Graphical Abstract
Summary: The genetic relationship between male reproduction and economically relevant traits is unknown. Here, we used genome-wide imputed genotypes to estimate genetic correlation between 6 estimates of male fertility and 57 dairy traits. We used allele substitution effects calculated with an additive regression approach for 11,804,781 genome-wide sequence variants. Correlations between male reproduction and other traits were small (r = -0.0681–0.0787). However, we observed correlations among various semen quality parameters and sire reproductive success (head anomalies and tail anomalies: r = 0.8380, head anomalies and motility: r = -0.7083, tail anomalies and motility: r = -0.7739, motility and volume: r = -0.1266, concentration and volume: r = -0.3503, nonreturn rate and motility: r = 0.1598, nonreturn rate and head anomalies: r = -0.1640, nonreturn rate and tail anomalies: r = -0.1580). To identify QTL responsible for the correlation among these traits, we conducted a multitrait meta-analysis; this revealed pleiotropy of a known QTL on chromosome 6 and identified a novel QTL for sperm head anomalies on chromosome 11.
Highlights
-
•
Correlations between male fertility and other economically relevant traits were low in Brown Swiss cattle.
-
•
Motility was correlated with anomalies and volume; volume and concentration were correlated.
-
•
Sire nonreturn rate was correlated with sperm anomalies and motility.
-
•
Correlation among male fertility traits suggests the presence of pleiotropic QTL.
-
•
Meta-analysis recovered QTL for male fertility on chromosomes 6 and 11.
Abstract
Undisturbed reproduction is key for successful breeding of beef and dairy cattle. Improving reproductive ability can be difficult because of antagonistic relationships with other economically relevant traits. In cattle, thorough investigation of female fertility revealed unfavorable genetic correlations with various production phenotypes. However, the correlation between male reproductive ability and production traits remains poorly understood. Here, we investigated the genetic relationships among and between male fertility characteristics and economically relevant traits in a population of Brown Swiss cattle. We performed GWAS with imputed genotypes at nearly 12 million sequence variants for semen quality (sperm head and tail anomalies, motility, concentration, and volume), male fertility, and 57 production phenotypes. Allele substitution effects were then correlated on a trait-by-trait basis to estimate genetic correlations. Correlations between male reproductive characteristics and traits of economic value were small and ranged from −0.0681 to 0.0787. Among the semen quality parameters, sperm motility was negatively correlated with anomalies (head: r = −0.7083 ± 0.0002; tail: r = −0.7739 ± 0.0002) and volume (r = −0.1266 ± 0.0003), whereas volume was negatively correlated with concentration (r = −0.3503 ± 0.0002). Sire nonreturn rate was negatively correlated with sperm anomalies (head: r = −0.1640 ± 0.0002; tail: r = −0.1580 ± 0.0002) and positively correlated with motility (r = 0.1598 ± 0.0002). A meta-analysis of male reproductive traits identified 2 quantitative trait loci: a previously described region on chromosome 6 showed pleiotropic effects and a novel region on chromosome 11 was associated with sperm head anomalies. In conclusion, our results suggest that selection for economically important dairy and production phenotypes has little impact on semen quality and fertility of Brown Swiss bulls.
Efficient and successful reproduction is crucial to both the dairy and beef industries. Repeated breedings incur costs and may interfere with the production scheme. However, improving cattle fertility by selection is challenging because of its low heritability (Berry et al., 2014; Butler et al., 2019). Moreover, pleiotropic QTL can cause antagonistic relationships between fertility and production (Kadri et al., 2014; de Souza Fonseca et al., 2018). This phenomenon is exemplified when considering female reproductive ability and production characteristics in dairy cattle: fertility and milk production demonstrate unfavorable relationships with moderate genetic correlations (Oltenacu et al., 1991; Roxström et al., 2001; Pryce et al., 2004). Indeed, antagonistic relationships between phenotypes have been thoroughly documented for dairy cows and heifers (VanRaden et al., 2004; Kadri et al., 2014; Miglior et al., 2017). However, the relationship between production and male fertility is not well understood, despite male reproductive ability having a significant economic impact on the dairy industry (Braundmeier and Miller, 2001).
Evidence that semen quality is correlated with production traits has been identified in species other than cattle (Oh et al., 2006; Wolf, 2009). In cattle, previous studies have identified genetic correlations between scrotal circumference (which is correlated with semen quality; Palasz et al., 1994) and both linear conformation characters and female fertility (Meyer et al., 1991; Martínez-Velázquez et al., 2003; Boligon et al., 2010). Kealey et al. (2006) found correlations between birth weight and semen characteristics in Hereford cattle. However, these studies were conducted in beef or indicine breeds and generally did not consider dairy traits. Thus, characterizing the relationship between dairy traits and male reproduction is warranted to improve dairy herd management.
Although the heritability of semen quality traits varies considerably by population (Butler et al., 2019), moderate heritability estimates of certain characteristics, such as sperm motility and concentration, have been repeatedly recovered (Berry et al., 2014). Several QTL that affect bull fertility and ejaculate characteristics have been identified in beef and dairy breeds (Peñagaricano et al., 2012; Rezende et al., 2018; Butler et al., 2019). In Brown Swiss (BSW) cattle, considerable heritable variation in male reproductive success has been identified between bulls (Hiltpold et al., 2020; Pacheco et al., 2021). Some of this variation is due to large-effect QTL for sperm motility, sperm anomalies, and bull fertility located on chromosomes 1,6, 18, 25, and 26 (Hiltpold et al., 2020, 2021). However, the largest proportion of heritable variation in semen quality and male fertility is due to unknown small-effect QTL. Previously, meta-analyses of multiple correlated phenotypes increased the power to detect QTL (Bolormaa et al., 2014; Fang and Pausch, 2019); thus, implementing a meta-analysis with male fertility characteristics may reveal undescribed small-effect QTL.
Here, we leverage genome-wide polymorphism data, consisting of imputed genotypes for nearly 12 million sequence variants, and an array of densely sampled phenotypes to explore the relationship between male fertility and production traits in BSW cattle. Specifically, we used allele substitution effects inferred for 63 phenotypes to assess genetic correlations between male reproduction and economically relevant traits. In addition, we performed a multitrait meta-analysis to detect QTL that influence semen quality and bull fertility.
We used the score for sperm head and tail anomalies, percent of motile sperm, sperm concentration (million sperm/mL), ejaculate volume, and insemination success (sire nonreturn rate) to describe male reproduction. Briefly, we considered only ejaculates that fulfilled minimum quality requirements for AI, which were collected from bulls between 400 and 1,000 d of age. The ejaculate phenotypes considered, including collection and filtration methods, are described in detail in Hiltpold et al. (2020, 2021). Ejaculate characteristics and reproductive success were assessed in 30,885 fresh ejaculates and 850,708 AIs, respectively, for 983 bulls. Each ejaculate was fitted with the linear model
| y = 1μ + ha + df + Ct + Sn + e, |
for all considered phenotypes to account for confounding variables, where y is a vector of trait-value for each ejaculate, μ is the intercept, h is a vector of bull's age (in days) at semen collection, d is a vector of number of days since the preceding ejaculate was collected, C is an incidence matrix relating the phenotype to the semen collector, and S is an incidence matrix relating the phenotype to collection date (4 seasons for each year); a, f, t, and n are the respective effects of the aforementioned variables, and e is a vector of random residuals. The random residuals were averaged for each individual and used as the input phenotype in subsequent analyses. We used R v4.0.5 (R Core Team, 2021) to construct the linear model and for subsequent analyses, unless otherwise specified.
Estimates of sire fertility were provided by Swissgenetics (Zollikofen, Switzerland). Insemination success was not available on an ejaculate level. Instead, a bull's reproductive ability was quantified as the 56-d nonreturn rate following the first insemination in both cows and heifers in the 12 successive months with the most inseminations. Sire nonreturn rate was computed using the linear mixed model proposed by Schaeffer (1993), which accounts for insemination month, parity (cow or heifer), cost of the semen dose, insemination technician, the combination of the bull's and cow's breed, and herd. Hiltpold et al. (2020, 2021) provides a detailed description of the model. Additionally, 57 phenotypes of economic importance were available from routine breeding value estimations in the form deregressed EBVs (drEBV) and belonged to the following categories: dairy (n = 7), linear conformation (n = 29), longevity (n = 1), slaughter–adult (n = 3), slaughter–calf (n = 3), calving (n = 8), cell count (n = 1), and female fertility (n = 5). A list of these traits can be found in Figure 1. The number of BSW sires considered for each production trait varied and ranged from 1,698 to 5,843.
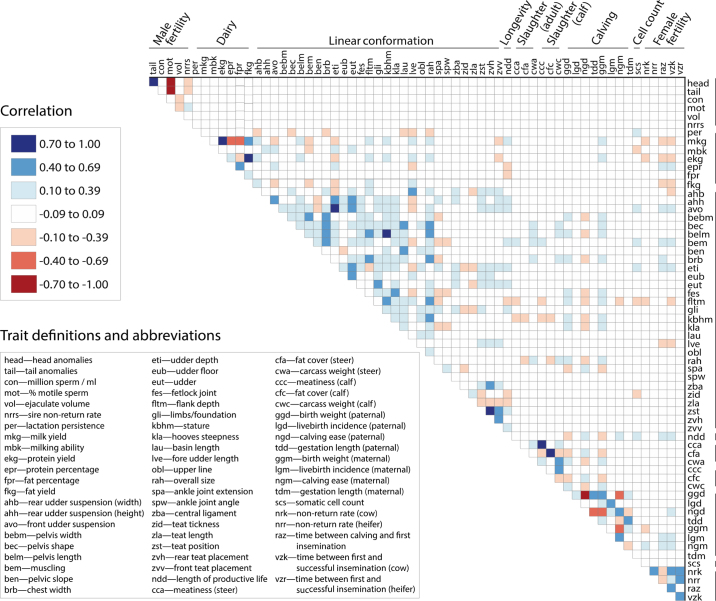
Figure 1.
Correlation matrix of the allele substitution effects estimated from single-trait GWAS. Traits considered and their abbreviations are listed in the bottom left key. Colors correspond to correlation category (high, moderate, low, and none) and direction (red: negative, blue: positive).
Procedures used to obtain sequence variant genotypes from the bulls are outlined in Lloret-Villas et al. (2021). In brief, sire genotypes were inferred with either low- to medium-density (Illumina 20k to 150k) or high-density (Illumina 777k) chips, and then partially imputed to 683,609 SNPs using 1,166 BSW animals that had 777k genotypes as a reference. Subsequently, a reference panel of 368 sequenced BSW individuals was used to further impute the partially imputed genotypes to 27,214,878 sequence variants with Beagle (version 5; Browning et al., 2018).
Genome-wide association studies were conducted for all traits using the GCTA-MLMA tool (Yang et al., 2011, 2014) and a standard linear mixed model that included a genomic relationship matrix (GRM) to model the random polygenic term. The model also included the top 4 principal components of the GRM as fixed effects to account for population stratification and weights to consider either the number of observations per individual for male fertility characteristics or the effective daughter contributions (EDC) for the deregressed proofs (drEBV). Weights for ejaculate characteristics were computed for each sire as (1/number of ejaculates), and weights for sire nonreturn rate were inferred as (1/number of inseminations). Weights for the drEBV were computed as (1/EDC). The GRM was constructed with 589,791 autosomal variants from the aforementioned partially imputed data set that had minor allele frequencies (MAF) >0.5%. Association tests were conducted for 11,804,781 sequence variants that had MAF >1% and model-based imputation accuracy >0.5. Variants with a P-value less than 4.24 × 10−9 were deemed significant, as informed by a 5% Bonferroni-corrected significance threshold. Heritabilities of ejaculate characteristics were inferred with the average information REML estimation algorithm in AIREMLF90 (Misztal et al., 2002) and followed the approach described in Hiltpold et al. (2020).
For each trait pair, we estimated genetic correlations using the allele substitution effects obtained from the single-trait GWAS. Following the approach proposed by Bolormaa et al. (2014), we inferred signed t-values for each trait with where b is the allele substitution effect and (se)b is the standard error. Variants for which the t-value deviated more than 5 standard deviations from the mean were removed. The filtered t-values were used to compute a Pearson correlation coefficient for each trait pair, thus producing a correlation matrix.
We administered a multitrait meta-analysis to identify QTL contributing to the correlation among semen quality and male fertility traits. Specifically, we performed a multitrait chi-squared (χ2) test with the test statistic described as in Bolormaa et al. (2014), where n is the number of traits considered, is a n × 1 matrix of signed t-values at the ith sequence variant across n traits, and is the inverse of the correlation matrix calculated as described above. The same significance threshold used in the single-trait GWAS (P < 4.24 × 10−9) was used to identify significant loci.
A correlation matrix constructed from the estimated substitution effects is visualized in Figure 1. Our analysis recovered multiple relationships previously described in BSW cattle; specifically, negative correlations between milk yield and content (e.g., milk yield and fat percentage, r = −0.6642 ± 0.0002; milk yield and protein yield, r = −0.8032 ± 0.0001), an antagonism between milk yield and female fertility (e.g., milk yield and cow nonreturn rate, r = −0.1415 ± 0.0002), and a positive correlation between stature and milk yield (r = 0.1418; de Haas et al., 2007; Gibson and Dechow, 2018; Martinez-Castillero et al., 2021), thereby supporting the general validity of our findings. Correlations between male reproductive characteristics and traits of economic value were small and ranged from −0.0681 to 0.0787. Both the most-positive and the most-negative relationships were observed with calving phenotypes (ejaculate volume and weight of calves, r = 0.0787 ± 0.0003; ejaculate volume and share of normal births, r = −0.0681 ± 0.0003). Other studies found that testis size (e.g., scrotal circumference, length, volume) is genetically correlated with calving interval, ADG, and daughter pregnancy rates (Toelle and Robinson 1985; Meyer et al., 1991; Raidan et al., 2017). However, testis measurements were not available for the BSW bulls considered in our study. In addition, Kealey et al. (2006) found that live birth weight is positively correlated with sperm motility and concentration. Although the lack of correlation between male fertility and other traits in our data set could be attributed to differences in beef and dairy breeds, it is still interesting to note that the strongest relationships we identified were with calving phenotypes.
Many of the relationships we observed within the ejaculate quality traits have been described in other dairy breeds (Butler et al., 2019). Correlation coefficients among ejaculate characteristics are reported in Table 1. Sperm head and tail anomalies were positively correlated (r = 0.8380 ± 0.0001) while being negatively correlated with motility (head: r = −0.7083 ± 0.0002; tail: r = −0.7739 ± 0.0002). Sperm motility was negatively correlated with ejaculate volume (r = −0.1266 ± 0.0003), and ejaculate volume was negatively correlated with sperm concentration (r = −0.3503 ± 0.0002). Most of these associations are in agreement with those of other studies, particularly the relationships between anomalies and motility, motility and volume, and volume and concentration (Ducrocq and Humblot, 1995; Druet et al., 2009; Karoui et al., 2011). We did not recover other frequently observed relationships, such as a positive correlation between motility and concentration (Berry et al., 2019; Burren et al., 2019; Olsen et al., 2020). However, a majority of these studies have investigated the relationships among semen quality parameters in small samples of AI bulls, and thus inferred correlation coefficients with large standard errors. Differences in semen quality phenotyping and the stringent filtering of raw data (cf. Hiltpold et al., 2020) may explain the differences between our results and those previously reported.
Table 1.
Phenotypic and genetic correlations among male fertility characteristics1
| Trait | Head anomalies | Tail anomalies | Motility | Concentration | Volume | Sire nonreturn rate |
|---|---|---|---|---|---|---|
| Head anomalies | 0 | 0.8380 | −0.7083 | −0.0018 | 0.0278 | −0.1640 |
| (0.0001) | (0.0002) | (0.0003) | (0.0003) | (0.0002) | ||
| Tail anomalies | 0.8363 | 0 | −0.7739 | −0.0131 | 0.0588 | −0.1580 |
| (0.0203) | (0.0002) | (0.0003) | (0.0003) | (0.0002) | ||
| Motility | −0.6767 | −0.7380 | 0.1160 | 0.0858 | −0.1271 | 0.1598 |
| (0.0273) | (0.0250) | (0.0410) | (0.0003) | (0.0003) | (0.0002) | |
| Concentration | NS2 | NS | NS | 0.2588 | −0.3503 | 0.0174 |
| (0.0517) | (0.0002) | (0.0003) | ||||
| Volume | 0.1044 | 0.1143 | −0.1671 | −0.3629 | 0.2102 | −0.0745 |
| (0.0368) | (0.0367) | (0.0364) | (0.0345) | (0.0436) | (0.0003) | |
| Sire nonreturn rate | −0.2344 | −0.2336 | 0.2340 | NS | −0.1392 | NA |
| (0.0360) | (0.0260) | (0.0360) | (0.0366) |
Genetic correlation was estimated from allele substitution effects from linear regression-based GWAS (upper diagonal) and phenotypic correlation was inferred with filtered observations (lower diagonal). Standard errors are reported in parentheses. Heritability for ejaculate quality traits can found on the diagonals. Heritability for sire nonreturn rate was not calculated (NA), because insemination success was not available on an ejaculate level.
Nonsignificant relationship (P > 0.05).
In accordance with other studies (Gredler et al., 2007; Gebreyesus et al., 2021), bull fertility was associated with several ejaculate characteristics. Correlation coefficients between ejaculate characteristics and male reproductive success are reported in Table 1. We observed a negative correlation between sperm anomalies and sire nonreturn rate (head: r = −0.1640 ± 0.0002; tail: r = −0.1580 ± 0.0002). Sperm motility was positively correlated with bull fertility (r = 0.1598 ± 0.0002). Ejaculate volume and sperm concentration were not correlated with sire nonreturn rate. The association between motility and sire nonreturn rate agrees with Gebreyesus et al. (2021), where they observed a positive genetic correlation between post-thaw motility and fertility. In addition, Gredler et al. (2007) identified a positive correlation between bull fertility and motility. Our results further support the possibility of motility—or motility-related measures such as head and tail anomalies—as a predictive measure of bull fertility. We further hypothesize that our study underestimates the correlations between semen quality and sire nonreturn rate, because semen doses from ejaculates of inferior quality are compensated for. Therefore, doses from lower-quality ejaculates contain more sperm. In addition, our stringent filtering removed low-quality ejaculates and large outliers from the data set, furthering the possibility that the inferred correlations are underestimated.
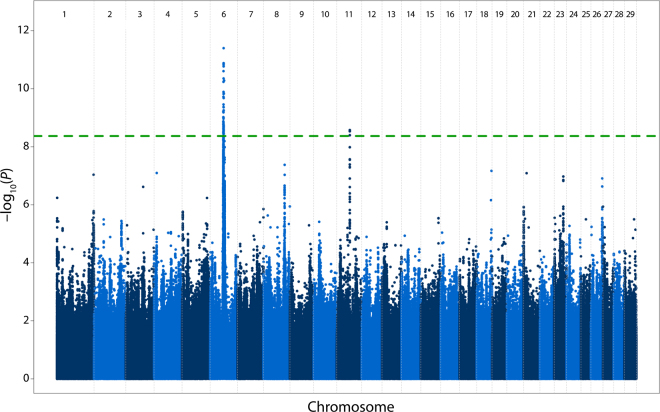
To identify QTL responsible for the correlated semen quality and fertility characteristics, we conducted a sequence-based multitrait meta-analysis. The meta-analysis recovered a total of 132 significant variants (P < 4.24 × 10−9) clustered in 2 distinct QTL regions on chromosomes 6 and 11 (Figure 2). A QTL on BTA 6 was delimited by 119 significantly associated markers (P < 4.22 × 10−9, MAF = 0.20–0.21) between positions 56643511 and 60068785 (ARS-UCD1.2; ftp://ftp.ncbi.nlm.nih.gov/genomes/refseq/vertebrate_mammalian/Bos_taurus/latest_assembly_versions/GCF_002263795.1_ARS-UCD1.2/), with the lead located at 57946201 bp (P = 4.00 × 10−12, MAF = 0.24). The QTL on BTA 6 was also identified in the individual trait GWAS for motility (P = 5.69 × 10−11), head anomalies (P = 8.35 × 10−12), and tail anomalies (P = 3.81 × 10−13), suggesting pleiotropic effects. This QTL harbors a putatively causal variant that activates cryptic splicing in WDR19 encoding WD repeat domain 19 (Hiltpold et al., 2020, 2021). The QTL on BTA 11 was located between positions 55996705 and 56001087 (P < 4.02 × 10−9, MAF = 0.09) with 2 leading SNPs that had identical P-values (56001085 and 56001087, P = 2.66 × 10−9). However, this region was only significant in the single-trait GWAS for sperm head anomalies (P = 3.42 × 10−11). The top variants were within an intron of CTNNA2 (encoding catenin α-2, a testis-expressed gene, http://cattlegeneatlas.roslin.ed.ac.uk/) that has yet to be associated with bull fertility or semen quality. The multitrait meta-analysis did not reveal associations at putative QTL underpinning male reproduction on chromosomes 1, 18, 25, and 26, which were reported in the same cohort by Hiltpold et al. (2021). These QTL act recessively, but their effect on male fertility traits is lower than that of the BTA 6 QTL, thus they likely remained undetected in our meta-analysis that assumed an additive mode of inheritance.
Figure 2.
Manhattan plot from the meta-analysis of male fertility and reproduction traits [percent of sperm anomalies–head, percent of sperm anomalies–tail, percent of motile sperm, concentration (million sperm/mL), ejaculate volume, and insemination success (sire nonreturn rate)]. The analysis considered 11,804,781 genome-wide variants, with the green line representing a significance threshold of P < 4.24 × 10−9.
In conclusion, we did not observe notable genetic correlations between male fertility and other economically relevant traits in the BSW cattle population. The genetic correlations were calculated using allele substitution effects estimated from an additive regression model; we did not consider other modes of inheritance that have been shown to contribute to variation in male reproduction (Peñagaricano et al., 2012; Nani et al., 2019; Hiltpold et al., 2021). Future studies that also consider recessive and dominant modes of inheritance may provide further insight into the genetic relationships between economically relevant traits and bull fertility. Moreover, our genetic correlation estimates were based on genome-wide markers. It is plausible that individual QTL exhibit pleiotropic effects on the traits studied and remain undetected by correlation coefficients estimated from genome-wide approaches. Our male reproductive trait GWAS, from which we obtained allele substitution effects to estimate the genetic correlations with production phenotypes, relied on 983 individuals. This small sample size may have hindered our ability to identify minor relationships between these trait categories. However, our recovery of other well-documented instances of genetic correlation (e.g., milk yield and cow nonreturn rate, stature and milk yield) demonstrates the possibility of identifying common relationships with small data sets. A large-effect QTL on BTA 6 with pleiotropic effects on male fertility characteristics did not exhibit notable effects on other traits of economic relevance. Large mapping cohorts with well-defined phenotypes may reveal further QTL underpinning male fertility. However, such QTL are likely either to be rare in BSW cattle or to have low-to-moderate effects on the traits studied, otherwise they would have been detected in the current study or in Hiltpold et al. (2020, 2021).
Notes
This study was financially supported by Swissgenetics (Zollikofen, Switzerland; https://swissgenetics.ch/), the Swiss National Science Foundation (Grant ID: 310030_185229; Bern, Switzerland), the European Union's Horizon 2020 research and innovation program under Grant Agreement No 815668 (BovReg; European Commission), and an ETH Research Grant (Grant ID: ETH-17 20-2; Zurich, Switzerland). The funding bodies were not involved in the design of the study and collection, analysis, interpretation of data, or writing the manuscript.
We thank Fritz Schmitz-Hsu (Swissgenetics, Zollikofen, Switzerland) for providing sire nonreturn rate estimates for the BSW bulls. We thank Jeremie Vandenplas (Wageningen University & Research, Wageningen, the Netherlands) for providing a modified version of the GCTA tool that allows for fitting weights while considering allele dosage data. We thank QualitasAG and Braunvieh Schweiz for providing genotype and phenotype data.
The authors have not stated any conflicts of interest.
References
- Berry D.P., Eivers B., Dunne G., McParland S. Genetics of bull semen characteristics in a multi-breed cattle population. Theriogenology. 2019;123:202–208. doi: 10.1016/j.theriogenology.2018.10.006. 30317043. [DOI] [PubMed] [Google Scholar]
- Berry D.P., Wall E., Pryce J.E. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal. 2014;8(Suppl. 1):105–121. doi: 10.1017/S1751731114000743. 24703258. [DOI] [PubMed] [Google Scholar]
- Boligon A.A., Silva J.A.V., Sesana R.C., Sesana J.C., Junqueira J.B., Albuquerque L.G. Estimation of genetic parameters for body weights, scrotal circumference, and testicular volume measured at different ages in Nellore cattle. J. Anim. Sci. 2010;88:1215–1219. doi: 10.2527/jas.2008-1719. 19966155. [DOI] [PubMed] [Google Scholar]
- Bolormaa S., Pryce J.E., Reverter A., Zhang Y., Barendse W., Kemper K., Tier B., Savin K., Hayes B.J., Goddard M.E. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004198. 24675618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braundmeier A.G., Miller D.J. The search is on: Finding accurate molecular markers of male fertility. J. Dairy Sci. 2001;84:1915–1925. doi: 10.3168/jds.S0022-0302(01)74633-4. 11573769. [DOI] [PubMed] [Google Scholar]
- Browning B.L., Zhou Y., Browning S.R. A one-penny imputed genome from next-generation reference panels. Am. J. Hum. Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. 30100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burren A., Joerg H., Erbe M., Gilmour A.R., Witschi U., Schmitz-Hsu F. Genetic parameters for semen production traits in Swiss dairy bulls. Reprod. Domest. Anim. 2019;54:1177–1181. doi: 10.1111/rda.13492. 31206856. [DOI] [PubMed] [Google Scholar]
- Butler M.L., Bormann J.M., Weaber R.L., Grieger D.M., Rolf M.M. Selection for bull fertility: A review. Transl. Anim. Sci. 2019;4:423–441. doi: 10.1093/tas/txz174. 32705001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haas Y., Janss L.L.G., Kadarmideen H.N. Genetic and phenotypic parameters for conformation and yield traits in three Swiss dairy cattle breeds. J. Anim. Breed. Genet. 2007;124:12–19. doi: 10.1111/j.1439-0388.2007.00630.x. 17302955. [DOI] [PubMed] [Google Scholar]
- de Souza Fonseca P.A., Id-Lahoucine S., Reverter A., Medrano J.F., Fortes M.S., Casellas J., Miglior F., Brito L., Carvalho M.R.S., Schenkel F.S., Nguyen L.T., Porto-Neto L.R., Thomas M.G., Cánovas A. Combining multi-OMICs information to identify key-regulator genes for pleiotropic effect on fertility and production traits in beef cattle. PLoS One. 2018;13 doi: 10.1371/journal.pone.0205295. 30335783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druet T., Fritz S., Sellem E., Basso B., Gérard O., Salas-Cortes L., Humblot P., Druart X., Eggen A. Estimation of genetic parameters and genome scan for 15 semen characteristics traits of Holstein bulls. J. Anim. Breed. Genet. 2009;126:269–277. doi: 10.1111/j.1439-0388.2008.00788.x. 19630877. [DOI] [PubMed] [Google Scholar]
- Ducrocq V., Humblot P. Genetic characteristics and evolution of semen production of young Normande bulls. Livest. Prod. Sci. 1995;41:1–10. doi: 10.1016/0301-6226(94)00029-7. [DOI] [Google Scholar]
- Fang Z.H., Pausch H. Multi-trait meta-analyses reveal 25 quantitative trait loci for economically important traits in Brown Swiss cattle. BMC Genomics. 2019;20:695. doi: 10.1186/s12864-019-6066-6. 31481029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebreyesus G., Lund M.S., Kupisiewicz K., Su G. Genetic parameters of semen quality traits and genetic correlations with service sire nonreturn rate in Nordic Holstein bulls. J. Dairy Sci. 2021;104:10010–10019. doi: 10.3168/jds.2021-20403. 34099302. [DOI] [PubMed] [Google Scholar]
- Gibson K.D., Dechow C.D. Genetic parameters for yield, fitness, and type traits in US Brown Swiss dairy cattle. J. Dairy Sci. 2018;101:1251–1257. doi: 10.3168/jds.2017-13041. 29174155. [DOI] [PubMed] [Google Scholar]
- Gredler B., Fuerst C., Fuerst-Waltl B., Schwarzenbacher H., Sölkner J. Genetic parameters for semen production traits in Austrian dual-purpose Simmental bulls. Reprod. Domest. Anim. 2007;42:326–328. doi: 10.1111/j.1439-0531.2006.00778.x. 17506813. [DOI] [PubMed] [Google Scholar]
- Hiltpold M., Kadri N.K., Janett F., Witschi U., Schmitz-Hsu F., Pausch H. Autosomal recessive loci contribute significantly to quantitative variation of male fertility in a dairy cattle population. BMC Genomics. 2021;22:225. doi: 10.1186/s12864-021-07523-3. 33784962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiltpold M., Niu G., Kadri N.K., Crysnanto D., Fang Z.H., Spengeler M., Schmitz-Hsu F., Fuerst C., Schwarzenbacher H., Seefried F.R., Seehusen F., Witschi U., Schnieke A., Fries R., Bollwein H., Flisikowski K., Pausch H. Activation of cryptic splicing in bovine WDR19 is associated with reduced semen quality and male fertility. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008804. 32407316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri N.K., Sahana G., Charlier C., Iso-Touru T., Guldbrandtsen B., Karim L., Nielsen U.S., Panitz F., Aamand G.P., Schulman N., Georges M., Vilkki J., Lund M.S., Druet T. A 660-kb deletion with antagonistic effects on fertility and milk production segregates at high frequency in Nordic red cattle: additional evidence for the common occurrence of balancing selection in livestock. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004049. 24391517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karoui S., Díaz C., Serrano M., Cue R., Celorrio I., Carabaño M.J. Time trends, environmental factors and genetic basis of semen traits collected in Holstein bulls under commercial conditions. Anim. Reprod. Sci. 2011;124:28–38. doi: 10.1016/j.anireprosci.2011.02.008. 21377297. [DOI] [PubMed] [Google Scholar]
- Kealey C.G., MacNeil M.D., Tess M.W., Geary T.W., Bellows R.A. Genetic parameter estimates for scrotal circumference and semen characteristics of Line 1 Hereford bulls. J. Anim. Sci. 2006;84:283–290. doi: 10.2527/2006.842283x. 16424254. [DOI] [PubMed] [Google Scholar]
- Lloret-Villas A., Bhati M., Kumar Kadri N., Fries R., Pausch H. Investigating the impact of reference assembly choice on genomic analyses in a cattle breed. BMC Genomics. 2021;22:363. doi: 10.1186/s12864-021-07554-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Castillero M., Pegolo S., Sartori C., Toledo-Alvarado H., Varona L., Degano L., Vicario D., Finocchiaro R., Bittante G., Cecchinato A. Genetic correlations between fertility traits and milk composition and fatty acids in Holstein-Friesian, Brown Swiss, and Simmental cattle using recursive models. J. Dairy Sci. 2021;104:6832–6846. doi: 10.3168/jds.2020-19694. 33773778. [DOI] [PubMed] [Google Scholar]
- Martínez-Velázquez G., Gregory K.E., Bennett G.L., Van Vleck L.D. Genetic relationships between scrotal circumference and female reproductive traits. J. Anim. Sci. 2003;81:395–401. doi: 10.2527/2003.812395x. 12643482. [DOI] [PubMed] [Google Scholar]
- Meyer K., Hammond K., Mackinnon M.J., Parnell P.F. Estimates of covariances between reproduction and growth in Australian beef cattle. J. Anim. Sci. 1991;69:3533–3543. doi: 10.2527/1991.6993533x. 1938640. [DOI] [PubMed] [Google Scholar]
- Miglior F., Fleming A., Malchiodi F., Brito L.F., Martin P., Baes C.F. A 100-Year Review: Identification and genetic selection of economically important traits in dairy cattle. J. Dairy Sci. 2017;100:10251–10271. doi: 10.3168/jds.2017-12968. 29153164. [DOI] [PubMed] [Google Scholar]
- Misztal I., Tsuruta S., Strabel T., Auvray B., Druet T., Lee D.H. Proc. 7th World Congress on Genetics Applied to Livestock Production. 2002. BLUPF90 and related programs (BGF90) pp. 743–744. [Google Scholar]
- Nani J.P., Rezende F.M., Peñagaricano F. Predicting male fertility in dairy cattle using markers with large effect and functional annotation data. BMC Genomics. 2019;20:258. doi: 10.1186/s12864-019-5644-y. 30940077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.H., See M.T., Long T.E., Galvin J.M. Estimates of genetic correlations between production and semen traits in boar. Asian-Australas. J. Anim. Sci. 2006;19:160–164. doi: 10.5713/ajas.2006.160. [DOI] [Google Scholar]
- Olsen H.B., Heringstad B., Klemetsdal G. Genetic analysis of semen characteristic traits in young Norwegian Red bulls. J. Dairy Sci. 2020;103:545–555. doi: 10.3168/jds.2019-17291. 31668445. [DOI] [PubMed] [Google Scholar]
- Oltenacu P.A., Frick A., Lindhé B. Relationship of fertility to milk yield in Swedish cattle. J. Dairy Sci. 1991;74:264–268. doi: 10.3168/jds.S0022-0302(91)78169-1. 2030172. [DOI] [PubMed] [Google Scholar]
- Pacheco H.A., Battagin M., Rossoni A., Cecchinato A., Peñagaricano F. Evaluation of bull fertility in Italian Brown Swiss dairy cattle using cow field data. J. Dairy Sci. 2021;104:10896–10904. doi: 10.3168/jds.2021-20332. 34304869. [DOI] [PubMed] [Google Scholar]
- Palasz A.T., Cates W.F., Barth A.D., Mapletoft R.J. The relationship between scrotal circumference and quantitative testicular traits in yearling beef bulls. Theriogenology. 1994;42:715–726. doi: 10.1016/0093-691X(94)90388-y. 16727577. [DOI] [PubMed] [Google Scholar]
- Peñagaricano F., Weigel K.A., Khatib H. Genome-wide association study identifies candidate markers for bull fertility in Holstein dairy cattle. Anim. Genet. 2012;43(Suppl. 1):65–71. doi: 10.1111/j.1365-2052.2012.02350.x. 22742504. [DOI] [PubMed] [Google Scholar]
- Pryce J.E., Royal M.D., Garnsworthy P.C., Mao I.L. Fertility in the high-producing dairy cow. Livest. Prod. Sci. 2004;86:125–135. doi: 10.1016/S0301-6226(03)00145-3. [DOI] [Google Scholar]
- R Core Team R: A language and environment for statistical computing. 2021. https://www.R-project.org/
- Raidan F.S.S., Tineo J.S.A., de Moraes M.M., Escarce T.C., de Araújo A.E.M., Gomes M. M. de C., Ventura H.T., Toral F.L.B. Associations among growth, scrotal circumference, and visual score of beef cattle in performance tests on pasture or in feedlots. Rev. Bras. Zootec. 2017;46:309–316. doi: 10.1590/s1806-92902017000400006. [DOI] [Google Scholar]
- Rezende F.M., Dietsch G.O., Peñagaricano F. Genetic dissection of bull fertility in US Jersey dairy cattle. Anim. Genet. 2018;49:393–402. doi: 10.1111/age.12710. 30109710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roxström A., Strandberg E., Berglund B., Emanuelson U., Philipsson J. Genetic and environmental correlations among female fertility traits and milk production in different parities of Swedish red and white dairy cattle. Acta Agric. Scand. A Anim. Sci. 2001;51:7–14. doi: 10.1080/090647001300004745. [DOI] [Google Scholar]
- Schaeffer L.R. Evaluation of bulls for nonreturn rates within artificial insemination organizations. J. Dairy Sci. 1993;76:837–842. doi: 10.3168/jds.S0022-0302(93)77409-3. [DOI] [Google Scholar]
- Toelle V.D., Robison O.W. Estimates of genetic correlations between testicular measurements and female reproductive traits in cattle. J. Anim. Sci. 1985;60:89–100. doi: 10.2527/jas1985.60189x. [DOI] [PubMed] [Google Scholar]
- VanRaden P.M., Sanders A.H., Tooker M.E., Miller R.H., Norman H.O., Kuhn M.T., Wiggans G.R. Development of a national genetic evaluation for cow fertility. J. Dairy Sci. 2004;87:2285–2292. doi: 10.3168/jds.S0022-0302(04)70049-1. 15328243. [DOI] [PubMed] [Google Scholar]
- Wolf J. Genetic correlations between production and semen traits in pig. Animal. 2009;3:1094–1099. doi: 10.1017/S1751731109004686. 22444838. [DOI] [PubMed] [Google Scholar]
- Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. 21167468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zaitlen N.A., Goddard M.E., Visscher P.M., Price A.L. Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 2014;46:100–106. doi: 10.1038/ng.2876. 24473328. [DOI] [PMC free article] [PubMed] [Google Scholar]