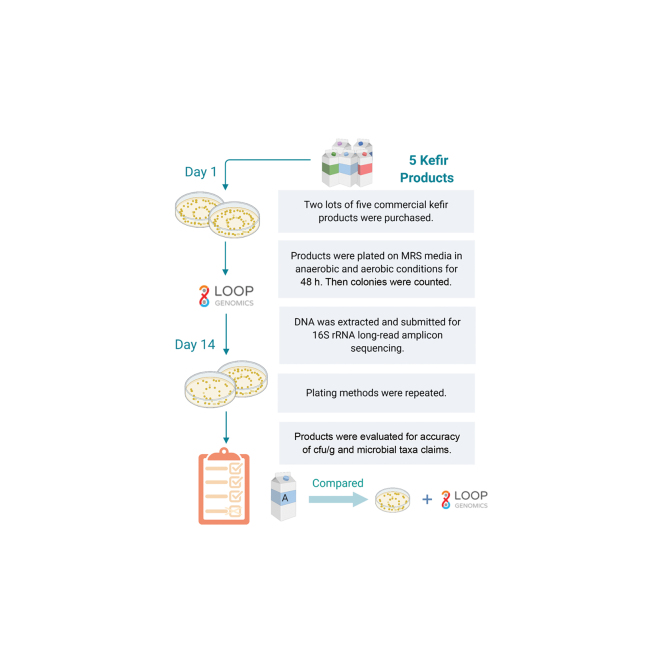
Graphical Abstract
Summary: In this study, label claims of commercial kefir products were subjected to bacterial enumeration and taxa integrity. Previous research suggests that claims of colony-forming units per gram and bacterial taxa on product labels are inaccurate and may be misleading to consumers. This project analyzed 5 commercial kefir products and revealed inaccuracies of label claims that are of great importance to everyday consumers.
Highlights
-
•
Using 16S rRNA gene sequencing combined with traditional plating techniques offers greater label accuracy testing of commercial fermented foods than plating alone.
-
•
The Loop sequencing method allows for high-throughput identification of species-level bacterial taxa present in fermented foods.
-
•
Although a moderate level of label accuracy was observed, some bacterial species, including Streptococcus salivarius and Lactobacillus paracasei, were detected in 4 of 5 products yet not claimed on labels.
Abstract
Kefirs are fermented beverages containing yeast and bacteria produced by the fermentation of water or milk with kefir grains. Because microorganism density may influence a product's health benefits, label accuracy regarding viable bacterial density and taxonomy of fermented foods is important. In this study, the microbiota of 5 commercial kefir products were measured quantitatively using standard plating techniques and characterized using high-resolution, long-read 16S rRNA gene amplicon sequencing. To enumerate viable lactic acid bacteria, 2 lots of each product were plated on de Man, Rogosa and Sharpe agar upon opening and following 14 d and incubated under anaerobic and aerobic conditions. Results revealed that 66% of products with a guaranteed count of colony-forming units per gram overstated microorganism density by at least 1 log, with only product E exceeding 1 × 109 cfu/g. Sequencing results demonstrated moderate product label accuracy in regard to taxonomy, yet several products contained bacterial species above the minimum detectable threshold (0.001% relative abundance) that were not included on the labels (e.g., Streptococcus salivarius, Lactobacillus paracasei). Our results demonstrate a moderate level of labeling accuracy for commercial kefir products intended for human consumption. Regulatory agencies and consumers must continue to scrutinize these products and demand a higher level of accuracy and quality.
The importance of the gut microbiome to human health has become increasingly relevant in fields pertaining to nutrition and nutrient quality. When considering the effect of microorganisms on human health, the International Scientific Association for Probiotics and Prebiotics states that several probiotic strains have documented health benefits, and probiotic fermented foods are defined as having documented health benefits due to the actions of the probiotic (Hill et al., 2014). Probiotic mechanisms can include modulation of immune function, production of antimicrobial compounds, interactions with resident microbiota, and improvement of barrier integrity and enzyme formation (Verruck et al., 2019; Marco et al., 2021), but these effects cannot be expected from consumption of fermented foods. Recently, many fermented foods such as kombucha, kefir, and kimchi have been marketed as beneficial to human health, yet no documented health benefit is necessary for sale (Hill et al., 2014; Marco, 2020). It is important for consumers to understand this difference and know the accurate contents of the fermented foods they consume.
Kefir is a common fermented dairy beverage that contains both yeast and bacteria produced by the fermentation of water or milk with kefir grains (Rosa et al., 2017). Kefir has been consumed for centuries, and due to its affordability and potential to improve health status, many consumers are incorporating it into their lifestyle as a fermented beverage that may contain live microorganisms (Stanton et al., 2001; de Sainz et al., 2020; Mitra and Ghosh, 2020). In addition to testing sensory acceptance (Larosa et al., 2021), several randomized human clinical trials have been conducted in recent years bolstering kefir's health effects (Rosa et al., 2017). These include lower fasting insulin levels, lower levels of serum zonulin, lower erythrocyte sedimentation rate, lower C-reactive protein levels in patients with Crohn's disease, and lower serum lipid profiles in premenopausal women (Fathi et al., 2017; Bellikci-Koyu et al., 2019; Yılmaz et al., 2019; Pražnikar et al., 2020). Each human trial used a different kefir product, which must be considered given the vast bacterial variability present between homemade and commercially produced kefir products (Bourrie et al., 2021). Because the benefits associated with added microorganisms are largely strain specific, verifying the bacterial taxa of kefir products used in clinical trials is of extreme importance when it comes to applying the results to human health (Kechagia et al., 2013).
The US Food and Drug Administration (FDA) considers microorganisms added to products as dietary supplements, and according to the FDA Code of Federal Regulations Title 21 (Code of Federal Regulations, 1975), kefir is categorized as a cultured milk that contains aroma- and flavor-producing microbial cultures. Under this title, kefir companies can make claims pertaining to colony-forming units per gram (cfu/g) and viable bacteria content without being closely monitored or regulated. Research by Merenstein et al. (2019) showed that 65% of the 93 commercial probiotic-containing products assessed lacked supporting clinical research. Correspondingly, research by Weese and Martin (2011) showed that many commercial pet products containing probiotics do not contain what their labeling suggests. Our recent assessment of pet kefir products showed similar inaccuracies in that segment of the market (Metras et al., 2020). Despite a surge in the market for fermented foods such as kefir, little research has been done to confirm the product claims of colony-forming units per gram and bacterial taxa. Regulatory agencies will need to identify core microbial components to impose novel regulations and labeling for fermented foods (Marco et al., 2021).
To address this potential gap in label accuracy, the objectives of this study were to measure viable microbial counts and characterize bacterial taxa of commercial kefir products intended for human consumption as well as compare enumeration and taxonomy data to label claims. Based on the results of previous studies (Weese and Martin, 2011; Merenstein et al., 2019; Metras et al., 2020), we hypothesized that commercially available kefir products would not closely match label claims of viable colony counts or bacterial taxonomy.
Kefir products were purchased from online vendors and local stores in the Urbana, Illinois, area. Products included the following: product A, Maple Hill Plain Kefir; product B, Siggi's Plain Filmjölk; product C, Redwood Hill Farm Plain Goat Milk Kefir; product D, CoYo Kefir; and product E, Lifeway Original Kefir. Chobani Plain Yogurt 0% Milk Fat (YOG) was included as a validation of techniques. For each product, 2 lots were obtained for analysis, and each lot was plated on d 1 and d 14 for thorough enumeration results. Lots were chosen at random, and products were outwardly labeled by their chosen product letter. Labels were evaluated for colony-forming units per gram claims and a clear description of microbial taxa; lot numbers and expiration dates were recorded as stated on the labels. All liquid samples were refrigerated at 4°C until analysis.
To begin, 2 lots of each product were thoroughly vortexed until homogenized, 1 g of product was added to 9 mL of PBS, and the product–PBS mixture was vortexed again. Serial 10-fold dilutions were prepared with PBS. A 50-μL volume of 105 dilutions and a 50-μL volume of 107 dilutions were both plated in duplicate via spiral plater (Eddy Jet Spiral Plater, Neutec Group Inc.) onto de Man, Rogosa and Sharpe (MRS) medium (BD Difco). The MRS plates were incubated anaerobically at 37°C for 48 h and aerobically at 30°C for 48 h to culture lactic acid bacteria. Colony counts were measured via enumeration software (Colony Counter, IUL Flash and Go, Neutec Group Inc.). No attempt to distinguish organisms was made, only enumeration. Fourteen days after product opening, enumeration was repeated for all product lots in identical conditions. Results at time of opening and at d 14 are averages from the duplicate plates of each product lot.
For DNA extraction, 250 mg of kefir product samples from the day of opening was used (DNeasy PowerLyzer PowerSoil Kit, Qiagen) following the manufacturer's instructions. The DNA quantity was determined using a Qubit 3.0 fluorometer (Life Technologies), and DNA quality was assessed via gel electrophoresis (E-Gel Power Snap and E-Gel EX 1% Agarose, Invitrogen). The DNA extracts were sequenced at the W. M. Keck Center for Biotechnology at the University of Illinois at Urbana–Champaign using the Loop Sequencing 16S Long Read Kit (Loop Genomics) at a 1 ng/μL concentration. All samples were processed following the standard Loop Genomics protocol for full-length 16S sequencing. Sequence results were processed through the Silva132 Large Subunit rRNA Database (https://www.arb-silva.de/documentation/release-132/). All taxa with a relative abundance greater than 0.001% were retained for analysis. For plating procedures and DNA extraction, researchers were not blinded to products. For sequencing methods, researchers were blinded to product identification.
Statistical analyses were conducted using R x64 version 4.0.2 (R Foundation for Statistical Computing). Data obtained from enumeration samples on d 1 and 14 and relative abundance lot 1 and relative abundance lot 2 were tested for significance via paired Student's t-test. A P-value <0.05 was considered statistically significant.
All 5 kefir products tested claimed the presence of specific microorganisms in their products, and 3 of 5 products guaranteed minimum counts in colony-forming units per gram (Table 1, Table 2). Analysis of product A specifically identified 5 out of the 10 species claimed on the label, with 7 species being detected in total. In addition, although the label claimed 3 different Lactococcus lactis subspecies, Loop sequencing was unable to distinguish and grouped them together as Lactococcus sp. Streptococcus thermophilus was the majority species detected at 84.50%, with Streptococcus salivarius second highest at 9.30%. These 2 species have very similar 16S rRNA gene sequence, and it is possible that small errors in sequence assembly resulted in misidentifying Strep. thermophilus sequence reads as Strep. salivarius. This product did not make colony-forming units per gram claims, but at opening we detected similar results between lots, with 2.03 × 108 and 1.66 × 108 cfu/g in lots 1 and 2, respectively, under anerobic conditions, and 1.02 × 109 and 1.36 × 109 cfu/g in lots 1 and 2, respectively, under anaerobic conditions. Product B fared well when comparing its label claims and Loop sequencing; 4 out of the 4 genera claimed were detected. As with product A, Loop sequencing was unable to distinguish the Lc. lactis subspecies and identified both Strep. thermophilus and Strep. salivarius. The top 3 genera were Streptococcus (53.60%), Lactococcus (24.15%), and Lactobacillus (3.75%). Although the label did not specify a Leuconostoc species, Loop sequencing identified Leuconostoc mesenteroides in both lots. This product did not make colony-forming units per gram claims, and enumeration values did not change from d 1 to d 14. When considering product C, sequencing specifically detected 4 of 11 species claimed on the label (Strep. thermophilus, Lactobacillus rhamnosus, Lactobacillus casei, and Lactobacillus acidophilus). In addition, Loop sequencing was unable to distinguish the 2 Lactobacillus delbrueckii subspecies and 3 Lc. lactis subspecies that were indicated on the label. No bifidobacteria or Leuconostoc were detected by sequencing. Lactococcus sp. was the second most abundant genus (1.65%) after Streptococcus sp. (79.35%). This product claimed 1 × 1011 cfu/g in its product, but our culturing methods never observed counts as high; lot 1 contained the most viable bacteria, 1.71 × 108 cfu/g at d 1 under anaerobic conditions. Product D contained Strep. thermophilus (88.05%) and Strep. salivarius (7.75%) as the majority taxa. The label claimed 2 different bifidobacteria, but Loop sequencing was able to identify only Bifidobacterium sp. In addition, Lb. acidophilus and Lb. rhamnosus were not specifically identified with sequencing but Lactobacillus sp. might include these 2 species. Interestingly, Pseudomonas was detected in both lots of product D. With claims of 1 × 109 cfu/g on its label, we detected this quantity on d 14 in anaerobic conditions in lot 2. No other culturing test achieved counts as high for product D. The last commercial kefir product we tested was product E, and sequencing confirmed 3 of 11 species claimed on its label (Lb. rhamnosus, Lb. acidophilus, and Lb. casei). Although the label claimed Streptococcus diacetylactis as the only streptococcus added, sequencing identified Strep. thermophilus, Strep. salivarius, and Streptococcus sp. In addition, sequencing did not identify any bifidobacteria or Leuconostoc despite the label claim for each. For product E, culturing results were consistent between lots, with counts exceeding 1 × 109 cfu/g, but none reaching 1 × 1010 cfu/g as its label claims. The YOG sequenced to validate our techniques specifically identified 4 of 5 species on its label, and counts matched the label claims of 1 × 1010 cfu/g. These results give confidence in our methods applied to the kefir products. The data from Table 1, Table 2 justify an expansion of testing to other kefirs and commercialized fermented foods. Out of the 3 products that guaranteed viable bacteria counts, none of those products met the colony-forming units per gram listed at the time of opening. Product D (lot 2) did meet claims when tested at d 14 in anaerobic conditions.
Table 1.
Comparison of bacterial cell count (cfu/g) label claims and enumeration results on de Man, Rogosa and Sharpe (MRS) medium in aerobic (AER) and anaerobic (ANA) conditions
| Product1 | Claimed | Lot | Day 1 |
Day 14 |
||
|---|---|---|---|---|---|---|
| MRS AER | MRS ANA | MRS AER | MRS ANA | |||
| A | NA2 | 1 | 2.03 × 108 ± 6.77 × 107 | 1.02 × 109 ± 6.62 × 107 | 7.53 × 108 ± 4.74 × 106 | 1.37 × 109 ± 2.97 × 108 |
| 2 | 1.66 × 108 ± 5.62 × 107 | 1.36 × 109 ± 2.22 × 108 | 8.71 × 109 ± 7.72 × 109 | 1.80 × 1010 ± 3.84 × 108 | ||
| B | NA | 1 | 4.74 × 108 ± 2.81 × 108 | <2.0 × 106 | 8.49 × 108 ± 1.66 × 107* | 9.92 × 108 ± 6.24 × 108 |
| 2 | 5.46 × 108 ± 3.08 × 108 | 5.84 × 108 ± 1.66 × 107 | 2.56 × 108 ± 5.62 × 107* | 8.81 × 108 ± 8.63 × 107 | ||
| C | 1.00 × 1011 | 1 | 2.24 × 107 ± 1.00 × 10−3 | 1.71 × 108 ± 1.73 × 108 | 1.43 × 107 ± 1.00 × 10−3 | 3.46 × 107 ± 1.15 × 107 |
| 2 | 2.04 × 107 ± 1.44 × 107 | 7.13 × 107 ± 8.65 × 106 | 2.55 × 107 ± 1.44 × 106 | 6.83 × 107 ± 1.44 × 106 | ||
| D | 1.00 × 109 | 1 | 1.89 × 108 ± 7.64 × 107 | 5.67 × 108 ± 1.18 × 107 | 2.69 × 108 ± 1.00 × 10−3 | 3.69 × 108 ± 8.99 × 106 |
| 2 | 8.97 × 107 ± 1.00 × 10−3 | 3.59 × 108 ± 2.31 × 107 | 1.86 × 108 ± 5.62 × 107 | 1.20 × 109 ± 6.24 × 107 | ||
| E | 3.00 × 1010 | 1 | 1.45 × 109 ± 3.95 × 108 | 1.10 × 109 ± 9.74 × 107 | 1.69 × 109 ± 4.07 × 107 | 1.67 × 109 ± 1.00 × 10−3 |
| 2 | 1.55 × 109 ± 1.44 × 108 | 1.28 × 109 ± 2.57 × 108 | 1.00 × 109 ± 7.10 × 106 | 9.36 × 108 ± 2.60 × 107 | ||
| YOG | 1.00 × 1010 | 1 | 5.12 × 109 ± 1.00 × 10−3 | 5.71 × 109 ± 2.51 × 108 | 3.14 × 109 ± 2.12 × 108 | 4.08 × 109 ± 7.94 × 107 |
| 2 | 9.00 × 109 ± 1.08 × 109 | 1.19 × 1010 ± 6.96 × 109 | 4.64 × 109 ± 1.01 × 109 | 1.09 × 1010 ± 1.30 × 109 | ||
Product A: Maple Hill Plain Kefir; product B: Siggi's Plain Filmjölk; product C: Redwood Hill Farm Plain Goat Milk Kefir; product D: CoYo Kefir; product E: Lifeway Original Kefir; YOG: Chobani Plain Yogurt 0% Milk Fat.
Not available.
Indicates a significant difference from d 1 to d 14 (P < 0.05 by paired Student's t-test).
Table 2.
Comparison of label claims and 16S rRNA sequencing results (relative abundance) of commercial kefir products
| Product1 | Claimed | Species | Lot 1 | Lot 2 |
|---|---|---|---|---|
| A | Streptococcus thermophilus | Streptococcus thermophilus | 0.843 | 0.847 |
| Lactobacillus acidophilus | Streptococcus salivarius | 0.090 | 0.096 | |
| Lactobacillus delbrueckii ssp. bulgaricus | Lactobacillus acidophilus | 0.024 | 0.021 | |
| Lactobacillus rhamnosus | Lactobacillus sp. | 0.016 | 0.011 | |
| Lactococcus lactis ssp. lactisbiovar diacetylactis | Streptococcus sp. | 0.013 | 0.013 | |
| Lactococcus lactis ssp. cremoris | Lactobacillus paracasei | 0.006 | 0.005 | |
| Lactococcus lactis ssp. lactis | Lactobacillus delbrueckii | 0.003 | 0.003 | |
| Lactobacillus casei | Lactobacillus rhamnosus | 0.002 | 0.001 | |
| Bifidobacterium lactis | Lactococcus sp. | 0.002 | 0.001 | |
| Leuconostoc cremoris | Lactobacillus casei | 0.001 | 0.001 | |
| B | Streptococcus thermophilus | Streptococcus thermophilus | 0.489 | 0.583 |
| Lactococcus lactis ssp. lactisbiovar diacetylactis | Lactococcus sp. | 0.329 | 0.154 | |
| Lactococcus lactis ssp. lactis | Lactobacillus sp. | 0.049 | 0.026 | |
| Lactococcus lactis ssp. cremoris | Streptococcus salivarius | 0.045 | 0.107 | |
| Lactobacillus delbrueckii ssp. bulgaricus | Lactococcus lactis | 0.043 | 0.051 | |
| Leuconostoc sp. | Leuconostoc mesenteroides | 0.010 | 0.004 | |
| Streptococcus sp. | 0.010 | 0.023 | ||
| Lactococcus raffinolactis | 0.007 | 0.005 | ||
| Lactobacillus rhamnosus | 0.006 | 0.016 | ||
| Lactobacillus delbrueckii | 0.005 | 0.011 | ||
| Lactobacillus paracasei | 0.005 | 0.012 | ||
| Lactobacillus casei | 0.002 | 0.006 | ||
| Leuconostoc pseudomesenteroides | 0.001 | 0.000 | ||
| C | Streptococcus thermophilus | Streptococcus thermophilus | 0.823 | 0.620 |
| Lactococcus lactis ssp. lactisbiovar diacetylactis | Streptococcus salivarius | 0.081 | 0.063 | |
| Lactococcus lactis ssp. cremoris | Lactococcus sp. | 0.032 | 0.001 | |
| Lactobacillus delbrueckii ssp. bulgaricus | Lactobacillus sp. | 0.029 | 0.023 | |
| Lactobacillus delbrueckii ssp. lactis | Streptococcus sp. | 0.014 | 0.007 | |
| Lactobacillus rhamnosus | Lactobacillus delbrueckii | 0.007 | 0.282 | |
| Lactobacillus casei | Lactobacillus rhamnosus | 0.004 | 0.001 | |
| Lactococcus lactis ssp. lactis | Lactobacillus casei | 0.003 | 0.001 | |
| Lactobacillus acidophilus | Lactobacillus paracasei | 0.003 | 0.001 | |
| Leuconostoc mesenteroides ssp. cremoris | Lactococcus lactis | 0.003 | 0.000 | |
| Bifidobacterium lactis | Lactobacillus acidophilus | 0.001 | 0.001 | |
| D | Streptococcus thermophilus | Streptococcus thermophilus | 0.884 | 0.877 |
| Lactobacillus delbrueckii ssp. bulgaricus | Streptococcus salivarius | 0.069 | 0.086 | |
| Bifidobacterium lactis | Streptococcus sp. | 0.011 | 0.012 | |
| Bifidobacterium bifidum | Lactobacillus delbrueckii | 0.011 | 0.009 | |
| Lactobacillus casei | Bifidobacterium sp. | 0.007 | 0.005 | |
| Lactobacillus acidophilus | Leuconostoc mesenteroides | 0.005 | 0.000 | |
| Lactobacillus rhamnosus | Pseudomonas sp. | 0.005 | 0.000 | |
| Pseudomonas lundensis | 0.003 | 0.003 | ||
| Lactobacillus sp. | 0.002 | 0.002 | ||
| Lactococcus sp. | 0.000 | 0.001 | ||
| Lactobacillus casei | 0.000 | 0.001 | ||
| E | Streptococcus diacetylactis | Streptococcus thermophilus | 0.648 | 0.680 |
| Lactobacillus rhamnosus | Lactobacillus delbrueckii | 0.119 | 0.108 | |
| Lactococcus lactis | Lactobacillus paracasei | 0.103 | 0.103 | |
| Lactobacillus acidophilus | Streptococcus salivarius | 0.065 | 0.055 | |
| Lactobacillus reuteri | Lactococcus sp. | 0.030 | 0.024 | |
| Lactobacillus plantarum | Lactobacillus casei | 0.014 | 0.010 | |
| Lactobacillus casei | Streptococcus sp. | 0.009 | 0.011 | |
| Bifidobacterium longum | Lactobacillus sp. | 0.006 | 0.001 | |
| Bifidobacterium lactis | Lactobacillus rhamnosus | 0.004 | 0.001 | |
| Bifidobacterium breve | Lactococcus lactis | 0.002 | 0.001 | |
| Leuconostoc cremoris | Lactobacillus acidophilus | 0.000 | 0.001 | |
| YOG | Streptococcus thermophilus | Streptococcus thermophilus | 0.775 | |
| Lactobacillus acidophilus | Streptococcus salivarius | 0.087 | ||
| Lactobacillus casei | Lactobacillus paracasei | 0.046 | ||
| Lactobacillus bulgaricus | Lactobacillus sp. | 0.044 | ||
| Lactobacillus bifidus | Lactobacillus acidophilus | 0.014 | ||
| Streptococcus sp. | 0.014 | |||
| Lactobacillus casei | 0.009 | |||
| Lactobacillus rhamnosus | 0.008 | |||
| Lactobacillus delbrueckii | 0.002 |
Product A: Maple Hill Plain Kefir; product B: Siggi's Plain Filmjölk; product C: Redwood Hill Farm Plain Goat Milk Kefir; product D: CoYo Kefir; product E: Lifeway Original Kefir; YOG: Chobani Plain Yogurt 0% Milk Fat.
Based on the results in Table 1, there should be more regulatory oversight on label accuracy for commercial kefir products to reduce the number of claims that can be misleading to consumers. Classification as a “cultured milk product” by the FDA requires disclosure of added microorganisms, yet regulation of ingredient quality and viability need to be better scrutinized. All 5 kefir products guaranteed specific bacterial species used in fermentation, yet no product matched its labeling completely. Incomplete labeling status can be defined by whether species are detected by sequencing and not listed on the label or when the label did not have enough DNA present of the species listed to be detected by 16S sequencing. Although our results demonstrated close matches across product labels and their stated contents, several products contained bacterial species above the minimum detectable threshold that were not included on the labels. For example, all products contained Strep. salivarius, and 4 out of 5 products contained Lactobacillus paracasei. Although there is regulatory oversight preventing companies from making unproven health claims, they should be held to higher standards of ingredient accuracy in their products (Marco et al., 2021). Previous research enumerating commercial pet probiotics and kefirs showed that label claims of colony-forming units per gram and microbial taxa are likely to be incorrect (Weese and Martin, 2011; Metras et al., 2020), and no other recent literature has investigated the efficacy of labels in this lucrative market.
Limitations of DNA-based sequencing methods could explain why taxa stated on labels were not detected and why unclaimed viable species were identified. Perhaps species were present but not in high enough abundance for Silva132 species-level classification; thus, they were classified under the bacterial genera (sp.) by the Loop Genomics pipeline [B. J. Callahan, D. Grinevich, and S. Thakur (North Carolina State University), M. A. Balamotis and T. B. Yehezkel (Loop Genomics, San Jose, CA); unpublished data]. For example, Lb. casei and Lb. paracasei both may exist in a sample, yet appear grouped as Lactobacillus sp. Databases also have limitations, as they are constantly updated with new species and nomenclature adjustments; running the data against updated databases may provide better categorization (Zheng et al., 2020). A limitation to our bacterial plating was that it was not specific to any taxa. The goal was to culture as much lactic acid bacteria as possible. Thus, the use of specific plating assays may yield different colony-forming units per gram results. It should also be noted that nonviable bacteria may exist in these products. If so, they might not have been cultured on MRS medium (Adams, 2010) and thus would not be included in final counts (cfu/g). Using methods such as sonication instead of vortexing the kefir products before plating might have broken up cell clumps better and resulted in more accurate counts. The YOG product was used to verify method accuracy. The results of the YOG enumeration and Loop sequencing in Table 1, Table 2 closely matched YOG label claims, giving confidence to our methods. The small sample size of 5 products can also be seen as a limitation in the ever-growing market of commercial kefir products. More fermented food research is needed so health professionals can identify which commercial products are truthful on their labels; then consumers might have more protection when purchasing from the vast market of commercial fermented foods.
In conclusion, our analysis of commercial kefir products via culture enumeration and sequencing revealed many deficiencies in label accuracy. Better quality control of these fermented foods is required to demonstrate and understand their potential health benefits for humans. Given the absence of regulatory scrutiny, consumers should be wary of this segment of the market and demand better quality commercial fermented foods.
Acknowledgments
This work was supported by USDA National Institute of Food and Agriculture (Hatch Grant #ILLU-538–937).
The authors have no conflicts of interest.
References
- Adams C.A. The probiotic paradox: Live and dead cells are biological response modifiers. Nutr. Res. Rev. 2010;23:37–46. doi: 10.1017/S0954422410000090. 20403231. [DOI] [PubMed] [Google Scholar]
- Bellikci-Koyu E., Sarer-Yurekli B.P., Akyon Y., Aydin-Kose F., Karagozlu C., Ozgen A.G., Brinkmann A., Nitsche A., Ergunay K., Yilmaz E., Buyuktuncer Z. Effects of regular kefir consumption on gut microbiota in patients with metabolic syndrome: A parallel-group, randomized, controlled study. Nutrients. 2019;11 doi: 10.3390/nu11092089. 31487797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourrie B.C., Ju T., Fouhse J.M., Forgie A.J., Sergi C., Cotter P.D., Willing B.P. Kefir microbial composition is a deciding factor in the physiological impact of kefir in a mouse model of obesity. Br. J. Nutr. 2021;125:129–138. doi: 10.1017/S0007114520002743. 32684173. [DOI] [PubMed] [Google Scholar]
- Code of Federal Regulations . US Food and Drug Administration; 1975. 21CFR1240.3. Control of communicable diseases: General definitions.https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?fr=1240.3 [Google Scholar]
- de Sainz I., Redondo-Solano M., Solano G., Ramírez L. Short communication: Effect of different kefir grains on the attributes of kefir produced with milk from Costa Rica. J. Dairy Sci. 2020;103:215–219. doi: 10.3168/jds.2018-15970. 31733842. [DOI] [PubMed] [Google Scholar]
- Hill C., Guarner F., Reid G., Gibson G., Merenstein D., Pot B., Morelli L., Canani R., Flint H., Salminen S., Calder P., Sanders M. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. 24912386. [DOI] [PubMed] [Google Scholar]
- Kechagia M., Basoulis D., Konstantopoulou S., Dimitriadi D., Gyftopoulou K., Skarmoutsou N., Fakiri E.M. Health benefits of probiotics: A review. ISRN Nutr. 2013;2013:1–7. doi: 10.5402/2013/481651. 24959545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larosa C.P., Balthazar C.F., Guimaraes J.T., Rocha R.S., Silva R., Pimentel T.C., Granato D., Duarte M.C.K.H., Silva M.C., Freitas M.Q., Cruz A.G., Esmerino E.A. Sheep milk kefir sweetened with different sugars: Sensory acceptance and consumer emotion profiling. J. Dairy Sci. 2021;104:295–300. doi: 10.3168/jds.2020-18702. 33162085. [DOI] [PubMed] [Google Scholar]
- Marco M.L. Defining how microorganisms benefit human health. Microb. Biotechnol. 2020 doi: 10.1111/1751-7915.13685. 33099885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco M.L., Sanders M.E., Gänzle M., Arrieta M.C., Cotter P.D., De Vuyst L., Hill C., Holzapfel W., Lebeer S., Merenstein D., Reid G., Wolfe B.E., Hutkins R. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on fermented foods. Nat. Rev. Gastroenterol. Hepatol. 2021 doi: 10.1038/s41575-020-00390-5. 33398112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenstein D., Guzzi J., Sanders M.E. More information needed on probiotic supplement product labels. J. Gen. Intern. Med. 2019;34:2735–2737. doi: 10.1007/s11606-019-05077-5. 31161565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metras B.N., Holle M.J., Parker V.J., Miller M.J., Swanson K.S. Assessment of commercial companion animal kefir products for label accuracy of microbial composition and quantity. J. Anim. Sci. 2020;98 doi: 10.1093/jas/skaa301. 32914845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S., Ghosh B.C. Quality characteristics of kefir as a carrier for probiotic Lactobacillus rhamnosus GG. Int. J. Dairy Technol. 2020;73:384–391. doi: 10.1111/1471-0307.12664. [DOI] [Google Scholar]
- Pražnikar Z.J., Kenig S., Vardjan T., Bizjak M.Č., Petelin A. Effects of kefir or milk supplementation on zonulin in overweight subjects. J. Dairy Sci. 2020;103:3961–3970. doi: 10.3168/jds.2019-17696. 32171508. [DOI] [PubMed] [Google Scholar]
- Rosa D.D., Dias M.M.S., Grześkowiak Ł.M., Reis S.A., Conceição L.L., Peluzio M.C.G. Milk kefir: Nutritional, microbiological and health benefits. Nutr. Res. Rev. 2017;30:82–96. doi: 10.1017/S0954422416000275. 28222814. [DOI] [PubMed] [Google Scholar]
- Stanton C., Gardiner G., Meehan H., Collins K., Fitzgerald G., Lynch P.B., Ross R.P. Market potential for probiotics. Am. J. Clin. Nutr. 2001;73(2 Suppl.):476S–483S. doi: 10.1093/ajcn/73.2.476s. 11157361. [DOI] [PubMed] [Google Scholar]
- Verruck S., Balthazar C.F., Rocha R.S., Silva R., Esmerino E.A., Pimentel T.C., Freitas M.Q., Silva M.C., da Cruz A.G., Prudencio E.S. Dairy foods and positive impact on the consumer's health. Adv. Food Nutr. Res. 2019;89:95–164. doi: 10.1016/bs.afnr.2019.03.002. 31351531. [DOI] [PubMed] [Google Scholar]
- Weese J.S., Martin H. Assessment of commercial probiotic bacterial contents and label accuracy. Can. Vet. J. 2011;52:43–46. [PMC free article] [PubMed] [Google Scholar]
- Yılmaz İ., Enver Dolar M., Ozpinar H. Effect of administering kefir on the changes in fecal microbiota and symptoms of inflammatory bowel disease: A randomized controlled trial. Turk. J. Gastroenterol. 2019;30:242–253. doi: 10.5152/tjg.2018.18227. 30662004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wittouck S., Salvetti E., Franz C.M.A.P., Harris H.M.B., Mattarelli P., O'Toole P.W., Pot B., Vandamme P., Walter J., Watanabe K., Wuyts S., Felis G.E., Gänzle M.G., Lebeer S. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. 32293557. [DOI] [PubMed] [Google Scholar]
Uncited Reference
- Fathi Y., Ghodrati N., Zibaeenezhad M.-J., Faghih S. Kefir drink causes a significant yet similar improvement in serum lipid profile, compared with low-fat milk, in a dairy-rich diet in overweight or obese premenopausal women: A randomized controlled trial. J. Clin. Lipidol. 2016;11:136–146. doi: 10.1016/j.jacl.2016.10.016. 28391880. [DOI] [PubMed] [Google Scholar]