Abstract
Objective(s):
Assess the impact of pretreatment high- and low-frequency drug-resistant human immunodeficiency (HIV) variants on long-term outcomes of 1st-line efavirenz-based antiretroviral therapy (ART).
Design:
Prospective observational study.
Methods:
Participants’ pretreatment plasma RNA had two sections of HIV pol encoding reverse transcriptase sequenced (Illumina, MiSeq) using unique molecular identifiers to detect wild-type (pretreatment drug-resistant variants <1% of viral quasispecies), low-frequency (1–9%) or high-frequency drug-resistant variants (10–100%). Associations between pretreatment drug resistance and virologic outcomes over 24 months of efavirenz-based ART were assessed for the number and frequency of mutations by drug class and other resistance parameters.
Results:
Virologic failure was detected in 30/352 (9%) and pretreatment drug-resistant variants were detected in the viral quasispecies of 31/352 (9%) participants prescribed efavirenz-based ART. Survival analyses revealed statistically significant associations between pretreatment drug-resistance at low (p<0.0001) and high (p<0.001) frequencies, at oligonucleotide ligation assay (OLA) (<0.00001) and non-OLA (<0.01) codons, to a single-antiretroviral class (<0.00001), and a shorter time to virologic failure of efavirenz-based ART. Regression analyses detected independent effects across resistance categories including both low- (<0.01) and high-(<0.001) frequency drug-resistant variants.
Conclusions:
We observed that (1) pretreatment HIV drug-resistance detected at low-frequencies increased the risk of virologic failure over 24 months of efavirenz-based ART, but that (2) most failures, regardless of drug-resistant variants’ frequencies, were detected within a year of ART-initiation. These observations suggest that when efavirenz-based ART is prescribed, screening for pretreatment drug resistance by an assay capable of detecting low-frequency variants, including OLA, may guide clinicians to prescribe more effective ART.
Keywords: Human immunodeficiency virus (HIV), Drug resistance, Low frequency variants, High frequency variants, Non-nucleoside reverse transcriptase inhibitors (NNRTI), Efavirenz
Introduction
The prevalence of human immunodeficiency virus (HIV) variants exhibiting resistance to antiretroviral drugs in Kenya and other low- and middle-income countries has increased with the expansion of antiretroviral uptake.1,2 Concerns that such resistance, especially to non-nucleoside reverse transcriptase inhibitors (NNRTIs), could compromise the effectiveness of antiretroviral therapy (ART) led the World Health Organization (WHO) to update ART treatment recommendations in 2019, designating dolutegravir-based ART as preferred in 1st- and 2nd-line regimens.3 However, some individuals may not tolerate dolutegravir, and efavirenz combined with two nucleoside reverse transcriptase inhibitors (NRTI) remains a recommended alternative regimen for adolescents and adults living with HIV and initiating ART treatment;3 indeed, in some countries efavirenz-based ART remains the most prevalent ART regimen.4 As ART regimens are prescribed for an individual’s lifetime, and few regimens exist without cross-resistance, understanding the effects of pretreatment HIV variants resistant to NRTIs and NNRTIs remains relevant in the age of dolutegravir.
Drug-resistant variants detected within an individual’s HIV quasispecies by consensus or Sanger sequencing (typically ≥15–25% of quasispecies) have long been associated with failure of ART to suppress viral replication.5 More sensitive genotypic assays can detect drug-resistant variants at frequencies below the limit of detection of consensus sequencing, yielding potential insights into the role of low-frequency variants in the success or failure of antiretroviral therapies.6–9 However, assay sensitivities for “low-frequency” pretreatment variants have varied across studies, significantly limiting the determination of clinically meaningful thresholds. A systematic review of 103 studies assessed the impact of “low-frequency” pretreatment drug-resistant variants on the efficacy of ART.10 The frequency cut-offs varied across a 4 log10 range (0.001%–10%). Pretreatment low-frequency drug-resistant variants and virologic failure were associated in 11/25 studies (44.0%) of populations taking NNRTI-based ART. Similarly, another study suggests a variant frequency cut-off of 2% may provide optimal specificity and sensitivity11 while a third suggests 5%.12 The recent “Winnipeg Consensus” described these inconsistencies as a major challenge to large-scale implementation of next-generation sequencing-based drug resistance genotyping, warranting further examination.13
We previously conducted a randomized clinical trial in Kenya to assess the value of a low-cost oligonucleotide ligation point mutation assay (OLA) for identifying pretreatment drug-resistance and whether guiding the selection of 1st-line ART regimens using this assay’s results improved outcomes.14 Among participants randomized to the OLA-guided arm, those with pretreatment drug-resistant variants detected at ≥10% of the individual’s viral population or quasispecies received a 2nd-line ART regimen consisting of two NRTIs and lopinavir-boosted with ritonavir, while those in the standard-of-care arm and those in the OLA arm with resistance between 0–10% were prescribed 1st-line NNRTI-based ART. As hypothesized, participants with pretreatment drug-resistant variants in the OLA-guided arm who were prescribed lopinavir-based ART experienced virologic failure at a significantly lower rate than those in the standard-of-care arm prescribed NNRTI-based ART. Among those with pretreatment resistance by OLA who were randomized to standard-of-care and received NNRTI-based ART, failure rates by month-12 of ART were higher in participants with drug-resistant frequencies ≥10%, and those with frequencies 2–9%; however, the latter group size was small and did not statistically differ from those without resistance. These findings suggested that the threshold of 10% resistance for recommending 2nd-line ART may have been too high, and raised the question as to whether participants with pretreatment drug-resistant variants at frequencies <10% across OLA and non-OLA codons might experience virologic failure at higher rates, particularly if followed for a longer duration of ART.
To evaluate the effects of low-frequency pretreatment drug-resistant variants on longer-term virologic outcomes of 1st-line NNRTI-based ART, study participants who had maintained virologic suppression at month-12 of ART at the largest of the three study sites were offered enrollment for a second year of follow-up. Pretreatment plasma HIV RNA specimens from these participants and those who had already experienced virologic failure by 12 months of NNRTI-based ART underwent genotyping by next-generation sequencing to assess the contribution of a broad array of codons with drug-resistant variants at low-frequencies on virologic failure over 24-months of efavirenz-based ART. (Note: Because nevirapine has been rendered largely obsolete following the introduction of efavirenz in sub-Saharan Africa, we limited our analyses to the larger group of participants prescribed efavirenz-based ART.) We aimed to determine if over two years of 1st-line-efavirenz-based ART (1) low frequency drug-resistant HIV variants comprising 1–9% of an individual’s quasispecies contributed to increased rates of virologic failure compared to rates in individuals with wild-type virus, (2) the time to virologic failure is longer in individuals with low-frequency variants (1–9%) compared to participants with high-frequency variants (10–100%), and (3) drug-resistance mutations at codons not assessed by the OLA contributed to virologic failure.
Methods
Study design and population
A randomized control trial (Clinicaltrials.gov # NCT01898754) was conducted in HIV-infected persons 2 or more years of age who qualified for 1st-line ART at three Coptic Hope Center clinics in Kenya between May 2013 and February 2016.14 Enrolled participants were assigned to receive either OLA testing prior to ART initiation or no pre-ART OLA testing (i.e., standard-of-care). Participants randomized to OLA-guided therapy who had drug-resistant variants detected initiated 2nd-line ART (TDF or ZDV + 3TC + LPV/r). A switch in antiretroviral drugs included in “1st-line ART” (from ZDV to TFD) occurred in the Kenyan ART Program during the study. In addition, individuals in the OLA arm who were resistant to either ZDV or TDF had the other drug recommended by the Study Team, however, the actual choice of ZDV or TDF was made by each participant’s clinician. Participants who tested negative for resistance by OLA as well as those randomized to standard-of-care were prescribed 1st-line NNRTI-based ART (TDF or ZDV + 3TC + NVP or EFV) as per Kenyan guidelines at the time of the trial. Participants’ clinical histories, demographic information, and blood specimens were collected at enrollment, and at 4, 8, and 12 months after ART initiation for plasma HIV-1 RNA quantification.
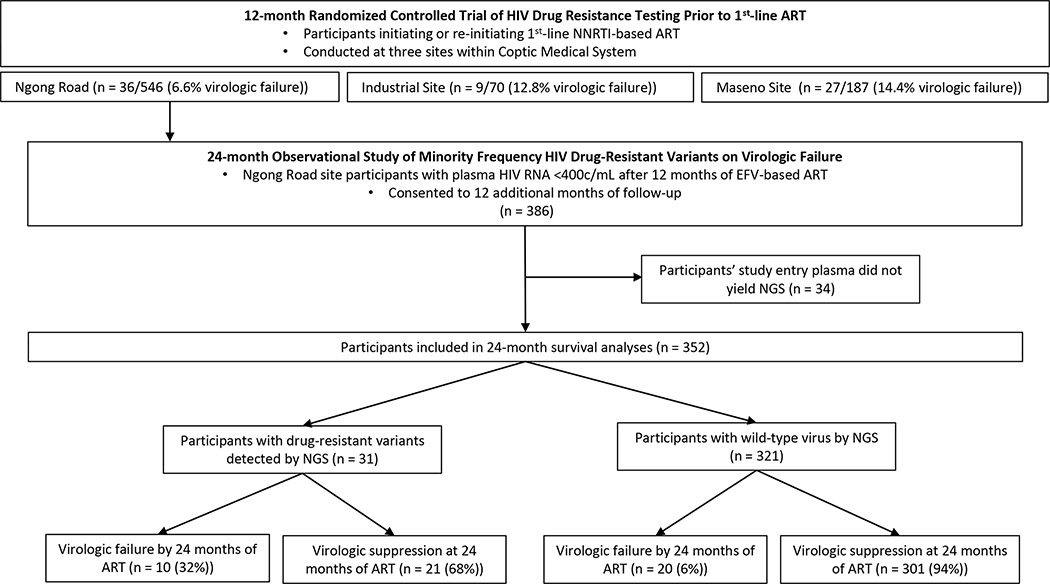
Participants with virologic suppression at month-12 of efavirenz-based ART at the Ngong Road clinic were invited to enroll for another 12 months of follow-up (Figure 1). Blood was collected at 16, 20 and 24 months after ART initiation for plasma HIV-1 RNA quantification. Pretreatment plasma specimens from participants followed for 24 months, as well as those who experienced virologic failure by month-12 of efavirenz-based ART, were assessed for pretreatment drug-resistant variants by next-generation sequencing.
Figure 1. Schema of participant enrollment, drug-resistance genotyping, and virologic outcomes.
Study outcomes
Outcomes of the 12-month study have been reported.14 The prespecified outcomes of this 24-month study included differences in (1) the rate of and (2) the time to virologic failure over 24 months of efavirenz-based ART between participants by frequency of drug-resistant variants detected by next-generation sequencing at enrollment, and (3) by drug-resistant codons that were or were not included in the OLA. Drug-resistance frequency was defined as the proportion of a nucleotide variant encoding a drug-resistant codon in specimens with ≥100 viral templates sequenced. Virologic failure was defined as plasma HIV RNA ≥400 c/mL in sequential specimens or at the final study visit in participants prescribed efavirenz-based ART.
Laboratory Methods
Plasma specimens were assessed for drug-resistant variants by next-generation sequencing using the Illumina platform. One mL blood plasma was centrifuged at 4°C and 20,000 relative centrifugal force for 90 minutes. Pelleted virus was extracted using the QIAamp Viral Mini RNA Kit (Qiagen, Hilden, Germany) per manufacturer’s protocol. Ten uL of extracted HIV RNA were reverse transcribed into cDNA separately in 20uL reactions using Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, California) and primers targeting two regions of the HIV pol gene encoding reverse transcriptase. Primers consisted of an HIV-specific sequence followed by an 8bp string of random nucleotides (a “unique molecular identifier”) and a universal 24bp Illumina reverse adapter sequence: RT1R: 5’-CTCGGAGATGTGTATAAGAGACAGNNNNNNNNACTAGGTATGGTRAATGCAGTATA-3’, HXB2 coordinates 2928–2951; RT2R: 5’-CTCGGAGATGTGTATAAGAGACAGNNNNNNNNAAYTTCTGTATATCATTGACAGTCCA -3’, HXB2 coordinates 3303–3328). cDNA was purified using a 1:1 ratio of cDNA to purification beads (Agencourt Ampure XP, Beckman-Coulter, Beverley, Massachusetts), eluted in 60uL of 10mM Tris-HCl pH 8.0, 0.1mM EDTA,, and divided equally between four separate 50uL PCR reactions per region. Separate single-round 45-cycle PCR amplified each of the two regions and added forward and reverse adapter sequences required by the Illumina sequencing platform. The PCR used a high-fidelity polymerase (FastStart, Roche Diagnostics, Mannheim, Germany) and primers RT1F: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAAACAATGGCCATTRACAGAAGA-3’; RT2F: 5’-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCACAGGGATGGAAAGGATCAC-3’; ILR_fullseq (Illumina Reverse Adapter): 5’-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3’). Amplicons underwent an 8-cycle indexing PCR according to the Illumina 16S Metagenomic library preparation guide and were then pooled together and bidirectionally sequenced across 300bp on an Illumina Miseq (MiSeq Reagent Kit v3, 600Cycles, Illumina, Inc., San Diego, California).
Sequence data processing
Sequences were processed and variants called as previously described,1 with additional steps to first generate consensus sequences for unique viral RNA templates. Three or more high-quality sequencing reads containing identical 8bp unique molecular identifiers were collapsed into a single consensus sequence and aligned to the HXB2 reference sequence using the Burrows–Wheeler algorithm.15 Collapsed consensus sequences containing nucleotide variants associated with resistance to NRTIs and NNRTIs were filtered for statistical significance and annotated. Sequencing datasets generated from fewer than 100 viral templates of each of a participant’s genomic regions were excluded as unrepresentative of a viral quasispecies. Drug-resistant variants were defined by Stanford’s HIV Drug Resistance Database version 8.9.16 Sequences were phylogenetically examined for potential cross-contamination. The variant calling pipeline is available at https://github.com/MullinsLab/drm-snp-calling.
Plasma HIV RNA loads at enrollment and longitudinal timepoints were quantified (Abbott Laboratories, Abbott Park, IL) in a Virology Quality Assurance Program compliant laboratory.
Statistical analysis
Participants’ demographic characteristics including age, sex, CD4+ T-cell count, and enrollment plasma HIV RNA load were compared between those who did and did not experience virologic failure using the Chi-square test for categorical variables and Kruskal Wallis test for continuous variables. Pretreatment drug-resistance identified by next-generation sequencing was classified as an ordinal variable in five different ways; 1) the largest (“peak”) drug-resistant variant frequency detected as a continuous variable, 2) the peak variant frequency as a categorical variable: wild-type (0%), low-frequency (1–9%), and high-frequency (10–100%); 3) the class of drug resistance: wild-type, single-class (NRTI or NNRTI), or dual-class resistance (1 or more variant encoding resistance to each of NRTIs and NNRTIs); 4) the presence of resistant variants at codons assayed by the OLA (K65R, K103N, Y181C, M184V, G190A) or at other codons; and 5) the total Stanford resistance penalty score encoded by all detected variants to individual participant’s ART regimens: wild-type, ≤60, and >60. The primary study outcome variable, virologic failure by month-24 of efavirenz-based ART, was compared between individuals with and without pretreatment drug-resistance using univariable and multivariable Cox Proportional Hazards regression. Time to virologic failure was compared among groups by Kaplan-Meier survival analysis with pairwise log-rank tests for statistical significance. All analyses were performed in R Studio (version 3.6.3).
Results
Demographics and clinical characteristics of study participants
Of the original 386 participants who consented to a second year of follow-up, baseline plasma specimens from 34 (9%) failed sequencing, yielding a total study population of 352 individuals for this analysis of the impact of pretreatment drug-resistant variants on virologic outcomes over 24 months of efavirenz-based ART (Figure 1). This included 332 (94%) participants who maintained virologic suppression through month-12 of ART and consented to a second year of follow-up, and 20 (6%) participants who experienced virologic failure by month-12. Prior to initiation of ART at study month 0, the 352 participants were predominantly female (n=228, 65%), antiretroviral naïve (n=327, 93%), and their median (interquartile range, IQR) age was 38 (32–45) years, enrollment CD4 T-cell count was 227 (104–304) cells/uL, and enrollment log viral load was 4.7 (4.1–5.3) log10 c/mL (Table 1). Participants in this study were largely similar to those in the original randomized controlled trial (Table 1).
Table 1.
Comparison of demographic characteristics between participants enrolled in initial 12-month randomized controlled trial and participants included in the extension of the study to 24 months
| Variable | Original RCT | 2nd Year Supplement | P-value |
|---|---|---|---|
| Total participants (N) | 759 | 352 | |
| Age (years), median (IQR) | 38 (31–45) | 38 (32–45) | 1 |
| Female, N (%) | 499 (65.7) | 228 (64.4) | 0.8 |
| CD4 count (cells/μL), median (IQR) | 235 (132–316) | 231 (109–309) | 0.65 |
| Pre-ART VL (log10 c/mL), median (IQR) | 4.7 (3.9–5.2) | 4.7 (4.1–5.3)* | 1 |
| History of antiretroviral exposure, N (%) | 0.17 | ||
| Antiretroviral-naïve | 679 (89.5) | 327 (92.9) | |
| Antiretroviral-experienced | 72 (9.5) | 25 (7.1) | |
| Virologic failure by month-12 of ART, N (%) | 65 (8.6) | 20 (5.7) | 0.12 |
1 missing
Detection of pretreatment drug-resistant variants
A total of 38 drug-resistant variants were detected by next-generation sequencing in the pretreatment plasma of 31 (9%) study participants; 15 (48%) participants had only low-frequency variants, and 16 (52%) had at least one drug-resistance mutation at a high-frequency. The median variant frequency (IQR) across all drug-resistant variants was 11.6% (3.4–70.6%), among participants harboring only low-frequency variants was 3.4% (1.5–4.2%), and in those with at least one high-frequency variant was 45.5% (23.0–97.0%).
K103N was the drug-resistant variant most commonly detected and was present at a median variant frequency (IQR) of 23.8% (2.9–83.1%) in plasma specimens from 12 (39%) of the 31 participants with pretreatment drug-resistance. No other drug-resistant variant was detected in more than three participants (Supplementary Table 1). Twenty-eight (90%) and three (10%) participants with pretreatment drug-resistance harbored variants resistant to a single antiretroviral class (NRTI or NNRTI) or dual-classes (NRTI and NNRTI), respectively. Among those with single-class resistance, 5 (18%) and 23 (82%) had resistance to NRTIs and NNRTIs, respectively. Finally, pretreatment drug-resistant variants encoding cumulative Stanford HIV Database penalty scores of ≤60 and >60 were detected in 28 (90%) and 3 (10%) participants, respectively.
Pretreatment drug resistant variants and virologic outcomes
In total, 30/352 (9%) participants experienced virologic failure by month-24 of efavirenz-based ART. Those who experienced virologic failure by month-24 of ART had similar demographic characteristics to those who maintained suppression (Table 2), except for younger age (mean (SD) 34.4 years (11.8) versus 38.9 (9.8), P = 0.021) and higher enrollment viral loads (mean (SD) 5.0 log10 copies (c)/mL(0.8) versus 4.6 (0.9), P = 0.031). Among the 30 participants who experienced virologic failure, 10 (33%) had pretreatment drug-resistant variants detected by next-generation sequencing, compared to 21/322 (7%) participants with virologic suppression (P<0.001). Peak mutant variant frequency also differed between those with virologic failure and suppression (mean (SD) 14.1% (32.5%) versus 4.2% (18.0%), P = 0.008).
Table 2.
Comparison of demographic characteristics and pretreatment drug-resistance genotypes detected by next-generation sequencing between study participants with and without virologic failure by month-24 of efavirenz-based ART.
| Variable | Suppression (N=322) | Virologic Failure (N=30) | P value |
|---|---|---|---|
|
| |||
| Age (years), mean (SD) | 38.8 (9.8) | 34.4 (11.8) | 0.022 |
|
| |||
| Sex, n (%) | 0.531 | ||
| female | 207 (64.3%) | 21 (70.0%) | |
| male | 115 (35.7%) | 9 (30.0%) | |
| CD4 count (cells/uL), mean (SD) | 219.1 (137.1) | 202.6 (128.5) | 0.525 |
|
| |||
| Enrollment Viral Load, log10 HIV RNA (copies/mL) plasma, mean (SD) | 4.6 (0.9) | 5.0 (0.8) | 0.031 |
|
| |||
| Peak Resistant Variant Frequency, mean (SD) | 4.2 (18.0) | 14.1 (32.5) | 0.008 |
|
| |||
| Resistant Variant Frequency, n (%) | <0.001 | ||
| wild-type | 301 (93.5%) | 20 (66.7%) | |
| low | 10 (3.1%) | 5 (16.7%) | |
| high | 11 (3.4%) | 5 (16.7%) | |
|
| |||
| OLA or Non-OLA codons, n (%) | <0.001 | ||
| wild-type | 301 (93.5%) | 20 (66.7%) | |
| non-OLA | 11 (3.4%) | 4 (13.3%) | |
| OLA | 10 (3.1%) | 6 (20.0%) | |
|
| |||
| Resistance Class Encoded by All Variants, n (%) | <0.001 | ||
| wild-type | 301 (93.5%) | 20 (66.7%) | |
| single | 19 (5.9%) | 9 (30.0%) | |
| dual | 2 (0.6%) | 1 (3.3%) | |
|
| |||
| Cumulative Stanford Resistance xPenalty Score for ART Regimen, n (%) | <0.001 | ||
| wild-type | 301 (93.5%) | 20 (66.7%) | |
| ≤60* | 20 (6.2%) | 8 (26.7%) | |
| >60 | 1 (0.3%) | 2 (6.7%) | |
includes 9 participants with K103N variant alone, 2/9 experienced virologic failure
In univariable Cox proportional hazard regression analyses, risk of virologic failure decreased with increasing age (HR = 0.96 (0.92–0.99), P=0.021) and increased with higher enrollment viral load (HR = 1.71 (1.06–2.74), P=0.027) (Table 3). In multivariable Cox Proportional Hazards regression analyses, younger age, higher enrollment viral load, and pretreatment drug-resistance coded as continuous and as multiple ordinal variables were all associated with increased risk of virologic failure by month-24 of ART (Table 3). Risk of virologic failure increased with the frequency of the drug-resistant variant (HR = 4.73 (1.53–14.59), P=0.007), with the detection of only low-frequency variants (HR = 5.35 (1.79–15.97), P=0.003), with detection of at least one high-frequency variant (HR = 6.60 (2.45–17.74) P<0.001), and with the detection of drug-resistant variants at both OLA (HR = 8.92 (3.43–23.22), P<0.001) and non-OLA (HR = 3.72 (1.10–12.53), P=0.034) codons. Finally, while sample size prohibited testing for statistical significance, increased risk of virologic failure was observed among participants with variants encoding resistance to both a single (NRTI or NNRTI) or to dual (NRTI and NNRTI) classes of antiretrovirals, as well as cumulative Stanford resistance scores both ≤60 and >60. However, among participants with variants encoding resistance to a single class of antiretrovirals, only 1/5 (20.0%) participants with resistance to NRTIs alone experienced virologic failure, compared to 8/23 (34.8%) participants with resistance to NNRTIs alone. Notably, the risk of virologic failure did not appear increased with the detection of K103N alone (Supplementary Table 1). We detected K103N in combination with mutations associated with resistance to NRTIs (M184V/I) in only one participant, who experienced virologic failure at study month-8.
Table 3.
Cox proportional hazards regression (univariable and multivariable) assessing associations between participant characteristics and virologic outcomes of efavirenz-based ART.
| Variable | Level | Value | Hazard Ratio (Univariable) | Hazard Ratio (Multivariable by Peak Variant Frequency) | Hazard Ratio (Multivariable by Variant Frequency Category) | Hazard Ratio (Multivariable by OLA Codon) | Hazard Ratio (Multivariable by Resistance Class) | Hazard Ratio (Multivariable by Stanford Resistance Score) |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Age, mean (SD) | 38.5 (10.1) | 0.96 (0.92–0.99, p=0.021) | 0.96 (0.92–1.00, p=0.039) | 0.96 (0.92–1.00, p=0.044) | 0.96 (0.92–1.00, p=0.038) | 0.96 (0.92–1.00, p=0.048) | 0.95 (0.91–0.99, p=0.015) | |
|
| ||||||||
| Sex, n (%) | female | 228 (64.8) | - | - | - | - | - | - |
| male | 124 (35.2) | 0.77 (0.35–1.68, p=0.507) | 0.80 (0.34–1.86, p=0.600) | 0.82 (0.35–1.93, p=0.649) | 0.86 (0.36–2.03, p=0.729) | 0.80 (0.34–1.89, p=0.610) | 0.76 (0.31–1.82, p=0.536) | |
|
| ||||||||
| CD4 Count, cells/uL, mean (SD) | 217.7 (136.2) | 1.00 (1.00–1.00, p=0.504) | 1.00 (1.00–1.00, p=0.580) | 1.00 (1.00–1.00, p=0.882) | 1.00 (1.00–1.00, p=0.990) | 1.00 (1.00–1.00, p=0.938) | 1.00 (1.00–1.00, p=0.973) | |
|
| ||||||||
| Enrollment Viral Load, Log HIV RNA Copies/mL Plasma, mean (SD) | 4.7 (0.9) | 1.71 (1.06–2.74, p=0.027) | 1.94 (1.14–3.32, p=0.015) | 2.00 (1.16–3.45, p=0.013) | 2.03 (1.16–3.57, p=0.013) | 2.05 (1.16–3.63, p=0.014) | 2.15 (1.26–3.65, p=0.005) | |
|
| ||||||||
| Peak Resistance Variant Frequency, mean (SD) | 0.0 (0.2) | 4.07 (1.36–12.14, p=0.012) | 4.73 (1.53–14.59, p=0.007) | - | - | - | - | |
|
| ||||||||
| Resistant Variant Frequency Category, n (%) | wt | 321 (91.2) | - | - | - | - | - | - |
| low | 15 (4.3) | 6.60 (2.47–17.61, p<0.001) | - | 5.35 (1.79–15.97, p=0.003) | - | - | - | |
| high | 16 (4.5) | 5.67 (2.13–15.12, p=0.001) | - | 6.60 (2.45–17.74, p<0.001) | - | - | - | |
|
|
||||||||
| Resistance at Codons Assayed by OLA, n (%) | wt | 321 (91.2) | - | - | - | - | - | - |
| non-ola | 15 (4.3) | 4.55 (1.55–13.30, p=0.006) | - | - | 3.72 (1.10–12.53, p=0.034) | - | - | |
| ola | 16 (4.5) | 7.91 (3.17–19.74, p<0.001) | - | - | 8.92 (3.43–23.22, p<0.001) | - | - | |
|
|
||||||||
| Class of Encoded Drug Resistance, n (%) | wt | 321 (91.2) | - | - | - | - | - | - |
| single | 28 (8.0) | 6.08 (2.76–13.37, p<0.001) | - | - | - | 6.38 (2.72–14.97, p<0.001) | - | |
| dual | 3 (0.9) | 6.26 (0.84–46.66, p=0.074) | - | - | - | 4.17 (0.52–33.34, p=0.178) | - | |
|
|
||||||||
| Cumulative Stanford Resistance Penalty Score for ART Regimen, n (%) | wt | 321 (91.2) | - | - | - | - | - | - |
| ≤60 | 28 (8.0) | 5.23 (2.30–11.88, p<0.001) | - | - | - | - | 4.82 (2.02–11.51, p<0.001) | |
| >60 | 3 (0.9) | 18.61 (4.32–80.21, p<0.001) | - | - | - | - | 48.03 (9.62–239.86, p<0.001) | |
Pretreatment drug-resistant variants and time to virologic failure
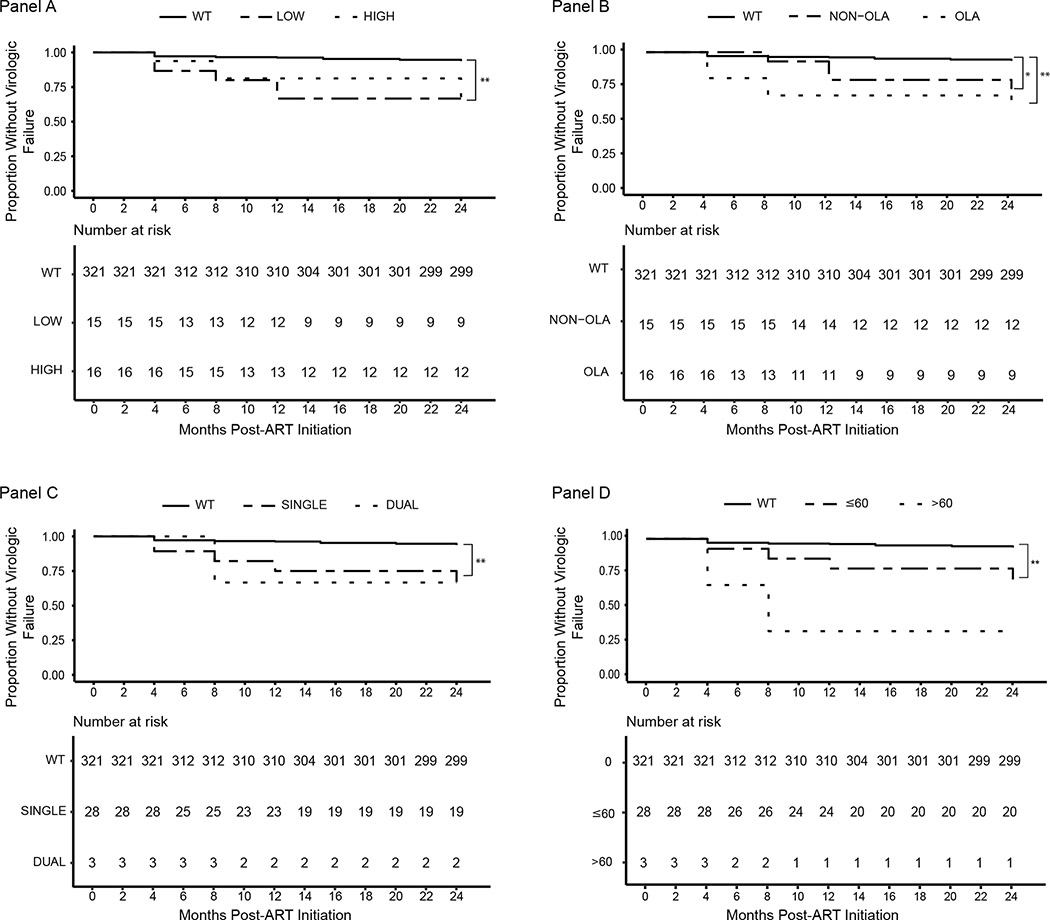
Of the 30 participants with virologic failure, 17 (57%) experienced failure by month 8 of ART, another three for a total of 20 (67%) by month 12, and another 10 (33%) between months 12 and 24. Among the 10 participants who experienced virologic failure between 12 and 24 months of ART, just two harbored pretreatment drug-resistant variants, with both at majority frequencies and both experienced virologic failure at month-24. Survival curves comparing the rates of virologic failure across participants with and without pretreatment drug-resistance are displayed in Figure 2, panels A-D. Overall, the detection of low-frequency variants alone (P<0.001) and the detection of any high-frequency variants (P<0.001) were associated with shorter time to failure (Figure 2, Panel A), as was the detection of variants at OLA (P<0.001) or non-OLA (P=0.003) codons (Figure 2, Panel B). Shorter times to virologic failure were also observed among participants with HIV variants encoding resistance to a single class or to dual-classes of antiretrovirals (Figure 2, Panel C), as well as with Stanford resistance penalty scores of ≤60 or >60 (Figure 2, Panel D), although the small sample sizes of groups in Panels C and D prohibited testing for statistical significance.
Figure 2. Kaplan Meier Survival Analysis of participants with and without pretreatment drug-resistant variants detected by next-generation sequencing.
Panel A: Shorter time to virologic failure was observed among study participants with low-frequency variants alone (P<0.001) or any high-frequency variants (P<0.001), compared to those with wild-type virus. Panel B: Participants with variants at OLA (P<0.001) or non-OLA codons alone (P=0.003) experienced shorter time to virologic failure. Panel C: Shorter time to virologic failure was observed among participants with variants encoding resistance to both a single class (NRTI or NNRTI) alone or dual-drug classes (at least one variant resistant to NRTIs and at least one variant resistant to NNRTIs), although sample size limited testing for statistical significance. Panel D: Shorter time to virologic failure was observed among participants with drug-resistant variants encoding Stanford resistance penalty scores of both less-than-or-equal-to 60 or greater-than 60, but sample size prohibited testing for statistical significance. * = P<0.01; ** = P<0.001
Discussion
Pretreatment HIV resistance to ART in sub-Saharan Africa has progressively increased with the uptake of ART and has been associated with virologic failure.1,2 However, the clinical relevance of low-frequency pretreatment drug-resistant variants appears inconsistent across studies.6–13 To our knowledge, this is the first study evaluating the impact of low-frequency drug-resistance variants on virologic outcomes over 24 months of efavirenz-based ART, with the goal of determining if virologic failure due to low frequency pretreatment drug-resistance occurs over a prolonged period of ART. Our detection of pretreatment drug-resistance in approximately 9% of study participants mirrored the prevalence in other reports.17 Our novel observations include that both majority and minority frequency drug-resistant variants were associated with both an increased rate and a shorter time to virologic failure compared to wild-type virus over 24 months of ART. Notably, two-thirds of the participants who experienced virologic failure failed by study month-12, and only two of the ten participants who failed between study months 12–24 harbored resistance, both with majority frequency pretreatment drug-resistant variants.
Our observation that low-frequency variants were associated with virologic failure confirms6,7,9 and refutes other8 studies examining the clinical relevance of low-frequency variants in antiretroviral-naïve individuals taking NNRTI-based ART. We previously reported findings for a larger population (participants from three Coptic Hope Center clinics) that included participants in the present study (Ngong Road clinic only) demonstrating that low-frequency variants at OLA codons were increased but not statistically significantly associated with virologic failure during the first 12 months of ART.18 This larger group included significantly more ART-experienced individuals and only examined resistance at OLA codons. Our observation that low-frequency variants are associated with virologic failure in the present study is likely due to a greater proportion of antiretroviral-naïve participants, the detection of a greater number of low-frequency pretreatment drug-resistant variants by next-generation sequencing, and a greater rate of ART-suppression among participants enrolled from this clinic. It seems likely that the overall lower rate of virologic failure at this clinic enhanced our ability to detect the effects of pretreatment low-frequency drug-resistant variants.
We speculate that the effects of pretreatment drug resistance on virologic failure are frequently overshadowed by other causes of virologic failure. For example, our larger aforementioned study found that while majority frequency pretreatment drug-resistance contributed to virologic failure, relatively few individuals with virologic failure had pretreatment drug-resistance and that deaths were a major cause of failure.14 Notably, the ADVANCE Trial reported that pretreatment NNRTI resistance was associated with virologic failure in participants randomized to integrase inhibitor-based ART.17 Given that NNRTI mutations do not confer cross-resistance to dolutegravir and next-generation sequencing in the ADVANCE Trial identified very few participants with pretreatment NRTI resistance, we speculate that poor adherence by participants to their first ART regimen may have persisted during the ADVANCE Trial and led to virologic failure despite dolutegravir-based ART. These observations suggest that delayed initiation of ART until the late stages of infection14 and poor ART adherence17 may pose greater threats to ART-suppression than pretreatment drug-resistance.
Our findings that drug-resistant variants at both OLA and non-OLA codons and at high and low frequencies were associated with virologic failure suggest a continued utility for both rapid point-mutation and ultrasensitive sequencing-based HIV drug-resistance genotyping assays to guide ART prescription. However, we also noted that the majority of participants harboring K103N alone, even when present at high frequencies in the viral quasispecies, maintained virologic suppression throughout 24 months of efavirenz-based ART. We and others have reported this phenomenon previously9,14,18,19, and while in the current study our sample size was limited, this finding suggests that resistance at this codon alone may pose little risk of virologic failure. In contrast, another study of efavirenz-based ART observed an association between pretreatment drug-resistance with K103N and virologic failure20, although the association in this study appears to be more frequent when efavirenz is combined with zidovudine compared to tenofovir; as was observed in our18 and others19 studies.
This study faces several limitations. First, the small number of participants with pretreatment majority and minority drug-resistance mutations limited nuanced analysis of the impact of variant frequency on the risk and timing of virologic failure. Second, effects of mutations at specific codons, single versus dual-class resistance, and Stanford resistance penalty score on the risk of virologic failure were similarly limited. Third, amplification of two separate regions within pol encoding reverse transcriptase prevented our assessment of linkages between some drug-resistant mutations, particularly between variants encoding resistance to multiple drug classes. Amplicon regions were assessed for variant frequencies separately, and while we may have been able to infer linkages for variants with majority frequencies, we could not always assess whether low-frequency variants were on the same viral templates. In a recently published study, linked dual-class pretreatment resistant variants on the same viral genome were associated with virologic failure of nevirapine-based ART; however, linked single class resistant variants were not associated with virologic failure.21 Inability to examine the effects of linked mutations was recognized as a major challenge for practical application of next-generation sequencing for drug-resistance genotyping during the recent Winnipeg Consensus.13 Finally, while we sought to mitigate PCR-induced errors as well as quantify the number of viral templates sequenced by use of unique molecular identifiers during the reverse transcription step of our workflow, we cannot dismiss the potential for nucleotide incorporation errors during reverse transcription or the early cycles of PCR, which could alter variant frequencies.
This study of a population with a low rate of virologic failure during the first two years of efavirenz-based ART revealed that either low or high frequency pretreatment drug-resistance contributes to virologic failure and usually within the first year of ART. Additionally, pretreatment resistance to a single antiretroviral class, with Stanford scores ≤60 or >60 and at OLA codons, or to a lesser degree at non-OLA codons were independently associated with virologic failure. These findings suggest that when treatment with efavirenz-based ART is considered, screening for pretreatment drug-resistance by an assay capable of detecting low-frequency variants, including OLA, may guide clinicians to prescribe more effective ART.
Supplementary Material
Supplemental Table 1. Pretreatment drug-resistant variants detected by next-generation sequencing among participants with and without virologic failure at 24-months of efavirenz-based ART.
Acknowledgements
The principal contributions made by each of the authors are as follows:
The study was conceived and led by LMF, MHC and facilitated by SRS and JNK; participants were enrolled and followed CK and NY; with drug resistance by OLA determined by IAB, ML, IS, NA, JK, RSM and NGS conducted by RSM; with NGS data derived by RSM; data analyzed by RSM, IAB, NP, WD, LMF; manuscript prepared by RSM, IAB and LMF; and reviewed by all authors.
Sources of Support:
This study was supported by National Institutes of Health awards R01 AI100037 (LMF) and R01 AI100037 Administrative Supplement (LMF), the President’s Emergency Plan for AIDS Relief cooperative agreement U62/CCU024512 (MHC) from the Centers for Disease Control and Prevention to fund Coptic Hope Center for Infectious Diseases, the Clinical and Retrovirology Research Core and the Molecular Profiling and Computational Biology Core of the University of Washington Fred Hutch Center for AIDS Research [P30 AI027757]. In addition, we appreciate the expertise of Richard D’Aquila, James Hughes, and Robert Shafer for participating on the study’s Data Safety and Monitoring Board and Joseph Fitzgibbon and Catherine Godfrey, National Institutes of Health Program Officers, for valuable contributions to the study.
Footnotes
Conflicts of Interest:
No author declared a conflict of interest.
References
- 1.Silverman RA, Beck IA, Kiptinness C, et al. Prevalence of Pre-antiretroviral-Treatment Drug Resistance by Gender, Age, and Other Factors in HIV-Infected Individuals Initiating Therapy in Kenya, 2013–2014. J Infect Dis. 2017;216(12):1569–1578. doi: 10.1093/infdis/jix544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gupta RK, Gregson J, Parkin N, et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis. 2018;18(3):346–355. doi: 10.1016/S1473-3099(17)30702-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Update of Recommendations on First- and Second-Line Antiretroviral Regimens. World Health Organization; 2019. [Google Scholar]
- 4.Kouamou V, Mavetera J, Manasa J, et al. Pretreatment HIV Drug Resistance Among Adults Initiating or Re-Initiating First-Line Antiretroviral Therapy in Zimbabwe: Fast-Tracking the Transition to Dolutegravir-Based First-Line Regimens? AIDS Res Hum Retroviruses. 2021. Feb 5. doi: 10.1089/AID.2020.0242. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 5.Kantor R, Smeaton L, Vardhanabhuti S, et al. Pretreatment HIV Drug Resistance and HIV-1 Subtype C Are Independently Associated With Virologic Failure: Results From the Multinational PEARLS (ACTG A5175) Clinical Trial. Clin Infect Dis Off Publ Infect Dis Soc Am. 2015;60(10):1541–1549. doi: 10.1093/cid/civ102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305(13):1327–1335. doi: 10.1001/jama.2011.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F, et al. Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother. 2015;70(3):930–940. doi: 10.1093/jac/dku426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nicot F, Sauné K, Raymond S, et al. Minority resistant HIV-1 variants and the response to first-line NNRTI therapy. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2015;62:20–24. doi: 10.1016/j.jcv.2014.10.020 [DOI] [PubMed] [Google Scholar]
- 9.Derache A, Iwuji CC, Baisley K, et al. Impact of Next-generation Sequencing Defined Human Immunodeficiency Virus Pretreatment Drug Resistance on Virological Outcomes in the ANRS 12249 Treatment-as-Prevention Trial. Clin Infect Dis Off Publ Infect Dis Soc Am. 2019;69(2):207–214. doi: 10.1093/cid/ciy881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mbunkah HA, Bertagnolio S, Hamers RL, et al. Low-Abundance Drug-Resistant HIV-1 Variants in Antiretroviral Drug-Naive Individuals: A Systematic Review of Detection Methods, Prevalence, and Clinical Impact. J Infect Dis. 2020;221(10):1584–1597. doi: 10.1093/infdis/jiz650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee ER, Parkin N, Jennings C, et al. Performance comparison of next generation sequencing analysis pipelines for HIV-1 drug resistance testing. Sci Rep. 2020;10(1):1634. doi: 10.1038/s41598-020-58544-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inzaule SC, Hamers RL, Noguera-Julian M, et al. Clinically relevant thresholds for ultrasensitive HIV drug resistance testing: a multi-country nested case-control study. Lancet HIV. 2018;5(11): e638–e646. doi: 10.1016/S2352-3018(18)30177-2 [DOI] [PubMed] [Google Scholar]
- 13.Ji H, Sandstrom P, Paredes R, et al. Are We Ready for NGS HIV Drug Resistance Testing? The Second “Winnipeg Consensus” Symposium. Viruses. 2020;12(6). doi: 10.3390/v12060586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung MH, McGrath CJ, Beck IA, et al. Evaluation of the management of pretreatment HIV drug resistance by oligonucleotide ligation assay: a randomised controlled trial. Lancet HIV. 2020;7(2):e104–e112. doi: 10.1016/S2352-3018(19)30337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu TF, Shafer RW(2006). Web Resources for HIV type 1 Genotypic-Resistance Test Interpretation. Clin Infect Dis 42(11):1608–18. Epub 2006 Apr 28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siedner MJ, Moorhouse MA, Simmons B, et al. Reduced efficacy of HIV-1 integrase inhibitors in patients with drug resistance mutations in reverse transcriptase. Nat Commun. 2020. Dec 1;11(1):5922. doi: 10.1038/s41467-020-19801-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beck IA, Levine M, McGrath CJ, et al. Pre-treatment HIV-drug resistance associated with virologic outcome of first-line NNRTI-antiretroviral therapy: A cohort study in Kenya. EClinicalMedicine. 2020. Jan 14;18:100239. doi: 10.1016/j.eclinm.2019.100239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodman DD, Zhou Y, Margot NA, McColl DJ, Zhong L, Borroto-Esoda K, et al. Low level of the K103N HIV-1 above a threshold is associated with virological failure in treatment-naive individuals undergoing efavirenz-containing therapy. AIDS 2011;25(3):325–33. [DOI] [PubMed] [Google Scholar]
- 20.Kantor R, Smeaton L, Vardhanabhuti S, et al. ; AIDS Clinical Trials Group A5175 Study Team. Pretreatment HIV drug resistance and HIV-1 subtype C are independently associated with virologic failure: results from the multinational PEARLS (ACTG A5175) clinical trial. Clin Infect Dis 2015; 60:1541–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boltz VF, Shao W, Bale MJ, et al. Linked dual-class HIV resistance mutations are associated with treatment failure. JCI Insight. 2019;4(19). doi: 10.1172/jci.insight.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Pretreatment drug-resistant variants detected by next-generation sequencing among participants with and without virologic failure at 24-months of efavirenz-based ART.