Abstract
Exposure to traffic-related pollutants, including diesel exhaust, is associated with increased risk of cardiopulmonary disease and mortality; however, the precise biochemical pathways underlying these effects are not known. To investigate biological response mechanisms underlying exposure to traffic related pollutants, we used an integrated molecular response approach that included high-resolution metabolomic profiling and peripheral blood gene expression to identify biological responses to diesel exhaust exposure. Plasma samples were collected from 73 non-smoking males employed in the US trucking industry between February 2009 and October 2010, and analyzed using untargeted high-resolution metabolomics to characterize association with shift- and week-averaged levels of elemental carbon (EC), organic carbon (OC) and particulate matter with diameter ≤ 2.5 μm (PM2.5). Metabolic associations with EC, OC and PM2.5 were evaluated for biochemical processes known to be associated with disease risk. Annotated metabolites associated with exposure were then tested for relationships with the peripheral blood transcriptome using multivariate selection and network correlation. Week-averaged EC and OC levels, which were averaged across multiple shifts during the workweek, resulted in the greatest exposure-associated metabolic alterations compared to shift-averaged exposure levels. Metabolic changes associated with EC exposure suggest increased lipid peroxidation products, biomarkers of oxidative stress, thrombotic signaling lipids, and metabolites associated with endothelial dysfunction from altered nitric oxide metabolism, while OC exposures were associated with antioxidants, oxidative stress biomarkers and critical intermediates in nitric oxide production. Correlation with whole blood RNA gene expression provided additional evidence of changes in processes related to endothelial function, immune response, inflammation, and oxidative stress. We did not detect metabolic associations with PM2.5. This study provides an integrated molecular assessment of human exposure to traffic-related air pollutants that includes diesel exhaust. Metabolite and transcriptomic changes associated with exposure to EC and OC are consistent with increased risk of cardiovascular diseases and the adverse health effects of traffic-related air pollution.
Keywords: Metabolomics, Exposomics, High-resolution mass spectrometry, Multi-omics, Traffic-related pollutants, Diesel exhaust
Graphical Abstract
INTRODUCTION
Exposure to traffic-related pollution has been linked to increased risk of all-cause mortality, cardiovascular disease (CVD), cardiopulmonary outcomes and lung cancer mortality, (Hart 2016; Pope et al. 2002; Pope et al. 2009) with 2010 estimates suggesting 3.1 million deaths were attributable to ambient particulate exposure alone. (Lim et al. 2012) Suspected mechanisms that contribute to disease risk include increased oxidative stress, endothelial dysfunction and inflammation; however, study in human populations has shown conflicting results. (Bates et al. 2015b; Brugge et al. 2013; Chuang et al. 2007; Donaldson et al. 2001; Sorensen et al. 2003)
Exhaust emissions consist of a complex mixture of particle-bound and free chemicals. Source, fuel type, combustion efficiency and distance from the source contribute to both the individual constituents and composition of particulate matter present in traffic-related air pollution. However, in general, combustion related particulate matter includes ultrafine particles with an elemental carbon core (EC) and a particulate organic carbon (OC) fraction that originates both from chemicals produced during incomplete combustion as well as condensation of organic volatile gases on the particle surface during atmospheric transport. Evidence suggests the composition of particulate matter, specifically EC, are a robust indicator of combustion-based exposures from traffic. (Janssen et al. 2012) EC is a major component of carbonaceous particulates arising from diesel exhaust lacking modern emission controls and is also present to a lesser extent in exhaust from other mobile sources, including spark-ignition vehicles. (Costantini et al. 2016; Grahame et al. 2014; Schauer 2003; Sheesley et al. 2009) Several studies have shown using EC as a measure of traffic-related exposures results in higher effect estimates associated with cardiopulmonary disease when compared to PM2.5 (particulate matter with diameter ≤ 2.5 μm); however, these findings have not been consistent across different populations and it is not clear if EC is independently associated with outcomes or an indirect measure of particle mass. (Beelen et al. 2008; Lipfert et al. 2006; Luben et al. 2017; Ostro et al. 2007; Sarnat et al. 2015) Cumulative EC exposure based upon 5- and 1-year lag periods has been associated with lung cancer mortality in trucking industry workers in the US, (Garshick et al. 2008) while the International Agency for Research on Cancer has classified diesel exhaust as a Group 1 carcinogen. (Benbrahim-Tallaa et al. 2012) Although health risks of EC have been documented, the contribution of EC and other traffic-related pollutants such as particle associated organic chemicals (assessed by measuring OC) to metabolic responses has not been well characterized.
The human metabolome is a functional measure of the interaction between the genome, diet, environment, and metabolism. (Wishart et al. 2013) Identifying metabolic changes associated with environmental exposures has the potential to improve understanding of response to exposures, providing new insight into the biological changes underlying exposure-related diseases. Untargeted chemical profiling techniques based upon high resolution mass spectrometry now make possible measurement of up to 20,000 chemical features, providing sufficient metabolic characterization for precision medicine and exposome research. (Liu et al. 2016) When combined with complementary response measures, such as gene expression, protein levels or epigenetic changes, it is possible to develop an in vivo, systems biology approach to investigate how environmental exposures contribute to disease risk in humans. Development of aggregated biological response patterns across multiple ‘omic layers represents a new paradigm for combining toxicology with molecular and environmental epidemiology.
In the present study, we used untargeted high-resolution metabolomics (HRM) and whole blood RNA gene expression to characterize the integrated molecular response of US trucking industry workers following occupational exposure to diesel exhaust. Using exposure levels measured at the workplace and blood samples collected at multiple time points throughout the workweek, we evaluated metabolic alterations associated with shift- and week-averaged EC and organic carbon (OC) in particles <1 μm in diameter (PM1.0), and fine particulate matter (PM2.5). Using metabolic changes associated with week-averaged exposures, we applied a data-driven network approach to identify correlating gene expression pathways. Our objective was to provide an integrated assessment of human molecular response to traffic-related pollutants and identify biological pathways that may underlie the observed effects of these exposures.
METHODS
Study population
Details on the study population, recruitment and sample collection have been reported previously. (Neophytou et al. 2013; Neophytou et al. 2014) Briefly, 95 participants were recruited from 10 unionized trucking terminals located throughout the northeastern US (Connecticut, Massachusetts, Maryland, New Jersey, New York and Pennsylvania), and classified based upon job duties with different patterns of roadway traffic and freight terminal exposures from working in proximity to diesel powered trucks. Job classifications include pickup and delivery drivers, freight dock workers, and office clerks. Participants were selected for enrollment in the study if they had at least two days off prior to the start of the study. Measurements and sample collection were completed between February 2009 and October 2010, with terminals sampled one at a time for up to 8 continuous days. To reduce confounding due to smoking and sex differences, the present analysis was restricted to 73 non-smoking, male workers. All participants provided informed consent, and the study protocol was approved by the Institutional Review Board of the Brigham and Women’s Hospital and the Human Subject Committee of the Harvard T.H. Chan School of Public Health.
Exposure assessment
Stationary samplers for exposure assessment were placed at indoor office spaces, terminal docks and within truck cabs for all 10 terminal locations during the full workweek (6–9 days). Exposure measures of traffic-related pollutants included PM2.5, and EC and OC in PM1.0. Collection protocols and detailed methodology on assigning exposures are described elsewhere. (Neophytou et al. 2013) PM2.5 was collected on a 37-mm Teflon filter (Gelman/Pall, Port Washington, NY, USA) after passing through a precision-machined cyclone pre-selector (GK2–5-SH, BGI, Inc., Waltham, MA USA) to remove particles greater than 2.5 μm in aerodynamic diameter. The method was consistent with the EPA PQ200 Federal Reference Method. (Tainio et al. 2005; Yanosky and MacIntosh 2001) EC and OC were measured by collecting PM1.0 on a 22-mm quartz tissue filter preceded by precision machined cyclone separator (SCC1.062 Triplex, BGI Inc., Waltham, MA USA), which was then analyzed by thermal-optical carbon analyzer using the NIOSH 5040 method. (NIOSH 2003) Individualized, personal exposures were then estimated for each participant using a weighted average of the reported time spent at each work location on each workday.
Biomarker sampling
A total of four blood samples was collected from each participant over the course of the workweek. The first blood sample was collected from each participant prior to the day’s work shift on their first day back after at least two days off, followed by a second blood sample at the end of the first work shift. Pre- and post-shift samples were collected again on the last day of the same workweek. Following each blood draw, blood tubes were stored at 4°C until processing. EDTA plasma samples for metabolomic analysis were centrifuged, aliquoted, and stored in the vapor phase of liquid nitrogen freezers at < −130°C. RNA was extracted from PaxGene tubes using the Qiagen RNAEasy kit and stored at −80°C.
High-resolution metabolomics
Untargeted HRM profiling was completed using verified protocols. (Accardi et al. 2016a; Go et al. 2015) Plasma aliquots were removed from storage and thawed on ice. A 65 μL aliquot of plasma was then added to 130 μL of acetonitrile containing a mixture of stable isotopic standards that included [13C6]-D-glucose, [15N]-indole, [2-15N]-L-lysine dihydrochloride, [13C5]-L-glutamic acid, [13C7]-benzoic acid, [3,4-13C2]-cholesterol, [15N]-L-tyrosine, [trimethyl-13C3]-caffeine, [15N2]-uracil, [3,3-13C2]-cystine, [1,2-13C2]-palmitic acid, [15N,13C5]-L-methionine, [15N]-choline chloride, and 2’-deoxyguanosine-15N2,13C10-5’-monophosphate, vortexed, and allowed to equilibrate for 30 minutes. (Soltow et al. 2013) Triplicate 10 μL aliquots were analyzed by reverse-phase C18 liquid chromatography (Targa 100 mm x 2.1mm x 2.6 μm, Higgins Analytical Inc) with detection by a Thermo Scientific Q-Exactive high-resolution mass spectrometer. Analyte separation was accomplished using water, acetonitrile and 2% [v/v] formic acid in water (solution A) mobile phases operating under the following gradient: initial 2 min period of 80% A, 5% water, 15% acetonitrile, followed by linear increase to 0% A, 5% water, 95% acetonitrile at 6 min and then held for an additional 4 min. Mobile phase flow rate was held at 0.35 mL/min for 6 min, and then increased to 0.5 mL/min. The high-resolution mass spectrometer was equipped with an electrospray ionization source operated in positive ion mode with spray voltage of 4.5 kV, probe, capillary temperature 275°C, sheath gas flow 45 (arbitrary units), auxiliary gas flow 5 (arbitrary units) and S-lens RF level of 69. Resolution was set at 70,000 (FWHM) and mass-to-charge (m/z) scan range 85–1275. Spectra were collected in full scan only without MSMS. Samples were analyzed in batches of 20, in addition to a quality control (QC) pooled reference sample included at the beginning and end of each batch of samples to evaluate batch effects and reproducibility of the detected metabolite features.
Upon injection of all study and quality control samples, mass spectral features with replicate coefficient of variation (CV) ≤ 100% were extracted and aligned using apLCMS (Yu et al. 2013) with modifications by xMSanalyzer (Uppal et al. 2013) and batch effect correction by ComBat (Johnson et al. 2007). Detected chemical signals were defined by accurate mass-to-charge ratio (m/z), retention time and intensity, referred to as metabolite features throughout. Prior to statistical analysis, replicate injections were averaged, and metabolite features not detected in >50% of the participants were removed, resulting in 7,042 and 7,035 metabolite features for the shift- and week-averaged analyses, respectively. All participant covariates and untargeted metabolomics data underlying this article are available under Study ID ST001930 in the Metabolomics Workbench at http://dx.doi.org/10.21228/M84Q3H.
Whole blood RNA gene expression
Gene expression associations with PM2.5, EC and OC in this cohort have been described previously. (Chu et al. 2016) Analysis was conducted using the Illumina HumanHT-12 v4 Expression BeadChip, with RNA labeling and array hybridization performed according to protocol. Image capture was performed using the Illumina BeadArray Reader. Standard QC and pre-processing procedures were applied, with background correction and quantile normalization procedures completed using the R package lumi. (Du et al. 2008) The final data set was log2 transformed prior to network integration and included information for 47,295 probes. Previous analysis of gene expression profiles from this cohort show no significant differences were present when comparing pre- and post-shift gene expression on day 1, or across post-shift samples on days 1 and 4. (Chu et al. 2016) Thus, to reduce adding any additional variability due to diurnal effects and simplify interpretation of correlation results only post-shift measurements from both days were considered.
Metabolome wide-association study of traffic-related pollutants
Due to the availability of repeat HRM and exposure measures in this study, we completed metabolome-wide association study (MWAS) of PM2.5, EC and OC separately for shift- and week-averaged workplace exposures. The association of each metabolite feature with PM2.5, EC and OC were determined using linear mixed effects regression models, which included a random intercept for each subject to account for baseline inter-individual differences. Fixed effects for both models included the interquartile range (IQR) normalized exposure measure, age, day (factor for first and last day of the workweek), and body mass index (BMI, calculated from measured height and weight). The effect of day as a continuous variable and race was also evaluated; however, accounting for time differences between blood sample collection or race as a fixed factor in the regression models did not impact MWAS results and were excluded as covariates. We first tested for daily associations with PM2.5, EC and OC using metabolite feature measured in post-shift blood samples collected on both days and the corresponding exposures measured over the course of that workday. Week-averaged effects were evaluated using the average exposure calculated using the first and last shift for each participant and metabolomic results measured in blood samples collected at all time points. To account for diurnal effects, we included sampling time (pre- or post-shift) as an additional covariate to allow metabolite peak intensity to vary with time irrespective of exposure measures. Regression analysis was performed using the R package lme4 and completed separately for each exposure and time combination. (Bates et al. 2015a) The likelihood ratio test of the complete model against the null model, which excluded the corresponding exposure measures as the independent variable, was used to obtain p for each metabolite feature-exposure pair. Model fits were evaluated by calculating the conditional and marginal r2. To account for multiple hypothesis testing and estimate the expected proportion of incorrectly rejected null hypotheses (i.e. false positives), a Benjamini-Hochberg false discovery rate (FDR) was applied to identify metabolite features associated with each pollutant. (Benjamini and Hochberg 1995) Associated metabolite features were identified using an FDR threshold of 20% to balance the rate of type I and II errors, which when combined with dose-response linear regression and metabolic pathway enrichment has been shown to improve detection of biological activity and reduce false positives. (Li et al. 2013; Walker et al. 2016)
Metabolite annotation and pathway enrichment
The metabolite features associated with each exposure were first matched using accurate m/z detected using MS1 and retention time to a database of 120 metabolites previously confirmed with MS2 and co-elution studies (Level 1 identification). (Accardi et al. 2016b; Liu et al. 2020; Schymanski et al. 2014) To generate additional biological insight into effects of exposure EC, OC and PM2.5, metabolite features not matching the database of 120 metabolites were annotated using MS1 detected accurate m/z based upon positive electrospray ionization adducts and the Human Metabolome Database (HMDB) release 3.0. (Wishart et al. 2013) Metabolite annotations were assigned using evidence scoring (Uppal et al. 2016a) and ± 10 parts-per-million (ppm) mass tolerance (Δmerror /mtheoretical × 106). While these annotations provide low confidence matches (Level 4 annotation) that predict chemical formulas only and do not inform on compound structures, combining low confidence annotations with functional activity patterns within the untargeted data improves the ability to uncover novel insight into metabolite response profiles to exposures. (Uppal et al. 2016b) Thus, we combined annotation of features with pathway-based functional enrichment analysis using Mummichog to characterize metabolic activity patterns associated with each exposure. (Li et al. 2013) Enriched metabolite pathways were selected using a Mummichog scoring threshold ≤ 0.05.
Metabolome x transcriptome exposure response pathways
To evaluate integrated metabolic and gene expression patterns associated with exposure to traffic-related pollutants, we performed a network-based correlation analysis using xMWAS to characterize relationships between annotated metabolites associated with week-averaged exposures and peripheral blood transcriptomic results. (Uppal et al. 2018) Correlations with |r| ≥ 0.4 and p< 0.05 were then selected for visualization in Cytoscape, with gene and metabolite pathway enrichment determined using MetaCore (Thomson Reuters) and Mummichog, respectively. An enrichment score of 0.1 was used to identify metabolite pathways and p <0.05 for gene expression pathways. Previously reported relationships between gene pathways present in the integrated network and exposure to traffic related pollutants were evaluated using the Comparative Toxicogenomics Database (CTD). (Davis et al. 2013) To identify previous gene-exposure relationships, we used the top two most connected gene nodes from each cluster for searching reported gene-chemical interactions within CTD.
RESULTS
Study population
A summary of participant characteristics is provided in Table 1. HRM profiling was limited to 73 individuals who were otherwise healthy, non-smoking males from all three-job categories with blood samples available on both sampling days. Comparison of exposure levels at the beginning and end of the workweek showed no differences between the two workdays (p >0.7). Week-averaged exposures showed a weak correlation between PM2.5 and EC (Pearson r= 0.39, p= 0.0007) and between PM2.5 and OC (Pearson r= 0.27, p= 0.021), while no correlation was present between EC and OC (Pearson r= 0.03, p= 0.78). Dockworkers and pickup and delivery drivers had higher PM2.5 and EC exposures compared to office clerks, while OC was lowest among dockworkers. (Neophytou et al. 2013; Neophytou et al. 2014) Participant shift-and week-averaged exposure levels for EC, OC and PM2.5 are provided in Supplementary Figure 1.
Table 1.
Population characteristics of 73 male unionized trucking industry workers included in HRM profiling
| Characteristic | Total |
|---|---|
| Number of individuals | 73 |
| Age (years, mean±SD) | 49.8 ± 8.3 |
| BMI (mean±SD) | 29.8 ± 4.5 |
| Race (n, (%)) | |
| White | 68 (93%) |
| Hispanic | 5 (7%) |
| Primary job title (n, (%)) | |
| Pick-up and delivery driver | 38 (52%) |
| Dockworker | 14 (19%) |
| Officeworker | 21 (29%) |
| Average first day of work exposures (mean±SD) | |
| PM2.5 (μg/m3) | 10.0 ± 5.7 |
| EC (μg/m3) | 0.6 ± 0.5 |
| OC (μg/m3) | 8.8 ± 2.9 |
| Average last day of work exposures (mean±SD) | |
| PM2.5 (μg/m3) | 9.7 ± 4.8 |
| EC (μg/m3) | 0.6 ± 0.4 |
| OC (μg/m3) | 8.6 ± 4.3 |
| Average work-week exposures (mean±SD) | |
| PM2.5 (μg/m3) | 10.0 ± 4.5 |
| EC (μg/m3) | 0.6 ± 0.4 |
| OC (μg/m3) | 8.7 ± 3.1 |
| Median work-week exposures (median, IQR) | |
| PM2.5 (μg/m3) | 9.7, 6.3 |
| EC (μg/m3) | 0.5, 0.6 |
| OC (μg/m3) | 8.9, 4.0 |
Shift-averaged exposure MWAS
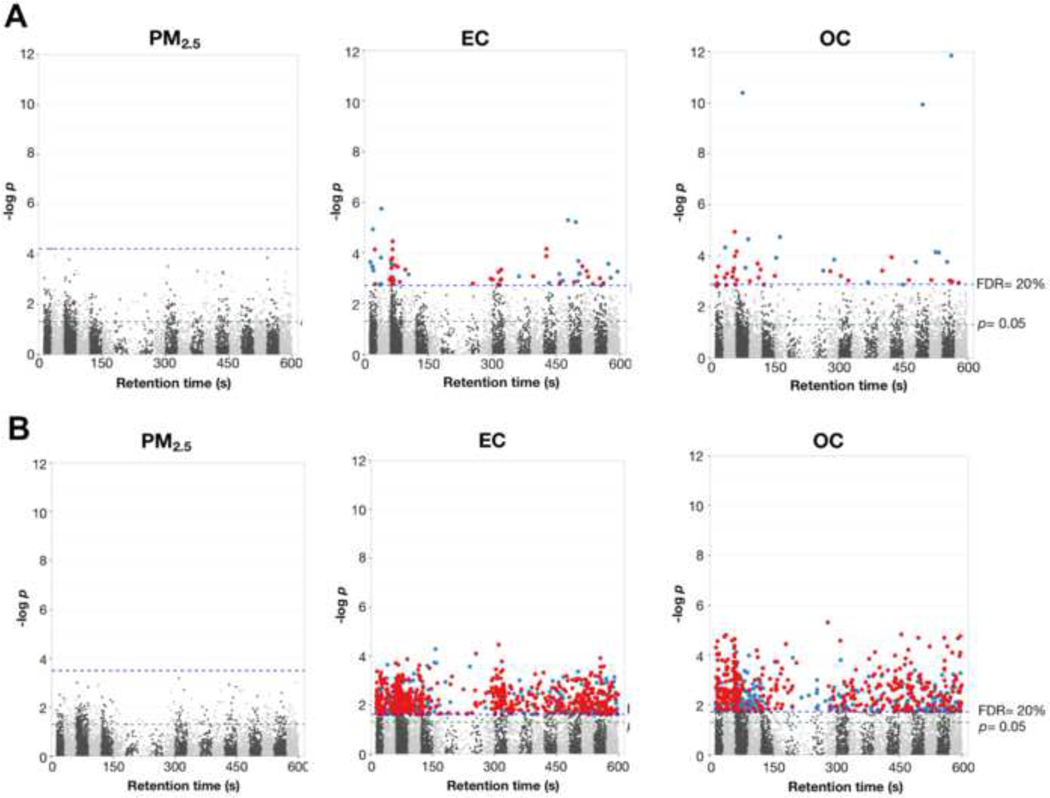
To evaluate acute effects of exposure to EC, OC and PM2.5, we used an MWAS framework to test the relationship between shift-averaged exposures and metabolite features measured in post-shift samples obtained on both workdays (Figure 1A). Using an FDR threshold of 20%, 68 and 48 unique m/z features were associated with EC and OC, respectively (Supplementary Tables 1 and 2). Only 3 metabolites were identified at level 1, the remaining were annotations based upon accurate mass matching only. Annotated metabolites associated with EC included metabolic intermediates related to inflammation, endothelial function, lipid peroxidation and co-factors (Table 2); enriched pathways included butanoate metabolism (p= 0.01) and urea cycle/amino group metabolism (p= 0.04). For OC, metabolite annotations were consistent with nucleotide metabolism, porphyrin metabolites and lipid peroxidation products (Table 2), and enriched pathways included changes in selenoamino acid metabolism (p= 0.003), glycerophospholipids (p= 0.003), pyruvate metabolism (p= 0.009), serine metabolism (p= 0.01), glycolysis/gluconeogenesis (p= 0.03) and arginine/proline metabolism (p= 0.03). None of the metabolite feature intensities were associated with shift PM2.5 at FDR<20%.
Figure 1:
Manhattan plot showing −log p as a function m/z feature retention time for A) daily and B) week-average exposure to PM2.5, EC and OC. MWAS was completed using linear mixed effects models and post-shift metabolite intensity only. Fixed effects included daily shift length exposure, age, day and BMI. Red= positive association; Blue= negative association
Table 2.
Annotated metabolites related to oxidative stress and endothelial function associated with shift-averaged exposure to elemental carbon (EC) and organic carbon (OC) detected in plasma from 73 male unionized trucking industry workers.
| m/z | Associated exposure | Identity | Number of matches1 | Annotation confidence2 | Exposure β (95% CI) | p-value |
|---|---|---|---|---|---|---|
| 155.1067 | EC | 4-oxo-2-Nonenal | 13 | 4 | −0.9 (−1.3, −0.5) | 4.88E-06 |
| 389.1350 | EC | 16-Hydroxy-4-carboxyretinoic acid | 15 | 4 | −2.4 (−3.7, −1.0) | 1.09E-03 |
| 105.0341 | EC | Benzoic acid | 1 | 1 | −3.7 (−5.8, −1.5) | 8.18E-04 |
| 167.9929 | EC | 2-Mercaptobenzothiazole | 1 | 4 | 2.3 (0.9, 3.7) | 1.61E-03 |
| 422.2357 | EC | Leukotriene E4 | 16 | 4 | 3.2 (1.4, 5.1) | 8.66E-04 |
| 223.1054 | EC | Dihydrobiopterin | 2 | 4 | 3.4 (1.5, 5.5) | 1.15E-03 |
| 854.373 | EC | Heme A | 1 | 4 | 3.9 (2.0,5.8) | 7.20E-05 |
| 130.0337 | OC | Methionine-sulfoxide | 1 | 1 | −1.8 (−2.2, −1.3) | 1.40E-12 |
| 311.1286 | OC | Porphyrin | 25 | 4 | −1.8 (−2.7, −1.0) | 7.10E-05 |
| 136.0622 | OC | Adenine | 2 | 4 | −2.4 (−3.8, −1.0) | 1.05E-03 |
| 157.1224 | OC | 4-hydroxynonenal | 28 | 4 | −2.8 (−4.2, −1.4) | 1.65E-04 |
| 589.2770 | OC | Mesoporphyrin | 2 | 4 | 2.7 (1.1, 4.3) | 1.35E-03 |
| 402.0927 | OC | Lactoylglutathione | 11 | 4 | 3.3 (1.6, 5.1) | 2.50E-04 |
| 107.0537 | OC | Serine | 1 | 1 | 3.5 (1.7, 5.3) | 2.00E-04 |
| 132.0657 | OC | L-Glutamate-5-semialdehyde | 19 | 4 | 3.9 (1.8, 6.1) | 3.70E-04 |
Total number of compounds matching accurate mass m/z using HMDB 3.0 at 10 ppm mass accuracy.
Identification confidence based upon Schymanski et al. (2014). Level 4 confidence levels are based upon accurate mass matching only, and do not inform on compound structures. The full list of metabolite matches from HMDB 3.0 are provided in Supplementary Table 1 (EC) and Supplementary Table 2 (OC).
Week-averaged exposure MWAS
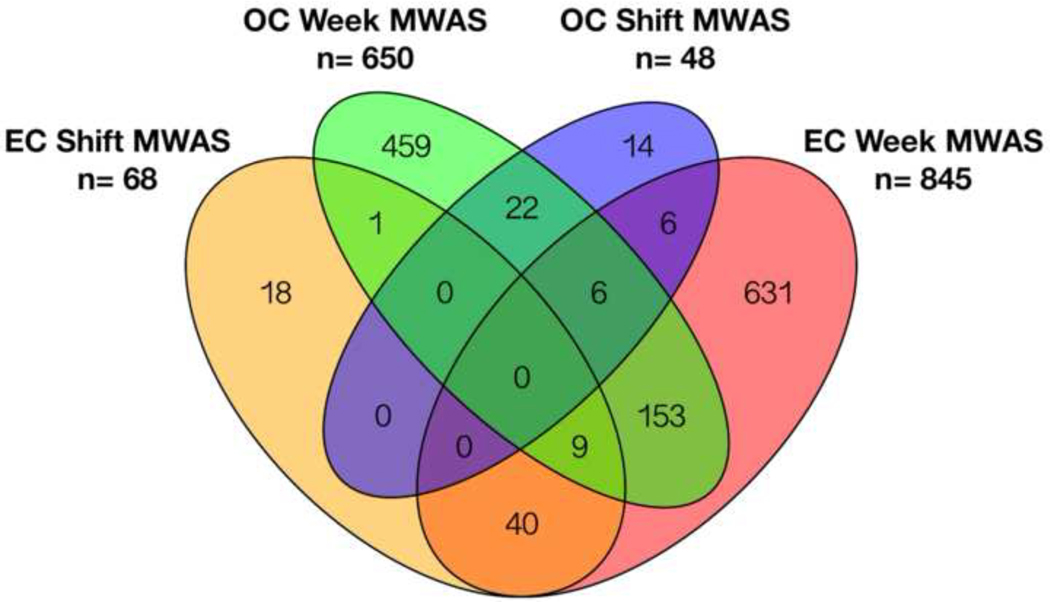
Previous results in this cohort have shown changes in gene expression, inflammatory markers, oxidative stress and urinary exposure biomarkers are largely dependent on week-averaged, rather than shift-averaged exposures. (Chu et al. 2016; Neophytou et al. 2013; Neophytou et al. 2014) Therefore, we next performed MWAS for PM2.5, EC and OC using plasma samples collected at all timepoints and averaged exposures for the workweek (Figure 1B). Using FDR <20%, 845 and 650 metabolite features were associated with week-averaged EC and OC levels, respectively; none met the FDR criteria for PM2.5 (Supplementary Tables 3 and 4). Comparison of the results for week-averaged MWAS showed 168 metabolite features were associated with both EC and OC. For EC 49 metabolite features were associated with both shift- and week-averaged exposure levels, while OC included 28 metabolite features associated with both exposure time periods. (Figure 2). The majority of the metabolite features showed a positive association with exposure at FDR<20%, including 634 for EC and 384 for OC that increased with increasing exposure levels.
Figure 2:
Overlapping features associated with shift-length and week averaged exposure to the two pollutants at false discovery rate threshold 20%.
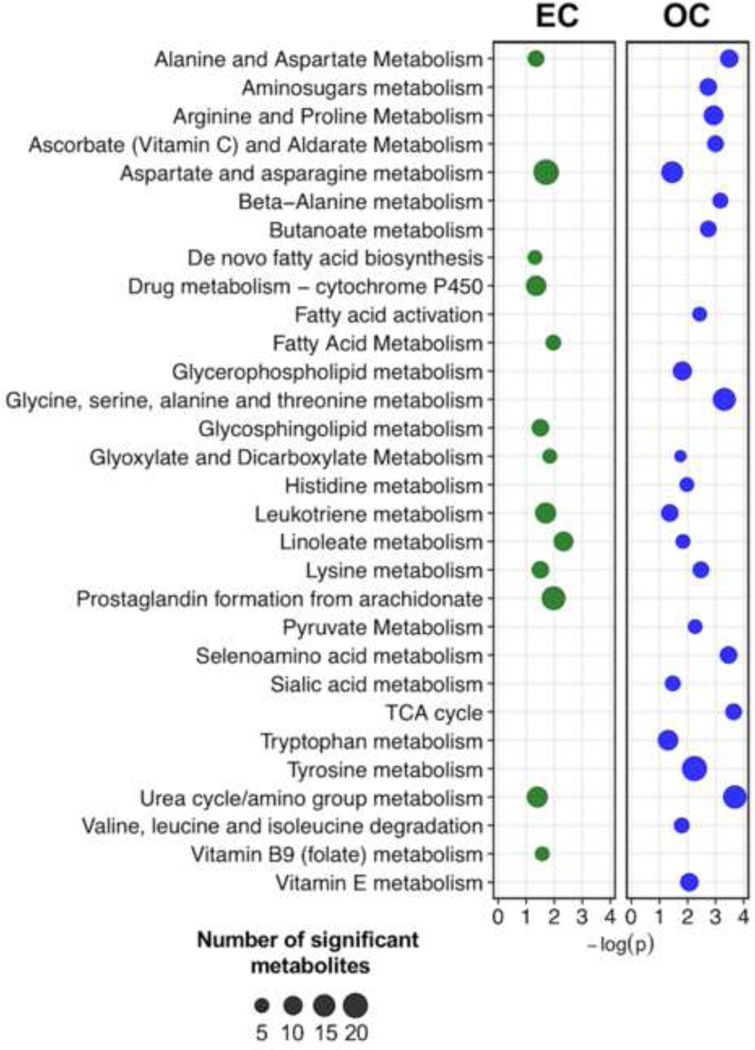
Metabolite identification for both exposures was first completed by comparison to a reference database of confirmed metabolites (Level 1 identification), which was then complemented with annotation by accurate mass matching and confidence scoring, which allows prediction of chemical formulas and potential metabolite structures (Level 4 annotation). Annotated endogenous metabolites associated with week-averaged EC included lipid peroxidation products, oxidative stress biomarkers, endothelial function-related metabolites and co-factors (Table 3). Lipid peroxidation products and oxidative stress metabolites included 4-oxo-2-nonenal, glutathione and 12-oxo-10E-octadecenoic acid. Pathway enrichment identified 13 metabolic pathways, including changes to fatty acid metabolism, pro-inflammatory lipid signaling, co-factor metabolism and nitrogen catabolism (Figure 3). Only urea cycle/amino group metabolism was also associated with shift-averaged EC.
Table 3.
Annotated metabolites related to oxidative stress, endothelial function and cofactor metabolism associated with week-averaged exposure to elemental carbon (EC) detected in plasma from 73 male unionized trucking industry workers.
| m/z | Identity | Number of Matches1 | Identification confidence2 | Change per IQR (95% CI) | p |
|---|---|---|---|---|---|
| 196.1335 | 4-oxo-2-nonenal | 3 | 4 | 0.5 (0.1, 0.9) | 1.5E-02 |
| 263.2366 | Linoleic acid | 1 | 1 | 1.6 (0.5, 2.8) | 6.9E-03 |
| 297.2410 | 12-oxo-10E-octadecenoic acid | 8 | 4 | 1.8 (0.6, 2.9) | 3.2E-03 |
| 371.1007 | Glutathione | 11 | 4 | 2.4 (0.7, 4.1) | 6.0E-03 |
| 177.0400 | Ascorbic acid | 1 | 1 | 3.9 (1.9, 5.9) | 2.3E-04 |
| 351.2144 | Prostaglandin I2 | 19 | 4 | 1.7 (0.3, 3.1) | 1.7E-02 |
| 335.2214 | Thromboxane B2 | 5 | 4 | 2.6 (.5, 4.8) | 1.5E-01 |
| 375.2141 | Thromboxane A2 | 6 | 4 | 2.7 (0.8, 4.5) | 5.8E-03 |
| 624.2953 | Hepoxilin A3-C | 7 | 4 | 2.9 (1.0, 4.8) | 3.6E-03 |
| 854.3730 | Heme A | 1 | 4 | 3.5 (1.7, 5.2) | 2.2E-04 |
| 227.1097 | Hydroxy-L-homoarginine | 5 | 4 | 2.6 (0.8, 4.4) | 6.3E-03 |
| 169.0592 | Glutamine | 1 | 1 | −0.5 (−1.0, −0.1) | 1.7E-02 |
| 156.0278 | Aspartic acid | 1 | 1 | 2.7 (0.6, 4.8) | 1.1E-02 |
| 445.1649 | Dihydrofolic acid | 1 | 1 | −2.2 (−3.7, −0.7) | 5.4E-03 |
| 459.1986 | Riboflavin | 12 | 4 | −3.8 (−6.0, −1.6) | 8.2E-04 |
Total number of compounds matching accurate mass m/z using HMDB 3.0 at 10 ppm mass accuracy.
Identification confidence based upon Schymanski et al. (2014). Level 4 confidence levels are based upon accurate mass matching only, and do not inform on compound structures. The full list of metabolite matches from HMDB 3.0 are provided in Supplementary Table 3.
Figure 3:
Metabolic pathways associated with week-averaged exposure to elemental and organic carbon. Dot size is a function of the number of metabolites from that pathway associated with the corresponding exposure measure. Only pathways with P<0.05 were considered.
Annotation of metabolite features associated with week-averaged OC show changes to key metabolic intermediates, including co-factor metabolism, increased oxidative stress, and disruption to nitric oxide (NO) production and endothelial function (Table 4). Pathway enrichment identified 23 pathways associated with week-averaged OC, which were related to endothelial function, co-factor metabolism, inflammatory signaling, nitrogen catabolism, mitochondrial bioenergetics and amino acid metabolism (Figure 3). Pathways including selenoamino acid metabolism, serine metabolism, arginine/proline metabolism, pyruvate metabolism and glycerphospholipid were associated with both shift- and week-averaged OC exposure.
Table 4.
Annotated metabolites related to oxidative stress, endothelial function and cofactor metabolism associated with week-averaged exposure to organic carbon (OC) detected in plasma from 73 male unionized trucking industry workers.
| m/z | Identity | Number of matches1 | Identification confidence2 | Change per IQR (95% CI) | p |
|---|---|---|---|---|---|
| 221.0940 | Hippuric acid | 1 | −2.5 (−4.2, − 0.8) | 5.2E-03 | |
| 204.0086 | Methionine sulfoxide | 1 | 1 | 0.8 (0.3, 1.4) | 3.5E-03 |
| 402.0921 | Lactoylglutathione | 11 | 4 | 3.4 (1.0, 5.7) | 5.1E-03 |
| 177.0691 | Homocysteine | 1 | 1 | −2.4 (−4.3, −0.4) | 1.7E-02 |
| 156.1021 | 4-hydroxyhexenal | 16 | 4 | −2.6 (−4.5, −0.6) | 9.3E-03 |
| 214.0578 | Citrulline | 1 | 1 | −0.7 (−1.2, −0.1) | 1.6E-02 |
| 303.1179 | Dihydrobiopterin | 5 | 4 | −3.8 (−5.6, −2) | 9.8E-05 |
| 335.2214 | Thromboxane B2 | 5 | 4 | −3.8 (−6.2, −1.4) | 2.4E-03 |
| 227.1097 | Hydroxy-Lhomoarginine | 5 | 4 | −3.9 (−5.9, 1.8) | 4.2E-04 |
| 169.0860 | Agmatine | 11 | 4 | −4.2 (−6.4, −1.9) | 3.8E-04 |
| 209.0081 | Ornithine | 4 | 4 | 1.3 (0.3, 2.2) | 1.1E-02 |
| 217.1045 | Acetylagmatine | 1 | 4 | 1.9 (0.4, 3.3) | 1.1E-02 |
| 229.0682 | N-(omega)-Hydroxyarginine | 8 | 4 | 2.7 (0.9, 4.4) | 2.9E-03 |
| 132.0657 | Lactate | 19 | 4 | 3.1 (1.2, 4.9) | 1.7E-03 |
| 195.9779 | Threonine | 1 | 1 | −3.7 (−5.6, −1.9) | 1.5E-04 |
| 156.0278 | Aspartic acid | 1 | 1 | −3.6 (−6, −1.2) | 4.2E-03 |
| 216.1960 | alpha-Tocopherol | 4 | 4 | 1.9 (0.6, 3.2) | 6.1E-03 |
| 134.0635 | Asparagine | 1 | 1 | 2.6 (1, 4.2) | 1.7E-03 |
| 107.0537 | Serine | 1 | 1 | 4.1 (2.2, 6) | 6.1E-05 |
| 202.1074 | Pantothenic acid | 17 | 4 | 3.9 (1.6, 6.3) | 1.4E-03 |
Total number of compounds matching accurate mass m/z using HMDB 3.0 at 10 ppm mass accuracy.
Identification confidence based upon Schymanski et al. (2014). Level 4 confidence levels are based upon accurate mass matching only, and do not inform on compound structures. The full list of metabolite matches from HMDB 3.0 are provided in Supplementary Table 4.
Network correlation of molecular response
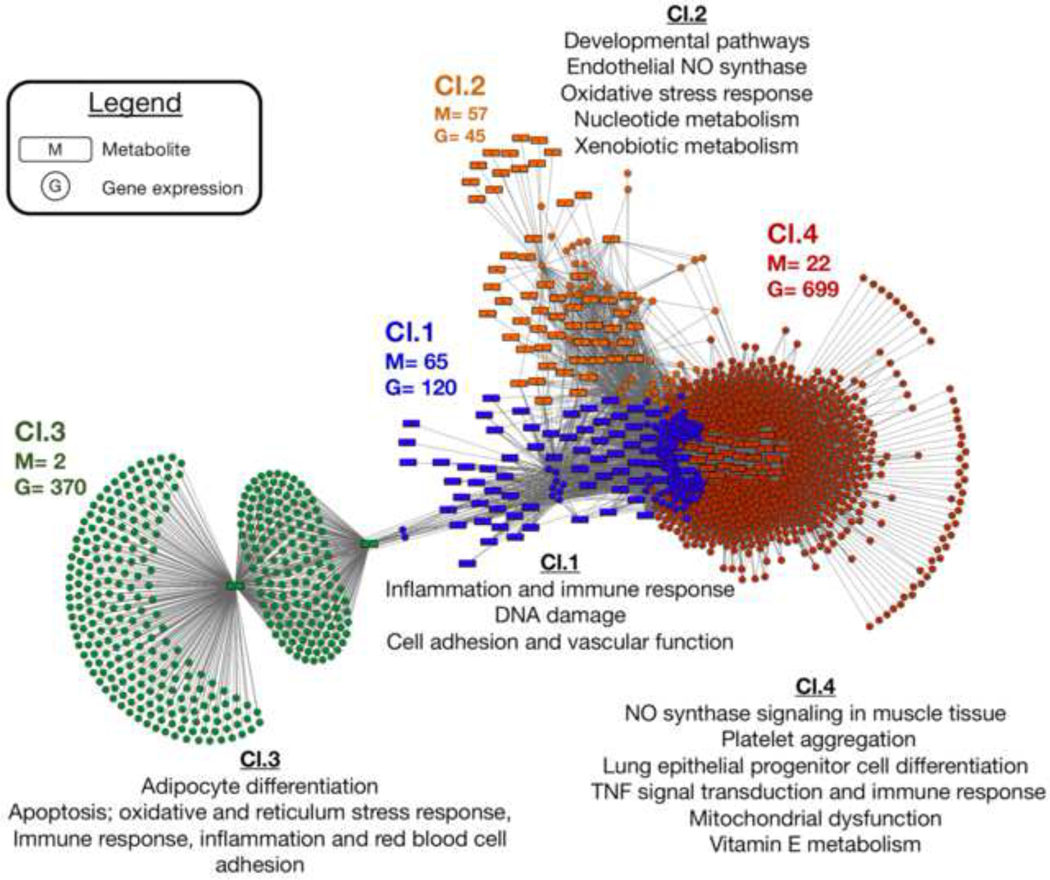
We used an integrative network approach developed for combining multi-omic datasets to identify correlated gene and metabolite pathways associated with week-averaged EC and OC exposure (Figure 4). A high degree of connectivity was observed for the two datasets, resulting in 1,234 molecular probes and 146 metabolites with |r| ≥ 0.4 and p≤ 0.05. Multi-level community detection identified four clusters in the molecular response network. Excluding cluster 2, the remaining were largely gene expression dominated, with clusters 1, 3 and 4 including 120, 370 and 699 genes, respectively, associated with 65, 2 and 22 metabolites. Cluster 2 contained 57 metabolites and 45 genes. To assess the biological functions associated with each of the clusters, we tested for the presence of metabolite and gene expression pathways enriched in each cluster. The complete results are provided in Supplementary Table 5. Cluster 1 showed association with 15 metabolic and 23 gene expression pathways, which included pathways related to inflammation and immune response, DNA damage, cell adhesion and vascular function. Metabolites present in cluster 2 included 26 metabolic and 13 gene expression pathways. These were consistent with developmental processes, endothelial NO synthases, nucleotide metabolism and oxidative stress processes. Oxidative stress pathways included changes to ascorbate/aldarate metabolism, sulfur amino acid metabolism and TNF-alpha-induced ROS-dependent Caspase-3 signaling, with the metabolites ascorbic acid, glutathione and cysteamine also present in the correlation cluster. Cluster 3 consists almost entirely of gene expression nodes, with the vitamin E metabolite 7’-carboxy-alpha-chromanol and pyrroline the only two identified metabolites present. Due to this, 60 gene expression pathways were enriched from a diverse range of processes, including adipocyte differentiation, apoptosis, immune response, oxidative and reticulum stress response, inflammation and red blood cell adhesion. Similarly, cluster 4 only showed enrichment in Vitamin E metabolism. The gene expression results showed 47 pathways, including NO synthase signaling in muscle tissue, platelet aggregation, TNF signal transduction, mitochondrial dysfunction, immune response and regulation of lung epithelial progenitor cell differentiation.
Figure 4:
Metabolite and gene expression integration network. Discriminatory features and network correlations were determined using the R package xMWAS. The top discriminatory genes and identified metabolites were first selected using multilevel sparse partial least squares regression analysis, which were then evaluated for correlation with |r|≥ 0.4 and p< 0.05. Clusters of gene expression and metabolite profiles were identified using a using multi-level community detection algorithm that groups nodes showing a high-degree of connection among each other and are less connected to other nodes in the network (cluster 1= blue nodes, cluster 2= orange nodes; cluster 3= green nodes; cluster 4= red nodes). Each cluster was then evaluated for biological activity and overlap between metabolite and gene expression pathways. Three of clusters that were identified included both metabolites and genes; cluster 3 was predominantly genes and only included two metabolites, suggesting a minimal degree of overlap between metabolic and gene expression related processes present in cluster 3. For each cluster, the main pathways linked to endothelial function, inflammation, oxidative stress and related are provided; a full list of gene expression and metabolite pathways present in the correlation network are provided in Supplementary Table 5. Cl= Cluster; M= metabolite node, G= gene expression node.
Integration of gene expression and metabolites associated with EC and OC identified four main clusters of gene-metabolite correlations. We next evaluated the top two most connected genes present in each cluster to see if these genes had previously been linked to traffic related pollutants by searching CTD for literature reported gene-chemical interactions. For cluster 1, MATR3 and PPM1A showed interaction with benzo[a]pyrene, and exposures listed as volatile organic chemicals and vehicle emissions. In addition, BPDE, dinitrotoluene and nanoparticle exposures had reported effects on expression of MATR3. Cluster 2 nodes included one gene providing no matches in CTD (LOC100131165). The second gene, SNORA14A, only showed interactions with 1-(4-methoxyphenyl)-2-(3,4,5-trimethoxyphenyl)ethane, a methylated analog of resveratrol that has been shown to reduce levels of induced NO synthase. Chemical interactions listed in CTD for the topmost connected gene expression nodes in cluster 3 included environmental pollutants originating from exhaust emissions. For KIDINS220, traffic-related pollutants included dinitrobenzene, benzene and PAH dipole epoxides, while dinitrotoluenes, PAHs and dipole epoxide metabolites, carbon nanotubes, tobacco smoke and vehicle emissions have documented interactions with expression of IVNS1ABP listed in CTD. Cluster 4 resulted in similar interactions, including dinitrotoluenes, dioxin and PAH-related chemicals for the topmost connected gene in that cluster (UBE2D2), while interactions with dinitrotoluene, dinitrobenzene, a PAH dipole epoxide, dioxins and nitroarenes were present for OTUB1.
DISCUSSION
The present study provides an integrated molecular characterization of the biological response to PM2.5, EC and OC based upon trucking industry workers whose jobs included operating and working in proximity to diesel powered trucks in trucking terminals and on roadways. Untargeted metabolomics was used to characterize metabolic changes associated with EC and OC from shift- and week-averaged exposures. The results showed week-averaged EC and OC were associated with the greatest number of metabolic alterations, and included metabolite changes consistent with oxidative stress, endothelial function and inflammation. Changes in these pathways were also present in gene expression profiles correlated with EC and OC-associated metabolites, providing new insight into biological effects underlying exposure to traffic-related pollutants and cardiovascular disease risk.
Metabolomic profiling of exposure to traffic-related pollutants
Due to the ability of metabolomics to characterize multiple key biological processes related to human health and disease, metabolomics approaches studying the effects of traffic related pollution include a wide range of exposure scenarios and timescales, which were recently detailed extensively by Jin et al. (2021). These studies include controlled exposure to ambient air pollution and specific constituents, (Cheng et al. 2018) observational studies that provide comprehensive assessment to traffic related pollutants, (Liang et al. 2018; Vlaanderen et al. 2017) natural and non-natural intervention studies, (Li et al. 2017; Mu et al. 2019) and long-term exposure assessment using location-based exposure estimates. (Menni et al. 2015; Walker et al. 2018; Yan et al. 2019) In several of these studies, how the metabolome mediates relationships between air pollution and other health phenotypes, including pregnancy, asthma and cardiovascular disease, has provided new insight into potential pathways underlying adverse effects of these exposures. (Inoue et al. 2020; Jeong et al. 2018; Liang et al. 2019) While reported metabolites associated with traffic-related pollution have varied depending on the study design and metabolomic platforms used, most studies have consistently shown exposure is associated with endothelial pathways that include critical intermediates for NO, alterations in vitamin E metabolites, antioxidants, sulfur-containing amino acids, fatty acids and lipid peroxidation products consistent with oxidative stress, and changes in inflammatory signaling pathways that include prostaglandins and leukotrienes. Many of these metabolites have also been identified in metabolomic studies of diseases linked to air pollution exposure, including lung cancer, chronic obstructive pulmonary disease (COPD) and cardiovascular disease. (Cruickshank-Quinn et al. 2018; Fitzpatrick et al. 2014; Seow et al. 2019) To date, these studies have largely focused on ambient exposures, which can include emissions from many sources. In the present study, we provide metabolomic characterization specific to workers with occupational exposures primarily from diesel exhaust. Metabolomic and gene expression changes in our study were consistent with many of the pathways described by others, suggesting diesel exhaust exposure results in similar biological response pathways. However, these associations were largely driven by exposure composition, measured as EC and OC, rather than PM2.5. These results were unexpected, since PM2.5 and other particulate exposures have been linked to metabolic changes and all-cause mortality. (Cohen et al. 2017; Walker et al. 2018) Lack of association with PM2.5 could be due to the focus on monitoring exposures during working hours, which would be expected to catch the majority of diesel-related EC and OC exposure, while PM2.5. levels could vary considerable throughout the day based upon other factors, such as residence location, home ventilation, commute, and cooking. These results highlight the importance of considering air pollution composition and levels outside of monitoring periods when evaluating metabolic effects of exposure.
Both EC and OC were associated with dose-associated alterations to central metabolic pathways, including amino acids, co-factors, lipids and fatty acids and consistent with changes in oxidative stress, endothelial dysfunction and inflammation. Due to the complexity and varied composition of EC and OC and associated exhaust constituents in particulate matter, findings on the health effects of these two pollutants have largely been inconsistent across different population studies. (Schauer 2003) However, both toxicological and epidemiological studies suggest EC as a marker of traffic exposure and important contributor to adverse health effects linked to traffic-related exposures. (Janssen et al. 2011) Only a small number of studies have evaluated metabolic effects associated with EC and diesel exhaust. (Liang et al. 2019; Liang et al. 2018) To measure acute response to biodiesel fuel exhaust exposure, Surowiec et al. (2016) evaluated human lung lavage fluids from different lung regions following a 1-hr controlled exposure to biodiesel exhaust emissions, and similar to the associations with EC and OC in the present study identified changes in amino acids, including aspartic acid, ornithine, fatty acids, benzoic acid metabolites and co-factor metabolism following exposure. Additional studies in humans show diesel exhaust results in altered DNA methylation profiles, (Jiang et al. 2014) gene expression, (Peretz et al. 2007) and changes to bioactive lipids in lung lavage fluids. (Gouveia-Figueira et al. 2017) Model systems provide additional evidence of metabolic changes associated with exposure to diesel emissions. Oeder et al. (2015) applied a multi-omic approach that included proteomic, transcriptomic and metabolomic assessment of exposure to emissions from diesel and heavy fuel oil using human epithelial lung cells. The results showed diesel fuel resulted in pathway alterations across multiple omic-layers that were not observed for the heavy fuel oil particulates. Metabolomic profiling of mouse models using mixed vehicle emissions that included both gasoline and diesel exhaust have identified similar metabolic changes in oxidative stress-related metabolites, inflammation and lipid peroxidation products, in addition to alterations in intermediates of NO production critical for proper vascular and endothelial function. (Brower et al. 2016)
Metabolomic response profiles of occupational EC exposure
Week-averaged EC levels were associated with antioxidant metabolites and biomarkers of increased oxidative stress, which have been linked to risk of CVD. Increased oxidative stress and formation of in vivo ROS has been well documented in exposure to particulate matter. (Bates et al. 2015b; Lodovici and Bigagli 2011; Neophytou et al. 2014) Previous HRM studies have also identified similar changes in oxidative stress-related metabolites associated with ultrafine particulates from near highway exposures, (Walker et al. 2018) including increased levels of lipid peroxidation products, which have been implicated in the pathogenesis of multiple chronic diseases. (Akude et al. 2010; Selley 1998) In addition, the two antioxidants GSH and ascorbic acid were identified. GSH and its corresponding oxidized form, glutathione disulfide (GSSG) are one of the primary thiol/disulfide redox couples and provide multiple critical functions, which include protecting against oxidative stress, chemical detoxification and arginine turnover during NO synthase. (Hofmann and Schmidt 1995) Ascorbic acid, which is a protective antioxidant with functions similar to GSH, contributes in multiple ways to endothelial functions, including chemical stabilization of tetrahydrobiopterin during NO synthesis, vascular endothelial function, and has beneficial effects on vascular dilation. (Brown and Hu 2001; Heller et al. 2001; Jackson et al. 1998; Padayatty et al. 2003) Increased oxidation, particularly of thiol couples, and subsequent shift towards a more positive steady state redox potential has been shown to regulate early events in atherosclerosis and is associated with CVD and endothelial dysfunction. (Ashfaq et al. 2008; Go and Jones 2005) A high burden of oxidative stress has also been associated with mortality in patients with coronary heart disease. (Patel et al. 2016)
Additional associations with EC exposure consistent with CVD risk include inflammatory and endothelial-related metabolites. Thromboxane A2 (TxA2) provides multiple physiological functions, which include signaling for activation of new platelets and increased aggregation, vasoconstriction, endothelial adhesion molecule expression, cell proliferation and acts as a hypertensive agent. (Ding and Murray 2005; Ishizuka et al. 1998; Smyth 2010) Elevated biosynthesis of TxA2 and increased receptor expression are elevated in CVD. (Smyth 2010) Additional inducers of TxA2 production and receptor expression include inflammation, oxidative stress and NO, with TxA2 and other prostaglandin formation almost entirely driven by NO following inflammatory events. (Mollace et al. 2005) This is consistent with the EC-associated increase in hydroxy-L-homoarginine, which is a minor metabolic byproduct of NO production by oxidation of the arginine (Arg) analogue, homoarginine. (Moali et al. 1998) Increased prostaglandin I2 (PGI2) and hepoxilin A3-C (HxA3-C) provides additional evidence of disruption to this pathway. The physiological functions of PGI2 include mediation of vasodilation and platelet inhibition, (Cheng et al. 2002; Whittle et al. 1978) and homeostatic control for the vasoactive effects of TxA2. HxA3-C is the glutathione adduct of hepoxilin A3, which is also an arachidonic acid metabolite with pro-inflammatory and vasodilation signaling properties. Taken together, these results provide strong evidence that exposure to EC results in changes to physiologically active signaling pathways that are implicated in a wide range of processes related to CVD and endothelial dysfunction.
Metabolomic response profiles of occupational OC exposure
Both shift- and week-averaged OC exposures resulted in changes to metabolites providing antioxidant effects and markers of oxidative stress. Metabolite changes suggesting increased oxidative stress include methionine sulfoxide, (Moskovitz et al. 1997) and LGSH, which is a byproduct of GSH dependent degradation of methylglyoxal (MG) by glyoxalate I. (Chang and Wu 2006) MG has been implicated in a number of disease processes, including hypertension, diabetes and formation of atherosclerotic lesions. (Chang and Wu 2006) Pathways consistent with antioxidant processes were also detected, including vitamin E metabolism and ascorbate/aldarate metabolism. Alpha-tocopherol was positively associated with OC, which has previously been observed to protect against decreasing lung function following PM2.5 exposure, (Menni et al. 2015) and has been associated with exposure to other traffic-related pollutants. (Liang et al. 2018)
OC exposure was associated with the greatest number of metabolites related to endothelial function. This included alterations to arginine and proline metabolism, and changes to metabolites with functions that include NO production from arginine, required co-factors, decreased thrombotic signaling molecules and endogenous inhibitors of NO. Many of these changes indicate OC results in disruption to NO production itself, while EC associations suggest increased platelet formation and aggregation. Changes consistent with possible disruption to NO included agmatine, acetylagmatine and n-(omega)-hydroxyarginine. Agmatine, which is an endogenously produced decarboxylation product of Arg, inhibits inducible NO synthase and acts as a modulator of NO production. (Raghavan and Dikshit 2004) Decreased levels suggest elevated rates of NO production, which is consistent with the changes in n-(omega)-hydroxyarginine but not citrulline. N-(omega)-hydroxyarginine is the first oxidative product of arginine by NO synthase and can as both an inhibitor of Mn dependent arginase enzymes and catalytic intermediate in NO production. (Cox et al. 2001) Taken together, these results suggest OC exposure contributes to metabolic alterations that have important implications for endothelial function.
Differential metabolomic response to EC and OC occupational exposures
While many of the underlying biological processes associated with OC were similar to those observed for EC, specific metabolite changes differed and overlapping metabolites showed opposing direction of change following exposure to the two pollutants. A possible explanation for this trend is different patterns of exposure and particulate origin, which is consistent with the lack of correlation between EC and OC. (Zhang et al. 2011) Prior characterization of this cohort has shown exposure patterns differ based upon job title and pollutant, with pickup/delivery drivers and dockworkers exhibiting highest exposure levels of EC, while office workers and drivers showed the highest OC levels. (Neophytou et al. 2013) The OC fraction of particulate matter consists of a complex mixture of polar and non-polar chemicals that can be formed after initial combustion, including organic matter condensation, binding of semi-volatile organics on the particulate surface and interaction with aerosols, in addition to atmospheric changes occurring during transport. Thus, while EC represents exposures from diesel fuel exhaust, the measured OC could arise from a variety of sources and may not be due to diesel emissions alone.
Gene expression by metabolome molecular correlation networks
Following characterization of the metabolome, we tested for additional associations with gene expression levels to characterize potential biological activity networks linking metabolite and gene expression pathways. Exposure to environmental chemicals can influence local and global changes in gene transcription and enzyme activity, resulting in varied biological changes that can influence metabolic changes and contribute to underlying toxicological mechanisms. Integrating gene expression results with MWAS has the potential to enhance understanding of how functional molecular mechanisms interact due to environmental exposures, providing a systems biology framework for studying toxicological effects of air pollution in vivo. Enriched gene pathways were consistent with the identified metabolic changes associated with EC and OC exposure, providing additional evidence that exposure to traffic-related pollution leads to changes in endothelial function, inflammation and oxidative stress. Most importantly, enriched metabolite and gene expression pathways within each cluster consisted of different biochemical processes, suggesting that the biological response to traffic-related air pollution is systemic and has important implications for homeostatic control and health. Comparison to curated data in CTD showed the most-connected genes for each cluster have previously been demonstrated to interact with pollutants present in exhaust emissions, including PAHs and diol epoxide metabolites, VOCs and nitroaromatics. Thus, not only were the in vivo identified biological changes from gene expression and the metabolome consistent, using CTD we show the most connected transcript in each cluster have been associated with gene changes due to traffic-related pollutants in vitro.
Limitations
We acknowledge several limitations of this work. First, this study was limited to an all-male, non-smoking, predominantly Caucasian cohort employed in the US unionized trucking industry. Therefore, we were unable to examine differences due to sex or other unaccounted for confounders. Although samples were collected over the course of a workweek, only exposures occurring at the workplace were considered, and we could not account for those that occurred during commuting or at home. Second, results showed week-averaged exposures resulted in the greatest molecular changes; however, these were estimated based upon one week only, and due to the length of the study we could not evaluate the effects of longer exposure periods or how levels varied over time. Third, the results from this study are correlative in nature. While we accounted for age, BMI and measurement day in our model, we could not account for unknown and uncharacterized confounders. In addition, we evaluated correlation between gene expression levels and the metabolome. Previous results in humans have shown signals in whole blood generally reflect organism wide processes; (Bartel et al. 2015) however, although the metabolome represents an integrated profile of multiple body compartments and processes, it is not possible to assess cell specific metabolite or gene changes. Fourth, only a small number of metabolites were identified by comparison to standards, and the majority of metabolite features were annotated using MS1 accurate mass only. Limited reference standards, low abundance of many of the metabolite features and lack of fragmentation spectra from MSMS render characterization and identification challenging. Finally, exposure was determined using aggregate measures that included PM2.5, EC and OC. We were unable to characterize other exposures associated with vehicle-exhaust exposures (e.g. nitrogen oxides, volatile or semi-volatile organics, or even individual PAHs or nitro-PAHs). Despite these limitations, we identified EC and OC exposure-associated changes consistent with increased oxidative stress, endothelial dysfunction and inflammation, which are common risk factors for diseases related to air pollution and diesel exhaust exposure. Integrating gene expression and metabolome profiles showed these changes were present across both molecular layers, providing additional evidence of biological response pathways underlying environmental exposure to traffic related pollutants. Continued application of these approaches in humans will provide improved understanding of how environmental exposures contribute to adverse health outcomes.
CONCLUSIONS
To elucidate the molecular response of occupational exposure to traffic-related pollutants in the US trucking industry, we used an MWAS framework to identify plasma metabolic changes in workers following shift- and week-averaged exposure to EC, OC and PM2.5. MWAS of EC and OC identified dose-associated metabolic alterations consistent with endothelial function, inflammation and oxidative stress. These include annotated metabolites that function as thrombotic signaling molecules, antioxidants, biomarkers of oxidative stress, lipid peroxidation products and intermediate metabolites in NO production. Week-averaged levels of both pollutants were associated with the greatest number of metabolite alterations observed in study participants, suggesting persistent workplace exposure influence biological changes that are not mediated by time off. Data-driven integration of gene expression levels with exposure-associated metabolites identified additional functional changes related to endothelial function, NO synthase, inflammation and immune response, including changes that have been reported as associated with traffic-related pollutants in other studies. Taken together, the results show exposure to traffic-related pollutants that includes diesel exhaust emissions influences metabolite and gene expression pathways implicated in cardiopulmonary disease risk and provide insight into the molecular pathways underlying the adverse effects of traffic-related pollution.
Supplementary Material
HIGHLIGHTS.
Diesel exhaust pollutants were evaluated for workers in the US trucking industry.
Metabolomic alterations were associated with elemental carbon and organic carbon.
No metabolites were associated with PM2.5 exposure.
Exposure-associated metabolites were related to oxidative stress and nitric oxide production
Gene and metabolite networks support associations with cardiopulmonary disease
ACKNOWLEDGEMENTS
This work was supported by funds received from the National Institute of Health, award numbers ES019776, ES025632, ES030859, ES013726, CA090792, ES016284, P30 ES000002 and OD018006.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Statement
Douglas I. Walker: Investigation, Formal analysis, Writing - Original Draft, Visualization
Jaime E. Hart: Conceptualization, Methodology, Data Curation, Writing - Review & Editing
Chirag J. Patel: Writing - Review & Editing
Ruthann Rudel: Writing - Review & Editing
Jen-hwa Chu: Investigation, Data Curation
Eric Garshick: Methodology, Writing - Review & Editing
Kurt D. Pennell: Writing - Review & Editing
Francine Laden: Conceptualization, Funding acquisition, Resources, Writing - Review & Editing
Dean P. Jones: Supervision, Writing - Review & Editing
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Conflicts of Interest: None to declare
REFERENCES
- Accardi CJ; Walker DI; Uppal K; Quyyumi AA; Rohrbeck P; Pennell KD; Mallon COLTM; Jones DP High-Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. Journal of Occupational and Environmental Medicine 2016a;58:S80–S88 [DOI] [PubMed] [Google Scholar]
- Accardi CJ; Walker DI; Uppal K; Quyyumi AA; Rohrbeck P; Pennell KD; Mallon CT; Jones DP High-Resolution Metabolomics for Nutrition and Health Assessment of Armed Forces Personnel. Journal of occupational and environmental medicine / American College of Occupational and Environmental Medicine 2016b;58:S80–88 [DOI] [PubMed] [Google Scholar]
- Akude E; Zherebitskaya E; Roy Chowdhury SK; Girling K; Fernyhough P. 4-Hydroxy-2-nonenal induces mitochondrial dysfunction and aberrant axonal outgrowth in adult sensory neurons that mimics features of diabetic neuropathy. Neurotox Res 2010;17:28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashfaq S; Abramson JL; Jones DP; Rhodes SD; Weintraub WS; Hooper WC; Vaccarino V; Alexander RW; Harrison DG; Quyyumi AA Endothelial function and aminothiol biomarkers of oxidative stress in healthy adults. Hypertension 2008;52:80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel J; Krumsiek J; Schramm K; Adamski J; Gieger C; Herder C; Carstensen M; Peters A; Rathmann W; Roden M; Strauch K; Suhre K; Kastenmuller G; Prokisch H; Theis FJ The Human Blood Metabolome-Transcriptome Interface. PLoS genetics 2015;11:e1005274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D; Mächler M; Bolker B; Walker S. Fitting Linear Mixed-Effects Models Using lme4. Journal of Statistical Software 2015a;67 [Google Scholar]
- Bates JT; Weber RJ; Abrams J; Verma V; Fang T; Klein M; Strickland MJ; Sarnat SE; Chang HH; Mulholland JA; Tolbert PE; Russell AG Reactive Oxygen Species Generation Linked to Sources of Atmospheric Particulate Matter and Cardiorespiratory Effects. Environmental science & technology 2015b;49:13605–13612 [DOI] [PubMed] [Google Scholar]
- Beelen R; Hoek G; van den Brandt PA; Goldbohm RA; Fischer P; Schouten LJ; Jerrett M; Hughes E; Armstrong B; Brunekreef B. Long-term effects of traffic-related air pollution on mortality in a Dutch cohort (NLCS-AIR study). Environmental health perspectives 2008;116:196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benbrahim-Tallaa L; Baan RA; Grosse Y; Lauby-Secretan B; El Ghissassi F; Bouvard V; Guha N; Loomis D; Straif K; International Agency for Research on Cancer Monograph Working, G. Carcinogenicity of diesel-engine and gasoline-engine exhausts and some nitroarenes. The Lancet Oncology 2012;13:663–664 [DOI] [PubMed] [Google Scholar]
- Benjamini Y; Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J Roy Stat Soc B Met 1995;57:289–300 [Google Scholar]
- Brower JB; Doyle-Eisele M; Moeller B; Stirdivant S; McDonald JD; Campen MJ Metabolomic changes in murine serum following inhalation exposure to gasoline and diesel engine emissions. Inhal Toxicol 2016;28:241–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AA; Hu FB Dietary modulation of endothelial function: implications for cardiovascular disease. Am J Clin Nutr 2001;73:673–686 [DOI] [PubMed] [Google Scholar]
- Brugge D; Lane K; Padro-Martinez LT; Stewart A; Hoesterey K; Weiss D; Wang DD; Levy JI; Patton AP; Zamore W; Mwamburi M. Highway proximity associated with cardiovascular disease risk: the influence of individual-level confounders and exposure misclassification. Environ Health 2013;12:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T; Wu L. Methylglyoxal, oxidative stress, and hypertension. Can J Physiol Pharmacol 2006;84:1229–1238 [DOI] [PubMed] [Google Scholar]
- Cheng W; Duncan KE; Ghio AJ; Ward-Caviness C; Karoly ED; Diaz-Sanchez D; Conolly RB; Devlin RB Changes in Metabolites Present in Lung-Lining Fluid Following Exposure of Humans to Ozone. Toxicological sciences : an official journal of the Society of Toxicology 2018;163:430–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y; Austin SC; Rocca B; Koller BH; Coffman TM; Grosser T; Lawson JA; FitzGerald GA Role of prostacyclin in the cardiovascular response to thromboxane A2. Science 2002;296:539–541 [DOI] [PubMed] [Google Scholar]
- Chu JH; Hart JE; Chhabra D; Garshick E; Raby BA; Laden F. Gene expression network analyses in response to air pollution exposures in the trucking industry. Environ Health 2016;15:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang KJ; Chan CC; Su TC; Lee CT; Tang CS The effect of urban air pollution on inflammation, oxidative stress, coagulation, and autonomic dysfunction in young adults. Am J Respir Crit Care Med 2007;176:370–376 [DOI] [PubMed] [Google Scholar]
- Cohen AJ; Brauer M; Burnett R; Anderson HR; Frostad J; Estep K; Balakrishnan K; Brunekreef B; Dandona L; Dandona R; Feigin V; Freedman G; Hubbell B; Jobling A; Kan H; Knibbs L; Liu Y; Martin R; Morawska L; Pope CA 3rd; Shin H; Straif K; Shaddick G; Thomas M; van Dingenen R; van Donkelaar A; Vos T; Murray CJL; Forouzanfar MH Estimates and 25-year trends of the global burden of disease attributable to ambient air pollution: an analysis of data from the Global Burden of Diseases Study 2015. Lancet 2017;389:1907–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini MG; Khalek I; McDonald JD; van Erp AM The Advanced Collaborative Emissions Study (ACES) of 2007- and 2010-Emissions Compliant Heavy-Duty Diesel Engines: Characterization of Emissions and Health Effects. Emission Control Science and Technology 2016;2:215–227 [Google Scholar]
- Cox JD; Cama E; Colleluori DM; Pethe S; Boucher JL; Mansuy D; Ash DE; Christianson DW Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase. Biochemistry 2001;40:2689–2701 [DOI] [PubMed] [Google Scholar]
- Cruickshank-Quinn CI; Jacobson S; Hughes G; Powell RL; Petrache I; Kechris K; Bowler R; Reisdorph N. Metabolomics and transcriptomics pathway approach reveals outcome-specific perturbations in COPD. Sci Rep 2018;8:17132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP; Murphy CG; Johnson R; Lay JM; Lennon-Hopkins K; Saraceni-Richards C; Sciaky D; King BL; Rosenstein MC; Wiegers TC; Mattingly CJ The Comparative Toxicogenomics Database: update 2013. Nucleic acids research 2013;41:D1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X; Murray PA Cellular mechanisms of thromboxane A2-mediated contraction in pulmonary veins. Am J Physiol Lung Cell Mol Physiol 2005;289:L825–833 [DOI] [PubMed] [Google Scholar]
- Donaldson K; Stone V; Seaton A; MacNee W. Ambient particle inhalation and the cardiovascular system: potential mechanisms. Environmental health perspectives 2001;109 Suppl 4:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P; Kibbe WA; Lin SM lumi: a pipeline for processing Illumina microarray. Bioinformatics 2008;24:1547–1548 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AM; Park Y; Brown LA; Jones DP Children with severe asthma have unique oxidative stress-associated metabolomic profiles. The Journal of allergy and clinical immunology 2014;133:258–261 e251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garshick E; Laden F; Hart JE; Rosner B; Davis ME; Eisen EA; Smith TJ Lung cancer and vehicle exhaust in trucking industry workers. Environmental health perspectives 2008;116:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go YM; Jones DP Intracellular proatherogenic events and cell adhesion modulated by extracellular thiol/disulfide redox state. Circulation 2005;111:2973–2980 [DOI] [PubMed] [Google Scholar]
- Go YM; Walker DI; Liang Y; Uppal K; Soltow QA; Tran V; Strobel F; Quyyumi AA; Ziegler TR; Pennell KD; Miller GW; Jones DP Reference Standardization for Mass Spectrometry and High-resolution Metabolomics Applications to Exposome Research. Toxicological sciences : an official journal of the Society of Toxicology 2015;148:531–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia-Figueira S; Karimpour M; Bosson JA; Blomberg A; Unosson J; Pourazar J; Sandstrom T; Behndig AF; Nording ML Mass spectrometry profiling of oxylipins, endocannabinoids, and N-acylethanolamines in human lung lavage fluids reveals responsiveness of prostaglandin E2 and associated lipid metabolites to biodiesel exhaust exposure. Analytical and bioanalytical chemistry 2017;409:2967–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grahame TJ; Klemm R; Schlesinger RB Public health and components of particulate matter: the changing assessment of black carbon. J Air Waste Manag Assoc 2014;64:620–660 [DOI] [PubMed] [Google Scholar]
- Hart JE Air pollution affects lung cancer survival. Thorax 2016;71:875–876 [DOI] [PubMed] [Google Scholar]
- Heller R; Unbehaun A; Schellenberg B; Mayer B; Werner-Felmayer G; Werner ER Lascorbic acid potentiates endothelial nitric oxide synthesis via a chemical stabilization of tetrahydrobiopterin. The Journal of biological chemistry 2001;276:40–47 [DOI] [PubMed] [Google Scholar]
- Hofmann H; Schmidt HH Thiol dependence of nitric oxide synthase. Biochemistry 1995;34:13443–13452 [DOI] [PubMed] [Google Scholar]
- Inoue K; Yan Q; Arah OA; Paul K; Walker DI; Jones DP; Ritz B. Air Pollution and Adverse Pregnancy and Birth Outcomes: Mediation Analysis Using Metabolomic Profiles. Curr Environ Health Rep 2020;7:231–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T; Kawakami M; Hidaka T; Matsuki Y; Takamizawa M; Suzuki K; Kurita A; Nakamura H. Stimulation with thromboxane A2 (TXA2) receptor agonist enhances ICAM-1, VCAM-1 or ELAM-1 expression by human vascular endothelial cells. Clin Exp Immunol 1998;112:464–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TS; Xu A; Vita JA; Keaney JF Jr. Ascorbate prevents the interaction of superoxide and nitric oxide only at very high physiological concentrations. Circ Res 1998;83:916–922 [DOI] [PubMed] [Google Scholar]
- Janssen N; Gerlofs-Nijland M; Lanki T; Salonen R; Cassee F; Hoek G; Fischer P; Brunekreef B; Krzyzanowski M. Health effects of black carbon. World Health Organization; 2012 [Google Scholar]
- Janssen NA; Hoek G; Simic-Lawson M; Fischer P; van Bree L; ten Brink H; Keuken M; Atkinson RW; Anderson HR; Brunekreef B; Cassee FR Black carbon as an additional indicator of the adverse health effects of airborne particles compared with PM10 and PM2.5. Environmental health perspectives 2011;119:1691–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong A; Fiorito G; Keski-Rahkonen P; Imboden M; Kiss A; Robinot N; Gmuender H; Vlaanderen J; Vermeulen R; Kyrtopoulos S; Herceg Z; Ghantous A; Lovison G; Galassi C; Ranzi A; Krogh V; Grioni S; Agnoli C; Sacerdote C; Mostafavi N; Naccarati A; Scalbert A; Vineis P; Probst-Hensch N; Consortium EX Perturbation of metabolic pathways mediates the association of air pollutants with asthma and cardiovascular diseases. Environment international 2018;119:334–345 [DOI] [PubMed] [Google Scholar]
- Jiang R; Jones MJ; Sava F; Kobor MS; Carlsten C. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol 2014;11:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L; Godri Pollitt KJ; Liew Z; Rosen Vollmar AK; Vasiliou V; Johnson CH; Zhang Y. Use of Untargeted Metabolomics to Explore the Air Pollution-Related Disease Continuum. Curr Environ Health Rep 2021;8:7–22 [DOI] [PubMed] [Google Scholar]
- Johnson WE; Li C; Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2007;8:118–127 [DOI] [PubMed] [Google Scholar]
- Li H; Cai J; Chen R; Zhao Z; Ying Z; Wang L; Chen J; Hao K; Kinney PL; Chen H; Kan H. Particulate Matter Exposure and Stress Hormone Levels: A Randomized, Double-Blind, Crossover Trial of Air Purification. Circulation 2017;136:618–627 [DOI] [PubMed] [Google Scholar]
- Li S; Park Y; Duraisingham S; Strobel FH; Khan N; Soltow QA; Jones DP; Pulendran B. Predicting network activity from high throughput metabolomics. PLoS computational biology 2013;9:e1003123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D; Ladva CN; Golan R; Yu T; Walker DI; Sarnat SE; Greenwald R; Uppal K; Tran V; Jones DP; Russell AG; Sarnat JA Perturbations of the arginine metabolome following exposures to traffic-related air pollution in a panel of commuters with and without asthma. Environment international 2019;127:503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D; Moutinho JL; Golan R; Yu T; Ladva CN; Niedzwiecki M; Walker DI; Sarnat SE; Chang HH; Greenwald R; Jones DP; Russell AG; Sarnat JA Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution. Environment international 2018;120:145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SS; Vos T; Flaxman AD; Danaei G; Shibuya K; Adair-Rohani H; Amann M; Anderson HR; Andrews KG; Aryee M; Atkinson C; Bacchus LJ; Bahalim AN; Balakrishnan K; Balmes J; Barker-Collo S; Baxter A; Bell ML; Blore JD; Blyth F; Bonner C; Borges G; Bourne R; Boussinesq M; Brauer M; Brooks P; Bruce NG; Brunekreef B; Bryan-Hancock C; Bucello C; Buchbinder R; Bull F; Burnett RT; Byers TE; Calabria B; Carapetis J; Carnahan E; Chafe Z; Charlson F; Chen H; Chen JS; Cheng AT; Child JC; Cohen A; Colson KE; Cowie BC; Darby S; Darling S; Davis A; Degenhardt L; Dentener F; Des Jarlais DC; Devries K; Dherani M; Ding EL; Dorsey ER; Driscoll T; Edmond K; Ali SE; Engell RE; Erwin PJ; Fahimi S; Falder G; Farzadfar F; Ferrari A; Finucane MM; Flaxman S; Fowkes FG; Freedman G; Freeman MK; Gakidou E; Ghosh S; Giovannucci E; Gmel G; Graham K; Grainger R; Grant B; Gunnell D; Gutierrez HR; Hall W; Hoek HW; Hogan A; Hosgood HD 3rd; Hoy D; Hu H; Hubbell BJ; Hutchings SJ; Ibeanusi SE; Jacklyn GL; Jasrasaria R; Jonas JB; Kan H; Kanis JA; Kassebaum N; Kawakami N; Khang YH; Khatibzadeh S; Khoo JP; Kok C; Laden F; Lalloo R; Lan Q; Lathlean T; Leasher JL; Leigh J; Li Y; Lin JK; Lipshultz SE; London S; Lozano R; Lu Y; Mak J; Malekzadeh R; Mallinger L; Marcenes W; March L; Marks R; Martin R; McGale P; McGrath J; Mehta S; Mensah GA; Merriman TR; Micha R; Michaud C; Mishra V; Mohd Hanafiah K; Mokdad AA; Morawska L; Mozaffarian D; Murphy T; Naghavi M; Neal B; Nelson PK; Nolla JM; Norman R; Olives C; Omer SB; Orchard J; Osborne R; Ostro B; Page A; Pandey KD; Parry CD; Passmore E; Patra J; Pearce N; Pelizzari PM; Petzold M; Phillips MR; Pope D; Pope CA 3rd; Powles J; Rao M; Razavi H; Rehfuess EA; Rehm JT; Ritz B; Rivara FP; Roberts T; Robinson C; Rodriguez-Portales JA; Romieu I; Room R; Rosenfeld LC; Roy A; Rushton L; Salomon JA; Sampson U; Sanchez-Riera L; Sanman E; Sapkota A; Seedat S; Shi P; Shield K; Shivakoti R; Singh GM; Sleet DA; Smith E; Smith KR; Stapelberg NJ; Steenland K; Stockl H; Stovner LJ; Straif K; Straney L; Thurston GD; Tran JH; Van Dingenen R; van Donkelaar A; Veerman JL; Vijayakumar L; Weintraub R; Weissman MM; White RA; Whiteford H; Wiersma ST; Wilkinson JD; Williams HC; Williams W; Wilson N; Woolf AD; Yip P; Zielinski JM; Lopez AD; Murray CJ; Ezzati M; AlMazroa MA; Memish ZA A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 2012;380:2224–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipfert FW; Baty JD; Miller JP; Wyzga RE PM2.5 constituents and related air quality variables as predictors of survival in a cohort of U.S. military veterans. Inhal Toxicol 2006;18:645–657 [DOI] [PubMed] [Google Scholar]
- Liu KH; Nellis M; Uppal K; Ma C; Tran V; Liang Y; Walker DI; Jones DP Reference Standardization for Quantification and Harmonization of Large-Scale Metabolomics. Analytical chemistry 2020; [DOI] [PMC free article] [PubMed]
- Liu KH; Walker DI; Uppal K; Tran V; Rohrbeck P; Mallon TM; Jones D. High resolution metabolomics assessment of military personnel. Journal of Occupational and Environmental Medicine 2016;Submitted; DoD Biomarkers Supplement [DOI] [PMC free article] [PubMed]
- Lodovici M; Bigagli E. Oxidative Stress and Air Pollution Exposure. Journal of Toxicology 2011;2011. [DOI] [PMC free article] [PubMed]
- Luben TJ; Nichols JL; Dutton SJ; Kirrane E; Owens EO; Datko-Williams L; Madden M; Sacks JD A systematic review of cardiovascular emergency department visits, hospital admissions and mortality associated with ambient black carbon. Environment international 2017;107:154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menni C; Metrustry SJ; Mohney RP; Beevers S; Barratt B; Spector TD; Kelly FJ; Valdes AM Circulating levels of antioxidant vitamins correlate with better lung function and reduced exposure to ambient pollution. Am J Respir Crit Care Med 2015;191:1203–1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moali C; Boucher JL; Sari MA; Stuehr DJ; Mansuy D. Substrate specificity of NO synthases: detailed comparison of L-arginine, homo-L-arginine, their N omega-hydroxy derivatives, and N omega-hydroxynor-L-arginine. Biochemistry 1998;37:10453–10460 [DOI] [PubMed] [Google Scholar]
- Mollace V; Muscoli C; Masini E; Cuzzocrea S; Salvemini D. Modulation of prostaglandin biosynthesis by nitric oxide and nitric oxide donors. Pharmacol Rev 2005;57:217–252 [DOI] [PubMed] [Google Scholar]
- Moskovitz J; Berlett BS; Poston JM; Stadtman ER The yeast peptide-methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci U S A 1997;94:9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu L; Niu Z; Blair RH; Yu H; Browne RW; Bonner MR; Fanter T; Deng F; Swanson M. Metabolomics Profiling before, during, and after the Beijing Olympics: A Panel Study of Within-Individual Differences during Periods of High and Low Air Pollution. Environmental health perspectives 2019;127:57010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou AM; Hart JE; Cavallari JM; Smith TJ; Dockery DW; Coull BA; Garshick E; Laden F. Traffic-related exposures and biomarkers of systemic inflammation, endothelial activation and oxidative stress: a panel study in the US trucking industry. Environ Health 2013;12:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neophytou AM; Hart JE; Chang Y; Zhang JJ; Smith TJ; Garshick E; Laden F. Short-term traffic related exposures and biomarkers of nitro-PAH exposure and oxidative DNA damage. Toxics 2014;2:377–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIOSH. Elemental Carbon (Diesel Particulate) 5040. in: Casinelli M, PF OC, eds. NIOSH Manual of Analytical Methods, 4th Ed. Cincinnati, OH: U.S Department of Helath and Human Services, Centers for Disease Control and Prevention, National Institute for Occupational Safety and Health, DHHS (NIOSH); 2003 [Google Scholar]
- Oeder S; Kanashova T; Sippula O; Sapcariu SC; Streibel T; Arteaga-Salas JM; Passig J; Dilger M; Paur HR; Schlager C; Mulhopt S; Diabate S; Weiss C; Stengel B; Rabe R; Harndorf H; Torvela T; Jokiniemi JK; Hirvonen MR; Schmidt-Weber C; Traidl-Hoffmann C; BeruBe KA; Wlodarczyk AJ; Prytherch Z; Michalke B; Krebs T; Prevot AS; Kelbg M; Tiggesbaumker J; Karg E; Jakobi G; Scholtes S; Schnelle-Kreis J; Lintelmann J; Matuschek G; Sklorz M; Klingbeil S; Orasche J; Richthammer P; Muller L; Elsasser M; Reda A; Groger T; Weggler B; Schwemer T; Czech H; Ruger CP; Abbaszade G; Radischat C; Hiller K; Buters JT; Dittmar G; Zimmermann R. Particulate matter from both heavy fuel oil and diesel fuel shipping emissions show strong biological effects on human lung cells at realistic and comparable in vitro exposure conditions. PloS one 2015;10:e0126536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostro B; Feng WY; Broadwin R; Green S; Lipsett M. The effects of components of fine particulate air pollution on mortality in california: results from CALFINE. Environmental health perspectives 2007;115:13–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padayatty SJ; Katz A; Wang Y; Eck P; Kwon O; Lee JH; Chen S; Corpe C; Dutta A; Dutta SK; Levine M. Vitamin C as an antioxidant: evaluation of its role in disease prevention. J Am Coll Nutr 2003;22:18–35 [DOI] [PubMed] [Google Scholar]
- Patel RS; Ghasemzadeh N; Eapen DJ; Sher S; Arshad S; Ko YA; Veledar E; Samady H; Zafari AM; Sperling L; Vaccarino V; Jones DP; Quyyumi AA Novel Biomarker of Oxidative Stress Is Associated With Risk of Death in Patients With Coronary Artery Disease. Circulation 2016;133:361–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretz A; Peck EC; Bammler TK; Beyer RP; Sullivan JH; Trenga CA; Srinouanprachnah S; Farin FM; Kaufman JD Diesel exhaust inhalation and assessment of peripheral blood mononuclear cell gene transcription effects: an exploratory study of healthy human volunteers. Inhal Toxicol 2007;19:1107–1119 [DOI] [PubMed] [Google Scholar]
- Pope CA 3rd; Burnett RT; Thun MJ; Calle EE; Krewski D; Ito K; Thurston GD Lung cancer, cardiopulmonary mortality, and long-term exposure to fine particulate air pollution. JAMA : the journal of the American Medical Association 2002;287:1132–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope CA 3rd; Ezzati M; Dockery DW Fine-particulate air pollution and life expectancy in the United States. The New England journal of medicine 2009;360:376–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan SA; Dikshit M. Vascular regulation by the L-arginine metabolites, nitric oxide and agmatine. Pharmacol Res 2004;49:397–414 [DOI] [PubMed] [Google Scholar]
- Sarnat SE; Winquist A; Schauer JJ; Turner JR; Sarnat JA Fine particulate matter components and emergency department visits for cardiovascular and respiratory diseases in the St. Louis, Missouri-Illinois, metropolitan area. Environmental health perspectives 2015;123:437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer JJ Evaluation of elemental carbon as a marker for diesel particulate matter. Journal of exposure analysis and environmental epidemiology 2003;13:443–453 [DOI] [PubMed] [Google Scholar]
- Schymanski EL; Jeon J; Gulde R; Fenner K; Ruff M; Singer HP; Hollender J. Identifying small molecules via high resolution mass spectrometry: communicating confidence. Environmental science & technology 2014;48:2097–2098 [DOI] [PubMed] [Google Scholar]
- Selley ML (E)-4-hydroxy-2-nonenal may be involved in the pathogenesis of Parkinson’s disease. Free radical biology & medicine 1998;25:169–174 [DOI] [PubMed] [Google Scholar]
- Seow WJ; Shu XO; Nicholson JK; Holmes E; Walker DI; Hu W; Cai Q; Gao YT; Xiang YB; Moore SC; Bassig BA; Wong JYY; Zhang J; Ji BT; Boulange CL; Kaluarachchi M; Wijeyesekera A; Zheng W; Elliott P; Rothman N; Lan Q. Association of Untargeted Urinary Metabolomics and Lung Cancer Risk Among Never-Smoking Women in China. JAMA Netw Open 2019;2:e1911970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheesley RJ; Schauer JJ; Garshick E; Laden F; Smith TJ; Blicharz AP; Deminter JT Tracking personal exposure to particulate diesel exhaust in a diesel freight terminal using organic tracer analysis. Journal of exposure science & environmental epidemiology 2009;19:172–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth EM Thromboxane and the thromboxane receptor in cardiovascular disease. Clin Lipidol 2010;5:209–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltow QA; Strobel FH; Mansfield KG; Wachtman L; Park Y; Jones DP High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics 2013;9:S132–S143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen M; Daneshvar B; Hansen M; Dragsted LO; Hertel O; Knudsen L; Loft S. Personal PM2.5 exposure and markers of oxidative stress in blood. Environmental health perspectives 2003;111:161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surowiec I; Karimpour M; Gouveia-Figueira S; Wu J; Unosson J; Bosson JA; Blomberg A; Pourazar J; Sandstrom T; Behndig AF; Trygg J; Nording ML Multi-platform metabolomics assays for human lung lavage fluids in an air pollution exposure study. Analytical and bioanalytical chemistry 2016;408:4751–4764 [DOI] [PubMed] [Google Scholar]
- Tainio M; Tuomisto JT; Hanninen O; Aarnio P; Koistinen KJ; Jantunen MJ; Pekkanen J. Health effects caused by primary fine particulate matter (PM2.5) emitted from buses in the Helsinki metropolitan area, Finland. Risk Anal 2005;25:151–160 [DOI] [PubMed] [Google Scholar]
- Uppal K; Ma C; Go YM; Jones DP xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 2018;34:701–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K; Soltow QA; Strobel FH; Pittard WS; Gernert KM; Yu T; Jones DP xMSanalyzer: automated pipeline for improved feature detection and downstream analysis of large-scale, non-targeted metabolomics data. BMC bioinformatics 2013;14:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K; Walker DI; Jones DP xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Analytical chemistry 2016a; [DOI] [PMC free article] [PubMed]
- Uppal K; Walker DI; Liu K; Li S; Go YM; Jones DP Computational Metabolomics: A Framework for the Million Metabolome. Chemical research in toxicology 2016b;29:1956–1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlaanderen JJ; Janssen NA; Hoek G; Keski-Rahkonen P; Barupal DK; Cassee FR; Gosens I; Strak M; Steenhof M; Lan Q; Brunekreef B; Scalbert A; Vermeulen RCH The impact of ambient air pollution on the human blood metabolome. Environ Res 2017;156:341–348 [DOI] [PubMed] [Google Scholar]
- Walker DI; Lane KJ; Liu K; Uppal K; Patton AP; Durant JL; Jones DP; Brugge D; Pennell KD Metabolomic assessment of exposure to near-highway ultrafine particles. Journal of exposure science & environmental epidemiology 2018; [DOI] [PMC free article] [PubMed]
- Walker DI; Uppal K; Zhang L; Vermeulen R; Smith M; Hu W; Purdue MP; Tang X; Reiss B; Kim S; Li L; Huang H; Pennell KD; Jones DP; Rothman N; Lan Q. High-resolution metabolomics of occupational exposure to trichloroethylene. International journal of epidemiology 2016;45:1517–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle BJ; Moncada S; Vane JR Comparison of the effects of prostacyclin (PGI2), prostaglandin E1 and D2 on platelet aggregation in different species. Prostaglandins 1978;16:373–388 [DOI] [PubMed] [Google Scholar]
- Wishart DS; Jewison T; Guo AC; Wilson M; Knox C; Liu Y; Djoumbou Y; Mandal R; Aziat F; Dong E; Bouatra S; Sinelnikov I; Arndt D; Xia J; Liu P; Yallou F; Bjorndahl T; Perez-Pineiro R; Eisner R; Allen F; Neveu V; Greiner R; Scalbert A. HMDB 3.0--The Human Metabolome Database in 2013. Nucleic acids research 2013;41:D801–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q; Liew Z; Uppal K; Cui X; Ling C; Heck JE; von Ehrenstein OS; Wu J; Walker DI; Jones DP; Ritz B. Maternal serum metabolome and traffic-related air pollution exposure in pregnancy. Environment international 2019;130:104872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanosky JD; MacIntosh DL A comparison of four gravimetric fine particle sampling methods. J Air Waste Manag Assoc 2001;51:878–884 [DOI] [PubMed] [Google Scholar]
- Yu T; Park Y; Li S; Jones DP Hybrid feature detection and information accumulation using high-resolution LC-MS metabolomics data. Journal of proteome research 2013;12:1419–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F; Zhao J; Chen J; Xu Y; Xu L. Pollution characteristics of organic and elemental carbon in PM2.5 in Xiamen, China. J Environ Sci (China) 2011;23:1342–1349 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.