Summary
Background
Homologous recombination repair (HRR) is the main mechanism of repair of DNA double-strand breaks. Its deficiency (HRD) is a common feature of epithelial ovarian cancers (EOCs). BRCA1/2 mutations and/or other aberrations in genes of HRR are well known causes of HRD and genomic instability. Poly ADP-ribose polymerase inhibitors (PARPi) have revolutionized the management of BRCA mutant EOCs and demonstrated activity in HRD tumor cells. Determining HRD status can provide informations on the magnitude of benefit for PARPi therapy. Myriad MyChoice CDx is a next generation sequencing- based in vitro diagnostic test that assesses the Genomic Instability Score (GIS) which is an algorithmic measurement of loss of heterozygosity, telomeric allelic imbalance, and large-scale state transitions using DNA isolated from formalin-fixed paraffin embedded tumor tissue specimens. However Myriad MyChoice CDx, is a centrally performed and costly assay, with no reimbursement scheduled, at least in Italy.
Methods
In this report, we described our experience in performing the HRD Focus AmoyDx (Amoy Diagnostics Ltd, Xiamen, Fujian, China) on the same samples of EOCs evaluated with Myriad MyChoiceCDx assay.
Results
The overall percent agreement between AmoyDx and Myriad was 87.8% (65 of 74 tumors tested). All the 36 AmoyDx negative cases were confirmed to be negative by Myriad (negative predictive value, 100%).
Conclusions
The concordance of the results with the gold standard Myriad MyChoice CDx assay suggest the feasibility and reliability of HRD testing in diagnostic laboratories with high-throughput NGS platforms and qualified personnel.
Key words: ovarian cancer, homologous recombination deficiency, genomic scar, HRD
Introduction
Ovarian cancer represents the 3% of all cancers occurring in female and is the sixth cause of cancer-related death in women worldwide, globally accounting for 294,000 new cases and 198,000 deaths per year 1 and 5000 new cases and more than 3000 deaths per year in Italy 2. The high grade serous ovarian carcinoma (HGSOC) is the most frequent (about 70%) and lethal ovarian cancer histotype. The large majority of HGSOC are diagnosed in advanced stage (FIGO stage III-IV) and despite cytoreductive surgery associated with platinum-based chemotherapy, will relapse within two years 3. The introduction of PARP inhibitors (PARPi) in first-line therapeutic regimens of women with platinum-sensitive ovarian cancers has dramatically changed clinical outcomes, both in terms of progression free and overall survival 4-7. Except for BRCA1/2 mutated cancers, which present the higher magnitude of clinical benefit for PARPi, this class of drugs show great efficacy also in BRCA wild type tumors with homologous recombination repair deficiency (HRD) 6-8. Clinically meaningful improvements reported in recent trials 6,7 have lead to the approval of PARPis alone or in combination with antiangiogenetic therapy for the maintenance treatment of patients with HRD-positive advanced ovarian cancer (Food and Drug Administration 9 and European Medicines Agency 10 in 2020, and Agenzia Italiana del Farmaco 11 in 2022). As recently reported in European expert consensus recommendations 12, BRCA1/2 tumor assessment should be associated with the evaluation of homologous recombination repair (HRR) status, as a pivotal step to extend effective PARPi treatment to the largest number of patients, considering that about 20-25% of HGSOCs harbor BRCA1/2 alterations and more than 50% are characterized by HRD 13.
The challenging topic is how to evaluate HRD in routine clinical practice. In the clinical trials PAOLA1 7, PRIMA 6 and VELIA 8, HRD assessment was performed with the FDA approved myChoiceCDx (Myriad) assay, which considered BRCA1 and BRCA2 status and HRD-induced genomic scar. However, this assay is centrally performed, costly and not reimbursed by the National Healthcare System. Next generation sequencing panels evaluating HRR genes, beyond BRCA1/2, may improve the detection rate of tumour with HRD by only 5-6% 13. Commercial assays applicable in diagnostic laboratories that screen for HRR genes along with genomic scar have been recently developed 14.
In this study, we report our first experience with in-house HRD testing, using the HRD Focus panel (AmoyDx), which evaluates both BRCA1/2 status and genomic instability. We performed a double-blind evaluation of HRD status in a consecutive series of high grade epithelial ovarian cancers that were analyzed in our laboratory with HRD Focus panel and sent to Myriad for MyChoiceCDx testing. We aimed: i) to evaluate the feasibility of HRD testing with an assay applicable in a diagnostic clinical setting; ii) to compare HRD assessment obtained with the HRD Focus panel and the reference assay myChoiceCDx.
Methods
STUDY COHORT
This single-institution study obtained specific Review Board approval (UID 2386). From the institutional electronic database, we selected all patients with high grade serous and endometrioid ovarian cancer treated at the European Institute of Oncology (IEO), Milan, Italy who underwent molecular analysis of HRR deficiency between April 2021 and April 2022. Demographic, clinicopathological and surgical characteristics were abstracted from clinical records. First line therapy indications were considered according with EMA criteria. Study data were collected and managed using REDCap (Research Electronic Data Capture) electronic data capture tools 15.
HRD TESTING
In-house HRD evaluation was performed with the HRD Focus Assay (CE-IVD) provided by AmoyDx (AmoyDx, Xiamen, China), following the manufacturer’s instructions. Briefly, 100 ng of DNA (50 ng minimum yield request) extracted from representative formalin-fixed paraffin-embedded (FFPE) tumor tissue blocks were used for library preparation, and then sequenced on Illumina NextSeq platform. This assay allowed the simultaneous analysis of SNVs and indels in the whole coding regions and exon-intron boundaries of BRCA1 and BRCA2, and estimated a genomic scar score (GSS) based on the analysis of 24,000 SNPs 16. A GGS equal or higher than 50 was indicative of HRD positivity. The bioinformatic algorithm applied for the NGS data analysis was the version 1.1.
The same FFPE tumor blocks used for AmoyDx evaluation were sent to Myriad, for performing myChoiceCDx assay. BRCA1 and BRCA2 status was evaluated along with HRD assessment, measured by a genomic instability score (GIS) score encompassing measured by loss of heterozygosity, telomeric allelic imbalance) and large-scale state transitions across the entire genome. A GIS equal or higher than 42 was indicative of HRD positivity.
STATISTICAL ANALYSIS
Statistical analysis was carried out using SPSS Statistic 25 software. Chi-Square test with Yates correction and t-test calculators were used for data comparison of categorical variables. p-values < 0.05 were considered statistically significant.
Results
Among the 101 ovarian cancer patients referred to the Clinical Unit of Oncogenomics for HRD analysis, 6 were excluded from the current analysis as they had already undergone BRCA testing externally and were evaluated with Myriad myChoiceCDx only. The remaining 95 cases reached the minimum tumor cellular content required for AmoyDx (above than 30%) and Myriad (above than 20%) and were included in the present study. The most representative FFPE tumor block was sent to Myriad after section cutting (6 sections 5 μm-thick) for AmoyDxHRD testing (Fig. 1). The clinicopathological characteristics of this population are reported in Table I.
Figure 1.
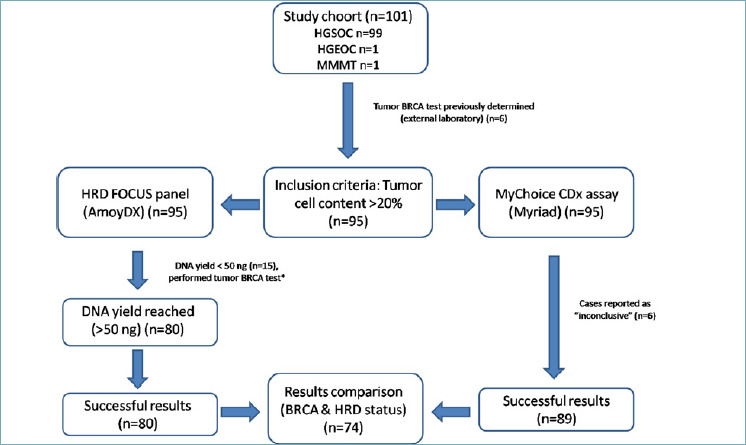
Study design.
*Next Generation sequencing panel: “Oncomine BRCA Research Assay” (ThermoFisher Scientific, Waltham, Massachussets, USA). HGSOC: High grade serous carcinoma; HGEOC: High grade endometrioid carcinoma; MMMT: Malignant Mixed Mullerian tumor.
Table I.
Clinico-pathological features of the study population.
| Clinico-pathological features | N (%) |
|---|---|
| Patients | 95 |
| Age at diagnosis | |
| Median (years), range | 62 (36-82) |
| Oncological treatment | |
| NACT | 41 (43.2%) |
| PCS | 54 (56.8%) |
| Family history for cancer in first and second degree relatives | |
| Positive | 58 (61.1%) |
| Negative | 37 (38.9%) |
| Histotype | |
| High grade serous carcinoma (HGSOC) | 93 (97.9%) |
| High grade endometrioid carcinoma (HGEOC) | 1 (1.1%) |
| Malignant mixed Mullerian tumor (MMMT) | 1 (1%) |
| tBRCA status | |
| BRCA1/2 wt | 87(91.6%) |
| BRCA1 pathogenic/BRCA2 wt | 3 (3.2%) |
| BRCA1 wt/BRCA2 pathogenic | 2 (2.1%) |
| NA | 3 (3.2%) |
NACT = neoadjuvant chemotherapy, PCS = primary cytoreductive surgery, tBRCA = tumor BRCA, wt = wild type.
AmoyDx HRD FOCUS PANEL RESULTS
95 cases underwent DNA extraction,obtaining a median concentration of 38.6 ng/μl (range 1.1-99.7 ng/μl).In 84 cases (88.4%) DNA was extracted from surgical specimens, while in 11 cases (11.6%) DNA was extracted from biopsies.15 of 95 (15.8%) samples, including 5 biopsies and 10 surgical specimens, were not adequate for analysis with AmoyDx HRD Focus panel due to the low DNA yield and were addressed to tumor BRCA test assessment only, as previously reported 17. The remaining 80 (84.2%) samples were subjected to HRD testing, giving a result in all the cases. The successful rate of HRD Focus assay in our cohort reached 84.2%. (Fig. 1). The Myriad MyChoiceCDx HRD evaluation was successfully performed in 89 of 95 cases (93.7%), and in the remaining 6 cases (surgical specimens) the analysis resulted inconclusive. The median turnaround time (TAT) from the test request to the available report was 7 days (range 5-9 days) for AmoyDx HRD Focus panel and 18 days (range 17-25 days) for Myriad MyChoiceCDx.
AmoyDx HRD Focus panel identified 38 (47.5%) HRD positive and 42 (52.5%) HRD negative tumors. HRD positive cases had a significant lower age at diagnosis, whereas no other significantly correlation with clinicopathological features was observed (Tab. II).
Table II.
AmoyDX HRD testing results according to clinicopathological characteristics.
| Clinico-pathological feature | HRD Positive (n = 38) | HRD Negative (n = 42) | p value | |
|---|---|---|---|---|
| Age at diagnosis median (range) | 59.5 (36-79) | 64 (43-82) | 0.03* | |
| Histotype | ||||
| HGSOC | 38 (100%) | 40 (95.2%) | ||
| HGEOC | 0 | 1 (2.4%) | 0.99 | |
| MMMT | 0 | 1 (2.4%) | ||
| Family history | ||||
| Positive | 26 (68.4%) | 21 (50%) | 0.09 | |
| Negative | 12 (31.6%) | 21 (50%) | ||
| tBRCA status | ||||
| Positive | 4 (10.5%) | 1 (2.4%) | 0.13 | |
| Negative | 34 (89.5%) | 41 (97.6%) | ||
* p value statistically significant.
HGSOC: High grade serous carcinoma; HGEOC: High grade endometrioid carcinoma; MMMT: Malignant mixed Mullerian tumor; tBRCA = tumor BRCA.
AmoyDx HRD FOCUS PANEL PERFORMANCE: COMPARISON WITH MYRIAD MyChoiceCDx RESULTS
The comparison between AmoyDx HRD focus panel and Myriad MyChoice results was performed including 74 cases which were successful analyzed with both assays.
AmoyDx and Myriad assays focused on BRCA status assessment along with HRD evaluation. BRCA pathogenic mutations were found in 5 cases with both the assays. Complete concordance was achieved in 72 (97.3%) samples, including 68 BRCA negative and 4 BRCA positive (pathogenic mutation) tumors, whereas 2 (2.7%) samples reported a discordant results. In one case a somatic BRCA1 large deletion from exon 14 to exon 18 was identified by Myriad assay only and another sample carried an alteration classified as pathogenic for AmoyDx test and VUS (Variant of Uncertain Significance) for Myriad evaluation (BRCA2 variant in exon11:c.4284_4285insT; p.Q1429Sfs*9).
Regarding HRD status, the overall percent agreement (OPA) between AmoyDx and Myriad was 87.8% (65 of 74 tumours tested) (Tab. III and Fig. 2). In detail, using AmoyDx assay, all the negative cases (36 of 36 tumors, 100%) were confirmed as negative by Myriad whereas among 38 cases identified as HRD positive by AmoyDx, 29 (76.3%) tumors resulted positive and 9 (23.7%) negative by Myriad. The positive predictive value (PPV) of AmoyDX test was 83.3% and the negative predictive value (NPV) was 100%. The clinicopathological characteristics of the discordant cases are reported in Table IV.
Table III.
HRD status comparison between AmoyDX and Myriad results.
| HRD Focus panel | AmoyDX | Positive (N = 38) | Negative (N = 36) |
|---|---|---|---|
| myChoiceCDx Myriad | |||
| Positive (N = 29) | 29 (76.3%) | - | |
| Negative (N = 45) | 9 (23.7%) | 36 (100%) | |
Figure 2.
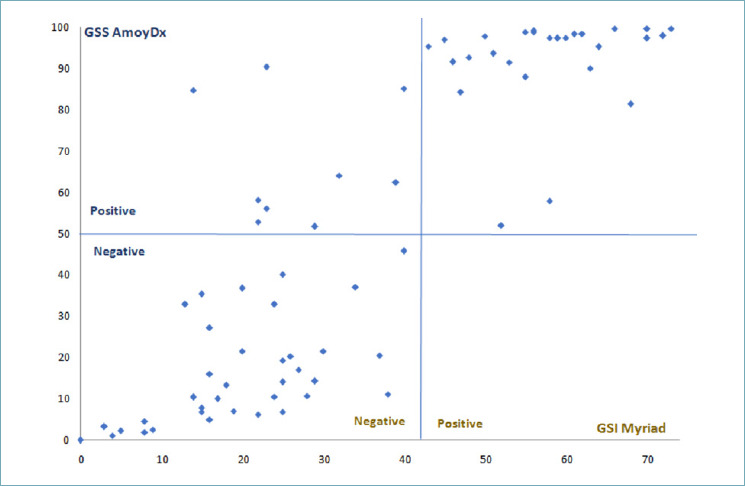
Comparison between HRD score: GSS AmoyDx (cut-off 50) and GSI Myriad (cut-off 42).
Table IV.
Clinicopathological characteristics of HRD discordant cases.
| GIS score Myriad | GSS score Amoy | Histotype | Age at diagnosis | Family history | FIGO stage | Surgery | Residual tumor | NACT | |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 22 | 58.2 | HGSOC | 40-45 | No | IIIC | IDS | Yes * | Yes |
| 2 | 29 | 51.7 | HGSOC | 65-70 | Yes | IVB | PCS | No | No |
| 3 | 14 | 84.8 | HGSOC | 55-60 | No | IIIC | IDS | No | Yes |
| 4 | 40 | 85.1 | HGSOC | 55-60 | No | IVB | PCS | Yes * | No |
| 5 | 32 | 64.1 | HGSOC | 50-55 | No | IVB | PCS | No | No |
| 6 | 22 | 52.9 | HGSOC | 65-70 | Yes | IIIC | PCS | No | No |
| 7 | 23 | 56.1 | HGSOC | 60-65 | No | IIIC | PCS | No | No |
| 8 | 23 | 90.4 | HGSOC | 60-65 | Yes | IIIC | IDS | Yes * | Yes |
| 9 | 39 | 62.5 | HGSOC | 65-70 | Yes | IIIC | IDS | No | Yes |
* ≤ 0.5 cm.
HGSOC: High grade serous carcinoma; IDS: interval debulking surgery; PCS: primary cytoreductive surgery; NACT = neoadjuvant chemotherapy.
Discussion
In the last years, the clinical management of women affected by ovarian cancers has been through a rapid evolution, prompted by progress in precision medicine. The recently introduced HRD assessment beyond BRCA1/2 status as a clinically relevant biomarker for therapeutic indications, poses a major challenge in hospital workflow as the gold standard assay for HRD evaluation. Myriad MyChoiceCDx is a centrally performed and costly assay, with no reimbursement scheduled, at least in Italy.
In this report, we describe our experience in with the HRD Focus AmoyDx (CE-IVD) in our diagnostic workflow, focusing on the feasibility and reliability of in-house HRD-testing. To our knowledge, this is one of the firsts report of feasibility of HRD-testing in a real-life diagnostic setting, evaluating a consecutive series of advanced carcinoma who may change their treatment indications. We observed a successful rate of 84.2%, with failures due to low extracted DNA yield, mainly related to small biopsies or DNA quality suboptimal, linked to formalin treatment and preanalytical condition that determined DNA deamination and fragmentation. Notably, the AmoyDx median TAT from the test request to the available report was 7 days, which was significantly shorter than the Myriad TAT of 18 days, the latter is also effected by logistic handling and transportation. Considering the clinical need to schedule the most effective therapy for the single patient in a short timeframe, both the successful rate and the TAT are crucial parameters to be taken under consideration to establish the clinical utility of an assay.
Applying the HRD Focus panel, we identified 47.5% HRD positive tumors, in line with the incidence reported in the PAOLA1 (48%) 7, PRIMA (50.9%) 6 and VELIA (50.1%) 8 trials. All tumors were evaluated with the gold standard Myriad MyChoiceCDx, obtaining an OPA of 87.8%. Our data were in accordance with the recent findings of Weichert and colleagues 18 that reported an OPA of 81.6% between AmoyDX and Myriad assays in HRD assessment.
Recently, other HRD assays (i.e., Oncomine Comprehensive Assay Plus, ThermoFisher Scientific or DDM HRD Solution, SOPHiA Genetics) have been placed on the market, aiming to provide HRD testing in diagnostic laboratories equipped with high throughput NGS systems. Moreover, great efforts have been made by European academic centers to develop a reliable and in-house feasible HRD test to replicate the Myriad MyChoiceCDx results 19. Exciting results have been recently reached by the Leuven HRD testing, a targeted next generation sequencing - capture based investigating about 90,000 genome wide SNPs and HRR involved genes running on Illumina NovaSeq instrument. This test has an OPA with Myriad MyChoice PLUS of 91%, based on the analysis of 468 samples from the PAOLA-1 study and remarkably showed a similar impact of olaparib on progression free survival as Myriad test 20. These very promising results may pave the way to the introduction of in-house academic-developed HRD testing, even if some criticisms have to be addressed, including the requirement of powerful NGS instruments and the need to obtain the European Certification required for in vitro diagnostic use (CE-IVD mark).
Our study presents some limitations. This is a feasibility study and the results obtained should be considered preliminary and need to be confirmed in a larger cohort. Moreover, most of the patients in this population are still undergoing adjuvant treatment and are waiting to start maintenance treatment. Maintenance treatment and follow-up are fundamental parameters, especially in AmoyDx-Myriad discordant cases, to assess the utility of the AmoyDx HRD test in predicting clinical outcomes or likely magnitude of benefit from PARPis.
In conclusion, we report on HRD assessment using the HRD Focus AmoyDx (CE-IVD) in a real-life diagnostic setting. The major limitation we faced was the DNA yield required in the HRD Focus test, which lowered the success rate. However, the turnaround time compatible with clinical needs and the high concordance with the gold standard Myriad MyChoice CDX assay suggest the feasibility and reliability of HRD testing in diagnostic laboratories.
ACKNOWLEDGEMENTS
We acknowledge the laboratory technicians of the Division of Pathology (European Institute of Oncology IRCCS, Milan, Italy) for sample collection and processing.
Figures and tables
Footnotes
CONFLICT OF INTEREST
NC has received honoraria for consulting from Roche (ongoing), PharmaMar (ended), AstraZeneca (ongoing), Tesaro/GSK (ended), Immunogen (ongoing), Pfizer (ended), Clovis Oncology (ongoing), Merck Sharp & Dohme Corp. (ongoing), BIOCAD (ended), Mersana (ended), Eisai (ongoing), Oncxerna (ended); for speakers’ bureau(s) from Clovis (ongoing), Novartis (ended), AstraZeneca (ongoing), Tesaro (ended), Merck Sharp & Dohme Corp. (ongoing), GSK (ongoing), Eisai (ongoing); grant/research support from AstraZeneca (ended), PharmaMar (ended), Roche (ended) and its Member Steering Committee ESMO Clinical Guidelines (ongoing) and Chair Scientific Committee ACTO Scientific onlus (ongoing). EG-R has received honoraria and/or advisory fees and/or research funding from AstraZeneca, Exact Sciences, Novartis, Roche, and ThermoFisher. MB has received honoraria for consulting, advisory role, speakers’ bureau, travel, accommodation, expenses from MSD Oncology, Roche/Genetech, AstraZeneca, Thermofisher Scientific and Illumina.
FUNDING
This work was partially supported by the Italian Ministry of Health with “RicercaCorrente”, “5 × 1000”.
ETHICAL CONSIDERATION
All information regarding human material was managed using anonymous numerical codes, and all samples were handled in compliance with the Helsinki Declaration. The study received the European Institute Review Board approval: UID 2386. Informed consent was obtained from all subjects involved in the study.
AUTHOR CONTRIBUTIONS
Conceptualization: CF, IB, MB, and EGR. Methodology: CF, and IB Data collection: A.Ranghiero., IB, A.Rappa, GB, DV. Statistical analyses: CF. Writing-original draft: CF, IB. and MB. Writing-review and editing: CF, A.Ranghiero, IB, EGR, GB, A.Rappa, DV, MB. All authors have read and agreed to the published version of the manuscript.
References
- 1.Global Burden of Disease 2019 Cancer Collaboration Kocarnik JM, Compton K, Dean FE, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 Cancer Groups From 2010 to 2019: a systematic analysis for the Global Burden of Disease Study 2019. JAMA Oncol 2022;8:420-444. https://doi.org/10.1001/jamaoncol.2021.6987 10.1001/jamaoncol.2021.6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I numeri del cancro in Italia 2021. – https://www.aiom.it/wp-content/uploads/2021/10/2021_NumeriCancro_web.pdf.
- 3.Lisio MA, Fu L, Goyeneche A, et al. High-grade serous ovarian cancer: basic sciences, clinical and therapeutic standpoints. Int J Mol Sci 2019;20:952. https://doi.org/10.3390/ijms20040952 10.3390/ijms20040952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee S, Moore KN, Colombo N, et al. Maintenance olaparib for patients with newly diagnosed advanced ovarian cancer and a BRCA mutation (SOLO1/GOG 3004): 5-year follow-up of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2021;22:1721-1731. https://doi.org/10.1016/S1470-2045(21)00531-3 10.1016/S1470-2045(21)00531-3 [DOI] [PubMed] [Google Scholar]
- 5.Coleman RL, Fleming GF, Brady MF, et al. Veliparib with first-line chemotherapy and as maintenance therapy in ovarian cancer. N Engl J Med 2019;381:2403-2415. https://doi.org/10.1056/NEJMoa1909707 10.1056/NEJMoa1909707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.González-Martín A, Pothuri B, Vergote I, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2019;381:2391-2402. https://doi.org/10.1056/NEJMoa1910962 10.1056/NEJMoa1910962 [DOI] [PubMed] [Google Scholar]
- 7.Ray-Coquard I, Pautier P, Pignata S, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416-2428. https://doi.org/10.1056/NEJMoa1911361 10.1056/NEJMoa1911361 [DOI] [PubMed] [Google Scholar]
- 8.Swisher EM, Aghajanian C, O’Malley DM, et al. Impact of homologous recombination status and responses with veliparib combined with first-line chemotherapy in ovarian cancer in the Phase 3 VELIA/GOG-3005 study. Gynecol Oncol 2022;164:245-253. https://doi.org/10.1016/j.ygyno.2021.12.003 10.1016/j.ygyno.2021.12.003 [DOI] [PubMed] [Google Scholar]
- 9.FDA Lynparza: https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/208558s013lbl.pdf (accessed on 10 May 2022)
- 10.EMA Lynparza: https://www.ema.europa.eu/en/documents/smop/chmp-post-authorisation-summary-positive-opinion-lynparza-ii-35-ii-36_en.pdf (accessed on 10 May 2022)
- 11.AIFA Lynparza: https://www.aifa.gov.it/documents/20142/961234/Determina_194-2022_Lynparza.pdf (accessed on 10 May 2022)
- 12.Vergote I, González-Martín A, Ray-Coquard I, et al. European experts consensus: BRCA/homologous recombination deficiency testing in first-line ovarian cancer. Ann Oncol 2022;33:276-287. https://doi.org/10.1016/j.annonc.2021.11.013 10.1016/j.annonc.2021.11.013 [DOI] [PubMed] [Google Scholar]
- 13.Konstantinopoulos PA, Ceccaldi R, Shapiro GI, et al. Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov 2015;11:1137-1154. https://doi.org/10.1158/2159-8290.Cd-15-071 10.1158/2159-8290.Cd-15-071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagener-Ryczek S, Merkelbach-Bruse S, Siemanowski J. Biomarkers for homologous recombination deficiency in cancer. J Pers Med 2021;11:612. doi:10.3390/jpm11070612 10.3390/jpm11070612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris PA, Taylor R, Thielke R, et al. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009;42:377-381. https://doi.org/10.1016/j.jbi.2008.08.010 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.AmoyDiagnostics. Available online: http://www.amoydiagnostics.com/productDetail_46.htlm
- 17.Fumagalli C, Betella I, Rappa A, et al. Tumor BRCA Testing in epithelial ovarian cancers: past and future-five-years’ single-institution experience of 762 consecutive patients. Cancers 2022;14:1638. https://doi.org/10.3390/cancers14071638 10.3390/cancers14071638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weichert W, Lukashchuk N, Yarunin A, et al. An evaluation of the performance of molecular assays to identify homologous recombination deficiency-positive tumours in ovarian cancer Int J Gynecol Cancer 2021;31:A366. [Google Scholar]
- 19.Pujade-Lauraine E, Christinat Y, D’incalci M, et al. 201 Homologous recombination deficiency testing in advanced ovarian cancer: description of the ENGOT HRD European initiative. Int J Gynecol Cancer 2021;31:A208. [Google Scholar]
- 20.SGO 2022 Annual Meeting| March 18-21, 2022. https://sgomtg22.us2.pathable.com/meetings/virtual/nRerYfh2bMwvczGuJ [Google Scholar]


