to the cutoff being applied.
Treat Modified Peptides as Different Peptides (ModPeptidesAreDifferent): This Search Task parameter requires the protein parsimony algorithm to consider modified peptides distinct from their unmodified form. This can potentially disambiguate protein groups by the presence of annotated PTMs.
Trim MS1 Peaks (TrimMs1Peaks): This parameter is present in all search task types (Search, XL Search, and Glyco Search) and enables the filtering of MS1 peaks as part of spectra pre-processing.
Trim MS2 Peaks (TrimMsMsPeaks): This parameter is present in all search task types (Search, XL Search, and Glyco Search) and enables the filtering of MS2 peaks as part of spectra pre-processing.
Use Delta Scores for FDR (UseDeltaScore): This Search Task parameter specifies whether the Delta Score, instead of the Score, should be used for ranking PSMs prior to statistical analysis. The Delta Score is the difference between the scores of the two best matching peptides for the same MS2 spectrum. If the Delta Score produces fewer PSMs at a 1% FDR, then the Score will be automatically used instead.
Use Provided Precursor (UseProvidedPrecursorInfo): Present in the GPTMD, Search, XL Search, and Glyco Search Tasks, this parameter indicates that the precursor mass reported in the spectra should be used as the observed precursor mass for the search. This can be used in addition to deconvoluted precursor masses.
Variable Modifications (ListOfModsVariable): Present in all tasks (Calibration, GPTMD, Search, XL Search, and Glyco Search), this parameter dictates which PTMS are “variable” and that modified and unmodified forms of all peptides should be generated. Variable modifications should be used with caution because they massively increase the search space and typically lead to high false-positive rates. With the exception of variable oxidation of methionine, all other potentially present variable modifications should be searched for using the GPTMD approach (see Subheading 3.6).
Write .mzID (WriteMzId): This Search Task parameter requires additional search result files to be written in .mzID format. This is the output file type defined by the Human Proteome Organization (HUPO) and was designed to be a standardized format for reporting search results across different searching platforms.
Write .pep.XML (WritePepXml): This XL Search Task parameter requires additional result files to be written in .pep.XML format. This file format is widely accepted for the output of proteomics search engines. This result file format can be used as input for ProXL (see Note 20) for visualization of crosslinking results.
Write Contaminants (WriteContaminants): This Search Task parameter specifies the inclusion of contaminant peptide identifications in the result files. Contaminant identifications are clearly annotated as contaminants.
Write Decoys (WriteDecoys): This Search Task parameter specifying the inclusion of decoy peptide identifications in the result files. Decoy identifications are clearly annotated as decoys.
Write Individual File Results (WriteIndividualFiles): This output option within the Search Task specifies that result files for each individual spectra file be written in addition to the cumulative result files.
Write Two Pruned Databases [Mod and Mod+Protein Pruned] (WritePrunedDatabase): This post-search analysis parameter within the Search Task triggers the construction and export of two custom pruned .XML databases (modification pruned and modification + protein pruned). Modification pruning limits the PTMs annotated in the database to either those that were confidently identified at 1% FDR, annotated in the original database, or both depending on the user’s specifications. The process of protein pruning restricts the database to protein entries that have peptide-level support at 1% FDR. These pruned databases can be beneficial for subsequent top-down and intact-mass proteoform analysis [17].
4. Notes
Versions of MetaMorpheus prior to 0.0.309 have a different GUI layout, but the operations remain the same, and instructions in this protocol can still be applied to older versions.
Conversion of spectra files to different formats can be achieved using the software program MSConvert. MSConvert is part of ProteoWizard and can be downloaded at http://proteowizard.sourceforge.net/download.html.
The traditional MetaMorpheus score was designed for the comparison of high-resolution MS2 spectra to theoretical spectra. The integer component of the score is the number of fragment ions that match between the observed and theoretical spectra. The decimal component is the fraction of the total ion current (TIC) of the experimental spectra that is represented by the matching fragment ions. This scoring approach is much less effective for low-resolution MS2 spectra. To adjust for this, MetaMorpheus has implemented an adapted XCorr [18] scoring algorithm for use with low-resolution CID fragmentation.
.XML-formatted protein databases contain more information than .FASTA format. One of the greatest advantages of the use of UniProt .XML databases is the presence of annotated PTMs. UniProt .XML databases for reference proteomes can be retrieved at https://www.uniprot.org/proteomes/.
The larger the proteomic dataset being analyzed, the more RAM the analysis will require.
Windows (https://dotnet.microsoft.com/download/dotnet-core/thank-you/runtime-desktop-3.1.3-windows-x64-installer). MacOS (https://dotnet.microsoft.com/download/dotnet-core/thank-you/runtime-3.1.3-macos-x64-installer). Linux (https://docs.microsoft.com/dotnet/core/install/linux-package-managers).
When files are added using the drag and drop approach, MetaMorpheus automatically detects the file type (spectra file, database file, etc.), based on the file extension, and places it in the proper location.
The inclusion of contaminant databases enables the correct identification of contaminant peptides, preventing their misidentification as false-positive target PSMs. MetaMorpheus has a custom contaminant database designed to include many common contaminants from basic sample preparation methods.
More than one file can be added at a time either by dragging and dropping a group of files or by selecting multiple files in the file explorer window.
If file-specific settings are not specified, parameters from the task’s settings are used. When file-specific settings are specified, task parameters are ignored in favor of the file-specific parameters.
Select multiple spectra files to alter file-specific parameters for several spectra at once.
Once added, tasks appear in the Tasks window in the order they will be run.
To edit the parameters of any task after it has been added to the workflow, right click the task and select Edit task or double click the task and the parameters window will reopen for adjustment. Once necessary adjustments have been made, simply select the Save [task name] task button.
To alter the default parameters for a task, make the necessary parameter adjustments in the task window and select the Save As Default button at the bottom right of the window.
The default GPTMD modifications have been curated to contain modifications empirically determined to be common biological modifications across multiple species, common metal adducts, and common chemical artifacts that can occur during sample preparation.
Modifications can be located and selected using the search bar in addition to the drop-down menus.
To create a custom PTM, select the Settings tab in the menu on the left side of the GUI window. Select Create new modification from the Settings window to open up a new window for PTM creation. Information in fields marked with a red asterisk are required for PTM generation. Provide a name for the modification, the category to place the modification under in the list (this can be an existing category or you can create a new one such as ‘Custom’), what amino acid, or amino acid motifs the modification is present on, the chemical formula, and finally if the modification is located on a specific peptide/protein terminus. Additionally, neutral losses and diagnostic ions can be provided for the PTM based on the dissociation type. Once all of the required information has been provided, click the Save Mod button. The program must be restarted for the new modification to appear in the GUI.
When files from multiple proteolytic digests are loaded and protein inference is applied, the multi-protease protein inference algorithm [11] is automatically triggered providing improved protein inference results.
To add a custom crosslinker, exit the XL Search Task, and select the Settings tab in the menu on the left side of the GUI. Select Create new crosslinker from the Settings window to open a new window containing fields for the required information for crosslinker generation. Select Save Crosslinker once the required information has been provided. MetaMorpheus must be restarted before the custom crosslinker will show up as an option within the XL Search Task.
ProXL is a web based software tool for analyzing, visualizing, and sharing protein crosslinking mass spectrometry data and can be accessed at http://proxl-ms.org/.
Custom glycan databases can be constructed and added to MetaMorpheus for use in the Glyco Search Task. To add a database, navigate to the Settings tab in the menu on the left side of the GUI. Select Open mods/data folder to open up file explorer to MetaMorpheus’ location. Select the Glycan_Mods folder followed by either the N- or O-glycan folder depending on what kind of database you are adding. Move the custom glycan database to this location and restart MetaMorpheus for its incorporation into the GUI. The format of the custom glycan databases should follow formatting of the existing gly- can databases. Briefly, the database entries should contain a composition based glycan description, a symbol based glycan description and the molecular weight of the glycan (e.g., HexNAc(1)Hex(1)Fuc(1) N1H1F1S0 511.1901). MetaMorpheus’ Glyco Search currently supports the interpretation of the following monosaccharides and modifications from glycan databases: Hex (H), HexNAc (N), NeuAc (A), NeuGc (G), Fuc (F), Phospho (P), Sulfo (S), Na (Y), Ac (C), Xylose (X), SuccinylHex (U), and Formylation (M).
A typical MetaMorpheus run will consist of a Calibration, GPTMD and Search Task.
When specifying a file path following an argument, it is preferable to use the absolute file path inside of quotes. This is required if there are spaces in the path name.
The experimental design for label-free quantification can be added to MetaMorpheus by selecting the SET EXPERIMEN TAL DESIGN button to the left of the SET FILE-SPECIFC SETTINGS button in the Spectra window. Specify the file condition, biological replicate number, fraction number and technical replicate number for each spectra file. This information enables FlashLFQ [10] to perform normalization and protein quantification on the provided data (see Chapter 13).
Custom mass tolerances can be created using the following syntax: name dot # PPM, #, # where the first # is the ppm error and the subsequent #s are the missed monoisotopics (e.g., test dot 4 PPM, 1, 2). You can also perform an interval search using the syntax name interval [#,#], where each # is either a min or max in Da with both numbers relative to the precursor mass.
It is recommended that the product mass tolerance be set in ppm when an Orbitrap mass spectrometer is used for data acquisition.
To add a custom protease to MetaMorpheus, select the Settings tab in the menu on the left side of the GUI. Select Open mods/data folder to open a file explorer window at MetaMorpheus’ location. Select the Proteolytic Digestion folder and open the proteases.tsv file. This file contains all the proteases, and their cleavage-specific information for MetaMorpheus. A new protease can be added by creating a new row in the file. For each new protease, provide the name and define its digestion motif using the following syntax: (a) specify the amino acids inducing cleavage by listing the single amino acid codes with the “—” character on the left for N-terminal cleavage and on the right for C-terminal cleavage, (b) provide any amino acids residue(s) that prevent proteolytic cleavage using “[ ]”, (c) Use “X” to denote any amino acid within a cleavage motif that is a wildcard (could be any amino acid), (d) use “” to define any exceptions to the wildcard character, and (e) Use “,” to separate multiple cleavage motifs for a single protease (e.g., Chymotrypsin (do not cleave before proline) F[P]—,W[P]—,Y [P]—). Once all necessary protease information has been added, save the edited protease.tsv file and restart MetaMorpheus for the custom protease to appear in the GUI.
In the classic search algorithm, a MS2 spectrum is compared to every theoretical spectrum that has the same precursor mass within the task defined precursor mass tolerance. The highest scoring match is reported.
The modern search algorithm creates an index of all theoretical peptide spectra which serves as a look-up table. During the comparison of theoretical and experimental spectra, experimental fragment ions can be rapidly compared to theoretical target and decoy peptide MS2 fragments in the look-up table. The theoretical target or decoy peptide with the most matching fragment ions is recorded in the results file. The modern search algorithm is much faster than the classic search when conducting an open-mass search.
The semi-specific search algorithm employs a novel digestion and search strategy to identify peptides where one of the termini may not follow the cleavage motif of the selected protease [19]. A separate q-value calculation prevents the semi-specific search from introducing significant false-positive identifications.
The non-specific search algorithm utilizes the same digestion and search strategy as the semi-specific approach [19], but differs because up to 2 of the peptide’s termini may not conform to the protease’s cleavage motif. If no protease was used for digestion of the sample (e.g., peptidomics), select the non-specific option for the protease parameter. A separate q-value calculation prevents the non-specific search from introducing significant false-positive identifications when a specific protease is selected.
Click the Add Isotope Label button that appears in the Search Task window when the SILAC/SILAM: Quantify peptides/protein with stable isotope labels parameter is selected. Specify the labeled amino acid in the top left corner. The remaining fields will be automatically filled with the information for the unlabeled amino acid. Alter the count of heavy isotope to match the composition of the label and the chemical formula and mass difference will be automatically updated. The final parameter specifies the type of labeling experiment performed (i.e., Multiplex or Turnover/Pulse). After completing the label design, select Save Label(s) to go back to the Search Task, or click Add Additional Labels to This Condition ifmore than one heavy labeled amino acid was used. For the command-line version, include a [[SearchParameters.SilacLabels]] section containing the following parameters, OriginalAminoAcid, AminoAcidLabel, LabelChemicalFormula, and MassDifference for each of the labeled amino acids.
Acknowledgements
The development and maintenance of MetaMorpheus is supported by NIH-NIGMS grant R35GM126914. Rachel M. Miller was supported in part by the NIH Chemistry-Biology Interface Training Grant (T32GM008505). Robert J. Millikin was supported by the NIH Genomic Sciences Training Program (5T32HG002760). Zach Rolfs was supported by NIH-NCI grant U24CA199347.
References
- 1.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR (1999) Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol 17(7):676–682. 10.1038/10890 [DOI] [PubMed] [Google Scholar]
- 2.Nesvizhskii AI (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J Proteomics 73(11):2092–2123. 10.1016/j.jprot.2010.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner OS, Kelleher NL (2015) Illuminating the dark matter of shotgun proteomics. Nat Biotechnol 33(7):717–718. 10.1038/nbt.3287 [DOI] [PubMed] [Google Scholar]
- 4.Chick JM, Kolippakkam D, Nusinow DP, Zhai B, Rad R, Huttlin EL, Gygi SP (2015) A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nat Biotechnol 33(7):743–749. 10.1038/nbt.3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griss J, Perez-Riverol Y, Lewis S, Tabb DL, Dianes JA, Del-Toro N, Rurik M, Walzer M, Kohlbacher O, Hermjakob H, et al. (2016) Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nate Methods 13(8):651–656. 10.1038/nmeth.3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesvizhskii AI, Roos FF, Grossmann J, Vogelzang M, Eddes JS, Gruissem W, Baginsky S, Aebersold R (2006) Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteom 5(4):652–670. 10.1074/mcp.M500319-MCP200 [DOI] [PubMed] [Google Scholar]
- 7.Solntsev SK, Shortreed MR, Frey BL, Smith LM (2018) Enhanced global post-translational modification discovery with metamorpheus. J Proteome Res 17(5):1844–1851. 10.1021/acs.jproteome.7b00873 [DOI] [PubMed] [Google Scholar]
- 8.Lu L, Millikin RJ, Solntsev SK, Rolfs Z, Scalf M, Shortreed MR, Smith LM (2018) Identification of MS-cleavable and noncleavable chemically cross-linked peptides with metamorpheus. J Proteome Res 17(7):2370–2376. 10.1021/acs.jproteome.8b00141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Riley NM, Shortreed MR, Bertozzi CR, Smith LM (2020) O-pair search with metamorpheus for o-glycopeptide characterization. bioRxiv 10.1101/2020.05.18.102327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millikin RJ, Solntsev SK, Shortreed MR, Smith LM (2018) Ultrafast peptide label-free quantification with FlashLFQ. J Proteome Res 17(1):386–391. 10.1021/acs.jproteome.7b00608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller RM, Millikin RJ, Hoffmann CV, Solntsev SK, Sheynkman GM, Shortreed MR, Smith LM (2019) Improved protein inference from multiple protease bottom-up mass spectrometry data. J Proteome Res 18(9):3429–3438. 10.1021/acs.jproteome.9b00330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong AT, Leprevost FV, Avtonomov DM, Mellacheruvu D, Nesvizhskii AI (2017) MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat Methods 14(5):513–520. 10.1038/nmeth.4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesnik AJ, Miller RM, Ibrahim K, Lu L, Millikin RJ, Shortreed MR, Frey BL, Smith LM (2020) Spritz: A proteogenomic database engine. bioRxiv 10.1101/2020.06.08.140681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giansanti P, Tsiatsiani L, Low TY, Heck AJ (2016) Six alternative proteases for mass spectrometry–based proteomics beyond trypsin. Nat Protocols 11(5):993–1006. 10.1038/nprot.2016.057 [DOI] [PubMed] [Google Scholar]
- 15.Varki A (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3(2):97–130. 10.1093/glycob/3.2.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta N, Pevzner PA (2009) False discovery rates of protein identifications: a strike against the two-peptide rule. J Proteome Res 8(9):4173–4181. 10.1021/pr9004794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai Y, Buxton KE, Schaffer LV, Miller RM, Millikin RJ, Scalf M, Frey BL, Shortreed MR, Smith LM (2019) Constructing human proteoform families using intact-mass and top-down proteomics with a multi-protease global post-translational modification discovery database. J Proteome Res 18(10):3671–3680. 10.1021/acs.jproteome.9b00339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howbert JJ, Noble WS (2014) Computing exact p-values for a cross-correlation shotgun proteomics score function. Mol Cell Proteom 13(9):2467–2479. 10.1074/mcp.O113.036327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolfs Z, Millikin RJ, Smith LM (2020) An algorithm to improve the speed of semi- and non-specific enzyme searches in proteomics. Curr Bioinf 15:1–9. 10.2174/1574893615999200429123334 [DOI] [PMC free article] [PubMed] [Google Scholar]