Abstract
Tobacco mild green mosaic virus (TMGMV) is a plant virus closely related to Tobacco mosaic virus (TMV), sharing many of its structural and chemical features. These rod-shaped viruses, comprised of 2130 identical coat protein subunits, have been utilized as nanotechnological platforms for a myriad of applications, ranging from drug delivery to precision agriculture. This versatility for functionalization is due to their chemically active external and internal surfaces. While both viruses are similar, they do exhibit some key differences in their surface chemistry, suggesting the reactive residue distribution on TMGMV should not overlap with TMV. In this work, we focused on the establishment and refinement of chemical bioconjugation strategies to load molecules into or onto TMGMV for targeted delivery. A combination of NHS, EDC, and diazo coupling reactions in combination with click chemistry were used to modify the N-terminus, glutamic/aspartic acid residues, and tyrosines in TMGMV. We report loading with over 600 moieties per TMGMV via diazo-coupling, which is a >3-fold increase compared to previous studies. We also report that cargo can be loaded to the solvent-exposed N-terminus and carboxylates on the exterior/interior surfaces. Mass spectrometry revealed the most reactive sites to be Y12 and Y72, both tyrosine side chains are located on the exterior surface. For the carboxylates, interior E106 (66.53%) was the most reactive for EDC-propargylamine coupled reactions, with the exterior E145 accounting for >15% reactivity, overturning previous assumptions that only interior glutamic acid residues were accessible. A deeper understanding of the chemical properties of TMGMV further enables its functionalization and use as a multifunctional nanocarrier platform for applications in medicine and precision farming.
Keywords: bioconjugation, drug delivery, nanocarriers, plant virus, precision agriculture
Graphical Abstract

Using the plant virus nanoparticle TMGMV, we systematically assessed which amino acids are solvent exposed and addressable by bioconjugation strategies. We determined that glutamic acids on the exterior and interior surfaces, tyrosines on the exterior surface as well as the exterior and solvent-exposed N-terminus are reactive but to varying degrees (Tyr>Glu>N-term.) Bioconjugation reactions and proteomics paired with structural analysis of TMGMV provided rationale for the underlying reactivity of target sites. A deeper understanding of the chemical properties of TMGMV further enables its functionalization and use as a multifunctional nanocarrier platform for applications in medicine and precision farming.
Introduction
Viruses are proteinaceous nanoparticles with a high degree of symmetry and polyvalency, imparting them with regularly spaced chemical units and handles for functionalization on their inner and exterior capsid surfaces.[1–4] Typically, viruses are expressed as rod-like or icosahedral protein capsid that protect their genome; more complex shapes have been achieved through post-harvest treatments and genetic modifications.[5,6] Viruses, in particularly those derived from plant viruses and bacteriophages, often exhibit high thermal and chemical stability when compared to other protein materials and they can be produced in high yields, allowing them to overcome many of the challenges of other protein-based materials.[7–9] Together, these features uniquely position viruses as nanomaterials where new functionalities can be introduced through bioconjugation reactions. Building on the development or repurposing of viruses as nanoparticles – also termed viral nanoparticles (VNPs) – for drug delivery targeting human health,[10–16] there also has been growing interest in using the application of VNPs in precision agriculture for delivery of agrochemicals.[17–19] In addition, viral nanotechnology is a powerful platform for specially controlled and programmed materials assembly for development of advanced biocatalytic materials.[20–25] Some prominent examples of plant viruses and bacteriophages utilized in viral nanotechnology include: virus-like particles (VLPs, which are devoid of the viral genome) of Qβ and Physalis mottle virus (PhMV)[26,27] and VNPs from plants such as Cowpea mosaic virus (CPMV), Potato virus X (PVX), Turnip mosaic virus (TuMV) and Cowpea chlorotic mottle virus (CCMV).[28–35] In particular, Tobacco mosaic virus (TMV) has been extensively analyzed for its reactivity and demonstrated as a robust platform in materials science, drug delivery, and structural biology.[36–40] Diverse applications have been made possible by the available chemical modification strategies.
We gained interested in application of Tobacco mild green mosaic virus (TMGMV) for precision farming. TMGMV is utilized as a herbicide[41,42] and approved by the United States Department of Agriculture; more recently TMGMV has been tested for precision agriculture applications. For example, we have demonstrated that TMGMV exhibits high motility within soil and that it can carry and deliver nematicides to effectively kill nematodes.[19,43] In those studies, the model pesticides were either infused and loaded through electrostatic interactions or via covalent coupling to solvent exposed amino acids – data indicate that active ingredients must be strongly bonded to facilitate co-delivery through soil. Therefore, to understand the full potential of TMGMV as a nanocarrier, it is important to elucidate the full array of amino acid residues available for conjugation reactions. Having a diverse toolset of available reactions and a structural understanding of sites modified provides structure-based design input for the formulation of next generation nanotechnologies based on TMGMV. While we previously demonstrated that active ingredients can be coupled to TMGMV,[43–45] its chemical reactivity remains largely unexplored.
The proteinaceous nature of VNPs allows for bioconjugation to address solvent-exposed amino acids. The most common targets are the side chains of lysines or the N-terminus, cysteines, aspartic/glutamic acids, and tyrosines.[46–49] Herein, we describe an effort to identify the active residues involved in TMGMV bioconjugation. Starting from the reported crystal structure of the coat protein, we identified three probable conjugation reactions via amino, carboxylate, amino, and tyrosine modifications. Using two fluorescent reagents and biotin, we confirmed successful bioconjugation and quantified the efficiency of each chemistry performed. We used a proteomic approach to delineate the specific bioconjugation handles.
Results and Discussion
Structural analysis to predict reactive residues
TMGMV is a rod-shaped virus with dimensions of 300 nm by 18 nm, with a 4 nm-wide channel. The VNP is closely related to TMV, with 71% amino acid sequence similarity of the coat proteins (Uniprot IDs: TMGMV- P03579 and TMV-P69687). While related, structural data indicates key differences in their surface accessible reactive amino acids. To systematically assess the reactive residues available on TMGMV, we used a combination of in silico analysis to identify tyrosine, N-terminus, and glutamic acids as target residues (Fig. 1A–D) – we then used bioconjugation reactions to probe the chemical reactivity of these sites. We chose three model cargos and conjugated sulfo-Cyanine5 (sulfo-Cy5), ATTO488 and biotin (Fig. 1E, F).
Fig. 1.
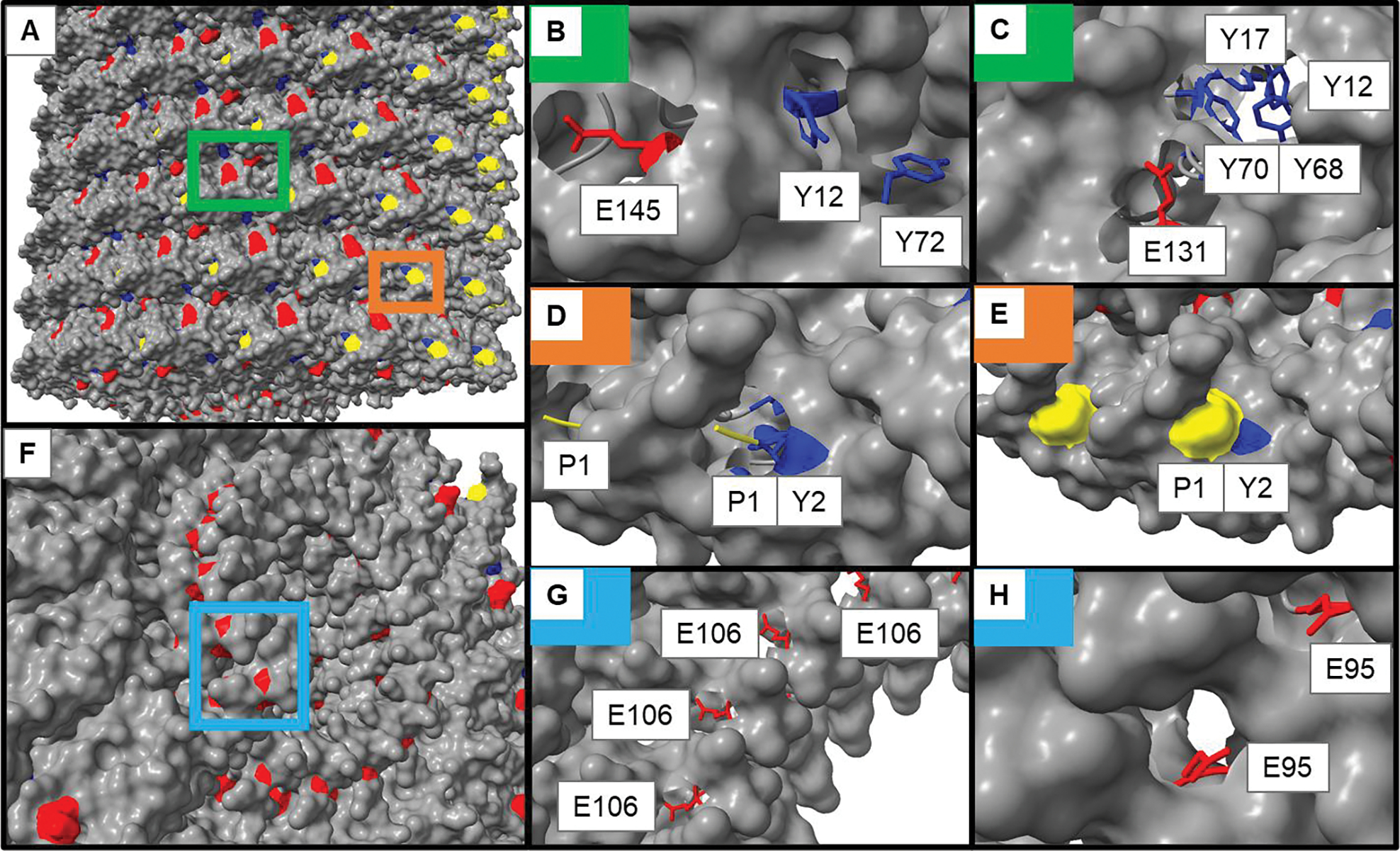
A structural model of TMGMV (PDB: 1VTM) showing different reactive residues (rendered in UCSF Chimera X). A) Exterior surface of TMGMV with areas of interest highlighted. B-C) exterior glutamic acid (red) and tyrosine (blue) residues. B) accessibility of E145 and Y12/Y72. C) accessibility of E131 and the tyrosine binding pocket (Y12/Y17/Y68/Y70). D-E) surface exposure of the N-terminus (yellow) and Y2. F) cross-section of the interior surface of TMGMV. G-H) accessibility of the interior glutamic acid residues G) E106 and H) E95. The colored boxes in (A) and (F) correspond to the areas with the matching insets in the remaining panels.
The TMV reactive sites have been mapped and are principally Y139 on the exterior surface, and Glu97 and Glu106 inside the channel[50]; to the best of our knowledge the reactivity of the N-terminal amines or lysines has not been reported. However, a lysine-added mutant termed TMV-Lys (amongst other engineered TMV nanoparticles) has been reported.[51] Using the model of TMGMV generated by Chimera X (UCSF, Version 1.2.5[52]), analysis of the surface accessible residues was conducted (Fig. 1). While this approach allows to identify solvent-exposed amino acid residues, it should be noted that the structural model is a static representation of a dynamic protein surface that may change based on the bathing conditions and reactant.[53] When comparing the structures of TMGMV and TMV, some key differences emerge:
Tyrosines:
Importantly, the known reactive tyrosine from TMV (Y139) is F139 on TMGMV.40 While both VNPs share Y2, Y70, and Y72, TMGMV has three additional Tyr side chains: Y12, Y17, and Y68 – all of which appear to be surface accessible but to varying degrees. Y12 and Y72 are relatively solvent-accessible with their meta positions similarly exposed, suggesting both may have similar reactivity.[54] Y2, Y20, Y17, and Y68 appear clustered which may suggest a good microenvironment for diazo coupling reactions by increasing the electron-withdrawing behavior of reactive tyrosine through π-stacking, though only Y12 and Y68 seem exposed enough to be reactive due to steric hindrance of the other side chains.[55,56] All identified solvent-exposed Tyr side chains are located on the exterior surface of TMGMV (Fig. 1A–C).
Glutamic and aspartic acids:
Carboxylate residues were analyzed next, with E145 (Fig. 1B) and E131 (Fig. 1C) shown to be relatively accessible on TMGMV’s exterior, suggesting these residues may be reactive.[57] Two exposed interior glutamate residues were identified in a cross-section of TMGMV (Fig. 1F–H, light blue box). In Fig. 1G, E106 is shown to have an exposed carboxylate group, suggesting this residue should be reactive. This is consistent with the results for TMV, where E106 has previously been shown to react in EDC-activated bioconjugations. The less-exposed E95, shown in Fig. 1H, does still have a solvent-accessible carboxylate group, suggesting this amino acid should also be reactive, though it may be less reactive than E106. Aspartic acid residues were considered putatively active on TMGMV by structural analysis, particularly in positions D19 and D66, where their carboxylate groups may be exposed enough to be reactive with EDC-based chemistries (Fig. S1).
N-terminus:
TMGMV does not have any Lys side chains – therefore we analyzed the N-terminus for chemical reactivity. The N-terminus of the TMGMV coat protein (Fig. 1D–E, orange box) is indeed solvent-exposed – the structural analysis indicates that the N-terminus may render the meta position on Y2 inaccessible. The N-terminal residue is shown to be proline, which does not retain a free amino group required for NHS bioconjugation. However, all organisms have a mix of N-terminal methionine retention and cleavage on their proteins. Thus, in a single TMGMV assembly with more than 2000 copies of identical coat proteins, we would expect around 400 N-terminal methionine residues retained on the protein, assuming approximately 80% cleavage of methionine by methionine aminopeptidase (MAP).[58] Because MAP is ubiquitous enzyme across all organisms, MAP1 from E. coli should perform similarly to MAP from the plant expression system used to make TMGMV, though the exact percentage of cleavage with proline in the penultimate position for plant expression systems has yet to be reported.
Reactivity of TMGMV and analysis of VNPs
To probe the reactivity of Tyr, Glu/Asp, and the N-terminus (Fig. 2A–D), a set of bioconjugation reactions was performed using fluorophores and biotin with appropriate ligation handles (Fig. 2E–F). Reactivity of the N-terminus was tested using NHS-active compounds for direct coupling. While Tyr and Glu/Asp reactivity was probed using two-step reactions: Tyr were converted to present an alkyne handle using diazonium coupling with a diazonium salt prepared from 4-ethynylaniline and nitric acid. Glu/Asp were modified with an alkyne using propargylamine and 1-ethyl-3-(-3-dimethylaminopropyl) carbodiimide (EDC) as the coupling reagent. TMGMV-alkyne was then modified using Cu(I)-catalyzed azide-alkyne cycloadditions[59] to give the final product (Fig. 2G).
Fig. 2.
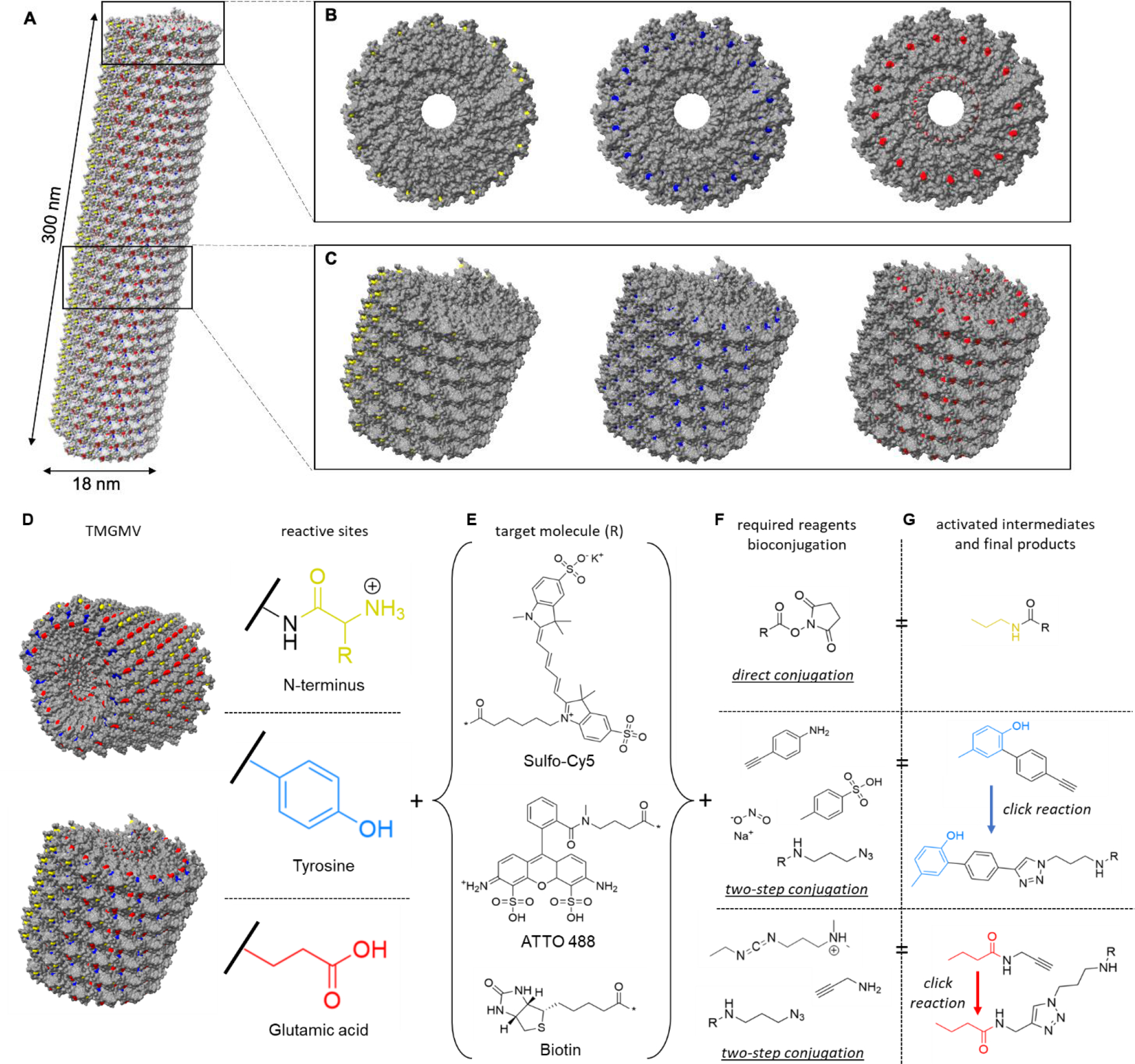
The structure of TMGMV. (A) The atomic model of TMGMV showing a helical structure assembled of 2130 CP subunits. N-terminus, tyrosine and glutamic acid residues are highlighted in yellow, blue and red, respectively. (B) Top view and (C) side view of a short segment of TMGMV displaying exposed N-terminus (yellow), tyrosine (blue) and glutamic acid (red) residues. (D) Reactive surface of TMGMV, N-terminus (yellow), tyrosine residues (blue) and glutamic acid residues (red), and a summary of the reactive moieties on the surface of TMGMV. (E) Model compounds for TMGMV bioconjugation (Sulfo-Cy5, ATTO488 and biotin), where * denotes the reactive position on the molecule. (F) Reactive groups required for bioconjugation using the residue in the same row, with NHS-amine coupling (upper) and with azide-alkyne cycloaddition via azo coupling of tyrosine and a diazonium salt (middle) and via glutamic acid activation by EDC (lower). (G) The reactive activated intermediate and the final compound structure for each type of bioconjugation. R denotes the target molecule (E).
First, conjugation reactions using fluorescent dyes (Sulfo-Cy5 and ATTO488) were performed for quantitative analysis by UV/vis spectroscopy. Conjugations were performed using a two-step diazo/click or EDC/click, or direct conjugation of NHS-functionalized Sulfo-Cy5 or ATTO488 at different molar ratios (dye:CP 1:1, 2:1, 5:1, 10:1, 20:1 and 50:1). After the reaction was allowed to proceed for one hour, the reaction mix was purified by ultracentrifugation, resuspended overnight, and further purified using size exclusion spin filters to remove any excess reagents. The degree of labeling was determined using absorbance measurements and the Beer-Lambert law for TMGMV and dyes (Sulfo-Cy5 or ATTO448) with their molar extinction coefficients (Fig. 3A, S2A).
Fig. 3.
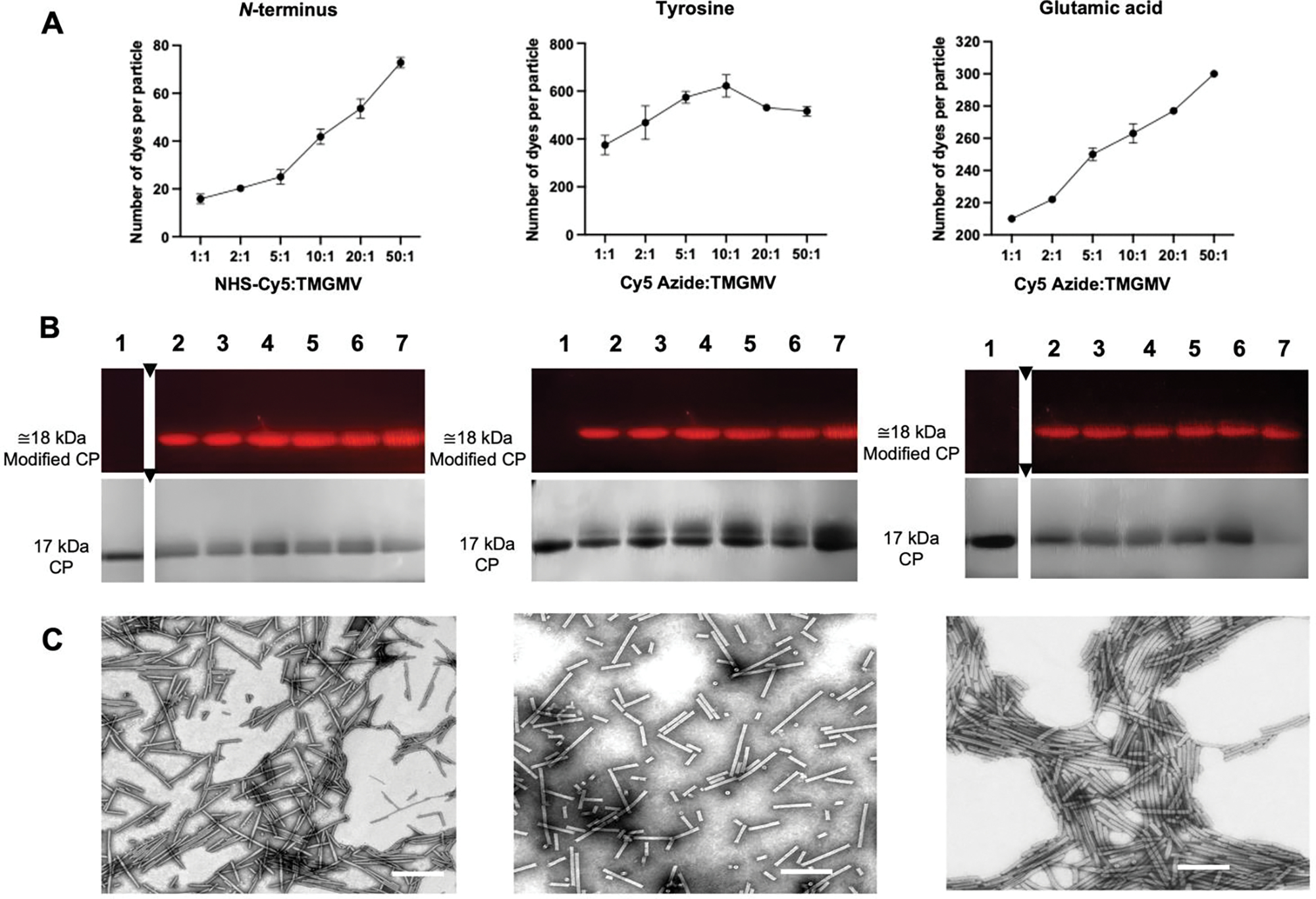
Characterization of TMGMV labeled with Sulfo-Cy5 (TMGMV-Cy5) at reactive amines from the N-terminus (left) and reactive alkynes from tyrosine residues (middle) and glutamic acid residues (right). (A) Quantification of Cy5 per TMGMV as determined by UV/vis spectroscopy. Averaged values from three independent experiments are shown; error bars represent the standard deviation. (B) SDS-PAGE of TMGMV-Cy5 visualized under UV light allowing for dye detection and one under white light after Gel Code staining allowing for protein detection. M: SeeBlue Plus2 Protein Standards, (1): non-modified TMGMV control, (2)–(7): increasing ratios of Cy5:CP: (2) 1:1, (3) 2:1, (4) 5:1, (5) 10:1, (6) 20:1, (7) 50:1. (C) TEM images of Cy5:CP ratio = 10:1 (scale bar: 200 nm).
Tyrosines:
For the diazo/click reaction targeting Tyr side chains, we achieved saturation: data indicate the maximum degree of labeling was achieved using 10:1 (dye:CP) ratio in the case of Sulfo-Cy5 (Fig. 3A) and at 20:1 (dye:CP) for ATTO488 (Fig. S2A). Over 600 Sulfo-Cy5 or ATTO488 dyes per TMGMV virion were attached when targeting tyrosine residues, i.e. 30% of the total number of coat proteins. This result is >3-fold higher than the previously reported degree of labeling applying this chemistry to TMGMV.[43] Based on prior results with TMV and copper-catalyzed azide-alkyne cycloadditions, it seems likely the limiting step is the incorporation of alkyne residues.[43] The Cy5 labeling was slightly more efficient compared to reactions with ATTO488, however comparable loading was achieved using a higher excess of ATTO488. The slightly lower reactivity of ATTO488 might be due to its relatively planar structure compared to Cy5 and its lower flexibility, which has been attributed to lower success in bioconjugation efficiency in previous studies.[60,61]
Glutamic and aspartic acids:
EDC/click conjugations exhibited a linear trend between efficiency and molar excess (Fig. 3A, S2A) – indicating saturation was not achieved. Labeling with ~310 Cy5 azide per TMVGMV (14.5% protein modification) or 285 ATT488 molecules per TMVGMV (13.3%) was achieved using a 50:1 dye:coat protein excess (Fig. 3A, S2A). Again, the reactivity of ATTO488 was slightly lower compared to reactions with Cy5. For EDC-propargylamine, we reported an increase of over 50 molecules to previously reported conjugation efficiencies.[43] Data indicate that saturation is not achieved; therefore, higher loading may be obtained when using more excess, performing multiple rounds of reactions or using more reactive chemistries.[61]
N-terminus:
NHS conjugations were the least efficient, reaching around 80 molecules per virion (3.5–4% protein modification, ATTO488 and Sulfo-Cy5 respectively) (Fig.3A and S2A). The lower reactivity might be explained by the distribution of proline and methionine residues expected to exist at the N-terminus, the former of which would not be receptive to NHS-coupling. However, it is important to note that maximum saturation was not reached indicating that labeling could be increased; for example, biorthogonal chemistries, modifications using pH control, enzymes or transamination, etc., could be used to increase the degree of labeling through the N-terminus.[62]
Covalent attachment of Sulfo-Cy5 or ATTO488 to the CP was determined by SDS-PAGE. Increased molecular weight bands and fluorescence signals from the modified coat proteins (visualized under white light after Coomassie staining) indicate covalent modification of the CPs (Fig. 3B, S2B). This was further validated my mass spectrometry (see below).
To verify structural integrity of the modified TMGMV particles, size exclusion chromatography (SEC) and transmission electron microscopy (TEM) were performed. SEC measurements showed no significant difference between native and conjugated TMGMV for any of the conjugation reactions, with all of them showing elution volumes around 9 mL and an A260:280 ratio of 1.2, indicative of intact TMGMV (Fig. S3). Likewise, TEM imaging showed no significant differences in the length of the virus particles between the amine-, diazo/click or EDC/click-conjugated TMGMV particles. In TEM TMGMV labeled via N-terminus measured 175±23 nm, via tyrosine residues 152±41 nm and via glutamic acid residues 177±17 nm (Fig. 3C), which are shorter than expected. While TMGMV forms 300 nm nucleoprotein complexes, the TMGMV preparations show shorter rods, even in untreated samples (191.4±21 nm, Fig. S4) - this could be due to grid preparation or sample processing.[63]
Next, addressability of the reactive sites using biotin tags was assessed. Biotin is a small vitamin that is easily detected through interaction with avidin/streptavidin conjugates[64] in biochemical assays such as Western blot. Also imaging techniques such as immunogold TEM allow to visualize the degree of labeling.62 Biotin-azide and NHS-biotin at different molar ratios (1:1, 2:1, 5:1, 10:1, 20:1 and 50:1) was reacted with TMGMV-alkyne (where the alkyne was introduced at Tyr or Glu) or TMGMV. Successful biotinylation was assessed by SDS-PAGE and Western Blot analysis (Fig. 4A,B). A shift in the coat protein band electrophoretic mobility, achieved by conjugation to biotin, was observed and confirmed by anti-biotin reaction with Western Blot. Then immunogold staining using gold-labeled anti-biotin antibodies and TEM imaging were performed (Fig. 4C). Consistent with the Western blot, all samples tested positive and gold nanoparticles tagged the biotinylated TMGMV – albeit at different degrees. The degree of gold labeling was proportional to the amount of dyes conjugated to TMGMV with reactivity being as follows: Tyr>Glu>N-terminus (Fig. 4C). Interestingly, immunogold TEM indicates that EDC/click reactions led to conjugation of the exterior surface (at least in part), while previous data suggested that only the interior channel had reactive sites for this chemistry.[43] Nevertheless, data are consistent with the structural analysis of TMGMV which suggests there should be some accessible exterior glutamic acids for EDC-activation reactions.
Fig. 4.
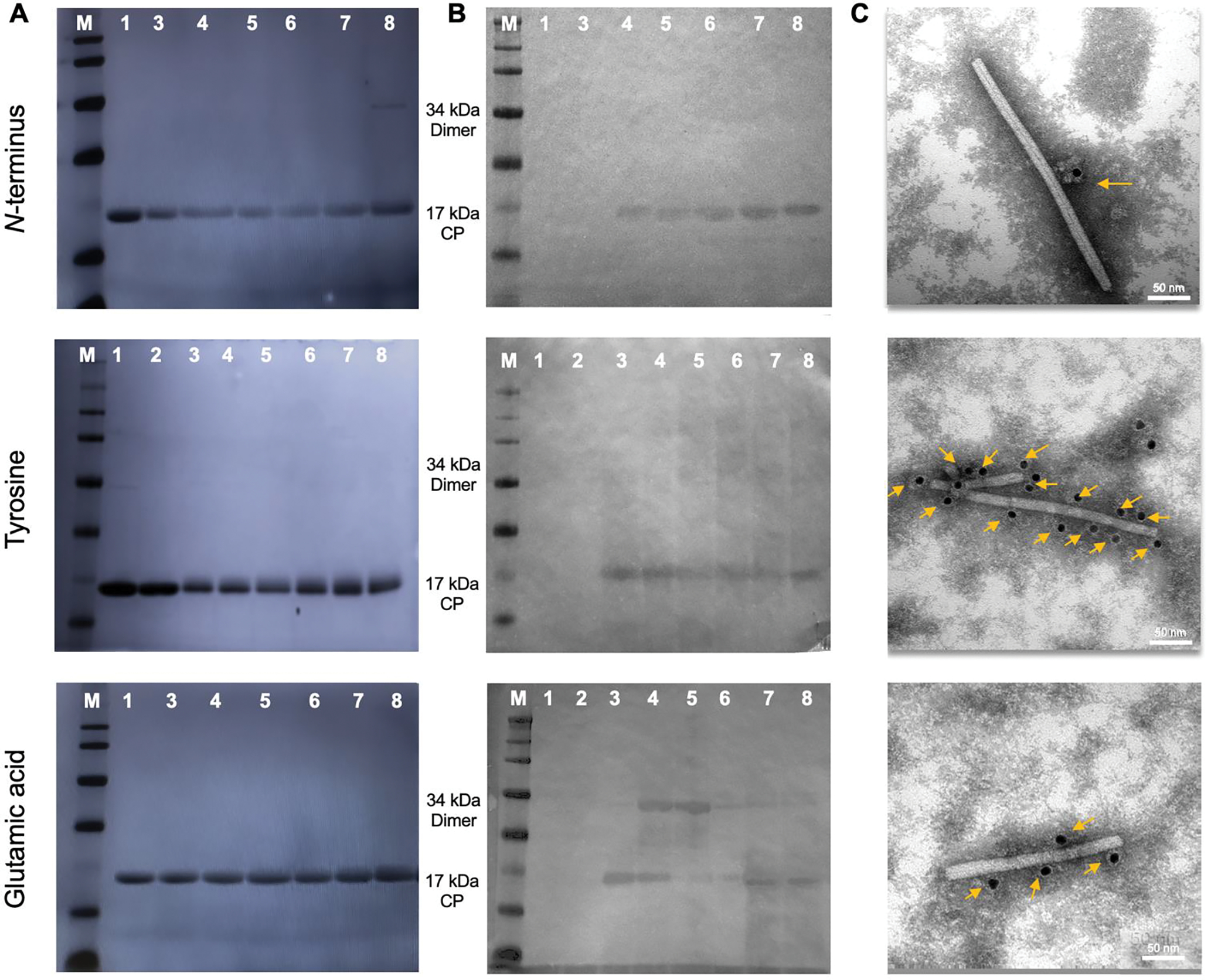
Characterization of biotin-labeled TMGMV at the N-terminus (top), tyrosine residues (middle) and or glutamic acid residues (lower). (A) SDS-PAGE (left) and (B) Western blot (right) of TMGMV-biotin. The gel was visualized after Gel Code Blue staining under white light and blots were probed with alkaline phosphatase-conjugated streptavidin. M: SeeBlue Plus2 Protein Standards, (1) non-modified TMGMV control, (2) non-modified TMGMV-alkyne (3)–(7): increasing ratios of Biotin:CP: (3) 1:1, (4) 2:1, (5) 5:1, (6) 10:1, (7) 20:1, (8) 50:1. (C) TEM of immunogold staining of TMGMV-biotin using gold-labeled anti-biotin antibodies. Yellow arrows show gold anti-biotin labeled nanoparticles.
Confirmation of biotin conjugation by LC-MS/MS
To confirm identity of the amino acids labeled, biotinylated TMGMV was subjected to tandem mass spectroscopy and subsequent proteomic analysis after gel electrophoresis and tryptic digest. The samples with modification at the carboxylates (EDC and EDC/click chemistry) and tyrosine side chains (diazo coupling and diazo/click) were submitted, as well as a control of untreated TMGMV. The samples modified at the N-terminus using NHS-biotin could not be analyzed by LC-MS/MS, because the technique cannot resolve N-terminal modifications.[65] As there are no surface exposed lysine in the viral capsid of TMGMV and NHS has a relatively high fidelity to reacting with primary amines, minimal reactivity with serine, cysteine, and methionine residues was expected.[66] The distribution of reacted residues for each type of reaction is shown in Table 1 with ‘modified alkyne’ percentage and percentage of biotin-conjugation (‘modified cycloaddition’) tabulated.
Table 1.
Modifications of glutamic acids and tyrosine residues and their relative abundance via LC-MS/MS of TMGMV after diazo coupling reaction and azide-alkyne cycloaddition with azide-conjugated biotin.
| Target Residue | Position | Modified Alkyne (%) | Modified Cycloaddltlon <%) | Position on Virus |
|---|---|---|---|---|
| Y | 12 | 37.79 | 52.38 | outside |
| Y | 17 | 2.91 | 1.59 | outside |
| Y | 67 | 20.35 | 19.05 | outside |
| Y | 71 | 38.95 | 26.98 | outside |
| E | 95 | 12.26 | 6.93 | inside |
| E | 106 | 66.53 | 60.14 | inside |
| E | 131 | 5.51 | 0.00 | outside |
| E | 145 | 15.70 | 32.93 | outside |
Tyrosines:
When analyzing the distribution of alkyne conjugation to tyrosine by diazo coupling reaction using the diazonium salt of 4-ethynylaniline, the highest levels of modification were observed for Y72 accounting for ~39% and Y12 accounting for 37.8%, followed by Y68 accounting for 20.4% of the modifications. Y17 showed negligible degree of modification accounting for ~3% of the modifications. The structural data (Fig. 1E) is in agreement indicating that steric hindrance of Y2 which is blocked by the N-terminus. Y12 and Y72 appear to be solvent exposed (Fig. 1B). Y68 is located in the tyrosine pocket (Fig. 1C) with Y68 being more exposed compared to Y17 and Y70. Therefore, the structural data and LC-MS/MS are in good agreement. For the subsequent biotin-azide conjugation, there is a shift in the distribution of reactive residues with Y12 being the most reactive site accounting for 52.4% of the modifications, followed by Y72 which account for 27% of the modifications. Y68, located inside the tyrosine pocket, accounted for 19% of the modifications. Y17 remained the least reactive with 1.6% of the modifications. The differences in reactivity can be explained by the varying microenvironments and conjugation of a small molecule (alkyne) vs. slightly larger, but highly polar biotin moiety.
Glutamic and aspartic acids:
A consistent trend in reactivity was observed when comparing the EDC coupling of alkyne and cycloaddition of biotin reactions with interior E106 being the most reactive side followed by E145 on the exterior; the degree of modification was 66.5% and 60.1% for E106 vs. 15.7% and 33% for E145 after the first and second conjugation respectively. Interior E95 showed some degree of modification accounting for 12.3% of alkynes and ~7% of biotins being conjugated to this site; the least reactive residue was E131 with 5.5% of alkynes located at this side chain; however, biotin conjugation could not be achieved. On the interior surface, E106 is 5.4-fold more reactive for the propargylamine addition, but around 10-fold more reactive for the biotin-azide conjugation. In the context of the structural data, E106 appears more solvent exposed compared to E95 (Fig. 1G+H) – and this is consistent with the LC-MS/MS analysis. Nevertheless, E95 is solvent-exposed – therefore steric hindrance is likely not the only reason explaining the difference in reactivity; differences in the microenvironment, i.e. the surrounding amino acids may explain the differences. These results are mirrored for TMV with E106 being more reactive compared to E95; TMGMV and TMV show a high degree of structural similarity in this region of the capsid.[67] For the exterior surface, to our knowledge, glutamic acid modifications for TMGMV or TMV have not been previously reported. Our results demonstrate that both E145 has some degree of reactivity and E131 has negligible reactivity. The latter is likely attributed to steric hindrance and/or a poor microenvironment with E131 being somewhat buried inside a pocket (Fig. 1D+E). After performing biotin-azide conjugation, >30% of the modifications occurred at external E145 and ~60% of the modification were determined at the internal E106, indicating exterior modifications during EDC-activation of TMGMV are an important consideration. This contrasts to TMV where data suggest that glutamic acid modifications only occur in the interior of the virus62 – therefore providing spatial control.
The aspartic acids D19 and D66, showed no modifications by proteomic analysis, suggesting these residues were not susceptible to activation by EDC. This may be due to steric hindrance of D19 or a poor microenvironment – we note also the proximal E22 did not show any modifications. D66 seems more buried than D19, so it is likely that it was unreactive due to steric effects (Fig S1).
Lastly, when analyzing the untreated TMGMV control group, three types of modifications are observed (Table S1). A mass addition of 57 Da to cysteine, 16 Da to methionine, and 16 Da to tryptophan was observed. Iodoacetamide, commonly used to reduce and cap cysteine during protein mapping, adds a mass of 57 Da.[68] The mass addition of 16 Da to methionine and tryptophan are previously reported oxidations of these amino acids observed in LC-MS/MS.[69] The results from the LC-MS/MS and proteomics algorithm were filtered to identify on the peptide fragments that matched TMGMV, and a sample of the data is shown in Table S2. As this method is highly sensitive, other protein fragments such as human keratin were identified in the sample, but were not considered for further analysis as they were present in negligible amounts.
The observed added masses and the expected added masses for each type of reaction are reported in Table S3, with distributions that span around 20 Da above and below the expected value for each kind of reaction. This is likely due to the ionized environment and mixture of reactive species in the LC-MS/MS causing unexpected fragmentation or addition of reactive groups on the peptide of interest.[70] This may also be attributed to errors in the fitting algorithm for the peptide fragments, which has previously been reported for proteomics fitting analyses involving bioconjugations and post-translational modifications. The expected mass additions of 128 Da and 572 Da for diazo/click reactions to install the alkyne handle followed by biotin, as well as the expected additions of 37 Da and 481 Da for the EDC/click reactions were observed. Importantly, no unreacted alkyne was observed in any sample that underwent azide-alkyne cycloaddition reactions, attesting to the high efficiency of the click chemistry protocol.
In summary, this work highlights the importance of cross-analyzing structural models with proteomics to gain detailed understanding of the reactivity of any virus or protein. Data suggest that multivalent modifications occur with a distribution of amino acids, located on both the interior (Glu) and exterior (Glu, Tyr) surfaces, targeted. Further optimization of the reaction conditions should be pursued to maximize coupling efficiency. The most reactive sites are E106 (interior), E145 (exterior), Y12 (exterior) and Y72 (exterior). Therefore, if complete functionalization was achieved, one could load >2,000 copies of active ingredient into the channel and display >6,000 cargos on the exterior surface. Alternate chemistries or multiple rounds of conjugation may allow to increase overall loading. For the latter, it should be noted that there would be a tradeoff in yield and particle integrity due to the increase in processing steps and the integrity of the particles, as well as the cost of preparation. All these factors should be considered together to create a technology that is effective, of high quality, and able to be translated into commercial applications.
Conclusion
TMGMV has high potential as a platform nanotechnology for cargo delivery as it offers multiple functionalization sites inside and out. The chemical addressability of TMGMV was established using fluorescence and biotin labeling, proteomic analysis, electron microscopy, and a structural model. The yield of modification and distribution of modified amino acid residues for three types of bioconjugations, targeting the N-terminus, tyrosine and glutamic acid residues using a combination of NHS, diazonium, EDC and click chemistry was determined, and the results were corroborated with analysis of the available TMGMV structure. While TMGMV and the well-established platform TMV share structural similarity, the reactive sites do not overlap. TMGMV offers a reactive N-terminus on its exterior surface, addressable surface tyrosines and glutamic acids on the interior and exterior surface with reactivity of multiple sites (the most reactive sites were E106 on the interior, and E145, Y12, Y72, N-terminus on the exterior). Highest loading was achieved by targeting tyrosine side chains resulting in ~600 cargos per TMV, followed by labeling glutamic acids (~300 cargos/TMGMV) and the N-terminus (~80 cargos/TMGMV). Knowledge about the native reactive sites and near-atomic resolution structural data enables structure-based engineering to impart new functionalities for nanotechnology applications.
Experimental Section
Materials and supplies
Unless otherwise specified, materials were purchased from Millipore Sigma and supplies were purchased from Avantor VWR International.
Preparation of TMGMV
TMGMV was obtained from BioProdex (Gainesville, FL, USA) and stored at −20 °C until use. The solution was thawed at 4 °C overnight and then dialyzed against potassium phosphate buffer (KP; 10 mM, pH 7.2) for 24 hours at 4 °C using 12–14 kDa dialysis tubing (Fisher Scientific S432700; Waltham, MA, USA). The buffer solution was replaced, and the dialysis continued for an additional 48 hours. The solution was then centrifuged at 10,000 × g for 20 min (Beckman Coulter Allegra or Avanti centrifuges). The supernatant was collected and ultracentrifuged at 42,000 rpm for 2.5 hours at 4 °C (Beckman Coulter Optima L-90k Ultracentrifuge with 50.2 Ti rotor; Brea, CA, USA). The pellet was resuspended under rotational mixing overnight at 4 °C in KP buffer. The sample concentration was then confirmed using a Nanodrop 2000 (Thermo Scientific; Waltham, MA, USA); the concentration was adjusted to 10 mg mL−1 in 10 mM KP before storing at 4 °C (for TMGMV CP, ε260 = 3 mL mg−1 cm−1).
NHS conjugation of TMGMV
To an ultracentrifugation tube (Beckman Coulter 357448, Indianapolis, IN, USA), 1 mg of TMGMV was added. The equivalences of the NHS-esters per TMGMV coat protein were varied, and the volume of 10 mM KP was adjusted for a final volume of 500 μL. The reaction was left to progress for 4 hours at 25 °C. To the bottom of the same tube, a 200 μL sucrose cushion (30% w/v) was added, and the sample was then ultracentrifuged at 50,000 rpm for 1 hour at 4 °C (Beckman Optima MAX-XP with TLA-55 rotor). The supernatant was removed, and the pellet was resuspended under rotational mixing at 4 °C overnight before further characterization.
Coupling of propargylamine to TMGMV
A 5 mL solution of 2 mg mL−1 TMGMV was prepared in 10 mM KP with 25 molar equivalents (0.73 mg) of propargylamine. Once mixed, 22.5 molar equivalents (25 mg) of EDC were added as a powder to the reaction and mixed immediately by inversion. The sample was left to rotate at room temperature for 4 hours, and then a second aliquot of 25 mg EDC (for a total of 45 molar equivalences) was added to the reaction and mixed. The solution was left to react in the dark for another 12 hours at room temperature. The solution was centrifuged at 50,000 rpm in the tabletop ultracentrifuge (Beckman Optima MAX-XP with TLA-55 rotor) for 1 hour on a sucrose cushion (30% w/v). The viral pellet was resuspended in 10 mM KP overnight at 4 °C on a rotary shaker.
Preparation of diazonium salt from 4-ethynylaniline
In a 5 mL tube, 298 mg of 4-ethynylaniline was dissolved in 2 mL methanol. In a 50 mL tube, 1.09 g p-toluenesulfonic acid was dissolved in 20 mL DIH2O. Both solutions were placed at −20 °C for 10 minutes to precool. A solution of 1.5 mL of 3M sodium nitrite (258 mg in 1.5 mL DIH2O) was prepared and placed at −20 °C for 5 minutes to precool. A 50 mL beaker was submerged in ice/water slurry on a stir plate. The solutions were removed from the freezer. A stir bar was added to the 20 mL of precooled acid in the submerged beaker. Once mixing, the methanol solution was added. The solution turned opaque and beige in color. The nitrite solution was gradually dropped into the acid solution and the mixture gradually turned yellow and eventually turned red after 30–60 minutes of reaction time. A sample of 1 mL of the diazonium slurry was collected and centrifuged for 2 minutes at 10,000 × g to isolate diazonium salts. On ice, the supernatant was removed and the diazonium salts were resuspended in 1 mL of precooled ethanol. The prepared diazonium salts were used immediately for tyrosine modification.
Coupling of diazonium to TMGMV
A solution of 962 μL of 2 mg mL−1 TMGMV in 100 mM borate buffer (pH 8.5) was prepared and precooled on ice. The diazonium salt solution was added to the TMGMV solution at a volume of 80 μL. The solution was mixed by inversion and reacted on ice for 30 minutes. The solution was centrifuged at 50,000 rpm in the tabletop ultracentrifuge (Beckman Optima MAX-XP with TLA-55 rotor) for 1 hour on a sucrose cushion (30% w/v). The viral pellet was resuspended in 10 mM KP overnight at 4 °C on a rotary shaker.
Copper-catalyzed azide-alkyne cycloaddition reaction
To an ultracentrifugation tube (Beckman Coulter 357448, Indianapolis, IN, USA), 1 mg of TMGMV was added. The reaction medium consisted of 1 mM copper sulfate, 2 mM aminoguanidine, 2 mM L-ascorbic acid, and 3.7 mM tris(benzyltriazolylmethyl)amine. The equivalences of the reactive azides (biotin, Cy5, and ATTO488) per TMGMV coat protein were varied, and the volume of 10 mM KP was adjusted for a final volume of 500 μL. The reaction was left to progress for 1 hour on ice. To the bottom of the same tube, a 200 μL sucrose cushion (30% w/v) was added, and the sample was then ultracentrifuged at 50,000 rpm for 1 hour at 4 °C (Beckman Optima MAX-XP with TLA-55 rotor). The supernatant was removed, and the pellet was resuspended under rotational mixing at 4 °C overnight before further characterization.
Virus model analysis
The position and microenvironment of various amino acids and the N-terminus was analyzed using the structural model of TMGMV generated by UCSF Chimera X (1.2.5). TMGMV coat protein (PDB: 1VTM) was used to generate a helically symmetrical model with a rise of 1.41 nm per subunit and an angle (rotation per subunit) of 22.04°. Both the surface accessibility and the identity of proximal residues were used in the analysis.
Characterization of chemically labelled TMGMV
SDS-PAGE:
Denatured biotinylated and fluorophore-conjugated TMGMV samples (10 μg) were loaded on a 12% NuPAGE gel (Life Technologies) and run on 1x MOPS Running Buffer (Life Technologies). Fluorescent proteins were visualized under UV light (for fluorophore-conjugated samples) and then Gel Code Blue stain (Life Technologies) was used to stain proteins and visualized under white light.
FPLC (Size exclusion chromatography):
Fluorophore-conjugated TMGMV samples (500 μL at 0.5 mg/mL) were analyzed using a Superose6 Increase 100 GL column and an ÄKTA Pure25 chromatography system (GE Healthcare) using a flow rate 0.5 mL/min in 10 mM KP (pH 7.4). The absorbance at 260 and 280 nm was recorded, as well as 488 nm and 647nm for ATTO448 and Cy5, respectively.
Western blot:
Biotinylated TMGMV protein separated by SDS-PAGE were transferred onto a nitrocellulose membrane (Thermo Scientific) using NuPAGE Transfer Buffer (Life Technologies). The membrane was then blocked overnight at 4°C using 0.1 M Tris-buffered saline (TBS, pH 7.6) containing 5% (w/v) skim milk powder. Anti-biotin peroxidase antibody (Sigma Aldrich, dilution 1:10000) was used to detect the biotinylated TMGMV. Peroxidase activity was detected using a fluorescent gel imager.
TEM imaging:
Samples were diluted to the concentration of 0.05 mg/mL and absorbed onto carbon-coated TEM grids (Electron Microscopy Sciences). The grids were then washed three times with pure water. Then, grids were stained by 2% (w/v) uranyl acetate for 2 min for imaging. TEM was conducted using a FEI Tecnai F30 transmission electron microscope operated at 300 kV.
Immunogold labeling:
Biotinylated TMGMV samples were diluted to the concentration of 0.5 mg/mL and absorbed onto the carbon-coated copper TEM grids (Electron Microscopy Sciences). Excess was removed by washing the grids using KP buffer. Grids were then blocked using one droplet of 1% (w/v) bovine serum albumin (BSA) in TBS pH 7.4 containing 0.1% (v/v) Tween 20 (TBST) for 30 min. Afterwards, they were equilibrated with 0.1% (w/v) BSA for 5 min and then stained with 10 nm-sized gold nanoparticle-labeled goat anti-biotin antibodies (AURION) diluted 5 x in KP buffer for 1 h. The grids were then washed by TBST and three washes of pure water, followed by the procedure stated before for TEM imaging.
Liquid chromatography and tandem mass spectrometry (LC-MS/MS) and proteomic analysis: Samples were run on SDS-PAGE and the bands of interest were excised using a razor after Coomassie staining. The gels were cut into multiple sections, reduced, alkylated, and digested by trypsin after processing using established protocols.[71] In brief, the digested bands were then run on liquid chromatography (Synergi C18 column) with tandem mass spectrometry using a 0.1% formic acids mobile phase of MeCN and deionized water. The mass spectrometer used was an LTQ Orbitrap Velos with tandem capabilities. The peaks were then extracted and analyzed using Mascot distiller. The data sheet was filtered to only analyze the peptide fragments which matched TMGMV coat protein. Modified residues unique from the untreated virus sample were identified and the range of added masses to the target residue was determined for each reaction. The peptide fragments were then grouped according to the modified residue, and the relative abundance of each type of modified residue was calculated to generate a distribution of residues.
Supplementary Material
Acknowledgements
This work supported in part by grants from USDA, NIFA-2020–67021-31255 (to NFS) and NIFA-2022–67012-36698 (to AAC), the NSF MRSEC at UCSD (DMR-2011924 to NFS), and a research contract through BASF (30034300). The authors thank the University of California, San Diego - Cellular and Molecular Medicine Electron Microscopy Core (UCSD-CMM-EM Core, RRID:SCR_022039) for equipment access and technical assistance. The UCSD-CMM-EM Core is supported in part by the National Institutes of Health Award number S10OD023527. Specifically, the authors thank Guillaume Castillon for training and technical support with the EM equipment. The authors would like to acknowledge Majid Ghassemian of the Biomolecular and Proteomics Mass Spectrometry Facility (BPMSF) at the University of California/San Diego for his assistance and use of facilities. The BPMSF is funded by the NIH under grants S10 OD016234 (Synapt-HDX-MS) and S10 OD021724 (LUMOS Orbi-Trap). The authors thank Matthew Shin, Dr. Krister Barkovich, and Dr. Soo Khim Chan (UC San Diego) for helpful discussions.
References
- [1].Müller TG, Sakin V, Müller B, Molecules 2019, 24, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pokorski JK, Steinmetz NF, Mol. Pharm. 2011, 8, 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhang Y, Dong Y, Zhou J, Li X, Wang F, Molecules 2018, 23, DOI 10.3390/molecules23092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mateu MG, Protein Eng. Des. Sel. 2011, 24, 53–63. [DOI] [PubMed] [Google Scholar]
- [5].Bruckman MA, VanMeter A, Steinmetz NF, ACS Biomater. Sci. Eng. 2015, 1, 13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Balaji S, Applied Plant Virology (Ed.: Awasthi LP), Academic Press, 2020, pp. 113–128. [Google Scholar]
- [7].Steinmetz NF, Evans DJ, Org. Biomol. Chem. 2007, 5, 2891–2902. [DOI] [PubMed] [Google Scholar]
- [8].Hefferon K, Biomedicines 2017, 5, DOI 10.3390/biomedicines5030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yusibov V, Rabindran S, Commandeur U, Twyman RM, Fischer R, Drugs in R & D 2006, 7, 203–217. [DOI] [PubMed] [Google Scholar]
- [10].Roberta Z, Annalisa B, Eva P, Elena B, Elena G, Elena R, George L, Yulia M, Luca S, Francesca I, Matilde M, Elisa T, Francesco D, Laura N, Guido S, Mario P, Alberto F, Linda A, Sci. Adv. 2022, 6, eaaz0295. [Google Scholar]
- [11].Eiben S, Koch C, Altintoprak K, Southan A, Tovar G, Laschat S, Weiss IM, Wege C, Adv. Drug Deliv. Rev. 2019, 145, 96–118. [DOI] [PubMed] [Google Scholar]
- [12].Chung YH, Cai H, Steinmetz NF, Adv. Drug Deliv. Rev. 2020, 156, 214–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Frías-Sánchez AI, Quevedo-Moreno DA, Samandari M, Tavares-Negrete JA, Sánchez-Rodríguez VH, González-Gamboa I, Ponz F, Alvarez MM, Trujillo-de Santiago G, Biofabrication 2021, 13, 035015. [DOI] [PubMed] [Google Scholar]
- [14].Herbert FC, Brohlin OR, Galbraith T, Benjamin C, Reyes CA, Luzuriaga MA, Shahrivarkevishahi A, Gassensmith JJ, Bioconjugate Chem. 2020, 31, 1529–1536. [DOI] [PubMed] [Google Scholar]
- [15].Aljabali AAA, al Zoubi MS, Al-Batanyeh KM, Al-Radaideh A, Obeid MA, al Sharabi A, Alshaer W, AbuFares B, Al-Zanati T, Tambuwala MM, Akbar N, Evans DJ, Beilstein J Nanotechnol. 2019, 10, 1983–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu Z, Zhou J, Nkanga CI, Jin Z, He T, Borum RM, Yim W, Zhou J, Cheng Y, Xu M, Steinmetz NF, v Jokerst J, ACS Appl. Mater. Interfaces 2022, 14, 13692–13702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chariou PL, Ortega-Rivera OA, Steinmetz NF, ACS Nano 2020, 14, 2678–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chariou PL, Dogan AB, Welsh AG, Saidel GM, Baskaran H, Steinmetz NF, Nat. Nanotechnol. 2019, 14, 712–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cao J, Guenther RH, Sit TL, Lommel SA, Opperman CH, Willoughby JA, ACS Appl. Mater. Interfaces 2015, 7, 9546–9553. [DOI] [PubMed] [Google Scholar]
- [20].Caparco AA, Dautel DR, Champion JA, Small 2022, n/a, 2106425. [DOI] [PubMed] [Google Scholar]
- [21].Edwardson TGW, Hilvert D, J. Am. Chem. Soc. 2019, 141, 9432–9443. [DOI] [PubMed] [Google Scholar]
- [22].Das S, Zhao L, Elofson K, Finn MG, Biochemistry 2020, 59, 2870–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fiedler JD, Brown SD, Lau JL, Finn MG, Angew. Chem., Int. Ed. 2010, 49, 9648–9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cuenca S, Mansilla C, Aguado M, Yuste-Calvo C, Sánchez F, Sánchez-Montero JM, Ponz F, Front. Plant Sci. 2016, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Poghossian A, Jablonski M, Koch C, Bronder TS, Rolka D, Wege C, Schöning MJ, Biosens. Bioelectron. 2018, 110, 168–174. [DOI] [PubMed] [Google Scholar]
- [26].Masarapu H, Patel BK, Chariou PL, Hu H, Gulati NM, Carpenter BL, Ghiladi RA, Shukla S, Steinmetz NF, Biomacromolecules 2017, 18, 4141–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hu H, Steinmetz NF, Biotechnol. J. 2020, 15, 2000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hu H, Yang Q, Baroni S, Yang H, Aime S, Steinmetz NF, Nanoscale 2019, 11, 9760–9768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Beatty PH, Lewis JD, Adv. Drug Deliv. Rev. 2019, 145, 130–144. [DOI] [PubMed] [Google Scholar]
- [30].Tian Y, Zhou M, Shi H, Gao S, Xie G, Zhu M, Wu M, Chen J, Niu Z, Nano Lett. 2018, 18, 5453–5460. [DOI] [PubMed] [Google Scholar]
- [31].Liu T, Zhao X, Wang L, Yang J, Metavarayuth K, Lin Y, Yuan J, Wang Q, J. Biomed. Nanotech. 2019, 15, 363–372. [DOI] [PubMed] [Google Scholar]
- [32].Lin Y-Y, Schuphan J, Dickmeis C, Buhl EM, Commandeur U, Fischer H, Adv. Hearthc. Mater. 2020, 9, 2001245. [DOI] [PubMed] [Google Scholar]
- [33].Yuste-Calvo C, González-Gamboa I, Pacios LF, Sánchez F, Ponz F, ACS Omega 2019, 4, 5019–5028. [Google Scholar]
- [34].Shukla S, Roe AJ, Liu R, Veliz FA, Commandeur U, Wald DN, Steinmetz NF, Biomater. Sci. 2020, 8, 3935–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].González-Gamboa I, Manrique P, Sánchez F, Ponz F, J. Biotechnol. 2017, 254, 17–24. [DOI] [PubMed] [Google Scholar]
- [36].Wu FC, Zhang H, Zhou Q, Wu M, Ballard Z, Tian Y, Wang JY, Niu ZW, Huang Y, Chem. Commun. 2014, 50, 4007–4009. [DOI] [PubMed] [Google Scholar]
- [37].Yi L, Shi J, Gao S, Li S, Niu C, Xi Z, Tetrahedron Lett. 2009, 50, 759–762. [Google Scholar]
- [38].Smith ML, Lindbo JA, Dillard-Telm S, Brosio PM, Lasnik AB, McCormick AA, Nguyen L. v., Palmer KE, Virology 2006, 348, 475–488. [DOI] [PubMed] [Google Scholar]
- [39].Bruckman MA, Kaur G, Lee LA, Xie F, Sepulveda J, Breitenkamp R, Zhang X, Joralemon M, Russell TP, Emrick T, Wang Q, ChemBioChem 2008, 9, 519–523. [DOI] [PubMed] [Google Scholar]
- [40].Harrison BD, Wilson TMA, Klug A, Philos. Trans. R. Soc. Lond., B, Biol. Sci. 1999, 354, 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Charudattan R, Hiebert E, Currey W, Elliott M, Devalerio J, Maia G, Weeds 2020. [Google Scholar]
- [42].Charudattan R, Hiebert E, Outlooks on Pest Manag. 2007, 18, 167–171. [Google Scholar]
- [43].Chariou PL, Steinmetz NF, ACS Nano 2017, 11, 4719–4730. [DOI] [PubMed] [Google Scholar]
- [44].Shin MD, Hochberg JD, Pokorski JK, Steinmetz NF, ACS Appl. Mater. Interfaces 2021, 13, 59618–59632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chariou PL, Wang L, Desai C, Park J, Robbins LK, von Recum HA, Ghiladi RA, Steinmetz NF, Macromol. Biosci. 2019, 19, 1800407. [DOI] [PubMed] [Google Scholar]
- [46].Smith MT, Hawes AK, Bundy BC, Curr. Opin. Biotechnol. 2013, 24, 620–626. [DOI] [PubMed] [Google Scholar]
- [47].Le DHT, Hu H, Commandeur U, Steinmetz NF, J. Struct. Biol. 2017, 200, 360–368. [DOI] [PubMed] [Google Scholar]
- [48].Bruckman MA, Steinmetz NF, Virus Hybrids as Nanomaterials: Methods and Protocols (Eds.: Lin B, Ratna B), Humana Press, Totowa, NJ, 2014, pp. 173–185. [Google Scholar]
- [49].Spicer CD, Davis BG, Nat. Commun. 2014, 5, 4740. [DOI] [PubMed] [Google Scholar]
- [50].Schlick TL, Ding Z, Kovacs EW, Francis MB, J. Am. Chem. Soc. 2005, 127, 3718–3723. [DOI] [PubMed] [Google Scholar]
- [51].Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C, Nanoscale 2013, 5, 3808–3816. [DOI] [PubMed] [Google Scholar]
- [52].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE, J. Comput. Chem. 2004, 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
- [53].Kuhlman B, Bradley P, Nat. Rev. Mol. Cell Biol 2019, 20, 681–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Wang L, Gruzdys V, Pang N, Meng F, Sun X-L, RSC Adv. 2014, 4, 39446–39452. [Google Scholar]
- [55].McGaughey GB, Gagné M, Rappé AK, J. Biol. Chem. 1998, 273, 15458–15463. [DOI] [PubMed] [Google Scholar]
- [56].Boutureira O, Bernardes GJL, Chem. Rev. 2015, 115, 2174–2195. [DOI] [PubMed] [Google Scholar]
- [57].Cammarata CR, Hughes ME, Ofner CM, Mol. Pharm. 2015, 12, 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T, Mol. Cell Proteomics 2006, 5, 2336–2349. [DOI] [PubMed] [Google Scholar]
- [59].Hong V, Presolski SI, Ma C, Finn MG, Angew. Chem. Int. Ed. 2009, 48, 9879–9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rajeshkumar V, Courté M, Fichou D, Stuparu MC, Eur. J. Org. Chem. 2016, 2016, 6010–6014. [Google Scholar]
- [61].Pickens CJ, Johnson SN, Pressnall MM, Leon MA, Berkland CJ, Bioconjug. Chem. 2018, 29, 686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Rosen CB, Francis MB, Nat. Chem. Biol. 2017, 13, 697–705. [DOI] [PubMed] [Google Scholar]
- [63].Pricet M, J. Virol. 1993. [Google Scholar]
- [64].Jain A, Cheng K, J. Control. Release 2017, 245, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sun M, Liang Y, Li Y, Yang K, Zhao B, Yuan H, Li X, Zhang X, Liang Z, Shan Y, Zhang L, Zhang Y, Anal. Chem. 2020, 92, 567–572. [DOI] [PubMed] [Google Scholar]
- [66].Pickens CJ, Johnson SN, Pressnall MM, Leon MA, Berkland CJ, Bioconjugate Chem. 2018, 29, 686–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Bruckman MA, Steinmetz NF, Methods Mol. Biol. 2014, 1108, 173–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Taylor SW, Fahy E, Murray J, Capaldi RA, Ghosh SS, J. Biol. Chem. 2003, 278, 19587–19590. [DOI] [PubMed] [Google Scholar]
- [69].Liu H, Ponniah G, Neill A, Patel R, Andrien B, Anal. Chem. 2013, 85, 11705–11709. [DOI] [PubMed] [Google Scholar]
- [70].Kim M-S, Zhong J, Pandey A, PROTEOMICS 2016, 16, 700–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Piersma SR, Warmoes MO, de Wit M, de Reus I, Knol JC, Jiménez CR, Proteome Sci. 2013, 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


