INTRODUCTION:
Several complications of decompensated cirrhosis are believed to result from increased intestinal permeability. However, little is known about the relationship between mucosal bacteria and epithelial permeability in cirrhosis. We aimed to assess epithelial permeability and associations with mucosal bacteria in patients with compensated cirrhosis.
METHODS:
We obtained duodenal tissue biopsies from patients with compensated cirrhosis and controls. Patients were excluded if they used antibiotics or immunosuppression. The composition of mucosal microbiota was determined by 16S rRNA gene sequencing and epithelial permeability by transepithelial electrical resistance (TEER) and tight junction protein expression.
RESULTS:
We studied 24 patients with compensated cirrhosis and 20 controls. Patients with cirrhosis were older than controls (62 vs 52 years, P = 0.02) but had a similar number of extrahepatic comorbidities (2.2 vs 1.4, P = 0.13). Patients with compensated cirrhosis had lower duodenal TEER (i.e., increased epithelial permeability; 13.3 Ω/cm2 ± 3.4 vs 18.9 Ω/cm2 ± 7.1; P = 0.004). Patients with compensated cirrhosis trended toward a distinct mucosal microbiota community structure relative to controls (P = 0.09). Clustering analysis identified two unique enterotypes. These enterotypes differed in bacterial composition and also TEER. A beta-binomial model found 13 individual bacteria associated with TEER, including Lactobacillus and Bifidobacterium taxa. Thirty-six taxa were associated with tight junction protein expression, including Lactobacillus and Bifidobacterium.
DISCUSSION:
Compensated cirrhosis is characterized by increased duodenal epithelial permeability with a distinct mucosal microbial community. Intriguingly, bacteria previously associated with health were protective of duodenal permeability.
INTRODUCTION
Decompensated cirrhosis is characterized by symptomatic complications of liver dysfunction, poor quality of life, and high mortality (1,2). Several manifestations of decompensated cirrhosis are believed to result from increased intestinal permeability including systemic infections, spontaneous bacterial peritonitis, hepatic encephalopathy, and acute-on-chronic liver failure.
There is no consensus on which gut segment is the most permeable in cirrhosis. However, prior research suggests that patients with cirrhosis have increased duodenal epithelial permeability (3). Tight junctions connect adjacent intestinal epithelial cells and regulate paracellular permeability. Patients with cirrhosis have diminished expression of some duodenal tight junction proteins, which in turn results in increased epithelial permeability (3–5).
Although there is evidence for increased duodenal epithelial permeability in cirrhosis, there are limited data regarding the role of microbiota. Microbiota influence intestinal epithelia and barrier function in several keyways including shared metabolism and immune regulation (6,7). The duodenum is where dietary nutrients, bile acids, pancreatic enzymes, and bacteria first coincide and engage in important metabolic activities, making it plausible that duodenal mucosal bacteria serve important functions. In patients with cirrhosis, fecal microbiota transplant using oral capsules led to a shift in duodenal mucosal microbiota composition, diminished proinflammatory cytokines, and increased intestinal barrier proteins (8).
The first step in assessing whether microbiota has a causative role in the pathogenesis of increased intestinal permeability is to establish whether intestinal microbiota is associated with permeability. In this study, we aimed to assess associations between duodenal mucosal bacteria and duodenal epithelial permeability in patients with compensated cirrhosis.
METHODS
Study design
We conducted a cross-sectional study collecting clinical, mucosal microbiome, and intestinal permeability data from human subjects at a single time point. We collected duodenum tissue biopsies from patients with compensated cirrhosis and controls undergoing routine outpatient endoscopies from October 2020 to December 2021. The clinical indication for upper endoscopy in patients with cirrhosis was esophageal variceal screening. The clinical indications for upper endoscopy in controls were globus sensation, prebariatric surgery, and anemia without iron deficiency. Four duodenal biopsy samples and duodenal aspirate were obtained from each patient. This study was approved by the University of Michigan Institutional Review Board, and all patients provided informed written consent for this study.
Patient selection
Patients and controls were excluded if they had inflammatory bowel disease or used an antibiotic (including rifaximin) or immunosuppressive medications in the preceding 4 weeks. Patients with a platelet count of <50/nL or international normalized ratio >1.5 were excluded to minimize bleeding risk, in accordance with an established research protocol. Cirrhosis was diagnosed by a hepatologist based on biopsy, imaging, or elastography. Compensated cirrhosis was defined as Child-Pugh class A and no recent symptomatic decompensation including variceal bleeding, ascites, or hepatic encephalopathy. Controls had no evidence of liver disease and no gastrointestinal symptoms. We also included data from a previously published cohort of 10 controls, collected and analyzed with the same protocols and instruments as described here (9). Controls were heterogeneous for nonhepatic comorbidities. We divided controls into 2 groups based on the Charlson Comorbidity Index (CCI): healthy controls (CCI 0–1) and comorbid controls (CCI ≥ 2).
Clinical data collection
Patients were asked about proton pump inhibitor, antibiotic, and probiotic use before endoscopy. Other clinical data including age, sex, model for end-stage liver disease score, Child-Pugh score, comorbidities, lactulose use, and etiology of liver disease were collected from a chart review.
Microbiome assessment
One duodenal biopsy sample per patient was analyzed for mucosal microbiota composition by 16S rRNA-encoding gene sequence analysis. Mucosal biopsies underwent DNA isolation using a Qiagen MagAttract PowerMag Microbiome DNA Isolation Kit (Germantown, MD). Barcoded dual-index primers specific to the V4 region of the 16S rRNA gene were used to amplify DNA using a “touchdown polymerase chain reaction” protocol given that duodenal mucosa is potentially of low biomass (10). Multiple negative controls were run in parallel. Libraries were prepared and sequenced using the Illumina 500-cycle MiSeq Reagent Kit V2 (San Diego, CA) (11). The mothur software package v1.45.3 was used to trim, screen, and align sequences; calculate distances; remove chimeras; assign sequences with ≥97% similarity to operational taxonomic units; and calculate alpha and beta diversities (11,12). Taxonomic classification was based on the Ribosomal Database Project (version 6/2020) (13). The SILVA rRNA reference alignment (release v132) was used to align the V4 region (14). The overall sequencing error rate, based on a comparison with a mock community, was 0.0017%.
Microbiome sequencing occurred in 2 separate runs. A Pseudomonas taxon was found in negative control samples in the first run. This same Pseudomonas was found in the second sequencing run in patients and not in negative controls—in other words, not as a contaminant in the second run. There was no difference in the abundance of this Pseudomonas taxon between the 2 sequencing runs (P = 0.98).
Epithelial permeability assessment
Two duodenal biopsies per patient were evaluated for epithelial permeability by transepithelial electrical resistance (TEER), a measure of paracellular permeability. After temperature and pH stabilization, duodenal biopsies underwent TEER analysis within 30 minutes of collection with a micro-Snapwell system with an EndOhm sural sensory nerve action potential electrode attached to an EVOM2 epithelial volt-ohm meter (World Precision Instruments) following published protocols (9). The results were reported as an average of the 2 biopsies and expressed in ohms per square centimeter (Ω/cm2).
One duodenal biopsy per patient was evaluated for tight junction protein gene expression. Each biopsy underwent real-time polymerase chain reaction to evaluate tight junction proteins claudin-1, occludin, and zonulin-1 as well as control gene glyceraldehyde-3-phosphate dehydrogenase expression using published protocols (CFX Connect Real-Time PCR Detection System, SYBR Green detection, Bio-Rad Laboratories, Hercules, CA) (9). Primers were obtained from Qiagen. Tight junction gene expression was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (15).
Statistical analysis
Descriptive statistics were reported as median and interquartile range for continuous variables and percentage for categorical variables. Wilcoxon rank-sum tests were used to compare continuous variables between 2 groups, and the Fisher exact test was used to compare 2 categorical variables. Unadjusted regression was used to identify covariates that influenced TEER. For microbiome data analysis, alpha diversity was assessed with inverse Simpson, as generated in mothur (12). Comparisons of the microbiota community structure between groups were based on analysis of molecular variance (AMOVA) of the Yue and Clayton dissimilarity index (θYC) and displayed as principal coordinate analysis of θYC distances (16). The partitioning around medoids (PAM) clustering algorithm was used to cluster samples into community clusters based on θYC distances (17). Beta-binomial regression models were used to associate the relative abundance of individual taxon with TEER, patient type, and microbiome community cluster. Unlike other modeling techniques, beta-binomial models allow for jointly assessing relative abundance and differential variability, the latter of which is valuable because some disease states manifest increased variability in bacterial abundance (18). R packages were used to analyze mothur outputs and other data and for figure creation (R Foundation for Statistical Computing, Vienna, Austria) (19).
Based on previous studies showing reduced TEER in patients with cirrhosis (17.0 [SD 0.8]) compared with controls (21.4 [SD 1.1]) (3), a sample size of 10 samples per patient group would be sufficient to provide 90% power, with an alpha of 0.05. Owing to a lack of available data on duodenal mucosal microbiota composition, we were unable to base power calculations on microbiome feature differences.
RESULTS
We sampled 24 patients with compensated cirrhosis and 10 controls and included data from a previously published cohort of 10 controls, without gastrointestinal symptoms (see Supplementary Figure 1, Supplementary Digital Content, http://links.lww.com/CTG/A855) (9). Therefore, we analyzed data from 24 patients with compensated cirrhosis and 20 controls (12 healthy controls with CCI 0–1 and 8 comorbid controls with CCI ≥2).
Clinical characteristics of patients studied
Patients with compensated cirrhosis were older than controls (median age 62 vs 52 years, P = 0.02) and had a similar number of extrahepatic comorbidities (CCI without points for liver disease; 2.2 vs 1.4, P = 0.13, Table 1). Patients with compensated cirrhosis were more likely to be male (62% vs 21%, P = 0.01). There was a trend toward a greater use of proton pump inhibitors in controls (41% vs 12%, P = 0.06). Six (25%) patients with cirrhosis and 7 (35%) controls underwent bowel preparation and colonoscopy on the same day as their upper endoscopy.
Table 1.
Patient characteristics
| Variable | Patient group | |
| Compensated cirrhosis, N = 24a | Control, N = 20a | |
| Age | 62 (56, 65) | 52 (38, 60) |
| Sex | ||
| Female | 9 (38%) | 15 (79%) |
| Male | 15 (62%) | 4 (21%) |
| Recent PPI | 3 (12%) | 7 (41%) |
| Recent probiotic | 3 (12%) | 2 (29%) |
| Recent lactulose | 2 (8.3%) | 0 (0%) |
| MELD | 7 (7,10) | |
| Viral cirrhosis | 5 (21%) | |
| NASH cirrhosis | 15 (62%) | |
| Alcohol cirrhosis | 6 (25%) | |
| Hepatocellular carcinoma | 1 (4.2%) | |
Double refers to combined upper endoscopy and colonoscopy in 1 session; these patients received bowel preparation fluid. Probiotic use was self-reported and referred to over-the-counter or prescribed probiotic supplements; specific probiotics were not recorded. Patients with 2 etiologies of liver disease are listed in both categories. Recent refers to the preceding 4 wk.
IQR, interquartile range; MELD, model for end-stage liver disease; NASH, nonalcoholic steatohepatitis; PPI, proton pump inhibitor.
Median (IQR); n (%).
Duodenal epithelial permeability
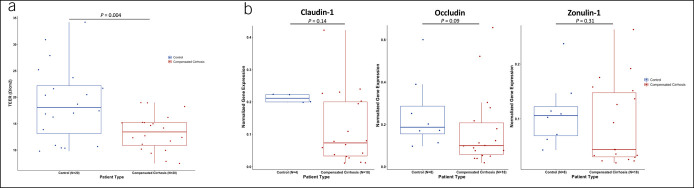
We evaluated the direct association between cirrhosis and duodenal epithelial permeability. Patients with compensated cirrhosis had lower duodenal TEER than controls (i.e., increased epithelial permeability; 13.3 Ω/cm2 ± 3.4 vs. 18.9 Ω/cm2 ± 7.1; P = 0.004; Figure 1a). Removing patients on proton pump inhibitors did not change this finding (cirrhosis: 13.7 vs controls: 20.5 Ω/cm2, P = 0.03). In unadjusted regression, duodenal TEER did not vary by age (r2 = 0.002, P = 0.79), sex (r2 = 0.04, P = 0.10), alcohol etiology of cirrhosis (r2 = 0.06, P = 0.31), presence/absence of colonoscopy preparation (r2 = 0.07, P = 0.09), model for end-stage liver disease (r2 = 0.01, P = 0.47), platelet count (r2 = 0.15, P = 0.47), or proton pump inhibitor use (r2 = 0.003, P = 0.77). Controls were heterogeneous for nonhepatic comorbidities. Patients with compensated cirrhosis had lower duodenal TEER than healthy controls (13.3 Ω/cm2 ± 3.4 vs 18.9 Ω/cm2 ± 7.2; P = 0.03; see Supplementary Figure 2, Supplementary Digital Content, http://links.lww.com/CTG/A856) and trended toward lower duodenal TEER than comorbid controls (13.3 Ω/cm2 ± 3.4 vs 19.0 Ω/cm2 ± 7.3; P = 0.07). Tight junction proteins claudin-1 (P = 0.14) and occludin (P = 0.09), although not zonulin-1 (P = 0.31), trended toward lower normalized gene expression in compensated cirrhosis compared with controls (Figure 1b).
Figure 1.
Duodenal epithelial permeability measures in patients with cirrhosis and noncirrhotic Controls. (a) Duodenal TEER between controls and compensated cirrhosis. (b) Tight junction protein gene expression between controls and compensated cirrhosis. TEER, transepithelial electrical resistance.
Next, we evaluated the association between tight junction proteins and duodenal epithelial permeability. Tight junction protein expression was divided into high vs low by the median value. TEER was associated with occludin expression (r2 = 0.16, P = 0.03)—but not claudin-1 (r2 = 0.05, P = 0.17) or zonulin-1 (r2 = 0.04, P = 0.18)—although the direction of the association was positive in all 3 cases.
Duodenal mucosal microbiota
We evaluated the association between cirrhosis and duodenal mucosal microbiota composition. Duodenal microbiota analyses were performed in 23 patients with cirrhosis and 5 controls. Patients with compensated cirrhosis trended toward lower alpha diversity (median inverse Simpson: 4.6 vs13.1, P = 0.14; Figure 2a) and may have a distinct mucosal microbiota community structure relative to controls based on AMOVA of the θYC dissimilarity index (P = 0.09; Figure 2b). Duodenal mucosa in compensated cirrhosis had the highest relative abundance of Proteobacteria, followed by Firmicutes and Bacteroides phyla, while controls had the highest relative abundance of Firmicutes, followed by Proteobacteria and Bacteroides (Figure 2c). Duodenal mucosa microbiota composition did not vary by sex or age.
Figure 2.
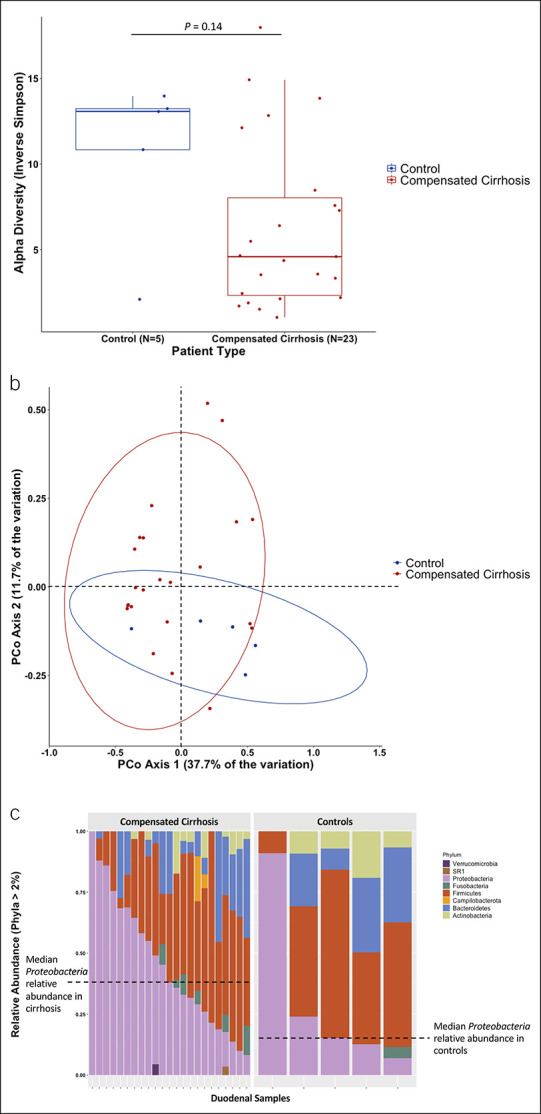
Duodenal mucosal microbiota may differ between patients with cirrhosis and noncirrhotic controls. (a) Alpha diversity as measured by inverse Simpson between controls and compensated cirrhosis. Higher inverse Simpson signifies greater alpha diversity. (b) PCoA comparing duodenal mucosal microbiota in controls vs compensated cirrhosis. (c) Comparison of relative abundance of phyla (those > 2% abundance) between compensated cirrhosis and controls. PCoA, principal coordinate analysis.
Enterotypes and association with duodenal epithelial permeability
We evaluated for the presence of enterotypes and their association with epithelial permeability. When duodenal mucosal microbiota data in patients with cirrhosis and controls were combined, PAM clustering analysis identified 2 unique clusters or enterotypes (Figure 3a) with distinct bacterial family composition (Figure 3b). Patients with cluster 1 enterotype had significantly lower TEER (12.3 Ω/cm2 ± 3.3, vs 15.8 Ω/cm2 ± 3.0, P = 0.01; Figure 3c). Cluster 1 included 15 patients with cirrhosis and 1 control while cluster 2 included 8 patients with cirrhosis and 4 controls (Fisher exact test: P = 0.13). The control patient with a cluster 1 enterotype was 70 years old, had a CCI of 4, and was the same outlier patient for alpha diversity (see Figure 3b). A beta-binomial model found 48 taxa that significantly differed between clusters 1 and 2 with a false discovery rate of 0.05.
Figure 3.
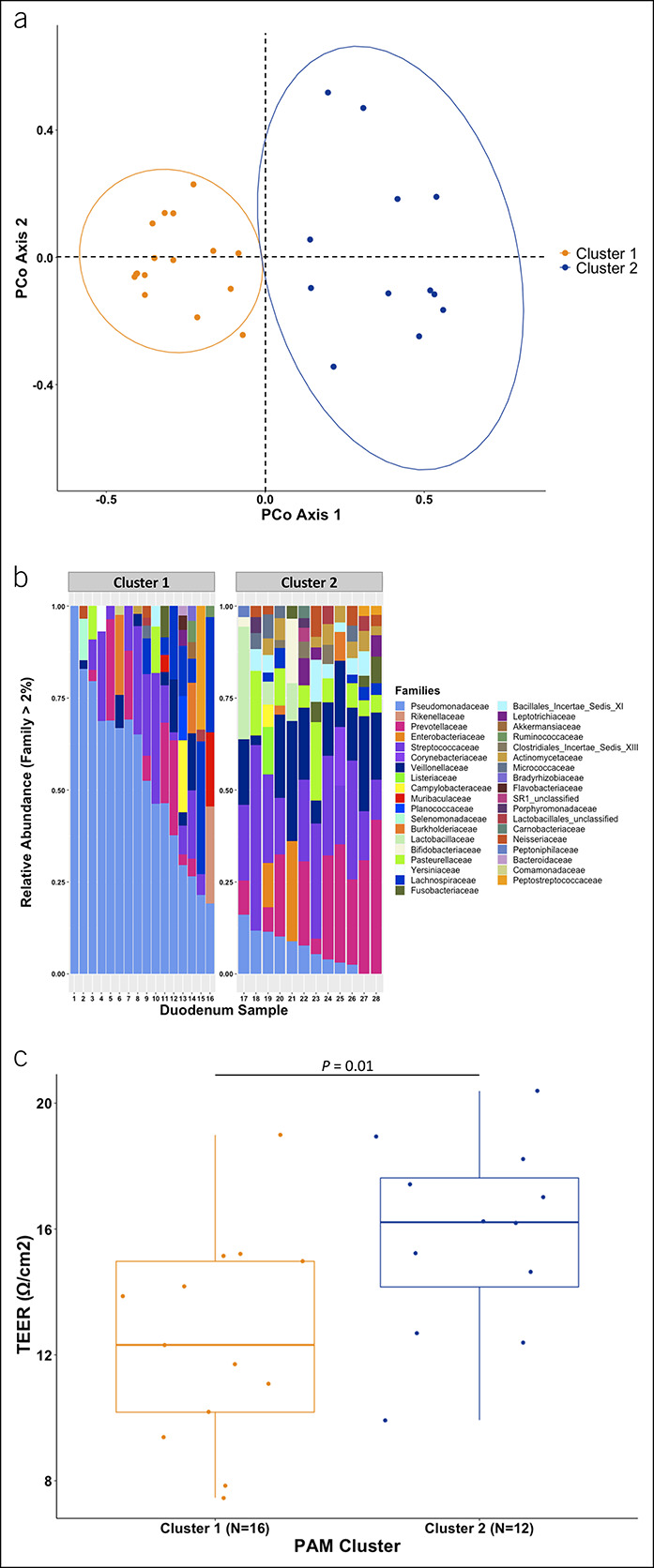
Duodenal mucosal microbiome enterotypes vary by epithelial permeability in the combined cohort of patients with cirrhosis and controls. (a) PCoA of θYC distances comparing duodenal mucosal microbiota in cluster 1 with cluster 2 enterotypes. (b) Comparison of relative abundance of families (those with >2% abundance) between cluster 1 and 2 enterotypes. (c) Duodenal transepithelial electrical resistance between those with a cluster 1 vs cluster 2 enterotype. PCoA, principal coordinate analysis.
We looked specifically at the Pseudomonas bacteria, which contaminated negative controls in the first sequencing run but appeared in patients in both sequencing runs. These Pseudomonas taxa were more abundant in cluster 1 than cluster 2 enterotypes (median: 52% vs 6%, P < 0.001). However, sequencing run did not significantly associate with cluster (P = 1.00) or Pseudomonas taxa abundance (P = 0.98), indicating that cluster identity was not driven by Pseudomonas contamination in the first sequencing run.
Association between duodenal microbiota and permeability in cirrhosis
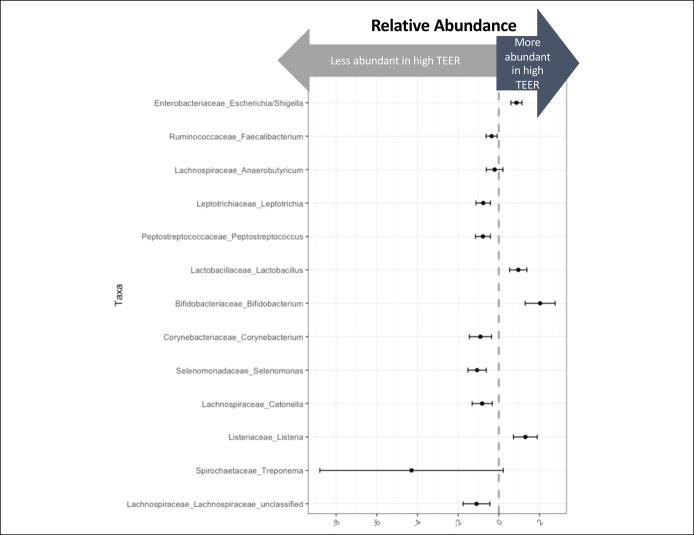
We evaluated whether specific taxa associated with epithelial permeability. A beta-binomial model found that 13 taxa associated with TEER with a false discovery rate of 0.05 (Figure 4; see Supplementary Table 1, Supplementary Digital Content, http://links.lww.com/CTG/A857). Relative abundance of Lactobacillus and Bifidobacterium bacteria positively associated with higher TEER, indicating that these 2 taxa are associated with a less permeable duodenal epithelium. Eight of these 13 taxa associated with TEER were also differentially abundant in cirrhosis, including Enterobacteriaceae Escherichia/Shigella, Lachnospiraceae Anaerobutyricum, Lactobacillaceae Lactobacillus, and Bifidobacteriaceae Bifidobacterium.
Figure 4.
Duodenal mucosal bacteria that associate with epithelial permeability. Thirteen taxa were significantly associated with TEER in a beta-binomial model with a false discovery rate of 0.05. TEER, transepithelial electrical resistance.
Next, we evaluated whether specific taxa associated with tight junction protein expression. A beta-binomial model found that 20 taxa associated with claudin-1, 13 taxa with occludin, and 20 taxa with zonulin-1, with a false discovery rate of 0.05. Four taxa positively associated with all 3 tight junction proteins, including Lachnospiraceae Blautia, Lactobacillaceae Lactobacillus, and Bifidobacteriaceae Scardovia.
Duodenal aspirate vs mucosal microbiota in cirrhosis
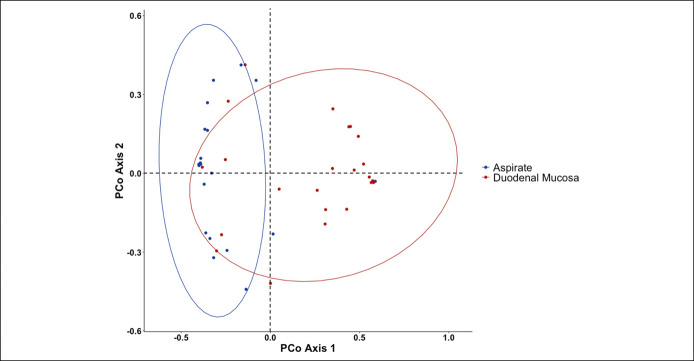
In patients with cirrhosis, duodenal aspirate microbiota composition differed from that in duodenal mucosa in AMOVA (P < 0.001; Figure 5). A beta-binomial model found that 6 taxa from duodenal aspirate associated with TEER, when using a false discovery rate of 0.05 (see Supplementary Table 2, Supplementary Digital Content, http://links.lww.com/CTG/A858). Two of these 6, Enterobacteriaceae Escherichia/Shigella and Bifidobacteriaceae Bifidobacterium, were also associated with higher TEER in the model with duodenal mucosal microbiota and were more abundant in controls vs patients with cirrhosis.
Figure 5.
Duodenal mucosa and aspirate microbiota differ in cirrhosis. PCoA of θYC distances comparing microbiota composition in duodenal mucosa and aspirate from patients with cirrhosis. PCoA, principal coordinate analysis.
DISCUSSION
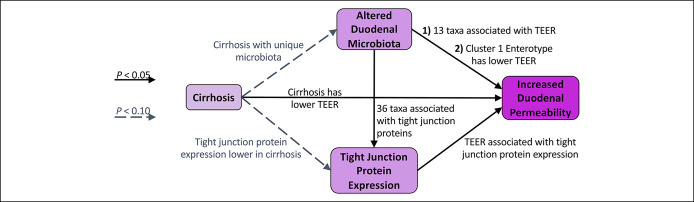
In this study, we investigated relationships between mucosal bacteria and epithelial permeability in patients with cirrhosis (Figure 6). We focused on the duodenum because previous literature shows diminished duodenal tight junction protein expression and high permeability in cirrhosis, and the duodenum is where enteric bacteria first engage in important metabolic activities. We confirmed that even patients with early-stage or compensated cirrhosis have increased permeability of their duodenal epithelia—in other words, leaky gut. We found that patients with compensated cirrhosis may have distinct duodenal mucosal bacterial community structure and that these bacterial communities and specific taxa are associated with intestinal permeability.
Figure 6.
Proposed causal diagram of how cirrhosis leads to increased duodenal permeability. Each arrow represents a hypothesized causal interaction, and the relationship between the variables at the base and tip of each arrow was evaluated through statistical analyses in the following sections. The figure summarizes the results of these analyses. Solid arrows represent analyses which found a P value of <0.05 while dashed arrows represent analyses which found a P value of <0.10.
Figure 6 shows the causal framework we used when approaching the analyses for this study. Each arrow represents a hypothesized causal interaction, and the relationship between the variables at the base and tip of each arrow was evaluated through statistical analyses in the following sections. The figure demonstrates the results of these analyses.
Our study confirms and expands on previous studies. Several previous studies have demonstrated increased intestinal permeability in cirrhosis through increased serum lipopolysaccharide (LPS), but LPS is influenced by Gram-negative bacterial abundance and not just intestinal permeability alone (3,4,20,21). One previous study demonstrated low duodenal TEER (i.e., high epithelial permeability) in cirrhosis, but that study included both compensated and decompensated patients with cirrhosis (3). Our study found high duodenal epithelial permeability in patients with compensated cirrhosis, indicating that small intestinal barrier function degrades even in early stages of cirrhosis, even when compared with older controls with comorbidities.
Our study also confirms previous studies demonstrating that intestinal microbial composition differs between cirrhosis and controls (22–24). This difference has been demonstrated in stool and sigmoid colon mucosa previously. Our study is the first to suggest that duodenal mucosal microbiota differs between controls and compensated cirrhosis. In our study, duodenal mucosa in cirrhosis was dominated by taxa in the phylum Proteobacteria while controls were dominated by Firmicutes. These alterations match those previously found in stool samples (22).
The novel findings of this study are the associations between mucosal bacteria and intestinal permeability. The PAM clustering algorithm was used to group patients into clusters (or enterotypes) based on similar bacterial composition. Two unique patient clusters were found. Cluster 1 contained more Pseudomonas, a genera with almost uniformly pathogenic qualities (25), whereas cluster 2 contained more Lactobacillus, Bifidobacterium, and Clostridial taxa, each known to have potentially beneficial effects on the host (26–31). Patients with the cluster 1 enterotype had significantly higher epithelial permeability (“leaky” gut) than patients with the cluster 2 enterotype. All controls except for 1 outlier were in cluster 2, lending further credence to this being the enterotype more associated with intestinal epithelial health. Several taxa including Lactobacillus and Bifidobacterium were differentially abundant and dispersed between the 2 clusters and also associated with TEER. Some Lactobacillus and Bifidobacterium strains are known to decrease intestinal permeability either through increased tight junction protein production or by providing short-chain fatty acids (SCFAs) as a nutritional source to intestinal epithelia (26–31). While SCFAs are known for their importance to colonic epithelial health, SCFAs are also produced in the small bowel (32,33), and SCFAs have decreased duodenal permeability in an animal model (34,35). It is unclear whether Lactobacillus and Bifidobacterium directly influence intestinal permeability in cirrhosis or simply represent a biomarker of intestinal health, but there is clearly an association between duodenal mucosal bacteria and intestinal permeability in compensated cirrhosis.
Tight junction protein expression may be one of the ways in which mucosal microbiota can influence intestinal permeability. Published data to date are mixed on claudin-1, occludin, and zonulin-1 expression in the cirrhosis duodenum (3–5). Our study found that occludin, in particular, trended toward lower expression in cirrhosis and was associated with TEER. We found 36 taxa which significantly associated with tight junction protein expression, including Lactobacillus whose abundance positively associated with the expression of all 3 tight junction proteins. Several Lactobacillus strains have been shown to increase tight junction protein production (26–28).
This study focused on the duodenum, which has biological and practical implications. Most prior investigations into intestinal microbiota and permeability in cirrhosis focused on the colon, in part, because this is the gut segment with the largest burden of intestinal bacteria. However, at least 1 previous study has found that manipulating the microbiome with oral fecal microbiota transplant capsules also influences the duodenal mucosal microbiome and antimicrobial peptides and is associated with cognitive improvement (e.g., systemic changes) in patients with cirrhosis (8). The duodenum is the place where enteral nutrients, pancreatic enzymes, and bile acids first coincide and engage in important metabolic activities. It is, therefore, biologically plausible that mucosal bacteria in the duodenum have important metabolic functions, which in turn influence the intestinal barrier. On a practical note, patients with cirrhosis frequently undergo routine upper endoscopy for gastroesophageal varices surveillance, so duodenal sampling is readily available for future clinical research.
Our results must be interpreted within the context of the study design. We were limited to ex vivo techniques (TEER and tight junction protein gene expression) for assessing epithelial permeability. We may, therefore, have missed some contribution of portal hypertension or other influences which could only be assessed in vivo. This was a cross-sectional study, and we were unable to assess stability of our findings over time or a cause-and-effect relationship. Bacterial composition may be a biomarker of liver disease, diet, environment, metabolism, or immune function, as opposed to direct influencers of barrier function. Owing to the small sample size, we were not able to compare the ability of duodenal mucosal microbiota with that of duodenal aspirate to predict TEER. With only 5 control microbiota samples, we were unable to draw conclusions about differences between control and cirrhosis duodenal microbiota. Finally, a limitation of this work was the Pseudomonas contamination in negative controls in the first sequencing run. Further analysis found that this Pseudomonas was also abundant in the second sequencing run (without contamination); this taxon did not predict TEER; and sequencing run did not determine PAM clustering.
In conclusion, this study confirms that duodenal epithelial permeability is increased (“leaky”) in compensated cirrhosis. It also suggests that the duodenal mucosal microbiome in cirrhosis differs from controls and is associated with intestinal permeability. This work moves beyond typical techniques in human studies to assess microbiota composition (stool samples) and intestinal permeability (serum LPS). The next step is to perform a longitudinal study to investigate the role of microbiota, especially Lactobacillus, Bifidobacterium, and occludin, in influencing intestinal permeability and the development of subsequent decompensating events, such as hepatic encephalopathy and spontaneous bacterial peritonitis. Specific pathogenic pathways will need to be confirmed in model systems to confirm a causal relationship. These findings may lead to microbial interventions such as therapy to prevent or treat decompensating events in patients with cirrhosis.
CONFLICTS OF INTEREST
Guarantor of the article: P.P. Bloom, MD.
Specific author contributions: P.P.B.: conception of the work, acquisition of data, analysis, interpretation of data, and drafting the work. K.R.: data analysis and interpretation and critical revisions. C.M.B.: data analysis and interpretation and critical revision. S.Y.Z.: data acquisition. B.N.: data acquisition and critical revisions. C.O.: conception of the work. V.B.Y.: conception of the work, analysis, interpretation of data, and critical revisions. A.S.L.: conception of the work, acquisition of data, analysis, interpretation of data, and critical revisions.
Financial support: P.P.B. received funding from the American College of Gastroenterology (ACG Junior Faculty Award). V.B.Y. was funded by NIH Award AI124255.
Potential competing interests: P.P.B. holds a research grant from Vedanta Biosciences. V.B.Y. serves as a consultant to Vedanta, Bio-K+ International, and Pantheryx.
Study Highlights.
WHAT IS KNOWN
✓ Cirrhosis is associated with increased intestinal permeability.
✓ Patients with cirrhosis have a different stool microbiome than healthy people.
WHAT IS NEW HERE
✓ Patients with compensated cirrhosis have increased duodenal permeability.
✓ Compensated cirrhosis is characterized by a distinct duodenal mucosal microbial community.
✓ Bacteria previously associated with health were protective of duodenal permeability.
Supplementary Material
ACKNOWLEDGEMENTS
This research was supported by work performed in the University of Michigan's Microbiome Core.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A855, http://links.lww.com/CTG/A856, http://links.lww.com/CTG/A857, http://links.lww.com/CTG/A858
Contributor Information
K. Rao, Email: krirao@med.umich.edu.
C.M. Bassis, Email: cbassis@med.umich.edu.
S.Y. Zhou, Email: zhousy@med.umich.edu.
B. Nojkov, Email: bnojkov@med.umich.edu.
C. Owyang, Email: cowyang@med.umich.edu.
V.B. Young, Email: youngvi@med.umich.edu.
A.S. Lok, Email: aslok@med.umich.edu.
REFERENCES
- 1.Rabiee A, Ximenes RO, Nikayin S, et al. Factors associated with health-related quality of life in patients with cirrhosis: A systematic review. Liver Int 2021;41(1):6–15. [DOI] [PubMed] [Google Scholar]
- 2.D'Amico G, Garcia-Tsao G, Pagliaro L. Natural history and prognostic indicators of survival in cirrhosis: A systematic review of 118 studies. J Hepatol 2006;44(1):217–31. [DOI] [PubMed] [Google Scholar]
- 3.Du Plessis J, Vanheel H, Janssen CEI, et al. Activated intestinal macrophages in patients with cirrhosis release NO and IL-6 that may disrupt intestinal barrier function. J Hepatol 2013;58(6):1125–32. [DOI] [PubMed] [Google Scholar]
- 4.Assimakopoulos SF, Tsamandas AC, Tsiaoussis GI, et al. Altered intestinal tight junctions' expression in patients with liver cirrhosis: A pathogenetic mechanism of intestinal hyperpermeability. Eur J Clin Invest 2012;42(4):439–46. [DOI] [PubMed] [Google Scholar]
- 5.Pijls KE, Koek GH, Elamin EE, et al. Large intestine permeability is increased in patients with compensated liver cirrhosis. Am J Physiol Gastrointest Liver Physiol 2014;306(2):G147–53. [DOI] [PubMed] [Google Scholar]
- 6.Sanders ME, Merenstein DJ, Reid G, et al. Probiotics and prebiotics in intestinal health and disease: From biology to the clinic. Nat Rev Gastroenterol Hepatol 2019;16(10):605–16. [DOI] [PubMed] [Google Scholar]
- 7.Visconti A, Le Roy CI, Rosa F, et al. Interplay between the human gut microbiome and host metabolism. Nat Commun 2019;10(1):4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bajaj JS, Salzman NH, Acharya C, et al. Fecal microbial transplant capsules are safe in hepatic encephalopathy: A phase 1, randomized, placebo-controlled trial [published correction appears in Hepatology]. Hepatology 2020;7270(45):15011690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nojkov B, Zhou SY, Dolan RD, et al. Evidence of duodenal epithelial barrier impairment and increased pyroptosis in patients with functional dyspepsia on confocal laser endomicroscopy and “ex vivo” mucosa analysis. Am J Gastroenterol 2020;115(11):1891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koenigsknecht MJ, Theriot CM, Bergin IL, et al. Dynamics and establishment of Clostridium difficile infection in the murine gastrointestinal tract. Infect Immun 2015;83(3):934–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozich JJ, Westcott SL, Baxter NT, et al. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79(17):5112–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75(23):7537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cole JR, Wang Q, Fish JA, et al. Ribosomal database project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42(Database issue):D633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 2013;41(Database issue):D590–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 16.Yue JC. A similarity measure based on species proportions. Commun Stat Theory Methods 2008;34:2123–31. [Google Scholar]
- 17.Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome [published correction appears in Nature. 2011 Jun 30;474(7353):666] [published correction appears in Nature. 2014 Feb 27;506(7489):516]. Nature 2011;473(7346):174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin BD, Witten D, Willis AD. Modeling microbial abundances and dysbiosis with beta-binomial regression. Ann Appl Stat 2020;14(1):94–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2020. https://www.R-project.org/. [Google Scholar]
- 20.Assimakopoulos SF, Tsamandas AC, Tsiaoussis GI, et al. Intestinal mucosal proliferation, apoptosis and oxidative stress in patients with liver cirrhosis. Ann Hepatol 2013;12(2):301–7. [PubMed] [Google Scholar]
- 21.Alexopoulou A, Agiasotelli D, Vasilieva LE, Dourakis SP. Bacterial translocation markers in liver cirrhosis. Ann Gastroenterol 2017;30(5):486–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bajaj JS, Heuman DM, Hylemon PB, et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J Hepatol 2014;60(5):940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bajaj JS, Hylemon PB, Ridlon JM, et al. Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation. Am J Physiol Gastrointest Liver Physiol 2012;303(6):G675–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513(7516):59–64. [DOI] [PubMed] [Google Scholar]
- 25.Laughlin RS, Musch MW, Hollbrook CJ, et al. The key role of Pseudomonas aeruginosa PA-I lectin on experimental gut-derived sepsis. Ann Surg 2000;232(1):133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahmadi S, Wang S, Nagpal R, et al. A human-origin probiotic cocktail ameliorates aging-related leaky gut and inflammation via modulating the microbiota/taurine/tight junction axis. JCI Insight 2020;5(9):e132055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou Q, Huang Y, Wang Y, et al. Lactobacillus casei LC01 regulates intestinal epithelial permeability through miR-144 targeting of OCLN and ZO1. J Microbiol Biotechnol 2020;30(10):1480–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W, Li H, Zhao N, et al. Lactobacillus johnsonii BS15 combined with abdominal massage on intestinal permeability in rats with nonalcoholic fatty liver and cell biofilm repair. Bioengineered 2021;12(1):6354–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Sadi R, Dharmaprakash V, Nighot P, et al. Bifidobacterium bifidum enhances the intestinal epithelial tight junction barrier and protects against intestinal inflammation by targeting the toll-like receptor-2 pathway in an NF-κB-Independent manner. Int J Mol Sci 2021;22(15):8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh CY, Osaka T, Moriyama E, et al. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol Rep 2015;3(3):e12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling X, Linglong P, Weixia D, Hong W. Protective effects of Bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of caco-2 monolayers and in a rat NEC model. Plos One 2016;11(8):e0161635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zoetendal EG, Raes J, van den Bogert B, et al. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 2012;6(7):1415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neis EP, van Eijk HM, Lenaerts K, et al. Distal versus proximal intestinal short-chain fatty acid release in man. Gut 2019;68(4):764–5. [DOI] [PubMed] [Google Scholar]
- 34.Akiba Y, Inoue T, Kaji I, et al. Short-chain fatty acid sensing in rat duodenum. J Physiol 2015;593(3):585–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan Saudi WS, Sjöblom M. Short-chain fatty acids augment rat duodenal mucosal barrier function. Exp Physiol 2017;102(7):791–803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.