INTRODUCTION:
Microscopic colitis is a relatively common cause of chronic diarrhea and may be linked to luminal factors. Given the essential role of the microbiome in human gut health, analysis of microbiome changes associated with microscopic colitis could provide insights into the development of the disease.
METHODS:
We enrolled patients who underwent colonoscopy for diarrhea. An experienced pathologist classified patients as having microscopic colitis (n = 52) or controls (n = 153). Research biopsies were taken from the ascending (ASC) and descending (DES) colon, and the microbiome was characterized with Illumina sequencing. We analyzed the associations between microscopic colitis and microbiome with a series of increasingly complex models adjusted for a range of demographic and health factors.
RESULTS:
We found that alpha diversity was significantly lower in cases with microscopic colitis compared with that in controls in the DES colon microbiome. In the DES colon, a series of models that adjusted for an increasing number of covariates found taxa significantly associated with microscopic colitis, including Proteobacteria that was enriched in cases and Collinsella that was enriched in controls. While the alpha diversity and taxa were not significantly associated with microscopic colitis in the ASC colon microbiome, the inference P values based on ASC and DES microbiomes were highly correlated.
DISCUSSION:
Our study demonstrates an altered microbiome in cases with microscopic colitis compared with that in controls. Because both the cases and controls experienced diarrhea, we have identified candidate taxa that could be mechanistically responsible for the development of microscopic colitis independent of changes to the microbial community caused by diarrhea.
INTRODUCTION
Microscopic colitis is a common cause of chronic diarrhea (1,2). The colon appears grossly normal during colonoscopy, but there is a thickened collagen band or lymphocytic infiltration microscopically. Although microscopic colitis was previously believed to be rare, the incidence has increased in Europe and North America (3–9). The incidence is comparable with Crohn's disease and ulcerative colitis, conditions that have received much more research attention.
The exact etiology of microscopic colitis is unknown, but the gut microbiome is considered to play an important role. It is widely accepted that the condition represents an abnormal immune reaction to luminal antigens in the predisposed host (10). The hypothesis of the involvement of a luminal factor is supported by resolution of the disease with diversion of the fecal stream but recurrence when continuity is restored (11,12). Fecal diversion also has a profound effect on the gut microbiome (13). The human gut microbiome therefore likely plays important roles in the development of microscopic colitis. Investigation of the changes in microbiome composition associated with microscopic colitis could contribute to our understanding of the etiology of microscopic colitis and provide insights on treatment.
Previous studies (14–19) on the microbiome associated with microscopic colitis have not always reported consistent results, and they have generally involved small numbers of patients, fecal samples, and comparison with healthy controls. Diarrhea in patients with microscopic colitis can affect the microbial composition of their fecal samples (20), and this difference could contribute to the differences between the fecal samples of patients with microscopic colitis and healthy controls.
To learn more about the possible roles of bacteria in microscopic colitis, we conducted a prospective study at a single academic medical center. We recruited study participants from patients who underwent colonoscopy for diarrhea. A research pathologist reviewed biopsies to determine whether the diarrhea was caused by microscopic colitis. We characterized the microbiomes of the colon biopsies with 16S ribosomal RNA (rRNA) sequencing and systematically collected detailed demographic and exposure information from patient interviews.
METHODS
Study population and sample collection
We identified patients who were referred to the University of North Carolina Hospitals for diarrhea between April 1, 2015, and December 22, 2020. Patients with known inflammatory bowel disease, Clostridioides difficile infection, or infectious diarrhea based on chart review and patients with gross evidence of inflammatory bowel disease were excluded. Patients had to report loose stools, as measured by the Bristol Stool Form Scale type 5–7 (21). Eligible patients signed informed consent, The Health Insurance Portability and Accountability Act of 1996, and Storing Biological Specimens with Identifying Information form. During colonoscopy, clinical biopsies were taken for standard pathologic review. Research biopsies were taken from the ascending (ASC) colon and descending (DES) colon and immediately frozen in liquid nitrogen for later microbial analysis. Specimens were taken to the laboratory where they were stored at −80 °C. Patients were classified as cases with microscopic colitis or controls by an experienced gastrointestinal pathologist (J.T.W.). Patients with microscopic colitis had increased numbers of intraepithelial or lamina propria lymphocytes or a thickened collagen band. The control group had neither increased lymphocytes nor thickened collagen band. Patients with nonlymphocytic colitis were excluded. Patients were interviewed by phone about demographic factors, diet, medications, symptoms, and autoimmune disease. Patients were asked whether they had taken antibiotics in the 3 months before their colonoscopy. An identical interview was offered to some patients to be self-completed over the Internet. The study was approved by the Institutional Review Board of the University of North Carolina at Chapel Hill (14-3156).
DNA extraction, PCR, and sequencing
Bacterial genomic DNA was extracted using previously described protocols (22,23). Normal colonic mucosal biopsies were placed in lysozyme-containing buffer for 30 minutes, followed by bead beating and DNA extraction with the Qiagen DNA Blood and Tissue Kit (Qiagen Cat# 69504). The purified DNA samples were stored in aliquots at −20 °C.
Illumina library preps were performed using previously published protocols (24,25). In brief, polymerase chain reaction (PCR) amplification was conducted in 2 separate reaction steps. The first PCR (PCR1) reaction contained the Phusion High-Fidelity Master Mix (Life Technologies, Carlsbad, CA) and primers targeting the V2 region of the 16S bacterial rRNA gene. The PCR1 product served as a template for the second PCR step (PCR2). PCR2 reaction mix contained primers with an Illumina index barcode sequence, Illumina adapter sequence, and a tag sequence. An equal volume of each sample library was pooled, followed by cleaning using AxyPrep Mag Beads (24). The pool was stored at −20 °C and shipped to the University of Maryland Institute for Genome Sciences for Illumina MiSeq sequencing (24). Positive and negative controls were included in all sample preparation steps. The sequencing data analyzed in this study are available at NCBI as BioProject PRJNA768799.
Sequence processing and statistical analysis
The sequencing reads were analyzed with QIIME2 and DADA2 following the instructions (26,27). The forward reads were first truncated to 250 bp and denoised with DADA2 with the chimera removed using DADA2 consensus method. The amplicon sequencing variants were then classified with the QIIME2 feature classifier classify-sklearn based on the SILVA database (release 132) (28). The taxonomic abundance tables were normalized as previously described to correct for the different sequencing depth across samples (29).
Statistical analyses and visualization were performed with R (version 4.0.5). The associations between the microbial community and case/control were analyzed with the permutational multivariate ANOVA (PERMANOVA) test using the R function adonis in the package vegan. The principal coordinate analysis (PCoA) of the microbiomes was based on the Bray-Curtis dissimilarity at the genus level and visualized with functions in the same package. Shannon diversity was calculated with the function diversity of the R package vegan and used to characterize the alpha diversity of the microbiome. We used multivariate-adjusted linear regression models with the R function lm to analyze the associations between case/control and Shannon diversity, PCoA coordinates, and individual taxa. The covariates were selected based on their known associations with microscopic colitis or gut microbiome (30–32). Missing data were not included. Continuous data were not grouped. We use 4 linear models: model 1 was adjusted for the covariate education, proton pump inhibitor (PPI) use, and batch effects; model 2 included additional covariates sex and antibiotics use; model 3 was additionally adjusted for age; model 4 was additionally adjusted for body mass index (BMI). Rare taxa (prevalence <10% participants) were not included to avoid overadjustment for false discovery rate (FDR). P values were adjusted for multiple hypotheses testing with the Benjamini-Hochberg method. Significant taxa were visualized on taxonomic trees using the function tree_view in the R package plotmicrobiome (https://github.com/ssun6/plotmicrobiome).
RESULTS
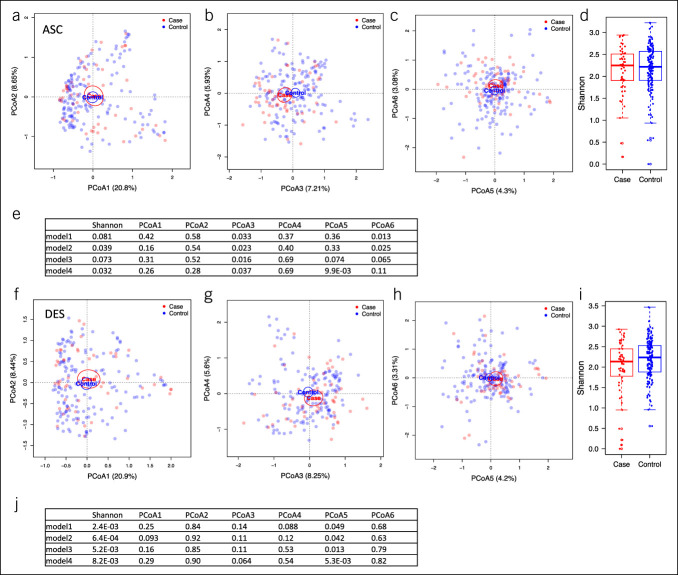
The characteristics of study participants are summarized in Table 1. The selection criteria flowchart for this analysis is shown in Supplementary Figure S1 (see Supplementary Digital Content 1, http://links.lww.com/CTG/A876). Cases with microscopic colitis were older than controls. Cases were more likely to be female and better educated. The BMI of cases was lower than controls. We characterized the ASC and DES colon microbiomes by Illumina sequencing technology to determine whether the microbiomes were different between cases with microscopic colitis and controls. We first analyzed the associations between case/control and the microbial community at the genus level with PCoA and univariate PERMANOVA tests. The case and control microbiomes were not separated at PCoA1 or PCoA2 for either ASC or DES microbiomes but showed better separation at PCoA 3–6 (Figure 1a–d and f–i). The PERMANOVA test indicated that the genus-level composition was not significantly associated with case/control in the ASC microbiome (P = 0.092) but was significant in the DES microbiome (R2 = 0.0087, P = 0.043). We used 4 models that were adjusted for different variables to analyze the associations between Shannon diversity, PCoA 1–6 (Figure 1e and j), and individual taxa (see Methods). Model 1 was adjusted for education, PPI use, and batch effects; model 2 included additional covariates sex and antibiotics use; model 3 was additionally adjusted for age, and model 4 was additionally adjusted for BMI. Shannon diversity of the ASC microbiomes was significantly different between cases and controls for models 2 and 4 (Figure 1d and e). However, in the DES microbiome, Shannon diversity was significantly lower in cases compared with that in controls in all 4 models (Figure 1i and j). In the ASC microbiome, PCoA3 was significantly associated with microscopic colitis with all 4 models (Figure 1e), and PCoA5 and PCoA6 were associated with microscopic colitis in some models (Figure 1e). In the DES microbiome, PCoA5 was significantly associated with microscopic colitis in all 4 models (Figure 1j).
Table 1.
Characteristics of study participants
| Participants characteristics | Case (n = 52) | Control (n = 153) | P valuea |
| Age, yr, mean (SD) | 62.2 (13.5) | 53.7 (11.8) | 9.0e-05 |
| Female, % | 90.4 | 75.2 | 0.019 |
| White, % | 96.2 | 85.6 | 0.53 |
| Education, % | 0.029 | ||
| Postgraduate degree | 28.8 | 23.9 | |
| College degree | 40.4 | 19.4 | |
| Some college | 23.1 | 35.1 | |
| GED | 1.9 | 1.5 | |
| High school degree | 5.8 | 15.7 | |
| Some high school | 0 | 3.0 | |
| Eighth grade or less | 0 Missing = 0 |
1.5 Missing = 19 |
|
| BMI, mean (SD) | 25.2 (6.2) Missing = 2 |
29.5 (6.9) Missing = 5 |
7.5e-05 |
BMI, body mass index; GED, general educational development.
For age and BMI, P value derived from the Wilcoxon rank sum test. For sex, race, and education, P value derived from the Fisher's exact test.
Figure 1.
PCoA plots and alpha diversity of the ASC and DES colon microbiomes of study participants. (a–c) PCoA plots of the case and control microbiomes in the ASC microbiome showing PCoA 1–2 (a), 3–4 (b) and 5–6 (c). (d) Boxplots of the Shannon diversity of cases and controls in the ASC microbiome. (e) P values of linear regression models analyzing the associations between case/control and Shannon diversity, PCoA1–6 adjusted for covariates (see Methods) in the ASC microbiome. (f–h) PCoA plots of case and control microbiomes in the DES microbiome. (i) Boxplots of the Shannon diversity of cases and controls for the DES microbiome. (j) The P values of linear regression models analyzing the associations between case/control and Shannon index, PCoA1–6 adjusted for covariates (see Methods) in the DES microbiome. The ellipses in PCoA plots indicate 95% confidence limits. The boxplots showed the median and 25th and 75th percentiles. ASC, ascending; DES, descending; PCoA, principal coordinate analysis.
We also used the 4 models to analyze the associations between individual taxa and case/control to identify the differential taxa associated with microscopic colitis. The taxa of the ASC colon microbiome were not significantly associated with case/control in any of the models after adjusting for multiple testing, whereas there were significantly associated taxa in the DES microbiome (Table 2, FDR <0.1). Model 1 (adjusted for batch, education, and PPI) and model 2 (additionally adjusted for sex and antibiotics use) revealed similar differential taxa (21 in common, 3 only in model 1, and 6 only in model 2) (Figure 2). However, model 3 revealed 10 taxa after being additionally adjusted for age, and model 4 revealed only 1 taxa after being additionally adjusted for BMI (Figure 2). Because age and BMI were significantly different between case and control participants, it is possible that age and BMI confounded the microbiome associations with case/control.
Table 2.
The P values of linear regression models analyzing the associations between individual taxa and case/control adjusted for covariates (see Methods) in the ASC and DES colon microbiome
| ASC colon | DES colon | |||||||||||||||
| Model 1 | Model 2 | Model 3 | Model 4 | Model 1 | Model 2 | Model 3 | Model 4 | |||||||||
| t value | FDR | t value | FDR | t value | FDR | t value | FDR | t value | FDR | t value | FDR | t value | FDR | t value | FDR | |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales | −3.22 | 0.26 | −3.11 | 0.18 | −3.64 | 0.12 | −3.64 | 0.12 | −4.31 | 0.009 | −4.14 | 0.017 | −3.80 | 0.07 | −3.77 | 0.08 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Burkholderiaceae | −2.96 | 0.26 | −2.89 | 0.2 | −3.36 | 0.16 | −3.40 | 0.14 | −3.87 | 0.024 | −3.76 | 0.037 | −3.33 | 0.09 | −3.30 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Burkholderiaceae—g__Sutterella—s__unclassified | −2.55 | 0.28 | −2.53 | 0.24 | −2.95 | 0.24 | −2.73 | 0.26 | −3.42 | 0.058 | −3.33 | 0.045 | −2.69 | 0.16 | −2.28 | 0.25 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Pasteurellales—f__Pasteurellaceae—g__Haemophilus—s__unclassified | −1.04 | 0.79 | −1.08 | 0.69 | −1.49 | 0.65 | −1.07 | 0.87 | −3.38 | 0.058 | −3.18 | 0.045 | −3.24 | 0.09 | −2.59 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Pasteurellales—f__Pasteurellaceae—g__Haemophilus | −1.04 | 0.79 | −1.08 | 0.69 | −1.49 | 0.65 | −1.07 | 0.87 | −3.38 | 0.058 | −3.18 | 0.045 | −3.23 | 0.09 | −2.59 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Burkholderiaceae—g__Sutterella | −2.31 | 0.35 | −2.32 | 0.31 | −2.70 | 0.24 | −2.50 | 0.29 | −3.2 | 0.063 | −3.16 | 0.045 | −2.51 | 0.18 | −2.10 | 0.31 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Pasteurellales | −1.14 | 0.71 | −1.17 | 0.64 | −1.55 | 0.65 | −1.16 | 0.87 | −3.04 | 0.063 | −2.83 | 0.081 | −2.57 | 0.18 | −2.11 | 0.31 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Pasteurellales—f__Pasteurellaceae | −1.14 | 0.71 | −1.17 | 0.64 | −1.55 | 0.65 | −1.16 | 0.87 | −3.04 | 0.063 | −2.83 | 0.081 | −2.57 | 0.18 | −2.11 | 0.31 |
| p__Proteobacteria—c__Gammaproteobacteria | −2.95 | 0.26 | −3.31 | 0.18 | −2.99 | 0.24 | −3.10 | 0.14 | −3.03 | 0.063 | −3.25 | 0.045 | −2.36 | 0.21 | −2.38 | 0.22 |
| p__Proteobacteria | −2.91 | 0.26 | −3.28 | 0.18 | −2.99 | 0.24 | −3.08 | 0.14 | −2.96 | 0.063 | −3.17 | 0.045 | −2.29 | 0.23 | −2.33 | 0.23 |
| p__Firmicutes—c__Bacilli—o__Lactobacillales—f__Streptococcaceae—g__Streptococcus—s__unclassified | −1.47 | 0.54 | −1.22 | 0.62 | −1.74 | 0.52 | −1.73 | 0.57 | −2.95 | 0.063 | −2.83 | 0.081 | −3.08 | 0.09 | −2.71 | 0.16 |
| p__Firmicutes—c__Bacilli—o__Lactobacillales | −1.62 | 0.52 | −1.39 | 0.56 | −1.82 | 0.51 | −1.85 | 0.57 | −2.94 | 0.063 | −2.83 | 0.081 | −3.06 | 0.09 | −2.76 | 0.16 |
| p__Firmicutes—c__Bacilli—o__Lactobacillales—f__Streptococcaceae | −1.46 | 0.54 | −1.22 | 0.62 | −1.74 | 0.52 | −1.73 | 0.57 | −2.93 | 0.063 | −2.82 | 0.081 | −3.07 | 0.09 | −2.68 | 0.16 |
| p__Firmicutes—c__Bacilli—o__Lactobacillales—f__Streptococcaceae—g__Streptococcus | −1.47 | 0.54 | −1.22 | 0.62 | −1.74 | 0.52 | −1.74 | 0.57 | −2.93 | 0.063 | −2.82 | 0.081 | −3.07 | 0.09 | −2.69 | 0.16 |
| p__Bacteroidetes—c__Bacteroidia—o__Bacteroidales—f__Marinifilaceae | −2.82 | 0.26 | −2.79 | 0.21 | −2.16 | 0.40 | −1.92 | 0.55 | −2.88 | 0.068 | −2.77 | 0.083 | −2.09 | 0.31 | −1.95 | 0.35 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Neisseriaceae—g__Neisseria | −1.96 | 0.4 | −1.68 | 0.51 | −2.55 | 0.25 | −2.24 | 0.35 | −2.74 | 0.094 | −2.42 | 0.16 | −2.56 | 0.18 | −2.58 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Neisseriaceae—g__Neisseria—s__unclassified | −1.96 | 0.4 | −1.68 | 0.51 | −2.55 | 0.25 | −2.24 | 0.35 | −2.74 | 0.094 | −2.42 | 0.16 | −2.56 | 0.18 | −2.58 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Betaproteobacteriales—f__Neisseriaceae | −2.05 | 0.37 | −1.78 | 0.51 | −2.65 | 0.24 | −2.41 | 0.29 | −2.73 | 0.094 | −2.41 | 0.16 | −2.56 | 0.18 | −2.57 | 0.16 |
| p__Bacteroidetes—c__Bacteroidia—o__Bacteroidales—f__Marinifilaceae—g__Odoribacter—s__unclassified | −2.08 | 0.37 | −2.34 | 0.31 | −2.09 | 0.40 | −2.21 | 0.36 | −2.52 | 0.15 | −2.71 | 0.09 | −2.37 | 0.21 | −2.62 | 0.16 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Enterobacteriales | −2.64 | 0.28 | −3.03 | 0.18 | −2.70 | 0.24 | −2.85 | 0.21 | −2.47 | 0.16 | −2.79 | 0.08 | −2.03 | 0.33 | −2.05 | 0.31 |
| p__Proteobacteria—c__Gammaproteobacteria—o__Enterobacteriales—f__Enterobacteriaceae | −2.64 | 0.28 | −3.03 | 0.18 | −2.70 | 0.24 | −2.85 | 0.21 | −2.47 | 0.16 | −2.79 | 0.08 | −2.03 | 0.33 | −2.05 | 0.31 |
| p__Firmicutes—c__Clostridia—o__Clostridiales—f__Lachnospiraceae—g__Blautia | 1.92 | 0.4 | 2.14 | 0.34 | 1.32 | 0.75 | 1.15 | 0.87 | 2.36 | 0.19 | 2.67 | 0.1 | 1.40 | 0.57 | 1.16 | 0.63 |
| p__Firmicutes—c__Clostridia—o__Clostridiales—f__Lachnospiraceae—g__Blautia—s__unclassified | 1.92 | 0.4 | 2.14 | 0.34 | 1.32 | 0.75 | 1.15 | 0.87 | 2.36 | 0.19 | 2.67 | 0.1 | 1.40 | 0.57 | 1.16 | 0.63 |
| p__Firmicutes—c__Negativicutes—o__Selenomonadales—f__Acidaminococcaceae—g__Phascolarctobacterium—s__unclassified | 2.09 | 0.37 | 2.47 | 0.26 | 2.66 | 0.24 | 2.43 | 0.29 | 2.62 | 0.12 | 2.86 | 0.08 | 2.70 | 0.16 | 2.46 | 0.19 |
| p__Actinobacteria | 2.19 | 0.36 | 2.32 | 0.31 | 1.99 | 0.42 | 1.73 | 0.57 | 2.88 | 0.07 | 3.05 | 0.06 | 2.45 | 0.20 | 2.26 | 0.26 |
| p__Actinobacteria—c__Coriobacteriia—o__Coriobacteriales—f__Coriobacteriaceae—g__Collinsella | 2.5 | 0.28 | 2.56 | 0.24 | 2.38 | 0.29 | 2.18 | 0.36 | 3.09 | 0.06 | 3.28 | 0.05 | 2.84 | 0.12 | 2.60 | 0.16 |
| p__Actinobacteria—c__Coriobacteriia—o__Coriobacteriales—f__Coriobacteriaceae—g__Collinsella—s__unclassified | 2.5 | 0.28 | 2.56 | 0.24 | 2.38 | 0.29 | 2.18 | 0.36 | 3.09 | 0.06 | 3.28 | 0.05 | 2.84 | 0.12 | 2.60 | 0.16 |
| p__Actinobacteria—c__Coriobacteriia—o__Coriobacteriales—f__Coriobacteriaceae | 2.53 | 0.28 | 2.58 | 0.24 | 2.40 | 0.29 | 2.20 | 0.36 | 3.1 | 0.06 | 3.3 | 0.05 | 2.86 | 0.12 | 2.62 | 0.16 |
| p__Actinobacteria—c__Coriobacteriia | 2.8 | 0.26 | 2.85 | 0.2 | 2.68 | 0.24 | 2.45 | 0.29 | 3.27 | 0.06 | 3.45 | 0.05 | 3.06 | 0.09 | 2.83 | 0.16 |
| p__Actinobacteria—c__Coriobacteriia—o__Coriobacteriales | 2.8 | 0.26 | 2.85 | 0.2 | 2.68 | 0.24 | 2.45 | 0.29 | 3.27 | 0.06 | 3.45 | 0.05 | 3.06 | 0.09 | 2.83 | 0.16 |
Bold numbers indicate significant differences (FDR <0.1).
ASC, ascending; DES, descending; FDR, false discovery rate.
Figure 2.
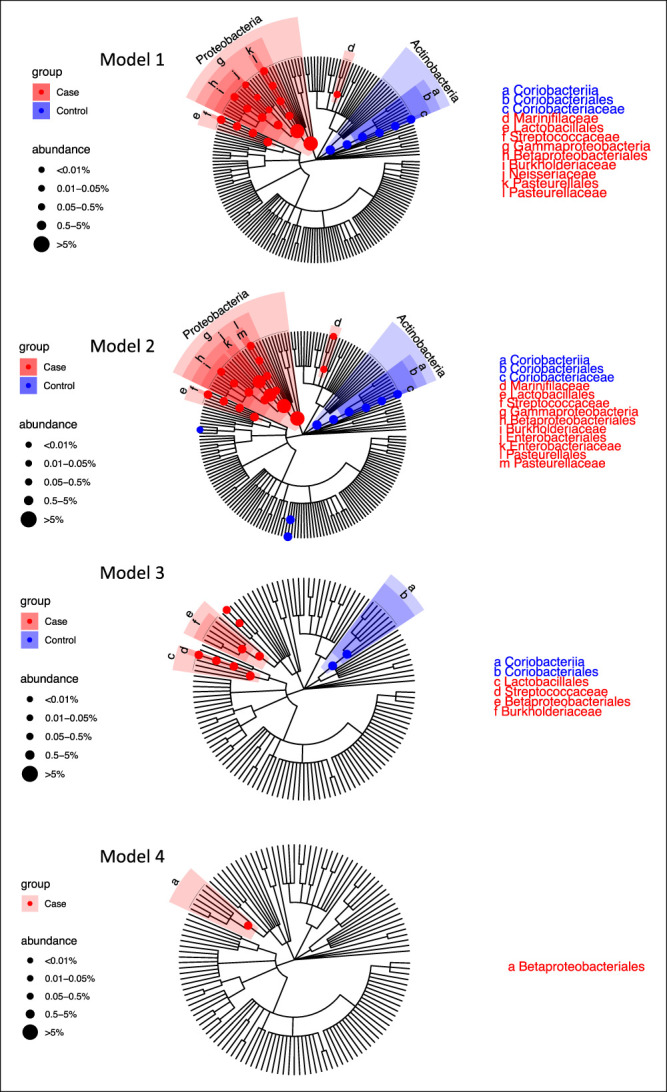
Significant differential taxa between microscopic colitis cases and controls highlighted in the taxonomic tree of the DES colon microbiome for the 4 linear regression models. Model 1 is adjusted for batch, education, and PPI use. Model 2 is additionally adjusted for sex and antibiotics. Model 3 is adjusted for age along with the covariates in model 2. Model 4 is adjusted for BMI along with the covariates in model 3. The branches of significant taxa from phylum to family level were highlighted and labeled. The node sizes are proportional to the overall relative abundance of the taxa. BMI, body mass index; DES, descending; PPI, proton pump inhibitor.
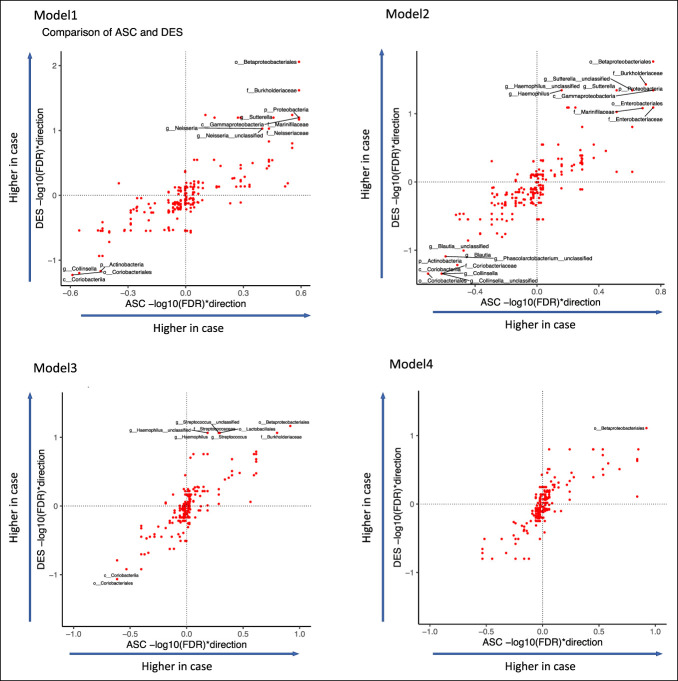
Among the 24 differential taxa revealed with model 1, 18 taxa were more abundant in cases and 6 were more abundant in controls, while among the 27 differential taxa revealed with model 2, 18 enriched in cases and 9 enriched in controls. The taxa enriched in the controls mostly belonged to Actinobacteria, mainly driven by the genus Collinsella and its higher taxonomic levels (Table 2 and Figure 2). However, these associations were not significant in models that included age and BMI (model 4). The taxa enriched in cases were mostly Gammaproteobacteria, including Sutterella, Haemophilus, Neisseria, Enterobacteriaceae, and Pasteurellaceae. Genus Streptococcus in phylum Firmicutes and family Marinifilaceae in phylum Bacteroidetes were also enriched in microscopic colitis cases (Table 2 and Figure 2). While the number of taxa associated with cases decreased with increasingly model complexity, there were taxa associated with cases in all 4 models, suggesting that confounding with age and BMI cannot explain all the associations we saw with microscopic colitis. In model 3, the taxa enriched in cases included genus Haemophilus and Streptococcus, family Burkholderiaceae, and order Betaproteobacteriales, and the taxa enriched in controls included order Coriobacteriales and class Coriobacteriia, while the taxa revealed by model 4 was Betaproteobacteriales that was enriched in cases. While we found no significant taxa associated with case/control in the ASC microbiome, the FDR-corrected P values estimated from the ASC and DES microbiomes were highly correlated (Figure 3), indicating that while the microbial features associated with microscopic colitis were stronger in DES than ASC, a similar pattern of difference was seen at both sampling sites. We also ran separate models in patients who did not take antibiotics, instead of adding antibiotics as a covariate, and these results are generally consistent, with significant taxa in DES microbiome but not in ASC microbiome (see Supplementary Figures S2 and S3, Supplementary Digital Content 1, http://links.lww.com/CTG/A876).
Figure 3.
Comparison of the associations of individual taxa and case/control in ASC and DES microbiome with the 4 models. The x and y axes show the −log10 (FDR) multiplied by +1/−1 to indicate the direction of changes. Significant taxa (FDR <0.1) were labeled. ASC, ascending; DES, descending; FDR, false discovery rate.
We have separately reported that stool frequency was greater in cases than in controls (30). When we included stool frequency in the model, there were no significant taxa associated with cases and control (data not shown). Because, as previously reported, the case control status and stool frequency are associated, however, having these terms in the same model might lead to unreliable statistical inferences. To address this, we created models stratified by the number of liquid stools in the week before the patients' colonoscopy to multiple levels and tested the associations between microbiome and case/control within each stratum. We found that there are taxa associated with case/control in some, but not all, of the strata (see Supplementary Table S1, Supplementary Digital Content 1, http://links.lww.com/CTG/A876). However, the smaller sample size from stratification reduced the statistical power of detecting the associated taxa, and fewer taxa were significant compared with the original results. Overall, these results suggest that stool frequency may have affected our results but is unlikely to explain all the relationships between case and control because some of these relationships can be observed in data stratified by stool frequency.
DISCUSSION
In this study of the microbiome of microscopic colitis cases and diarrhea controls, we found that alpha diversity was significantly lower in cases than in controls in the DES colon microbiome. We also found microorganisms that are associated with microscopic colitis, including taxa in phylum Proteobacteria that are potentially inflammation related. These differential taxa remained significant after adjusting the models for demographic factors and medicines, including sex, education, PPI, and antibiotic use. Some taxa were not significant after adjusting for age and BMI. The ASC and DES colon microbiome showed consistent microbial changes, but the changes were not significant in the ASC colon microbiome.
While the etiology of microscopic colitis remains unknown, there are some clues. In 1995, Järnerot et al. (11) described the experience of 20 patients with microscopic colitis with severe diarrhea and increased thickness of the subepithelial collagen layer. Following fecal diversion, the diarrhea stopped in all patients and the collagen layer thinned. Symptoms recurred when intestinal continuity was restored. A more recent study used Ussing chambers to measure intestinal permeability in a single patient before and after fecal diversion (33). Diversion of the fecal stream decreased inflammation of the mucosa and normalized epithelial degeneration and mucosal permeability. The permeability changes recurred when the diversion was removed. These studies suggested a luminal factor for the onset of microscopic colitis. The gut microbiota is a logical target of investigation, given the important roles of bacteria in the intact gut and the profound change when the fecal stream is diverted.
Previous studies have reported alterations in the gut microbiome associated with microscopic colitis using fecal samples and that alpha diversity of the gut microbiome was often decreased in patients with microscopic colitis. Rindom Krogsgaard et al. (17) studied stool findings from 10 patients with lymphocytic colitis, 10 with collagenous colitis, and 10 healthy controls and reported that the bacterial diversity of the cases was lower from the controls at baseline but not after treatment with budesonide. Similarly, in a study by Hertz et al. (18), the stool microbiota in 15 patients with microscopic colitis was less diverse than in 21 healthy controls. Morgan et al. (16) compared 20 patients with microscopic colitis with 20 healthy controls and 20 patients with functional diarrhea and found that alpha diversity was lower in the active cases compared with that in healthy controls and diarrhea controls, but the results were not significant.
While changes in alpha diversity were broadly consistent between studies, the microorganisms associated with microscopic colitis reported in previous studies have not been always consistent, potentially because of small sample sizes and different ways that different studies define control groups. Helal et al. (34) cultured biopsies from 20 patients and 10 normal controls and reported an association between Escherichia coli and lymphocytic colitis. In another study, Fischer et al. (15) examined fecal samples in 10 female patients with microscopic colitis and compared them with 7 healthy controls and observed that the patients with microscopic colitis had lower amounts of Akkermansia. Millien et al. (19) studied 20 cases of microscopic colitis and 20 healthy controls and reported that the cases had an increase in proinflammatory sulfur-reducing bacteria with a significant decrease in the Coriobacteriaceae family that was abundant in the healthy gut. Morgan et al. (16) found that the relative abundance of Haemophilus parainfluenzae, Veillonella parvula, and Veillonella unclassified was higher in microscopic colitis cases, and the abundance of Alistipes putredinis was higher in healthy controls.
In this study, we collected ASC and DES colon biopsy samples of study participants and compared the cases with microscopic colitis and controls with a diarrhea history. Our approach minimizes the potential influence of diarrhea on microbial composition of fecal samples. With our relatively large sample size of 52 cases and 153 controls, we observed a decreased Shannon diversity in the DES colon microbiome associated with microscopic colitis, which verified some previous findings (17,18). In this study, we also reported some differences in individual taxa that are consistent with previous studies such as enrichment of Gammaproteobacteria with microscopic colitis (18). This consistent signal associated with Gammaproteobacteria is especially interesting because Gammaproteobacteria are known to be inflammation related (35–37). Consistent with previous studies (14,19), the relative abundance of family Coriobacteriaceae and genus Collinsella was higher in controls compared with that in cases with microscopic colitis. Our study also had some differences from previous studies. For example, we did not observe a significant difference in mucin-degrading Akkermansia that has been previously reported (15), which might be explained by our unique approach to defining controls. We have previously reported that stool frequency was greater in cases than in controls (30), but we do not think that stool frequency can explain all the observed relationships between taxa and case/control because we see some significant taxa in models stratified by stool frequency.
A number of factors distinguish our study from previous studies. Our study is one of the largest studies analyzing the microbiomes of patients with microscopic colitis. Our choice of controls with diarrhea is an important strength. Diarrhea can alter the gut microbiota (20), and thus comparing microscopic colitis cases with diarrhea with healthy controls, as conducted in many previous studies, risks not considering the influence of diarrhea on microbial composition. In our study, we examined adherent bacteria by obtaining biopsies from 2 locations in the colon rather than stool specimens, as has been conducted in most previous studies. The human gut microbiota consists of resident (adherent) and transient (fecal) bacteria. We selected adherent bacteria obtained by biopsy in this study for the following reasons. First, the adherent bacteria are more permanent than bacteria in feces and could interact more directly with host tissues, so they are potentially more relevant to disease etiology. Second, we have data using fluorescence in situ hybridization showing that there are abundant bacteria from many species densely adherent to the mucosa, even after a purge (23). The bacteria can be influenced by fasting and purge before colonoscopy, but all subjects were fasted and prepped, so comparisons between groups are valid. Finally, it has been shown that adherent bacteria are better predictors of disease activity than fecal samples in Crohn's disease (38). Our biopsies were flash frozen, compared with some other studies using formalin-fixed and paraffin-embedded tissues, which while easily obtained can result in altered microbial composition (19). In our study, we also adjusted for a number of covariates including age and BMI that potentially obscure the signal of microbial associations with microscopic colitis. Finally, our study finds a consistent signal in the ASC and DES colon, although the signal in the DES colon is much stronger.
There are a few limitations to our study. The ages and BMI of patients are different for cases and controls. While we controlled for age and BMI in the linear models, this may have decreased the statistical power for detecting taxa associated with cases. We used 16 rRNA sequencing techniques because low biomass makes generation of shotgun metagenome sequencing data very challenging. We therefore cannot provide information on functional genes that are related to the roles of the microbiome in microscopic colitis. Another potential limitation of our study was that we combined lymphocytic and collagenous colitis. Although these subtypes are often considered separately in the literature, similarities in risk factors, histology, and response to treatment would suggest that they are subtypes of the same entity (39). Our study is cross-sectional, and we therefore do not know whether the observed differences in microbial communities are a cause or a consequence of microscopic colitis. Future studies with a larger sample size and using shotgun metagenome sequencing will likely provide further insights on the potential roles of the microbial community in the development of microscopic colitis.
In conclusion, altered microbial communities were associated with microscopic colitis, including increased alpha diversity, increase in inflammation-related taxa, and decrease in Collinsella. The changes of microbial features were consistent between ASC and DES colon microbiomes but not significant in the ASC colon microbiome. The altered microbial communities in patients with microscopic colitis raise the possibility that live biotherapeutic products could someday play a role in the management of patients with microscopic colitis.
CONFLICTS OF INTEREST
Guarantor of the article: Robert S. Sandler, MD, MPH.
Specific author contributions: R.S.S., A.A.F., T.O.K., J.T.W., and A.F.P.: contributed to the conception, design, data acquisition, analysis, and supervision of the work. S.S. and I.C.B.: contributed to the analysis and interpretation of the data. S.S., R.S.S., and A.A.F.: drafted the manuscript. I.C.B., T.O.K., J.T.W., and A.F.P.: contributed to revision of the manuscript. All authors have read and approved the final manuscript.
Financial support: This research was supported by fundings from the National Institutes of Health R01 DK 105114 and P30 DK 034987.
Potential competing interests: None to report.
Ethics approval: The study met the standards for the ethical treatment of participants and was approved by the Institutional Review Board of the University of North Carolina at Chapel Hill (14-3156).
Study Highlights.
WHAT IS KNOWN
✓ Microscopic colitis is a common cause of chronic diarrhea.
✓ The exact etiology of microscopic colitis is unknown, but the gut microbiome is considered to play an important role.
WHAT IS NEW HERE
✓ The descending colon biopsies microbiome was altered in patients with microscopic colitis.
✓ Lower alpha diversity, increase in inflammation-related taxa, and decrease in Collinsella was associated with microscopic colitis.
✓ The changes of microbial features were consistent between ascending and descending colon microbiomes.
Supplementary Material
ACKNOWLEDGMENT
We thank Ms. Amber Nicole McCoy and Ms. Alondra Rodriguez for technical assistance with DNA extractions and library preparation.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A876
Contributor Information
Ivory C. Blakley, Email: ieclabau@uncc.edu.
Anthony A. Fodor, Email: afodor@uncc.edu.
Temitope O. Keku, Email: temitope_keku@med.unc.edu.
John T. Woosley, Email: John_woosley@med.unc.edu.
Anne F. Peery, Email: anne_peery@med.unc.edu.
Robert S. Sandler, Email: robert_sandler@med.unc.edu.
REFERENCES
- 1.Pardi DS, Smyrk TC, Tremaine WJ, et al. Microscopic colitis: A review. Am J Gastroenterol 2002;97(4):794–802. [DOI] [PubMed] [Google Scholar]
- 2.Williams JJ, Beck PL, Andrews CN, et al. Microscopic colitis—A common cause of diarrhoea in older adults. Age Ageing 2010;39(2):162–8. [DOI] [PubMed] [Google Scholar]
- 3.Weimers P, Ankersen DV, Lophaven S, et al. Incidence and prevalence of microscopic colitis between 2001 and 2016: A Danish nationwide cohort study. J Crohns Colitis 2020;14(12):1717–23. [DOI] [PubMed] [Google Scholar]
- 4.Maye H, Safroneeva E, Godat S, et al. Increasing incidence of microscopic colitis in a population-based cohort study in Switzerland. Clin Gastroenterol Hepatol 2021;19(10):2205–6. [DOI] [PubMed] [Google Scholar]
- 5.Bergman D, Clements MS, Khalili H, et al. A nationwide cohort study of the incidence of microscopic colitis in Sweden. Aliment Pharmacol Ther 2019;49(11):1395–400. [DOI] [PubMed] [Google Scholar]
- 6.Davidson S, Sjoberg K, Engel PJH, et al. Microscopic colitis in Denmark and Sweden: Incidence, putative risk factors, histological assessment and endoscopic activity. Scand J Gastroenterol 2018;53(7):818–24. [DOI] [PubMed] [Google Scholar]
- 7.Fumery M, Kohut M, Gower-Rousseau C, et al. Incidence, clinical presentation, and associated factors of microscopic colitis in Northern France: A population-based study. Dig Dis Sci 2017;62(6):1571–9. [DOI] [PubMed] [Google Scholar]
- 8.Pardi DS, Loftus EV, Smyrk TC, et al. The epidemiology of microscopic colitis: A population based study in Olmsted County, Minnesota. Gut 2007;56(4):504–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams JJ, Kaplan GG, Makhija S, et al. Microscopic colitis–defining incidence rates and risk factors: A population-based study. Clin Gastroenterol Hepatol 2008;6(1):35–40. [DOI] [PubMed] [Google Scholar]
- 10.Beaugerie L, Pardi DS. Review article: Drug-induced microscopic colitis: Proposal for a scoring system and review of the literature. Aliment Pharmacol Ther 2005;22(4):277–84. [DOI] [PubMed] [Google Scholar]
- 11.Järnerot G, Tysk C, Bohr J, et al. Collagenous colitis and fecal stream diversion. Gastroenterology 1995;109(2):449–55. [DOI] [PubMed] [Google Scholar]
- 12.Veress B, Lofberg R, Bergman L. Microscopic colitis syndrome. Gut 1995;36(6):880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Sluis WB, Bouman M-B, Mullender MG, et al. The effect of surgical fecal stream diversion of the healthy colon on the colonic microbiota. Eur J Gastroenterol Hepatol 2019;31(4):451–7. [DOI] [PubMed] [Google Scholar]
- 14.Carstens A, Dicksved J, Nelson R, et al. The gut microbiota in collagenous colitis shares characteristics with inflammatory bowel disease-associated dysbiosis. Clin Transl Gastroenterol 2019;10(7):e00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fischer H, Holst E, Karlsson F, et al. Altered microbiota in microscopic colitis. Gut 2015;64(7):1185–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan DM, Cao Y, Miller K, et al. Microscopic colitis is characterized by intestinal dysbiosis. Clin Gastroenterol Hepatol 2020;18(4):984–6. [DOI] [PubMed] [Google Scholar]
- 17.Rindom Krogsgaard L, Kristian Munck L, Bytzer P, et al. An altered composition of the microbiome in microscopic colitis is driven towards the composition in healthy controls by treatment with budesonide. Scand J Gastroenterol 2019;54(4):446–52. [DOI] [PubMed] [Google Scholar]
- 18.Hertz S, Durack J, Kirk KF, et al. Microscopic colitis patients possess a perturbed and inflammatory gut microbiota. Dig Dis Sci 2022;67(6):2433–43. [DOI] [PubMed] [Google Scholar]
- 19.Millien V, Rosen D, Hou J, et al. Proinflammatory sulfur-reducing bacteria are more abundant in colonic biopsies of patients with microscopic colitis compared to healthy controls. Dig Dis Sci 2019;64(2):432–8. [DOI] [PubMed] [Google Scholar]
- 20.Vandeputte D, Falony G, Vieira-Silva S, et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 2016;65(1):57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 1997;32(9):920–4. [DOI] [PubMed] [Google Scholar]
- 22.McCoy AN, Araujo-Perez F, Azcarate-Peril A, et al. Fusobacterium is associated with colorectal adenomas. PLoS One 2013;8(1):e53653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen XJ, Rawls JF, Randall TA, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010;1(3):138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014;2(1):1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones RB, Fodor AA, Peery AF, et al. An aberrant microbiota is not strongly associated with incidental colonic diverticulosis. Sci Rep 2018;8(1):4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37(8):852–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13(7):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 2013;41(Database issue):D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones RB, Zhu X, Moan E, et al. Inter-niche and inter-individual variation in gut microbial community assessment using stool, rectal swab, and mucosal samples. Sci Rep 2018;8(1):4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sandler RS, Keku TO, Woosley JT, et al. Medication use and microscopic colitis. Aliment Pharmacol Ther 2021;54(9):1193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen EF, Pokhrel B, Bianchi LK, et al. Decreased colorectal cancer and adenoma risk in patients with microscopic colitis. Dig Dis Sci 2012;57(1):161–9. [DOI] [PubMed] [Google Scholar]
- 32.Keszthelyi D, Jansen SV, Schouten GA, et al. Proton pump inhibitor use is associated with an increased risk for microscopic colitis: A case–control study. Aliment Pharmacol Ther 2010;32(9):1124–8. [DOI] [PubMed] [Google Scholar]
- 33.Munch A, Soderholm JD, Wallon C, et al. Dynamics of mucosal permeability and inflammation in collagenous colitis before, during, and after loop ileostomy. Gut 2005;54(8):1126–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helal TE, Ahmed NS, El Fotoh OA. Lymphocytic colitis: A clue to bacterial etiology. World J Gastroenterol 2005;11(46):7266–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawls JF. Enteric infection and inflammation alter gut microbial ecology. Cell Host Microbe 2007;2(2):73–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mukhopadhya I, Hansen R, El-Omar EM, et al. IBD—What role do Proteobacteria play? Nat Rev Gastroenterol Hepatol 2012;9(4):219–30. [DOI] [PubMed] [Google Scholar]
- 37.Winter SE, Bäumler AJ. Dysbiosis in the inflamed intestine: Chance favors the prepared microbe. Gut Microbes 2014;5(1):71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 2014;15(3):382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen MA, Munck LK. Systematic review: Are lymphocytic colitis and collagenous colitis two subtypes of the same disease: Microscopic colitis? Aliment Pharmacol Ther 2012;36(2):79–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.