Graphical Abstract
Summary: Wooden boards were obtained from 3 cheese-processing facilities and assessed for bacterial diversity. Samples were obtained from the surface and at depths of 0.2 and 0.9 cm. DNA was extracted and amplified by PCR and 16S rRNA analysis was performed to identify bacterial communities present on the surface and within the boards. Although there was board-to-board variation in both diversity and richness, all boards contained bacteria in the phyla Actinobacteria, Firmicutes, and Proteobacteria. In general, the boards selected for saltand cold-tolerant bacteria, with 7 operational taxonomic units (Staphylococcus, Brevibacterium, Psychrobacter, Brachybacterium, Jeotgalicoccus, Nocardiopsaceae, and Yaniella) forming a core microbiota found on all boards.
Highlights
-
•
The phyla Actinobacteria, Firmicutes, and Proteobacteria dominated the microbiota of the boards.
-
•
The boards displayed differences in both diversity and richness.
-
•
We identified 288 total operational taxonomic units (OTU), with 7 OTU forming a core microbiota across all boards.
-
•
The boards appeared to select for salt- and cold-tolerant bacteria.
Abstract
Wooden boards are commonly used for aging artisan cheeses. Although considered critical to the development of desired flavors and aromas, knowledge about the microbial communities associated with these boards is limited. To begin to address this need, we performed a 16S ribosomal RNA analysis of the bacterial communities present on the surface and within 5 wooden boards used for cheese ripening that were obtained from 3 cheese-processing facilities. The 5 boards were dominated by bacteria in the phyla Actinobacteria, Firmicutes, and Proteobacteria and displayed differences in both diversity and richness. Analysis of these boards also identified significant board-to-board variation. A total of 288 operational taxonomic units were identified across all samples, with 7 operational taxonomic units forming a core microbiota across all boards. Taken together, these data reflect the cheese-ripening environment, which appears to select for salt- and cold-tolerant bacteria.
Wooden surfaces have been used for centuries for cheese-making and ripening. Although cheese-makers believe the porous structure of wood promotes a rich microbial community that contributes to the desirable organoleptic characteristics of cheese (as reviewed by Lortal et al., 2014), food safety concerns regarding the use of wood have periodically been raised due to perceived difficulty in maintaining cleanliness. However, previous studies of wood food-contact surfaces suggest that this might not be the case. For example, Ak et al. (1994a,b) compared the recovery of foodborne pathogens (Salmonella and Listeria monocytogenes) from wooden and plastic boards and recovered lower quantities of pathogens from wood than from plastic. A possible explanation was provided by Gough and Dodd (1998), who reported that bacterial cells were trapped and possibly inactivated more strongly in wood than in plastic boards.
Although cheese aged on wood had not been incriminated in any foodborne disease outbreaks (as reviewed in Aviat et al., 2016, and Lortal et al., 2014), in 2014 the United States Food and Drug Administration temporarily prohibited the centuries-old practice of aging cheese on wooden boards. This was met with strong resistance from cheese-makers across the United States, and the decision was soon reversed. However, questions remain regarding the use of wooden boards in aging cheese, in part because we have little detailed information about the microbiota residing on these wooden boards.
Several sampling methods have been used to explore the microflora on wooden cheese-ripening boards. These include the use of a sonicator to remove biofilms from the surface of the board (Mariani et al., 2007), swabs (Mounier et al., 2006), agar plates pressed with wooden blocks derived from ripening boards (Ak et al., 1994a), and sampling from ground or planned ripening board wood (Ismaïl et al., 2015). Importantly, the latter study found that grinding was the most reliable method for recovering bacteria from poplar, pine, and spruce wood surfaces, with an average recovery of 30% for Lis. monocytogenes on spruce and Escherichia coli on poplar. However, none of the above studies sampled wood at various depths or used modern RNA sequencing technologies to characterize the resident microbiota of wooden cheese-ripening boards.
The advent of high-throughput sequencing technologies makes it possible to explore the entire microbiota at various stages or environmental sites of the cheese-making process (Bokulich and Mills, 2012; Wolfe and Dutton, 2013). Mounier et al. (2006) investigated the microbiota on the hands of cheese-making personnel and on wooden and stainless-steel shelves used for cheese ripening. These authors found that Corynebacterium spp. and Staphylococcus saprophyticus were the dominant bacteria on both types of shelving and on the skin of cheese-making personnel. Quigley et al. (2012, 2013) used 16S rRNA sequencing to characterize the microbiota of raw and pasteurized milk and found several bacterial genera not previously associated with milk (Faecalibacterium, Prevotella, and Catenibacterium). More recently, Guzzon et al. (2017) and Quijada et al. (2018) included swab samples of wooden shelves in 16S rRNA gene analyses of the microbiota during production of specific cheeses.
Bokulich and Mills (2013) compared 2 cheese-processing facilities and found that in one plant the microbiota was dominated by gram-positive bacteria in the genera Brevibacterium, Psychrobacter, Brachybacterium, Corynebacterium, and Staphylococcus, whereas in the second plant the microbiota was dominated by gram-negative bacteria in the genera Pseudoalteromonas, Psychrobacter, and Vibrio. Moreover, certain microbial communities were associated with specific areas of the processing plants. For example, in both facilities, cheese-maturation rooms were dominated by bacteria in the genera Brevibacterium, Staphylococcus, Corynebacterium, and Halomonas. These genera were also recovered from cheese produced by the plants, leading the authors to suggest a possible “house microflora” characteristic of cheese-making plants (Bokulich and Mills, 2013).
Because wooden boards are a popular choice for the ripening of cheeses, there is a need to characterize their microbiota. To begin to address this issue, we used 16S ribosomal RNA targeted amplicon sequence analysis to explore the hypothesis that wooden boards used for cheese ripening have a complex microbiota that differs on the surface relative to microbiota within the wood. Gaining an understanding of the wooden board cheese-ripening microbiota will provide new insights into the potential roles these communities play in both the safety of artisanal cheeses and the development of their desirable characteristics.
Five wooden boards were obtained from 3 Wisconsin cheese-making facilities immediately after removal of cheese and transported to our laboratory. Boards A (cedar) and B (pine) came from one facility, board C (spruce) came from a second facility, and boards D and E (both spruce) came from a third facility. The dimensions of each board (length × breadth × height, in cm) were as follows: A, 104.5 × 14.0 × 2.5; B, 92.5 × 18.0 × 1.8; C, 122.6 × 29.0 × 1.9; D, 31.5 × 29.0 × 1.9; and E, 44.5 × 18.5 × 1.8. Boards A and B were used for aging surface-ripened cheese, as was board C. Board D was used for cave-ripening cheese, and board E was used for smear-ripened cheese.
Samples were collected from (1) the surface of all 5 boards; (2) the surface to a depth of 0.2 cm and at a depth of 0.7 to 0.9 cm from boards A, B, and C; and (3) from the surface to a depth of 0.2 cm for boards D and E. Surface samples were collected from a 64-cm2 area using a sterile cell scraper (Biologix Technologies Inc.) and cotton-tipped applicators (Fisher Scientific). Wood shavings from the surface to 0.2 cm and from a depth of 0.7 to 0.9 cm were collected using a sanitized 12.7-mm drill bit attached to a cordless drill (Dewalt, 20-Volt Max lithium-ion). The bit was sanitized with 70% EtOH between each sample collection. All samples were suspended in 25 mL of sterile PBS and stored at −80°C.
Samples were thawed at room temperature before total DNA extraction as described previously (Stevenson and Weimer, 2007; Li et al., 2019). Briefly, 1 mL of each sample (wood shavings with sterile PBS) was transferred to a 2-mL screw-cap tube with 0.5 g of 0.1-mm zirconium beads (Biospec Products). A total of 50 μL of 20% SDS and 700 μL of cold equilibrated phenol were added to each tube and subjected to bead beating for 2 min on a tabletop bead beater (Biospec Products). The mixture was then heated in a 60°C water bath for 10 min followed by 2 min of additional bead beating. Tubes were then centrifuged for 10 min at 4°C on a tabletop centrifuge (Beckman Coulter) at 9,350 × g for 10 min at 4°C. The aqueous layer was washed 2 to 4 times with 500 μL of cold equilibrated phenol:chloroform:isoamyl alcohol (25:24:1) to remove the lipid layer, and then DNA was precipitated overnight with isopropanol and 2 M sodium acetate at −20°C. The DNA was then pelleted, washed twice with 70% ethanol, and dried overnight in a fume hood. Pellets were resuspended in 20 µL of elution buffer (Thermo Fisher Scientific), quantified using a Qubit 2.0 Fluorometer (Invitrogen), and stored at 4°C.
Universal primers flanking variable region 4 of the bacterial 16S rRNA coding region, along with Illumina-specific sequencing adapters and dual-index barcodes, were used to perform PCR for each sample (Kozich et al., 2013). The PCR reactions were conducted with the following composition: 5 ng of template DNA, 0.5 µL of each primer (forward and reverse), 12.5 μL of 2× Hot Start Ready Mix (Kapa Biosystems), and nuclease-free water were added for a total volume of 25 µL. The PCR cycle conditions were as follows: initial denaturation of 95°C for 3 min; 25 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. Negative controls consisting of the same PCR reactions with water substituted for the DNA template were also performed with each PCR run. The PCR products were then assessed by gel electrophoresis on a 1.0% low-melt agarose gel (National Diagnostics). Bands at 350 bp were cut from each gel, placed into a 96-well collection plate, and purified using a Zymoclean Gel DNA Recovery Kit (Zymo Research). Purified DNA was quantified using a Qubit fluorometer, equimolar pooled, and combined with a 10% PhiX control. Samples were then sequenced at the University of Wisconsin–Madison Biotechnology Center on an Illumina MiSeq using a 2 × 250 v2 sequencing kit (Illumina) with custom sequencing primers as described by Li et al. (2019) and Kozich et al. (2013). Raw sequences were deposited into the NCBI's Short Read Archive and are publicly available under BioProject accession PRJNA648954.
All sequences were demultiplexed on the Illumina MiSeq and processed using mothur v.1.40.5 (Schloss et al., 2009) following a protocol adapted from Kozich et al. (2013). Briefly, paired-end sequences were assembled into continuous segments and poor-quality sequences were removed. The remaining sequences were aligned to the SILVA 16S rRNA gene reference database v132 (Pruesse et al., 2007), and sequences that did not align to variable region 4 were removed. Preclustering was performed (diffs = 2) to reduce error and computational load, and chimeric sequences were detected using UCHIME (Edgar et al., 2011) and removed. Sequences were taxonomically classified to the SILVA database with a bootstrap value cutoff of 80. Sequences classified to Cyanobacteria, mitochondria, eukarya, or archaea were removed. Bacterial sequences were grouped into operational taxonomic units (OTU) at a 97% sequence similarity using the opticlust method and taxonomically classified using the SILVA database. Sample coverage, as determined using Good's index (Good and Toulmin, 1956), was calculated and samples with <95% coverage were removed. The OTU counts were normalized to 7,336 sequences per sample, which represents the lowest number of sequences in our sample set.
All statistical analyses were performed in R (version 3.6.1; https://www.r-project.org/). α-Diversity was determined using Shannon's diversity and Chao's species richness estimators for all samples after validating the normal distribution of each sample using the Shapiro-Wilk test. Pairwise comparisons of α-diversity were performed using Tukey's honestly significant difference test. For β-diversity, Bray-Curtis dissimilarity (community structure) and Jaccard (community composition) metrics were calculated and visualized using metaMDS (vegan package; Oksanen et al., 2018).
A total of 17 samples were collected from the 5 wooden boards used in our study: 7 from the surface, 7 at a depth of 0.2 cm, and 3 at a depth of 0.9 cm. In total, we generated 443,768 raw bacterial sequences, of which 369,876 were considered high quality after filtering. As a result, we obtained an average of 7,336 ± 5.9 sequences per sample, which were grouped into 288 OTU. Satisfactory coverage was reached for all samples (>99%), as assessed using Good's index. The greatest number of OTU was recovered from the surface of board C (sample C.s), and the greatest number of subsurface OTU was recovered from board C at a depth of 0.2 cm. Shannon's diversity index ranged from 0.87 (board E at 0.2 cm) to 3.34 (board C at the surface), and the Chao1 richness estimator ranged from 24 (board A surface) to 213 (board C surface).
Surface samples from board C had the most diverse phyla and the greatest relative abundance of Proteobacteria. Actinobacteria was the dominant phylum at all depths for boards A and B. Firmicutes and Actinobacteria were the most abundant phyla for boards D and E. Figure 1 illustrates the most abundant genera among all samples collected. For board A, Brevibacterium (⁓63%) was the predominant genus, followed by Brachybacterium (⁓11%), Staphylococcus (⁓16%), Jeotgalicoccus (<1%), Nocardiopsis (<1%), and Corynebacterium (<1%). For board B (obtained from the same facility as board A), Brevibacterium (⁓46%), Brachybacterium (⁓11%), and Nocardiopsis (⁓30%) were the most abundant genera. Board C differed substantially from boards A and B as the most abundant surface genera were Psychrobacter (⁓42%), Staphylococcus (⁓11%), Brevibacterium (⁓23%), Brachybacterium (⁓23%), and Corynebacterium (<1%).
Figure 1.
Relative abundance of the 10 most abundant genera across all samples collected from wooden cheese-ripening boards as assessed using 16S rRNA sequencing. The x-axis labels indicate board (A–E), depth [surface (s), 0.2 cm, or 0.9 cm], and sample number (1 or 2).
In general, surface samples had greater diversity than those at depths of 0.2 cm or 0.7 to 0.9 cm; the exception was board D, where diversity was slightly higher at 0.2 cm (Shannon's diversity index of 2.01 vs. 1.82 at the surface). Surface diversity was greatest at the surface versus 0.2 or 0.9 cm for board C, where Shannon's diversity index ranged from 3.34 (surface) to 1.00 (0.9 cm). For boards D and E, Staphylococcus (⁓50%) was the most abundant genus, with Corynebacterium being more abundant on board E (12%) than on board D (<1%; Figure 1). Other abundant genera from board D included Brachybacterium (⁓11%), Nocardiopsis (⁓11%), Jeotgalicoccus (<1%), and Brevibacterium (⁓21%). Other genera identified from board E included Psychrobacter (⁓2%), Weissella (⁓6%), and Brevibacterium (⁓1%). Besides the most abundant genera illustrated in Figure 1, the genus Haererehalobacter was also abundant on the surface of board C (⁓10%). Overall, these data support the existence of a “house” microflora in different cheese-making facilities, as has been suggested in previous studies (Mounier et al., 2006; Bokulich and Mills, 2013).
To better understand the microbiota of these boards, we pooled samples collected from the same board (surface, 0.2 cm, and 0.7–0.9 cm) for further analysis. Overall, the boards did not differ significantly from one another with respect to both diversity and richness (P > 0.05). α-Diversity was lowest for board A and greatest for board C (not shown). Variation among samples, assessed using both Bray-Curtis dissimilarity and Jaccard metrics, indicated significant differences (P < 0.05) among boards (Figure 2). Pairwise comparisons using an Adonis analysis found boards A and D, A and E, B and D, B and E, C and E, and D and E to be significantly different from each other (P < 0.05).
Figure 2.

β-Diversity of boards (A–E) as assessed using (a) Bray-Curtis dissimilarity and (b) Jaccard similarity analysis and visualized using nonmetric multidimensional scaling (NMDS). Ellipses represent standard error of all samples collected for each respective cheese-ripening board.
We next sought to determine whether a core microbiota existed among all boards and samples in our study, and we identified a core bacterial community of 7 OTU (Figure 3). Six of these OTU were relatively abundant among all samples (Staphylococcus, Brevibacterium, Psychrobacter, Brachybacterium, Nocardiopsaceae, and Jeotgalicoccus); the exception was Yaniella (<1% relative abundance among all samples). We also identified 2 OTU (Dietzia and Prevotella) on boards collected from all 3 cheese-processing facilities, although not necessarily on all boards from the same facilities.
Figure 3.
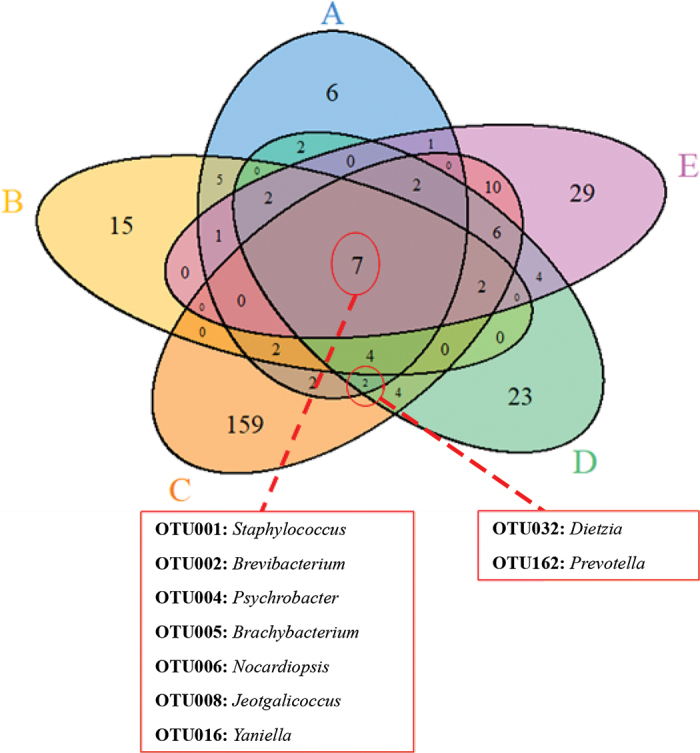
Venn diagram of the 288 operational taxonomic units (OTU) recovered from the 5 wooden boards. A core microbial community of 7 genera was common to all samples as indicated. Two additional OTU were found on at least one board from the 3 facilities (boards A, C, and D).
The results from this study agree with previous reports of prevalent bacterial genera identified in other cheese-processing facilities (Montel et al., 2014; Guzzon et al., 2017; Frétin et al., 2018; Quijada et al., 2018; Falardeau et al., 2019). Brevibacterium was part of the core microbiota in our study. This is expected because Brevibacterium is often abundant on cheese rind as it is involved in flavor, aroma, and color development during cheese ripening (Frétin et al., 2018). In a separate 16S rRNA gene study, Brevibacterium was abundant on the rind of an artisanal washed-rind hard cheese, although it was less abundant on wood shelves later in the aging process (Quijada et al., 2018).
We also found Staphylococcus in relatively high abundance on all 5 wooden boards. Comparison of a 253-bp sequence for this OTU against NCBI's GenBank revealed 100% sequence identity with the reference genome for Staphylococcus equorum KS1039 (data not shown) (Jeong et al., 2016). Staphylococcus equorum is frequently found in fermented food products and the food-processing environment, where it is thought to contribute to the formation of aroma during cheese ripening (Irlinger et al., 2012), particularly for smear-ripened cheese (Place et al., 2002). Other genera identified in our analysis, such as Haererehalobacter and Salinicoccus (Figure 1; Hyun et al., 2013; Birdilla Selva Donio et al., 2018), are halotolerant microorganisms, whose relative abundance in surface samples is commensurate with the high salt concentrations found on cheeses during the ripening process.
Finally, we found the psychrophilic genus Psychrobacter (Figure 1) present on all 5 wooden boards. Previous work has identified this genus in the cheese-making environment (Falardeau et al., 2019), particularly during cheese aging, which is characterized by cooler temperatures. The greatest abundance of Psychrobacter was found on board D, which was obtained from a cave used for cheese ripening that is consistent with cool temperature.
Bacteria from the genera Carnobacterium, Lactobacillus, Lactococcus, Leuconostoc, Nocardiopsis, and Yaniella were found on most boards in our study (Figure 1). The function of Nocardiopsis and Yaniella in the cheese-making environment is unknown, but these genera have been reported to be present in other food environments (Wolfe et al., 2014; Frétin et al., 2018). Traditionally, bacteria in the genera Lactobacillus and Lactococcus are associated with starter culture during the cheese-making process, and their presence on these boards is not unexpected. Carnobacterium spp. is known to have some ability to reduce the growth of a common cheese contaminant (i.e., Pseudomonas) when inoculated into ricotta cheese (Spanu et al., 2018).
Our results are similar to those of 2 previous studies that also performed 16S rRNA gene analyses of the surface of wooden shelves. Guzzon et al. (2017) found a very diverse microbiota on spruce shelves used to age raw milk Fontina cheese, in which Brevibacterium, Corynebacterium, and Aequorivita were in greatest relative abundance. These authors also demonstrated that cleaning boards with hot water or wet ozone greatly reduced the bacterial and fungal microbiota. Quijada et al. (2018) assessed the microbiota of an Austrian raw cow milk salt-brined cheese and sampled the environment at various stages in the cheese-making and aging process. Proteobacteria were in greater relative abundance on wooden shelves than cheese rind, where Actinobacteria were in greater relative abundance. The microbiota of shelves changed with time, as initially Brevibacterium, Staphylococcus (including Staph. equorum), and Psychrobacter were in greatest relative abundance. As cheese aged in the cellar, Comamonas and Pseudomonas came to predominate the shelf microbiota.
In conclusion, this study represents an initial survey of the bacterial communities on wooden boards used for cheese ripening. We acknowledge that the small sample size of our study is a limitation. However, we believe our findings provide a framework to guide future work in this field. Our study identified both abundant genera known previously to be present in cheese facilities and less common genera (e.g., Dietzia and Prevotella) that currently have no known roles in the cheese-making process. Future work should include greater and more detailed bacterial (16S rRNA) and fungal (18S rRNA) analyses. Such efforts will provide the cheese industry a more comprehensive view of the microbial communities present on wooden boards during cheese ripening. Finally, we note that we neither detected 16S rRNA signal for the foodborne pathogen Lis. monocytogenes nor recovered Listeria colonies on MOX (modified Oxford agar) plates, for any of our samples. Although we did not perform enrichment and cannot completely exclude the possible presence of low numbers of Lis. monocytogenes, our findings raise the possibility that the microbial communities present on wooden boards might be deleterious to the survival and multiplication of Lis. monocytogenes. Additional work is required to investigate this hypothesis.
Notes
This work was supported by a USDA National Institute of Food and Agriculture (Washington, DC) Hatch grant WIS02007 to G. Suen, the University of Wisconsin–Madison Food Research Institute, and the Walter and Martha Renk Endowed Laboratory for Food Safety (University of Wisconsin–Madison) to C. Czuprynski.
The authors have not stated any conflicts of interest.
References
- Ak N.O., Cliver D.O., Kaspar C.W. Cutting boards of plastic and wood contaminated experimentally with bacteria. J. Food Prot. 1994;57:16–22. doi: 10.4315/0362-028X-57.1.16. 31113021. [DOI] [PubMed] [Google Scholar]
- Ak N.O., Cliver D.O., Kaspar C.W. Decontamination of plastic and wooden cutting boards for kitchen use. J. Food Prot. 1994;57:23–30. doi: 10.4315/0362-028X-57.1.23. 31113026. [DOI] [PubMed] [Google Scholar]
- Aviat F., Gerhards C., Rodriguez-Jerez J., Michel V., Bayon I.L., Ismail R., Federighi M. Microbial safety of wood in contact with food: A review. Compr. Rev. Food Sci. Food Saf. 2016;15:491–505. doi: 10.1111/1541-4337.12199. 33401823. [DOI] [PubMed] [Google Scholar]
- Birdilla Selva Donio M., Chelladurai Karthikeyan S., Michaelbabu M., Uma G., Raja Jeya Sekar R., Citarasu T. Haererehalobacter sp. JS1, a bioemulsifier producing halophilic bacterium isolated from Indian solar salt works. J. Basic Microbiol. 2018;58:597–608. doi: 10.1002/jobm.201800056. 29775199. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Mills D.A. Next-generation approaches to the microbial ecology of food fermentations. BMB Rep. 2012;45:377–389. doi: 10.5483/BMBRep.2012.45.7.148. 22831972. [DOI] [PubMed] [Google Scholar]
- Bokulich N.A., Mills D.A. Facility-specific “house” microbiome drives microbial landscapes of artisan cheesemaking plants. Appl. Environ. Microbiol. 2013;79:5214–5223. doi: 10.1128/AEM.00934-13. 23793641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C., Haas B.J., Clemente J.C., Quince C., Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. 21700674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau J., Keeney K., Trmčić A., Kitts D., Wang S. Farm-to-fork profiling of bacterial communities associated with an artisan cheese production facility. Food Microbiol. 2019;83:48–58. doi: 10.1016/j.fm.2019.04.002. 31202418. [DOI] [PubMed] [Google Scholar]
- Frétin M., Martin B., Rifa E., Isabelle V.-M., Pomiès D., Ferlay A., Montel M.-C., Delbès C. Bacterial community assembly from cow teat skin to ripened cheeses is influenced by grazing systems. Sci. Rep. 2018;8:200. doi: 10.1038/s41598-017-18447-y. 29317671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good I.J., Toulmin G.H. The number of new species, and the increase in population coverage, when a sample is increased. Biometrika. 1956;43:45–63. doi: 10.1093/biomet/43.1-2.45. [DOI] [Google Scholar]
- Gough N.L., Dodd C.E.R. The survival and disinfection of Salmonella typhimurium on chopping board surfaces of wood and plastic. Food Control. 1998;9:363–368. doi: 10.1016/S0956-7135(98)00127-3. [DOI] [Google Scholar]
- Guzzon R., Carafa I., Tuohy K., Cervantes G., Vernetti L., Barmaz A., Larcher R., Franciosi E. Exploring the microbiota of the red-brown defect in smear-ripened cheese by 454-pyrosequencing and its prevention using different cleaning systems. Food Microbiol. 2017;62:160–168. doi: 10.1016/j.fm.2016.10.018. 27889144. [DOI] [PubMed] [Google Scholar]
- Hyun D.-W., Whon T.W., Cho Y.-J., Chun J., Kim M.-S., Jung M.-J., Shin N.R., Kim J.Y., Kim P.S., Yun J.H., Lee J., Oh S.J., Bae J.W. Genome sequence of the moderately halophilic bacterium Salinicoccus carnicancri type strain CrmT (= DSM 23852T) Stand. Genomic Sci. 2013;8:255–263. doi: 10.4056/sigs.3967649. 23991257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irlinger F., Loux V., Bento P., Gibrat J.-F., Straub C., Bonnarme P., Landaud S., Monnet C. Genome sequence of Staphylococcus equorum ssp. equorum Mu2, isolated from a French smear-ripened cheese. J. Bacteriol. 2012;194:5141–5142. doi: 10.1128/JB.01038-12. 22933766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismaïl R., Le Bayon I., Michel V., Jequel M., Kutnik M., Aviat F., Fédérighi M. Comparative study of three methods for recovering microorganisms from wooden surfaces in the food industry. Food Anal. Methods. 2015;8:1238–1247. doi: 10.1007/s12161-014-0008-3. [DOI] [Google Scholar]
- Jeong D.W., Na H., Ryu S., Lee J.H. Complete genome sequence of Staphylococcus equorum KS1039 isolated from Saeu-jeotgal, Korean high-salt-fermented seafood. J. Biotechnol. 2016;219:88–89. doi: 10.1016/j.jbiotec.2015.12.025. 26718562. [DOI] [PubMed] [Google Scholar]
- Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. 23793624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Edwards A., Riehle C., Cox M.S., Raabis S., Skarlupka J.H., Steinberger A.J., Walling J., Bickhart D., Suen G. Transcriptomics analysis of host liver and meta-transcriptome analysis of rumen epimural microbial community in young calves treated with artificial dosing of rumen content from adult donor cow. Sci. Rep. 2019;9:790. doi: 10.1038/s41598-018-37033-4. 30692556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lortal S., Licitra G., Valence F. Wooden tools: Reservoirs of microbial biodiversity in traditional cheesemaking. Microbiol. Spectr. 2014;2:167–176. doi: 10.1128/9781555818593.ch8. 26068076. [DOI] [PubMed] [Google Scholar]
- Mariani C., Briandet R., Chamba J.F., Notz E., Carnet-Pantiez A., Eyoug R.N., Oulahal N. Biofilm ecology of wooden shelves used in ripening the French raw milk smear cheese Reblochon de Savoie. J. Dairy Sci. 2007;90:1653–1661. doi: 10.3168/jds.2006-190. 17369205. [DOI] [PubMed] [Google Scholar]
- Montel M.-C., Buchin S., Mallet A., Delbes-Paus C., Vuitton D.A., Desmasures N., Berthier F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014;177:136–154. doi: 10.1016/j.ijfoodmicro.2014.02.019. 24642348. [DOI] [PubMed] [Google Scholar]
- Mounier J., Goerges S., Gelsomino R., Vancanneyt M., Vandemeulebroecke K., Hoste B., Brennan N., Scherer S., Swings J., Fitzgerald G., Cogan T. Sources of the adventitious microflora of a smear-ripened cheese. J. Appl. Microbiol. 2006;101:668–681. doi: 10.1111/j.1365-2672.2006.02922.x. 16907817. [DOI] [PubMed] [Google Scholar]
- Oksanen J., Blanchet F.G., Friendly M., Kindt R., Legendre P., McGlinn D., Minchin P.R., O'Hara R.B., Simpson G.L., Solymos P., Henry M., Stevens H., Szoecs E., Wagner H. vegan: Community ecology package. R package version 2.4-6. 2018. https://cran.r-project.org/package=vegan
- Place R.B., Hiestand D., Burri S., Teuber M. Staphylococcus succinus ssp. casei ssp. nov., a dominant isolate from a surface ripened cheese. Syst. Appl. Microbiol. 2002;25:353–359. doi: 10.1078/0723-2020-00130. 12421073. [DOI] [PubMed] [Google Scholar]
- Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., Glöckner F.O. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. 17947321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley L., McCarthy R., O'Sullivan O., Beresford T.P., Fitzgerald G.F., Ross R.P., Stanton C., Cotter P.D. The microbial content of raw and pasteurized cow milk as determined by molecular approaches. J. Dairy Sci. 2013;96:4928–4937. doi: 10.3168/jds.2013-6688. 23746589. [DOI] [PubMed] [Google Scholar]
- Quigley L., O'Sullivan O., Beresford T.P., Ross R.P., Fitzgerald G.F., Cotter P.D. High-throughput sequencing for detection of subpopulations of bacteria not previously associated with artisanal cheeses. Appl. Environ. Microbiol. 2012;78:5717–5723. doi: 10.1128/AEM.00918-12. 22685131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada N.M., Mann E., Wagner M., Rodríguez-Lázaro D., Hernández M., Schmitz-Esser S. Autochthonous facility-specific microbiota dominates washed-rind Austrian hard cheese surfaces and its production environment. Int. J. Food Microbiol. 2018;267:54–61. doi: 10.1016/j.ijfoodmicro.2017.12.025. 29291459. [DOI] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: Open-source, platform-independent, community supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. 19801464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanu C., Piras F., Mocci A.M., Nieddu G., De Santis E.P.L., Scarano C. Use of Carnobacterium spp protective culture in MAP packed Ricotta fresca cheese to control Pseudomonas spp. Food Microbiol. 2018;74:50–56. doi: 10.1016/j.fm.2018.02.020. 29706337. [DOI] [PubMed] [Google Scholar]
- Stevenson D.M., Weimer P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007;75:165–174. doi: 10.1007/s00253-006-0802-y. 17235560. [DOI] [PubMed] [Google Scholar]
- Wolfe B.E., Button J.E., Santarelli M., Dutton R.J. Cheese rind communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–433. doi: 10.1016/j.cell.2014.05.041. 25036636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe B.E., Dutton R.J. Towards an ecosystem approach to cheese microbiology. Microbiol. Spectr. 2013;1:311–321. doi: 10.1128/microbiolspec.CM-0012-12. 26184819. [DOI] [PubMed] [Google Scholar]




