Abstract
Small extracellular vesicles (sEVs) play a crucial role in local and distant cell communication. The intrinsic properties of sEVs make them compatible biomaterials for drug delivery, vaccines, and theranostic nanoparticles. Although sEV proteomics have been robustly studied, a direct instantaneous assessment of sEV structure dynamics remains difficult. Here, we use the high‐speed atomic force microscopy (HS‐AFM) to evaluate nanotopological changes of sEVs with respect to different physicochemical stresses including thermal stress, pH, and osmotic stress. The sEV structure is severely altered at high‐temperature, high‐pH, or hypertonic conditions. Surprisingly, the spherical shape of the sEVs is maintained in acidic or hypotonic environments. Real‐time observation by HS‐AFM imaging reveals an irreversible structural change in the sEVs during transition of pH or osmolarity. HS‐AFM imaging provides both qualitative and quantitative data at high spatiotemporal resolution (nanoscopic and millisecond levels). In summary, our study demonstrates the feasibility of HS‐AFM for structural characterization and assessment of nanoparticles.
1. INTRODUCTION
An extracellular vesicle (EV) is a collective term that includes apoptotic bodies, plasma membrane‐origin EVs (ectosomes), and endosome‐origin EVs (exosomes) (Théry et al., 2018). Exosomes are known as small EVs (sEVs) because they are generally smaller than 200 nm. sEVs are biologically versatile, where they can be nanocarriers, ligand/receptor‐presenting nanoparticles, antigen‐presenting nanoparticles, and more. Currently, rather than synthetic nanoparticles, the intrinsic properties of sEVs have made them suitable biomaterials for nanomedicines and vaccines. In addition, sEVs in biological samples contain diagnostic values for various diseases. Therefore, as well as the biochemical contents, the physical characterization of sEVs is indeed important. Conventional atomic force microscopy (AFM) has been used to directly visualize the nanotopology of sEVs, and to measure their membrane stiffness (Hardij et al., 2013; Szatanek et al., 2017; Whitehead et al., 2015; Yurtsever et al., 2021). Nonetheless, the images are rather static snapshots, and the stiffness is measured at a relatively low temporal resolution. The limitation in conventional AFM impedes the real‐time observation and measurement of millisecond dynamic changes in the sEV nanotopology. High‐speed atomic force microscopy (HS‐AFM) is a powerful nano‐imaging tool with high spatiotemporal resolution. A gentle cantilever tapping force allows HS‐AFM to perform millisecond imaging without sample damage (Ando, 2019). To date, we have directly visualized conformational dynamics of viral proteins (Lim et al., 2021, 2020, b), DNA wrapping on a histone (Nishide et al., 2021), and eukaryotic organelles (nuclear pores (Mohamed et al., 2020; Mohamed et al., 2017) and sEVs (Lim et al., 2020, 2021)) by using HS‐AFM. In this study, we report the dynamic change of the sEV nanotopology in response to various physicochemical stresses including temperature, pH, and osmotic pressure. The findings may provide valuable insights for sEV biology and applications of sEVs in the near future.
2. MATERIALS AND METHODS
2.1. Cell line
HEK293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, Nacalai Tesque, Kyoto, Japan) supplemented with 10% foetal bovine serum (FBS; Life Technologies, CA, USA) and 1% Penicillin‐Streptomycin (Nacalai Tesque, Kyoto, Japan). Cells were kept at 37°C in a humidified CO2 incubator.
2.2. Preparation, purification, and characterization of sEVs
sEVs of the HEK293T cell were isolated according to the protocol that was reported previously (Lim et al., 2020, 2021). After 4 days of culturing, cell‐conditioned medium was collected and subjected to differential centrifugation to remove cells, debris, and large EVs. Then, the supernatant was filtered through a 0.22‐μm Millex‐GV filter (Merck Millipore, MA, USA) to generate a final pre‐cleared cell conditioned medium, known as the 10K sup. After that, a MagCapture Exosome Isolation Kit PS (Wako, Osaka, Japan) was used following to the manufacturer's manual to purify the sEVs from the 10K sup. The purified sEVs were kept in a dialysis membrane (3500 MWCO) for overnight dialysis in PBS. NanoSIGHT LM10 (Malvern Panalytical, Malvern, UK) was used to perform nanoparticle tracking analysis (NTA) to measure the sEV concentration. The sEV purification process follows the Minimal Information for Studies of Extracellular Vesicle 2018 (MISEV2018) guidelines (Théry et al., 2018) because we uploaded the protocol to the EV‐TRACK knowledgebase in our earlier study (Lim et al., 2021) (EV‐TRACK ID: EV210256).
2.3. Nanofabrication of cantilever tip by using electron beam deposition (EBD)
A BL‐AC10DS‐A2 cantilever (Olympus, Tokyo, Japan) was used as a scanning probe to scan the sEVs. The spring constant (k) and resonance frequency (f) of the cantilever were 0.1 N/m and 0.6 MHz in water (1.5 MHz in air), respectively. The dimension of the cantilever was 9 μm (length), 2 μm (width), and 0.13 μm (thickness). Short cantilever tip length can affect the sensitivity of tip‐sample interaction. This issue can be solved by depositing amorphous carbon on top of the cantilever tip using EBD to increase cantilever tip length. First, the cantilever was cleaned by UV/O3, and then the cantilever was soaked in piranha solution (containing sulfuric acid and hydrogen peroxide). After that, EBD was conducted on the cantilever at 30‐kV accelerating voltage and 2 min of irradiation using a field emission scanning electron microscope, ELS‐7500 (Tokyo, Japan). The typical tip radius range of an EBD‐ed cantilever tip is 6–8 nm.
2.4. HS‐AFM imaging
HS‐AFM images were acquired by using our laboratory‐built HS‐AFM microscope, as previously mentioned (Lim et al., 2020, 2021). Briefly, a laser beam (λ = 670 nm) was irradiated through a 20× objective lens (CFI S Plan Fluor ELWD, Nikon, Tokyo, Japan) and focused on an electron beam deposited cantilever tip. A position‐sensing two‐segmented photodiode was used to detect the dynamic cantilever deflection. Free oscillation amplitude of the cantilever (A0) at 1.5–2.5 nm and the set point at 80%–90% of the free amplitude were required to generate a gentle tapping force to protect sample integrity. A muscovite mica layer (∼0.1‐mm thickness) was affixed on a glass stage, and the glass stage was then mounted on a HS‐AFM scanner. The mica layer was then coated with 0.1% w/v poly‐L‐lysine (PLL) to serve as a substrate to immobilize the sEVs.
To visualize the nanotopology of the sEVs, the sEVs were scanned under a physiological buffer (50 mM Tris‐HCl, 150 mM NaCl, pH 7.50). To study the effect of temperature on sEV nanotopology, the sEVs were incubated in a scanning buffer at 4°C, 37°C, 60°C, or 100°C for 4 h prior to HS‐AFM scanning. To determine the effect of pH on sEV nanotopology, sEVs were incubated in a scanning buffer at pH 4.00, pH 7.50, or pH 10.00 for 3 h followed by HS‐AFM imaging. For real‐time observation of sEV nanotopology in response to pH change, an sEV was initially scanned at pH 7.50, then the neutral scanning buffer was changed to an acidic scanning buffer (pH 4.00), and eventually to a basic scanning buffer (pH 10.00). To elucidate the effect of osmolarity on sEV topology, sEVs were incubated in a physiological buffer (50 mM Tris‐HCl, pH 7.50) containing 0 M, 0.15 M, or 1.8 M salt (NaCl) for 3 h before HS‐AFM imaging. For real‐time visualization of the sEV nanotopology in response to an osmolarity change, an sEV was first scanned in a buffer with 0.15 M NaCl, then the salt concentration was increased to 1.8 M, and finally to 0 M. All HS‐AFM scans of the sEV nanotopology in response to temperature, pH, or osmolarity were repeated at least three times.
2.5. Analysis of HS‐AFM images
ImageJ software (https://imagej.nih.gov/ij/) was used to process and analyse all HS‐AFM images. First, the HS‐AFM images were filtered by a fit polynomial filter (order for both x‐ and y‐directions: 1), follow by a low‐pass filter (Gaussian blur) to suppress noise and enhance the image clarity. Three‐dimensional (3D) images of the HS‐AFM images were computed by using the freeware Gwyddion (http://www.gwyddion.net). The parameters of spatial dimension analysis including cross‐sectional height, area, and circularity were also measured by using ImageJ. A circularity value of 1.0 indicates a perfect circle whereas a value close to zero indicates an increasingly elongated polygon. The sEV circularity is given by 4πSL−2, where L and S are the perimeter and the area surrounded by the object, respectively. The volume of sEV was calculated by multiplying area and height of the sEV as previously mentioned (Lim et al., 2020b). The aspect ratio (AR) was calculated using this formula, AR = h/davg (h is height of sEV; davg is the average of major (d1) and minor (d2) diameters), to measure sEV stretchability as previously reported (Yokota et al., 2019). The processed images were converted to videos in AVI format, and further edited and compiled using the Adobe Creative Cloud suite (https://www.adobe.com/creativecloud.html).
2.6. Statistical analysis
Graphs for descriptive statistics were drawn using the GraphPad Prism version 7 (GraphPad, CA, USA). Comparative statistical analysis was performed using SPSS version 22 (IBM Group, NY, USA). Mann‐Whitney U test and Kruskal‐Wallis H test were conducted to compare means between independent groups. The statistically significant level was fixed at p < 0.05 with a confidence interval of 95%.
3. RESULTS
3.1. Nanoscopic conformational dynamics of native and formalin‐fixed sEVs
Differential ultracentrifugation and ultrafiltration of EVs are insufficient to separate exosomes from other EVs with similar size (<200 nm). We believe that the Tim‐4 isolation protocol (Figure 1a) is suitable for exosome purification because Tim‐4 has a high affinity for phosphatidylserine (PS), which is enriched in exosomes (Nakai et al., 2016). Furthermore, the distribution of PS is exclusively located at the cytoplasmic site of the plasma membrane (Tsuji et al., 2019). As a result, Tim‐4 isolation could spare EVs originating from the plasma membrane (ectosomes) from the cell conditioned media. In addition, we have previously detected exosome markers, particularly the endosome‐origin marker TSG101, by using this protocol (Lim et al., 2021). By abiding to the MISEV guideline, we continue to mention exosomes as sEVs. The setup of HS‐AFM is illustrated in Figure 1b, and we have elaborated the technical details in the Materials and Method section. The optimal tapping force and high scanning speed allow HS‐AFM to scan an sEV at high spatiotemporal resolution without deteriorating the sEV. Therefore, we can manipulate several physicochemical stresses including temperature, pH, and osmolarity, to study the dynamic response of sEVs either in a real‐time manner or in a premix setting. Given that Tim‐4‐isolated sEVs are PS‐enriched sEVs, the sEV surface should be negatively charged. Therefore, we coated mica with PLL to provide a positively charged substrate to adsorb sEVs. This adsorption strategy has proven effective to adsorb the nuclear envelope (NE) (Mohamed et al., 2020; Mohamed et al., 2017) for HS‐AFM imaging because the NE layer also contains PS (Tsuji et al., 2019). Nanotopological features of sEV were evaluated either in native or in formalin‐fixed form (Figure 1c–d, Figure S1, and Movie S1). Native sEVs appeared to be ellipsoidal rather than spherical, probably due to the adsorption effect that pulled the lipid layer wider. In contrast, formalin‐fixed sEV presented as a sphere. We found that the height fluctuation of a formalin‐fixed sEV was less compared with a native sEV (Figure 1e). These findings suggest that formalin fixation could increase the sEV membrane rigidity. Real‐time imaging of an sEV revealed its smooth fluid‐filled lipid vesicle nanotopology, and the sharp cantilever did not damage the sEV structure (Figure 1f, and Movie S1). Results of spatial dimension analysis (Figure 1g) demonstrated the size heterogeneity of the sEVs. Altogether, these data indicate that use of HS‐AFM is feasible for recording the native spatiotemporal dynamics of HEK293T‐derived sEVs.
FIGURE 1.
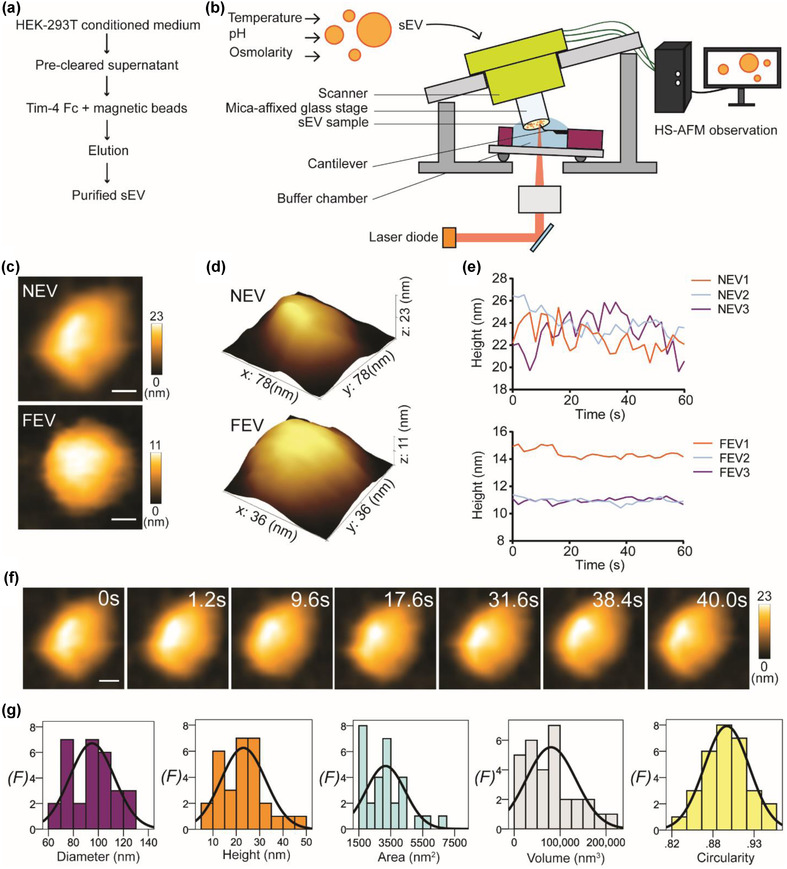
Real‐time visualization of native conformation of sEV in a physiological buffer. (a) A flow chart shows the TIM‐4 affinity purification of HEK293T‐derived sEVs. (b) A schematic diagram illustrates the setup of HS‐AFM for sEVs imaging under different conditions. (c) HS‐AFM images demonstrate the nanotopology of native (NEV) and formalin‐fixed (FEV) sEVs under physiological environment (scale bar, NEV: 18 nm; FEV: 10 nm). (d) 3D structure of NEV and FEV. (e) Line graphs delineate real‐time height of NEV and FEV. Results indicate that formalin fixation could increase the rigidity of sEVs as the height fluctuation is less for FEV compared with NEV. (f) Real‐time image sequence of an NEV. Fluidity of NEV layer is visible during HS‐AFM imaging (scale bar: 18 nm). (g) Spatial dimension analysis of NEV. Results are presented in histograms with normal distribution curves (n = 30; F, frequency)
3.2. Evaluation of thermal stress on sEV nanotopology by using HS‐AFM
Thermal stress exerts a major impact on sEV stability, and subsequently alters its structure and function. To study the outcome at the nanoscopic level, we performed HS‐AFM imaging to visualize sEVs which had been incubated at different temperatures. We noticed that high temperature (60°C and 100°C) destabilized the sEV structure (Figure 2 a–c, Figure S2, and Movie S2). Intriguingly, the proportion of deformed sEVs was similar at both 60°C and 100°C (Figure 2c), suggesting that the sEV pool could contain a proportion of heat‐resistant sEVs. This prediction is sensible given that an sEV is heterogeneous in nature. High temperature also reduced the sEV membrane elasticity (Figure 2d) as the height change was less dynamic in sEV60°C and sEV100°C compared with sEV4°C and sEV37°C. Although the height of sEV37°C was significantly lower than that of sEV4°C (Figure 2e), both diameter and area of sEV37°C were comparable with sEV4°C (Table S1). In addition, a spherical nanotopology was preserved in most of sEV37°C. These findings indicated that 37°C could promote the leakage of the sEV contents without eminently disrupting its membrane, as shown in sEV60°C and sEV100°C. Yokota et al. used the aspect ratio (AR) as an indicator to indicate the deformability of sEVs due to adsorption on a silicon dioxide (SiO2) substrate (Yokota et al., 2019). We also included this indicator in our study (Figure 2f‐g), but we rather called it as an indicator for stretchability to avoid confusion with sEV deformation by high temperature. AR was lowest in both sEV37°C and sEV60°C, suggesting that these two groups were strongly stretched on the PLL‐coated mica substrate. A negative surface charge (‐ζ potential) of sEV was previously found to increase as a function of temperature (Maroto et al., 2017), which subsequently increased the adsorption of sEVs (low AR) on the substrate. In accordance with this finding, our result depicted both sEV37°C and sEV60°C had lower AR (higher stretchability) compared with sEV4°C. At an extreme temperature (100°C), disruption of the sEV layer could eliminate the sEV100°C stretchability on the PLL‐coated mica substrate. Content loss (low height) and failed to expand after adsorption (small diameter) made sEV100°C had higher AR than sEV37°C and sEV60°C. The sEV4°C was also maintained at high AR suggesting that the increment of diameter (denominator of AR) after adsorption did not drastically reduce the AR because sEV height (numerator of AR) was maintained as lesser content loss at 4°C.
FIGURE 2.
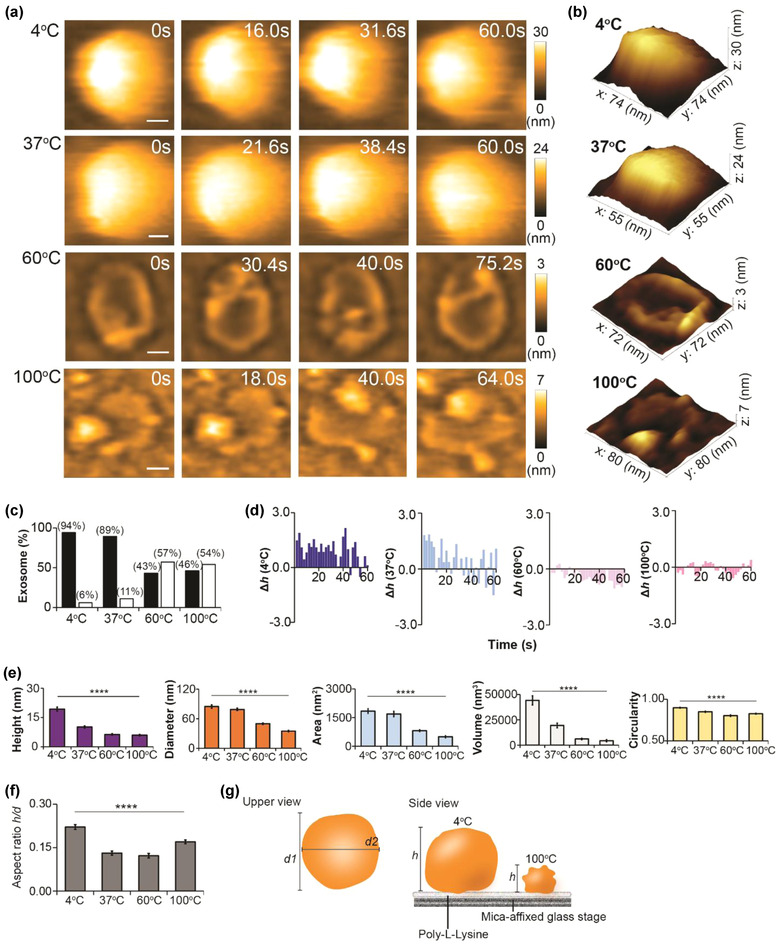
Temperature‐induced nanotopological change in sEV. (a) Direct visualization of sEV after exposure to 4°C, 37°C, 60°C, or 100°C for 4 h (scale bar, sEV4°C: 17 nm; sEV37°C: 11 nm; sEV60°C: 20 nm; sEV100°C: 20 nm). (b) 3D topology of respective sEVs in panel (a). (c) Bar graph demonstrating the percentage of normal and deformed sEVs in four sEV groups (black bar, normal; white bar, deformed). (d) Bar graphs illustrating the real‐time height change (Δh) of four sEV groups (n of each group: 1). The magnitudes of height change for sEV4°C and sEV37°C are greater than those for sEV60°C and sEV100°C. (e) Temperature significantly altering the spatial dimension and circularity of sEV. Comparison is performed using Kruskal‐Wallis H Test, and data are presented as mean ± SEM (****p < 0.0001). p‐values of detailed pairwise comparison are reported in Table S1. (f) Aspect ratio (AR) computed to determine the stretchability of an sEV on PLL‐coated mica substrate. Data are depicted as mean ± SEM (****p < 0.0001). (g) A schematic diagram showing the nanotopology of an sEV after exposure to low or high temperatures (d1, major diameter; d2, minor diameter). Sample sizes for (c), (e), and (f): sEV4°C, 80; sEV37°C, 82; sEV60°C, 92; sEV100°C, 104
3.3. Evaluation of acid‐base effect on sEV nanotopology by using HS‐AFM
A previous study reported that sEVs stored at pH 4.00 or pH 10.00 experienced protein loss without affecting their sizes (Cheng et al., 2019). By using HS‐AFM, we intended to observe the structural changes of sEVs in response to an acidic or basic environment. Results showed that the sEV shape was maintained at pH 4.00, but it became deformed (flattened or shrunken) at pH 10.00 (Figure 3a–c, Figure S3a, and Movie S3). Spatial dimension analysis revealed that height, diameter, area, and volume were significantly reduced in sEVpH 4.00 compared with sEVpH 7.50 (Figure 3d). These findings could imply that low pH only induced content loss in sEVpH 4.00 but spared its spherical shape. In contrast, sEVpH 10.00 lost its round shape, and spatial dimension parameters were greatly reduced when compared with sEVpH 7.50 (Figure 3d). Basic buffer (pH 10.00) could disrupt the sEV membrane stability and subsequently the membrane elasticity diminished, which were indicated by the low real‐time height change (Figure 3e) and low height fluctuation (Figure S3b) in sEVpH 10.00. The sEV structure deteriorated at pH 10.00 and stretched irreversibly on PLL‐coated substrate, depicted by the lowest AR value (Figure 3f). In a real‐time observation, we noticed that the sEV shrunk but remained round when the pH had changed from 7.50 to 4.00 (Figure 3g, and Movie S4), which was indicated by a rapid drop followed by subtle changes in area, diameter, and height (Figure 3h and Figure S3c). Moreover, the AR result illustrated a constant trend at pH 4.00 (Figure 3i). However, at pH 10.00, the sEV became unstable (Figure 3g‐i and Movie S4), and the diameter and area of the sEV were increased together with persistent drops in height and circularity, which suggested that the sEV could start to deform and widely spread on substrate. Interestingly, during the transition from pH 4.00 to pH 10.00, we did not find a reversion trend in the real‐time height (Figure S3d), which suggested that the rapid drop of height induced by pH 4.00 was irreversible (Figure 3j).
FIGURE 3.
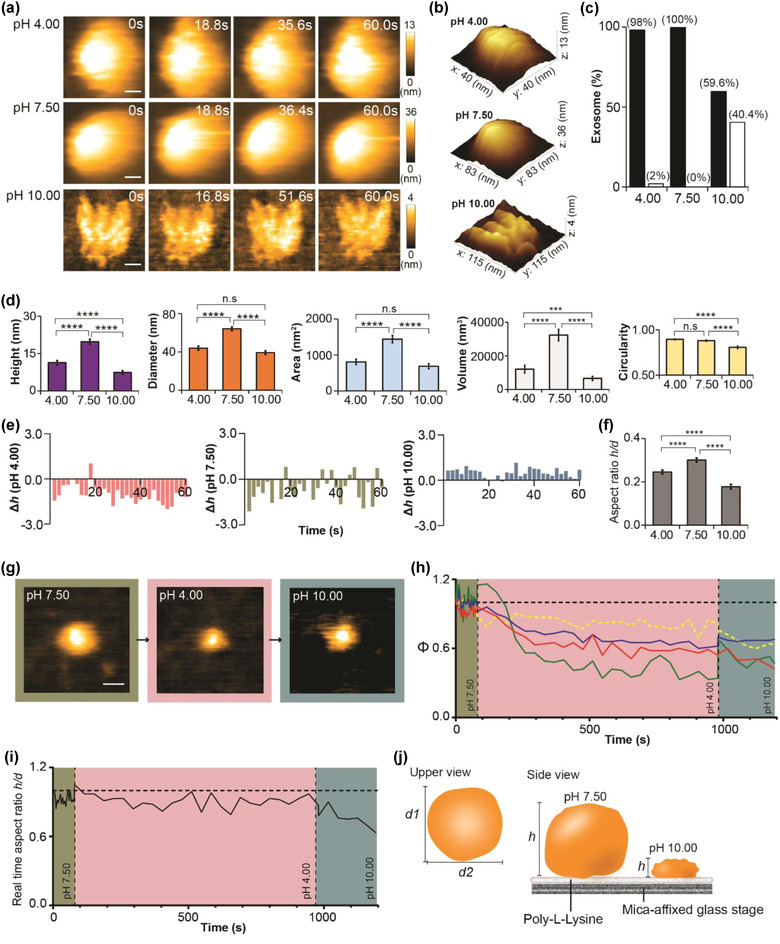
Nanotopological changes of sEVs in response to pH manipulation. (a) sEVs incubated in acidic (pH 4.00), neutral (pH 7.50), or basic (pH 10.00) buffers for 3 h before HS‐AFM imaging. sEV spherical shape is retained at pH 4.00 but deformed at pH 10.00 (scale bar, sEVpH 4.00: 8 nm; sEVpH 7.50: 17 nm; sEVpH 10.00: 23 nm). (b) 3D structure of sEVpH 4.00, sEVpH 7.50, and sEVpH 10.00. (c) Bar graph showing the proportion of deformed sEV after exposure to three different pH levels (black bar, normal; white bar, deformed). (d) pH level significantly alters spatial dimension parameters and circularity of sEV. Data are presented as mean ± SEM (***p < 0.001, ****p < 0.0001, n.s: not significant). (e) Bar graphs presenting the real‐time height changes (Δh) of an sEV pretreated at pH 4.00, pH 7.50, or pH 10.00. (f) sEVpH 10.00 is greatly stretched on PLL‐coated substrate compared with sEVpH 4.00 and sEVpH 7.50. Values are shown as mean ± SEM (****p < 0.0001). (g) Real‐time imaging of sEV nanotopology during pH transition (neutral‐acidic‐basic). sEV shrinks at pH 4.00 and later deforms at pH 10.00 (scale bar: 30 nm). (h) Line graphs demonstrating the real‐time measurement of height, diameter, area, and circularity in response to pH change. The values are illustrated as ratio relative to the initial state (t = 0) (red: height; blue: diameter; green: area; yellow dotted: circularity; green area: pH 7.50; red area: pH 4.00; purple: pH 10.00). (i) Real‐time change in aspect ratio of sEV at different pH levels. The aspect ratio drops in the basic environment, which is consistent to the result in a premix setting (green area: pH 7.50; red area: pH 4.00; purple area: pH 10.00). (j) A schematic diagram delineating the sEV nanotopology at different pH levels. Sample size for (c), (d), and (f): sEVpH 4.00, 52; sEVpH 7.50, 51; sEVpH 10.00, 52
3.4. Evaluation of osmotic stress on sEV nanotopology by using HS‐AFM
Osmotic pressure is regulated to maintain the viable cell structure and cell physiology. For example, haemolysis occurs when red blood cells are exposed to a hypotonic environment. Therefore, we postulated that osmotic stress could exert a similar effect on the sEV structure. First, we scanned sEVs after 3‐h incubation in Tris‐HCl buffer containing 0 M NaCl, 0.15 M NaCl, or 1.8 M NaCl. HS‐AFM images depicted that sEV1.8 M was contorted whereas the spherical shape remained intact in sEV0 M but with less bulging than sEV0.15 M (Figure 4a–b, Figure S4a, and Movie S5). The deformation rate was comparable for sEV0 M and sEV1.8 M (Figure 4c). Parameters of spatial dimension and circularity of sEV1.8 M were significantly lower than those of sEV0.15 M and sEV0 M (Figure 4d). Both diameter and area of sEV0 M were comparable with those of sEV0.15 M, but sEV0 M had much lower height, volume, and circularity than sEV0.15 M (Figure 4d). The sEV membrane became less elastic in buffer with zero or high salt (Figure 4e, and Figure S4b) and failed to resist stretching after the sEV was adsorbed on a PLL‐coated substrate (Figure 4f). Collectively, after 3‐h incubation, Tris‐HCl with 1.8 M NaCl was hypertonic to the sEV as shrinkage, membrane disruption, and content loss occurred in many sEVs. However, Tris‐HCl with zero salt was hypotonic to the sEV, and this condition led to an osmotic burst in many sEV without exaggeratedly disrupting the sEV nanotopology. Tris HCl with 0.15 M NaCl was isotonic to the sEV and hence the sEV nanotopology was well preserved even after 3‐h incubation in this condition. We further investigated the real‐time response of the sEV to osmotic stress (Figure 4g–i, and Movie S6). An sEV was initially scanned in an isotonic buffer, and then the osmotic pressure was manipulated by adjusting the NaCl concentration (Figure 4g). The sEV shrunk in hypertonic buffer, which was demonstrated by the rugged appearance and declination in height, diameter, area, and circularity (Figure 4h). The shrinkage was irreversible because the sEV did not expand in hypotonic buffer (Figure 4h–j), which implied that the hypertonic environment had irreversibly damaged the sEV membrane and the membrane failed to retain hydrostatic pressure for expansion in the hypotonic environment. Surprisingly, prolonged incubation of the sEV in hypotonic buffer promoted sEV fusion and formed large sEV aggregates (Figure S5, and Movie S7).
FIGURE 4.
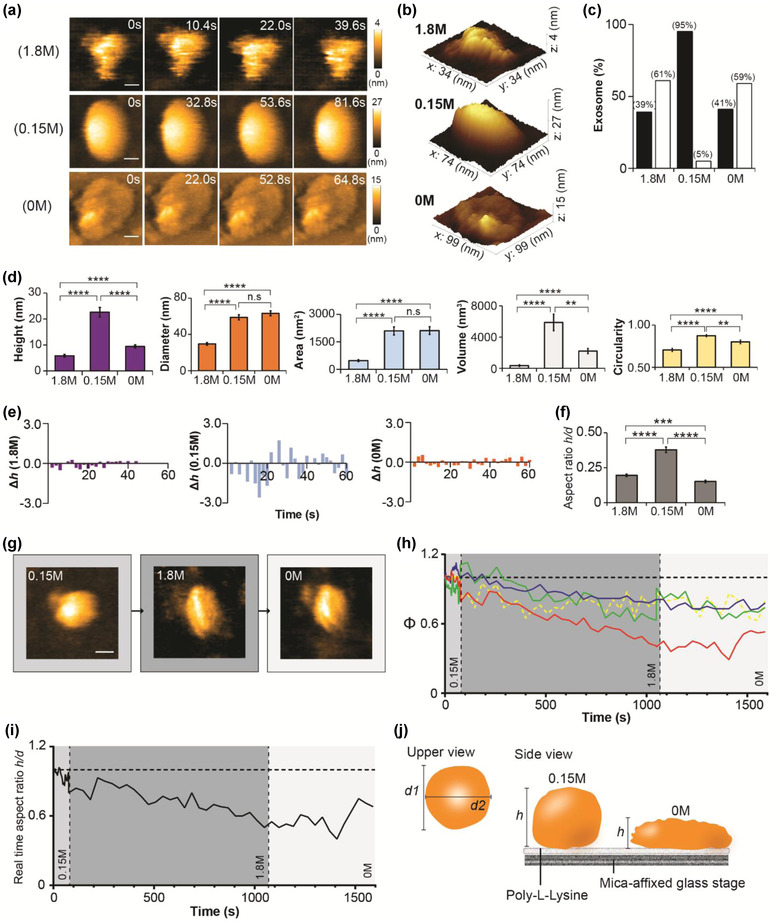
Osmotic stress on nanotopology of sEV. (a) sEVs incubated in a scanning buffer (50 mM Tris‐HCl pH 7.50) with different sodium chloride (NaCl) concentrations (0 M, 0.15 M, and 1.8 M) for 3 h before HS‐AFM observation. sEV structure is clearly deteriorated in hypertonic buffer (1.8 M NaCl) but its spherical shape remains intact in a hypotonic buffer (0 M NaCl) (scale bar, sEV1.8 M: 8 nm; sEV0.15 M: 16 nm; sEV0 M: 22 nm). (b) 3D structures of sEV1.8 M, sEV0.15 M, and sEV0 M. (c) Bar graph showing the percentage of deformed sEVs after exposure to different levels of osmotic stress (black bar, normal; white bar, deformed). (d) Osmotic stress significantly changes the spatial dimension and circularity of sEV. Mann‐Whitney U test is performed, and data are presented as mean ± SEM (**p < 0.01, ***p < 0.001, ****p < 0.0001, n.s: not significant). (e) Bar graphs illustrating the real‐time height change (Δh) in sEVs after treated with hypo‐, iso‐, or hypertonic buffer. Results show that both hypertonic and hypotonic buffers reduce the height change of an sEV. (f) Bar graph demonstrating the aspect ratio is significantly reduced in sEV1.8 M and sEV0 M. Data are depicted as mean ± SEM. (g) Real‐time tracing of nanotopological change in an sEV in response to different levels of osmotic stress (scale bar: 40 nm). (h‐i) Hypertonic condition induces an irreversible change in the sEV structure, and hence the sEV fails to revert to its initial bulging shape in hypotonic condition. The values are illustrated as ratio relative to the initial state (t = 0) (red: height; blue: diameter; green: area; yellow dotted: circularity; medium grey area: 0.15 M; dark grey: 1.8 M; white area: 0 M). (j) A schematic diagram showing nanotopology of an sEV at isotonic or hypotonic conditions. Sample size of (c), (d), and (f): sEV1.8 M, 51; sEV0.15 M, 40; sEV0 M, 44
4. DISCUSSION
Here, we have introduced a new paradigm for nanoscopic assessment of sEVs in response to various types of physicochemical stress, in both a premix setting and a real‐time manner (Figure 5). The advent of HS‐AFM has brought a revolutionary improvement to nanoimaging of biomolecules and organelles (Ando et al., 2001; Kodera et al., 2010). It supersedes other imaging tools in two respects: sample preparation and method of data acquisition. Additional sample preparation, fixation, and permeabilization, for example, are not necessary prerequisites for HS‐AFM imaging. Furthermore, the imaging environment of HS‐AFM is relatively physiological compared with the vacuum environment used in electron microscopy (EM). Therefore, we are able to observe and study the native conformation of sEV and life processes using HS‐AFM. Data acquisition is important for obtaining valuable information about sEV structure and function. EM methods, especially cryo‐EM, could potentially provide a comprehensive set of sEV structural data but only within the confines of static snapshots. Computational simulations to evaluate sEV dynamic properties are therefore needed. Alternatively, fluorescent markers can be used to tag sEVs, but the acquired data only reflect dynamic behaviour of the tag and not the sEVs themselves. Various AFM imaging modes have been established, such as Bio‐AFM, DM‐AFM, and Opto‐AFM (reviewed in (Dufrêne et al., 2017)), and AFM is frequently used to measure the membrane stiffness of sEVs (Ye et al., 2021; Yurtsever et al., 2021). Recently, 3D images of sEVs were generated using 3D‐AFM (Fukuma & Garcia, 2018; Yurtsever et al., 2021). In contrast to HS‐AFM, other AFM methods have lower temporal resolution, and it is not feasible to measure rapid changes in sEV nanotopology in response to stresses, such as those that occur during transition in pH or osmolarity. Although the spatiotemporal resolution of HS‐AFM is advantageous for capturing sEV conformational dynamics, it must be complemented with biochemical analysis to determine protein compositions in sEVs. Without a functionalized cantilever tip coated with antibodies, HS‐AFM is unable to detect exosomal markers such as CD9 and CD81 in sEVs. Unless sEVs are ruptured or deformed, HS‐AFM is only able to scan sEV surfaces. Indeed, we have identified exosomal contents from ruptured sEVs such as DNA strands and peptide filaments (Figure S6 and Movie S8), yet we still need additional tools to investigate exosomal content. Moreover, HS‐AFM is an in vitro approach, despite the scanning buffer providing near‐physiological conditions.
FIGURE 5.
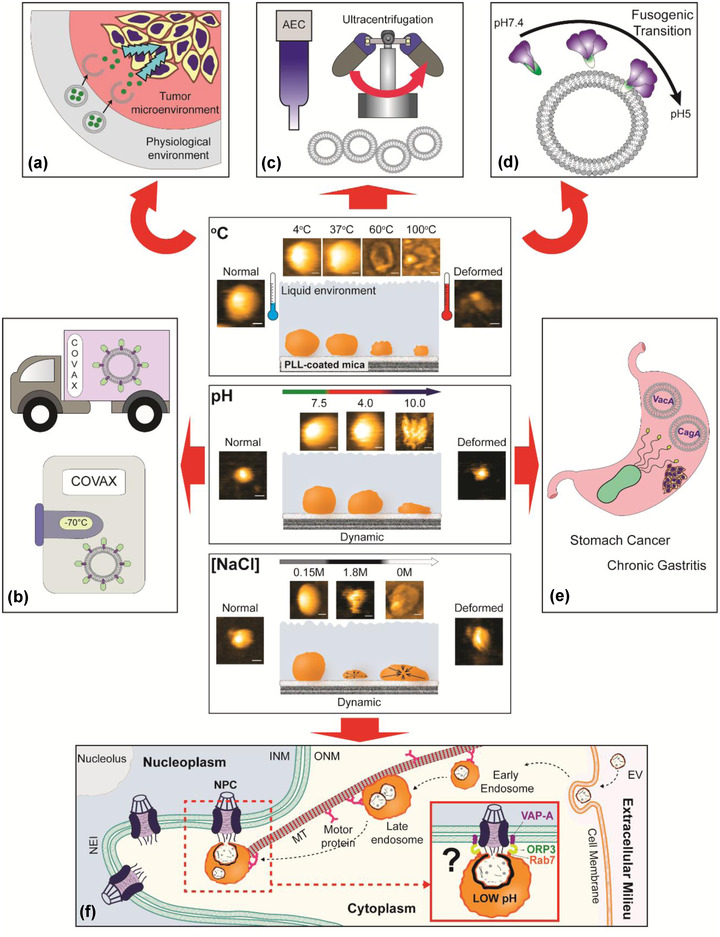
New paradigm of nanoparticle assessment using HS‐AFM. HS‐AFM imaging represents the continuity of nanotopological dynamics of sEV in response to different physicochemical parameters. The information gained is essential for sEV study in various aspects: (a) Structural dynamics of sEV‐based nanocarrier in drug release. (b) Material selection to produce sEV‐based products with high thermal stability (ex: sEV‐based COVID‐19 vaccine) for people in rural area with limited freezers and/or delivery service. (c) Quality control of sEV after isolation before downstream processes. (d) Real‐time observation of interaction between viral fusion protein and sEV layer during viral entry. sEV can be used to mimic endosome to provide a docking platform for viral fusion proteins during fusogenic transition triggered by low pH. (e) Nanoscopic assessment of pathogenic EVs. Helicobacter pylori EVs contain oncoproteins CagA and VacA, which are the etiological factors of chronic gastritis and gastric cancer. (f) Discovery of new nuclear transport mechanism. Non‐canonical nuclear transport of sEV contents (Corbeil et al., 2020) could be investigated using HS‐AFM in the near future. (AEC: anion exchange chromatography; INM: inner nuclear membrane; ONM: outer nuclear membrane; NPC: nuclear pore complex; NEI: nuclear envelope invagination; MT: microtubule; VAP‐A: (VAMP)‐associated protein A; ORP3: oxysterol‐binding protein (OSBP)‐related protein‐3; Rab7: late endosome‐associated small GTPase Rab7)
Our study provides insights into a wide range of sEV topics. First, our findings suggest that HS‐AFM is suitable for studying the relationship between storage conditions and sEV nanotopology, with temperature and pH being common storage parameters. The intrinsic properties of sEVs make them suitable for use as nanocarriers, vaccines, and theranostic nanoparticles. Robust HS‐AFM scanning of sEV nanotopology in response to pH and temperature manipulations allows the resilience of sEVs against unfavourable storage conditions to be evaluated. For example, sEV‐based COVID‐19 vaccines with high thermal stability allow people living in poor rural areas to receive protection (Figure 5b). Material selection of sEVs is not limited to biomaterial storage. For example, assessment of conformational dynamics of sEVs in response to different physicochemical parameters allows the drug release patterns of sEV‐based nanocarriers to be studied, which subsequently improves understanding of pharmacokinetic profile of drugs (Figure 5a).
Quality control of isolated sEVs is a routine procedure to ensure they are fit for downstream processes, such as nanoformulation of sEV‐based drugs and bioengineering of sEVs for vaccines (Figure 5c). Certain isolation techniques could affect the structural integrity of sEVs. For instance, EVs are exposed to a high salt concentration (e.g., 1 M NaCl) during their rapid isolation and enrichment using anion exchange chromatography (Heath et al., 2018). Our results illustrated that the nanotopology of sEVs was rapidly affected during an increase in salt concentration from 0.15 to 1.8 M. Images obtained using cryo‐EM may not represent the native conformation of EVs isolated using anion exchange chromatography. As a result, confirmation of the isolated sEV structure using HS‐AFM is beneficial.
The pH change during endocytosis, from early endosome to late endosome, can be readily established by changing the scanning buffer pH from 7 to 4. Interestingly, the nanotopology of sEVs was resistant to acidic conditions, presumably because both sEVs and endosomes originate from multivesicular bodies. This condition is crucial to several viral fusion proteins for which low pH triggers their fusogenic transition to orchestrate viral entry (Figure 5d). We have previously characterized the influenza A virus hemagglutinin fusion protein (Lim et al., 2020a) or the SARS‐CoV‐2 spike protein (Lim et al., 2021) docking on an sEV layer using HS‐AFM. We can also use HS‐AFM to study the nanotopology and structural dynamics of EVs released by Helicobacter pylori (Figure 5e), the pathogen that is able to survive in extremely acidic environments, and usually secretes urease to increase stomach acidity (Rektorschek et al., 1998). H. pylori EVs contain oncoproteins CagA and VacA, which induce chronic gastritis and gastric cancer (Choi et al., 2017). Choi et al. reported that H. pylori EVs accumulate in the stomach of gastric cancer patients (Choi et al., 2017). Using our methodology, we can investigate the pH‐dependent nanotopology of these EVs using HS‐AFM and relate the findings to the pathogenesis of chronic gastritis or gastric cancer. In the near future, we may also be able to perform HS‐AFM imaging to trace the non‐canonical nuclear transport of sEV contents (Corbeil et al., 2020; Santos et al., 2021), during which low pH induces membrane fusion of an endosome and sEV to release sEV contents near the nucleus before nuclear translocation (Corbeil et al., 2020) (Figure 5f). Further effort is needed to determine whether this uncommon transport pathway is essential for intercellular communication or nuclear transport of oncogenes/oncoproteins (Hashizume et al., 2010; Hazawa et al., 2018), and viral proteins (Sajidah et al., 2021).
In conclusion, we have presented HS‐AFM as a versatile nanoimaging tool that, by manipulating physicochemical stresses, is capable of answering a broad range of sEV‐ and EV‐related questions.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
We thank all members of the Richard Wong laboratory. We also thank Prof. Noriyuki Kodera, Dr. Azuma Taoka, Dr. Yousuke Kikuchi, and Ms Phuong Doan N. Nguyen for providing insightful HS‐AFM advice and technical support in this study. This project was funded by grants from the MEXT/JSPS KAKENHI (19K23841 to Keesiang Lim, 20K16262 to Keesiang Lim, 21H05744 to Richard W. Wong, 21K19043 to Richard W. Wong, 22H02209 to Richard W. Wong, and 22H05537 to Richard W. Wong), from LocSEES MEXT Japan (to Elma Sakinatus Sajidah), the TDRP grant (to Keesiang Lim) from WPI NanoLSI, from the JST CREST (No. JPMJCR18H4 to Rikinari Hanayama), from the Kobayashi International Scholarship Foundation (to Richard W. Wong), from the Shimadzu Science Foundation (to Richard W. Wong), and from the Takeda Science Foundation (to Richard W. Wong).
Sajidah, E. S. , Lim, K. , Yamano, T. , Nishide, G. , Qiu, Y. , Yoshida, T. , Wang, H. , Kobayashi, A. , Hazawa, M. , Dewi, F. R. P. , Hanayama, R. , Ando, T. , & Wong, R. W. (2022). Spatiotemporal tracking of small extracellular vesicle nanotopology in response to physicochemical stresses revealed by HS‐AFM. Journal of Extracellular Vesicles, 11, e12275. 10.1002/jev2.12275
Contributor Information
Keesiang Lim, Email: limkeesiang@staff.kanazawa-u.ac.jp.
Richard W. Wong, Email: rwong@staff.kanazawa-u.ac.jp.
REFERENCES
- Ando, T. (2019). High‐speed atomic force microscopy. Current Opinion in Chemical Biology, 51, 105–112. 10.1016/j.cbpa.2019.05.010 [DOI] [PubMed] [Google Scholar]
- Ando, T. , Kodera, N. , Takai, E. , Maruyama, D. , Saito, K. , & Toda, A. (2001). A high‐speed atomic force microscope for studying biological macromolecules. Proceedings of the National Academy of Sciences of the United States of America, 98, 12468–12472. 10.1073/pnas.211400898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Y. , Zeng, Q. , Han, Q. , & Xia, W. (2019). Effect of pH, temperature and freezing‐thawing on quantity changes and cellular uptake of exosomes. Protein & Cell, 10, 295–299. 10.1007/s13238-018-0529-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, H. I. , Choi, J. P. , Seo, J. , Kim, B. J. , Rho, M. , Han, J. K. , & Kim, J. G. (2017). Helicobacter pylori‐derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Experimental & Molecular Medicine, 49, e330. 10.1038/emm.2017.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbeil, D. , Santos, M. F. , Karbanová, J. , Kurth, T. , Rappa, G. , & Lorico, A. (2020). Uptake and fate of extracellular membrane vesicles: Nucleoplasmic reticulum‐associated late endosomes as a new gate to intercellular communication. Cells, 9, 1931. 10.3390/cells9091931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrêne, Y. F. , Ando, T. , Garcia, R. , Alsteens, D. , Martinez‐Martin, D. , Engel, A. , Gerber, C. , & Müller, D. J. (2017). Imaging modes of atomic force microscopy for application in molecular and cell biology. Nature Nanotechnology, 12, 295–307. 10.1038/nnano.2017.45 [DOI] [PubMed] [Google Scholar]
- Fukuma, T. , & Garcia, R. (2018). Atomic‐ and molecular‐resolution mapping of solid‐liquid interfaces by 3D atomic force microscopy. ACS Nano, 12, 11785–11797. 10.1021/acsnano.8b07216 [DOI] [PubMed] [Google Scholar]
- Hardij, J. , Cecchet, F. , Berquand, A. , Gheldof, D. , Chatelain, C. , Mullier, F. , Chatelain, B. , & Dogné, J. M. (2013). Characterisation of tissue factor‐bearing extracellular vesicles with AFM: Comparison of air‐tapping‐mode AFM and liquid Peak Force AFM. Journal of Extracellular Vesicles, 2, 21045–53. 10.3402/jev.v2i0.21045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume, C. , Nakano, H. , Yoshida, K. , & Wong, R. W. (2010). Characterization of the role of the tumor marker Nup88 in mitosis. Molecular Cancer, 9, 119. 10.1186/1476-4598-9-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa, M. , Lin, D. C. , Kobayashi, A. , Jiang, Y. Y. , Xu, L. , Dewi, F. R. P. , Mohamed, M. S. , Hartono, Nakada, M. , Meguro‐Horike, M. , Horike, S. ‐. I. , Koeffler, H. P. , & Wong, R. W. (2018). ROCK‐dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Reports, 19, 73–88. 10.15252/embr.201744523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, N. , Grant, L. , De Oliveira, T. M. , Rowlinson, R. , Osteikoetxea, X. , Dekker, N. , & Overman, R. (2018). Rapid isolation and enrichment of extracellular vesicle preparations using anion exchange chromatography. Scientific Reports, 8, 5730. 10.1038/s41598-018-24163-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodera, N. , Yamamoto, D. , Ishikawa, R. , & Ando, T. (2010). Video imaging of walking myosin V by high‐speed atomic force microscopy. Nature, 468, 72–76. 10.1038/nature09450 [DOI] [PubMed] [Google Scholar]
- Lim, K. , Kodera, N. , Wang, H. , Mohamed, M. S. , Hazawa, M. , Kobayashi, A. , Yoshida, T. , Hanayama, R. , Yano, S. , Ando, T. , & Wong, R. W. (2020). High‐speed AFM reveals molecular dynamics of human influenza a hemagglutinin and its interaction with exosomes. Nano Letters, 20, 6320–6328. 10.1021/acs.nanolett.0c01755 [DOI] [PubMed] [Google Scholar]
- Lim, K. , Nishide, G. , Yoshida, T. , Watanabe‐Nakayama, T. , Kobayashi, A. , Hazawa, M. , Hanayama, R. , Ando, T. , & Wong, R. W. (2021). Millisecond dynamic of SARS‐CoV‐2 spike and its interaction with ACE2 receptor and small extracellular vesicles. Journal of Extracellular Vesicles, 10, e12170. 10.1002/jev2.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, K. S. , Mohamed, M. S. , Wang, H. , Hazawa, M. , Kobayashi, A. , Voon, D. C. , Kodera, N. , Ando, T. , & Wong, R. W. (2020). Direct visualization of avian influenza H5N1 hemagglutinin precursor and its conformational change by high‐speed atomic force microscopy. Biochimica et Biophysica Acta. General Subjects, 1864, 129313. 10.1016/j.bbagen.2019.02.015 [DOI] [PubMed] [Google Scholar]
- Maroto, R. , Zhao, Y. , Jamaluddin, M. , Popov, V. L. , Wang, H. , Kalubowilage, M. , Zhang, Y. , Luisi, J. , Sun, H. , Culbertson, C. T. , Bossmann, S. H. , Motamedi, M. , & Brasier, A. R. (2017). Effects of storage temperature on airway exosome integrity for diagnostic and functional analyses. Journal of Extracellular Vesicles, 6, 1359478. 10.1080/20013078.2017.1359478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed, M. S. , Hazawa, M. , Kobayashi, A. , Guillaud, L. , Watanabe‐Nakayama, T. , Nakayama, M. , Wang, H. , Kodera, N. , Oshima, M. , Ando, T. , & Wong, R. W. (2020). Spatiotemporally tracking of nano‐biofilaments inside the nuclear pore complex core. Biomaterials, 256, 120198. 10.1016/j.biomaterials.2020.120198 [DOI] [PubMed] [Google Scholar]
- Mohamed, M. S. , Kobayashi, A. , Taoka, A. , Watanabe‐Nakayama, T. , Kikuchi, Y. , Hazawa, M. , Minamoto, T. , Fukumori, Y. , Kodera, N. , Uchihashi, T. , Ando, T. , & Wong, R. W. (2017). High‐speed atomic force microscopy reveals loss of nuclear pore resilience as a dying code in colorectal cancer cells. ACS Nano, 11, 5567–5578. 10.1021/acsnano.7b00906 [DOI] [PubMed] [Google Scholar]
- Nakai, W. , Yoshida, T. , Diez, D. , Miyatake, Y. , Nishibu, T. , Imawaka, N. , Naruse, K. , Sadamura, Y. , & Hanayama, R. (2016). A novel affinity‐based method for the isolation of highly purified extracellular vesicles. Scientific Reports, 6, 33935. 10.1038/srep33935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishide, G. , Lim, K. , Mohamed, M. S. , Kobayashi, A. , Hazawa, M. , Watanabe‐Nakayama, T. , Kodera, N. , Ando, T. , & Wong, R. W. (2021). High‐speed atomic force microscopy reveals spatiotemporal dynamics of histone protein H2A involution by DNA inchworming. The Journal of Physical Chemistry Letters, 12, 3837–3846. 10.1021/acs.jpclett.1c00697 [DOI] [PubMed] [Google Scholar]
- Rektorschek, M. , Weeks, D. , Sachs, G. , & Melchers, K. (1998). Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology, 115, 628–641. 10.1016/s0016-5085(98)70142-8 [DOI] [PubMed] [Google Scholar]
- Sajidah, E. S. , Lim, K. , & Wong, R. W. (2021). How SARS‐CoV‐2 and other viruses build an invasion route to hijack the host nucleocytoplasmic trafficking system. Cells, 10, 1424. 10.3390/cells10061424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, M. F. , Rappa, G. , Karbanová, J. , Fontana, S. , Bella, M. A. D. , Pope, M. R. , Parrino, B. , Cascioferro, S. M. , Vistoli, G. , Diana, P. , & Lorico, A. (2021). Itraconazole inhibits nuclear delivery of extracellular vesicle cargo by disrupting the entry of late endosomes into the nucleoplasmic reticulum. Journal of Extracellular Vesicles, 10, e12132. 10.1002/jev2.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatanek, R. , Baj‐Krzyworzeka, M. , Zimoch, J. , Lekka, M. , Siedlar, M. , & Baran, J. (2017). The methods of choice for extracellular vesicles (EVs) characterization. International Journal of Molecular Sciences, 18, 1153. 10.3390/ijms18061153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry, C. , Witwer, K. W. , Aikawa, E. , Alcaraz, M. J. , Anderson, J. D. , Andriantsitohaina, R. , Antoniou, A. , Arab, T. , Archer, F. , Atkin‐Smith, G. K. , Ayre, D. C. , Bach, J ‐M. , Bachurski, D. , Baharvand, H. , Balaj, L. , Baldacchino, S. , Bauer, N. N. , Baxter, A. A. , Bebawy, M. , & Zuba‐Surma, E ‐K. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. Journal of Extracellular Vesicles, 7, 1535750. 10.1080/20013078.2018.1535750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji, T. , Cheng, J. , Tatematsu, T. , Ebata, A. , Kamikawa, H. , Fujita, A. , Gyobu, S. , Segawa, K. , Arai, H. , Taguchi, T. , Nagata, S. , & Fujimoto, T. (2019). Predominant localization of phosphatidylserine at the cytoplasmic leaflet of the ER, and its TMEM16K‐dependent redistribution. Proceedings of the National Academy of Sciences of the United States of America, 116, 13368–13373. 10.1073/pnas.1822025116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead, B. , Wu, L. , Hvam, M. L. , Aslan, H. , Dong, M. , Dyrskjøt, L. , Ostenfeld, M. S. , Moghimi, S. M. , & Howard, K. A. (2015). Tumour exosomes display differential mechanical and complement activation properties dependent on malignant state: Implications in endothelial leakiness. Journal of Extracellular Vesicles, 4, 29685. 10.3402/jev.v4.29685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, S. , Li, W. , Wang, H. , Zhu, L. , Wang, C. , & Yang, Y. (2021). Quantitative nanomechanical analysis of small extracellular vesicles for tumor malignancy indication. Advanced Science (Weinheim, Baden‐Wurttemberg, Germany), 8, e2100825. 10.1002/advs.202100825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, S. , Kuramochi, H. , Okubo, K. , Iwaya, A. , Tsuchiya, S. , & Ichiki, T. (2019). Extracellular vesicles nanoarray technology: Immobilization of individual extracellular vesicles on nanopatterned polyethylene glycol‐lipid conjugate brushes. PloS One, 14, e0224091. 10.1371/journal.pone.0224091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurtsever, A. , Yoshida, T. , Badami Behjat, A. , Araki, Y. , Hanayama, R. , & Fukuma, T. (2021). Structural and mechanical characteristics of exosomes from osteosarcoma cells explored by 3D‐atomic force microscopy. Nanoscale, 13, 6661–6677. 10.1039/d0nr09178b [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Supporting Information


