Abstract
The rapid and accurate quantification of peptides is a critical element of modern proteomics that has become increasingly challenging as proteomic data sets grow in size and complexity. We present here FlashLFQ, a computer program for high-speed label-free quantification of peptides and proteins following a search of bottom-up mass spectrometry data. FlashLFQ is approximately an order of magnitude faster than established label-free quantification methods and can quantify data-dependent analysis (DDA) search results from any proteomics search program. It is available as a graphical user interface program, a command line tool, a Docker image, and integrated into the MetaMorpheus search software.
Keywords: Label-free quantification, post-translational modifications, quantitative proteomics, software
1. Introduction
Modern bottom-up proteomics workflows involve digesting a protein sample with a protease, followed by online separation of the resultant peptides and subsequent analysis by tandem mass spectrometry (MS/MS). In data-dependent acquisition (DDA), peptides are first observed in a survey (MS1) scan, then identified by their fragmentation (MS2) spectra. The MS1 scan contains isotopic envelopes of intact peptide ions, the signal intensity of which is a proxy for abundance of that ion in the mass spectrometer. This intensity can be used for quantification of the peptide species. There are many complex factors that determine how well a peptide ionizes. Accordingly, a relative quantification strategy is typically employed in which the signal intensity of a peptide is compared to the same peptide’s signal in another sample, rather than attempting absolute quantification. The trace of MS1 signal across time for a particular ion is called the extracted ion chromatogram (XIC). The absence of an added chemical label to assist in quantification gives this strategy its name: label-free quantification (LFQ).
Given the large amount of raw data afforded by these experiments (typically on the order of thousands of peptides in a single run), bioinformatics software is used for automatic data interpretation to ease the burden of manual annotation and quantification. FlashLFQ [1] is a software program that was created to quickly quantify peptides and proteins in LC-MS/MS data through XIC tracing. It was specifically designed to be a free, open-source LFQ tool that is agnostic of upstream and downstream software. One important motivation for creating FlashLFQ was the desire to quantify post-translational modification (PTM)-containing peptides discovered by MetaMorpheus and its global PTM discovery [2] (G-PTM-D) engine.
At its core, FlashLFQ is an algorithm for rapid XIC extraction/peakfinding. Several features have been added since its initial debut as a peptide quantification program, including match-between-runs, intensity normalization, and Bayesian protein quantification and hypothesis testing [3].
The Bayesian hypothesis testing option is intended to replace the typical workflow of importing peptide or protein intensities into a separate program to perform the Student’s t-test and multiple testing correction. While the t-test is a perfectly valid option for the interpretation of proteomics data, FlashLFQ’s Bayesian solution aggregates intensity-dependent uncertainty for each peptide into a protein-level uncertainty, increasing the selectivity of the results (see Note 1).
FlashLFQ ( https://github.com/smith-chem-wisc/FlashLFQ ) is available as a command-line interface (CLI) and graphical user interface (GUI) program. It is also integrated into the MetaMorpheus search software program. The CLI version can be run in Microsoft Windows, Apple macOS, and Linux; a CLI FlashLFQ/Linux Docker image is also available on Docker Hub ( https://hub.docker.com/r/smithchemwisc/flashlfq ). The GUI version of FlashLFQ is currently only available on Microsoft Windows.
FlashLFQ requires spectral data in .mzML or .raw file format, along with a list of peptide identifications from a proteomics search program (e.g., MetaMorpheus [2], Morpheus [4], Andromeda [5], SearchGUI [6], etc.).
2. Material
2.1. Data Inputs
FlashLFQ requires a list of peptide identifications (peptide spectral matches, PSMs) as well as LC-MS/MS data. Each peptide identification is required to have a(n):
spectra file name,
amino acid sequence,
string representation of the peptide containing any potential modifications (such as phosphorylation),
monoisotopic mass,
retention time,
charge,
protein identifier (e.g., protein accession).
The required data values are provided to FlashLFQ as a tab-delimited text file(s).
2.2. Accepted Data Formats
Several data formats from common search engines are recognized in their native format without modification:
MetaMorpheus .psmtsv,
Morpheus .tsv,
MaxQuant msms.txt,
PeptideShaker .tabular.
Output from other search engines can be modified to be compatible with FlashLFQ. The columns:
“File Name”,
“Base Sequence”,
“Full Sequence”,
“Peptide Monoisotopic Mass”,
“Scan Retention Time”,
“Precursor Charge”,
“Protein Accession”
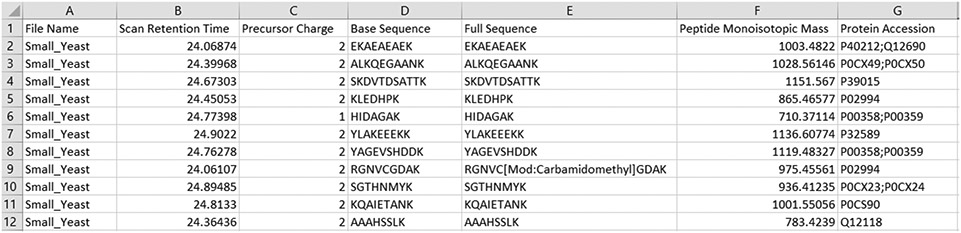
must be defined if this generic format is used, as described in the Data Inputs section above (see 2.1; see also https://github.com/smith-chem-wisc/FlashLFQ/wiki/Identification-Input-Formats ). See Figure 1 for an example of the generic PSM input format.
Fig. 1.
Example of a “generic” identification input file. The columns are tab-delimited. FlashLFQ automatically reads MetaMorpheus, Morpheus, MaxQuant, and PeptideShaker output, but output from other search programs can be adapted to this generic format for use by FlashLFQ.
The .mzML and Thermo .raw spectra file formats are valid inputs for the LC-MS/MS data. Other file formats can be converted to .mzML using ProteoWizard’s free MSConvert software [7] (see https://github.com/smith-chem-wisc/FlashLFQ/wiki/Converting-spectral-data-files-with-MSConvert ).
2.3. Hardware Requirements
There are no formal requirements for CPU speed or number of cores, but faster processors with additional cores will result in a faster processing time. FlashLFQ is a multithreaded program and uses all but one core by default. We advise that the user have at least 4 GB of RAM, plus 500 MB for each spectra file if match-between-runs is enabled.
2.4. Software Requirements
The operating system (OS) must be Microsoft Windows, Apple macOS, or Linux for the command-line version. For the GUI version of FlashLFQ, the OS must be Microsoft Windows.
.NET Core 3.1 must be installed (see Note 2).
To use the Docker image (see Note 3) of FlashLFQ, the software requirements above can be ignored; only a Docker installation is required, as the Docker image includes Alpine Linux and .NET Core 3.1.
2.5. Installation
GUI (Microsoft Windows only). The GUI (graphical user interface) version of FlashLFQ can be downloaded from GitHub by navigating to the “releases” page ( https://github.com/smith-chem-wisc/FlashLFQ/releases/latest ) and clicking “FlashLFQ.zip”. After extracting the archive to a folder, open FlashLFQ.exe.
- Command-line.
- Microsoft Windows: Download FlashLFQ.zip and extract it to a folder following the same steps as the GUI version. Running CMD.exe in the terminal will start FlashLFQ and display valid parameters.
- Linux and Apple macOS: Download FlashLFQ_DotNetCore.zip and extract it to a folder following the same steps as the GUI version. Running the command dotnet CMD.dll in the terminal will start FlashLFQ and display valid parameters.
-
Docker. A Docker image of FlashLFQ is hosted on DockerHub ( https://github.com/smith-chem-wisc/FlashLFQ/wiki/Docker-Image ). It can be pulled with the docker pull command; for example,
docker pull smithchemwisc/flashlfq:1.1.1
will download the Docker image containing FlashLFQ version 1.1.1. The most recent version of FlashLFQ is always tagged with latest (i.e., smithchemwisc/flashlfq:latest).
3. Methods
3.1. Adding identification files
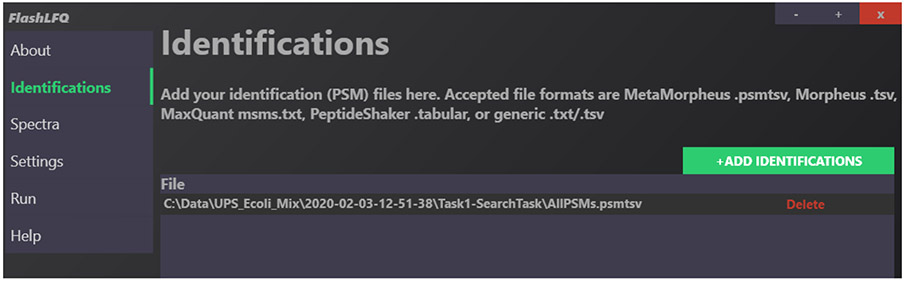
Identification (PSM) files can be added to FlashLFQ’s GUI interface by drag-and-drop or by clicking the “Add Identifications” button in the “Identifications” tab. In the command-line version, the “--idt” flag is used to specify an identification file. An example of a MetaMorpheus PSM file added in the GUI is shown in Figure 2.
Fig. 2.
Example of the “Identifications” page in the FlashLFQ GUI. A MetaMorpheus .psmtsv output file has been added.
3.2. Adding spectra files
Spectra files can be added by drag-and-drop or by clicking the “Add Spectra” button in the GUI in the “Spectra” tab. In the command-line version, the “--rep” flag is used to specify a folder containing the spectra files.
Each spectra file is associated with metadata containing:
the run’s experimental condition (e.g., normal or treatment),
sample number,
fraction number,
replicate number.
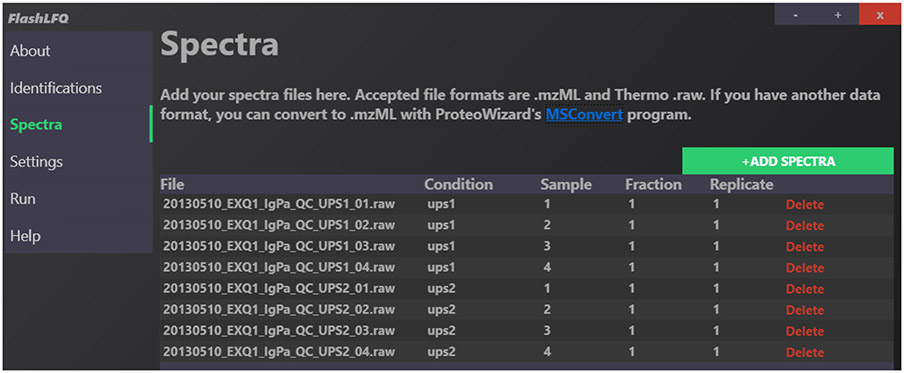
This collection of metadata is called the experimental design, which is saved to a file called ExperimentalDesign.tsv. This information is used to normalize intensities, perform statistical analysis, and improve match-between-runs. If you simply want to quantify peptides without performing these extra functions, the experimental design can be ignored. The Condition can be any text whereas the Sample, the Fraction and the Replicate must be an integer value. The Sample, Fraction and Replicate values must begin with 1 and there can be no missing values. An example from the GUI is shown in Figure 3, where the experimental design has been defined for an 8-file run.
Fig. 3.
Example of the “Spectra” page in the FlashLFQ GUI. Several Thermo .raw data files containing the Universal Proteomics Standard (UPS) proteins have been added, with UPS1 and UPS2 files divided between two experimental conditions, 4 samples per condition. The samples are unfractionated.
Command-line users must build the ExperimentalDesign.tsv file manually. You can find a detailed description of the contents of the file at https://github.com/smith-chem-wisc/FlashLFQ/wiki/Experimental-Design, along with an example file available for download.
3.3. Settings
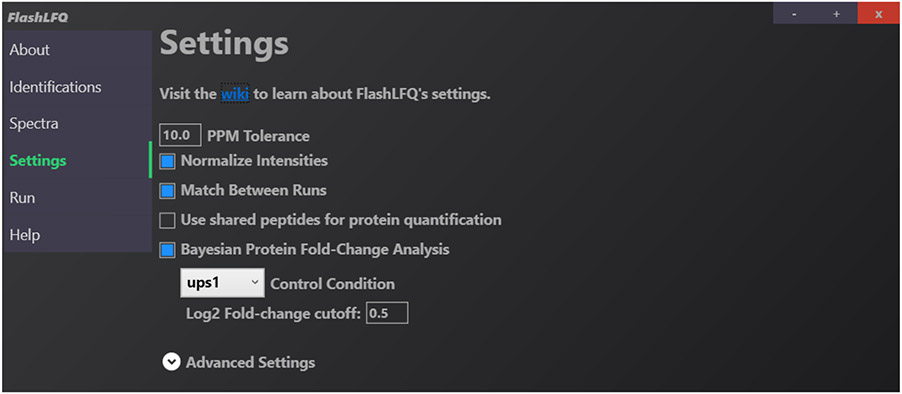
Settings are specified in the “Settings” tab in the GUI. Most settings can be enabled or disabled via a checkbox. In the command-line version, each setting has its own flag. An example in the GUI is shown in Figure 4.
Fig. 4.
Example of the “Settings” page in the FlashLFQ GUI. Normalization, match-between-runs, and the Bayesian protein quantification have been enabled.
PPM tolerance (see Note 4). Specifies the mass-error tolerance for the XIC peakfinding algorithm, in parts per million (ppm). The default is 10ppm.
Intensity normalization (see Note 5). Specifies whether or not intensity normalization should be performed. FlashLFQ’s intensity normalization algorithm uses a median normalization, which means that the median peptide intensity difference between samples is assumed to be zero (i.e., not changing). This normalization process rests on the assumption that most proteins are not changing in abundance between samples.
Match-between-runs (see Note 6). Specifies whether or not match-between-runs (MBR) should be performed. MBR attempts to identify peptides that were not fragmented and identified in some runs, using retention-time alignment between runs.
Using shared peptides for protein quantification (see Note 7). Specifies whether or not shared peptides should be used for protein quantification. If this is disabled, only peptides unique to a protein will be used for quantification (see Note 8).
Bayesian protein quantification (see Note 9). Specifies whether or not FlashLFQ’s Bayesian protein quantification system should be used. A control condition (see Note 10) must be specified, along with a fold-change cutoff (see Note 11).
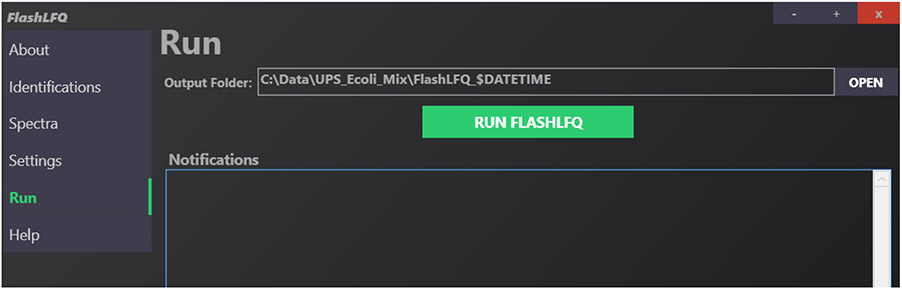
3.4. Run
On the “Run” tab, you can specify an output directory. A default directory will already be populated for you; this default directory is where your spectra files are located. The field “$DATETIME” is included in this name, which will be automatically replaced with the date and time of the FlashLFQ run. After you are satisfied with your settings, click “Run FlashLFQ”. The command-line flag to (optionally) specify an output directory is “--out”. An example in the GUI is shown in Figure 5, with the output folder being left as default.
Fig. 5.
Example of the “Run” page in the FlashLFQ GUI. The default output directory is shown.
3.5. Output
FlashLFQ writes several files as output: QuantifiedPeaks.tsv, QuantifiedPeptides.tsv, QuantifiedProteins.tsv, FlashLfqSettings.toml, and (optionally) BayesianFoldChange-Analysis.tsv.
QuantifiedPeaks.tsv reports each chromatographic peak along with information about that peak, such as its begin and end time, apex intensity, etc. Some peaks list 0 for their intensity, which indicates an unquantifiable peak (i.e., an identification was passed in to FlashLFQ, but that identification could not be quantified). Some peaks list “∣” as part of their base sequence or full sequence fields, which indicates multiple identifications were associated with the same chromatographic peak (i.e., the identity of the chromatographic peak is ambiguous).
QuantifiedPeptides.tsv reports all peptide sequences along with their intensity in all spectra files and their method of quantification (by MS/MS, by match-between-runs, etc.) in all files. For fractionated data, each fraction’s intensity is reported separately; these fraction intensities can be summed to get a sample intensity.
QuantifiedProteins.tsv reports protein intensities per spectra file, similar to QuantifiedPeptides.tsv. This file sums the 3 most intense peptides for that protein in each sample. It is a rough estimate of a protein’s intensity in a given file (see Note 12).
FlashLfqSettings.toml reports the settings used for the FlashLFQ run, so that results can be reproduced easily.
BayesianFoldChangeAnalysis.tsv reports output from the Bayesian protein quantification and hypothesis testing. It lists each protein, along with a fold-change (relative to the control condition specified in the settings) and the probability that the protein’s fold-change is less than the fold-change cutoff (the posterior error probability, or PEP).
3.6. Using the Docker image
Once a Docker image has been pulled (downloaded) from Docker Hub, you can run FlashLFQ within the Docker container with the docker run command. For example:
docker run [Docker args] smithchemwisc/flashlfq:latest [FlashLFQ args]
Typical usage will look something like this:
docker run --rm -v C:/MyDataFolder:/mnt/ smithchemwisc/flashlfq:latest --idt ./mnt/AllPSMs.psmtsv --rep ./mnt/ --out ./mnt/FlashLFQ _Output
The --rm flag tells Docker to clean up/remove the image after FlashLFQ is done executing. The -v C:/MyDataFolder:/mnt/ part "mounts" the C:/MyDataFolder directory to the Docker container, i.e., lets the container access your hard drive and gives that directory an alias of /mnt/. The rest of the line runs FlashLFQ with the passed arguments.
To ease the use of the Docker image, we recommend the following (see Note 13):
Sometimes Docker has trouble getting permission from your computer to access the hard disk; for Windows users, the C: drive is usually easiest to access and you might get a popup asking for permission.
Try not to use spaces in your path names if you can help it (in the Dockerfile, terminal commands, input/output files, etc.). It usually makes things harder than they need to be.
Notes
1. There were a few motivations for creating this system. Most protein quantification algorithms report only one protein-level intensity per sample, which is then used in downstream statistics. However, this throws out quite a lot of information; each peptide is an independent measurement of a protein’s abundance in a sample, and we wanted to use this information. Additionally, uncertainty in peptide measurements increases with lower intensity, and these peptide-level uncertainty measurements can be aggregated into a protein-level uncertainty; again, this information would be lost in most statistical pipelines. FlashLFQ takes a peptide-centric approach to bottom-up protein quantification and measures peptide fold-changes across experimental conditions, as well as peptide and protein uncertainty within a condition to estimate quantitative false-discovery rates. See Ref [3] for details.)
2. To use the Windows GUI, the .NET Core Desktop Runtime must be installed. This framework allows both GUI and CLI .NET Core programs to be run. This is in contrast to the .NET Core Runtime, which only runs CLI .NET Core programs.
3. Docker images are "containers" which encapsulate an operating system and pre-installed software. These containers can be run independent of one’s own operating system or other software requirements. The purpose of using a Docker container over a more "traditional" installation is usually to avoid installing software prerequisites, either out of convenience or inability, such as on a server in which the user does not have permission to install software.
4. CMD flag: “--ppm”.
5. CMD flag: “--nor”.
6. CMD flag: “--mbr”.
7. CMD flag: “--sha”.
8. Currently, if shared peptides are used for protein quantification, shared peptides are treated the same as unique peptides for protein quantification.
9. CMD flag: “--bay”.
10. Currently, one condition must be specified as the control to compare the other conditions to. For example, if you were to have 3 conditions of samples, “Normal”, “Tumor1”, and “Tumor2”, with “Normal” specified as the control, then the “Tumor1” and “Tumor2” conditions would be quantified relative to the “Normal” condition.
11. A change in a given protein’s relative abundance below which you are not interested.
12. Generally, the top-3 method is not as reliable as the Bayesian protein quantification, or importing FlashLFQ’s peptide intensities into other software for protein quantification. Future improvements are coming to the non-Bayesian method of protein quantification.
13. More documentation is given on the FlashLFQ wiki: https://github.com/smith-chem-wisc/FlashLFQ/wiki/Docker-Image.
Acknowledgements
This work was supported by grant R35GM126914 from the National Institute of General Medical Sciences. R.J.M. was supported by an NHGRI training grant to the Genomic Sciences Training Program 5T32HG002760. We thank the software development team of the Smith lab (Stefan K. Solntsev, Anthony J. Cesnik, Khairina Ibrahim, Lei Lu, Rachel M. Miller, Zach Rolfs, and Leah V. Schaffer), who contributed daily input and guidance for FlashLFQ’s improvement.
References
- [1].Millikin RJ, Solntsev SK, Shortreed MR, Smith LM (2018) Ultrafast peptide label-free quantification with flashlfq. Journal of proteome research 17(1):386–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li Q, Shortreed MR, Wenger CD, Frey BL, Schaffer LV, Scalf M, Smith LM (2017) Global post-translational modification discovery. Journal of proteome research 16(4):1383–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Millikin RJ, Shortreed MR, Scalf M, Smith LM (2020) A bayesian null interval hypothesis test controls false discovery rates and improves sensitivity in label-free quantitative proteomics. Journal of Proteome Research 19(5):1975–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wenger CD, Coon JJ (2013) A proteomics search algorithm specifically designed for high-resolution tandem mass spectra. Journal of proteome research 12(3):1377–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the maxquant environment. Journal of proteome research 10(4):1794–1805 [DOI] [PubMed] [Google Scholar]
- [6].Barsnes H, Vaudel M (2018) Searchgui: A highly adaptable common interface for proteomics search and de novo engines. Journal of proteome research 17(7):2552–2555 [DOI] [PubMed] [Google Scholar]
- [7].Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) Proteowizard: open source software for rapid proteomics tools development. Bioinformatics 24(21):2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]