Abstract
Background
Human and animal cases of Rift Valley fever (RVF) are typically only reported during large outbreaks. The occurrence of RVF cases that go undetected by national surveillance systems in the period between these outbreaks is considered likely. The last reported cases of RVF in Tanzania occurred during a large outbreak in 2007–2008.
Methods
Samples collected between 2017 and 2019 from livestock suffering abortion across northern Tanzania were retrospectively tested for evidence of RVF virus infection using serology and reverse transcription quantitative polymerase chain reaction (RT-qPCR).
Results
A total of 14 RVF-associated cattle abortions were identified among dairy cattle in a peri-urban area surrounding the town of Moshi. RVF cases occurred from May to August 2018 and were considered to represent an undetected, small-scale RVF outbreak. Milk samples from 3 of 14 cases (21%) were found to be RT-qPCR positive. Genotyping revealed circulation of RVF viruses from two distinct lineages.
Conclusions
RVF outbreaks can occur more often in endemic settings than would be expected on the basis of detection by national surveillance. The occurrence of RVF cases among peri-urban dairy cattle and evidence for viral shedding in milk, also highlights potentially emerging risks for RVF associated with increasing urban and peri-urban livestock populations.
Keywords: Africa; food microbiology; milk; population surveillance; Rift Valley fever, zoonoses
Introduction
Rift Valley fever (RVF) is a mosquito-borne disease caused by the Rift Valley fever virus (RVFV) that affects people and animals across Africa and the Arabian Peninsula. Previous RVF outbreaks have been major public health emergencies in affected countries and the disease is considered to be a priority for research and intervention by the World Health Organization (WHO).1 Ruminant livestock are highly susceptible to RVFV, with disease in cattle, goats and sheep associated with abortion and mortality in young animals.2
The epidemiology of RVF in East Africa is characterised by infrequent epidemics that are triggered by the emergence of large numbers of floodwater mosquitoes following periods of unusually heavy rainfall.3 In Tanzania, RVF epidemics have been reported every 10–15 y, with the last human or animal cases identified in the country in the year 2007.4 While clinical disease in people and animals is typically not reported outside epidemics in most countries in East Africa, an increasing number of serological surveys provide evidence for regular circulation of RVFV during this interepidemic period.2,5,6
Surveillance for RVF in most endemic countries, including Tanzania, is passive, requiring the reporting of suspicious illness in people or livestock. However, detection and confirmation of RVF cases during outbreaks is likely to be challenging for several reasons. In people and animals, presenting symptoms and signs of RVF are typically non-specific.2 Evidence-based thresholds for reporting and investigating unusually high rates of abortion or mortality in livestock have also generally not been established. Where suspect human or animal cases are reported, confirmation requires RVF-specific laboratory tests, which can have limited availability in endemic countries. These issues are compounded by medical and veterinary infrastructures being particularly weak in the grassland areas that tend to be at highest risk for RVF.3 Such infrastructure can be further weakened during periods of flooding, which are also the periods when risk of RVFV transmission is greatest.7 Passive surveillance for RVF in these circumstances is likely to have very low sensitivity for detecting disease events, particularly events with small numbers of cases or with limited geographic range.
Given regular circulation of RVFV and expected low sensitivity of current surveillance for disease detection, the occurrence of small-scale outbreaks with undetected human and animal RVFV infections during periods of heavy rainfall is likely.2,7 Such disease events could occur regularly: in the period 2013–2019, meteorological models predicted multiple areas of northern Tanzania to be at high risk for RVF outbreaks in all years except 2014.8
Here we describe a previously unreported outbreak of RVF among dairy cattle near the town of Moshi, Tanzania that occurred in 2018. Infections were detected retrospectively as a result of RVF testing performed on samples collected as part of a research study investigating the infectious aetiology of cattle, goat and sheep abortions across northern Tanzania. The outbreak provides an example of the type of small-scale RVF disease event that may go undetected by national animal health surveillance systems.
Methods
Prospective cohort study of livestock abortion
A prospective cohort study of livestock abortions was conducted from November 2017 through October 2019 in three administrative regions of northern Tanzania. This study and its principal findings are described in detail elsewhere.9 Briefly, the study population included all livestock-keeping households in 13 wards in Arusha, Manyara and Kilimanjaro Regions. Livestock keepers in these regions have been classified as engaging in a mixture of agropastoral, pastoral and smallholder-based livelihoods.10
Each study ward was served by a livestock field officer (LFO), a government-employed veterinary technician providing basic veterinary services. Community meetings were held in study wards and livestock keepers were asked to report any cattle, goat and sheep abortions to their LFO. Within 72 h of a reported abortion, the LFO collected blood, milk and vaginal swab samples from the aborting dam. Placental tissue samples and foetal surface swabs were also collected where available. Tissue, milk and swab samples were preserved in DNA/RNA shield (Zymo Research, Irvine, CA, USA). Basic individual animal- and household-level data, including breed, service date, animal demographics and vaccination history, were recorded.
All households were revisited 4–6 weeks after the investigated abortion and a convalescent-phase blood sample was collected from the affected dam.
RVF livestock abortion case definition
Based on World Organisation for Animal Health (OIE) guidelines,11 we used a combination of molecular and serological methods to identify RVF cases. A case of RVF-associated livestock abortion was defined as an abortion event with RVFV detected by one-step reverse transcription quantitative polymerase chain reaction (RT-qPCR) in any of the vaginal swab, foetal swab or placental tissues and evidence of RVFV antibodies by enzyme-linked immunosorbent assay (ELISA) on a blood sample collected on an acute or convalescent serum sample from the dam.9
RVF molecular and serological testing was performed retrospectively, with diagnoses made approximately 12 months after abortion events occurred. Households in which RVF cases were identified were revisited shortly after detection (i.e. around 12 months post-abortion) and livestock keepers informed of positive test results. During this third visit, a blood sample was collected from any animals that had been confirmed as RVF cases and were still present.
Serological testing for RVFV
Sera were separated from clotted whole blood samples by centrifugation (≤1300 g for 10 min). Sera from acute, convalescent and 12-month blood samples were tested for RVFV immunoglobulin G antibodies using a multispecies competitive ELISA (ID Screen, IDVet, Paris, France).
RT-qPCR
RNA was extracted from swab, tissue and milk samples using a RNeasy Mini Kit (Qiagen, Hilden, Germany) as described by Thomas et al.9 Details on extraction from milk samples are given in the supplementary materials. Detection of RVFV by RT-qPCR on extracted RNA was performed using published protocols.12 Briefly, a 94 base pair (bp) target of the RVFV G2 gene was amplified using the primer pair RVS and RVA and a fluorescent-labelled probe (RVP). A negative extraction control, negative PCR control (PCR grade water) and positive control (RNA extracted from cells experimentally infected with RVFV-MP1213) were included in each PCR run. Samples were tested in duplicate, with crossover threshold (Ct) values <40 on both wells considered positive.
Genotyping of RVFV
RNA from positive RT-qPCR samples was shipped to the University of Glasgow for viral genotyping. An approximately 900-bp target of the S genome segment was amplified using previously described primers.14 Reactions were performed using the Qiagen OneStep RT-PCR Kit using 8 μL of extracted RNA in a final reaction volume of 25 μL. Initial cycling conditions were reverse transcription at 50°C for 60 min followed by an initial PCR activation step of 95°C for 15 min and then 45 cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1.5 min. A final extension step of 72°C for 10 min was performed. For samples that failed to amplify under these initial conditions, the RT-qPCR was repeated using an extended RT step of 90 minutes at 50°C to increase the amount of complementary DNA template available for amplification. A negative control (PCR grade water) and positive control (RVFV-MP12) were included in each run. PCR products were visualised by gel electrophoresis. For sequencing, PCR products from positive samples were purified using the QIAquick Gel Extraction Kit (Qiagen). Sanger sequencing was performed using both forward and reverse primers by Source Bioscience (Nottingham, UK) using deoxyguanosine triphosphate technology.
Molecular phylogenetic analysis was performed in MEGA 7.0.15 Sequences were aligned using the Clustal W algorithm.15 The evolutionary history was inferred using the maximum likelihood method based on the Kimura two-parameter model with a discrete gamma distribution.16 RVFV sequences were compared with sequences from a diverse set of RVFV strains through BLAST searches of the GenBank nucleotide database to identify the infecting haplotype.17 The tree with the highest log likelihood value was selected and drawn to scale with branch lengths measured in the number of substitutions per site. All positions containing gaps or missing data were deleted.
In our analysis, particular attention was paid to viral sequences that fell into the same haplotype clade as the live vaccine used in the region (Smithburn RVFV modified vaccine strain, GenBank accession number DQ380157), as this can induce abortions in pregnant animals.18 We therefore sought to rule out vaccination as a cause of abortion among animals meeting our case definition. For this, phylogenetic analysis was first performed to estimate the genetic distance between samples obtained from case animals and the reference Smithburn vaccine strain. Next, the nucleotide sequences from field samples were translated into amino acid sequences using National Center for Biotechnology Information (NCBI) BLAST tools,19,20 aligned and compared with reference amino acid sequences from the Smithburn vaccine strain (DQ380157). We determined whether nucleotide substitutions in field-derived viral strains were synonymous mutations, which could indicate a modified vaccine strain, or non-synonymous mutations, which would be more consistent with a wild-type virus infection.
Results
Prospective cohort study of livestock abortion
Samples were collected from 215 abortion events from November 2017 through October 2019.9 Of the investigated abortions, 71 (33%) involved cattle, 100 (47%) goats and 44 (21%) sheep. Additional details on the breeds of affected animals are given in the supplementary materials. Both RT-qPCR and acute or convalescent phase serological testing for RVF were performed on samples from 212 of 215 (99%) abortion events. The number of abortion events investigated per ward was highly variable, with a minimum of 1 and maximum of 84 abortion events investigated (Supplementary Table S1).
Detection of RVF livestock abortion cases
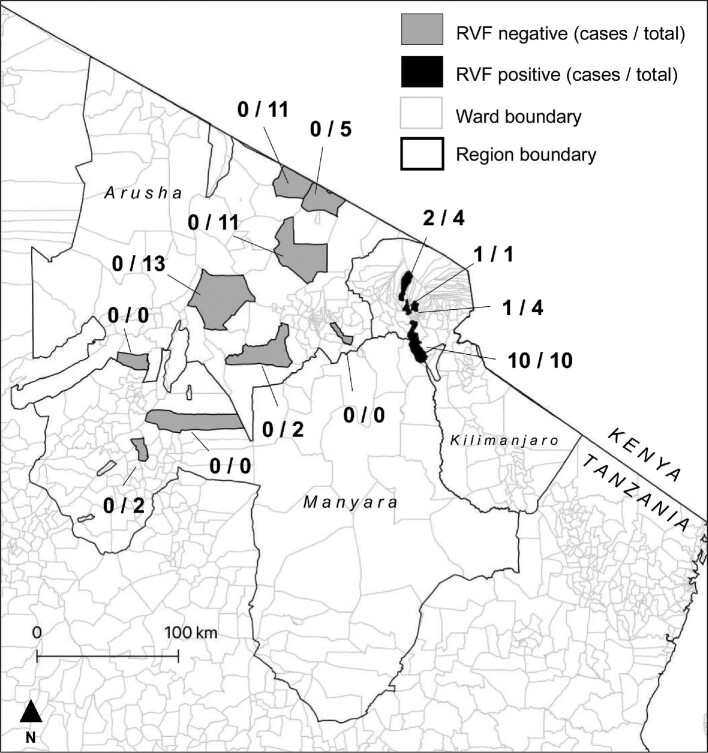
A total of 14 abortion events met our case definition for RVF-associated abortion. All occurred in cattle between 16 May and 11 August 2018 in 14 households. These 14 cases represented 23% of the 63 abortion investigations conducted as part of the prospective cohort study over this time period (i.e. the apparent outbreak period) (Figure 1). Case animals were all found in four wards in three districts surrounding the town of Moshi in the Kilimanjaro Region (Figure 2). These were Arusha Chini and Kindi Wards in Moshi Rural District, Rau Ward in Moshi Municipality and Machame Mashariki Ward in Hai District.
Figure 1.
RVFV RT-qPCR status of livestock abortions in northern Tanzania by week, November 2017–October 2019.
Figure 2.
Location of wards included in the prospective cohort study of livestock abortion in northern Tanzania, 2017– 2019. The number of RVF cases identified out of total abortions investigated during the apparent outbreak period (16 May–11 August 2018) are shown.
Table 1 shows the samples collected from case animals and the results of RT-qPCR and serological testing performed. Further details on Ct values for RT-qPCR and percentage competition ELISA values are given in the Supplementary Tables S2 and S3. Milk samples and vaginal swabs were available from all 14 cases, of which 3 (21%) and 11 (79%) were positive by RT-qPCR, respectively. Ten (91%) of 11 foetal swabs and all 6 (100%) placental samples from confirmed cases were positive for RVFV by RT-qPCR. Of the 14 RVF abortion cases, 13 (93%) dams were seropositive on the basis of samples collected within 72 h of abortion and 1 (7%) case demonstrated seroconversion between acute and convalescent sera (Table 1). All seven case animals that could be sampled at 12 months post-abortion remained seropositive (Table 1 and Supplementary Table S2).
Table 1.
Results of RT-qPCR and serological testing performed on samples from confirmed cattle RVF cases in northern Tanzania, May–July 2018
| RT-qPCR | Serology | |||||||
|---|---|---|---|---|---|---|---|---|
| Abortion date (2018) | District | Milk | Vaginal swab | Placental sample | Foetal swab | Acute serum | Convalescent serum | 12-month serum |
| 16 May | Moshi Rural | − | + | NA | +a | − | + | + |
| 29 May | Moshi Rural | + | + | NA | + | + | + | + |
| 29 May | Moshi Rural | + | + | NA | + | + | + | + |
| 12 June | Moshi Rural | − | + | NA | + | + | + | + |
| 11 June | Moshi Rural | − | + | + | + | + | NA | + |
| 17 June | Moshi Rural | + | − | +b | − | + | + | NA |
| 24 June | Moshi Rural | − | − | + | + | + | + | NA |
| 29 June | Moshi Rural | − | + | NA | NA | + | + | + |
| 3 July | Moshi Municipality | − | + | NA | +c | + | + | + |
| 3 July | Moshi Rural | − | + | NA | NA | + | NA | + |
| 15 July | Hai | − | − | + | NA | + | + | NA |
| 22 July | Moshi Rural | − | + | NA | + | + | + | + |
| 26 July | Hai | − | + | + | + | + | + | + |
| 11 August | Moshi Rural | − | + | + | NA | + | + | NA |
+: positive sample; −: negative sample; NA: sample type that was not available.
Samples used for genotyping (individual identifiers): aSEBI-051, bSEBI-079, cSEBI-094.
One cow that aborted in August 2018 in Hai District and one cow that aborted in July 2019 in Moshi Rural District were also seropositive for RVFV on acute serum samples (and the one available convalescent sample from these animals). Both animals were negative by RT-qPCR testing of milk, vaginal swabs and foetal swabs and so did not meet the case definition for RVF-associated abortion.
Characteristics of RVF cases and affected households
Of the 14 RVF cattle, 1 (7%) was an indigenous breed, 7 (50%) were European dairy breeds and 6 (43%) were cross-breeds. The service date was known for 12 case animals, with abortions occurring at a median of 181 d of gestation (range 128–251). No RVF vaccination was reported in any of the affected herds in the previous 24 months. The median number of adult female cattle kept by households with RVF cases was 5 (range 1–14). Seven of the affected households reported keeping goats in addition to cattle, with a median of 1 (range 0–30) adult female goat. No other livestock were kept by affected households.
Data on cattle grazing management were available for 12 (71%) of the 14 households with RVF cases. Of the households with these data, five (42%) managed cattle under a zero grazing system (permanently housed with fodder cut and brought to animals), three (25%) managed cattle under a tethered grazing system (unhoused but restrained by long rope and moved between grazing areas near the households) and four (33%) managed cattle under a herded grazing system (unhoused and moved between grazing areas by a herdsperson). One (8%) household reported using a mix of zero and herded grazing systems for cattle.
RVFV genotypes identified
An 802-bp fragment of RVFV genome segment S sequence was obtained and analysed from three RVF cases. Positive cattle came from Moshi Rural (SEBI-051 and SEBI-079) and neighbouring Moshi Municipality (SEBI-094) Districts.
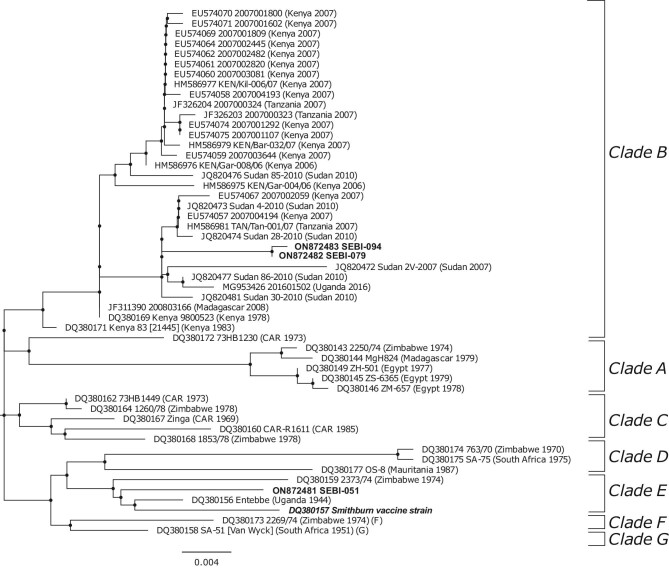
Sequences obtained from SEBI-079 (GenBank accession no. ON872482) and SEBI-094 (GenBank accession no. ON872483) were highly similar (99.9%), with only a single nucleotide substitution between the two sequences. Based on phylogenetic analysis, these two sequences fell within the clade B haplotype17 alongside numerous sequences obtained from previous RVF outbreaks in livestock in Kenya, Tanzania and Sudan (Figure 3). In contrast, the third sequence (SEBI-051, ON872481) fell within the clade E haplotype and was more similar to sequences obtained from a South African outbreak in 2009 than sequences previously described in East Africa (Figure 3). Despite falling into the same clade as the Smithburn vaccine strain, a total of 11 nucleotide substitutions were detected between the Smithburn vaccine strain and the field sample SEBI-051 in the coding region of the partial sequence of the S segment used for phylogenetic analysis. In the translated amino acid sequences, seven variable sites were identified (amino acid numbers 16, 67, 180, 208, 222, 242 and 250) between the attenuated Smithburn vaccine strain and wild-type viruses within the same RVFV clade. When compared with the sequence obtained from SEBI-051, all nucleotide substitutions were non-synonymous mutations resulting in a change of amino acids at all sites. Therefore the virus obtained from SEBI-051 was considered to be a wild-type virus.
Figure 3.
Molecular phylogenetic analysis of partial sequence (802 bp) of segment S of RVFV strains obtained from Tanzanian cattle, northern Tanzania, 2018. The tree with the highest log likelihood (−2414.72) is shown. Sequences are labelled with GenBank accession number and designated strain names with country and year of origin shown in parentheses. Sequences from this study are highlighted in bold and labelled with GenBank accession numbers and unique animal individual identifiers. Clades of viral lineages as designated by Bird et al. 17 are labelled by brackets. Sequence from the Smithburn vaccine strain is highlighted in bold italics.
Discussion
We describe an apparently small outbreak of RVF occurring among dairy cattle in a peri-urban area of northern Tanzania in 2018. Detection of livestock cases was accomplished through active, abortion-based sampling triggered by individual animal abortions rather than through a passive surveillance response to reports of unusually high levels of abortion. The retrospective detection of this previously unreported RVF outbreak provides evidence that RVF-associated livestock abortions are occurring more regularly in this endemic setting than would be expected on the basis of national surveillance alone. The occurrence of small-scale RVF outbreaks that go undetected by surveillance systems in endemic settings is not unexpected, but such events have rarely been described in detail.
The outbreak we describe has several notable features. First, all RVF cases were detected in animals reared in a peri-urban area. Transmission in urban and peri-urban areas is a key public health concern for many arboviruses21 but has been infrequently described for RVFV. Indeed, the WHO reports that there is no evidence for RVF outbreaks in urban areas,22 which could be taken to imply that RVF does not pose a threat to urban populations. In Tanzania, the peri-urban livestock sector is undergoing rapid expansion to meet the growing demand for milk and meat.23 Growing livestock populations within and surrounding high-density human population areas may pose new risks for zoonotic disease transmission,24 including for RVF.
These zoonotic risks are highlighted by the detection of RVFV nucleic acids in milk from multiple aborting dams sampled in this study. All RVF-associated abortions we identified were dairy cattle reared in an area that supplies the majority of milk to the town of Moshi.25 To our knowledge, the only published report of RVFV detection in milk was from experimentally infected animals in the 1950s.26 Our samples were preserved in a viricidal solution, so we were unable to assess the infectivity of milk from RVF cases, but milk-borne RVFV transmission is supported by literature-based reports of significant positive associations between RVFV seropositivity in people and a history of unboiled milk consumption.27 Few, if any, data exist on the effectiveness of heat treatment for inactivation of RVFV in milk and milk products. RVFV viability using culture-adapted virus has been detected following short periods of heat treatment at 56°C,28 raising concerns about the potential thermal stability of RVFV. In heat treatment experiments performed by our group using the RVFV-MP12 vaccine strain spiked in milk, we found no viral survival after incubation at 72°C for 15 min (further detail in the supplementary materials). Other authors report complete inactivation of RVFV after heating solutions containing the virus for 1 h at 60°C.29 These data suggest heat treatment is likely to reduce the risk of RVFV transmission via milk, but further work is needed to explore the effectiveness of standard milk pasteurisation and other milk preparation procedures on inactivation of the virus. Milk can be an important source of nutrition for food insecure households in RVF-endemic settings and interventions aimed at promoting milk safety should be carefully designed to avoid unintended consequences such as reduced consumption or inequitable access to safe products.25
A second notable feature of this outbreak is that despite its apparently limited spatial and temporal extent, we identified the circulation of viruses from two distinct lineages. The outbreak around Moshi occurred at the same time as outbreaks involving human and livestock populations were occurring in Kenya, Rwanda and Uganda.30 It is therefore possible that there were repeated virus introductions via livestock movements from neighbouring countries, which could explain the co-occurrence of these two viral haplotypes. This would also suggest that the Moshi outbreak was part of a more widespread, regional-level RVFV transmission rather than a small, isolated RVF outbreak arising autochthonously in northern Tanzania. Alternatively, the apparent viral diversity observed could be explained by repeated emergence from the low-level endemic viral cycling among livestock that has been described in Tanzania2,5,6 or spillover from unknown sylvatic cycles. It is worth noting that the town of Moshi sits on the edge of Kilimanjaro National Park and one of the wards in which cases were identified (Machame Mashiriki) directly borders this forested area. While limited data exist, it has been suggested that RVFV may circulate in unidentified sylvatic cycles in the forests of East Africa.31 Attempts to confirm and characterise these cycles in northern Tanzania would represent a valuable area for future research. At the time of writing, we are not aware of viral sequence data from the outbreaks in the wider East African region in 2018 that could help address questions around virus origin. These unanswered questions, and the diversity of viral strains we observed during this small outbreak, clearly demonstrate the value of genotyping during RVF oubreaks.32
It is also noteworthy that these two distinct viral lineages were identified in samples from only 3 of 14 RVF cases, raising the possibility of greater diversity than observed here. We performed genotyping on RT-qPCR products from placental and foetal samples, which tended to have the lowest Ct values (and therefore the highest concentration of viral RNA). Of the 12 available placental or foetal samples, insufficient sample was available for three after testing had been completed on the available aliquot at the University of Glasgow for the range of abortigenic pathogens evaluated as part of the wider research study9 (an additional aliquot has been retained in Tanzania). Among the six aliquots in which genotyping was not successful, two (SEBI-064 and SEBI-076) had Ct values in placental and/or foetal samples that were equivalent to the three samples in which we were able to amplify sufficient PCR product for genotyping (i.e. a Ct of ≤26). Although numbers are too small to draw firm conclusions, a hypothesis for this failure is primer mismatch with these viral haplotypes. The primers used in this study were adapted from an assay developed for working with the MP12 (lab modified) strain of RVFV rather than wild-type viruses. This may have resulted in a higher proportion of samples with amplification success of some haplotypes rather than others. Of note, a longer RT step was required for SEBI-051, which aligned with RVFV clade E, compared with SEBI-079 and SEBI-094, which aligned with RVFV clade B, which would support this hypothesis. A priority for future work will be to perform whole genome sequencing to overcome these potential issues with primer mismatch and to further enhance our understanding of the RVFV viral diversity associated with this outbreak.
We show that RVFV nucleic acids can be detected in a range of sample types collected within 72 h of cattle abortion. The number of available RVF cases was small and all sample types were not available for all animals, but placental material was most consistently RT-qPCR positive among the RVF cases, followed by swabs from the foetal surface. Encouragingly, vaginal swabs also appear to have good sensitivity for RVFV detection. Sampling directly from the dam has considerable advantages over collecting swabs or material from the foetus or placenta, which are frequently scavenged by dogs and wildlife or can become rapidly autolysed in hot climates. A surveillance system in which vaginal swabs are collected from recently aborted dams by livestock field officers or other trained people living in livestock-keeping communities and submitted for PCR-based testing at a regional laboratory, particularly during high-risk periods,8 would enable the early detection of RVF outbreaks, including small-scale outbreaks such as that described here. This could allow ring vaccination around an outbreak and the implementation of targeted livestock movement restrictions, reducing the risks of onward spread.33 Although there are likely to be many logistic and economic challenges with such a system, particularly given the historically low levels of investment in veterinary surveillance in many RVF-endemic countries,34 these costs and challenges may be offset if early detection of RVF cases and targeted interventions can contribute to preventing RVF outbreaks from becoming established as national and regional epidemics. In Tanzania, previous large-scale epidemics have resulted in widespread human illness and mortality, losses of large numbers of livestock and major impacts on the livestock sector through regional movement restrictions and slaughter bans.4,35
The small number of cases detected in this study, and the apparent geographic isolation of cases, suggest the occurrence of a small-scale RVF outbreak. However, there are important limitations to the prospective cohort study in which RVF cases were detected that create uncertainty about the true size of the outbreak we describe. In particular, there were substantial differences in the number of abortions investigated in each study ward, with reporting likely to be strongly biased by levels of engagement of community members with their LFO. A particular source of this bias that is reflected in our data is the large number of European dairy breeds and their crosses in the study sample and among cases. These animals are typically higher value than local breeds and can therefore be expected to receive higher levels of veterinary care. European-breed dairy cattle and their crosses are primarily reared in smallholder, peri-urban areas in northern Tanzania and are relatively rare across rural areas of the region.10 Smallholder dairy systems should therefore be considered to be overrepresented in our sample. Given this selection bias, we cannot rule out a larger RVF outbreak or multiple separate outbreaks occurring over a wider geographic area or longer time period. All RVF abortions were detected during a high-rainfall period in which multiple areas of northern Tanzania were considered to be at high risk for RVF outbreaks.8,30
Conclusions
Testing of samples collected as part of a research study investigating the aetiology of livestock abortion allowed us to retrospectively identify a small outbreak of RVF occurring among livestock in Tanzania in 2018 that involved RVF viruses from two distinct lineages. Our data provide a rare example of the type of small-scale RVF outbreak that can occur below the surveillance detection threshold in endemic countries. The outbreak we describe occurred among cattle in the emerging dairy sector in the peri-urban area surrounding the town of Moshi. Importantly, we identified shedding of RVFV nucleic acids in milk from affected animals, supporting growing evidence for milk as a potential source of human RVFV infection, potentially including urban consumers. Promotion of milk safety measures, including pasteurisation and home boiling, may therefore contribute to reduced zoonotic RVF risk in endemic settings during high-risk periods. Our data suggest that RT-qPCR performed on vaginal swabs collected within 72 h of a bovine abortion has good sensitivity for the detection of RVFV nucleic acids from RVF abortion cases. This simple and low-cost sampling approach has the potential to be integrated into active abortion-based surveillance, potentially enabling early detection and response to RVF outbreaks.
Supplementary Material
Acknowledgements
We would like to thank the participating livestock-keepers, as well as village, ward, district and regional authorities. Our particular thanks go to the LFOs who collected data for this study. We are also very grateful to Rigobert Tarimo, Fadhili Mshana, Matayo Melubo, Hassan Hussein, Ephrasia Hugho, Nelson Amani, Elizabeth Kasagama, Victor Mosha and Rosanne de Jong for their support with field and/or laboratory work.
Contributor Information
William A de Glanville, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK; University of Global Health Equity, Kigali 6955, Rwanda.
Kathryn J Allan, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK; School of Veterinary Medicine, University of Glasgow, Glasgow G61 1QH, UK.
James M Nyarobi, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK; Nelson Mandela African Institution of Science and Technology, Arusha 255, Tanzania.
Kate M Thomas, Centre for International Health, University of Otago, Dunedin 9054, New Zealand; Kilimanjaro Clinical Research Institute, Moshi 2236, Tanzania.
Felix Lankester, Paul G. Allen School for Global Health, Washington State University, Pullman, WA 99164, USA; Global Animal Health Tanzania, Arusha 1642, Tanzania.
Tito J Kibona, Nelson Mandela African Institution of Science and Technology, Arusha 255, Tanzania.
John R Claxton, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK.
Benjamin Brennan, MRC-University of Glasgow Centre for Virus Research, University of Glasgow, Glasgow, G61 1QH, UK.
Ryan W Carter, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK.
John A Crump, Centre for International Health, University of Otago, Dunedin 9054, New Zealand; Division of Infectious Diseases and International Health, Duke University Medical Center, Durham, NC 27710, USA; Duke Global Health Institute, Duke University, Durham, NC 27710, USA; Kilimanjaro Christian Medical University College, Tumaini University, Moshi 3010, Tanzania.
Jo E B Halliday, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK.
Georgia Ladbury, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK.
Blandina T Mmbaga, Kilimanjaro Clinical Research Institute, Moshi 2236, Tanzania; Division of Infectious Diseases and International Health, Duke University Medical Center, Durham, NC 27710, USA; Kilimanjaro Christian Medical University College, Tumaini University, Moshi 3010, Tanzania.
Furaha Mramba, Tanzania Veterinary Laboratory Agency, Dar es Salaam 9254, Tanzania.
Obed M Nyasebwa, Ministry of Livestock and Fisheries, Dodoma 40487, Tanzania.
Matthew P Rubach, Division of Infectious Diseases and International Health, Duke University Medical Center, Durham, NC 27710, USA; Duke Global Health Institute, Duke University, Durham, NC 27710, USA; Programme in Emerging Infectious Diseases, Duke-National University of Singapore, Singapore 169857, Singapore.
Melinda K Rostal, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK; EcoHealth Alliance, New York, NY 10018, USA.
Paul Sanka, Tanzania Veterinary Laboratory Agency, Dar es Salaam 9254, Tanzania.
Emmanuel S Swai, Ministry of Livestock and Fisheries, Dodoma 40487, Tanzania.
Agnieszka M Szemiel, MRC-University of Glasgow Centre for Virus Research, University of Glasgow, Glasgow, G61 1QH, UK.
Brian J Willett, MRC-University of Glasgow Centre for Virus Research, University of Glasgow, Glasgow, G61 1QH, UK.
Sarah Cleaveland, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, G12 8QQ, UK.
Authors’ contributions
SC conceived the study. WAdG, KJA, KMT, FL, TJK, JRC, OMN and SC designed the study protocol. WAdG, KMT, FL, TK, OMN and PS carried out data collection. JMN and RWC carried out serological analysis. KJA, JMN, KMT, BB, RWC, AMS and BJW carried out molecular analysis. AMS and BJW performed the milk heat treatment experiment. WAdG, KJA and JMN performed data analysis. WAdG drafted the manuscript. All authors revised and approved the final manuscript.
Funding
This research was supported by the Supporting Evidence Based Interventions project, University of Edinburgh (R83537) and the Zoonoses and Emerging Livestock Systems program (funded through the Biotechnology and Biological Sciences Research Council, Department for International Development, Economic and Social Research Council, Medical Research Council, Natural Environment Research Council and Defence Science and Technology Laboratory; BB/L018926/1, BB/L017679/1, BB/N503563/1). Additional financial support was provided through the Biotechnology and Biological Sciences Research Council (BB/J010367) and the National Institutes of Health (R01TW009237).
Competing interests
None declared.
Ethical approval
The study protocols, questionnaires and consent documents were approved by the Kilimanjaro Christian Medical Centre (832) and National Institute of Medical Research (2028) Research Ethics Committees, the University of Otago Ethics Committee (H17/069) and the University of Glasgow Medical, Veterinary and Life Sciences Ethics Committee (200140152 and 20017006). Permission to carry out the study in Tanzania was provided by the Tanzania Commission for Science and Technology (2014-244-ER-2005-141).
Data availability
The data underlying this article are available in the article and in its online supplementary material. The sequences generated by this study are available through NCBI GenBank (accession numbers ON872481–872483).
References
- 1. World Health Organization . An R&D blueprint for action to prevent epidemics. Plan of action. Geneva: World Health Organization; 2016. [Google Scholar]
- 2. Kariuki Njenga M, Bett B. Rift Valley fever virus—how and where virus is maintained during inter-epidemic periods. Curr Clin Micro Rep. 2019;6:18–24. [Google Scholar]
- 3. Anyamba A, Linthicum KJ, Mahoney Ret al. Mapping potential risk of Rift Valley fever outbreaks in African savannas using vegetation index time series data. Photogramm Eng Remote Sensing. 2002;68(2):137–45. [Google Scholar]
- 4. Sindato C, Karimuribo ED, Pfeiffer DUet al. Spatial and temporal pattern of Rift Valley fever outbreaks in Tanzania; 1930 to 2007. PLoS One. 2014;9(2):e88897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matiko MK, Salekwa LP, Kasanga CJet al. Serological evidence of inter-epizootic/inter-epidemic circulation of Rift Valley fever virus in domestic cattle in Kyela and Morogoro, Tanzania. PLoS Negl Trop Dis. 2018;12(11):e0006931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sumaye RD, Abatih EN, Thiry Eet al. Inter-epidemic acquisition of Rift Valley fever virus in humans in Tanzania. PLoS Negl Trop Dis. 2015;9(2):e0003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Labeaud AD, Ochiai Y, Peters CJet al. Spectrum of Rift Valley fever virus transmission in Kenya: insights from three distinct regions. Am J Trop Med Hyg. 2007;76(5):795–800. [PMC free article] [PubMed] [Google Scholar]
- 8. United States Department of Agriculture, Agriculture Research Service . Rift Valley Fever monitor, June 2018. Available from: https://www.ars.usda.gov/ARSUserFiles/60360500/rvf/archives/0618.pdf [accessed 10 August 2022].
- 9. Thomas KM, Kibona T, Claxton JRet al. Prospective cohort study reveals unexpected aetiologies of livestock abortion in northern Tanzania. Sci Rep. 2022;12(1):11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. de Glanville WA, Davis A, Allan KJet al. Classification and characterisation of livestock production systems in northern Tanzania. PLoS One. 2020;15(12):e0229478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Organization for Animal Health. Rift Valley fever (infection with Rift Valley fever virus). In: OIE Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Available from: https://www.woah.org/en/what-we-do/standards/codes-and-manuals/terrestrial-manual-online-access/ [accessed 3 November 2019]. [Google Scholar]
- 12. Drosten C, Göttig S, Schilling Set al. Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR. J Clin Microbiol. 2002;40(7):2323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ikegami T, Hill TE, Smith JKet al. Rift Valley fever virus MP-12 vaccine is fully attenuated by a combination of partial attenuations in the S, M, and L segments. J Virol. 2015;89(14):7262–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennan B, Welch SR, Elliott RM.. The consequences of reconfiguring the ambisense S genome segment of Rift Valley fever virus on viral replication in mammalian and mosquito cells and for genome packaging. PLoS Pathog. 2014;10(2):e1003922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar S, Stecher G, Tamura K.. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16(2):111–20. [DOI] [PubMed] [Google Scholar]
- 17. Bird BH, Khristova ML, Rollin PEet al. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J Virol. 2007;81(6):2805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Faburay B, LaBeaud AD, McVey DSet al. Current status of Rift Valley fever vaccine development. Vaccines (Basel). 2017;5(3):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul SF, Gish W, Miller Wet al. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 20. Ye J, McGinnis S, Madden TL.. BLAST: improvements for better sequence analysis. Nucleic Acids Res. 2006;34:W6–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Weaver SC. Urbanization and geographic expansion of zoonotic arboviral diseases: mechanisms and potential strategies for prevention. Trends Microbiol. 2013;21(8):360–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. World Health Organization . Rift Valley fever. Available from: https://www.who.int/news-room/fact-sheets/detail/rift-valley-fever [accessed 3 November 2019].
- 23. Katjiuongua H, Nelgen S.. Tanzania smallholder dairy value chain development. Situation analysis and trends. Project report. Nairobi:International Livestock Research Institute; 2014. [Google Scholar]
- 24. Ahmed S, Dávila JD, Allen Aet al. Does urbanization make emergence of zoonosis more likely? Evidence, myths and gaps. Environ Urban. 2019;31(2):443–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ladbury G. From cow to consumer: using value chain approaches to evaluate infectious disease risk along dairy value chains serving urban consumers in Moshi Municipality, northern Tanzania. PhD dissertation, University of Glasgow, 2018. [Google Scholar]
- 26. Alexander RA. Rift Valley fever in the union. J S Afr Vet Assoc. 1951;22:105–12. [Google Scholar]
- 27. Grossi-Soyster EN, Lee J, King CHet al. The influence of raw milk exposures on Rift Valley fever virus transmission. PLoS Negl Trop Dis. 2019;13(3):e0007258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daouam S, Fakri FZ, Ennaji MMet al. Heat stability of the Rift Valley fever virus clone 13 live vaccines. Trials Vaccinol. 2014;3:61–4. [Google Scholar]
- 29. Saluzzo JF, Leguenno B, Van der Groen G.. Use of heat inactivated viral haemorrhagic fever antigens in serological assays. J Virol Methods. 1988;22(2–3):165–72. [DOI] [PubMed] [Google Scholar]
- 30. Anyamba A, Soebiyanto RP, Small JLet al. Rift Valley fever outbreak in East Africa: signature of climate extremes. American Geophysical Union, Fall Meeting, Washington, DC, 10–14 December 2018; abstract GH14A-07. [Google Scholar]
- 31. Shoemaker TR, Nyakarahuka L, Balinandi Set al. First laboratory-confirmed outbreak of human and animal Rift Valley fever virus in Uganda in 48 years. Am J Trop Med Hyg. 2019;100(3):659–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maluleke MR, Phosiwa M, van Schalkwyk Aet al. A comparative genome analysis of Rift Valley fever virus isolates from foci of the disease outbreak in South Africa in 2008–2010. PLoS Negl Trop Dis. 2019;13(3):e0006576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaters GL, Johnson PCD, Cleaveland Set al. Analysing livestock network data for infectious disease control: an argument for routine data collection in emerging economies. Philos Trans R Soc Lond B Biol Sci. 2019;374(1776):20180264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Auty H, Swai E, Virhia Jet al. How can we realise the full potential of animal health systems for delivering development and health outcomes. Rev Sci Tech. 2021;40(2):483–95. [DOI] [PubMed] [Google Scholar]
- 35. Sindato C, Karimuribo E, Mboera LEG.. The epidemiology and socio-economic impact of rift valley fever epidemics in Tanzania: a review. Tanzan J Health Res. 2011;13(5 Suppl 1):305–18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. The sequences generated by this study are available through NCBI GenBank (accession numbers ON872481–872483).