Abstract
Male sex is a major risk factor for SARS-CoV-2 infection severity. To understand the basis for this sex difference, we studied SARS-CoV-2 infection in a young adult cohort of United States Marine recruits. Among 2,641 male and 244 female unvaccinated and seronegative recruits studied longitudinally, SARS-CoV-2 infections occurred in 1,033 males and 137 females. We identified sex differences in symptoms, viral load, blood transcriptome, RNA splicing, and proteomic signatures. Females had higher pre-infection expression of antiviral interferon-stimulated gene (ISG) programs. Causal mediation analysis implicated ISG differences in number of symptoms, levels of ISGs, and differential splicing of CD45 lymphocyte phosphatase during infection. Our results indicate that the antiviral innate immunity set point causally contributes to sex differences in response to SARS-CoV-2 infection. A record of this paper’s transparent peer review process is included in the supplemental information.
Keywords: SARS-CoV-2, sex differences, interferon-stimulated genes, viral infection, COVID-19, differential expression, alternative splicing, causal mediation
Sex differences across many levels of SARS-CoV-2 infection response have been observed globally. In a longitudinal study of young adults, we identify significant clinical and immune differences between males and females. We further show that differences in antiviral response immune pathways mediate sex-specific responses to SARS-CoV-2 infection in young adults.
Introduction
COVID-19, which has led to 4.25 million deaths as of August 2021, has a worse outcome in males (Takahashi et al., 2020; Ren et al., 2021; Scully et al., 2020). Innate and adaptive immune responses are different in males and females during infection by SARS-CoV-2, the virus that causes COVID-19 (Takahashi et al., 2020). These differences in response and outcome could be influenced by pre-existing factors that either worsen the disease in males, such as androgen effects or the prevalence of comorbidities, or that improve the outcome in females, such as immune system effectiveness (Klein and Flanagan, 2016; Bienvenu et al., 2020; Schultze and Aschenbrenner, 2021; Brodin, 2021; Meng et al., 2020). Because the basis for sex differences in clinical outcomes of SARS-CoV-2 has therapeutic implications, it is important to study whether aspects of the intrinsic immune state in females compared with males alter the response to SARS-CoV-2. However, without well-controlled studies, including pre-infection data and minimizing the confounding effect of comorbidities, it is impossible to tease apart which of these hypotheses contribute to the clinical differences observed during infection.
The prospective COVID-19 Health Action Response for Marines (CHARM) study enrolled 3,326 new Marine recruits beginning military training. A total of 2,641 males and 244 females were initially SARS-CoV-2 seronegative and were followed longitudinally with symptom screening, serial nares swab PCR for SARS-CoV-2, and blood sampling for molecular analyses (see STAR Methods). The cohort was physically fit, predominantly Caucasian, and had an average age of 19.1 ± 1.9 years (Table S1). During the 12 weeks after study entry, which included 2 weeks of supervised quarantine and 10 weeks of basic training, a total of 1,033 males and 137 females tested positive for SARS-CoV-2 by PCR (Figure S1). The study was conducted prior to FDA-approved vaccines and treatments specifically directed at SARS-CoV-2, and none of these participants were involved in any clinical trials at the time. All symptomatic participants were treated as outpatients, and none received any type of medication beyond symptomatic treatment such as non-steroidal anti-inflammatory drugs or acetaminophen. This longitudinal study of a unique cohort permitted a causal analysis of pre-existing immune system differences and their significance in molecular and clinical sex differences observed during SARS-CoV-2 infection.
Results
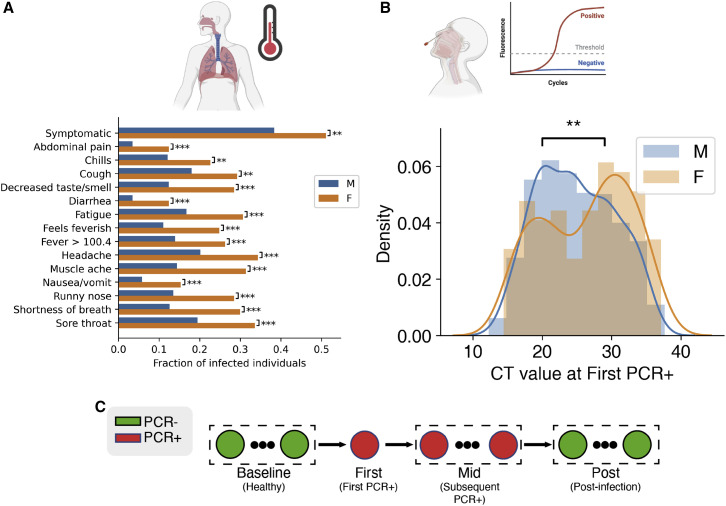
All SARS-CoV-2 infected participants were asymptomatic or had mild infection, with none requiring hospitalization or resulting in death. Infected females were more often symptomatic than males (OR: 1.68, 95% CI: 1.17–2.41, p = 0.004, Wald test). In addition, infected females were more likely to have a fever, defined as a recorded temperature greater than 100.4. Females reported a higher rate of each of 13 symptoms surveyed, including shortness of breath and decreased smell/taste, and had a higher total number of unique symptoms (3.3 ± 4.3 in females, 1.6 ± 2.8 in males, p = 1.72e−5, Mann-Whitney U test; Figure 1 A). The differences in fever rate suggest that the relationship of sex to SARS-CoV-2 clinical outcomes are not due to reporting bias. We previously performed a phylogenetic analysis of SARS-CoV-2 sequences recovered from more than 600 infected participants and found that most recruits were infected by one of 5 monophyletic clusters (Letizia et al., 2020, Lizewski et al., 2022). Higher symptomatic rates in females were consistent across these virus clusters, suggesting that the symptom-sex association did not result from viral genetic variability (Figure S2).
Figure 1.
Analysis of sex divergence in clinical measurements
(A) The fraction of each reported symptom for infected females (n = 137) and males (n = 1,033). Statistical significance was tested using the chi2 independence test. Statistical significance: ∗∗∗p < 0.001, ∗∗p <0.01, ∗p < 0.05.
(B) Initial viral load, measured by probe CT and averaged across the S, N, and ORF1ab genes, between females and males. Statistical significance was tested by the Kolmogorov-Smirnov test.
(C) Clinical data and biospecimens were collected longitudinally for each subject. Samples were labeled “baseline” if they tested negative for SARS-CoV-2, the first PCR-positive sample from each infected individual was labeled “first,” and any subsequent PCR-positive sample was labeled “mid.” Subjects were followed throughout infection, and any PCR-negative samples taken after infection were labeled “post.”
Viral load when subjects were first identified as SARS-CoV-2 PCR positive by quantitative RT-PCR as previously described (Letizia et al., 2021) was estimated by probe cycle threshold (CT) levels. Females showed an average 2.6-fold lower viral load than males, having an average CT value of 26.3 ± 6.343 compared with 24.9 ± 5.633 in males (p = 0.007, Mann-Whitney U test; p = 0.015 ANOVA after controlling for appropriate covariates; see method details and Figure 1B). Beyond the initial PCR-positive sample, viral load measurements were too sparse and too heterogeneous in time for reasonable comparison.
To explore the relationship of host states and sex differences in the effects of SARS-CoV-2, we analyzed 839 whole-blood samples from 310 participants (255 male and 55 female) by RNA-sequencing for gene expression and alternative splicing, and 397 serum samples from 106 participants (93 male, 13 female) by a proteomics inflammation panel of 92 analytes (Olink assay). Assays were performed at time points both before (baseline) and during (first/mid) SARS-CoV-2 infection, providing the opportunity to investigate sex differences in immune states both preceding and during acute infection (STAR Methods; see Figure 1C). We determined the significant multi-omic changes observed during infection, grouping them according to whether they were observed in both sexes (sex-independent), significantly greater in one sex (M/F-biased), or observed only in one sex (M/F-specific).
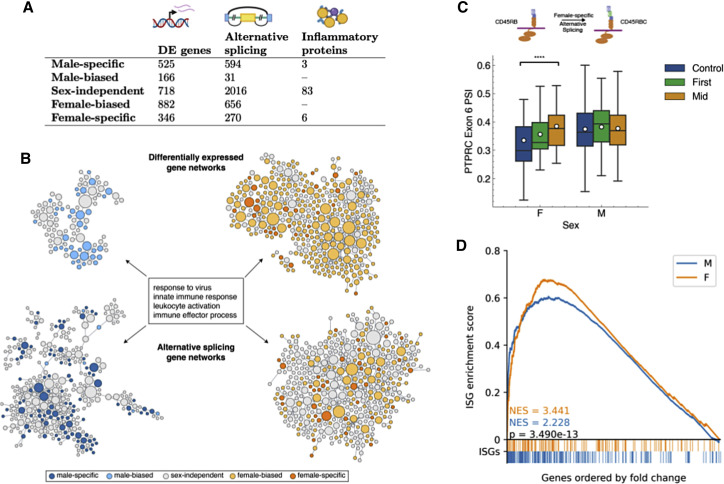
We identified sex-specific molecular signatures for gene expression, alternative splicing, and immunoproteomics (Figure 2 A). In particular, differential expression of 525 genes and differential splicing of 594 sites was found during infection only in males, whereas 346 genes and 270 sites were modulated only in females. Among 92 immune mediators assayed by proteomic analysis, 3 were regulated only in males and 6 only in females (see Figure S3 and Table S2). Both the gene expression and the alternative splicing sex-specific signatures were strongly enriched in an immune response pathway module when analyzed in the context of the HumanBase functional network (Wong et al., 2018; Greene et al., 2015) (Figure 2B; see method details). Many of these immune response genes are sex-biased or sex-specific, and there are more genes in the enriched immune response set in females than males across both the differential expression (females n = 360; males n = 98) and alternative splicing (females n = 391; males n = 325) analyses, with an especially marked difference in the differential expression networks. This suggests a broadly stronger transcriptional and post-transcriptional immune response to acute SARS-CoV-2 infection in females.
Figure 2.
Multi-omic integrative analysis of the sex-dependent molecular responses to SARS-CoV-2 infection
(A) The number of sex-dependent molecular variations upon SARS-CoV-2 infections discovered using gene expression, alternative splicing, and proteomics.
(B) Sex divergence of gene expression and alternative splicing visualized in the virus response module of a tissue-specific functional network. Each node represents a gene, with its size proportional to its connectedness in the network. Clusters shown here represent the immune response genes from males (left) and females (right) with differential expression (top) and differential alternative splicing events (bottom) during SARS-CoV-2 infection. This network is constructed by probabilistically integrating a compendium of thousands of public omics datasets to provide functional maps of biological processes and pathways. Projecting each sex-specific signature onto this network reveals significant enrichment of the immune response module in both gene expression and alternative splicing.
(C) Box-and-whisker plot of exon inclusion changes of PTPRC exon 6 upon SARS-CoV-2 infection in females (n = 55) and males (n = 255) throughout the infection course. The white circles represent the mean, the center line shows the median, and the upper and lower edges of the boxes represent the upper and lower quartiles, respectively. The differential splicing of PTPRC will alter the protein product of PTPRC, known as CD45, in females.
(D) ISGs are significantly enriched in both males and females during COVID-19 infection but significantly more so in females as shown by the higher normalized enrichment score (NES) from gene set enrichment analysis. Colored bars below the x axis represent ISG locations within the list of all differentially expressed genes, ordered by fold change from baseline samples. p value calculated using the Mann-Whitney U test.
Sex-based divergence in the immune responses to SARS-CoV-2 infection was seen in the alternative splicing of the protein tyrosine phosphatase receptor Type C (PTPRC) gene, a key immune cell signaling gatekeeper (Courtney et al., 2019). The isoform variations of CD45 (the protein product of PTPRC) are specific markers for immune cell groups (Charbonneau et al., 1988). Compared with pre-infection, females had a significantly higher induced percent spliced in (PSI) of PTPRC exon 6 during infection (baseline PSI = 0.335 ± 0.130, mid PSI = 0.385 ± 0.078, FDR = 3.765e−5), a regulatory event that was not observed in males (Figure 2C). Exon 6 inclusion is necessary for two CD45 isoforms, CD45RABC and CD45RBC, which identify functionally distinct subsets of B cells and T cells, respectively (Rodig et al., 2005; Fukuhara et al., 2002; McNeill et al., 2004). Thus, this molecular splicing regulation of PTPRC, consistent with alterations in the immune cell repertoire in response to SARS-CoV-2 infection, is restricted to females.
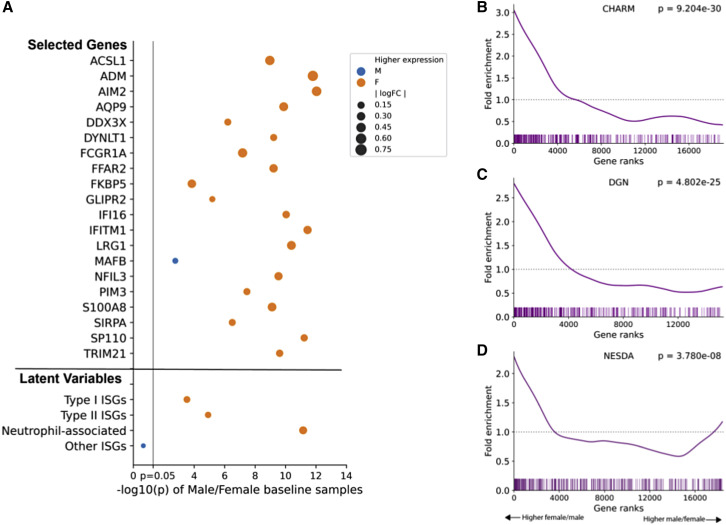
Transcriptional differences between the sexes were also widespread, and given that the strength of the interferon response correlates inversely with the severity of SARS-CoV-2 infections (Del Valle et al., 2020; Hadjadj et al., 2020; Gadotti et al., 2020), we analyzed the differential expression of interferon-stimulated genes (ISGs) as a potential source of sex divergence in SARS-CoV-2 infection responses. Using a published list of ISGs (Schoggins et al., 2011), we found higher ISG expression in females versus males during SARS-CoV-2 infection (first p = 6.88e−09, mid p = 1.87e−14, Mann-Whitney U test). Furthermore, although ISGs were significantly induced in both sexes during infection (GSEA normalized enrichment scores [NESs] > 2.2 and p values < 1e−4 for all analyses), the extent of induction was significantly higher for females compared with males (first p = 0.0207, mid p = 3.49e−13, Mann-Whitney U test; Figure 2D). No pathway from the Kyoto Encyclopedia of Genes and Genomes (KEGG) database had higher enrichment scores than ISGs in any of the comparisons, indicating ISGs play a key role in the transcriptional response to SARS-CoV-2 infection (see Table S3 for details).We also investigated interferon response levels before infection. Although serum levels of interferons were undetectable before infection by Luminex assays in most participants, we observed baseline ISG expression differences between uninfected males and females. Of the 172 differentially expressed ISGs, over 75% (n = 131) are overexpressed in females (FDR = 0.05 Mann-Whitney test with Benjamini-Hochberg multiple hypothesis correction). Even after correcting for cell type proportion differences between the sexes, a robust signal of increased ISG expression in females remained (examples in Figure 3 A; full table in Table S4). Overall baseline ISG expression was also significantly higher in females (rank-based enrichment p value p = 9.203e−30; Figure 3B). We replicated this observation in two independent studies of cohorts including a broad range of ages, comprising blood RNA-seq datasets from 922 (Battle et al., 2014) and 1,848 individuals (Wright et al., 2014) (Figures 3C and 3D). Both cohorts also showed significant enrichment of ISGs among genes with higher expression in females (p = 4.802e−25 and p = 3.780e−8, respectively), confirming higher baseline ISG expression levels in females across cohorts.
Figure 3.
Higher baseline ISG expression observed in females in CHARM and in reference datasets
(A) The majority of ISGs are significantly different between sexes in baseline samples. A subset of ISGs that remain statistically significant after cell type correction is shown (see Table S3 for full list), colored by the sex with higher expression. An orange marker represents genes with higher expression in females and blue represents genes with higher expression among males. Point size is proportional to the log fold change of expression between sexes.
(B–D) (B) ISGs are more highly expressed in females in CHARM baseline samples and two independent studies, (C) Depression Genes and Networks (DGN) (n = 922), and (D) Netherlands Study of Depression and Anxiety (NESDA) (n = 1,848). Vertical bars represent ISG locations among gene lists sorted by log fold change between females and males. Fold enrichment is a measure of the observed versus expected number of ISGs present at each gene rank. p values are computed from rank-based enrichment.
To functionally characterize these baseline sex-based differences in ISGs, we performed factor analysis identifying reproducible latent variables (LVs), four of which showed high correlation (Pearson’s R > 0.7) across all three cohorts. These LVs correspond to the Type I interferon response (top enriched pathway: Hallmark interferon alpha response), Type II (top enriched pathway: Hallmark interferon gamma response), neutrophil-associated (highly correlated with PLIER [Mao et al., 2019] estimated neutrophil proportion), and other (no clear association with known interferon groups) (details in Figure S4 and method details). Three of these LVs, representing Type I, Type II, and neutrophil-associated ISGs, were significantly more expressed in females (p < 5e–4 for all; Figure 3A) prior to SARS-CoV-2 infection. Thus, in addition to a stronger transcriptional and post-transcriptional immune response during SARS-CoV-2 infection, females show higher baseline levels of ISG expression, specifically Type I, Type II, and neutrophil-associated ISGs.
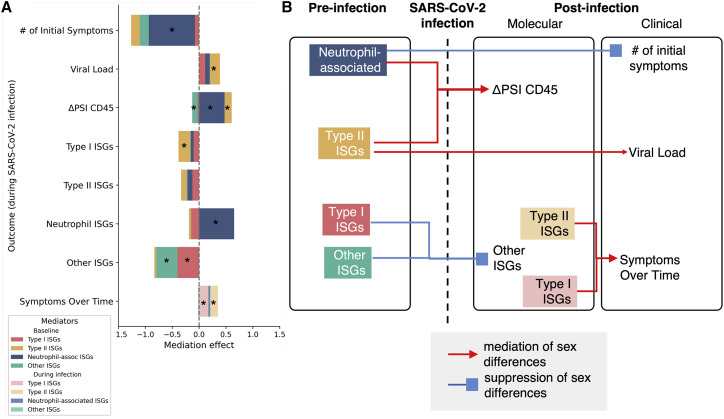
To identify which molecular mediators before and during infection underlie sex differences in the responses to SARS-CoV-2 infection, we performed mediation analyses (Imai et al., 2010) (Figure 4 ). Mediation analysis is a causal inference framework to test whether variables, such as pre-infection antiviral ISG signatures, contribute significantly to sex-specific differences in infection response, such as number of symptoms (see STAR Methods for details; mediation diagram in Figure S5, full list of tested mediators in Table S5). Under this framework, the independent variable is male or female sex, and we test the mediation of the four ISG LVs on several different molecular and clinical outcomes, including CD45 splicing, viral load, and number of symptoms, while controlling for race and ethnicity as potential confounders (multiple-mediator analysis results in Figure S6). This analysis found that higher pre-infection levels of the LV-representing neutrophil-associated ISGs in females mediated the higher levels of these immune response genes seen in females during infection. The pre-infection neutrophil-associated LV also mediated a suppression of the sex bias in the number of symptoms reported at the time of the first positive PCR test, indicating the presence of additional baseline mediating factors. Sex bias in symptoms was significantly mediated by the levels of Type I and Type II ISGs during infection, consistent with inflammatory responses triggered by interferons (McNab et al., 2015; Goel et al., 2021). The relationship of ISGs during infection and viral load is unclear because of feedback loops that violate the assumptions of mediation analysis (Sa Ribero et al., 2020). However, we did detect a trend of baseline Type II ISGs mediating sex differences in viral loads (p = 0.040, FDR 0.128). Finally, the higher induced CD45 isoform splicing in response to infection in females was mediated by pre-infection neutrophil and Type II ISGs and was suppressed by the “other” ISG LVs. Overall, the mediation analysis demonstrates for the first time that sex differences in molecular and clinical responses to acute SARS-CoV-2 infection can be causally attributed to pre-infection immunological sex differences. This suggests that exterior factors such as comorbidity levels and social behaviors are not enough to explain the persistent, significant differences between the response to SARS-CoV-2 infection between males and females, but inherent immune system differences that are evident in healthy adults are a significant contributing factor.
Figure 4.
Mediation analysis of the relationship between sex, ISGs, symptoms, and CD45 splice variant pre- and post-infection
(A) Causal mediation analysis was performed to test the mediation effects between pre-infection ISGs to post-infection molecular and clinical responses, as well as post-infection molecular responses to clinical responses. A stacked bar chart summarizes the full mediation results, with stars marking statistically significant mediation results (average causal mediation effect [ACME] p < 0.05).
(B) Pre-infection molecular and clinical measurements were separated from post-infection measurements. Selected significant results of mediation analysis were presented as a directed graph. Edges represent mediation or suppression effects.
Discussion
Although females show a better overall outcome after SARS-CoV-2 infection, we find higher symptom rates in females which echoes the higher female rate of side effects to SARS-CoV-2 vaccination (Gee et al., 2021). Females having a lower viral load suggests that their immune response to SARS-CoV-2 infection is more effective, consistent with the widely observed better COVID-19 outcomes among females (Scully et al., 2020). The increased symptoms seen in females in this young, healthy cohort may reflect a more proactive innate response that lessens the spread of infection. These observations parallel those in other viral infections, with lower viral load observed in females infected with human immunodeficiency virus (HIV) (Meier et al., 2009) and hepatitis B virus (Klein and Flanagan, 2016) and better female outcomes observed for SARS-CoV and Middle East respiratory syndrome-related coronavirus (MERS-CoV) (Matsuyama et al., 2016; Karlberg et al., 2004).
Previous studies of sex differences in immunology and SARS-CoV-2 have focused on changes observed after infection. Sex differences in both innate and adaptive immune responses during infection have been reported (Takahashi et al., 2020; Ren et al., 2021). Taking advantage of longitudinal sampling commencing before infection on a large number of participants, we are able to detect baseline immune sex differences that contribute to the sex bias in SARS-CoV-2 infection responses. The differences in pre-infection interferon signaling state implicated in the response sex bias are consistent with the relationship previously observed between disease severity and innate immune system variability (Schultze and Aschenbrenner, 2021), although they have never been causally linked before. In order to draw strong causal conclusions from our analysis, they assumption of sequential ignorability is required (Imai et al., 2010). Sequential ignorability has two components: first, we assume that the exposure assignment is statistically independent from the outcomes, given the mediators and confounders, and second, we assume that the mediator is conditionally independent of the outcome, given the exposure and confounders, i.e., there are no unmeasured mediator-outcome confounders (VanderWeele, 2016). The first piece, essentially randomization of exposure assignments, is easily satisfied in our analysis because sex has not been assigned based on any measured factor. The next piece of sequential ignorability is a much stronger assumption, and in general, it is difficult to know for certain whether the ignorability of the mediator holds even after researchers collect as many pretreatment confounders as possible. Such an assumption is often referred to as nonrefutable because it cannot be directly tested from the observed data (Manski, 2007).
Study of sex differences in COVID-19 is challenging because of variations in age, basal health and physical fitness, disease progression, and heterogeneity of diagnosis timing and treatment. Although around 90% of the participants in our study were male, the total number of females still allowed for meaningful comparisons and sufficient statistical power. Our study is limited to young healthy adults and did not include severe COVID-19, allowing us to tightly control for baseline health levels but limiting our ability to draw conclusions about severe disease. This large relatively homogeneous cohort of young Marine recruits exposed to the virus under similar basic training conditions minimizes the influence of age, comorbidities, race, ethnicity, and environmental confounders which allowed identification of the causal contribution of baseline immunological sex differences to the molecular responses and symptoms caused by SARS-CoV-2 infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Critical commercial assays | ||
| RNA sequencing | Illumina | N/A |
| Proximity Extension Assay | Olink | Target 96 Inflammation panel |
| PCR testing for SARS-CoV-2 | Thermo-Fisher | TaqPath COVID-19 Combo Kit |
| Deposited data | ||
| RNA-sequencing | This paper | GEO: GSE198449 |
| Software and algorithms | ||
| Mediation package | Tingley et al., 2014 | https://cran.r-project.org/web/packages/mediation/ |
| Other | ||
| Proximity Extension Assay | Soares-Schanoski et al., 2022 | Table S5 |
| DGN RNA-sequencing dataset | Battle et al., 2014 | Available by application through NIMH Center for Collaborative Genomic Studies on Mental Disorders |
| NESDA RNA-sequencing dataset | Wright et al., 2014 | Available by application through dbGaP |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Olga G. Troyanskaya (ogt@genomics.princeton.edu).
Materials availability
This study did not generate new materials.
Experimental model and subject details
Human subjects
Details of the human subjects who participated in this study can be found in this paper’s study design section of the method details, and in supplemental information (Figure S1). This study was approved by the Naval Medical Research Center (NMRC) institutional review board (IRB), protocol number NMRC.2020.0006, in compliance with all applicable U.S. federal regulations governing the protection of human subjects.
Method details
Study design
The COVID-19 Health Action Response for Marines (CHARM) prospective study enrolled 3,326 Marine recruits entering basic training at Parris Island, South Carolina from May to November, 2020. All study participants were tested for SARS-CoV-2 by PCR, had serum drawn to assess antibody status, and were administered a symptom questionnaire as well as demographic information at enrollment, and approximately 7, 14, 28, 42, and 56 days afterward. The majority of this cohort has been described in previous studies (Letizia et al., 2020, 2021).
Individuals who were seropositive or missing serology data upon enrollment were removed from this analysis. Among the remaining 2,885 consented participants, 2,641 were males and 297 were females (details in Figure S1). Intersex, transgender, and nonbinary individuals are not represented in this study. Biospecimens were sequenced by Illumina short read sequencing at an average depth of 25 million paired-end reads of 101 bp in length. In total, an average of n=3.17 RNA-seq samples per person were sequenced.
SARS-CoV-2 quantitative PCR testing was performed in mid-turbinate nares swabs and were performed within 48 h of sample collection at high complexity Clinical Laboratory Improvement Amendments-certified laboratories using the US Food and Drug Administration-authorized Thermo Fisher TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific, Waltham, MA, USA). Lab24Inc (Boca Raton, FL, USA) performed PCR testing from study initiation (May 11, 2020) until Aug 24, 2020, and the Naval Medical Research Center (Silver Spring, MD, USA) from Aug 24, 2020, until the conclusion of the study (Nov 2, 2020).
Depending on the infection status determined by PCR testing, RNA-seq samples were annotated to four distinct stages. RNA-seq samples from initial enrollment with negative PCR tests (PCR-) were annotated as Baselines. The RNA-seq samples at the first time a participant turned PCR+ were annotated as first-time infection, or first. Following the initial PCR+, all subsequent RNA-seq samples while the participant remained PCR+ were annotated Mid. Finally, the RNA-seq samples from the participants that turned PCR- after infection were annotated as Post.
Clinical feature analysis
Study participants self-reported symptoms and had temperatures checked at each study time point, regardless of PCR test result as part of the longitudinal testing. Participants were asked to report any symptoms experienced since the previous visit or during the previous two weeks. Symptom reporting fell into 14 total categories: abdominal pain, chills, cough, decreased taste or smell, diarrhea, fatigue, feeling feverish, headache, muscle ache, nausea/vomiting, runny nose, shortness of breath, sore throat, and other. A participant was considered “Symptomatic” if fever over 100.4 was measured or at least one symptom was reported within 2 weeks of the first positive PCR test. Statistical significance in the different proportions of symptom incidence between males and females was computed with a Chi-square test of independence.
Viral load measured by probe cycle threshold (CT) when each participant first turned PCR+ was analyzed by ANOVA for sex divergence. The statistical significance for biological sex was determined by Mann-Whitney U test, as well as by ANOVA when controlling for results obtained from two different CLIP/CLIA certified laboratories running the ThermoFisher Emergency Use Authorization SARS-CoV-2 assay and different months the participants were infected.
RNA-seq preprocessing
A uniform pipeline was used to process all RNA-seq fastq files. STAR (v2.7.4) (Dobin et al., 2013) was used to align the reads to hg38 genome build with Gencode v34 index. To quantify gene expression levels, kallisto (v0.46.0) (Bray et al., 2016) was used to pseudo-align RNA-seq reads to Gencode v34 transcripts (Harrow et al., 2012). Throughout this study, Gencode v34 genome annotation was used as the reference gene annotations wherever applicable.
Differential gene expression analysis
Due to the large difference in sample sizes of males and females in this study, differential gene expression analysis was performed within each sex separately. Transcript expression quantifications from kallisto were aggregated to gene level with the tximport (v1.14.2) (Charlotte Soneson et al., 2015) package, and differential gene expression analysis was carried out with DESeq2 (v1.26) (Love et al., 2014), comparing samples of each sex in the First category to Baseline and Mid to Baseline. All participants with seropositive results or missing serology data at baseline were removed prior to analysis. The study design controlled for plate number to minimize batch effects from the RNA-seq processing.
Gene set enrichment analysis was performed with a published list of interferon stimulated genes (ISGs) (Schoggins et al., 2011) through GSEApy (v0.10.4) (Fang). For each sex and time annotation, the prerank module was used to compute the ISG enrichment within the background of all differentially expressed genes (DEGs) for the given sex and time comparison (e.g. First vs. Baseline in Female samples). DEGs were defined by an adjusted p-value (according to the Benjamini-Hochberg method) below 0.05 and ordered by their log fold change. The p-values comparing enrichment scores between males and females were computed by randomly selecting n=10,000 sets of genes of the same length as the ISG list, computing their enrichment scores in males and females, and counting the number of times there was an equal or greater enrichment score difference as seen in the ISG comparisons.
Differential alternative splicing analysis
Two complementary approaches leveraging different aspects of RNA-seq reads to quantify percent spliced in (PSI) were employed to reduce potential counting bias. Using genome read alignment generated by STAR as input, the junction read counts for alternative splicing events were counted by DARTS/rMATS-turbo (Zhang et al., 2019; Shen et al., 2014). Using the transcript quantifications generated by kallisto as input, the ratio between long and short isoforms were computed by SUPPA2 (Trincado et al., 2018). We analyzed four basic types of alternative splicing events, i.e., skipped exons, alternative 5’ splice sites, alternative 3’ splice sites, and retained introns.
To identify the alternative spliced exons upon SARS-CoV-2 infection, we employed a linear mixed model (LMM) regression model implemented in Python library statsmodels (v0.11.1) (Seabold and Josef Perktold, 2010). Similar to DEG analysis, splicing analysis was performed within each sex separately. The LMM regressed the logit-transformed exon usage measured by PSI to disease stage and potential confounding factors. To control for potential batch effects, the RNA-seq plate numbers were included in the regression model.
Fixed-effect regression coefficients and random-effect variance components were estimated by statsmodels. Statistical significance was determined by Wald tests and p-values were multiple testing corrected by Benjamini-Hochberg False Discovery Rate (FDR). Alternative splicing events with FDR<0.05 were called as statistically significant.
O-link processing and analysis
For proteomic analysis we used the OLINK commercially available assay, as described previously (Lundberg et al., 2011). We heat-inactivated serum samples and the OLINK analysis were performed at the Human Immuno Monitoring Core at the Icahn School of Medicine, Mount Sinai, New York. For the assay, paired cDNA-tagged antibodies against different analytes were used to target the proteins in the sera. The protein level was quantified by real-time PCR, after DNA oligonucleotides hybridization and extension. Finally, the results were submitted to rigorous quality control.
Sex divergence was computed for each protein by comparing the NPX values in males and females in the Baseline, First, and Mid categories via a Mann-Whitney test. Any protein with a significant difference between sexes after multiple hypothesis testing at FDR=0.05 with the Benjamini-Hochberg method was considered sex-specific. Among these sex-specific proteins, each was assigned to male-specific or female-specific groups based on which sex had a higher median NPX value in the significant comparison. A list of all 9 sex-specific proteins is provided in Table S1.
Cell type proportion correction
Cell type proportions were estimated from the bulk RNA-seq data using CIBERSORTx (Newman et al., 2019), with the built-in LM22 reference covering 22 immune cell types. Baseline ISG expression differences between the sexes were determined by linear regression analysis with each of the 22 cell types as covariates to control for their confounding effects. FDR was controlled at a level of 0.05 with the Benjamini-Hochberg procedure.
Sex-biased and sex-specific labeling
For the creation of tisue- and sex-specific networks from the DEG analysis, genes were first filtered down to only those with an adjusted p-value below 0.05 and log2 fold change greater than 0.5 in at least one of the four DEG analyses (Male/Female, First/Mid vs Baseline). For each gene in this group, normalized TPM values were compared between males and females for each of Baseline, First, and Mid categories with a Mann-Whitney test, and then multiple hypothesis testing was performed with the Benjamini-Hochberg procedure at an FDR=0.05. Genes that did not pass this statistical testing for any of the three time categories (Control, First, and Mid) were considered sex-independent. Those that showed significant differences in expression between the sexes were further categorized into sex-biased and sex-specific groups. The sex-specific groups include genes that are only significantly differentially expressed (adjusted p-value < 0.05) in one of the two sexes. Genes that were significant in both groups were categorized as sex-biased based on which sex had higher log fold change (First/Mid) or TPM (Baseline) values in the category with significant sex differences. This resulted in many more genes in the female-biased group than the male-biased group, a pattern which persisted even with down-sampling the male samples to the same number as the females.
We adopted a similar approach to partition sex-dependent differentially spliced exons. The exons were filtered down to adjusted p-values below 0.05 and absolute delta PSI values larger than 1% in at least one of the four DAS analyses (Male/Female, First/Mid vs Baseline). Mann-Whitney test was used to compare the PSI values for each tested exon between males and females for each of Baseline, First and Mid categories and p-values were corrected by FDR. Exons were divided into sex-independent, sex-biased, and sex-specific similarly to DEGs, with the following modification: exons were assigned to sex-biased group based on which group had the larger absolute delta PSI (First/Mid) instead of log fold change. To determine the sex dependency on the gene level with multiple differentially spliced exons, the hierarchy was followed as sex-specific exons > sex-biased exons > sex-independent exons. That is, a gene was sex-specific as long as at least one of its exons was sex-specific, regardless of the definition of other exons. Thus, it is possible that a gene was defined as both female- and male-specific differentially spliced due to the existence of female- and male-specific exons.
Functional network analysis
Functional networks in HumanBase (Wong et al., 2018; Greene et al., 2015) were used to analyze the tissue-specific network modules and functional annotation enrichment for male- and female- differentially expression and spliced genes, i.e., the union of sex-biased, sex-specific, and sex-independent spliced gene lists. Detected modules for male and female expression and splicing signatures were analyzed for the enriched Gene Ontology (GO) terms. In each of the four analyses corresponding to each sex with DEGs and DAS events, there was a functional module related to virus response and immune activation, with representative GO terms such as “response to virus”, “innate immune response”, and “leukocyte activation”. We focused on this module from each network, comparing the numbers of sex-independent and sex-dependent genes.
Independent cohorts: DGN and NESDA
The DGN cohort includes a transcriptomic study of whole blood samples from 922 individuals (Battle et al., 2014). The cohort consists of 274 male and 648 female European individuals with an average age of 44.70, ranging from 21 to 60. We directly used the “trans” normalized data as described in Battle et al. (2014) which were normalized for known technical factors and genotype principal components. The NESDA (Netherlands Study of Depression and Anxiety) cohort (Wright et al., 2014) was available from dbGAP (phs000486.v1) including 1,848 transcriptomic samples. This cohort includes 1241 female and 607 male adults from 18 to 65 years old, with an average age of 41.78. The NESDA RNA-seq data was normalized for available technical factors and first three genotype principal components.
Interferon-stimulated gene (ISG) analysis
A list of 362 ISGs was acquired from a previous study characterizing interferon response (Schoggins et al., 2011). One gene (FLJ23556) was unable to be mapped to our data, therefore all ISG analysis is performed with the 361 remaining genes. Baseline expression differences between males and females were computed using TPM values normalized by the counts function of DESeq2 (Love et al., 2014). For each participant with multiple samples, normalized TPM values were averaged to give only one baseline value per gene per study participant. Statistical testing was performed with the Mann-Whitney test, and multiple hypothesis tests were performed with an FDR of 0.05 and the Benjamini-Hochberg procedure.
Gene set enrichment analysis was performed with the prerank function of the gseapy package (Fang). The 2021 KEGG pathway list was used for comparison with ISGs. The KEGG pathway analysis showed that female comparisons tended to result in higher normalized enrichment scores (NES), so to control for this bias we performed a permutation test for the NES difference observed with ISGs. For both First and Mid, we randomly selected n=10,000 sets of 361 genes and computed NES values for males and females with these random gene sets. The NES difference p-value is therefore computed as the fraction of times we observed a F-M difference greater than or equal to that of the ISG difference for the given annotation (First p = 0.2235; Mid p = 0.0328).
In order to determine the relative enrichment of ISGs in genes with significant sex differences at baseline, we computed the log fold change of all genes between females and males. Genes were then ranked by log fold change, and we compared the rankings of ISGs to the background of all genes with significant differences between the sexes, excluding ISGs. ISGs were significantly enriched for increased expression in females across all three cohorts (CHARM p=9.203e-30; DGN p=4.802e-25; NESDA p=3.780e-8).
We performed matrix factorization using the ‘simpleDecomp’ function from the PLIER package (Mao et al., 2019) on the ISGs from three blood datasets (CHARM, DGN, and NESDA) to identify 10 ISG latent variables (LVs). In order to find ISG patterns that were robust across datasets we computed the Pearson correlation of the loadings for each CHARM LV with the loadings of the other two datasets, retaining the maximum value. LVs that had a mean inter-dataset correlation above 0.7 were considered highly reproducible across datasets (Figure S4).
The four highly reproducible LVs were annotated based on pathway enrichment and the top-ranked genes with respect to the corresponding loading values. LV6 is highly enriched with the “Hallmark interferon gamma response” pathway (p=3.816e-16), and interferon gamma associated genes, e.g, GBP1, GBP4 and GBP5, so it was annotated as the Type II interferon cluster. LV10 shows a similar expression pattern as LV6 with a distinct list of genes including IFI6, MX1, IFIT1, ISG15, and was most enriched for the “Hallmark interferon alpha response” pathway (p = 2.217e-19). LV10 was therefore annotated as the Type I interferon cluster. Top-ranked genes of LV3 include known neutrophil genes, such as IL1RN, IL1R1, and MAP3K5, and it is highly correlated with neutrophil proportion estimated from bulk RNA-seq using PLIER (Mao et al., 2019). We called LV3 the Neutrophil-associated cluster. LV5 has no clear association with known interferon groups, and we annotated it as Other.
Mediation analysis
The relationships between biological sex, symptoms, and molecular variations were tested by a causal mediation statistical framework implemented in the R package ‘mediation’ (33). Briefly, the mediation analysis leveraged a potential outcome framework to partition the total effect from exposure variable (in this case, sex) to outcome variables as the sum of causal mediation effect and direct effect. When the mediation effects had opposite signs as the total effect, the tested mediation variable was deemed as having suppression effect to the outcome. Average causal mediation effects (ACME) were estimated by a model-based approach that sampled counterfactual outcomes; statistical inference was drawn against the null hypothesis where the average causal mediation effect was zero. In this study, mediators with ACME p-value < 0.05 were considered as statistically significant.
We performed this statistical analysis for the following combinations of outcomes and mediators: 1) outcome is number of symptoms at the time of first PCR positive and mediator is each of the pre-infection ISG latent variables (LVs); 2) outcome is the initial viral load and mediator is each of the pre-infection ISG latent variables (LVs); 3) outcome is the induced CD45 splice level and mediator is each of the pre-infection ISG LVs; 4) outcome is each of the post-infection ISG LVs and mediator is each of the pre-infection ISG LVs; 5) outcome is the number of symptoms throughout infection and mediator is each of the post-infection ISG LVs.
If the dependent variable (i.e., outcome and mediator) was continuous, a linear regression was used to model the relationships; if the dependent variable was discrete counts, a Poisson regression was used. For test conditions 4 and 5 described above, we included the number of days since each participant initial infection as a covariate to adjust for the time of sample collections after infection. Furthermore, to model multiple samples from the same participant, random effects for each participant were introduced to both the linear regression of post-infection ISG LVs and the Poisson regression of symptoms.
Quantification and statistical analyses
Statistical tests used for computational tests are explained in each subsection of the STAR Methods. Inclusion and exclusion criteria are described in the study design section of the method details.
Acknowledgments
We thank the Human Immuno Monitoring Core (HIMC) at Icahn School of Medicine at Mount Sinai for performing the proteomics OLINK assays. We thank the many US Navy corpsmen who assisted in the logistics and sample acquisition and the devoted Marine recruits who volunteered for this study. This study was approved by the Naval Medical Research Center (NMRC) institutional review board (IRB), protocol number NMRC.2020.0006, in compliance with all applicable US federal regulations governing the protection of human subjects. This study was funded by Defense Advanced Research Projects Agency contract number N6600119C4022 (S.C.S.), Defense Healthy Agency grant 9700130 through the Naval Medical Research Center (A.G.L.), National Institutes of Health grant R01GM071966 (O.G.T.), and Simons Foundation grant 395506 (O.G.T.).
Author contributions
Conceptualization, Z.Z., N.S., O.G.T., and S.C.S.; investigation: I.R., A.S.S., V.D.N., M.A.S.A., M.V., N.S., K.G., N.M., C.M.M., S.V., R.S.G.S., H.A., A.S.G.-R., A.v.d.G., C.W.G., R.A.L., S.E.L., G.N., M.A.S., M.S.T., D.L.W., E.Z., and H.V.B.; resources: S.C.S.; data curation: Y.G.; methodology, Z.Z., N.S., and Y.N.W.; formal analysis, Z.Z., N.S., W.M., and M.C.; funding acquisition: A.G.L., S.C.S., and O.G.T.; project administration, A.G.L., S.C.S., and O.G.T.; supervision: S.C.S. and O.G.T.; writing – original draft, Z.Z., N.S., O.G.T., and S.C.S.; writing – review & editing, Z.Z., N.S., O.G.T., S.C.S., A.G.L., A.S.S., Y.G., W.M., and I.R.
Declaration of interests
C.W.G., R.A.L., S.E.L., M.A.S., M.S.T., D.L.W., and A.G.L. are military service members or government service employees. This work was prepared as part of their official duties. Title 17, US Code §105 provides that copyright protection under this title is not available for any work of the US Government. Title 17, US code §101 defines a US Government work as a work prepared by a military service member or employee of the US Government as part of that person’s official duties. The views expressed in the article are those of the authors and do not necessarily express the official policy and position of the US Navy, the Department of Defense, and the US Government or the institutions affiliated with the authors. O.G.T. is on the advisory board of Cell Systems.
Published: November 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cels.2022.10.005.
Supplemental information
All normalized enrichment scores (NES) and p values for KEGG pathways in males and females at first and mid time points
Full results (p values and log fold changes) on the comparison between males and females of baseline (pre-infection) expression of ISG genes
This table includes the full results of the mediation analysis for all mediator-outcome relationships tested
Data and code availability
-
•
Source data statement: RNA sequencing data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/7xmbpz6p2f.1
-
•
Code statement: This paper does not report original code.s
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Battle A., Mostafavi S., Zhu X., Potash J.B., Weissman M.M., McCormick C., Haudenschild C.D., Beckman K.B., Shi J., Mei R., et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienvenu L.A., Noonan J., Wang X., Peter K. Higher mortality of COVID-19 in males: sex differences in immune response and cardiovascular comorbidities. Cardiovasc. Res. 2020;116:2197–2206. doi: 10.1093/cvr/cvaa284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- Charbonneau H., Tonks N.K., Walsh K.A., Fischer E.H. The leukocyte common antigen (CD45): a putative receptor-linked protein tyrosine phosphatase. Proc. Natl. Acad. Sci. USA. 1988;85:7182–7186. doi: 10.1073/pnas.85.19.7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney A.H., Shvets A.A., Lu W., Griffante G., Mollenauer M., Horkova V., Lo W.L., Yu S., Stepanek O., Chakraborty A.K., Weiss A. CD45 functions as a signaling gatekeeper in T cells. Sci. Signal. 2019;12:eaaw8151. doi: 10.1126/scisignal.aaw8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Huang H.H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T.H., Madduri D., Stock A., et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, Z.Q. GSEApy: gene set enrichment analysis in Python (Github; https://github.com/zqfang/GSEApy). [DOI] [PMC free article] [PubMed]
- Fukuhara K., Okumura M., Shiono H., Inoue M., Kadota Y., Miyoshi S., Matsuda H. A study on CD45 isoform expression during T-cell development and selection events in the human thymus. Hum. Immunol. 2002;63:394–404. doi: 10.1016/s0198-8859(02)00379-8. [DOI] [PubMed] [Google Scholar]
- Gadotti A.C., de Castro Deus M., Telles J.P., Wind R., Goes M., Garcia Charello Ossoski R., de Padua A.M., de Noronha L., Moreno-Amaral A., Baena C.P., Tuon F.F. IFN-γ is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020;289:198171. doi: 10.1016/j.virusres.2020.198171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gee J., Marquez P., Su J., Calvert G.M., Liu R., Myers T., Nair N., Martin S., Clark T., Markowitz L., et al. First month of COVID-19 vaccine safety monitoring – United States December 14, 2020-January 13, 2021. MMWR Morb. Mortal. Wkly. Rep. 2021;70:283–288. doi: 10.15585/mmwr.mm7008e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel R.R., Kotenko S.V., Kaplan M.J. Interferon lambda in inflammation and autoimmune rheumatic diseases. Nat. Rev. Rheumatol. 2021;17:349–362. doi: 10.1038/s41584-021-00606-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C., et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015;47:569–576. doi: 10.1038/ng.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., et al. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K., Keele L., Tingley D. A general approach to causal mediation analysis. Psychol. Methods. 2010;15:309–334. doi: 10.1037/a0020761. [DOI] [PubMed] [Google Scholar]
- Karlberg J., Chong D.S.Y., Lai W.Y.Y. Do men have a higher case fatality rate of severe acute respiratory syndrome than women do? Am. J. Epidemiol. 2004;159:229–231. doi: 10.1093/aje/kwh056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- Letizia A.G., Ge Y., Vangeti S., Goforth C., Weir D.L., Kuzmina N.A., Balinsky C.A., Chen H.W., Ewing D., Soares-Schanoski A.S., et al. SARS-CoV-2 seropositivity and subsequent infection risk in healthy young adults: a prospective cohort study. Lancet Respir Med. 2021;9:712–720. doi: 10.1016/S2213-2600(21)00158-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letizia A.G., Ramos I., Obla A., Goforth C., Weir D.L., Ge Y., Bamman M.M., Dutta J., Ellis E., Estrella L., et al. SARS-CoV-2 Transmission among Marine recruits during quarantine. N. Engl. J. Med. 2020;383:2407–2416. doi: 10.1056/NEJMoa2029717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizewski Rhonda A., Sealfon Rachel S.G., Park Sang Woo, Smith Gregory R., Porter Chad K., Gonzalez-Reiche Ana S., Ge Yongchao, Miller Clare M., Goforth Carl W., Pincas Hanna, et al. SARS-CoV-2 Outbreak Dynamics in an Isolated US Military Recruit Training Center With Rigorous Prevention Measures. Epidemiology. 2022;33:797–807. doi: 10.1097/EDE.0000000000001523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg M., Eriksson A., Tran B., Assarsson E., Fredriksson S. Homogeneous antibody-based proximity extension assays provide sensitive and specific detection of low-abundant proteins in human blood. Nucleic Acids Res. 2011;39:e102. doi: 10.1093/nar/gkr424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manski C.F. Harvard University Press; 2007. Identification for Prediction and Decision. [Google Scholar]
- Mao W., Zaslavsky E., Hartmann B.M., Sealfon S.C., Chikina M. Pathway-level information extractor (PLIER) for gene expression data. Nat. Methods. 2019;16:607–610. doi: 10.1038/s41592-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama R., Nishiura H., Kutsuna S., Hayakawa K., Ohmagari N. Clinical determinants of the severity of Middle East respiratory syndrome (MERS): a systematic review and meta-analysis. BMC Public Health. 2016;16:1203. doi: 10.1186/s12889-016-3881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNab F., Mayer-Barber K., Sher A., Wack A., O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill L., Cassady R.L., Sarkardei S., Cooper J.C., Morgan G., Alexander D.R. CD45 isoforms in T cell signalling and development. Immunol. Lett. 2004;92:125–134. doi: 10.1016/j.imlet.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D., et al. Sex differences in the toll-like receptor-mediated response of plasmacytoid dendritic cells to HIV-1. Nat. Med. 2009;15:955–959. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Wu P., Lu W., Liu K., Ma K., Huang L., Cai J., Zhang H., Qin Y., Sun H., et al. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: A retrospective study of 168 severe patients. PLoS Pathog. 2020;16:e1008520. doi: 10.1371/journal.ppat.1008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F., et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913.e19. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodig S.J., Shahsafaei A., Li B., Dorfman D.M. The CD45 isoform B220 identifies select subsets of human B cells and B-cell lymphoproliferative disorders. Hum. Pathol. 2005;36:51–57. doi: 10.1016/j.humpath.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Sa Ribero M.S., Jouvenet N., Dreux M., Nisole S. Interplay between SARS-CoV-2 and the type I interferon response. PLoS Pathog. 2020;16:e1008737. doi: 10.1371/journal.ppat.1008737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins J.W., Wilson S.J., Panis M., Murphy M.Y., Jones C.T., Bieniasz P., Rice C.M. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultze J.L., Aschenbrenner A.C. COVID-19 and the human innate immune system. Cell. 2021;184:1671–1692. doi: 10.1016/j.cell.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully E.P., Haverfield J., Ursin R.L., Tannenbaum C., Klein S.L. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA. 2014;111:E5593–E5601. doi: 10.1073/pnas.1419161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold, Skipper, and Josef Perktold, 2010. statsmodels: Econometric and statistical modeling with python. Proceedings of the 9th Python in Science Conference.
- Charlotte Soneson, Michael I. Love, Mark D. Robinson, 2015. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences, F1000Research, 4:1521. [DOI] [PMC free article] [PubMed]
- Soares-Schanoski Alessandra, Sauerwald Natalie, Goforth Carl W., Periasamy Sivakumar, Weir Dawn L., Lizewski Stephen, Lizewski Rhonda, Ge Yongchao, Kuzmina Natalia A., Nair Venugopalan D., et al. Asymptomatic SARS-CoV-2 Infection Is Associated With Higher Levels of Serum IL-17C, Matrix Metalloproteinase 10 and Fibroblast Growth Factors Than Mild Symptomatic COVID-19. Frontiers in Immunology. 2022;13 doi: 10.3389/fimmu.2022.821730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Ellingson M.K., Wong P., Israelow B., Lucas C., Klein J., Silva J., Mao T., Oh J.E., Tokuyama M., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tingley Dustin, Yamamoto Teppei, Hirose Kentaro, Keele Luke, Imai Kosuke. mediation: R package for causal mediation analysis. Journal of Statistical Software. 2014;59(5) [Google Scholar]
- Trincado J.L., Entizne J.C., Hysenaj G., Singh B., Skalic M., Elliott D.J., Eyras E. SUPPA2: fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018;19:40. doi: 10.1186/s13059-018-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderWeele T.J. Mediation analysis: a practitioner’s guide. Annu. Rev. Public Health. 2016;37:17–32. doi: 10.1146/annurev-publhealth-032315-021402. [DOI] [PubMed] [Google Scholar]
- Wong A.K., Krishnan A., Troyanskaya O.G. GIANT 2.0: genome-scale integrated analysis of gene networks in tissues. Nucleic Acids Res. 2018;46:W65–W70. doi: 10.1093/nar/gky408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F.A., Sullivan P.F., Brooks A.I., Zou F., Sun W., Xia K., Madar V., Jansen R., Chung W., Zhou Y.H., et al. Heritability and genomics of gene expression in peripheral blood. Nat. Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Pan Z., Ying Y., Xie Z., Adhikari S., Phillips J., Carstens R.P., Black D.L., Wu Y., Xing Y. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods. 2019;16:307–310. doi: 10.1038/s41592-019-0351-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All normalized enrichment scores (NES) and p values for KEGG pathways in males and females at first and mid time points
Full results (p values and log fold changes) on the comparison between males and females of baseline (pre-infection) expression of ISG genes
This table includes the full results of the mediation analysis for all mediator-outcome relationships tested
Data Availability Statement
-
•
Source data statement: RNA sequencing data have been deposited at the Gene Expression Omnibus and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Additional Supplemental Items are available from Mendeley Data: https://doi.org/10.17632/7xmbpz6p2f.1
-
•
Code statement: This paper does not report original code.s
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.