SUMMARY
Apolipoprotein E4 (APOE4) is the greatest known genetic risk factor for developing sporadic Alzheimer’s disease. How the interaction of APOE4 microglia with neurons differs from microglia expressing the disease-neutral APOE3 allele remains unknown. Here, we employ CRISPR-edited induced pluripotent stem cells (iPSCs) to dissect the impact of APOE4 in neuron-microglia communication. Our results reveal that APOE4 induces a lipid-accumulated state that renders microglia weakly responsive to neuronal activity. By examining the transcriptional signatures of APOE3 versus APOE4 microglia in response to neuronal conditioned media, we established that neuronal cues differentially induce a lipogenic program in APOE4 microglia that exacerbates pro-inflammatory signals. Through decreased uptake of extracellular fatty acids and lipoproteins, we identified that APOE4 microglia disrupts the coordinated activity of neuronal ensembles. These findings suggest that abnormal neuronal network-level disturbances observed in Alzheimer’s disease patients harboring APOE4 may in part be triggered by impairment in lipid homeostasis in non-neuronal cells.
In brief
Tsai and colleagues explored the impact of the Alzheimer’s disease-associated risk gene APOE4 onto the cellular communication of neurons and microglia. Through combinatorial experiments with cells derived from CRISPR-edited APOE isogenic lines, this work defines the functional consequence of impaired microglial lipid metabolism induced by APOE4 onto neuronal network activity.
Graphical Abstract
INTRODUCTION
Recent findings that microglia can actively regulate neuronal activity (Badimon et al., 2020; Cheadle et al., 2020; Cserep et al., 2020; Merlini et al., 2021) have led many to the speculate that altered neuronal excitability, which is typical of many neurological diseases, may partly manifest through early deficits in microglial surveillance and regulation of neuronal networks. This is particularly interesting in the context of Alzheimer’s disease (AD), where a large number of risk variants have been identified in genes that are highly or solely expressed by non-neuronal cells (Kunkle et al., 2019). Nevertheless, it remains unclear how disease-associated genetic variants enriched in glia impact neuronal network dynamics.
Apolipoprotein E (APOE) is a polymorphic gene with three common alleles: ε2, ε3 and ε4. While APOE ε3 (APOE3) is the most common allele and deemed to be neutral with respect to disease, APOE ε4 (APOE4) is the greatest known genetic risk factor for developing late-onset AD (Liu et al., 2013; Yamazaki et al., 2019). Within the central nervous system, APOE is predominantly expressed by glia, although neuronal expression of APOE has been reported in stress-induced conditions (Xu et al., 2006; Zalocusky et al., 2021). By leveraging CRISPR/Cas9 genome-editing with induced pluripotent stem cells (iPSCs), we have previously generated isogenic lines that are genetically identical except for the modification in the APOE allele (Lin et al., 2018). We previously reported that iPS-derived neurons, astrocytes, and microglia that harbored the APOE4 genotype displayed distinct cell type-dependent phenotypes (Lin et al., 2018). In this study, we sought to build upon these findings to test the impact of APOE4 onto the bi-directional communication between neurons and microglia using an iPSCs.
RESULTS
iPSC-derived microglia-like cells respond to soluble factors secreted by neurons in an activity-dependent manner
Purinergic signaling is a powerful modulator of microglia chemotaxis, phagocytosis, and pro-inflammatory cytokine production (Davalos et al., 2005; Koizumi et al., 2007; Monif et al., 2009). Within the brain, the purinergic receptor P2RY12 is highly expressed by microglia (Cserep et al., 2020), enabling the sensing of extracellular adenosine di- or tri-phosphate (ADP or ATP) (Calovi et al., 2019). In addition, many other soluble neuronal factors are thought to maintain microglia in surveillance states, such as the CX3CL1-CX3CR1 signaling axis (Finneran and Nash, 2019). Interestingly, microglia also express numerous neurotransmitter receptors (Szepesi et al., 2018) (Figure 1A). We sought to determine if microglia derived from iPSCs expressed receptors thought to mediate neuron-microglia communication. We generated human microglia-like cells (iMGLs) (line #2191; see Key Resources Table) using established protocols (Abud et al., 2017; McQuade et al., 2018). iMGLs stained positive for microglial specific-markers such as IBA1 and P2RY12 and display electrophysiological properties typical of ex vivo microglia (Figures 1B and S1A–S1E). Additionally, immunostaining of iMGLs revealed the expression of canonical ion channels P2X1 and THIK-1, voltage-gated channels such as KCNE3 and CACNA2D4, as well as the glutamate receptor GLUR7 (Izquierdo et al., 2021; Koizumi et al., 2013; Madry et al., 2018) (Figure 1B). Collectively these findings suggest that iMGL recapitulate expression patterns of receptors governing surveillance of neuronal activity.
Figure 1. Neuronal activity evokes Ca2+ transients in iPSC-derived microglia-like cells.
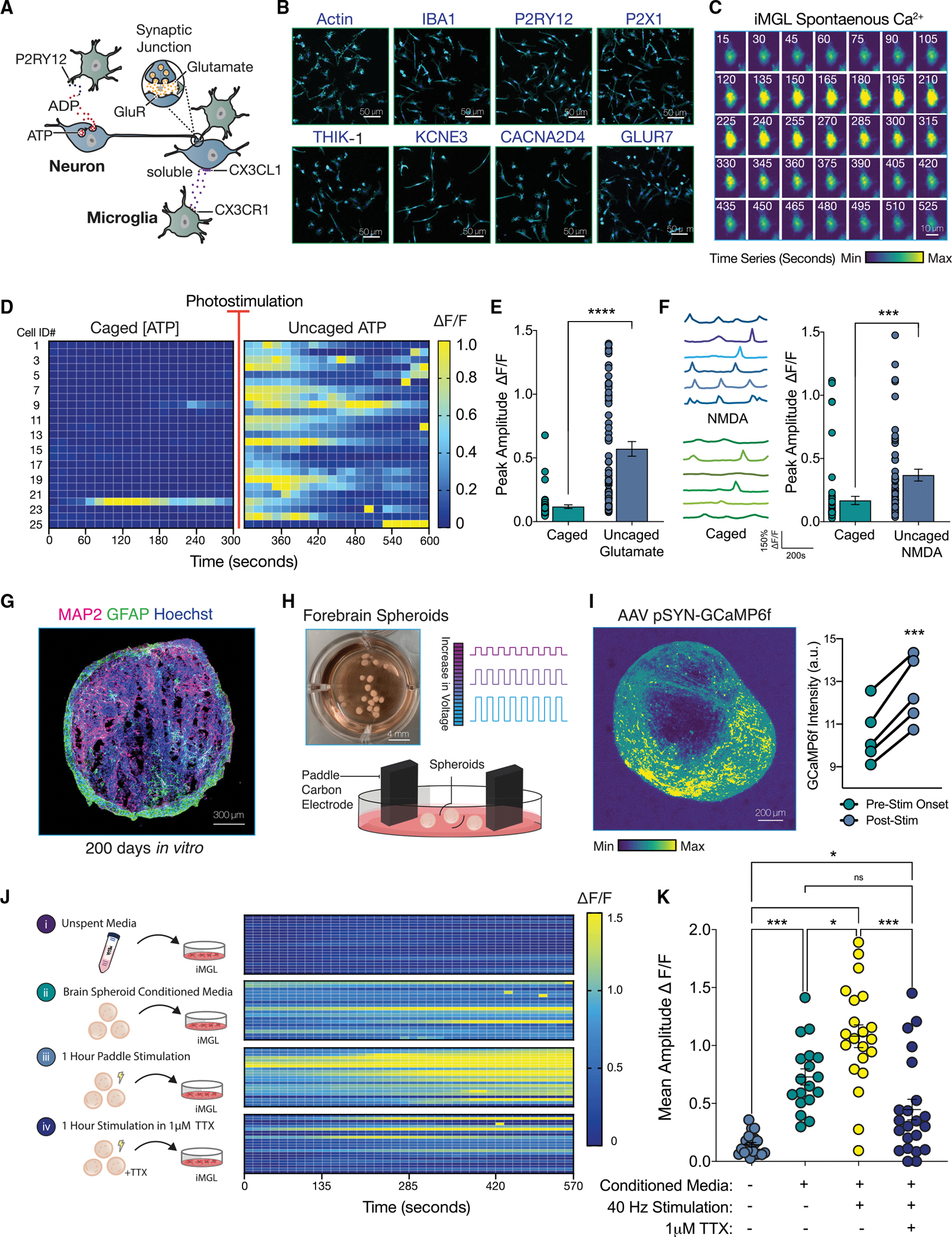
(A) Diagram depicting neuron-microglia communication.
(B) iMGL (line #2191, see Key Resources Table) express canonical microglial receptors.
(C) Time series (15 s) of a spontaneous calcium transient in iMGL with Fluo-4 AM. Relative minimum and maximum amplitudes are shown.
(D) Heatmap of iMGL upon 1 mM ATP uncaging. Change in fluorescence shown as Delta F/F.
(E) Change in fluorescence upon uncaging of 5 mM glutamate plotted as peak amplitude for baseline (caged) and post photostimulation (uncaged). (Paired t test; n = 64 cells.)
(F) Representative calcium traces from 6 iMGLs before and after uncaging of 1 mM NMDA (paired t test; n = 60 cells).
(G) Immunostaining of a forebrain spheroid 200 DIV (days in vitro). Neuronal marker MAP2 (magenta) and astrocyte marker, GFAP (green), nuclei stained with Hoechst (blue).
(H) Forebrain spheroids stimulated using paddle carbon electrodes.
(I) Live-imaging of pSYN-GCaMP6f-infected spheroids before and after electrical stimulation. Representative image of maximum projection across time frames. (Paired t test; n = 5 spheroids.)
(J) Calcium imaging of iMGL in monoculture following 2 h of incubation in spheroid conditioned media (CM).
(K) Peak amplitude of calcium transients from (J) is quantified and plotted (one-way ANOVA with post-hoc Tukey test; n = 18–21 cells per group).
For all statistical tests, n.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Chicken Anti-MAP2 (used at 1:10,000) | BioLegend | Cat#PCK-554P (replaced with Cat#822501) RRID AB_2564858 |
| Guinea Pig Anti-IBA1 (used at 1:500) | Synaptic Systems | Cat#234004 (replaced with 234 308) RRID:AB_2493179 |
| Mouse Anti-GLUR7 (used at 1:200) | Thermo Scientific | Cat#MA5-31743 RRID AB_2787366 |
| Mouse Anti-KCNE3 (used at 1:100) | Santa Cruz Biotechnology | Cat#sc-393841 |
| Mouse Anti-NEUROFILAMENT (used at 1:1000) | BioLegend | Cat#SMI-311R (replaced with #837801) RRID AB_2565383 |
| Mouse Anti-SYNAPTOPHYSIN (used at 1:200) | Milipore Sigma | Cat#S5768 RRID:AB_477523 |
| Mouse Anti-Tubulin β-III (used at 1:2000) | BioLegend | Cat#MMS-435 (replaced with 801213) RRID AB_2728521 |
| Rabbit Anti-CACNA2D4 (used at 1:50) | EpiGentek | Cat#A65974-020 |
| Rabbit Anti-KCNJ9 (GIRK3) (used at 1:100) | Atlas Antibodies | Cat#HPA070478 RRID:AB_2686270 |
| Rabbit Anti-P2RY12 (used at 1:100) | Milipore Sigma | Cat#HPA014518 RRID:AB_2669027 |
| Rabbit Anti-P2X1 (used at 1:100) | Thermo Scientific | Cat#PA5-77662 RRID AB_2736289 |
| Rabbit Anti-THIK-1 (used at 1:100) | Abcam | Cat#ab237624 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| AAV9 hSYN-EGFP | Addgene | Cat#50465-AAV9 |
| AAV1 SYN-GCaMP6f-WPRE-SV40 | Addgene | Cat#100843-AAV1 |
| Lentiviral vector pLKO5-sgRNA-EFS-tRFP | Addgene | Cat#57823 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Accutase | Life Technologies | Cat#A11105-01 |
| ActinGreen ReadyProbes | Invitrogen | Cat#R37110 |
| ATP | Sigma-Aldrich | Cat#A9187 |
| B-27 Supplement | Gibco | Cat#17504-044 |
| BODIPY 493/503 | Cayman Chemical | Cat#25892 |
| BODIPY-Cholesterol | Cayman Chemical | Cat#24618 |
| BrainPhys Neuronal Medium | StemCell Technologies | Cat#05790 |
| BsmBI | New England Biolabs | Cat#R0580S |
| DMEM/F12 | Thermo Fisher Scientific | Cat#11330-057 |
| DMNPE-Caged ATP | Fisher Scientific | Cat#1049 |
| Dulbecco’s PBS | Thermo Fisher Scientific | Cat#14190235 |
| FBS | GEMINI BIO PRODUCTS | Cat#100-106 |
| FLUO4-AM | Invitrogen | Cat# F14201 |
| HBSS | Thermo Fisher Scientific | Cat#14175-095 |
| Hoechst 33342 | Thermo Fisher Scientific | Cat#H3570 |
| IL-34 | PeproTech | Cat#200-34 |
| Isolectin GS-IB4 Alexa Fluor 594 | Invitrogen | Cat#I21413 |
| Matigel hESC-Qualified Matrix | Corning | Cat#354277 |
| MatTek plates | MatTek Corporation | Cat#P06G-0-14-F |
| m-CSF | PeproTech | Cat#300-25 |
| MNI-Caged L-Glutamate | Tocris | Cat#1490 |
| MNI-Caged NMDA | Tocris | Cat#2224 |
| mTeSRI medium | STEMCELL Technologies | Cat#85850 |
| N-2 Supplement | Gibco | Cat#17502-048 |
| Neurobasal media | Gibco | Cat#21103049 |
| Oleic Acid | Sigma-Aldrich | Cat#03008 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#15714-S |
| pHrodo-LDL | Invitrogen | Cat#L34356 |
| Poly-D-Lysine | Sigma-Aldrich | Cat#P6407-10X5MG |
| Ponceau S | CST | Cat#59803 |
| ReLeSR | STEMCELL Technologies | Cat#05872 |
| RIPA lysis buffer | Sigma-Aldrich | Cat#R0278 |
| RNA to cDNA EcoDry Premix | Takara | Cat#639549 |
| ROCK inhibitor | BioVision | Cat#2342-25 |
| SsoFast EvaGreen Supermix | Bio-Rad | Cat#1725202 |
| Tetrodotoxin | Trocris | Cat#1078 |
| Tissue-Tek OTC | Sakura | Cat#4583 |
| Triacsin C | Cayman Chemical | Cat#10007448 |
| Vybrant Alexa Fluor 594 Lipid Raft Labeling Kit | Invitrogen | Cat##V34405 |
| Water-soluble cholesterol | Sigma-Aldrich | Cat#C4951 |
| WesternBright Quantum HRP substrate | Advansta | Cat#K-12042 |
| AR-C 66096 Tetrasodium Salt | Tocris | Cat#3321 |
| (+)-MK 801 Maleate | Tocris | Cat#0924 |
| PPADS Tetrasodium Salt | Tocris | Cat#0625 |
| MPEP Hydrochloride | Tocris | Cat#1212 |
| L-AP4 | Tocris | Cat#0103 |
| YM 58483 (BTP2) | Tocris | Cat#3939 |
| CNQX Disodium Salt | Tocris | Cat#1045 |
| D-AP5 (APV) | Tocris | Cat#0106 |
| PTX (Picrotoxin) | Tocris | Cat#1128 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Pierce BCA Protein Assay Kit | Thermo Fisher Scientific | Cat#23225 |
| Trans Blot Turbo Transfer System | Bio-rad | Cat#1704150EDU |
| Cholesterol Assay Kit | Abcam | Cat#ab65390 |
| Apolipoprotein E Human ELISA kit | Invitrogen | Cat#EHAPOE |
| RNeasy Plus Mini Kit | Qiagen | Cat#74134 |
| NEBNext Ultra II RNA Library Prep Kit | New England Biolabs | Cat#E7770S |
| Quick Ligase Kit | New England Biolabs | Cat#M2200S |
| STEMdiff Hematopoietic Kit | STEMCELL Technologies | Cat#05310 |
|
| ||
| Deposited data | ||
|
| ||
| APOE3 and APOE4 iMGL RNA-seq | NCBI/GEO | GSE203019 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| iPS Line 2191 (87 years old healthy male control) Genotyped in house: APOE3/3 | Reprogrammed in house | |
| CRISPRi iPSC line(dCas9-KRAB) – Genotyped in house and determined to harbor APOE3/3 genotype. | Allen Institute for cell Science | Cat#AICS-0090 cl.391 |
| APOE 3/3 isogenic line (Parental – Unaffected, Coriell #AG09173, female, Age 75) | Lin et al., 2018 | Cat#ADE3/3 |
| APOE 4/4 isogenic line (CRISPR-edited #AG09173) | Lin et al., 2018 | Cat#ADE4/4 |
| sAPOE 4/4 isogenic line (Parental - Sporadic AD Patient, Coriell #AG10788, female, Age 87) | Lin et al., 2018 | Cat#sADE4/4 |
| sAPOE 3/3 isogenic line (CRISPR-edited #AG10788) | Lin et al., 2018 | Cat#sADE3/3 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| NEB Stabl3 competent E. coli | New England Biolabs | Cat#C3040H |
| HEK-293T cell line | ATCC | Cat#CRL-3216 |
|
| ||
| Oligonucleotides | ||
|
| ||
| PCR Primers for amplifying APOE locus in CRISPRi line 5’–3’ (Forward) | This Paper | ATGGACGAGACCATGAAGG |
| PCR Primers for amplifying APOE locus in CRISPRi line 5’–3’ (Reverse). | This Paper | CTGCCCATCTCCTCCATCC |
| Sanger Sequencing primer for determination of APOE genotype 5’–3’ (Forward). | This Paper | GCACGGCTGTCCAAGGAG |
| Sanger Sequencing primer for determination of APOE genotype 5’–3’ (Reverse). | This Paper | CAGCTCCTCGGTGCTCTG |
| KCNJ9 (GIRK3) 5’–3’ sgRNA: 1 | Horlbeck et al., 2016 | GCCCCCACGGGCCCCCCGAA |
| KCNJ9 (GIRK3) 5’–3’ sgRNA: 2 | Horlbeck et al., 2016 | GCACGGGCCCCCCGAAGGGT |
| KCNJ9 (GIRK3) 5’–3’ sgRNA: 3 | Horlbeck et al., 2016 | GTGTAGCGGCAGCTCTGACT |
| For qPCR Primers see Table S4 | This Paper | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pLKO5-sgRNA-EFS-tRFP | (Heckl etal., 2014) | Cat# 57823; Addgene |
| AAV9 hSYN-EGFP | Roth Lab DREADDS (Unpublished) | Cat#50465-AAV9; Addgene |
| AAV1 SYN-GCaMP6f-WPRE-SV40 | (Chen etal., 2013) | Cat#100837-AAV1; Addgene |
|
| ||
| Software and algorithms | ||
|
| ||
| EnrichR Appyter | EnrichR | https://maayanlab.doud/Enrichr/ |
| GraphPad Prism | GraphPad Software | https://www.graphpad.com/ |
| ImageJ cell | https://maayanlab.doud/Enrichr/ | |
| IMARIS | Oxford Instruments | https://imaris.oxinst.com/products/imaris-for-cell-biologists?gclid=EAIaIQobChMIi4qN2JXp9wIVyTizAB3d6QuLEAAYASAAEgLHyvD_BwE_ |
| Linear Stack Alignment with SIFT | ImageJ | https://imagej.net/plugins/linear-stack-alignment-with-sift |
| Neural Metrics Tool | Axion Biosystems | https://www.axionbiosystems.com/products/software/neural-module |
| pCLAMP 11 | Molecular Devices | https://www.moleculardevices.com/products/axon-patch-clamp-system/acquisition-and-analysis-software/pclamp-software-suite#gref |
| ShinyGO 0.76 | Bioinformatics.sdstate | http://bioinformatics.sdstate.edu/go/ |
| SnapGene | SnapGene | https://www.snapgene.com/ |
|
| ||
| Other | ||
|
| ||
| 70 μM strainer | VWR International | Cat #21008-952 |
| AggreWell 800 | STEMCELL technologies | Cat#34815 |
| Automated Cell Counter | Invitrogen | Countess II |
| C-Dish | Ionoptix | Cat#CLD6WFC |
| ChemiDoc MP Imaging System | Bio-Rad | Cat#12003154 |
| C-Pace EM | Ionoptix | Cat#TCI100 |
| Cryostat | Leica | Cat#CM3050s |
| CytoView MEA 48-well plate | Axion BioSystems | Cat#M768-tMEA-48B |
| EnSpire plate reader | Perkin Elmer | Cat#2300-9040 |
| EVOS cell imaging system | Thermo Fisher Scientific | AMF4300 |
| Nitrocellulose membranes | Bio-Rad | Cat#1704159 |
| Real-Time PCR | Bio-Rad | CFX96 Touch Real-Time PCR Detection System |
Next, we examined calcium signaling in iMGLs in response to extracellular cues. We reasoned that non-viral mediated approaches to visualize calcium transients may be better suited to avoid evoking microglial activation. Thus, we labeled monocultures of iMGLs with the membrane-permeant calcium indicator Fluo-4 AM. At baseline, we observed sparse microglial calcium transients (Figure 1C). Because iMGLs have been shown to respond to extracellular ATP (Abud et al., 2017; Konttinen et al., 2019), we next attempted to evoke calcium transients by exposing iMGLs to a biologically inactive analog of ATP that can be photostimulated with ultraviolet light for rapid activation (caged ATP versus uncaged ATP). As expected, ATP uncaging elicited a robust increase in calcium transients (Figures 1D and Video S1). Importantly, photostimulation alone did not elicit a response (Figure S1G). We also determined that iMGLs sensed extracellular ATP with whole-cell patch-clamp (Figure S1F). Similarly to how microglia are known to respond to excitatory neurotransmitters in vivo (Eyo et al., 2014), we found that uncaging NMDA or glutamate increased the amplitude of calcium transients (Figures 1E and 1F). This suggested that iMGLs may sense secreted neuronal soluble factors, such as nucleotides or neurotransmitters.
To test the possibility that iMGLs can respond to a more physiological neuronal stimulus, we next generated forebrain spheroids following previously established protocols (Gordon et al., 2021; Sloan et al., 2018; Yoon et al., 2019). Extended culture times of greater than 120 days yield a small but growing population of GFAP-positive astrocytes among MAP2-positive neurons (Figures 1G and S1H) (Sloan et al., 2017). We exclusively conducted our studies on spheroids between 60 and 90 days to restrict our findings to neuronal factors. Because high levels of neuronal activity is associated with a larger number of microglial calcium transients in vivo (Umpierre et al., 2020), we postulated that stimulating spheroids would yield greater release of neuronal soluble factors. We adapted paddle carbon electrodes built onto a tissue culture multi-well plate typically employed in iPSC-derived cardiomyocytes (Figure 1H) (Ronaldson-Bouchard et al., 2018). Stimulation with paddle electrodes evoked a robust and sustained neuronal response (Figure 1I).
Next, we performed media carry-over experiments in four experimental groups (Figure 1J). Unspent neuronal media evoked no response in iMGLs, while neuronal media conditioned for 24 h with forebrain spheroids significantly increased the number of calcium transients (Figures 1J and 1K). When iMGLs were treated with conditioned media from spheroids that had been stimulated with paddle electrodes, we observed increased calcium transients (Figures 1J and 1K). Importantly, when the stimulation was performed in the presence of tetrodotoxin (TTX), a potent inhibitor of neuronal activity, the increase in iMGL calcium transients was reduced to levels of non-stimulated spheroid media (Figure 1K).
Modeling neuron-microglia communication with CRISPR-edited APOE3 and APOE4 iMGLs
To determine the functional impact of APOE4 on neuron-microglia communication, we applied APOE3 spheroid conditioned media (CM) to monocultures of APOE3 or APOE4 iMGLs and measured their calcium transients (Figure 2A and 2B). Although we did not observe any differences in spontaneous calcium transients, application of neuronal CM evoked fewer calcium transients in APOE4 than APOE3 iGMLs (Figures 2C and 2D). This was not dependent on the APOE status of the spheroids (Figure S2A). To elucidate the signaling pathways that must be impaired in APOE4 iMGLs, we targeted broad classes of channels with co-application of selective antagonists to APOE3 iMGLs in the presence of CM (Figure 2E). We observed a dramatic decrease in calcium transients when iMGLs were treated with a selective antagonist of P2RY12, AR-C 66096 (Figures 2F and Video S2). Blocking other purinergic receptors with the antagonist PPADS (namely P2Xs, P2Y2, and P2Y4 receptors) did not induce similar decreases in calcium transients. In addition, we also observed little to no effect with synaptic depressants of metabotropic glutamate receptors (MPEP and L-AP4), AMPA/Kainate receptors (CNQX), NMDA receptors ([+] MK 801, and APV), GABA A receptors (Pricrotoxin or PTX), or the non-selective K+ channel blocker TEA. Interestingly, blocking store-operated Ca2+ entry (SOCE) with the inhibitor YM 58483 was also sufficient to suppress CM-evoked calcium transients. In addition, APOE4 iMGLs displayed a blunted response compared to APOE3 iMGL controls to ATP uncaging (Figure 2G). iMGLs derived from a distinct donor parental line showed similar responses (Figure 2H). Contrastingly, we observed no deficit in APOE4 iMGLs in response to uncaged glutamate (Figure 2I). This suggests that purinergic signaling might be particularly impaired in APOE4 iMGLs. Given that downregulation of purinergic receptors, namely P2RY12, is associated with microglial activation status, we reasoned that homeostatic surveillance state is shifted in APOE4 iMGLs.
Figure 2. APOE4 iMGL are weakly attuned to neuronal-derived ATP.
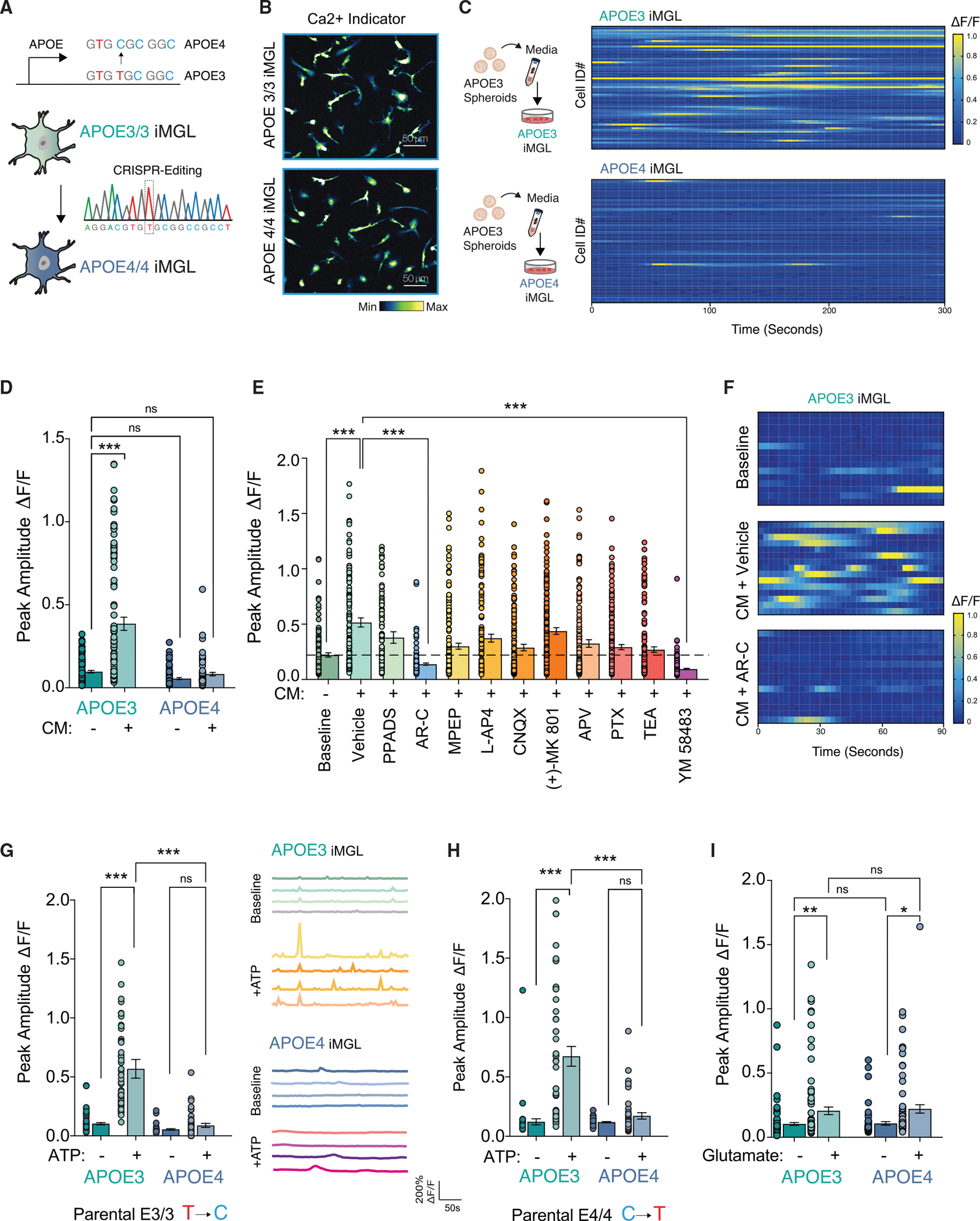
(A) Diagram of CRISPR-generated isogenic APOE3 and APOE4 iPSCs differentiated into iMGLs.
(B) Maximum intensity projection of calcium imaging of APOE3 and APOE4 iMGLs with Fluo-4 AM.
(C and D) Heatmap from isogenic pairs incubated with conditioned media (CM) from APOE3 forebrain spheroids (C), and quantified as peak amplitude change (D) in fluorescent intensity (n = 73–101 cells in each group).
(E) Pharmacological blockade of APOE3 iMGLs reveals CM-evoked calcium transients are mediated via P2RY12 (See Key Resources table for full descriptions of drugs) (n = 112–217 cells in each group).
(F) Representative heat maps from CM-evoked Ca2+ transients and AR-C treatment.
(G and H) ATP uncaging in APOE3 and APOE4 iMGLs in multiple isogenic lines. (G) APOE3 parental line (n = 39–45 cells per group) with representative traces and (H) APOE4 parental line (n = 41–65 cells per group).
(I) Glutamate uncaging (n = 61–101 cells per group).
All statistical testing was performed using ANOVA with post-hoc Tukey test. n.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
Neuronal conditioned media evokes distinct transcriptional responses in APOE3 versus APOE4 iMGLs
To dissect the mechanisms whereby APOE4 reduces microglial surveillance of neuronal activity, we probed the transcriptional profile of these cells at baseline and in response to spheroid conditioned media: APOE3 iMGLs versus APOE4 iMGLs and when exposed to APOE3 spheroid conditioned media (+CM) (Figure 3A). To understand how spheroid CM affects iMGLs, we first performed differential analysis comparing APOE3 iMGL versus APOE3 iMGL + CM. We identified 604 downregulated and 884 upregulated DE-Gs (Figures 3B and Table S1). Gene ontology (GO) analysis of transcripts in APOE3 iMGLs + CM revealed a strong signature of secondary signaling cascades including cAMP signaling (FDR 7.19 × 10−3), Phospholipase D signaling (FDR 9.14 × 10−4), and MAPK signaling (FDR 1.33 × 10−4) (Figure S3D and Table S2). In fact, one of the most highly enriched genes induced by CM was the cAMP response element modulator CREM (APOE3 iMGL versus APOE3 iMGL + CM; FDR 4.08 × 10−67). In contrast, APOE4 iMGL exposure to spheroid CM (APOE4 iMGL versus APOE4 iMGL + CM) evoked a larger transcriptional response with 1,305 down and 1,702 upregulated DEGs (Table S2). APOE4+CM iMGLs were significantly enriched for HIF-1 signaling (FDR 2.26 × 10−2), JAK-STAT signaling (FDR 2.15 × 10−2), and Cytokine-cytokine receptor interaction (FDR 2.59×10−3), suggesting a strong pro-inflammatory response (Figure S3D and Table S2). We observed decreased induction of several targets of intracellular Ca2+ signaling in APOE4 iMGLs + CM in direct comparison to APOE3 iMGLs + CM, congruent with our observation of decreased calcium transients in APOE4 iMGLs (Figure S3F).
Figure 3. APOE4 shifts iMGL into a metabolic distinct cell state that is marked by impaired lipid catabolism.
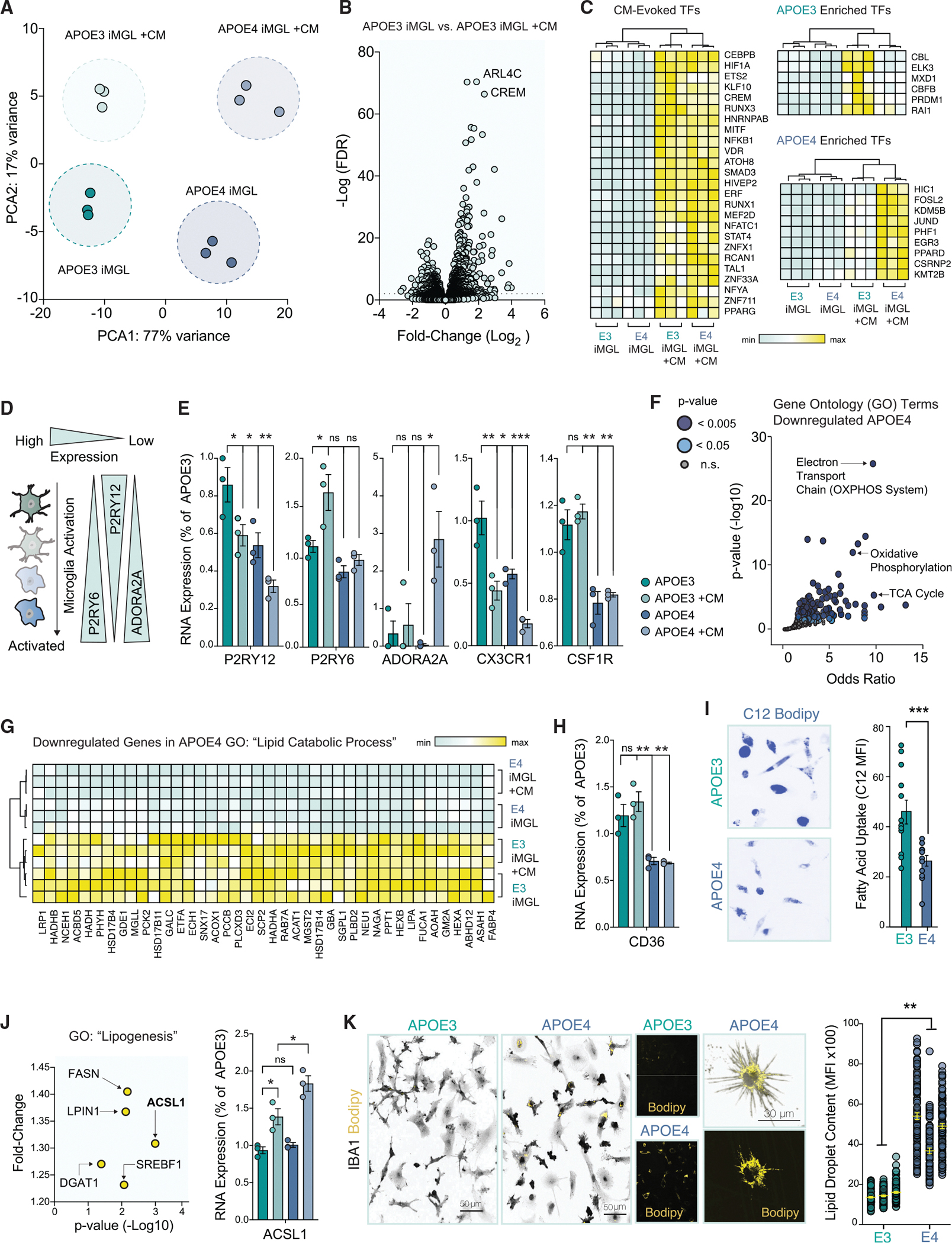
(A) Principal component analysis (PCA) of biological triplicates for four groups: APOE3 iMGL and APOE4 iMGL, with or without APOE3 spheroid conditioned media (CM).
(B) Volcano plot of differentially expressed genes (DEGs) evoked by CM in APOE3 iMGLs. Dotted line indicates FDR cut-off of 0.01.
(C) Heatmap of differentially expressed transcription factors (TFs) evoked by CM. Relative minimum and maximum values per row are shown as a gradient.
(D) Diagram depicting microglial activation-associated changes in purinergic signaling.
(E) Read counts from RNA-seq DEGs normalized to a single biological replicate of APOE3 for P2RY12, P2RY6, ADORA2A, CX3CR1, and CSF1R (ANOVA with post-hoc Tukey test).
(F) Gene ontology (GO) pathways for down-regulated DEGs in APOE4.
(G) Heatmap for DEGs associated with lipid catabolism.
(H) Levels of CD36 shown from normalized read counts from RNA-seq DEGs (ANOVA with post-hoc Tukey test).
(I) DEGs associated with lipogenesis, upregulated in APOE4 +CM. ACSL1 levels across all groups (ANOVA with post-hoc Tukey test).
(J) BODIPY staining (yellow), counterstained with IBA1 (gray) in APOE3 versus APOE4 (unpaired t test, n = 73–107 cells per group in three separate experiments; averages from the three groups were used).
(K) Twenty-four hours of incubation with the green fluorescent fatty acid, C12 Bodipy (shown in blue). Unpaired t test n = 12 separate replicates in each group with 16–33 cells quantified per replicate.
For all statistical tests, n.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
To dissect the regulatory landscape that governs the distinct inflammatory transcriptional response by APOE4 iMGLs, we identified transcription factors (TFs) induced by spheroid CM (Figure 3C). Evoked TFs included several master regulators of inflammation, including NFKB1 and STAT4. We also observed a distinct set of inducible TFs that bifurcated in a genotype-dependent manner on the known role to mitigate or exacerbate inflammatory processes (Figure 3C). In APOE3 iMGLs + CM, we observed the induction of inflammatory repressors: CBL (Lu et al., 2021; Shamim et al., 2007; Zhang et al., 2003), PRDM1 (Keller and Maniatis, 1991), and ELK3 (Chen et al., 2003; Tsoyi et al., 2015). Neuronal inducible-TFs unique to APOE4 iMGLs + CM have been linked with promoting inflammation (e.g., HIC1 and FOSL2) (Burrows et al., 2017; Renoux et al., 2020) or have been induced in macrophages by pro-inflammatory stimuli (e.g. CSRNP2, JUND, and EGR3) (Eichelbaum and Krijgsveld, 2014). Failure to evoke these inducible-TFs suggests that this immune checkpoint is left unchecked in APOE4 iMGLs, perhaps leading to overactivation of downstream immune effectors.
Since our assays indicated deficits in purinergic signaling, we next examined the levels of P2RY12 and found significantly lower levels were expressed in APOE4 iMGLs (Figure 3E). While exposure to spheroid CM significantly decreased levels of P2RY12 in both genotypes, P2RY6 expression levels increased only in APOE3 iMGLs + CM. Upregulation of P2RY6 in microglia is associated with a hypervigilant microglial state (Koizumi et al., 2013). Interestingly, APOE4 iMGLs + CM dramatically increased the expression levels of adenosine receptor A2A (encoded by the gene ADORA2A) (Figure 3E). In the brain, ATP can be hydrolyzed into adenosine, which is a potent activator of microglia (Colella et al., 2018). Moreover, we detected reduction in multiple homeostatic genes, including CX3CR1 and CSF1R (Figure 3E) (Krasemann et al., 2017). While our transcriptional profiling provided molecular insight into the decreased surveillance of APOE4 iMGLs, we next sought to identify the cellular mechanism by which APOE4 genotype shifts microglial status.
Microglial energetics and lipid processing are altered in APOE4 iMGLs
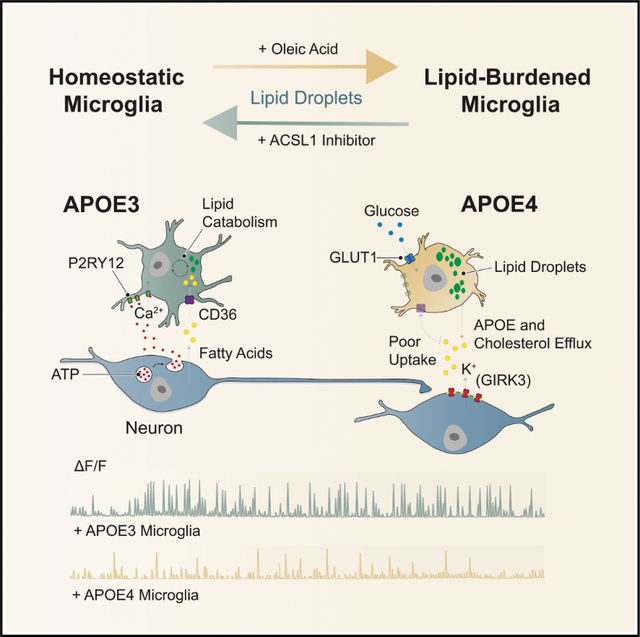
At baseline and upon spheroid CM, we observed a dramatic decrease in DEGs relating to mitochondrial oxidative phosphorylation (OXPHOS) in APOE4 iMGLs (Figures 3F and S3G). Pro-inflammatory stimuli are known to induce a metabolic switch in microglia from OXPHOS to glycolysis, modifying the rate of fatty acid catabolism for the production of ATP by instead relying on the consumption of glucose (Lauro and Limatola, 2020). In alignment with this, we observed a small but significant upregulation of the glucose transporter GLUT3 in APOE4 iMGLs (Log2 fold-change = 0.37, FDR 0.0315), while GLUT1 dramatically increased in expression (Log2 fold-change = 6.01, FDR 1.67 × 10−23) relative to APOE3 iMGLs (Figure S3H). Moreover, through our transcriptional analysis we observed that HIF-1 signaling is enriched in APOE4 iMGLs. HIF-1α is a master transcriptional regulator of glycolysis (Cheng et al., 2014). These results suggest that APOE4 expression in microglia induces a metabolic reprogramming in energy production that is associated with a pro-inflammatory state.
Energy depletion and impairment of fatty acid oxidation has been associated with intracellular lipid accumulation in activated microglia (Loving and Bruce, 2020). Concurrent with downregulation of genes related to OXPHOS, we also observe a dramatic decrease in DEGs related to lipid catabolic processes in APOE4 iMGLs in relation to APOE3 (Figure 3G). Moreover, we also detected a significant downregulation of the membrane fatty-acid transporter CD36 (also known as FAT) in APOE4 iMGLs in comparison to APOE3 iMGLs (Figure 3H). Defective fatty acid uptake has been previously observed in APOE4 astrocytes (Qi et al., 2021), and CD36 has been shown to be downregulated in AD-associated proinflammatory microglia (Dobri et al., 2021). Mirroring our RNA-seq analysis of decreased CD36 expression in APOE4 iMGLs, we observed decreased uptake of the green-fluorescent fatty-acid C12 BODIPY (C12 BODIPY) by APOE4 iMGLs (Figure 3I).
Interestingly, upon exposure to neuronal CM, we further detected the differential upregulation of a subset of genes in APOE4 iMGLs involved in the de novo production of lipids or in the regulation of its cellular storage, with the Acyl-Coa synthethase ACSL1, showing the most robust enrichment (Figure 3J). Upregulation of ACSL1 has been previously reported in the postmortem human brain of APOE4 carriers and to be similarly enriched in iPSC-derived astrocytes harboring the APOE4 allele (Sienski et al., 2021). Of note, snRNA-seq of postmortem human brains from (Mathys et al., 2019) did not show significant enrichment for ACSL1 in microglia of AD subjects, but rather significantly higher expression levels of ACSL5 (Figure S4A).
We next determined the abundance of intracellular lipids in APOE4 iMGLs. Staining for intracellular neutral lipid stores known as lipid droplets with the fluorescent dye BODIPY reveals significantly greater lipid droplet content in APOE4 iMGLs in comparison to APOE3 iMGLs (Figure 3K). Nonetheless, it remains unclear how impairment in purinergic signaling and thus reduced downstream Ca2+ activation impact lipid accumulation in APOE4 iMGLs. Interestingly, we found that blocking P2YR12 signaling significantly increased lipid droplet content in APOE3 iMGLs + CM (Figures S4A and S4B). Calcium signaling downstream of G-protein activation is thought to be an important mechanism of lipid break-down through activation of protein kinase A and C (PKA and PKC), which in turn increases the activity of lipases. Congruent with this body of work, we observed reduced expression of genes that encode the catalytic and regulatory domains of PKA and PKC in APOE4 iMGLs + CM (Figures S4C and S4D). This data suggests that Ca2+ mobilization may be a critical component in the different regulation of lipid storage between APOE3 and APOE4 microglia.
The accumulation of lipid droplets in microglia have been reported to represent a dysfunctional and pro-inflammatory state in the aging brain (Marschallinger et al., 2020). However, the functional repercussions of a lipid burdened microglial state to the activity of neuronal circuits remains unknown.
APOE4 Microglia impair the highly coordinated neuronal activity of APOE3 spheroids
To assess how APOE4 iMGLs impact the activity of neurons, we began by dissociating spheroids grown in 3D after 60 days and plating these cultures onto coverslips. After 4 weeks in culture, we found that dissociated spheroids displayed mature neuronal morphology with extensive neurite networks and were free of GFAP-positive cells (Figure S5A). To track microglia in mixed cultures with neurons, we pre-labeled iMGLs with the microglia-specific dye Isolectin IB4 (Boscia et al., 2013) (Figure 4A). We found that iMGLs persisted in these mixed cultures for at least 4 weeks (Figure 4B).
Figure 4. APOE4 iMGL disrupts the synchronized activity of neuronal ensembles.
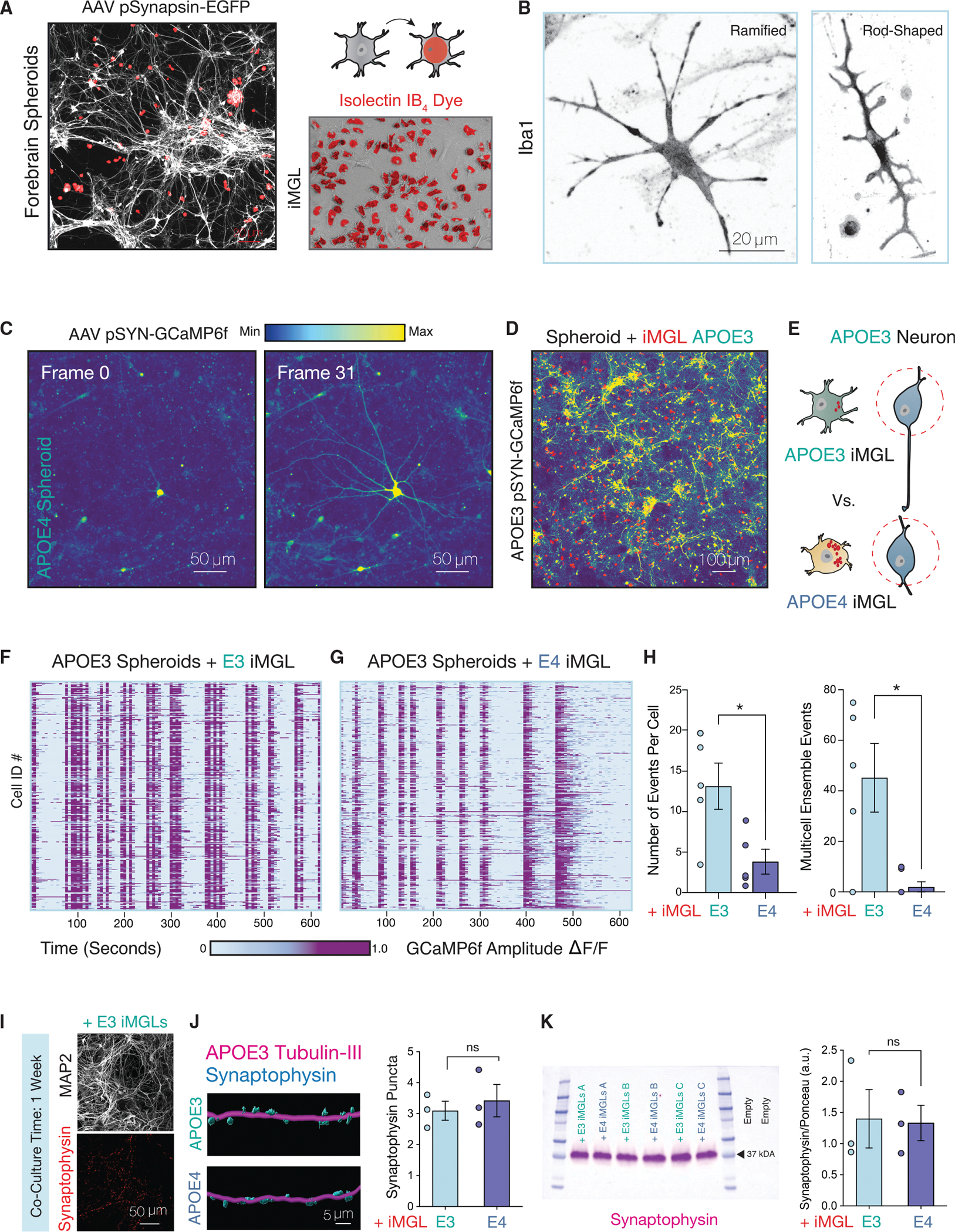
(A) APOE3 spheroids infected with AAV pSYN-EGFP (gray) and iMGL pre-labeled with Alexa Fluor 594 isolectin GS-IB4 conjugate (red).
(B) iMGLs in co-culture with spheroids immunostained with microglia marker IBA1.
(C) Activity of a single neuron from APOE4 spheroid cultures shown over 31 frames of 3 s each. Relative change in fluorescence amplitude is shown as a gradient.
(D) Maximum intensity projection of GCaMP6f spheroids infected with AAV pSYN-GCaMP6f in co-culture with iMGLs.
(E) Diagram depicting experimental approach.
(F) Rasterplots for spontaneous APOE3 neuronal network events in co-culture for 1 week with either APOE3 or (G) APOE4 iMGLs.
(H) Average number of events per cell and number of coordinated calcium transients with greater than 60% co-active cells indicative of ensemble events were quantified (n = 50–106 cells per group for 5–6 distinct experiments).
(I) Co-culture of iMGLs with APOE3 neurons. MAP2-positive neuronal cultures exhibit synaptic puncta as evident by synaptophysin staining.
(J) IMARIS 3D reconstruction of confocal z-stacks from neuron-microglia co-cultures from APOE3 neurons with either APOE3 and APOE4 iMGLs immunostained for neuronal marker Tubulin-III (magenta) and synaptic marker Synaptophysin (blue). Quantification for number of synaptophysin-positive puncta per 100 μm of tubulin-positive neuronal filament (n = 3 biological replicates, three field-of-views per replicate).
(K) Western blot analysis of APOE3 spheroids in co-culture with APOE3 or APOE4 iMGL for 1 week for synaptophysin (n = 3 separate experiments).
All statistical testing was performed using unpaired t test. N.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
Dissociated spheroids from APOE3 or APOE4 were transduced with AAV pSyn-GCaMP6f for calcium dynamics visualization (Figures 4C and S5E). Given the critical role of microglia in sculpting neural dynamics, we sought to determine the impact of APOE4 iMGLs to neuronal network activity (Figures 4D and 4E). By quantifying spontaneous calcium events of APOE3 neurons in co-culture for 1 week with either APOE3 (Figure 4F) or APOE4 iMGLs (Figure 4G), we found that APOE4 iMGLs decreased the overall number of calcium transients in APOE3 spheroid cultures (Figure 4H). Additionally, we observed that APOE4 iMGLs disrupted coordinated neuronal ensemble events in APOE3 neurons (Figure 4H). We did not detect differences in the size or number of synapses via immunostaining (Figures 4I and 4J) or western blotting (Figure 4K). Because we observed changes to neuronal calcium dynamics at a point in which we did not detect robust changes to synapses, we postulated that a non-phagocytic mechanism may mediate the contribution of APOE4 iMGLs to impaired neuronal network dynamics.
Imbalance in the net flux of lipids by APOE4 iMGLs
To investigate if microglial secreted factors impact neuronal activity, we conducted media carry-over experiments from APOE3 or APOE4 iMGL monocultures to APOE3 spheroid cultures labeled with AAV pSyn-GCaMP6f (Figure 5A). We observed a robust decrease in neuronal calcium transients in cultures that were exposed to APOE4 iMGL CM, while cultures treated with APOE3 iMGL CM continued to display highly synchronized calcium transients (Figures 5B and 5C and Video S3). This effect seemed to be independent of the APOE genotype of the neurons (Figures S2C–S2E). We also confirmed these results with neurons seeded onto multielectrode arrays (MEA) (Figures 5E and S6A–C). Interestingly, APOE3 spheroids can partially recover after withdrawal of APOE4 iMGL CM (Figure S6D).
Figure 5. Conditioned Media from APOE4 iMGLs is sufficient to disrupt neuronal activity via potentiation of neuronal lipid-gated K + currents.
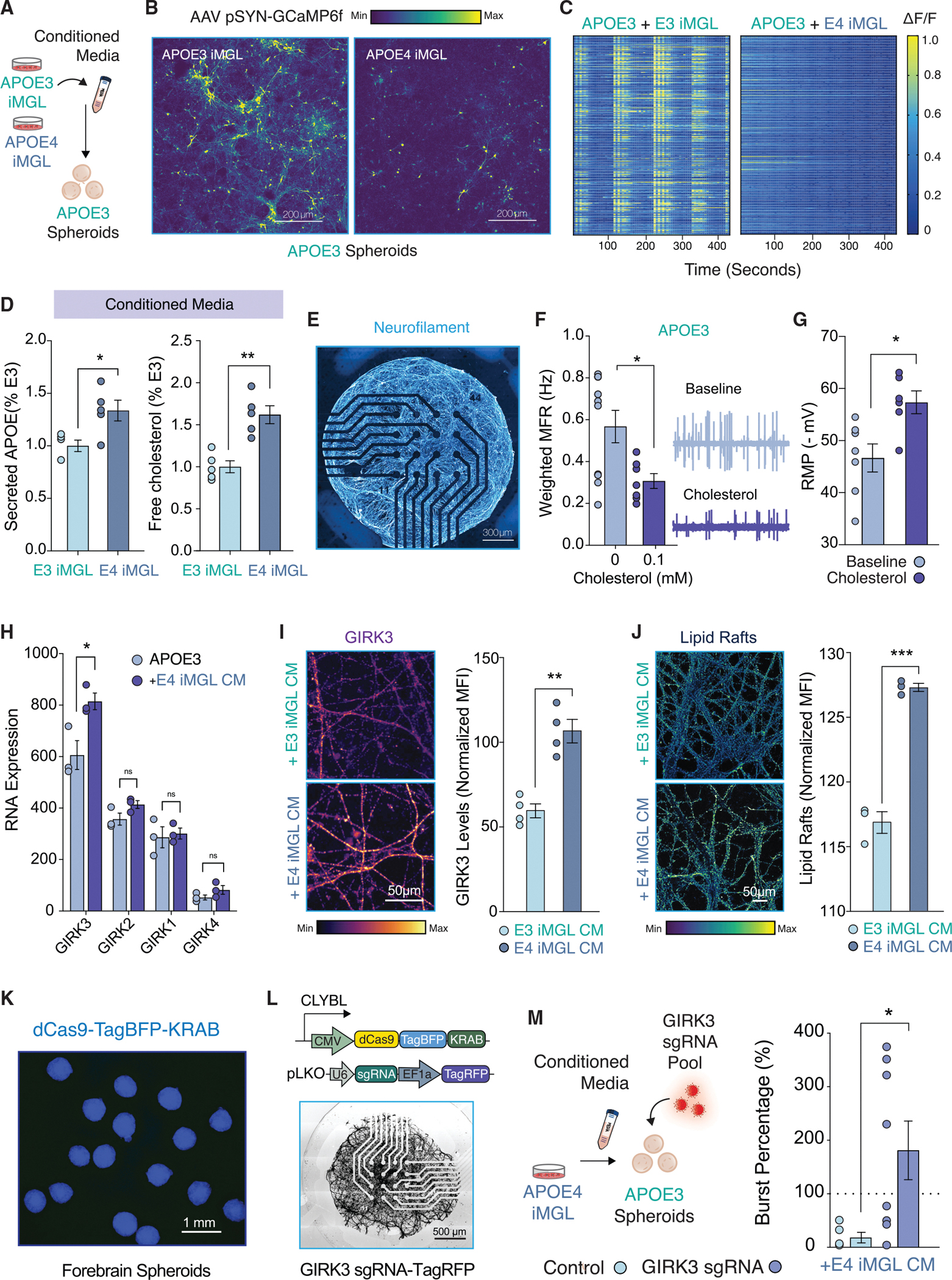
(A) Experimental approach.
(B) Calcium imaging of GCAMP6f-infected spheroid cultures treated with iMGL conditioned media (CM).
(C) Heatmap for changes in GCaMP6f fluorescence for APOE3 or APOE4 iMGL + CM.
(D) ELISA for APOE and Cholesterol (n = 5 biological replicates).
(E) NGN2-induced neurons on a multielectrode array (MEA) stained for neurofilament.
(F) Weighted mean firing rate (MFR) for APOE3 NGN2-neurons in presence of cholesterol. MEA traces for a random 120 s (n = 7–10 wells).
(G) Patch-clamp electrophysiology; hyperpolarization of the resting membrane potential (RMP) with cholesterol treatment (n = 6–7 per group).
(H) RNA expression of lipid-gated inwardly rectifying K+ channel family (GIRK) (n = 3 biological replicates).
(I–J) GIRK3 immunostaining (I) and membrane lipid rafts detection in APOE3 spheroids treated with iMGL CM (J). Mean fluorescence intensity (MFI) normalized for total arbor length in the field of view (n = averages from 3 fields of view from 3 biological replicates).
(K) CRISPRi spheroids.
(L and M) GIRK3 CRISPRi or APOE3 NGN2 neurons infected with a non-targeting sgRNA control were seeded on MEAs and exposed to APOE4 iMGL CM for 48 h. Dotted line represents baseline recording (n = 5–8 biological replicates).
All statistical testing was performed using unpaired t tests and (H) was corrected for multiple comparisons. N.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
As the major transporter of cholesterol in the brain, APOE mediates the delivery of cholesterol and other lipids between neurons and glia (Ioannou et al., 2019). Thus, we decided to examine levels of APOE and cholesterol in the supernatant of iMGL monocultures. We found that the supernatant of APOE4 iMGL cultures were enriched in both APOE and cholesterol (Figure 5D). We repeated this experiment using the fluorescent cholesterol analog BODIPY-cholesterol, which also revealed an increase in cholesterol in the media of APOE4 iMGL (Figure S4D and S4E). Although our transcriptional profiling is in alignment with recent suggestions that APOE4 glia exhibits reduced lipid transport (TCW et al., 2019) (Figure S4F), we reasoned that extracellular lipid accumulation could also be the net product of a relatively greater deficit in lipid influx. To test this idea, we exposed APOE3 or APOE4 iMGLs to low-density lipoprotein (LDL) isolated from human plasma. We observed a dramatic reduction in cellular uptake of LDL by APOE4 iMGLs in comparison to APOE3 controls (Figures S4G–S4I).
Neuronal accumulation of cholesterol-enriched membrane microdomains modify K+ currents
Cholesterol is essential for proper neuronal physiology, and cholesterol depletion is known to impair neurotransmission (Linetti et al., 2010; Liu et al., 2010). On the other hand, increased cholesterol is well-established to enhance amyloidogenesis (Di Paolo and Kim, 2011), and decreasing cholesterol biosynthesis in astrocytes was found to alleviate pathology in AD mouse models (Wang et al., 2021). Because we detect extracellular accumulation of cholesterol in APOE4 iMGLs, we wondered what the impact of exogenous cholesterol treatment would be to these neuronal cultures. Treatment of neurons seeded onto MEAs with cholesterol APOE4 iMGL CM treatment (Figure 5F). We next assessed iPSC-derived neurons by patch-clamp electrophysiology. Recordings demonstrated physiological properties akin to mature neurons in these cultures (Figures S5B and S5C). We found that addition of exogenous cholesterol significantly hyperpolarized the resting membrane potential (RMP) of neurons (Figure 5G). Moreover, we observed a changes in the I-V (current-voltage) curve indicative of greater inwardly rectifying potassium (Kir) currents (Figure S5D). Potentiation of Kir currents is aligned with our observations, as strong inwardly rectifying potassium currents are known to hyperpolarize resting membrane potentials and decrease neuronal excitability (Hodge, 2009).
We decided to isolate mRNA from APOE3 spheroids for bulk RNA-sequencing to define the expression of Kir channels. We detected abundant expression of Kir3.3 channel, which is encoded by the gene KCNJ9 (Figures 5H and S5F). Kir3.3 is a G-protein-gated inwardly rectifying K+ channel (GIRK3) that regulate neuronal excitability similarly to other Kir channels, with gain of function reducing neuronal activity and loss of function increasing neuronal activity (Luscher and Slesinger, 2010). Notably, GIRK channels are known to be lipid-gated (Huang et al., 1998). In neurons, cholesterol enhances its activity and thus decreases neuronal excitability (Bukiya et al., 2017, 2019; Mathiharan et al., 2021). Based on our RNA-seq analysis, application of APOE4 iMGL CM significantly upregulated transcript levels of GIRK3 in APOE3 spheroids (Figure 5H). We next assessed expression of GIRK3 in APOE3 spheroids exposed to either APOE3 or APOE4 iMGL CM by immunostaining. Protein levels of GIRK3 were significantly increased in neurons treated with APOE4 iMGL CM (Figure 5I). Interestingly, GIRK channels are known to localize to cholesterol-rich microdomains at neuronal membranes, often referred to as lipid rafts (Delling et al., 2002). Indeed, we detected an increased prevalence of lipid rafts in neurons treated with APOE4 iMGL CM (Figure 5J).
To determine if the potentiation of GIRK3 is necessary for the suppression of neuronal activity in APOE4 iMGL CM treated neurons, we targeted GIRK3 with CRISPR-interference (CRISPRi) using an iPSC line previously engineered by the Allen Institute (Figure 5K) (see Key Resources Table). Gene repression in iPSC-derived neurons with this CRISPRi vector has been previously reported (Tian et al., 2021). Neurons derived from these cultures were seeded onto MEAs and transduced with a lentiviral vector carrying three distinct sgRNAs targeting KCNJ9 (Figure 5L). Interestingly, we observed that GIRK3 knockdown prevented the repression of neuronal bursts by APOE4 iMGL condition media (Figure 5M). Of note, we did not observe a significant rescue in the number of spikes between GIRK3 knockdown and control (unpaired t-test, p-value = 0.1758), suggesting GIRK currents may be particularly important to regulate neuronal burst firing. Collectively, these results demonstrate that extracellular cholesterol accumulation, at least in part due to poor lipid re-uptake by APOE4 iMGLs, can suppress neuronal activity via potentiation of GIRK currents. We sought to determine the expression levels of GIRK channels in excitatory neurons from AD patients from published snRNA-seq data (Mathys et al., 2019). Indeed, levels of GIRK2 and GIRK3 were significantly enriched in excitatory neurons of AD patients in comparison to no pathology control subjects (Figure S4A).
Modulation of intracellular lipid content can reversibly drive purinergic signaling in microglia
To determine if APOE4-induced lipid accumulation is necessary and sufficient to drive microglia activation status away from homeostatic surveillance, we attempted to bidirectionally modulate lipid content iMGLs. We began by inducing lipid droplet accumulation in APOE3 iMGLs by exposing the cells to the mono-unsaturated fatty acid oleic acid (OA) (Figure 6A). Fatty acid overload is a potent inducer of lipid droplet formation, and as such IBA1-positive cells treated with OA accumulated intracellular BODIPY-positive lipid droplets (Figures 6B and 6C). The rise in lipid droplet content was also linked with a decrease in the cell size (Figure 6D), which resembles ameboid-like morphologies adopted by activated microglia. In fact, levels of the pro-inflammatory MHC-II marker CD74 were upregulated in OA-treated iMGLs (Figure 6E). In addition, OA treatment was sufficient to blunt calcium transients evoked by ATP uncaging relative to untreated APOE3 iMGLs (Figure 6F). A key mechanism mediating lipid storage into intracellular droplets is the activation of fatty acids by the Acyl-Coa synthethase ACSL1 (Ellis et al., 2010; Stremmel et al., 2001). ACSL1 expression has been reported to be modulated by lipogenic conditions (Li et al., 2006), and as such we observed that OA significantly induced the expression of ACSL1 (Figure 6G; see Table S4 for a list of qPCR primers). This is of particular interest because we also uncovered ACSL1 as the most enriched gene governing lipogenesis in APOE4 iMGLs through our RNA-seq analysis. Collectively, these results suggest that increasing lipid accumulation is sufficient to shift microglia away from homeostatic surveillance and phenocopies key aspects of the APOE4 iMGL state.
Figure 6. Bidirectional manipulation of lipid content can reversibly drive purinergic signaling in iMGLs.
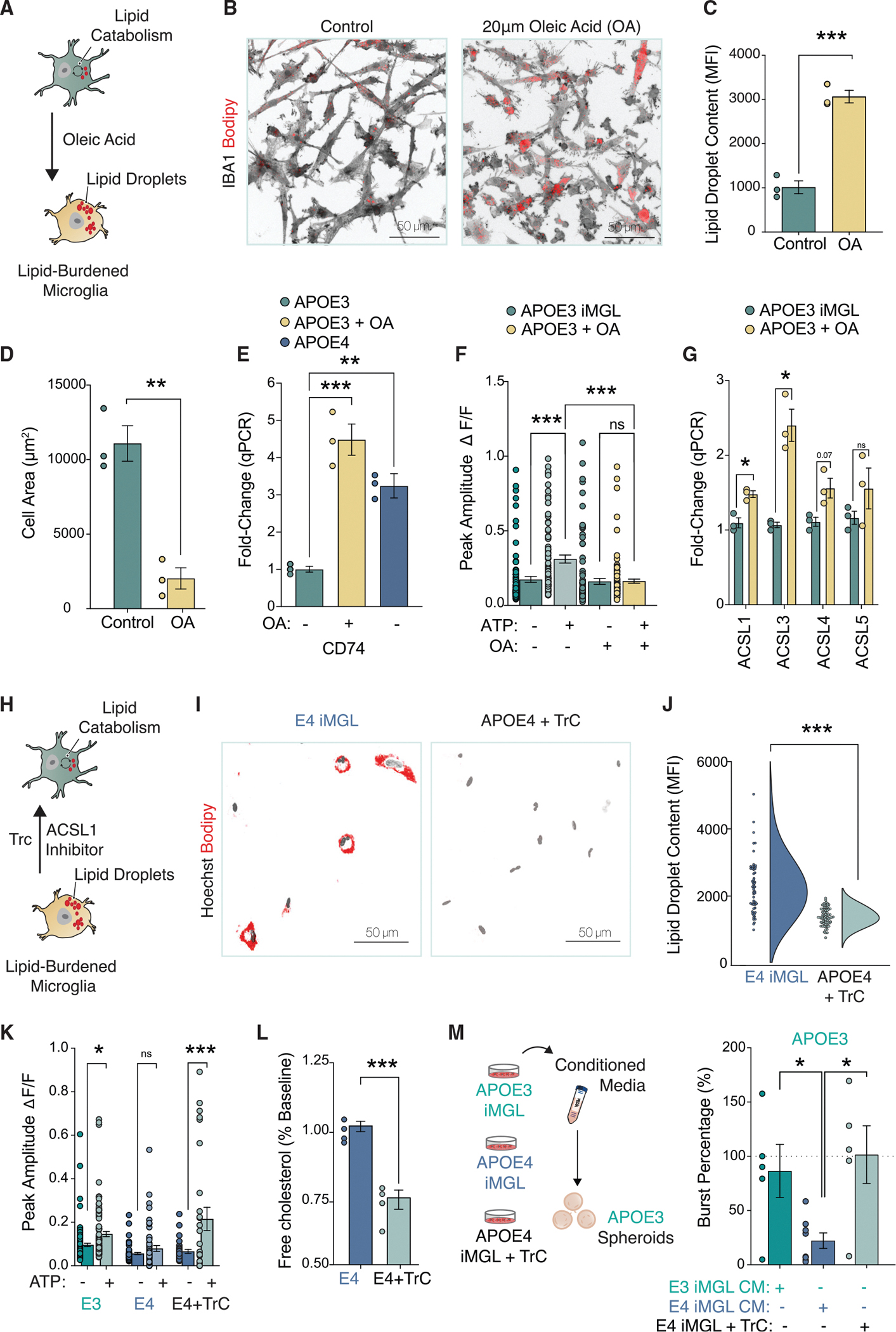
(A) iMGLs activation after 20 μM oleic acid (OA).
(B) BODIPY staining shown in red, IBA1 in gray (n = 3 biological replicates with 20–30 cells per replicate). MFI = mean fluorescence intensity.
(C and D) Lipid droplet accumulation and changes to cell size following OA (n = 3 biological replicates, 20–30 cells per replicate).
(E) qPCR analysis for CD74 (n = 3 biological replicates). Data normalized to GAPDH and shown relative to APOE3 control.
(F) OA blunts iMGL response to ATP (n = 85–110 cells quantified per group). Experiment repeated three times.
(G) qPCR for long-chain-fatty-acid-CoA ligases (ASCL1 – 5) in IMGLs.
(H–J) Treatment of iMGLs with 1 μM of Triacsin C depletes lipid droplets.
(K) 1 mM ATP uncaging upon Triacsin C treatment. n = 29–52 cells per group.
(L) Triacsin C reduces extracellular cholesterol accumulation via ELISA. n = 4 biological replicates.
(M) MEA recording conducted 48 h post CM incubation onto APOE3 neurons. n = 5–8 biological replicates.
Statistical testing performed using unpaired t test (C, D, J, and L).
All others used ANOVA with post-hoc Tukey test. N.s. = not significant; *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001; ****p-value < 0.0001. Error bars reflect SEM.
Having induced lipid formation in APOE3 iMGLs, we next attempted to deplete APOE4 iMGLs of lipid droplets to test whether this would alleviate APOE4 phenotypes. We turned to the ACSL1 inhibitor Triacsin C (TrC), a pharmacological intervention that has been shown to prevent lipid accumulation (Marschallinger et al., 2020; Qi et al., 2021) (Figure 6H). Treatment of APOE4 iMGLs with TrC was sufficient to dramatically reduce BODIPY-positive lipid droplets relative to DMSO-treated control cells (Figures 6I and 6J). Furthermore, purinergic signaling was restored after lipid droplet depletion in APOE4 IMGLs (Figure 6K). We observed a significant decrease in the levels of accumulated cholesterol in the media via ELISA, in line with previous reports that TrC can inhibit cholesterol biosynthesis (Igal et al., 1997; Marschallinger et al., 2020) (Figure 6L). Of note, we did not detect a similar reduction in the accumulated levels of APOE by TrC treatment (Figure S4J). Nevertheless, we reasoned that a decrease in extracellular cholesterol accumulation in APOE4 iMGLs treated with TrC was likely to also relieve the suppression of neuronal activity. While APOE4 iMGL CM decreased neuronal bursts in APOE3 neurons, as we previously observed, neural activity remained intact with pre-treatment of APOE4 iMGLs with TrC (Figure 6M). These results establish that the maintenance of lipid homeostasis in microglia sustains surveillance homeostatic states required to support proper neuronal network function.
DISCUSSION
Neuronal excitability and circuit dysfunction
In our studies, we observed that APOE4 microglia exhibit reduced capacity to uptake lipids, resulting in the net accumulation of lipids extracellularly, which we found to be detrimental to neuronal activity. Recently, it has been reported that co-culture of LPS activated microglia or exposure to its CM impaired the mitochondrial function of neurons (Park et al., 2020). Neuronal mitochondrial metabolism is critical for activity-dependent calcium buffering, synaptic transmission, and the generation of action potentials (Harris et al., 2012). Although our findings might be related to neurons exposed to LPS-activated microglia, it remains to be determined how lipids secreted by APOE4 iMGL impact neuronal mitochondrial function.
Although astrocytes are the primary cellular buffers of extracellular lipids and are thought to prevent the accumulation of toxic free fatty acids that are generated from neurons in an activity-dependent manner, microglia are also critical players in the regulation of lipid transport and consumption (Ioannou et al., 2019). In the context of APOE, which is predominantly expressed by astrocytes and microglia, APOE4-disruption of lipid homeostasis across both cell types is likely a double-hit on the buffering capacity of lipids within the brain, rendering neurons more susceptible to lipotoxicity. Mechanisms that decrease excitability (such as the potentiation of GIRKs) and thus the production of fatty acids to fuel the rise in metabolism associated with increased firing rates, may be neuroprotective but at the detriment of neural computations critical for learning and memory.
Searching for a common lipid-burdened transcriptional signature
In the mouse hippocampus, aging microglia have been shown to accumulate lipid droplets, a cellular state that has been termed lipid-droplet accumulating microglia (LDAM) (Marschallinger et al., 2020). Phenotypically, LDAM is marked by reduced phagocytosis and increased secretion of pro-inflammatory cytokines, which is similar to the cellular state we have previously characterized in APOE4 bearing iPSC-derived microglia (Lin et al., 2018). Contrary to our findings with human APOE4 iMGLs, mouse LDAMs have been reported to exhibit increased mitochondrial fatty acid oxidation. Through gene set activity analysis, we see poor convergence of transcriptional signatures between human APOE4 iMGL and mouse LDAM (Figure S3I).
Cholesterol homeostasis
Cellular pathways that govern the regulation of lipid and cholesterol homeostasis have emerged as a central node in the pathogenesis of AD. Similarly to our findings in microglia, we have previously reported that in astrocytes derived from APOE4 iPSCs cholesterol accumulates extracellularly (Lin et al., 2018). Although the mechanism by which cholesterol and other lipids may accumulate extracellularly in APOE4 glia is not clear, astrocytes activated with pro-inflammatory stimuli have recently been reported to secrete saturated lipids contained within APOE and APOJ lipoproteins that are toxic to neurons (Guttenplan et al., 2021). Moreover, dysfunctional lipid metabolism in astrocytes have been well-established in mice harboring the human APOE4 allele (Farmer et al., 2019). Evidence exists that microglia also secrete higher levels of APOE under pro-inflammatory conditions (Lanfranco et al., 2021). Because the metabolic profile of microglia is associated with its pro-inflammatory state, therapies aimed at reprogramming microglial metabolism may prove to be imperative in curbing inflammation and halting neurodegeneration in AD.
Limitations of the study
Lipid homeostasis is a tightly regulated process that involves many cell types, particularly through lipid exchange between cells (Ioannou et al., 2019). Thus, our study omitted key producers and consumers of lipids, such as astrocytes, which are also critically involved in modulating neuronal activity (Chung et al., 2015). In addition, because our analysis was performed in microglia that did not face pathology, such as Amyloid-β or Tau, it remains unclear to what degree the lipid-burdened transcriptional signatures of APOE4 microglia would change in a more complex model with multiple cell types and in the presence of pathology.
STAR★METHODS
RESOURCE AVAILABLITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Li-Huei Tsai (lhtsai@mit.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
RNA-seq data has been deposited at NCBI/GEO #GSE203019.
This paper does not report original code.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Culture of iPSC lines
All human iPSCs were maintained at 37°C and 5% CO2, in feeder-free conditions in mTeSR1 medium (Cat #85850; STEMCELL Technologies) on Matrigel-coated plates (Cat # 354277; Corning; hESC-Qualified Matrix). iPSCs were passaged at 60–80% confluence using ReLeSR (Cat# 05872; STEMCELL Technologies) and reseeded 1:6 onto Matrigel-coated plates. APOE isogenic lines were derived from a 75-year-old female (AGO9173) with an APOE3/3 genotype edited to harbor APOE4/4. A second distinct APOE isogenic line was derived from a 70-year-old female sporadic AD patient (AG10788) with APOE4/4 genotype (sADE4/4) and CRISPR-edited to APOE3/3 (sADE3/3). The iPSC lines were generated by the Picower Institute for Learning and Memory iPSC Facility as first described (Lin et al., 2018). CRISPRi iPSCs were acquired via the Allen Institute for Cell Science https://www.allencell.org and maintained similarly. APOE genotype for the CRISPRi line was determined by amplifying the APOE locus with PCR primers Forward: 5’- ATGGACGAGACCATGAAGG -3’ Reverse: 5’-CTGCCCATCTCCTCCATCC -3’ followed by Sanger sequencing with primer Forward: 5’- GCACGGCTGTCCAAGGAG-3’ and Reverse: 5’CAGCTCCTCGGTGCTCTG-3’.
METHOD DETAILS
Spheroid induction protocol
Dorsal forebrain spheroids were generated using a previously established protocol (Sloan et al., 2018) with an adapted iPS seeding strategy (Marton et al., 2019). Briefly, 60–80% confluent iPSCs were dissociated into a single-cell suspension after incubation in ReLeSR (Cat# 05872; STEMCELL Technologies) for three minutes at room temperature, followed by a dry incubation at 37°C for five more minutes. iPSC colonies were then scraped in mTeSR1 medium (Cat # 85850; STEMCELL Technologies) and dissociated into a single-cell suspension by mechanical pipetting. The cell suspension was centrifuged at 300 g for 5 min, resuspended in 1 mL of mTeSR1 medium supplemented with ROCK inhibitor, Rockout at 1:1000 (Cat# 2342–5; BioVision) and counted with an automated cell counter (Countess II; Invitrogen). 3 × 106 cells were then plated onto AggreWell 800 microwells (Cat# 34815; STEMCELL Technologies) for embryoid body induction. After 48 h, embryoid bodies were moved onto non-tissue culture treated Petri dishes for neural induction following forebrain spheroid dissociation protocol.
Spheroid dissociation and 2D plating
After growing in suspension for at least 60 days, spheroids were dissociated into a single cell suspension for plating onto coverslips to generate 2D cultures. Adapting a previously described protocol, spheroids were incubated in StemPro Accutase (Cat# A11105–01; Life Technologies) for 30 min at 37°C. Following Accutase aspiration, spheroids were mechanically dissociated by pipetting in 10% FBS (Cat# 100–106; Gemini Bio-Products) in Hank’s Balanced Salt Solution containing (HBSS) (Cat#14175-095; Thermo Scientific). The cell suspension was centrifuged at 300 × g and washed in warm Neurobasal media (Cat# 21103049; Gibco) supplemented with B-27 (Cat#17504-044; Gibco) and N-2 (Cat#17502–048; Gibco). This was then passed through a 70 μM strainer (Cat # 21008–952; VWR International) and plated in 24-well Poly-D-Lysine (Cat#P6407-10X5MG; Sigma-Aldrich) coated No. 0 glass coverslips in 6-well MatTek plates (Cat#P06G-0-14-F; MatTek) at a ratio of 1 spheroid per 3 wells. Cells were allowed to recover for one month before experiments in neuronal media, half-feeding every 3–4 days.
Microglia induction protocol
Embryoid bodies (EBs) were generated using the same protocol as the spheroid induction protocol and seeded onto Matrigel-coated 6-well tissue culture plates at a density of 15–30 EBs per well. EBs were first differentiated into hematopoietic progenitor cells (HPCs) using the STEMdiff Hematopoietic Kit (Cat#05310; STEMCELL Technologies). Following a previously established protocol, non-adherent HPCs were collected, centrifuged at 300 × g, and resuspended in 1 mL of microglia differentiation media (MDM) containing a mixed composition of half DMEM/F12 (Cat#11330–057; Thermo Fisher Scientific) and half Neurobasal media (Cat# 21103049; Gibco) supplemented with IL-34 (Cat#200–34; PeproTech) and m-CSF (Cat#300–25; PeproTech) (McQuade et al., 2018). Cells were plated in 6-well tissue culture plates at 200,000 cells per well and maintained in MDM for at least two weeks before experiments.
Microglia-neuron co-cultures
Dissociated neuronal cultures were switched from Neurobasal Media to BrainPhys Neuronal Medium (Cat#05790; STEMCELL Technologies) 1 month after dissociation and before iMGL seeding. Neurons were infected with 12 μL of AAV9 hSYN-EGFP (Cat#50465-AAV9; Addgene) at titer ≥7×1012 vg/mL or 12 μL AAV1 SYN-GCaMP6f-WPRE-SV40 (Cat#100837-AAV1; Addgene) at titer ≥1×1013 vg/mL per 24 wells. BrainPhys Neuronal Medium was reduced to 300 μL overnight during transduction. The following day, fresh media was added to reach a final culture volume of 500 μL. Upon harvesting and adding iMGLs, in suspension, to neuronal cultures, BrainPhys was supplemented with m-CSF (Cat#300–25; PeproTech). Co-cultures were half-fed every 3–4 days and were ready to be used for experiments after a minimum of 1 week.
Calcium imaging
Live-imagining was performed with Zeiss LSM900 equipped with a heated chamber kept at 37°C with humidity and CO2 control. Fluo-4 AM (Cat#F14201; Thermo Fisher Scientific) was applied to iMGLs in monoculture at a final concentration of 1 μM in DMSO and incubated at 37°C for 30–45 min. Images were acquired at 488 nm, and compound uncaging was done at 405 nm for 30 s post-baseline acquisition, followed immediately by post-stimulation image acquisition. For uncaging experiments, cells were pre-incubated in 1 mM DMNPE-Caged ATP (Cat# 1049; Fisher Scientific), 1 mM MNI-Caged NMDA (Cat#2224; Tocris) or 1 mM MNI-Caged L-Glutamate (Cat#1490; Tocris). Images were first stabilized to account for drift in the x-y direction; we used the ImageJ plugin “Linear Stack Alignment with SIFT”. Calcium traces from motion-corrected time series were manually segmented on ImageJ into individual cells based on threshold intensity, variance, and upper and lower limits for cell size. Image segmentation results were separately inspected for quality control. Fluorescence signal time series (ΔF/F: change in fluorescence divided by baseline fluorescence) were calculated for each segment. The baseline fluorescence for each cell was determined as the minimum fluorescence signal in the baseline recording epoch. For GCaMP-tagged neurons (Cat#100843-AAV1; Addgene), The onset of a calcium transient occurred when ΔF/F exceeded two standard deviations above the baseline fluorescence. The termination of a calcium transient was identified as occurring when ΔF/F fell below 0.5 standard deviations above the baseline fluorescence. To test for changes in functional connectivity between cells in the presence of APOE3 versus APOE4 microglia, we quantified multicellular ensembles. A multicellular ensemble event occurred when the number of simultaneously active cells exceeded 60% of all cells. Data was generated from distinct cultures recorded in separate experiments and plotted as averages per group for neuronal calcium dynamics. In contrast, for iMGLs, data was plotted from individual cells in one experiment, although the experiments were repeated at least three times. In experiments were iMGLs were exposed to spheroid conditioned media (CM), cells were pre-incubated in unspent neuronal media to acclimate to media condition for at least 24 h. Then, iMGLs were half-fed with CM for 2 h before recording session. We found that this was the length of time necessary to avoid confounding elevated calcium transients due to media manipulation. Controls were half-fed unspent media. For media carry-over experiments, and to avoid carrying-over TTX to the microglia (Related to Figure 1), stimulation was followed by three washes in warm media and a complete media switch free of TTX that was allowed to condition for 24 h. Non TTX-treated stimulated controls were handled identically to control for drug washout manipulation. Heatmaps were generated using GraphPad Prism (GraphPad Software).
Staining with non-antibody probes
Staining with non-antibody probes was performed following the manufacturer’s guidelines. ActinGreen ReadyProbes (Cat#R37110; Invitrogen) was used to visualize morphology, FLUO4-AM (Cat# F14201; Invitrogen) for microglia calcium imaging, BODIPY 493/503 (Cat#25892; Cayman Chemical) for lipid droplet staining, Isolectin GS-IB4 Alexa Fluor 594(Cat# I21413; Invitrogen) to label microglia before co-culture and Vybrant Alexa Fluor 594 Lipid Raft Labeling Kit (Cat# V34405; Invitrogen) for lipid raft staining. Microscopy was performed using a Zeiss LSM880 confocal system, and fluorescent Z stack images were quantified using IMARIS (Oxford Instruments).
Electrophysiology
Whole-cell patch-clamp recordings of neurons were performed at 6 to 8 weeks after spheroid dissociation and 2D plating or for iMGLs after 2 to 4 weeks of iMGL differentiation. Intracellular recordings were performed at room temperature using an Axon CV-7B headstage, Multiclamp 700B amplifier, and Digidata 1440A digitizer (Molecular Devices). Electrode pipettes were pulled from borosilicate glass on a Model P-97 Flaming/Brown micropipette puller (Sutter Instrument) and ranged between 4–7 MΩ resistance. Intrinsic neuronal properties were studied using the following solutions (in mM): Extracellular: 125 NaCl, 2.5 KCl, 1.2 NaH2PO42H2O, 1.2 MgCl26H2O, 2.4 CaCl22H2O, 26 NaHCO3, 11 glucose (pH 7.4). Intracellular: 135 K-gluconate, 5 KCl, 2 MgCl26H2O, 10 HEPES, 2 Mg-ATP, 0.2 Na2GTP (pH 7.2). Membrane potentials were typically kept between − 50 mV to −70 mV depending on whether neurons or iMGLs were recorded. In voltage-clamp mode, currents were recorded with voltage steps ranging from −160 mV to +80 mV. In current-clamp mode, action potentials were elicited by injection step currents from −50 pA to +50 pA. For experiments aimed at determining the impact of cholesterol on neuronal properties, one mM water-soluble cholesterol (Cat#C4951; Sigma-Aldrich) was supplemented with the external solution. ATP-evoked currents were recorded by local application of 100 μM (Cat#A9187; Sigma-Aldrich). Data was first collected and analyzed using pCLAMP 11 software (Molecular Devices). Further analysis was done in GraphPad Prism (GraphPad Software).
Spheroid electrical stimulation
Culture pacing system, C-Pace EM, (Cat#TCI100; Ionoptix) equipped with a 6-well carbon electrode dish, C-Dish (Cat#CLD6WFC; Ionoptix) was used to deliver electrostimulation to spheroids at 12V with a biphasic pulse train frequency of 40 Hz. Paddle carbon electrodes were scrubbed clean with ethanol following the manufacturer’s recommended procedure and allowed to fully dry before use. To prevent the effect of media hydrolysis, washes and full media switches immediately followed stimulation.
MEA
Dissociated spheroids or NGN2-induced neurons were plated as a 10 μL droplet in Poly-D-Lysine (Cat#P6407–10X5MG; Sigma-Aldrich) coated wells of a CytoView MEA 48-well plate (Cat#M768-tMEA-48B; Axion BioSystems). Typically, 50,000–75,000 cells were plated per well that each contained 16 low-impedance PEDOT electrodes 50 μm in diameter and arranged at a pitch of 350 μm. Intact spheroids were plated and covered in a Matrigel droplet (Cat # 354277; Corning) to anchor the spheroid. After 15–30 min in 37°C, droplets were flooded with warm BrainPhys Medium (Cat#05790; STEMCELL Technologies) and allowed to recover for at least four weeks before recording sessions. A recording session preceded the media treatment for conditioned media carry-over experiments, denoted as baseline recording. iMGL media was added to compose half of the final volume of the well and allowed to incubate for 24 h before a second recording was performed. All extracellular recordings were performed using the Axion Maestro Pro MEA system (Axion Biosystems). Spontaneous neural activity was recorded for 30 min at a sampling rate of 12.5 kHz, and an adaptive threshold set at 5.5 times the standard deviation of baseline noise was used for spike detection. Bursts were detected at each electrode using an inter-spike interval (ISI) threshold set to at least five spikes with a maximum ISI of 100 ms. Electrodes were defined as active if neuronal firing occurred at a minimal rate of 5 spikes/min. For MEA data analysis, only wells containing a minimum of 3 active electrodes were included. Neuronal firing metrics were exported as the averages from each well from Axion Biosystems’ Neural Metrics Tool and plotted with Prism GraphPad (GraphPad Software).
Drug treatment
To block voltage-gated sodium channels, 1 μM Tetrodotoxin (Cat#1078; Tocris) was applied to the media before electrostimulation of spheroids. Control cells were treated with DMSO as vehicle. The following drugs and final concentrations were applied for pharmacological blockade on iMGLs exposed to spheroid conditioned media: 100 μM AR-C 66096 Tetrasodium Salt (Cat#3321; Tocris), 200 μM (+)-MK 801 Maleate (Cat#0924; Tocris), 200 μM PPADS Tetrasodium Salt (Cat#0625; Tocris), 200 μM MPEP Hydrochloride (Cat#1212; Tocris), 50 μM L-AP4 (Cat#0103; Tocris), 10 μM YM 58483 (BTP2) (Cat#3939; Tocris), 10 μM CNQX Disodium Salt (Cat#1045; Tocris), 50 μM D-AP5 (APV) (Cat#0106; Tocris), 100 μM PTX (Picrotoxin) (Cat#1128; Tocris). To induce lipid accumulation, 20 μM Oleic Acid (Cat# 03008; Sigma-Aldrich) was applied overnight to iMGLs, control cells were treated with 0.1% BSA as vehicle. 1 μM Triacsin C (Cat#10007448; Cayman Chemical) was applied to iMGLs overnight to deplete lipid accumulation.
Western blot
Spheroids transplanted with APOE3 or APOE4 iMGLs for 10 days were washed once with cold 1x PBS and homogenized in RIPA lysis buffer (Cat#R0278; Sigma-Aldrich) containing Halt protease/phosphatase inhibitor cocktail (Cat#78440; Thermo Fisher Scientific) and EDTA. Supernatants were collected after centrifugation at 14,500 RPM for 15 min at four °C and stored at −80°C for later use. Total protein levels were quantified using the Pierce BCA Protein Assay Kit (Cat#23225; Thermo Fisher Scientific;), and five mg of protein were loaded from each sample per lane onto precast 4–20% polyacrylamide gels (Cat#4561094; Bio-Rad). Denatured/reduced samples were run at 150V for 75 min. Proteins were transferred from the gel to 0.2 μm nitrocellulose membranes (Cat#1704159; Bio-Rad) using the Trans-Blot Turbo Transfer System (Cat#1704150EDU; Bio-Rad) set to the mixed molecular weight program. Membranes were stained with Ponceau S (Cat#59803; CST) and subsequently blocked with 5% non-fat milk in 1x TBST (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.05% Tween 20) for 1 h before incubating with primary antibody. Membranes were incubated with rabbit anti-synaptophysin (Cat#S5768; Milipore Sigma) overnight at four °C, and the secondary antibody was later incubated at room temperature for 2 h. Wash buffer was 1x TBST. Proteins were detected by Western Bright Quantum HRP substrate (Cat#K-12042; Advansta) and visualized using the ChemiDoc MP Imaging System (Cat#12003154; Bio-Rad). Western blot densitometry was conducted using ImageJ. Synaptophysin levels were normalized to Ponceau S.
ELISA
Cholesterol levels from iMGLs in monoculture were measured using the Cholesterol Assay Kit (Cat#ab65390; Abcam), following the manufacturer’s instructions for fluorometric detection. Cells were grown in 6-well plates, and samples were either assayed immediately or frozen at −80°C Celsius and thawed once for cholesterol measurements. To obtain total cholesterol levels, cholesterol esterase was added to samples. For free cholesterol measurements, samples were used directly without adding the enzyme. Measurements were made using an EnSpire plate reader (Cat#2300–9040; Perkin Elmer). APOE levels were similarly processed and quantified from iMGL conditioned media using the Apolipoprotein E Human ELISA kit (Cat#EHAPOE; Invitrogen).
Lipid cellular assays
The fluorescently tagged cholesterol analogue, BODIPY-Cholesterol (Cholesterol with BODIPY at carbon-24 of the side chain) (Cat#24618; Cayman Chemical), was used to assay the extracellular accumulation of cholesterol in monocultures of APOE3 or APOE4 iMGLs. Cells were incubated with BODIPY-Cholesterol for 48 h to saturate cellular uptake, washed three times and further incubated for an additional 24 h before media was collected, centrifuged at 300 g for 5 min and assayed for fluorescence at 488 nm with an EnSpire plate reader (Cat#2300–9040; Perkin Elmer). Low-Density Lipoprotein (LDL) from human plasma complexed to pHrodo red (pHrodo-LDL) (Cat#L34356; Invitrogen) was used to determine LDL uptake in APOE3 and APOE4 iMGLs. Monocultures were treated with 5 μg/mL of pHrodo-LDL and incubated for 1 h before live cells were imaged with an EVOS cell imaging system (Cat# AMF4300; Thermo Fisher Scientific). Images were processed in IMARIS (Oxford Instruments) to reconstruct cellular boundaries and quantify intracellular content as mean fluorescence intensity.
RNA analysis of iMGLs and spheroids
RNA extraction from biological replicates (n = 3) for the isogenic pair (APOE3 and APOE4) of iMGLs exposed to spheroid conditioned media or unspent neuronal media was achieved with RNeasy Plus Mini Kit (Cat# Cat#74134; Qiagen). RNA integrity number (RIN) scores were above 9 before library preparation. MIT BioMicro Center prepared libraries using the NEBNext Ultra II RNA Library Prep Kit (Cat#E7770S; New England Biolabs) and performed 75 bases single-end run NextSeq 500 Illumina sequencing. FASTQ reads were aligned using STAR to GRCh37 reference genome (GENCODE 19)(Dobin et al., 2013). Transcripts were quantified using HTSeq, data was normalized utilizing RUV-seq and differential gene expression analysis was performed through DESeq2 as previously described (Meharena et al., 2022). Significant differentially expressed genes (DEGs) were called with an FDR < 0.05 with unrestricted log2 fold-change cut-offs. Gene ontology analysis was performed using ShinyGO 0.76 and EnrichR Appyter. Gene set activity scores related to the LDAM signatures (Supplementary Table T2–1 (Marschallinger et al., 2020), p < 0.05) were computed on the iPSC RNA-sequencing normalized counts matrix, as previously implemented in the R package GSVA (Hanzelmann et al., 2013). RNA extraction for qPCR analysis was performed similarly, and reverse transcription was performed with RNA to cDNA EcoDry Premix (Cat# Cat#639549; Takara) according to the manufacturer’s instruction. Gene expression was analyzed with Real-Time PCR (CFX96; Bio-Rad) and SsoFast EvaGreen Supermix (Cat#1725202; Bio-Rad). Expression data was normalized to housekeeping gene GAPDH using the 2−ΔΔCT relative quantification method. A list of qPCR primers used in this study can be found in Table S4.
Plasmids, cloning and lentivirus production
To target KCNJ9 (GIRK3) in CRISPRi iPS-derived neurons, three distinct gene target sequences were picked from the sgRNA/gene published library for CRISPRi (Horlbeck et al., 2016). Protospacer sequences (note that 5′G is added independent if it exists in the genome): sgRNA 1: 5′-GCCCCCACGGGCCCCCCGAA-3′, sgRNA 2: 5′-GCACGGGCCCCCCGAAGGGT-3′, sgRNA 3: 5’ -GT GTAGCGGCAGCTCTGACT-3′. 5ug of the lentiviral vector pLKO5-sgRNA-EFS-tRFP (Cat# 57,823; Addgene) was linearized with BsmBI (Cat#R0580S New England Biolabs) and oligos annealed with adapters to the sense 5’ -CACC and antisense 5′-AAAC were ligated using Quick Ligase Kit (Cat#M2200S; New England Biolabs). The ligated product was transformed into stabl3 competent E. coli (Cat#C3040H; New England Biolabs) and screened for insert via Sanger sequencing) and aligned to plasmid (SnapGene software). Clones were then transfected into HEK-293T cell line (Cat#CRL-3216; ATCC) with lentiviral packaging and envelope vectors to generate lentivirus following the previously published protocol (Victor et al., 2014). Lentiviral supernatant was collected 48 h after transfection and centrifuged for 2 h by ultracentrifugation at 25,000 RPM at 4°C. Pellets were resuspended in DPBS and frozen in 80at until used. All three sgRNA viruses were mixed equally to deliver a pool ttotalling12ul per well of a 48-well MEA plate. Virus was top-loaded and allowed to incubate overnight in neurons that had been seeded within the past 48 h. Within 5–7 days, tRFP (turbo Red Fluorescent Protein) expression was visible, and by three weeks, the vast majority of cells in the culture expressed high levels of RFP.
QUANTIFICATION AND STATISTICAL ANALYSES
Statistical analyses were performed in GraphPad Prism using a two-tailed Student’s t test or a one-way ANOVA followed by a post hoc Tukey’s test with *p < 0.05 considered significant. Multiple comparisons were corrected with the Dunnett method as described in the figure legends. Studies were performed blindly and automated whenever possible with IMARIS or ImageJ cell, and multiple investigators confirmed quantification results. Data in graphs are expressed as mean, and error bars represent the standard error of the mean (SEM). Outliers were detected and excluded with Grubbs’ test for alpha levels of 0.05. In the entirety of this study, only MEA data exhibited high variability and, therefore, had data points excluded based on this criterion. All experiments reporting values of single cells instead of population averages were performed at least three times.
Supplementary Material
Highlights.
Microglia-like cells (iMGLs) respond to soluble factors secreted by neurons
APOE4 renders iMGLs weakly responsive to neuronal activity
APOE4 iMGLs disrupts the coordinated activity of neuronal ensembles
Microglial lipid homeostasis is critical to sustain surveillance states
ACKNOWLEDGMENTS
We thank Y.T. Lin and T. Ko for experimental assistance with iPSCs; Y. Zhou, H. Cam, M. Mazzanti, and T. Garvey for administrative support. In addition to P. Narayan, L. Akay and all Tsai laboratory members for helpful discussions. Our work in the Tsai lab is only possible through the generous support of The Robert A. and Renee E. Belfer Family Foundation, Carol and Eugene Ludwig Family Foundation, the Cure Alzheimer’s Fund, The JPB Foundation, Joseph P. DiSabato and Nancy E. Sakamoto, Donald A. and Glenda G. Mattes, Lester A. Gimpelson, The Halis Family Foundation, The Dolby Family, David Emmes, and Alan and Susan Patricof. This work was supported by NIH grants R01-AG058002 and RF1-AG062377 to L.-H.T. M.B.V. is supported by the Howard Hughes Medical Institute Hanna H. Gray Postdoctoral Fellowship.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in science. One or more of the authors of this paper self-identifies as a member of the LGBTQ+ community. One or more of the authors of this paper received support from a program designed to increase minority representation in science. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at 10.1016/j.stem.2022.07.005.
REFERENCES
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, et al. (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron 94, 278–293.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badimon A, Strasburger HJ, Ayata P, Chen X, Nair A, Ikegami A, Hwang P, Chan AT, Graves SM, Uweru JO, et al. (2020). Negative feedback control of neuronal activity by microglia. Nature 586, 417–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscia F, Esposito CL, Casamassa A, de Franciscis V, Annunziato L, and Cerchia L (2013). The isolectin IB4 binds RET receptor tyrosine kinase in microglia. J. Neurochem. 126, 428–436. [DOI] [PubMed] [Google Scholar]
- Bukiya AN, Blank PS, and Rosenhouse-Dantsker A (2019). Cholesterol intake and statin use regulate neuronal G protein-gated inwardly rectifying potassium channels. J. Lipid Res. 60, 19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukiya AN, Durdagi S, Noskov S, and Rosenhouse-Dantsker A (2017). Cholesterol up-regulates neuronal G protein-gated inwardly rectifying potassium (GIRK) channel activity in the hippocampus. J. Biol. Chem. 292, 6135–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows K, Antignano F, Bramhall M, Chenery A, Scheer S, Korinek V, Underhill TM, and Zaph C (2017). The transcriptional repressor HIC1 regulates intestinal immune homeostasis. Mucosal Immunol. 10, 1518–1528. [DOI] [PubMed] [Google Scholar]
- Calovi S, Mut-Arbona P, and Sperlá gh B (2019). Microglia and the purinergic signaling system. Neuroscience 405, 137–147. [DOI] [PubMed] [Google Scholar]
- Cheadle L, Rivera SA, Phelps JS, Ennis KA, Stevens B, Burkly LC, Lee WCA, and Greenberg ME (2020). Sensory experience engages microglia to shape neural connectivity through a non-phagocytic mechanism. Neuron 108, 451–468.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Layne MD, Chung SW, Ejima K, Baron RM, Yet SF, and Perrella MA (2003). Elk-3 is a transcriptional repressor of nitric-oxide synthase 2. J. Biol. Chem. 278, 39572–39577. [DOI] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JHA, Rao NA, Aghajanirefah A, et al. (2014). mTOR- and HIF-1alpha-mediated aerobic glycolysis as metabolic basis for trained immunity. Science 345, 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WS, Allen NJ, and Eroglu C (2015). Astrocytes control synapse formation, function, and elimination. Cold Spring Harb. Perspect. Biol. 7, a020370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella M, Zinni M, Pansiot J, Cassanello M, Mairesse J, Ramenghi L, and Baud O (2018). Modulation of microglial activation by adenosine A2a receptor in animal models of perinatal brain injury. Front. Neurol. 9, 605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cserép C, Pósfai B, Lénárt N, Fekete R, László ZI, Lele Z, Orsolits B, Molnár G, Heindl S, Schwarcz AD, et al. (2020). Microglia monitor and protect neuronal function through specialized somatic purinergic junctions. Science 367, 528–537. [DOI] [PubMed] [Google Scholar]
- Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, Littman DR, Dustin ML, and Gan WB (2005). ATP mediates rapid microglial response to local brain injury in vivo. Nat. Neurosci. 8, 752–758. [DOI] [PubMed] [Google Scholar]
- Delling M, Wischmeyer E, Dityatev A, Sytnyk V, Veh RW, Karschin A, and Schachner M (2002). The neural cell adhesion molecule regulates cell-surface delivery of G-protein-activated inwardly rectifying potassium channels via lipid rafts. J. Neurosci. 22, 7154–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, and Kim TW (2011). Linking lipids to Alzheimer’s disease: cholesterol and beyond. Nat. Rev. Neurosci. 12, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobri AM, Dudău M, Enciu AM, and Hinescu ME (2021). CD36 in Alzheimer’s disease: an overview of molecular mechanisms and therapeutic targeting. Neuroscience 453, 301–311. [DOI] [PubMed] [Google Scholar]
- Eichelbaum K, and Krijgsveld J (2014). Rapid temporal dynamics of transcription, protein synthesis, and secretion during macrophage activation. Mol. Cell. Proteomics 13, 792–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Li LO, Wu PC, Koves TR, Ilkayeva O, Stevens RD, Watkins SM, Muoio DM, and Coleman RA (2010). Adipose acyl-CoA synthetase-1 directs fatty acids toward beta-oxidation and is required for cold thermogenesis. Cell Metab. 12, 53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, and Wu LJ (2014). Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J. Neurosci. 34, 10528–10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer BC, Kluemper J, and Johnson LA (2019). Apolipoprotein E4 alters astrocyte fatty acid metabolism and lipid droplet formation. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finneran DJ, and Nash KR (2019). Neuroinflammation and fractalkine signaling in Alzheimer’s disease. J. Neuroinflammation 16, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon A, Yoon SJ, Tran SS, Makinson CD, Park JY, Andersen J, Valencia AM, Horvath S, Xiao X, Huguenard JR, et al. (2021). Long-term maturation of human cortical organoids matches key early postnatal transitions. Nat. Neurosci. 24, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan KA, Weigel MK, Prakash P, Wijewardhane PR, Hasel P, Rufen-Blanchette U, Münch AE, Blum JA, Fine J, Neal MC, et al. (2021). Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature 599, 102–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, and Guinney J (2013). GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 14, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JJ, Jolivet R, and Attwell D (2012). Synaptic energy use and supply. Neuron 75, 762–777. [DOI] [PubMed] [Google Scholar]
- Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, and Ebert BL (2014). Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nature biotechnology 32, 941–946. 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge JJL (2009). Ion channels to inactivate neurons in Drosophila. Front. Mol. Neurosci. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, and Weissman JS (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. Elife 5, e19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CL, Feng S, and Hilgemann DW (1998). Direct activation of inward rectifier potassium channels by PIP2 and its stabilization by Gbetagamma. Nature 391, 803–806. [DOI] [PubMed] [Google Scholar]
- Igal RA, Wang P, and Coleman RA (1997). Triacsin C blocks de novo synthesis of glycerolipids and cholesterol esters but not recycling of fatty acid into phospholipid: evidence for functionally separate pools of acyl-CoA. Biochem. J. 324 (Pt 2), 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannou MS, Jackson J, Sheu SH, Chang CL, Weigel AV, Liu H, Pasolli HA, Xu CS, Pang S, Matthies D, et al. (2019). Neuron-astrocyte metabolic coupling protects against activity-induced fatty acid toxicity. Cell 177, 1522–1535.e14. [DOI] [PubMed] [Google Scholar]
- Izquierdo P, Shiina H, Hirunpattarasilp C, Gillis G, and Attwell D (2021). Synapse development is regulated by microglial THIK-1 K(+) channels. Proc. Natl. Acad. Sci. USA. 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCW J, Liang SA, Lu Q, Pipalia NH, Chao MJ, Shi Y, Bertelsen SE, Kapoor M, Marcora E, Sikora E, Holtzman DM, et al. (2019). Cholesterol and matrisome pathways dysregulated in human APOE ε4 glia. Preprint at bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller AD, and Maniatis T (1991). Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 5, 868–879. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Ohsawa K, Inoue K, and Kohsaka S (2013). Purinergic receptors in microglia: functional modal shifts of microglia mediated by P2 and P1 receptors. Glia 61, 47–54. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, Joshi BV, Jacobson KA, Kohsaka S, and Inoue K (2007). UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature 446, 1091–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konttinen H, Cabral-da-Silva MEC, Ohtonen S, Wojciechowski S, Shakirzyanova A, Caligola S, Giugno R, Ishchenko Y, Hernández D, Fazaludeen MF, et al. (2019). PSEN1DeltaE9, APPswe, and APOE4 confer disparate phenotypes in human iPSC-derived microglia. Stem Cell Rep. 13, 669–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O’Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, et al. (2019). Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat. Genet. 51, 414–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco MF, Sepulveda J, Kopetsky G, and Rebeck GW (2021). Expression and secretion of apoE isoforms in astrocytes and microglia during inflammation. Glia 69, 1478–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro C, and Limatola C (2020). Metabolic reprograming of microglia in the regulation of the innate inflammatory response. Front. Immunol. 11, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li LO, Mashek DG, An J, Doughman SD, Newgard CB, and Coleman RA (2006). Overexpression of rat long chain acyl-coa synthetase 1 alters fatty acid metabolism in rat primary hepatocytes. J. Biol. Chem. 281, 37246–37255. [DOI] [PubMed] [Google Scholar]
- Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, Cam HP, Gjoneska E, Raja WK, Cheng J, et al. (2018). APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human ipsc-derived brain cell types. Neuron 98, 1141–1154.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linetti A, Fratangeli A, Taverna E, Valnegri P, Francolini M, Cappello V, Matteoli M, Passafaro M, and Rosa P (2010). Cholesterol reduction impairs exocytosis of synaptic vesicles. J. Cell Sci. 123, 595–605. [DOI] [PubMed] [Google Scholar]
- Liu CC, Liu CC, Kanekiyo T, Xu H, and Bu G (2013). Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat. Rev. Neurol. 9, 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Trotter J, Zhang J, Peters MM, Cheng H, Bao J, Han X, Weeber EJ, and Bu G (2010). Neuronal LRP1 knockout in adult mice leads to impaired brain lipid metabolism and progressive, age-dependent synapse loss and neurodegeneration. J. Neurosci. 30, 17068–17078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving BA, and Bruce KD (2020). Lipid and lipoprotein metabolism in microglia. Front. Physiol. 11, 393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Chen L, Mansour AG, Yu MJ, Brooks N, Teng KY, Li Z, Zhang J, Barr T, Yu J, and Caligiuri MA (2021). Cbl-b is upregulated and plays a negative role in activated human NK cells. J. Immunol. 206, 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüscher C, and Slesinger PA (2010). Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat. Rev. Neurosci. 11, 301–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Kyrargyri V, Arancibia-Cárcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial ramification, surveillance, and interleukin-1beta release are regulated by the two-pore domain K(+) channel THIK-1. Neuron 97, 299–312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschallinger J, Iram T, Zardeneta M, Lee SE, Lehallier B, Haney MS, Pluvinage JV, Mathur V, Hahn O, Morgens DW, et al. (2020). Lipid-droplet-accumulating microglia represent a dysfunctional and proinflammatory state in the aging brain. Nat. Neurosci. 23, 194–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, Huguenard JR, and Pașca SP (2019). Differentiation and maturation of oligodendrocytes in human three-dimensional neural cultures. Nat. Neurosci. 22, 484–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiharan YK, Glaaser IW, Zhao Y, Robertson MJ, Skiniotis G, and Slesinger PA (2021). Structural insights into GIRK2 channel modulation by cholesterol and PIP2. Cell Rep. 36, 109619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. (2019). Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade A, Coburn M, Tu CH, Hasselmann J, Davtyan H, and Blurton-Jones M (2018). Development and validation of a simplified method to generate human microglia from pluripotent stem cells. Mol. Neurodegener. 13, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meharena HS, Marco A, Dileep V, Lockshin ER, Akatsu GY, Mullahoo J, Watson LA, Ko T, Guerin LN, Abdurrob F, et al. (2022). Down-syndrome-induced senescence disrupts the nuclear architecture of neural progenitors. Cell Stem Cell 29, 116–130.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini M, Rafalski VA, Ma K, Kim KY, Bushong EA, Rios Coronado PE, Yan Z, Mendiola AS, Sozmen EG, Ryu JK, et al. (2021). Microglial Gi-dependent dynamics regulate brain network hyperexcitability. Nat. Neurosci. 24, 19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monif M, Reid CA, Powell KL, Smart ML, and Williams DA (2009). The P2X7 receptor drives microglial activation and proliferation: a trophic role for P2X7R pore. J. Neurosci. 29, 3781–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park GH, Noh H, Shao Z, Ni P, Qin Y, Liu D, Beaudreault CP, Park JS, Abani CP, Park JM, et al. (2020). Activated microglia cause metabolic disruptions in developmental cortical interneurons that persist in interneurons from individuals with schizophrenia. Nat. Neurosci. 23, 1352–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi G, Mi Y, Shi X, Gu H, Brinton RD, and Yin F (2021). ApoE4 Impairs Neuron-Astrocyte Coupling of Fatty Acid Metabolism. Cell Rep. 34, 108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renoux F, Stellato M, Haftmann C, Vogetseder A, Huang R, Subramaniam A, Becker MO, Blyszczuk P, Becher B, Distler JHW, et al. (2020). The AP1 Transcription Factor Fosl2 Promotes Systemic Autoimmunity and Inflammation by Repressing Treg Development. Cell Rep. 31, 107826. [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, and Vunjak-Novakovic G (2018). Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 556, 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamim M, Nanjappa SG, Singh A, Plisch EH, LeBlanc SE, Walent J, Svaren J, Seroogy C, and Suresh M (2007). Cbl-b regulates antigen-induced TCR down-regulation and IFN-gamma production by effector CD8 T cells without affecting functional avidity. J. Immunol. 179, 7233–7243. [DOI] [PubMed] [Google Scholar]
- Sienski G, Narayan P, Bonner JM, Kory N, Boland S, Arczewska AA, Ralvenius WT, Akay L, Lockshin E, He L, et al. (2021). APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 13, eaaz4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Andersen J, Pașca AM, Birey F, and Pașca SP (2018). Generation and assembly of human brain region-specific three-dimensional cultures. Nat. Protoc. 13, 2062–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, and Paşca SP (2017). Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 95, 779–790.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremmel W, Pohl L, Ring A, and Herrmann T (2001). A new concept of cellular uptake and intracellular trafficking of long-chain fatty acids. Lipids 36, 981–989. [DOI] [PubMed] [Google Scholar]
- Szepesi Z, Manouchehrian O, Bachiller S, and Deierborg T (2018). Bidirectional Microglia-Neuron Communication in Health and Disease. Front. Cell. Neurosci. 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian R, Abarientos A, Hong J, Hashemi SH, Yan R, Dräger N, Leng K, Nalls MA, Singleton AB, Xu K, et al. (2021). Genome-wide CRISPRi/a screens in human neurons link lysosomal failure to ferroptosis. Nat. Neurosci. 24, 1020–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsoyi K, Geldart AM, Christou H, Liu X, Chung SW, and Perrella MA (2015). Elk-3 is a KLF4-regulated gene that modulates the phagocytosis of bacteria by macrophages. J. Leukoc. Biol. 97, 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umpierre AD, Bystrom LL, Ying Y, Liu YU, Worrell G, and Wu LJ (2020). Microglial calcium signaling is attuned to neuronal activity in awake mice. Elife 9, e56502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor MB, Richner M, Hermanstyne TO, Ransdell JL, Sobieski C, Deng PY, Klyachko VA, Nerbonne JM, and Yoo AS (2014). Generation of human striatal neurons by microRNA-dependent direct conversion of fibroblasts. Neuron 84, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Kulas JA, Wang C, Holtzman DM, Ferris HA, and Hansen SB (2021). Regulation of beta-amyloid production in neurons by astrocyte-derived cholesterol. Proc. Natl. Acad. Sci. USA. 118. e2102191118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, and Huang Y (2006). Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J. Neurosci. 26, 4985–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Zhao N, Caulfield TR, Liu CC, and Bu G (2019). Apolipoprotein E and Alzheimer disease: pathobiology and targeting strategies. Nat. Rev. Neurol. 15, 501–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SJ, Elahi LS, Pașca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, et al. (2019). Reliability of human cortical organoid generation. Nat. Methods 16, 75–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalocusky KA, Najm R, Taubes AL, Hao Y, Yoon SY, Koutsodendris N, Nelson MR, Rao A, Bennett DA, Bant J, et al. (2021). Neuronal ApoE upregulates MHC-I expression to drive selective neurodegeneration in Alzheimer’s disease. Nat. Neurosci. 24, 786–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Shao Y, Fang D, Huang J, Jeon MS, and Liu YC (2003). Negative regulation of T cell antigen receptor-mediated Crk-L-C3G signaling and cell adhesion by Cbl-b. J. Biol. Chem. 278, 23978–23983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
RNA-seq data has been deposited at NCBI/GEO #GSE203019.
This paper does not report original code.


