Graphical Abstract
Summary: The Australian Feed Saved estimated breeding value (EBV) includes 2 components: (1) body weight (BW) as an estimate of maintenance requirements, and (2) residual feed intake to identify metabolically efficient cows. We have updated the residual feed intake EBV using an expanded reference population of 3,711 Holstein cows using a multivariate model. Residual feed intake is combined with the BW EBV to calculate Feed Saved EBV. In 2015, Feed Saved was included in the Australian national selection indices. The updated 2020 model for the Feed Saved EBV has improved the reliability of Feed Saved by 10% compared with the 2015 model.
Highlights
-
•
The Feed Saved (FS) estimated breeding value (EBV) was updated by doubling the number of Australian and overseas cows.
-
•
The reliability of the residual feed intake component of FS has increased from 11% (2015 model) to 20% (current model).
-
•
The mean reliability of FS EBV in Holstein bulls that were born in the last 10 years has improved by 10%.
-
•
The genetic trend of FS EBV has been stabilizing since 2015.
Abstract
Although selection for increased milk production traits has led to a genetic increase in body weight (BW), the genetic gain in milk production has exceeded the gain in BW, so gross feed efficiency has improved. Nonetheless, greater gains may be possible by directly selecting for a measure of feed efficiency. Australia first introduced Feed Saved (FS) estimated breeding value (EBV) in 2015. Feed Saved combines residual feed intake (RFI) genomic EBV and maintenance requirements calculated from mature BW EBV. The FS EBV was designed to enable the selection of cows for reduced energy requirements with similar milk production. In this study, we used a reference population of 3,711 animals in a multivariate analysis including Australian heifers (AUSh), Australian cows (AUSc), and overseas cows (OVEc) to update the Australian EBV for lifetime RFI (i.e., a breeding value that incorporated RFI in growing and lactating cows) and to recalculate the FS EBV in Australian Holstein bulls (AUSb). The estimates of genomic heritabilities using univariate (only AUSc or AUSh) to trivariate (including the OVEc) analyses were similar. Genomic heritabilities for RFI were estimated as 0.18 for AUSc, 0.27 for OVEc, and 0.36 for AUSh. The genomic correlation for RFI between AUSc and AUSh was 0.47 and that between AUSc and OVEc was 0.94, but these estimates were associated with large standard errors (range: 0.18–0.28). The reliability of lifetime RFI (a component of FS) in the trivariate analysis (i.e., including OVEc) increased from 11% to 20% compared with the 2015 model and was greater, by 12%, than in a bivariate analysis in which the reference population included only AUSc and AUSh. By applying the prediction equation of the 2020 model, the average reliability of the FS EBV in 20,816 AUSb that were born between 2010 and 2020 improved from 33% to 43%. Previous selection strategies—that is, using the predecessor of the Balanced Performance Index (Australian Profit Ranking index) that did not include FS—have resulted in an unfavorable genetic trend in FS. However, this unfavorable trend has stabilized since 2015, when FS was included in the Balanced Performance Index, and is expected to move in a favorable direction with selection on Balanced Performance Index or the Health Weighted Index. Doubling the reference population, particularly by incorporating international data for feed efficiency, has improved the reliability of the FS EBV. This could lead to increased genetic gain for feed efficiency in the Australian industry.
Feed costs make up a large proportion of the variable and total costs on a dairy farm; therefore, improving production efficiency remains a key breeding objective. The dairy industry has seen tremendous gains in milk yield without a substantial increase in maintenance requirements, leading to an improvement in lifetime per cow gross efficiency (Pryce et al., 2018).
Feed efficiency in dairy cattle is often considered a measure of converting feed into additional volumes of milk solids (Berry and Crowley, 2013). This is often presented as a ratio trait, such as output (milk yield or weight gain) per unit of feed consumed. However, ratio traits may lead to unstable selection results (Gunsett, 1984). An alternative efficiency trait is residual feed intake (RFI), which is often defined as the difference in an animal's actual and predicted DMI, after adjusting for its body size, growth, and productivity. The RFI trait is of importance because it captures variation in activity, protein turnover, digestibility, and heat increment of fermentation (Herd et al., 2004). Pryce et al. (2015) proposed that combining maintenance requirements (through BW EBV) and metabolic efficiency (measured as RFI) might be a more suitable means to select for feed efficiency. This was defined as Feed Saved (FS).
The reference population used for the genomic prediction of RFI (that became part of the FS EBV) in 2015 was comparatively small (n = 2,036), including only 234 Australian cows, with the remainder being overseas cows. Since then, the number of Australian cows with genotypes and phenotypes for RFI has more than doubled. Access to a larger data set of overseas cows (OVEc) was enabled by participating in collaborations such as the Efficient Dairy Genome Project (EDGP; an international database including research herds from Europe and North America). This has provided an opportunity to further increase the size of the reference population and hence to update the FS EBV.
Compared with milk production traits [DataGene (https://datagene.com.au/) typically reports reliabilities of milk production traits of 78% for individuals without progeny or records], the reliability of FS is still low. The reliability of a new trait, such as feed efficiency, is likely to be lower than that of established traits because of data availability. However, achieving reliabilities of over 50% is desirable. To predict a priori the accuracy of genomic EBV for a specified reference population size, several studies (Goddard, 2009; Daetwyler et al., 2010) proposed deterministic approaches. MacLeod et al. (2014) demonstrated that the deterministic prediction of accuracy of genomic prediction closely matched the accuracies using genomic BLUP (GBLUP) in simulated data.
DataGene released the Australian FS EBV to the Australian dairy industry in 2015, representing the first implementation of this trait breeding value worldwide. The FS EBV was incorporated in 2 Australian national selection indices (the Balance Performance Index, BPI; and the Health Weighted Index, HWI), and these indices include traits aligned to profitability and farmer preferences (Byrne et al., 2016). Selecting animals based on FS EBV, especially as part of the BPI or HWI, is expected to reduce energy requirements for similar amounts of milk production. The concept of FS is now becoming popular in other countries (Van Raden et al., 2017; de Jong et al., 2019; Lidauer et al., 2019). For example, as a proxy for identifying metabolically efficient cows, the saved feed cost for maintenance index has been incorporated in the Dutch-Flemish total merit index (NVI; de Jong et al., 2019).
The main objectives of this study were (1) to estimate SNP effects for RFI from an enlarged reference population of 3,711 animals including Australian growing heifers (AUSc), Australian lactating cows (AUSc), and overseas cows (OVEc); (2) to recalculate the FS EBV and its reliability and compare with the 2015 EBV; and (3) to evaluate the correlated response of FS to selection in the current 21k genotyped AUSb.
The animals used in this study included 584 AUSc, 2,440 lactating OVEc [United States (n = 671) Canada (CAN, n = 473), Netherlands (NLD, n = 597), Denmark (n = 425), United Kingdom (GBR, n = 211), and Switzerland (CHE, n = 63)], and 824 AUSh (137 also recorded as cows and included in AUSc).
The phenotypes and genotypes of the OVEc, except NLD and GBR, were downloaded from the EDGP database; the NLD and GBR data were a part of the original data set used in the development of the 2015 FS EBV (Pryce et al., 2015) using the same data set as reported by de Haas et al. (2012a).
Full details of management of animals, diets fed, and collection of DMI and other data for NLD, GBR, AUSc, and AUSh were described in de Haas et al. (2012b), Williams et al. (2011), and Macdonald et al. (2014). The RFI phenotypes for AUSh were previously calculated as means of the difference in actual and predicted DMI that was measured over a 6- to 7-wk period of growth around 6 mo of age (Pryce et al., 2015):
| RFI = DMI – (mean + cohort + age + age2 + MBW + ΔBW), |
where DMI is the daily DMI, mean is the overall mean across population, cohort is cohort group, MBW is the means of BW, and ΔBW is change in BW. All AUSc had milk production traits (milk, fat, and protein yields), ECM, and DMI data available on most days over a 28-d period, starting at 5 DIM. The RFI phenotypes for AUSc were calculated as the average DMI over the 28-d experimental period using the following model as described in Pryce et al. (2015):
| RFI = DMI – (mean + cohort + parity + DIM + ECM + MBW + ΔBW), |
where ECM, MBW, and ΔBW are individual records of ECM, mean BW, and change in BW for DIM during the trial period, respectively; measurements were taken in the spring or summer months. The phenotypes of RFI for OVEc were calculated as the residual of the following model:
| RFI = DMI – (mean + parityST + DIM + HYS + AGEcalv + trial + ECM + BW + ΔBW), |
where parityST is parity by stage of lactation, HYS is herd-year-season of calving, AGEcalv is age of cows at calving fitted as a second-order orthogonal polynomial, trial is diet treatment, BW is body weight measurements recorded over the test period, and all other terms are as previously defined. Daily BW change (ΔBW) was calculated by fitting a fifth-order orthogonal polynomial regression on DIM (5 to 306 DIM) to daily BW, and then ΔBW was calculated as the difference in predicted BW between consecutive days. The classification effects in the OVEc were parityST, HYS, and trial (diet treatment). ParityST is a combination of parity (3 levels: 1, 2, and 3+) and stage of lactation (4 levels: ≤30, 31–100, 101–200, and >200 d), and HYS is a combination of herd, year, and season of calving (2 herds for CAN and CHE, and the remaining countries had animals from 1 herd, and 4 seasons: autumn (October 1–December 31), winter (January 1–March 31), spring (April 1–June 30), and summer (July 1–September 30). There were 3 diet treatments for CHE, and 1 diet treatment for other countries.
The 824 Australian heifers were genotyped using the Illumina Bovine HD (high density, ~600K) SNP chip. The 584 Australian cows were genotyped on a variety of low-density (8.5-25K SNP) to 50K chips, and these genotypes were imputed to 50K genotypes using FImpute (Sargolzaei et al., 2014). The genotypes of each overseas country in the EDGP database were from a variety of medium- to high-density chips, ranging in size from 55,647 to 777,961 SNP. The genotypes were recoded to 0, 1, and 2 copies of the B/A allele from the top/top Illumina format, and were mapped to the UMD 3.1 build of the bovine genome sequence assembled by the Center for Bioinformatics and Computational Biology at University of Maryland (CBCB; http://www.cbcb.umd.edu/research/bos_taurus_assembly.shtml). The genotypes of each data set were then prepared according to the ARS-UDC1.2 sequence map of the bovine genome (Rosen et al., 2020) from Run 7 of the 1000 Bull Genome Project, and all unknown chromosomes and positions were removed. Sporadic missing genotypes were imputed using FImpute (Sargolzaei et al., 2014) with a reference population of 2,700 Australian animals that were genotyped directly with HD SNP. Before merging each country's genotypes, the allele frequency of each SNP in each country was checked to ensure that the homozygote allele codes were likely to be in concordance. Additionally, the real and imputed 50K genotypes of 20,816 Australian Holstein bulls (AUSb) born between 2010 and 2020 were provided by DataGene. The imputation of the bulls' genotypes was done using the standard DataGene process where FImpute was used for all imputations. This study used 41,276 SNP overlapped between cow, heifer, and bull genotype sets.
A trivariate genomic-based REML (GREML) analysis was used to calculate genetic correlations between RFI traits and to predict GEBV, where the traits were RFI in AUSc, AUSh, and OVEc. The genomic relationship matrix (GRM) used in GREML analysis was built based on the genotypes with 41,276 SNP using the method of Yang et al. (2010). The variance components were estimated using ASReml (Gilmour et al., 2009).
Prediction of SNP effects for AUSc RFI and AUSh RFI was obtained by
where Z is the n × 41,276 matrix of the genotypes of 3,711 animals in the reference set, and is the descaled GEBV (GEBV multiplied by SD of RFI traits) for RFI of AUSc or AUSh in the reference population. The SNP effects from the prediction equations were applied to calculate the direct genomic breeding values (DGV) of 21K AUSb. To produce a genomic breeding value of lifetime RFI covering both growth and lactation stages, the RFI DGV for the cows and heifers were combined as described in Pryce et al. (2015):
| Lifetime RFI = (DGV AUSc × 305 × 4 + DGV AUSh × 700)/6, |
where lifetime RFI was expressed in kilograms of feed per year over a 6-yr period and assumed a rearing period of 2 yr and lactating period of 4 yr. Following Pryce et al. (2015), the FS EBV was then calculated by subtracting lifetime RFI from the amount of the feed required for maintenance (Feed_BW_kg): FS = Feed_BW_kg – lifetime RFI. Feed_BW_kg is a function of BW and calculated as EVBW × (EBVBW − 100)/(feedcost × MJME), where EVBW (economic value of maintenance) is A$5.14, feedcost (the cost of feed per MJ) is A$0.032/MJ, and MJME (the energy content of feed) is 11.9 MJ/kg of DMI (where A$1.00 = US$0.76). To calculate the reliabilities for RFI in the AUSb, the GREML was performed based on GRM including all cows and additionally 3,413 AUSb that overlapped with the data used in 2015. The calculation of reliabilities for lifetime RFI and FS followed the formulas described in Pryce et al. (2015). However, we slightly adjusted the weights on the contributions of heifers and cows considering the relative contribution of heifer and cow intake across a productive lifetime:
| RELlifetime RFI = 0.20 × RELheifer RFI + 0.80 × RELcow RFI, and |
| RELFS = 1 - PEV/SD2FS, |
where REL is reliability of RFI traits and FS, and PEV is the prediction error variance calculated as
| PEV =(1-RELlifetimeRFI)× SD2RFI + (1 - RELBW) × SD2BW. |
The response to selection for FS to achieve a 100-unit (A$) gain in the selection index was calculated by regressing the FS EBV of 21K AUSb on the selection index (BPI):
| Selection responseFS = r(index,FS) × SDFS × (100/SDindex), |
where r(index,FS) is a correlation between selection index (i.e., BPI or HWI) and FS EBV.
Heterozygosity predicted from the GRM was compared with mean observed heterozygosity per country and heterozygosity assuming Hardy-Weinberg equilibrium (calculated as described Bolormaa et al., 2013), using the 50K genotypes of 3,711 animals. There was good concordance between these population measures, with all genotype groups displaying a similar range of heterozygosity between 0.32 and 0.34. This shows that the GRM constructed using animals from different groups is a true representation of the relationships between and within groups of animals.
Standard deviations (SD) of RFI phenotypes were 0.42 kg of DM/d for AUSh, 1.28 kg of DM/d for AUSc, and 1.82 kg of DM/d for OVEc. The heritability estimates for RFI (h2 ± SE) using trivariate analysis were 0.18 ± 0.086 in AUSc, 0.36 ± 0.086 in AUSh, and 0.27 ± 0.034 in OVEc data sets. The single, bivariate, and trivariate analyses provided similar genomic heritability estimates. Because of the small size of the data set, the standard errors of the h2 estimates were large. However, standard errors were much smaller than the comparable estimates obtained using the data available in 2015, particularly for AUSc. The genetic correlations (rg ± SE) of RFI were 0.47 (±0.274) between AUSc and AUSh, 0.94 (±0.297) between AUSc and OVEc, and 0.20 (±0.175) between OVEc and AUSh data sets. The rg between AUSc RFI and OVEc RFI using the updated model was greater than the estimate in 2015 of 0.76 (±0.60). However, the estimates are associated with large standard errors.
The SD of FS EBV for the 3,413 AUSb in common between 2015 and the current data set was 79 kg (DM)/yr (Table 1), which was 14 kg/yr higher than the equivalent SD estimated in 2015. Cows with FS EBV that are 1 SD above the mean of 0 (i.e., +79 kg/yr) could save 1.3% of annual feed costs assuming a cow eats 20 kg of DM/d (3.5% of 570 kg of BW, estimated as the mean BW for AUSc). The correlations of lifetime RFI and FS EBV between the prediction equations of 2015 and 2020 were 0.65 and 0.80, respectively.
Table 1.
Mean, SD, and range of EBV and reliabilities for cow residual feed intake (RFI), heifer RFI, lifetime RFI, and Feed Saved (FS) in 3,413 Australian Holstein bulls
| Item | Cow RFI (kg×10/d) | Heifer RFI (kg×10/d) | Lifetime RFI (kg/yr) | Feed_BW_kg1 (kg/yr) | FS (kg/yr) | BW (kg/yr) |
|---|---|---|---|---|---|---|
| EBV | ||||||
| Mean | 1 | 0.23 | 23.2 | 8.7 | −14.4 | 99.4 |
| SD | 2.88 | 0.89 | 65.6 | 46.8 | 79.2 | 3.5 |
| Maximum | 11.7 | 3.1 | 262.9 | 195.7 | 250.1 | 113.9 |
| Minimum | −6.86 | −2.38 | −154.4 | −141.7 | −268.3 | 85.6 |
| Reliability | ||||||
| Mean | 0.22 | 0.12 | 0.20 | 0.47 | 0.71 | |
| SD | 0.038 | 0.035 | 0.036 | 0.039 | 0.062 | |
| Maximum | 0.45 | 0.29 | 0.39 | 0.69 | 0.99 | |
| Minimum | 0.07 | 0.01 | 0.06 | 0.28 | 0.45 |
Feed_BW_kg = feed required for maintenance.
The genetic variances of lifetime RFI and feed required for maintenance (Feed_BW_kg) per year were 30,318 and 33,325 kg2/yr, respectively; hence, the variance of FS was 63,643 kg2/yr. The mean reliabilities for lifetime RFI and FS EBV in the 3,413 AUSb were 0.20 and 0.47, respectively (Table 1). The mean reliability of FS for 21K AUSb that were born from 2010 onward using the updated 2020 equation was 0.43 (SD = 0.045), ranging from 0.15 to 0.61. This was 10% higher than using the prediction equation from 2015, when the reference population was almost half the size. The reliability of AUSc RFI using the bivariate model with RFI AUSc and RFI AUSh applied to 3,413 AUSb was low (0.08) compared with the reliability using the trivariate model (including the OVEc), showing a large benefit of using overseas data. Interestingly, the benefit of overseas data was more evident when we estimated the empirical accuracy of genomic prediction. For example, the accuracy using the trivariate model (including international cow data) increased by at least 8% compared with just using AUS cow and heifer data (unpublished results). Thus, international collaborations are of high importance for traits that are expensive to measure, such as feed intake.
Figure 1 shows the predicted reliability of lifetime RFI at 2 h2 values (0.18 and 0.27) achieved by expanding the size of the reference population through the addition of cows with phenotypes and genotypes. Using the deterministic equation described in MacLeod et al. (2014), 20,000 cows and heifers are needed to achieve a reliability of 0.50 for lifetime RFI assuming an h2 of 0.18, an effective population size (Ne) of 210 and a constant reliability of 0.12 for RFI heifer (assuming no more additional data are added at the growth stage). If a reliability of 0.50 for lifetime RFI is achieved, then the reliability for FS would be around 0.58 (Figure 1). With a greater h2, such as 0.27, which was estimated for OVEc, 13K and 24K cows would be sufficient to achieve reliabilities of 0.50 and 0.60 for lifetime RFI and FS, respectively (Figure 1). Expanding the heifer population has a small impact unless the weight on it is changed (because the weight on heifer RFI in the current model is only 20%). Because feed efficiency is expensive to measure, collecting such a large amount of data is probably only achievable through international collaborations. It is worth noting that because combining RFI heifer and RFI cow into lifetime RFI is an approximate approach, studies into how selection affects lifetime efficiency are worthwhile.
Figure 1.
Deterministically derived reliability for lifetime residual feed intake (RFI) and Feed Saved as a function of the size of reference population and heritability (h2).
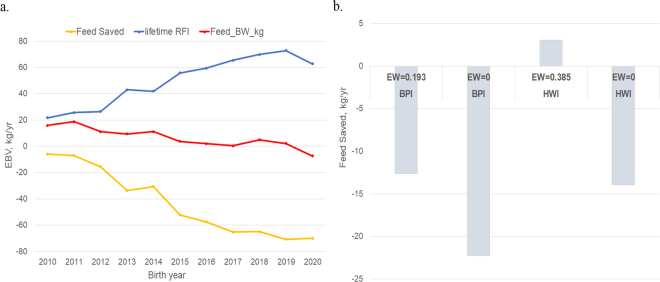
The genetic trends of lifetime RFI, Feed_BW_kg, and FS using EBV calculated for ~21K genotyped AUSb that were born from 2010 onward is shown Figure 2a. Since 2010, there has been an increase in lifetime RFI and a decrease in feed required for BW, and a negative (i.e., unfavorable) trend in EBV for FS. The unfavorable change in FS has slowed from 1/2 genetic SD to <1/4 SD of FS EBV when comparing the 5 yr before and after the inclusion of the FS EBV as part of BPI in 2015. This shows that including FS in the BPI has been effective in reducing the unfavorable genetic trend in FS.
Figure 2.
(a) Genetic trend by birth year of EBV (mean ± SE) for lifetime residual feed intake (RFI), feed required for maintenance (Feed_BW_kg), and Feed Saved (FS) in ~21K genotyped Holstein bulls. (b) Responses to selection in FS through 100-unit gain (A$) in the Balanced Performance Index (BPI) and Health Weighted Index (HWI) in the 21K Holstein bulls achieved through different economic weights (EW) on FS.
The FS EBV using the updated 2020 model was released by DataGene in December 2020 and was included in the BPI and HWI. The correlation between BPI and HWI using the ~21K AUSb was 0.92 and the SD of BPI and HWI were 105.3 and 118.4, respectively. The economic weight of FS from the economic model (Byrne et al., 2016) used to derive weights for the BPI was halved to A$0.1927/kg, the rationale being that low reliability traits hinder genetic improvement. As the reliability of FS continues to increase, the full economic value of FS in the BPI should be considered. However, the full value (A$0.385/kg) has already been applied in the health trait–focused HWI, which is expected to result in a better response to selection in FS. Therefore, it is not surprising that the predicted responses in FS were −12.63 kg/yr and 3.08 kg/yr as a result of selection on BPI and HWI, respectively, using a selection intensity to achieve 100 units (A$) gain (typically 5–10 yr of genetic gain) of BPI and HWI using the 21K AUSb data (Figure 2b). The response in FS to selection in BPI has a negative value, but the relative weight of FS in BPI is small (5.50%). Thus, the overall economic merit will increase while improving the feed efficiency. When the economic value of FS is set to zero in selection indices (e.g., BPI), FS falls more rapidly (Figure 2b). For breeds other than Holsteins, FS is calculated using only the BW component, as RFI has only been measured in a Holstein reference population currently. As reported by Lidauer et al. (2019), the Nordic Cattle Genetic Evaluation NAV (Denmark, Finland, and Sweden) is developing an index for FS, but it currently includes only the metabolic BW component, which is the same as we use in Australia for breeds other than Holstein.
In conclusion, the updated 2020 model for the FS EBV has improved the reliability of FS by 10% compared with the 2015 model. Feed Saved has been included in the BPI and HWI since 2015, which appears to have had a favorable effect on the genetic trend. Implementation of the updated FS EBV and its inclusion in BPI and HWI is expected to further improve the genetic trend of FS in AUSb and AUSc in the future and to improve feed efficiency in the Australian dairy industry. The current reference population based on Australian animals is small; therefore, international collaborations are crucial to achieve higher reliabilities of the FS EBV across dairy populations. For other breeds, RFI is not available; therefore, FS for non-Holsteins will continue to be calculated using only the BW EBV.
Notes
We thank DairyBio, jointly funded by Dairy Australia (Melbourne, Australia), The Gardiner Foundation (Melbourne, Australia), and Agriculture Victoria (Melbourne, Australia), for funding this project.
The authors extend special thanks to Roel Veerkamp and Yvette de Haas (Wageningen University, the Netherlands) for provision of genotypes and phenotypes on animals from the Netherlands, and to Mike Coffey and Eileen Wall for provision of genotypes and phenotypes on UK animals. The authors gratefully acknowledge use of data from Ellinbank Research Centre (belonging to Agriculture Victoria Research) and the Efficient Dairy Genome Project (EDGP) and its participants. EDGP was funded by Genome Canada (Ottawa, ON, Canada), Genome Alberta (Calgary, AB, Canada), Ontario Genomics (Toronto, ON, Canada), Alberta Ministry of Agriculture (Edmonton, AB, Canada), Ontario Ministry of Research and Innovation (Toronto, ON, Canada), Ontario Ministry of Agriculture, Food and Rural Affairs (Guelph, ON, Canada), Canadian Dairy Network (Guelph, ON, Canada), GrowSafe Systems (Airdrie, AB, Canada), Alberta Milk (Edmonton, AB, Canada), Victoria Agriculture (Australia), Scotland's Rural College (Edinburgh, UK), USDA Agricultural Research Service (Washington, DC), Qualitas AG (Zug, Switzerland), and Aarhus University (Aarhus, Denmark). The staff caring for the cows used in the study are gratefully acknowledged.
The authors have not stated any conflicts of interest.
References
- Berry D.P., Crowley J.J. Cell Biology Symposium: Genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 2013;91:1594–1613. doi: 10.2527/jas.2012-5862. 23345557. [DOI] [PubMed] [Google Scholar]
- Bolormaa S., Pryce J.E., Kemper K., Savin K., Hayes B.J., Barendse W., Zhang Y., Reich C.M., Mason B.A., Bunch R.J., Harrison B.E., Reverter A., Herd R.M., Tier B., Graser H.-U., Goddard M.E. Accuracy of prediction of genomic breeding values for residual feed intake, carcass and meat quality traits in Bos taurus, Bos indicus and composite beef cattle. J. Anim. Sci. 2013;91:3088–3104. doi: 10.2527/jas.2012-5827. 23658330. [DOI] [PubMed] [Google Scholar]
- Byrne T.J., Santos B.F.S., Amer P.R., Martin-Collado D., Pryce J.E., Axford M. New breeding objectives and selection indices for the Australian dairy industry. J. Dairy Sci. 2016;99:8146–8167. doi: 10.3168/jds.2015-10747. 27522425. [DOI] [PubMed] [Google Scholar]
- Daetwyler H.D., Pong-Wong R., Villanueva B., Woolliams J.A. The impact of genetic architecture on genome-wide evaluation methods. Genetics. 2010;185:1021–1031. doi: 10.1534/genetics.110.116855. 20407128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas Y., Calus M.P.L., Veerkamp R.F., Wall E., Coffey M.P., Daetwyler H.D., Hayes B.J., Pryce J.E. Improved accuracy of genomic prediction for dry matter intake of dairy cattle from combined European and Australian data sets. J. Dairy Sci. 2012;95:6103–6112. doi: 10.3168/jds.2011-5280. 22863091. [DOI] [PubMed] [Google Scholar]
- de Haas Y., Calus M.P.L., Veerkamp R.F., Wall E., Coffey M.P., Daetwyler H.D., Hayes B.J., Pryce J.E. Improved accuracy of genomic prediction for dry matter intake of dairy cattle from combined European and Australian data sets. J. Dairy Sci. 2012;95:6103–6112. doi: 10.3168/jds.2011-5280. 22863091. [DOI] [PubMed] [Google Scholar]
- de Jong D., Bouwmeester-Vosman J.J., van der Linde C., de Haas Y., Schopen G.C.B., Veerkamp R.F. Feed intake genetic evaluation: Progress and an index for saved feed cost. Interbull Bull. 2019;55:1–4. [Google Scholar]
- Gilmour A.R., Gogel B.J., Cullis B.R., Thompson R. VSN International Ltd; 2009. ASReml User Guide Release 3.0. [Google Scholar]
- Goddard M. Genomic selection: Prediction of accuracy and maximisation of long term response. Genetica. 2009;136:245–257. doi: 10.1007/s10709-008-9308-0. 18704696. [DOI] [PubMed] [Google Scholar]
- Gunsett F.C. Linear index selection to improve traits defined as ratios. J. Anim. Sci. 1984;59:1185–1193. doi: 10.2527/jas1984.5951185x. [DOI] [Google Scholar]
- Herd R.M., Oddy V.H., Richardson E.C. Biological basis for variation in residual feed intake in beef cattle. 1. Review of potential mechanisms. Aust. J. Exp. Agric. 2004;44:423–430. doi: 10.1071/EA02220. [DOI] [Google Scholar]
- Lidauer M.H., Leino A.-M., Stephansen R.S., Pösö J., Nielsen U.S., Fikse W.F., Aamand G.P. Genetic evaluation for maintenance–Towards genomic breeding values for saved feed in Nordic dairy cattle. Interbull Bull. 2019;55:21–25. [Google Scholar]
- Macdonald K.A., Pryce J.E., Spelman R.J., Davis S.R., Wales W.J., Waghorn G.C., Williams Y.J., Marett L.C., Hayes B.J. Holstein-Friesian calves selected for divergence in residual feed intake during growth exhibited significant but reduced residual feed intake divergence in their first lactation. J. Dairy Sci. 2014;97:1427–1435. doi: 10.3168/jds.2013-7227. 24377796. [DOI] [PubMed] [Google Scholar]
- MacLeod I.M., Hayes B.J., Goddard M.E. The effects of demography and long-term selection on the accuracy of genomic prediction with sequence data. Genetics. 2014;198:1671–1684. doi: 10.1534/genetics.114.168344. 25233989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce J.E., Gonzalez-Recio O., Nieuwhof G., Wales W.J., Coffey M.P., Hayes B.J., Goddard M.E. Hot topic: Definition and implementation of a breeding value for feed efficiency in dairy cows. J. Dairy Sci. 2015;98:7340–7350. doi: 10.3168/jds.2015-9621. 26254533. [DOI] [PubMed] [Google Scholar]
- Pryce J.E., Nguyen T.T.T., Axford M., Nieuwhof G., Shaffer M. Symposium review: Building a better cow—The Australian experience and future perspectives. J. Dairy Sci. 2018;101:3702–3713. doi: 10.3168/jds.2017-13377. 29454697. [DOI] [PubMed] [Google Scholar]
- Rosen B.D., Bickhart D.M., Schnabel R.D., Koren S., Elsik C.G., Tseng E., Rowan T.N., Low W.Y., Zimin A., Couldrey C. De novo assembly of the cattle reference genome with single-molecule sequencing. GigaScience. 2020;9 doi: 10.1093/gigascience/giaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., Chesnais J., Schenkel F. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 2014;15:478. doi: 10.1186/1471-2164-15-478. 24935670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raden P.M., Wright J.R., Connor E.E., Vandehaar M., Tempelman R.J., Liesman J., Weigel K. Preliminary genomic predictions of feed saved for 1.4 million Holsteins. J. Dairy Sci. 2017;100(Suppl. 2):200–201. (Abstr.) [Google Scholar]
- Williams Y.J., Pryce J.E., Grainger C., Wales W.J., Linden N., Porker M., Hayes B.J. Variation in residual feed intake in Holstein-Friesian dairy heifers in southern Australia. J. Dairy Sci. 2011;94:4715–4725. doi: 10.3168/jds.2010-4015. 21854946. [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy N.P., Gordon S., Henders A.K., Nyholt D.R., Madden P.A., Heath A.C., Martin N.G., Montgomery G.W., Goddard M.E., Visscher P.M. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 2010;42:565–569. doi: 10.1038/ng.608. 20562875. [DOI] [PMC free article] [PubMed] [Google Scholar]