INTRODUCTION:
Variants in SMAD4 or BMPR1A cause juvenile polyposis syndrome, a rare autosomal dominant condition characterized by multiple gastrointestinal hamartomatous polyps. A phenotype of attenuated adenomatous polyposis without hamartomatous polyps is rare.
METHODS:
We describe a retrospective cohort of individuals with SMAD4 or BMPR1A heterozygous germline variants, having ≥10 cumulative colorectal adenomas and/or colorectal cancer without hamartomatous polyps. All individuals had multigene panel and duplication/deletion analysis to exclude other genetic syndromes.
RESULTS:
The study cohort included 8 individuals. The pathogenic potential of the variants was analyzed. Variants detected included 4 missense variants, 1 nonsense variant, 1 splice site variant, and 2 genomic deletions. Features of pathogenicity were present in most variants, and cosegregation of the variant with polyposis or colorectal cancer was obtained in 7 of the 8 families. Three of 8 individuals had colorectal cancer (age less than 50 years) in addition to the polyposis phenotype. Two individuals had extraintestinal neoplasms (pancreas and ampulla of Vater).
DISCUSSION:
The clinical phenotype of SMAD4 and BMPR1A variants may infrequently extend beyond the classical juvenile polyposis syndrome phenotype. Applying multigene panel analysis of hereditary cancer-related genes in individuals with unexplained polyposis can provide syndrome-based clinical surveillance for carriers and their family members.
INTRODUCTION
Juvenile polyposis syndrome (JPS) is a rare autosomal dominant condition, affecting approximately 1:100,000 individuals (1,2). It is characterized by multiple gastrointestinal (GI) hamartomatous polyps. Individuals with JPS are at increased risk of colorectal, gastric, and small bowel cancers (3,4). JPS is clinically diagnosed in an individual with any of the following: (i) ≥5 colorectal juvenile polyps, (ii) juvenile polyps in other parts of the GI tract, or (iii) any number of juvenile polyps and ≥1 affected family member.
Up to 60% of individuals with clinically defined JPS carry variants in SMAD4 or BMPR1A genes (5,6), encoding 2 members of the transforming growth factor beta (TGF-β) superfamily signaling pathway. Approximately 20%–50% of JPS cases have no family history and may harbor de novo variants (7–10). On the other hand, approximately 80%–90% of the individuals having multiple colorectal adenomas (20–100 cumulative colorectal adenomas) remain genetically unsolved by testing the major high predisposition genes APC and MUTYH (11,12).
Interestingly, variants in SMAD4 and BMPR1A are also associated with a clinical picture of hereditary mixed polyposis syndrome characterized by adenomatous, serrated, and juvenile polyps (7,13–15). Sporadic observations suggest that individuals with SMAD4 or BMPR1A variants may also have otherwise unexplained attenuated adenomatous polyposis with no juvenile polyps (16,17). We describe the clinical and molecular phenotype of a small cohort of individuals with variants in SMAD4 or BMPR1A genes and adenomatous polyposis without juvenile polyps.
METHODS
This retrospective cohort study included individuals heterozygous for genetic variants in SMAD4 or BMPR1A, with multiple (≥10) cumulative colorectal adenomas or colorectal cancer (CRC), but without juvenile polyps. Individuals were followed over a period of 13 years (2009 until 2021) in 3 tertiary academic centers in Israel. Institutional review board approval was obtained in each center.
Individuals underwent upper and lower GI endoscopy, and polypectomy was performed as indicated. Polyp histology was reviewed by experienced GI pathologists in each of the medical centers, and juvenile polyp pathology was excluded in all cases.
Genetic evaluation was performed in all individuals. Genomic DNA was extracted from whole blood using a dedicated kit (5 PRIME, Gaithersburg, MD, ArchivePure) as instructed by manufacturer guidelines. Multigene panel analysis of hereditary cancer-related genes (testing 31–123 genes including APC, MUTYH, MSH2, MSH6, PMS2, MLH1, BMPR1A, SMAD4, STK11, PTEN, POLD1, POLE, GREM1, MSH3, NTHL1) was performed in certified genetic laboratories. The genes included in the panels are listed in Supplementary Table 1 (see Supplementary Digital Content, http://links.lww.com/CTG/A870). Segregation analysis results were available for 7 of the 8 families.
Each genetic variant was evaluated and classified according to the American College of Medical Genetics and Genomics (ACMG) guidelines (18). In addition, missense variants were introduced to the SWISS-MODEL algorithm (www.swissmodel.expasy.org/interactive), to evaluate their effect on the protein secondary structure. This effect was evaluated by comparing the predicted protein structure of the variants to those of the canonic wild types (NP_005350.1 for SMAD4 and NP_004320.2 for BMPR1A).
RESULTS
Clinical characteristics
Registries from 3 medical centers of individuals with variants in SMAD4 or BMPR1A revealed 8 individuals fulfilling inclusion criteria. Table 1 lists demographic data, clinical phenotypes, genetic findings, and variant classification.
Table 1.
Demographic, clinical phenotype, and genetic findings
| Patient no. (sex) | Origin | Age of onset (yr) | Clinical phenotype | Extraintestinal involvement | Surgical treatment | Genetic findings | Family history of colorectal cancer and colorectal polyposis | Familial cosegregation | Classification Verdict by: Franklina; VarSomeb Commercial laboratory |
| 1 (female) | Ashkenazi Jewish | 40–50 | 10 adenomas (right colon) + small gastric hyperplastic polyps | Pancreas SB-IPMN | None | SMAD4 (NM_005359.6):c.403C>T p.R135* | No | De novo | Pathogenic; PathogenicPathogenic |
| 2 (female) | Sephardic Jewish | 40–50 | Left-sided CRC + <10 adenomas | None | Left hemicolectomy | BMPR1A (NM_004329.3):c.675G>A p.L225= | First degree: CRC, polyposis Second degree: CRC |
Yes | Likely pathogenic; Pathogenic Likely pathogenic |
| 3 (male) | Sephardic Jewish | 20–30 | ∼40 adenomas (whole colon, advanced) | None | None |
BMPR1A (NM_004329.3): del ex1-13 Chr10: 86875867–89041308[hg19] |
First degree: CRC, polyposis Second degree: N/A |
Yes | Pathogenic; Pathogenic Pathogenic |
| 4 (female) | Sephardic Jewish | 20–30 | ∼20 adenomas (right colon) | None | None |
BMPR1A (NM_004329.3):del ex3-13 chr10:88611882-89041308[hg19] |
First degree: CRC, polyposis Second degree: CRC, polyposis |
Unknown | Pathogenic; Likely pathogenic Pathogenic |
| 5 (male) | Ashkenazi Jewish | 30–40 | Left-sided CRC + ∼20 adenomas (whole colon, advanced) | None | Total proctocolectomy with IPAA | SMAD4 (NM_005359.6):c.746A>C p.Q249P | Second degree: CRC, gastric carcinoma | Yes | VUS; Benign VUS |
| 6 (female) | Ashkenazi Jewish | 10–20 | ∼20 adenomas (whole colon, advanced) | None | Total proctocolectomy with IPAA | BMPR1A (NM_004329.3):c.388T>C p.C130R | First degree: CRC Second degree: CRC |
Yes | Likely pathogenic; VUS VUS |
| 7 (male) | Ashkenazi Jewish | 40–50 | Right-sided CRC + 10 adenomas (ascending and transverse colon, advanced) | None | Subtotal colectomy | BMPR1A (NM_004329.3):c.760C>T p.R254C | First degree: CRC Second degree: CRC |
Yes No |
VUS; VUS VUS |
| FANCI (MN_001376911.1):c.1856T>A p.L619Q | |||||||||
| VUS; VUS VUS | |||||||||
| 8 (female) | Ashkenazi Jewish | 50–60 | ∼20 adenomas (whole colon, advanced) | Papilla of Vater carcinoma | Whipple procedure | BMPR1A (NM_004329.3):c.676G>T p.V226F | First degree: CRC Second degree: CRC |
Unknown | VUS; VUS VUS |
CRC, colorectal cancer; IPAA, ileal pouch anal anastomosis; SB-IPMN, side branch intraductal papillary mucinous neoplasm; VUS, variant of unknown significance.
Franklin: gene-specific artificial intelligence-based variant search engine, by Genoox.
VarSome: the human genomic variant search engine.
Clinically, 3 individuals had early-onset (age less than 50 years) CRC; 2 were left-sided and 1 right-sided. Two of these individuals had first-degree and second-degree relatives with CRC. In the 5 individuals without a personal history of CRC, 4 had ≥1 first-degree relative with CRC. Extraintestinal neoplasms were detected in 2 individuals and included an ampulla of Vater carcinoma in a BMPR1A variant carrier (individual 6) and a pancreatic side-branch intraductal papillary mucinous neoplasm (SB-IPMN) in a SMAD4 variant carrier (individual 2). The phenotypes and segregation analysis of each family are described in detail in the Supplementary File (see Supplementary Digital Content, http://links.lww.com/CTG/A869).
Genetic findings
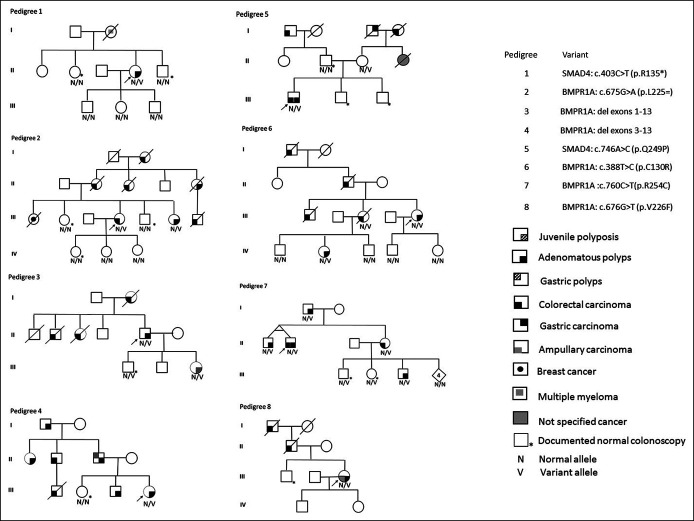
Six individuals carried a heterozygous single-base variant: 2 in SMAD4 (1 missense and 1 nonsense) and 4 in BMPR1A (3 missense variants and 1 splice site variant). Two individuals had genomic deletions encompassing BMPR1A exons 1–13 and exons 3–13, respectively. The 8 pedigrees are presented in Figure 1.
Figure 1.
Pedigrees.
All detected missense variants replace a conserved nucleic acid (with positive phyloP100way scores). Seven of the 8 individuals had no genetic variant in any other hereditary cancer-related gene. One individual (pedigree 7, Supplementary File [see Supplementary Digital Content, http://links.lww.com/CTG/A869]) carried an additional genetic variant in FANCI (c.1856T>A, p.L619Q), which is associated with Fanconi anemia, a recessive syndrome not associated with a risk for colorectal polyps. This variant did not cosegregate with the affected individuals in the extended family, and we, therefore, considered it to be an incidental finding.
The SMAD4 c.403C>T, p.R135* variant (individual number 1) causes a premature stop codon (PVS1 by the ACMG criteria) and, therefore, is classified as a pathogenic variant. In addition, this variant was not found among 141,456 normal adult exome and genome sequencing samples in the Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org).
BMPR1A: c.675G>A, p.L225= (individual number 2) is a synonymous inherited variant, located in the donor canonic splice site (−1) of exon 8. Therefore, it is predicted to affect the splicing process and is accordingly classified by varSEAK (https://varseak.bio) as class 4. This tool also recognizes a potential alternative splicing site in the intron, 30 nucleotides downstream of the 5' splice site position 675 + 1. In accordance, the combined annotation-dependent depletion (https://cadd.gs.washington.edu/snv) scaled score is 22, placing this variant among the 1% most deleterious variants in the human genome. This is further supported by a Transcript-inferred Pathogenicity score of 0.998 of 1. As expected, this nucleotide location is very conserved (genetic resources and enhancement program score = 5.51). All this in silico evidence fulfills PP3 ACMG criteria. In addition, this variant is not found in any healthy population (gnomAD), supporting PM2 criteria. It was recently reported in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar) by just a single submitter with no linked publication. Segregation of this variant in the family fulfills PP1 ACMG criteria for pathogenicity.
Individuals 3 and 4 were carriers of inherited copy number variations in BMPR1A. Individual number 3 had a deletion of exons 1–13 (with no boundaries defined). Individual number 4 had a deletion of exons 3–13 as part of the 429 kb deletion, encompassing 8 additional genes, downstream to BMPR1A, which has previously been reported as a common founder variant occurring in 1 of 124 Israelis of Jewish Bukharan origin, with a wide spectrum of clinical features (19).
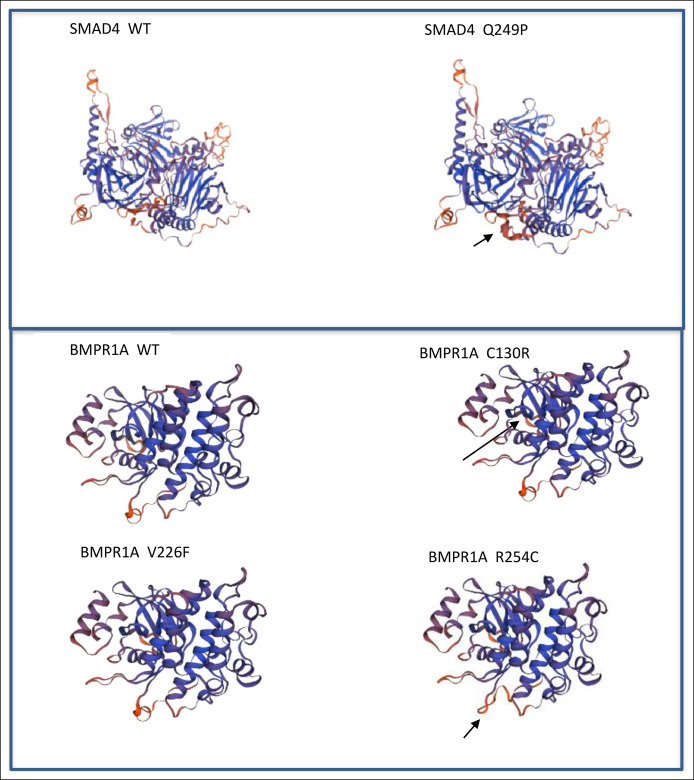
Individuals 5–8, a were each found to carry a missense variant. The SMAD4: c.746A>C, p.Q249P variant (individual number 5) is classified by search engines Franklin (https://www.genoox.com) and VarSome (https://varsome.com) as a variant of unknown significance (VUS) and as a benign variant, respectively. However, this variant is rare in Ashkenazi population (0.18%) and causes a change in conserved amino acid (phyloP100way score 5.006) (https://gnomad.broadinstitute.org, https://genome.ucsc.edu). Furthermore, the SWISS-MODEL predicted an apparent difference in the protein structure between the wild type and the variant protein (Figure 2), supporting the suspected pathogenicity of this variant.
Figure 2.
Comparison between local structure around the missense variant site and wild type protein, generated by Swiss-model repository online software.
The BMPR1A: c.388T>C, p.C130R variant (individual number 6) is not found in the gnomAD database of healthy populations, fulfilling the PM2 criteria of ACMG. Cysteine at position 130 is located in a disulfide bond domain. Cysteine is a sulfur-containing nucleophilic amino acid, frequently involved in bonds with molecules. Thus, its replacement by arginine, which has different biologic characteristics, is predicted to affect protein function. The BMPR1A secondary structure is also predicted by the SWISS-MODEL to be affected (Figure 2). Indeed, the Rare Exome Variant Ensemble Learner (REVEL) is a method for predicting pathogenicity of missense variants . REVEL provides this variant with an almost maximal combined score of 0.973 (range 0–1). In addition, the individual's family exhibited cosegregation of this variant with colonic polyposis/carcinomas. Therefore, based on this bioinformatic analysis, we predict the BMPR1A: c.388T>C, p.C130R variant as a likely pathogenic variant.
The BMPR1A: c.760C>T, p.R254C variant (individual number 7) is classified as VUS by both VarSome and Franklin engines. However, it is a rare alteration (0.086% in Ashkenazi Jews) that qualifies for ACMG PS4 criteria. Moreover, the variant is in the protein kinase domain, and its REVEL score is 0.721, which predicts pathogenicity. The SWISS-MODEL algorithm (included in the REVEL score ensemble) demonstrates an effect of this variant on the BMPR1A protein structure (Figure 2). These in silico tools fulfill the PP3 ACMG criteria. In addition, we found it to cosegregate with the polyposis and colorectal carcinoma phenotype in the family, thus fulfilling the PP1-supporting ACMG criteria for pathogenicity. Based on the combination of all these criteria, we predict this variant to be a likely pathogenic variant.
The BMPR1A: c.676G>T, p.V226F missense variant (individual number 8) did not affect the BMPR1A protein structure according to the SWISS-MODEL (Figure 2). There up-to-date 6 submissions of this variant to ClinVar, are all classified as VUS. However, 676 guanine is the first nucleotide in exon 9 and is highly conserved (Genetic Resources and Enhancement Program score 5.63). Its location within the acceptor splice site (+1) suggests a potential interruption of the splicing process by facilitating skipping of exon 9, which is highly expressed (proportion expressed across transcript score 0.80). Moreover, the distal 3' nucleic acid in exon 9 is included in the codon of the proximal 5' amino acid of exon 10; therefore, skipping exon 9 is expected to cause a frameshift in exon 10 (PM4 criteria). Accordingly, the varSEAK score of this variant is 4, indicating a likely splicing effect. In addition, this variant is extremely rare (8.36e-6 by gnomAD), supporting the PS4 criteria. Based on this interpretation, this variant is considered to be likely pathogenic.
DISCUSSION
Juvenile polyposis is characterized by colorectal hamartomas with or without adenomatous polyps. The small cohort presented here, with an atypical phenotype of multiple colorectal adenomas without juvenile polyps, further broadens the known clinical spectrum of SMAD4 and BMPR1A variations. This phenotype is unique and dictates a thorough genetic workup to exclude other etiologies such as familial adenomatous polyposis (FAP) and MUTYH-associated polyposis. Bioinformatic analysis of the genetic variations in this cohort, some of which were novel, raises a suspicion for their pathogenicity based on their conservation, rarity, alteration of the protein structure or the splicing process, along with familial polyposis/CRC segregation analysis.
A review of the literature revealed rare reports of SMAD4 or BMPR1A variant carriers presenting with multiple colorectal adenomas without evidence of juvenile polyps (14,16). Rohlin et al. (16) described 4 cases of FAP-like phenotype lacking an APC variant but with BMPR1A variants (3 cases) and a SMAD4 variant (1 case). A retrospective cohort study of 221 patients with JPS from 10 European centers described adenomas or serrated polyps, in addition to multiple juvenile polyps, in 42%–50% of the cases (20). O'Riordan et al. (13) found that ∼7%–14% of JPS polyps harbored adenomatous changes; however, adenomas as the sole histology of multiple colorectal polyps were not mentioned in any of these patients. Gao et al. (8) reported a family with the BMPR1A variant and multiple juvenile polyps misdiagnosed as multiple adenomas. In our study, polyp histology in all 8 patients, initially reported to be adenomas, was revised by GI pathologists, and the diagnosis of adenoma was confirmed in all cases.
A retrospective study on a large European JPS cohort reported that 15.4% of the patients were diagnosed with cancer at a median age of 41 years (20). In our cohort of 8 individuals, there were 3 cases of CRC (ages 39 years in a SMAD4 variant carrier and 43 years and 47 years in BMPR1A variant carriers). Interestingly, we report 1 BMPR1A variant carrier with an ampulla of Vater carcinoma at age 55 years. Whether the BMPR1A variant served as a driver variant or whether the cancer occurred sporadically is unclear because we were unable to perform somatic tumor genetic testing in this case. We also report a SMAD4 variant carrier with pancreatic side-branch intraductal papillary mucinous neoplasm in addition to multiple right-sided colorectal adenomas. Because of these observations, we reviewed the medical history of additional patients in the JPS registry who did not fulfill the inclusion criteria for this study (57 patients). These patients did not have any pancreatic and/or ampullary abnormalities. In the European JPS cohort (20), a single case of pancreatic cancer was reported. A retrospective review of JPS from Japan reported extracolonic tumors in the stomach, jejunum/ileum, breast, and thyroid (21), and a prospective JPS study in the United States reported extracolonic tumors in the stomach, upper GI tract, and testicles (22). These studies (21,22) did not report pancreatic or ampullary cancers.
The TGF-β pathway is important in CRC progression. Bone morphogenetic proteins (BMPs) are a subgroup within the TGF-β superfamily. BMPR1A signals, through cell-surface serine-threonine kinase receptors, to the intracellular SMAD4, which accumulates in the nucleus to regulate gene expression. BMP acts as a tumor suppressor that promotes apoptosis of mature colonic epithelial cells; therefore, perturbations in BMP signaling may lead to increased tumorigenesis (23). Selective transgenic inhibition of BMP signaling in mice intestinal epithelium leads to epithelial branching and budding, crypt dilatation, and reactive inflammatory changes and the subsequent development of dysplastic foci and adenomatous change (15).
TGF-β signaling in humans plays a role in both adenoma and hamartoma formation and their progression to cancer. In the hamartoma-carcinoma sequence, BMP signaling affects the first step in the formation of dysplastic aberrant crypts (23), namely, the transition from normal epithelium to hamartomatous polyps, which is further transformed into an adenoma by a somatic APC variant. In the adenoma-carcinoma sequence, loss of BMP signaling correlates tightly with progression to cancer and occurs in the transition from early adenomas to advanced adenomas, with BMPR1A variants associated with early adenoma transformation and SMAD4 variants associated with intermediate and advanced adenoma transformation (22). It remains unclear why most individuals with BMPR1A or SMAD4 pathogenic variants exhibit a clinical phenotype of hamartomas while some develop adenomatous polyposis, even in the same family (Figure 1, Pedigree 4).
Strengths of this study are the use of multigene panel testing to rule out other unrecognized variants which might cause polyposis syndromes and the thorough clinical follow-up the individuals had at a tertiary care center. Limitations of this study are the small sample size, which is expected given the rarity of JPS and, more specifically, the unusual adenomatous polyposis phenotype in JPS. An additional limitation is the subset of individuals carrying a missense variant because pathogenicity was not entirely established. It is possible that a pathogenic variant in a gene yet to be discovered may contribute to the adenomatous polyposis phenotype and cancer described in these cases.
In conclusion, we describe an atypical clinical phenotype of SMAD4 or BMPR1A variants, mimicking attenuated adenomatous polyposis syndrome. The use of multigene panel analysis of hereditary cancer-related genes in clinical practice helps achieve an accurate genetic diagnosis and allows specific syndrome-based clinical surveillance for variant carriers and their family members.
CONFLICTS OF INTEREST
Guarantor of the article: Guy Rosner, MD.
Specific author contributions: G.R. planned the study; acquired, analyzed, and interpreted study data; performed clinical follow-up of the study cohort; and wrote the manuscript. Y.P-.G. acquired, analyzed, and interpreted study data and wrote the manuscript. I.L., Z.L., R.K., H.S., and O.G. acquired study data and performed clinical follow-up of the study cohort. N.G. planned the study; acquired, analyzed, interpreted study data; and wrote the manuscript. All authors reviewed the manuscript and approved the final version.
Financial support: None to report.
Potential competing interests: None to report.
Patient consent for publication: Obtained.
Ethics approval: This study was approved by the local Ethics Committee at all participating medical centers (IRB Approval # 0557-21TLV). All participants provided informed consent for the performed genetic studies. All the procedures were performed under the Declaration of Helsinki and relevant policies.
Data availability statement: All data analyzed during this study are included in this published article and its supplementary information files.
Study Highlights.
WHAT IS KNOWN
✓ Juvenile polyposis syndrome (JPS) is a rare autosomal dominant condition caused by SMAD4 or BMPR1A mutations and characterized by multiple gastrointestinal hamartomatous polyps.
✓ A clinical phenotype of attenuated adenomatous polyposis without hamartomatous polyps is rarely described in JPS.
WHAT IS NEW HERE
✓ A small cohort of JPS individuals with a phenotype of attenuated familial adenomatous polyposis is described.
✓ Analysis of BMPR1A and SMAD4 variations, including novel variants, suggests their pathogenicity based on a combination of bioinformatic and clinical tools.
✓ Applying multigene panel analysis of hereditary cancer-related genes provides syndrome-based clinical surveillance for the carriers and their family members.
Supplementary Material
ACKNOWLEDGEMENT
We thank the patients and their families for participation in this study and for allowing us to publish their data.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A869, http://links.lww.com/CTG/A870
Contributor Information
Yael Petel-Galil, Email: yaelgal@tlvmc.gov.il.
Ido Laish, Email: idolaish@gmail.com.
Zohar Levi, Email: Zoharl@clalit.org.il.
Revital Kariv, Email: revitalk@tlvmc.gov.il.
Hana Strul, Email: hanas@tlvmc.gov.il.
Ophir Gilad, Email: ophirg@tlvmc.gov.il.
Nathan Gluck, Email: nathang@tlvmc.gov.il.
REFERENCES
- 1.Burt RW, Bishop DT, Lynch HT, et al. Risk and surveillance of individuals with heritable factors for colorectal cancer. WHO Collaborating Centre for the Prevention of Colorectal Cancer. Bull World Health Organ 1990;68:655–65. [PMC free article] [PubMed] [Google Scholar]
- 2.Chen HM, Fang JY. Genetics of the hamartomatous polyposis syndromes: A molecular review. Int J Colorectal Dis 2009;24:865–74. [DOI] [PubMed] [Google Scholar]
- 3.Latchford AR, Neale K, Phillips RK, et al. Juvenile polyposis syndrome: A study of genotype, phenotype, and long-term outcome. Dis Colon Rectum 2012;55:1038–43. [DOI] [PubMed] [Google Scholar]
- 4.McColl I, Busxey HJ, Veale AM, et al. Juvenile polyposis coli. Proc R Soc Med 1964;57:896–7. [PubMed] [Google Scholar]
- 5.Syngal S, Brand RE, Church JM, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 2015;110:223–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calva-Cerqueira D, Chinnathambi S, Pechman B, et al. The rate of germline mutations and large deletions of SMAD4 and BMPR1A in juvenile polyposis. Clin Genet 2009;75:79–85. [DOI] [PubMed] [Google Scholar]
- 7.Cheah PY, Wong YH, Chau YP, et al. Germline bone morphogenesis protein receptor 1A mutation causes colorectal tumorigenesis in hereditary mixed polyposis syndrome. Am J Gastroenterol 2009;104:3027–33. [DOI] [PubMed] [Google Scholar]
- 8.Gao XH, Li J, Zhao ZY, et al. Juvenile polyposis syndrome might be misdiagnosed as familial adenomatous polyposis: A case report and literature review. BMC Gastroenterol 2020;20:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayed MG, Ahmed AF, Ringold JR, et al. Germline SMAD4 or BMPR1A mutations and phenotype of juvenile polyposis. Ann Surg Oncol 2002;9:901–6. [DOI] [PubMed] [Google Scholar]
- 10.Aretz S, Stienen D, Uhlhaas S, et al. High proportion of large genomic deletions and a genotype phenotype update in 80 unrelated families with juvenile polyposis syndrome. J Med Genet 2007;44:702–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grover S, Kastrinos F, Steyerberg EW, et al. Prevalence and phenotypes of APC and MUTYH mutations in patients with multiple colorectal adenomas. JAMA 2012;308:485–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanich PP, Pearlman R, Hinton A, et al. Prevalence of germline mutations in polyposis and colorectal cancer-associated genes in patients with multiple colorectal polyps. Clin Gastroenterol Hepatol 2019;17:2008–15. [DOI] [PubMed] [Google Scholar]
- 13.O'Riordan JM, O'Donoghue D, Green A, et al. Hereditary mixed polyposis syndrome due to a BMPR1A mutation. Colorectal Dis 2010;12:570–3. [DOI] [PubMed] [Google Scholar]
- 14.Cao X, Eu KW, Kumarasinghe MP, et al. Mapping of hereditary mixed polyposis syndrome (HMPS) to chromosome 10q23 by genomewide high-density single nucleotide polymorphism (SNP) scan and identification of BMPR1A loss of function. J Med Genet 2006;43:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haramis AP, Begthel H, van den Born M, et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science 2004;303:1684–6. [DOI] [PubMed] [Google Scholar]
- 16.Rohlin A, Rambech E, Kvist A, et al. Expanding the genotype-phenotype spectrum in hereditary colorectal cancer by gene panel testing. Fam Cancer 2017;16:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cragun D, Radford C, Dolinsky JS, et al. Panel-based testing for inherited colorectal cancer: A descriptive study of clinical testing performed by a US laboratory. Clin Genet 2014;86:510–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman S, Beeri R, Walsh T, et al. Variable features of juvenile polyposis syndrome with gastric involvement among patients with a large genomic deletion of BMPR1A. Clin Transl Gastroenterol 2019;10:e00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blatter R, Tschupp B, Aretz S, et al. Disease expression in juvenile polyposis syndrome: A retrospective survey on a cohort of 221 European patients and comparison with a literature-derived cohort of 473 SMAD4/BMPR1A pathogenic variant carriers. Genet Med 2020;22:1524–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishida H, Ishibashi K, Iwama T. Malignant tumors associated with juvenile polyposis syndrome in Japan. Surg Today 2018;48:253–63. [DOI] [PubMed] [Google Scholar]
- 22.Aytac E, Sulu B, Heald B, et al. Genotype-defined cancer risk in juvenile polyposis syndrome. Br J Surg 2015;102:114–8. [DOI] [PubMed] [Google Scholar]
- 23.Cross W, Kovac M, Mustonen V, et al. The evolutionary landscape of colorectal tumorigenesis. Nat Ecol Evol 2018;2:1661–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.