Graphical Abstract
Summary: Houseflies in dairy farms can be a concern since they could play a crucial role in increased bacterial counts in milk and the potential occurrence of transmitted diseases that affect public and animal health. This study evaluated the bacterial communities associated with houseflies captured on a dairy farm in New York State. A total of 101 flies were collected at 3 farm sites: hospital pen, calf hutches, and milking parlor. Each housefly was tested by bacteriological analysis for microbial identification, and a total of 304 bacterial isolates were obtained. Twelve percent of the identified organisms are well known to affect dairy farms. We identified 26 bacterial species with implications for animal health since these are mastitis-causing pathogens; 5 bacteria are considered foodborne pathogens and, for this reason, represent a concern for human health, and last, but not least, we identified 5 milk spoilage bacteria species that affect the quality of dairy products.
Highlights
-
•
We obtained 101 houseflies from 3 different sites of one commercial dairy farm.
-
•
Bacteria culture was performed from external surfaces and internal parts of houseflies.
-
•
We identified 26 mastitis-causing pathogens, 5 foodborne pathogens, and 5 milk-spoiling organisms.
Abstract
Houseflies (Musca domestica) are nonbiting muscoids of importance because they can be mechanical vectors of many kinds of pathogens such as bacteria, protozoa, viruses, and helminth eggs. This study aimed to evaluate the bacterial communities associated with houseflies captured in 3 different areas on a dairy farm located in New York State. Variations in the bacterial community were also evaluated based on the flies' sex and external or internal location where the bacteria were isolated. A total of 101 flies were collected: 27 flies from the sick pen, 42 from calf hutches, and 32 from the milking parlor. A total of 485 organisms were isolated, 233 (48.0%) from 53 female flies and 252 (52.0%) from 48 male flies. Most (74%) bacteria were found in the internal parts of the flies, with only 26% isolated from the external surfaces. The number of isolates detected per fly ranged between 1 and 11. A total of 392 bacteria were identified at the species level. We isolated 26 species reported to be bovine contagious or environmental mastitis pathogens. Within the group of organisms considered contagious, we isolated Staphylococcus aureus and Mycoplasma arginini. This was the first time that a Mycoplasma species was isolated from houseflies. We identified 5 organisms considered foodborne pathogens that affect human health: Salmonella spp., Escherichia coli, Staph. aureus, Bacillus cereus, and Bacillus subtilis. Four of the organisms isolated in this study were also linked with milk spoilage, including Pseudomonas aeruginosa, Bacillus cereus, Bacillus licheniformis, and Paenibacillus lactis. This study confirmed that houseflies carry a high bacterial diversity, including organisms associated with animal infections, organisms that could be a concern for public health, or organisms that could negatively affect milk quality.
Houseflies (Musca domestica) live in close association with humans and domestic animals because both urban and rural environments enable their development (Gopal et al., 2015); human excrement and garbage, as well as animal manure and bedding, represent the main sources for their nutrition and oviposition (West, 1951). This insect, having a dispersal range of 5 to 32 km, can carry bacteria both on the surface of its exoskeleton and in the alimentary canal, and disperse them through mechanical translocation from the exoskeleton or by defecation and regurgitation (Nazni et al., 2005). The presence of bristles and glandular hairs on housefly legs enhances bacteria adhesion to their exterior surface (Graczyk et al., 2001). Feces viscosity increases the efficiency of flies' bristles and hair in trapping bacteria suspended in the manure (Graczyk et al., 1999). Bacteria can be stored in the fly's crop (Doud and Zurek, 2012) where they can multiply and be regurgitated or pass through the gut, leading to the concept of “bioenhanced transmission” (Onwugamba et al., 2018). Flies can contribute to the dissemination of bacteria of public health importance, such as Escherichia coli (Alves et al., 2018), Campylobacter jejuni (Bahrndorff et al., 2014), Salmonella spp. (Holt et al., 2007), and Staphylococcus spp. (Almeida et al., 2014), including strains resistant to antibiotics (Macovei et al., 2008; Akhtar et al., 2009). Flies are also discussed as reservoirs of mastitis pathogens such as Staphylococcus aureus. Still, most of the data associated with the dairy environment are related to horn flies (Haematabia irritans; Anderson et al., 2012; Ryman et al., 2013). Otherwise, in other studies conducted on dairies, the fly species are not mentioned (Roberson et al., 1994). To the best of our knowledge, published data of Staph. aureus isolated from houseflies are not specifically associated with dairy environments (Nayduch et al., 2013).
Surveillance of pathogens in flies can identify persistent pathogens in farm environments and their potential association with public and animal health. Houseflies can also act as vectors of multidrug-resistant bacteria, contributing to the dissemination of antimicrobial resistance between farm animals and humans (Zurek and Ghosh, 2014; Usui et al., 2015). Despite the importance of houseflies in disease transmission, little is known about the prevalence of pathogens on houseflies within different parts of a dairy, which could affect udder health, human health, and food shelf life (Geden et al., 2021). This study aimed to identify bacterial species associated with houseflies captured in different locations on a dairy farm structured by fly sex and bacterial presence in internal parts or external surfaces of flies.
This study was conducted on a commercial dairy farm in New York State that belongs to the client base of Quality Milk Production Services, Cornell University, Ithaca, New York. Approximately 1,300 lactating Holstein cows were housed in freestall pens, bedded with manure solids, milked 3 times per day, and fed a TMR consistent with National Research Council requirements (NRC, 2001).
The farm was selected based on a client list of the daily milk sample pick-up and 24-h result program of Quality Milk Production Services but also for a variety of pathogens isolated including Staph. aureus, Mycoplasma spp., Prototheca spp., and Lactococcus spp. Housefly collection was conducted upon approval by the farm owner.
All flies were collected during one farm visit (September 5, 2019), and investigators were trained for fly sampling by one experienced author (JCF). Flies were captured by a handheld sweep net from 3 farm sites: sick pen, calf hutch area, and milking parlor. The calf area was approximately 0.5 km apart from the main farm building.
Flies were individually transferred from the net into sterile vials (Capital Vial Corp.) and kept on ice during transport to the laboratory where they were stored at −30°C until further processing. To minimize cross-contamination between flies, the sweeping net was immersed into a sodium chlorite solution and air-dried after each fly was captured. Species and sex were determined through external morphology observations under steromicroscope by an experienced entomologist (JCF). All study personnel were trained in animal handling, and no endangered species were threatened.
Bacterial isolation and identification were performed from both external surfaces and internal parts of each fly. For isolation of bacteria from the external surface, 2 mL of sterile 1X PBS (Hardy Diagnostics) was added to each vial and gently shaken by inversion for 2 min. Culture enrichment was performed by transferring 0.5 mL of washing suspension to the following media: 2 mL of Todd Hewitt Broth (Northeast Laboratory) and incubated in an aerobic incubator at 37°C for 6 h; and 1 mL of Mycoplasma Broth (Hardy Diagnostics) and incubated for 72 h at 35°C to 38°C with 5 to 10% CO2. After enrichment, each sample was inoculated using cotton swabs on the following culture media: (1) Trypticase Soy Agar with 5% sheep blood and 0.1% esculin (Northeast Laboratory), (2) MacConkey Agar (Northeast Laboratory), (3) Edwards with 5% sheep blood agar (Northeast Laboratory), (4) Vogel Johnson Agar (Northeast Laboratory), (5) Prototheca Isolation Media (Animal Health Diagnostic Center, Cornell University), and (6) Mycoplasma Hayflick Agar (Northeast Laboratory). Based on bacterial growth requirements, agar plates were incubated as follows: 48 h at 37°C for media (1), (2), (3), and (4); 72 h at 37°C for medium (5); and 7 d at 35 to 38°C with 5 to 10% CO2 for medium (6).
For the isolation of bacteria from the internal part of flies, the first step was to sterilize the external surface. For this purpose, the remaining PBS was removed from each vial, and flies were washed by inversion for 5 min with 70% isopropyl alcohol (VWR International). Subsequently, the alcoholic solution was removed, and flies were exposed to UV light in a CLASS II biological safety cabinet (The Baker Company) for 10 min. To verify the decontamination efficiency, PBS was added to each vial containing the flies, and it was streaked on medium (1) and incubated at 37°C for 48 h. Flies were transferred into 2-mL sterile disruption tubes containing five 1.7-mm zirconium beads and 1 mL of PBS. Homogenates were obtained with 2 cycles on a bead beater (HT24, OPS Diagnostics), 1 min each at 7,000 rpm. Homogenates were centrifuged at 2,000 × g for 5 min at room temperature and cultured according to the procedure outlined above for bacterial isolation of the external surface.
Bacteria were identified by MALDI-TOF using MALDI Biotyper Microflex LT (Bruker Daltonics). Adaption of the MALDI-TOF library and sample preparation were performed as described by Randall et al. (2015). For all bacteria except NAS, a result was considered accurate if the score value was ≥1.7 for genus level identification, and a score ≥2.0 was set for a match at the species level. For NAS, a score value ≥1.7 was chosen (Cameron et al., 2018) to be indicative of a valid result at the species level (no scores <1.7 were obtained). Mycoplasma growth was detected by visual inspection of culture plates under an illuminated stage stereomicroscope. Mycoplasma colonies were confirmed by a colony PCR specific for Mycoplasma and Acholeplasma followed by Sanger sequencing for species identification (Gioia et al., 2016).
A total of 143 flies were caught from the 3 farm sites; 101 were identified as houseflies, and the remaining 42 were stable flies. Only houseflies were used for further investigation and from those, 53 (52.5%) flies were female and 48 (47.5%) were male. Out of the 101 flies, a total of 27 (26.7%) were found in the calf area, 42 (41.6%) were collected in the sick pen, and 32 (31.7%) were collected in the milking parlor. All colonies grown on plates were subjected to MALDI. A total of 485 isolates were obtained; 484 were bacterial and 1 was fungal. Based on MALDI scores, 91 (18.8%) were identified at the genus level and 392 (80.8%) at the species level. Multiple colonies of the same bacterial genus and species, if isolated from the same fly body's location, were reported only once. Two isolates grew on Mycoplasma medium; both were submitted for molecular confirmation and speciation. The average (mean ± SD) number of isolates detected per fly was 4.8 ± 2.4 ranging between 1 and 11. Figure 1 illustrates the taxonomic classification of 485 isolates (A) and their distribution divided by farm sites where houseflies were collected (B), flies' sex (C), and flies' body site from where the isolates were obtained (D). From the total 485 isolates, 278 (57.3%) were gram positive, 206 (42.5%) were gram negative, and 1 isolate (0.2%) was identified as a fungus. From the 2 isolates growing on Mycoplasma agar, one was identified as Mycoplasma arginini and one as Acholeplasma laidlawii. Both cases represent bacterial species classified as mollicutes and for the present study were included in the gram-positive cluster according to Razin et al. (1998). The isolate classified as a fungus was identified by MALDI as Candida krusei and it was obtained from the external surface of a male fly collected in the sick pen. Based on the site of collection (Figure 1B), a total of 211 (43.5%) isolates were obtained from 42 flies captured in the sick pen. Out of those, 126 (59.7%) were gram positive, 84 (39.8%) were gram negative, and 1 isolate (0.4%) was identified as a fungus. We obtained a total of 148 (30.5%) isolates from 27 flies of the calf hutch area; 70 (47.3%) were gram positive and 78 (52.7%) were gram negative. From 32 flies collected in the milking parlor, a total of 126 (25.9%) isolates were obtained; 82 (65%) were gram positive, and 44 (34.9%) were gram negative. The number of bacteria isolated from male or female flies was similar (Figure 1C). A total of 252 (51.9%) isolates were from 48 male flies, 150 (59.5%) were gram positive, 101 (40%) were gram negative, and 1 (0.4%) was a fungus. In 53 female flies we found 233 (48%) isolates, of which 128 (54.9%) were gram positive and 105 (45%) were gram negative. The majority, 349 (71.9%), of isolates were retrieved from internal parts, of which 170 (48.7%) were gram positive and 179 (51.3%) were gram negative. Only 136 (28%) isolates were obtained from the external part of the flies, of which 108 (79.4%) were gram positive, 27 (19.8%) were gram negative, and 1 (0.7%) was a fungus (Figure 1D).
Figure 1.
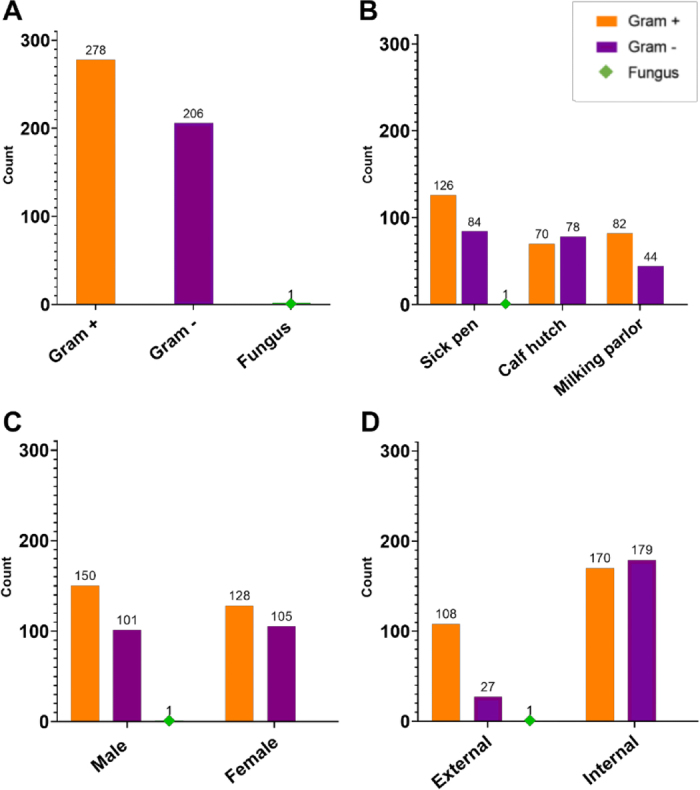
Bacterial isolate counts found in 101 houseflies obtained from 3 sites of 1 dairy farm. (A) Taxonomic classification of the isolates. Bacteria are classified as gram positive, gram negative, and fungus. (B) Bacteria count distribution between farm locations where flies were collected. (C) Bacteria count distribution between female and male flies. (D) Bacteria count distribution based on the flies' body site.
Infestations of houseflies on a dairy farm represent a concern due to possible microorganisms they could carry. Some of those microorganisms are harmless, but others could cause an increase of infectious diseases in cows or negatively affect the milk quality. Mastitis is one of the most common diseases of dairy cattle with high economic impact (Halasa et al., 2007). It has been demonstrated that different species of flies, including head flies (Hydrotaea irritans) and horn flies, could act as a possible vector for mastitis pathogens, particularly summer mastitis in heifers (McDougall et al., 2009). Implications on milk quality could be associated with foodborne pathogens that represent a hazard for human health or spoilage microorganisms that compromise milk's quality, flavor, and texture. From the total 392 organisms identified at the species level, 304 were selected based on their importance and possible implications in a dairy farm. Table 1 depicts the frequency distribution of 304 bacteria species isolated from external surfaces and internal parts of flies collected at different farm sites and their potential impact on milk quality and health of both humans and animals. We divided the isolates into 3 groups based on (1) mastitis-causing pathogens, (2) foodborne pathogens, and (3) milk-spoiling organisms.
-
(1)
In the group of organisms that can invade the teat canal and cause mastitis, we found NAS, Enterococcus spp., Klebsiella spp., and E. coli present in flies from all 3 farm locations. Citrobacter spp., Enterobacter cloacae, and Pseudomonas aeruginosa were only found in flies from calf hutches and the sick pen. Lactococcus spp. and Staph. aureus were found in flies from calf hutches and milking parlor. Only flies found in the sick pen carried Corynebacterium stationis and Mycoplasma arginini. Streptococcus uberis was only found in flies from the milking parlor. Except for Enterococcus spp., Klebsiella pneumoniae, E. coli, Enterobacter cloacae, Staph. aureus, and most of the NAS species, all other bacteria here classified as mastitis pathogens have not been reported to be isolated from houseflies in dairy farms before this study to our knowledge. Both Staph. aureus and Mycoplasma spp. are considered contagious mastitis pathogens and require timely management to prevent new infections within a dairy herd. It is well known that flies may be carriers of Staph. aureus, but no previous reports exist on the possibility to isolate Mycoplasma spp. from flies. Mycoplasma arginini is not one of the most pathogenic species, but it can persist for a long time under different environmental conditions (Nagatomo et al., 2001).
-
(2)
In the group of organisms previously classified as potential foodborne pathogens, we found B. cereus, B. subtilis, and E. coli carried by flies collected from all 3 farm locations. Other foodborne pathogens were Salmonella spp. found in the milking parlor, and Staph. aureus found in the calf hutches and the milking parlor. In our study, we found only 1 Salmonella isolate compared with Almeida et al. (2014) where Salmonella was found in 9.5% of muscoids.
-
(3)
Some of the bacterial species found in the present study have been previously classified as milk-spoiling organisms (Scheldeman et al., 2004; Scatamburlo et al., 2015; Awasti et al., 2019), and these included B. licheniformis and Lysinibacillus sphaericus, which were found in flies collected in the milking parlor and the sick pen, Pseudomonas aeruginosa found in the calf hutch and the sick pen, and Paenibacillus lactis isolated from 1 fly of the milking parlor. Before this study, none of these 3 organisms have been isolated from houseflies collected in dairies. In most previous publications, these organisms were found in water, soil, or milking equipment.
Table 1.
Selected bacterial species isolated from 101 houseflies in which bacterial selection was based on effects on udder health, human health, and milk quality
| Genus | Species | Effect1 | Farm site2 | Location3 | n (%)4 |
|---|---|---|---|---|---|
| Citrobacter | freundii5 | M | H, C | E, I | 4 (1.0) |
| amalonaticus5 | M | P | I | 1 (0.3) | |
| Enterobacter | cloacae5 | M | H, C, P | E, I | 25 (6.4) |
| Escherichia | coli | M, F | H, C, P | E, I | 59 (15.1) |
| Klebsiella | pneumoniae | M | H, C, P | E, I | 26 (6.6) |
| variicola5 | M | H, P | E, I | 7 (1.8) | |
| oxytoca5 | M | C, P | E, I | 4 (1.0) | |
| Salmonella | spp. | F | P | I | 1 (0.3) |
| Pseudomonas | aeruginosa5 | M, S | H, C | E, I | 7 (1.8) |
| Enterococcus | faecalis | M | H, C, P | E, I | 67 (17.1) |
| hirae | M | C | I | 1 (0.3) | |
| faecium | M | H, C, P | E, I | 18 (4.6) | |
| gallinarum5 | M | H | I | 2 (0.5) | |
| Lactococcus | garvieae5 | M | C, P | E, I | 6 (1.5) |
| lactis5 | M | C | E, I | 2 (0.5) | |
| Streptococcus | uberis5 | M | P | I | 1 (0.3) |
| Staphylococcus | aureus | M, F | C, P | I | 2 (0.5) |
| capitis5 | M | P | E | 1 (0.3) | |
| chromogenes | M | H, P | I | 6 (1.5) | |
| haemolyticus | M | H, C, P | E, I | 10 (2.6) | |
| lugdunensis | M | H | E, I | 2 (0.5) | |
| saprophyticus | M | H, C, P | E, I | 10 (2.6) | |
| sciuri | M | H, C, P | E | 10 (2.6) | |
| succinus | M | H, P | E | 2 (0.5) | |
| xylosus | M | C | E | 1 (0.3) | |
| Corynebacteriun | stationis5 | M | H | E | 1 (0.3) |
| Bacillus | cereus5 | F, S | H, C, P | E, I | 16 (4.1) |
| subtilis | F | H | E | 1 (0.3) | |
| licheniformis5 | S | H, P | E | 7 (1.8) | |
| Lysinibacillus | sphaericus5 | S | H, P | I | 3 (0.8) |
| Paenibacillus | lactis5 | S | P | E | 1 (0.3) |
| Mycoplasma | arginini5 | M | H | I | 1 (0.3) |
Effect on udder health (M), human health (F), and milk quality (S).
Farm site (sick pen, H; calf area, C; and milking parlor, P).
Location (external surface, E; internal parts, I)
Number and percentage (calculated from all isolates).
Species has not been previously identified from houseflies at dairies.
We detected a total of 17 species of pathogens that, to our knowledge, have not been previously reported from houseflies at dairies (Table 1); most of these are mastitis pathogens and some are milk-spoiling organisms. Species of pathogens that we and others have detected in houseflies from dairies include Enterococcus faecalis, hirae, and faecium (Ahmad et al., 2011), E. coli (Bahrndorff et al., 2017), and Klebsiella pneumoniae (Ranjbar et al., 2016).
A limitation of the sampling technique used in this study was the possibility of contaminating the external surfaces through bacteria that could have been captured from the air after air-drying the nets. However, we believe that it would have been minor if such contamination existed, or it would be seen as background from flies caught in the same area of the farm. In this study, the bacteria species found with higher prevalence were Enterococcus faecalis (17.1%), E. coli (15.1%), Klebsiella pneumoniae (6.6%), and Enterobacter cloacae (6.4%); these bacteria were isolated from internal parts and external surfaces of both female and male flies obtained at all 3 farm sites. The major pathogens that we found are all considered enteric bacteria that live in the digestive tracts of healthy animals, humans, and even insects. In a dairy farm, these organisms are typically found in feces, manure, and bedding; all are places where houseflies habitually go for feeding. Bacteria present in the organic matter stick on flies' body surfaces and can be carried around. The constant movement of houseflies back and forth from feces to food and drinking water places humans and animals at risk of infection. Enterococcus faecalis, E. coli, Klebsiella pneumoniae, and Enterobacter cloacae can be opportunistic invaders of the mammary glands and are considered environmental mastitis-causing pathogens. Mastitis caused by E. coli and Klebsiella spp. can be severe and occasionally fatal. The predominance of a single strain of E. coli or Klebsiella spp. in a dairy farm could indicate contagious transmission or exposure of multiple cows to an environmental point source (Munoz et al., 2007). For the aim of the present study, all isolates were identified at the genus and some at the species level; none of them were further tested by strain typing. For future studies, it would be interesting to see if there are matches between strains found in flies and strains of the same organism found in other sources such as milk samples from cows with clinical mastitis. The identification at the strain level could also be relevant to point out the presence of critical foodborne pathogens such as Shiga toxin-producing E. coli; this is important mainly for dairy farms that sell unpasteurized milk (Murinda et al., 2019). To our knowledge, this is the first study describing the distribution of the bacterial communities carried by flies in different areas of the same farm. Our results showed that the pathogens found differ by the farm location where flies were collected. Although 9 of the pathogens were detected in all 3 collection sites, 13 pathogens were found in only one of the locations within the facility. Considering that flies travel 5 to 32 km, our data give a snapshot of what these flies carried in the given location at sampling time. For future research, it would be interesting to collect flies on specific sites over time to see if there are circadian or seasonal changes or if there is a concentration of certain organisms on one site despite the flight radius of the fly. Indeed, the observations of this study need to be specifically associated with the farm selected where flies were collected. Because bacterial communities might be highly variable from farm to farm, it would be necessary for future studies to conduct the same kind of investigation in more than one farm, even located in different areas.
Our results are consistent with those from previous studies (Bahrndorff et al., 2017), suggesting that houseflies can be an effective vector of bacteria such as Salmonella spp., Staph. aureus, Pseudomonas aeruginosa, Klebsiella pneumoniae, Enterococcus faecalis, and E. coli. However, contrary to our study, Bahrndorff et al. (2017) used a culture-independent amplicon sequencing approach to characterize the bacterial communities of houseflies on dairy farms.
The large number of pathogens detected in this study, several reported for the first time, illustrate that surveys of pathogens on houseflies can help to understand and ultimately reduce disease spread.
Notes
This project was in part funded by Quality Milk Production Services (QMPS) internal program 2019.
The authors have not stated any conflicts of interest.
References
- Ahmad A., Ghosh A., Schal C., Zurek L. Insects in confined swine operations carry a large antibiotic resistant and potentially virulent enterococcal community. BMC Microbiol. 2011;11:23. doi: 10.1186/1471-2180-11-23. 21269466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar M., Hirt H., Zurek L. Horizontal transfer of the tetracycline resistance gene tetM mediated by pCF10 among Enterococcus faecalis in the house fly (Musca domestica L.) alimentary canal. Microb. Ecol. 2009;58:509–518. doi: 10.1007/s00248-009-9533-9. 19475445. [DOI] [PubMed] [Google Scholar]
- Almeida J.L., Giuffrida R., Andrade R.P.A., Chaves M.P. Muscoid Diptera as potential vectors of bacterial agents on dairy farms in the northern region of Paraná, Brazil. Semin. Cienc. Agrar. 2014;35:3127–3138. doi: 10.5433/1679-0359.2014v35n6p3127. [DOI] [Google Scholar]
- Alves T.D.S., Lara G.H.B., Maluta R.P., Ribeiro M.G., Leite D.D.S. Carrier flies of multidrug-resistant Escherichia coli as potential dissemination agent in dairy farm environment. Sci. Total Environ. 2018;633:1345–1351. doi: 10.1016/j.scitotenv.2018.03.304. 29758886. [DOI] [PubMed] [Google Scholar]
- Anderson K.L., Lyman R., Moury K., Ray D., Watson D.W., Correa M.T. Molecular epidemiology of Staphylococcus aureus mastitis in dairy heifers. J. Dairy Sci. 2012;95:4921–4930. doi: 10.3168/jds.2011-4913. 22916896. [DOI] [PubMed] [Google Scholar]
- Awasti N., Anand S., Djira G. Sporulating behavior of Bacillus licheniformis strains influences their population dynamics during raw milk holding. J. Dairy Sci. 2019;102:6001–6012. doi: 10.3168/jds.2018-15613. 31103302. [DOI] [PubMed] [Google Scholar]
- Bahrndorff S., de Jonge N., Skovgård H., Nielsen J.L. Bacterial communities associated with houseflies (Musca domestica L.) sampled within and between farms. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169753. 28081167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrndorff S., Gill C., Lowenberger C., Skovgård H., Hald B. The effects of temperature and innate immunity on transmission of Campylobacter jejuni (Campylobacterales: Campylobacteraceae) between life stages of Musca domestica (Diptera: Muscidae) J. Med. Entomol. 2014;51:670–677. doi: 10.1603/ME13220. 24897861. [DOI] [PubMed] [Google Scholar]
- Cameron M., Perry J., Middleton J.R., Chaffer M., Lewis J., Keefe G.P. Short communication: Evaluation of MALDI-TOF mass spectrometry and a custom reference spectra expanded database for the identification of bovine-associated coagulase-negative staphylococci. J. Dairy Sci. 2018;101:590–595. doi: 10.3168/jds.2017-13226. 29102131. [DOI] [PubMed] [Google Scholar]
- Doud C.W., Zurek L. Enterococcus faecalis OG1RF:pMV158 survives and proliferates in the house fly digestive tract. J. Med. Entomol. 2012;49:150–155. doi: 10.1603/ME11167. 22308783. [DOI] [PubMed] [Google Scholar]
- Geden C.J., Nayduch D., Scott J.G., Burgess E.R., IV, Gerry A.C., Kaufman P.E., Thomson J., Pickens V., Machtinger E.T. House fly (Diptera: Muscidae): Biology, pest status, current management prospects, and research needs. J. Integr. Pest Manag. 2021;12:39. doi: 10.1093/jipm/pmaa021. [DOI] [Google Scholar]
- Gioia G., Werner B., Nydam D.V., Moroni P. Validation of a mycoplasma molecular diagnostic test and distribution of mycoplasma species in bovine milk among New York State dairy farms. J. Dairy Sci. 2016;99:4668–4677. doi: 10.3168/jds.2015-10724. 27016831. [DOI] [PubMed] [Google Scholar]
- Gopal N., Hill C., Ross P.R., Beresford T.P., Fenelon M.A., Cotter P.D. The prevalence and control of Bacillus and related spore-forming bacteria in the dairy industry. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.01418. 26733963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graczyk T.K., Cranfield M.R., Bixler H., Fayer R. House flies (Musca domestica) as transport hosts of Cryptosporidium parvum. Am. J. Trop. Med. Hyg. 1999;61:500–504. doi: 10.4269/ajtmh.1999.61.500. 10497998. [DOI] [PubMed] [Google Scholar]
- Graczyk T.K., Knight R., Gilman R.H., Cranfield M.R. The role of non-biting flies in the epidemiology of human infectious diseases. Microbes Infect. 2001;3:231–235. doi: 10.1016/S1286-4579(01)01371-5. 11358717. [DOI] [PubMed] [Google Scholar]
- Halasa T., Huijps K., Østerås O., Hogeveen H. Economic effects of bovine mastitis and mastitis management: A review. Vet. Q. 2007;29:18–31. doi: 10.1080/01652176.2007.9695224. 17471788. [DOI] [PubMed] [Google Scholar]
- Holt P.S., Geden C.J., Moore R.W., Gast R.K. Isolation of Salmonella enterica serovar Enteritidis from houseflies (Musca domestica) found in rooms containing Salmonella serovar Enteritidis-challenged hens. Appl. Environ. Microbiol. 2007;73:6030–6035. doi: 10.1128/AEM.00803-07. 17675422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macovei L., Miles B., Zurek L. Potential of houseflies to contaminate ready-to-eat food with antibiotic-resistant enterococci. J. Food Prot. 2008;71:435–439. doi: 10.4315/0362-028X-71.2.435. 18326202. [DOI] [PubMed] [Google Scholar]
- McDougall S., Parker K.I., Heuer C., Compton C.W.R. A review of prevention and control of heifer mastitis via non-antibiotic strategies. Vet. Microbiol. 2009;134:177–185. doi: 10.1016/j.vetmic.2008.09.026. 18986782. [DOI] [PubMed] [Google Scholar]
- Munoz M.A., Welcome F.L., Schukken Y.H., Zadoks R.N. Molecular epidemiology of two Klebsiella pneumoniae mastitis outbreaks on a dairy farm in New York State. J. Clin. Microbiol. 2007;45:3964–3971. doi: 10.1128/JCM.00795-07. 17928424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murinda S.E., Ibekwe A.M., Rodriguez N.G., Quiroz K.L., Mujica A.P., Osmon K. Shiga toxin-producing Escherichia coli in mastitis: an international perspective. Foodborne Pathog. Dis. 2019;16:229–243. doi: 10.1089/fpd.2018.2491. 30624967. [DOI] [PubMed] [Google Scholar]
- Nagatomo H., Takegahara Y., Sonoda T., Yamaguchi A., Uemura R., Hagiwara S., Sueyoshi M. Comparative studies of the persistence of animal mycoplasmas under different environmental conditions. Vet. Microbiol. 2001;82:223–232. doi: 10.1016/S0378-1135(01)00385-6. 11470544. [DOI] [PubMed] [Google Scholar]
- Nayduch D., Cho H., Joyner C. Staphylococcus aureus in the house fly: temporospatial fate of bacteria and expression of the antimicrobial peptide defensin. J. Med. Entomol. 2013;50:171–178. doi: 10.1603/ME12189. 23427667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazni W.A., Luke H., Wan Rozita W.M., Abdullah A.G., Sa'diyah I., Azahari A.H., Zamree I., Tan S.B., Lee H.L., Sofian M.A. Determination of the flight range and dispersal of the house fly, Musca domestica (L.) using mark release recapture technique. Trop. Biomed. 2005;22:53–61. 16880754. [PubMed] [Google Scholar]
- NRC . 7th rev. ed. The National Academies Press; 2001. Nutrient Requirements of Dairy Cattle; p. 408. [Google Scholar]
- Onwugamba F.C., Fitzgerald J.R., Rochon K., Guardabassi L., Alabi A., Kühne S., Grobusch M.P., Schaumburg F. The role of ‘filth flies' in the spread of antimicrobial resistance. Travel Med. Infect. Dis. 2018;22:8–17. doi: 10.1016/j.tmaid.2018.02.007. 29482014. [DOI] [PubMed] [Google Scholar]
- Randall L.P., Lemma F., Koylass M., Rogers J., Ayling R.D., Worth D., Klita M., Steventon A., Line K., Wragg P., Muchowski J., Kostrzewa M., Whatmore A.M. Evaluation of MALDI-ToF as a method for the identification of bacteria in the veterinary diagnostic laboratory. Res. Vet. Sci. 2015;101:42–49. doi: 10.1016/j.rvsc.2015.05.018. 26267088. [DOI] [PubMed] [Google Scholar]
- Ranjbar R., Izadi M., Hafshejani T.T., Khamesipour F. Molecular detection and antimicrobial resistance of Klebsiella pneumoniae from house flies (Musca domestica) in kitchens, farms, hospitals and slaughterhouses. J. Infect. Public Health. 2016;9:499–505. doi: 10.1016/j.jiph.2015.12.012. 26876433. [DOI] [PubMed] [Google Scholar]
- Razin S., Yogev D., Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 1998;62:1094–1156. doi: 10.1128/MMBR.62.4.1094-1156.1998. 9841667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson J.R., Fox L.K., Hancock D.D., Gay J.M., Besser T.E. Ecology of Staphylococcus aureus isolated from various sites on dairy farms. J. Dairy Sci. 1994;77:3354–3364. doi: 10.3168/jds.S0022-0302(94)77277-5. 7814712. [DOI] [PubMed] [Google Scholar]
- Ryman V.E., Nickerson S.C., Hurley D.J., Berghaus R.D., Kautz F.M. Influence of horn flies (Haematobia irritans) on teat skin condition, intramammary infection, and serum anti-S. aureus antibody titres in holstein heifers. Res. Vet. Sci. 2013;95:343–346. doi: 10.1016/j.rvsc.2013.04.017. 23664017. [DOI] [PubMed] [Google Scholar]
- Scatamburlo T.M., Yamazi A.K., Cavicchioli V.Q., Pieri F.A., Nero L.A. Spoilage potential of Pseudomonas species isolated from goat milk. J. Dairy Sci. 2015;98:759–764. doi: 10.3168/jds.2014-8747. 25497792. [DOI] [PubMed] [Google Scholar]
- Scheldeman P., Goossens K., Rodriguez-Diaz M., Pil A., Goris J., Herman L., De Vos P., Logan N.A., Heyndrickx M. Paenibacillus lactis sp. nov., isolated from raw and heat-treated milk. Int. J. Syst. Evol. Microbiol. 2004;54:885–891. doi: 10.1099/ijs.0.02822-0. 15143040. [DOI] [PubMed] [Google Scholar]
- Usui M., Shirakawa T., Fukuda A., Tamura Y. The role of flies in disseminating plasmids with antimicrobial-resistance genes between farms. Microb. Drug Resist. 2015;21:562–569. doi: 10.1089/mdr.2015.0033. 26061440. [DOI] [PubMed] [Google Scholar]
- West L.S. Comstock Publ. Co; 1951. The Housefly: Its Natural History, Medical Importance, and Control. [Google Scholar]
- Zurek L., Ghosh A. Insects represent a link between food animal farms and the urban environment for antibiotic resistance traits. Appl. Environ. Microbiol. 2014;80:3562–3567. doi: 10.1128/AEM.00600-14. 24705326. [DOI] [PMC free article] [PubMed] [Google Scholar]