Abstract
Background
The usefulness of comprehensive genomic profiling (CGP) panels for thoracic malignancies after completion of the standard treatment is unclear.
Methods
The results of CGP panels for malignant thoracic diseases performed at our hospital between December 2019 and June 2022 were collected. We examined whether CGP panel results led to new treatment, correlated with the effectiveness of immune checkpoint inhibitors (ICIs), or revealed secondary findings related to hereditary tumors.
Results
A total of 60 patients were enrolled, of which 52 (86.6%) had lung cancer. In six (10%) patients, the panel results led to treatment with insurance‐listed molecular‐targeted agents; four patients had EGFR mutations not detected by the real‐time polymerase chain reaction assay and two had MET ex.14 skipping mutations. In small‐cell lung cancer, the tumor mutation burden was high in 4/6 (66.7%) patients and pembrolizumab was available. Another MET ex.14 skipping mutation was detected in two cases with EGFR‐tyrosine kinase inhibitor resistance. ICI efficacy was ≤1 year in patients with STK‐11, KEAP1, and NEF2L2 mutations. A BRCA2 mutation with a high probability of germline mutation was detected in one patient. A thymic carcinoma with no detectable oncogenic mutation responded to second‐line treatment with Tegafur‐Gimeracil‐Oteracil Potassium (TS‐1) for ≥9 years.
Conclusions
CGP panels are useful in thoracic malignancies, especially lung cancer, because they can detect overlooked driver mutations and genetic alterations. We believe that the significance of conducting a CGP panel prior to treatment may also exist, as it may lead to the prediction of ICI treatment efficacy.
Keywords: comprehensive genomic profiling, lung cancer, next‐generation sequencing, thoracic malignancy
The usefulness of comprehensive genomic profiling (CGP) panels for thoracic malignancies is unclear. A total of 60 patients were enrolled and in six (10%) patients the panel results led to treatment with insurance‐listed molecular‐targeted agents. Immune checkpoint inhibitor efficacy was poor in patients with STK‐11, KEAP1, and NEF2L2 mutations. CGP panels are useful because they can detect overlooked driver mutations and genetic alterations, and lead to the prediction of ICI treatment efficacy.

INTRODUCTION
A genetic test in which multiple regions of multiple genes are simultaneously analyzed using next‐generation sequencing (NGS) is called a cancer gene panel test. 1 , 2 , 3 Conventional genetic testing can analyze a limited area at a time, but NGS allows the analysis of several to several hundred genes at a time. The cancer gene panel test can simultaneously detect base substitution/insertion/deletion mutations, gene amplification/deletion, and gene fusion in all or part of the carried genes. In addition, there are gene panel tests that can estimate tumor mutation burden (TMB) and microsatellite instability. 4 The number of target genes and type of nucleic acids (DNA and RNA) used for analysis are different for each cancer gene panel test. Furthermore, some tests use only tumor‐derived nucleic acids, while others use nucleic acids derived from normal specimens, such as peripheral blood, as controls. 3 , 5 In addition, a test method that analyses tumor‐derived free DNA in blood using peripheral blood is under development. 6 , 7 This test is expected to be implemented in clinical practice in the future because of its low invasiveness for specimen collection.
Similar to conventional gene tests, gene panel tests include a companion‐diagnosis function to determine the appropriateness of administering molecular‐targeting drugs, 8 , 9 in addition to comprehensive genomic profiling (CGP) to determine the genetic abnormalities involved for appropriate treatment selection. The former does not require interpretation of the results because the results obtained are positive/negative for a specific genetic biomarker. However, the latter requires decisions regarding the pathological significance of the genetic abnormality detected and the availability of the corresponding candidate drug. Therefore, a review by an “expert panel” or “molecular tumor board” is required for insurance purposes. 10 Although cancer gene panel tests have been used for >2 years in Japan, many problems persist, including how to make the best use of gene panel test results for treatment. According to domestic and international reports, the percentage of patients who receive treatment after a gene panel test is currently about 10–20%. 3
Lung cancer is a malignant disease with a poor prognosis and is the leading and second leading cause of death among men and women in Japan, respectively. 11 However, among solid tumors, lung cancer has the highest number of identified druggable driver mutations. 12 In advanced‐stage lung cancer, it is recommended to identify epidermal growth factor receptor (EGFR), ALK fusion, ROS1 fusion, BRAF, RET fusion, and MET ex.14 skipping mutations before starting treatment. 13 To identify these mutations simultaneously, an NGS‐based gene panel (Oncomine Dx target test) is recommended. 13 , 14 Lung cancer is the second most likely solid tumor after malignant melanoma to respond to immune checkpoint inhibitors (ICIs), 15 and genetic mutations associated with ICI efficacy are being identified. 16 , 17 However, the usefulness of CGP panel tests in advanced‐stage lung cancer after standard treatment has been completed is unclear.
In Japan, CGP panels are covered by health insurance only after completion of the standard treatment defined by each guideline. Therefore, this study aimed to examine the impact of CGP panels conducted after the completion of standard treatment on actual clinical practice at our center for malignant thoracic diseases and the current usefulness of such panels.
PATIENTS AND METHODS
Patients and analysis procedure for CGP panels
All patients with malignant thoracic disease who underwent CGP panel tests at the Osaka International Cancer Institute between December 2019, when the CGP panel was approved for reimbursement in Japan, and June 2022 were included in this study. Patients' age, sex, disease, and number of lines of treatment at the time of CGP panel evaluation were collected. All participants were asked whether they wished to disclose the results of CGP panel analysis to parties other than themselves and whether they wished to disclose information related to any hereditary tumors prior to test submission. When tumor tissue was used, the attending physician decided whether to perform a Foundation One panel (F1 panel) or an OncoGuide NCC oncopanel (NCC panel). When tissue specimens were used, after obtaining consent, a pathologist determined whether they could be submitted for CGP panel testing based on the tumor area, tumor content, and specimen storage period. The percentage of patients whose specimens could not be submitted because of the pathologist's decision and whether these patients subsequently underwent re‐examination were investigated. The sampling method of tumor specimens was also investigated. For patients treated after August 2021, when F1 liquid was introduced, if tissue specimens were not available, we proposed the use of F1 liquid, and CGP panel testing was performed using F1 liquid for consenting patients. The time between obtaining consent and disclosing the CGP panel results after expert panel review to the patient was calculated as the turnaround time.
Expert panel for CGP panels
The results of all CGP panel analyses were reviewed by an expert panel within the Osaka International Cancer Institute and then explained to the patients. The expert panel consisted of an oncologist for each organ, a clinical geneticist, a genetic counselor, a pathologist, a clinical trial coordinator, and a pharmacist. For detected alterations, oncogenicity was annotated based on the reports of each gene panel and Center for Cancer Genomics and Advanced Therapeutics (C‐CAT) guidelines, 18 and treatment for oncogenic mutations was recommended based on the results of clinical trials mainly in Japan and the recommendation level (A to F) in C‐CAT. 3 , 8 The possibility of drug treatment through the patient offer system was also proposed. We considered genetic mutations that may be associated with hereditary tumors as secondary findings, and if these were disclosed we considered referring the patient for genetic counseling. All considerations were based on individual patients' medical history, with a focus on treatment history.
Heat map of reported oncogenic mutations
Reported oncogenic alterations with a frequency of >5% in the cohort were included in the heatmap. The heat map was created using custom R programming scripts with graphics modules of ggplot2 v.3.3.6 and cowplot v.1.1.1. Cluster classification was performed for each malignant thoracic disease.
Correlation of STK11, KEAP1, and NEF2L2 mutations with effects of ICI
Cases with STK11, KEAP1, and NEF2L2 mutations were extracted based on the results of the CGP panel, and in patients with a history of ICI administration the effect was evaluated in terms of progression‐free survival (PFS). PFS was defined as the point from the start of ICI administration to its discontinuation due to tumor progression or toxicity, based on medical records.
RESULTS
Patient characteristics
During the study period, 63 patients consented to CGP panel testing, of whom eight (8/63, 12.7%) were determined to have insufficient specimens; of these, four (50%) patients underwent re‐biopsy for CGP panel, one (12.5%) specimen was submitted in F1 liquid, and three (37.5%) patients declined to resubmit tests. Finally, 60 (95.2%) results were available for analysis (Figure 1). The clinical characteristics of the patients are shown in Table 1. The participants included 38 (63.3%) men and 22 (36.7%) women, with a median age of 69 (range 44–82) years. Histopathologically, there were 33 (55.0%) lung adenocarcinomas, 10 (16.6%) lung squamous cell carcinomas, three (5.0%) nonsmall‐cell lung carcinomas (NSCLC)‐not otherwise specified, six (10.0%) small‐cell lung carcinomas (SCLC), six (10.0%) thymic carcinomas, one (1.7%) thymoma, and one (1.7%) malignant pleural mesothelioma. The median number of treatment lines at the time of CGP panel submission was three. Submitted specimens included 21 (35.0%) surgical biopsies, 12 (20.0%) computed tomography‐guided biopsies, two (3.3%) pleural biopsies, nine (15.0%) bronchoscopic specimens, six (10.0%) endobronchial ultrasound‐guided transbronchial needle aspiration specimens, and 10 (16.7%) plasma samples. The F1 panel, NCC panel, and F1 liquid were used to analyze 47, three, and 10 specimens, respectively. The median turnaround time from obtaining consent to explaining the results was 48 (range 33–118) days. Eight (13.3%) patients did not want the results of the CGP panel to be disclosed to anyone other than themselves and three (5.0%) did not want the results of inherited tumor‐associated mutations to be disclosed. No case could be registered in a clinical trial, based on the genetic alterations detected in the CGP panel.
FIGURE 1.

Patient flow chart. Flow chart of 63 patients who consented to CGP panel evaluation
TABLE 1.
Patient characteristics
| Total n = 60 | ||
|---|---|---|
| Age | ||
| Median (range) | 69 | (44–82) |
| Sex, n (%) | ||
| Male | 38 | (63.3) |
| Female | 22 | (36.7) |
| Disease | ||
| LDA | 33 | (55.0) |
| LSq | 10 | (16.6) |
| NSCLC‐NOS | 3 | (5.0) |
| SCLC | 6 | (10.0) |
| Thymic carcinoma | 6 | (10.0) |
| Thymoma | 1 | (1.7) |
| MPM | 1 | (1.7) |
| Treatment lines | ||
| Median (range) | 3 | (1–12) |
| Sampling methods | ||
| Surgical | 21 | (35.0) |
| CT‐guided | 12 | (20.0) |
| Pleural biopsy | 2 | (3.3) |
| TBB | 9 | (15.0) |
| EBUS‐TBNA | 6 | (10.0) |
| Liquid | 10 | (16.7) |
| CGP panel | ||
| Foundation One | 47 | (78.3) |
| NCC oncopanel | 3 | (5.0) |
| Foundation One Liquid | 10 | (16.7) |
| Turn around time, days | ||
| from obtaining consent to result explanation | ||
| Median (range) | 48 | (33–118) |
Abbreviations: CGP, comprehensive genome profiling; CT, computed tomography; EBUS‐TBNA, endobronchial ultrasound‐guided transbronchial needle aspiration; LDA, lung adenocarcinoma, LSq, lung squamous carcinoma; MPM, malignant pleural mesothelioma; NCC, national cancer center; NOS, not other specified; NSCLC, non‐small‐cell lung carcinoma; SCLC, small‐cell lung carcinoma; TBB, transbronchial biopsy.
Landscape of genomic alterations in 60 patients
Of the mutations detected in the gene‐panel analyses of 60 cases, only those mutations or copy‐number alterations that were considered oncogenic mutations in the report and found in >5% of cases are shown in the heatmap image (Figure 2). The top 10 alterations detected were TP53 (30%), CDKN2A (27.3%), EGFR (23.6%), RB1 (23.6%), CDKN2B (16.4%), MTAP (14.5%), ERBB2 (12.7%), ARID1A (9.1%), NEF2L2 (9.1%), and MET (7.3%). In six (10%) cases, genetic mutations that were indications for insurance‐approved molecularly targeted drugs (Evidence level A) were first detected using CGP panel testing. Of these, four patients had major activating EGFR mutations and two had MET ex.14 skipping mutations. The median TMB was 4/Mb (range 0–24). Except for six cases with microsatellite instability, all tumors were stable. Notably, high TMB (≥10/Mb) was observed in four of six (66.7%) patients with SCLC. HER2 mutations eligible for trastuzumab deruxtecan therapy included three cases of A775_G776insYVMA and one case each of G776>VC and S310F.
FIGURE 2.

Heatmap of CGP panel in 60 cases. Heatmap of the mutation pattern of oncogenic alterations with frequency >5%. Each row represents a gene and each column represents a case. On the left side of the heatmap oncogenic alterations are listed in order of frequency, with gray bars representing the frequency. The heatmap shows oncogenic alterations in different colors. The horizontal axis shows clinical data for each case, including sex, histological type, and tumor mutation burden
EGFR mutation cases not detected by the first RT‐PCR test but detected by the CGP panel
All four patients with major activating EGFR mutations detected using the CGP panel had undergone RT‐PCR EGFR‐detection tests at diagnosis, but no EGFR mutation was detected and they were treated as EGFR‐mutation‐negative cases. The clinical courses and characteristics of the four cases are presented in Table 2. In all cases, CGP panels were performed after at least 2 years of chemotherapy. In case 1, the RT‐PCR test for EGFR gene mutation was submitted using bronchoscopy‐forceps washout. In case 2, RT‐PCR was performed using a section from a paraffin‐embedded block of tumor‐tissue specimen from a bronchial biopsy. EGFR L858R in case 2 was a two‐base substitution mutation of EGFR c.2572 _ 2573 CT>AG (Figure 3). EGFR gene mutations detected in cases 3 and 4 (Ex.20 ins A763_Y764 ins FGEA and Ex.20 S768_V769 > IL, respectively) were likely not detected because they were variants not covered by RT‐PCR testing.
TABLE 2.
Patient list of EGFR mutations detected by the CGP panel
| Case no. | Case 1 | Case 2 | Case 3 | Case 4 |
|---|---|---|---|---|
| EGFR mutation | E746_A750 del | L858R | Ex.20 ins A763_Y764 ins FQEA | Ex.20 S768_V769 > IL |
| Age at CGP panel | 76 years | 71 years | 71 years | 66 years |
| Sex | Female | Female | Female | Male |
| Smoking history | Light | Never | Never | Never |
| Treatment line at CGP panel | 6th line | 8th line | 3rd line | 12th line |
| Years of treatment at the time of CGP panel | 3 years | 8 years | 2 years | 8 years |
Abbreviations: CGP, comprehensive genome profiling; EGFR, epithelial growth factor receptor.
FIGURE 3.
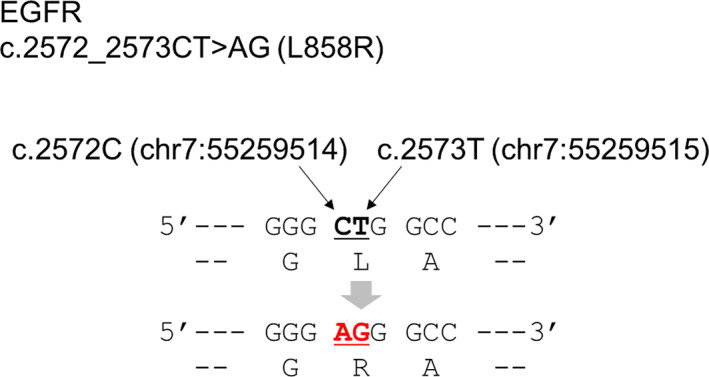
A case of EGFR L858R mutation caused by two‐base substitution. A two‐base substitution changes the codon encoding amino acid 858th L from “CFG” to “AGG”. “AGG” codes R
CGP panel testing for resistance mutations after treatment with tyrosine kinase inhibitor
A total of six patients underwent CGP panel testing to search for resistance mutations after tyrosine‐kinase inhibitor (TKI) treatment. Four patients had major EGFR mutations and two had ALK fusion; all six patients underwent TKI therapy for each mutation. MET ex.14 skipping mutation was detected as a resistance mutation in two of the four patients with EGFR mutations. In one patient, G724S mutation was detected as a new compound mutation in addition to the original Ex.19 deletion, leading to a change in TKI, based on EGFR structure. 19 In one case of ALK fusion, after first‐, second‐, third‐, and fourth‐line treatment with alectinib, lorlatinib, ceritinib, and a combination of CBDCA, paclitaxel, bevacizumab, and atezolizumab, respectively, and a fifth‐line lorlatinib rechallenge, CGP panel was performed, and BRAF‐KIAA1549 fusion was detected in addition to multiple ALK‐resistant mutations of ALK G1269A, L1196M and F1174C.
ICI‐resistant mutations
We studied STK11, KEAP1, and NEF2L2 mutations as ICI‐resistant gene mutations and the effect of ICIs on tumors with these mutations. Of the 60 patients, eight had oncogenic mutations of these three genes and seven had received ICI treatment. The age, sex, TMB, PD‐L1 tumor proportion score (%), and ICI and ICI treatment line administered were evaluated, and their correlation with PFS in these patients is summarized in Table 3. PFS was <1 year in all patients receiving ICIs, and in patients with KEAP1 and NEF2L2 mutations, PFS was <3 months despite administration of the first‐line therapy, indicating primary resistance to ICI.
TABLE 3.
List of patients with STK11, KEAP1, and NEF2L2 mutations who received ICI
| Age (years) | Sex | TMB (/Mb) | PD‐L1 TPS (%) | STK11, KEAP1, NEF2L2 | ICI treatment line | ICI treatment | PFS |
|---|---|---|---|---|---|---|---|
| Mutation or alteration | |||||||
| 80 | F | 10 | 0% | STK11 Q100fs*63 | 2nd | Atezolizumab | 6 months |
| 63 | F | 8 | 90% | STK11 loss | 1st | Pembrolizumab | 11 months |
| 73 | M | 3 | 30% | KEAP1 Q92* | 1st | Pembrolizumab | 2 months |
| 66 | F | 1 | 60% | KEAP1 W252* | 2nd | Pembrolizumab | 3 months |
| 48 | M | 3 | 80% | NEF2L2 D29H | 3rd | Pembrolizumab | 2 months |
| 55 | M | 4 | 1% | NEF2L2 E79K | 1st | CBDCA + PTX + Niv + Ipi | 2 months |
| 53 | M | 21.7 | 1% | NEF2L2 G31R | 1st | CBDCA + PEM + Pembrolizumab | 2 months |
| 76 | F | CBD | 20% | NEF2L2 G81S | NA | NA | NA |
Abbreviations: CBD, cannot be determined; F, female; ICI, immune checkpoint inhibitor; M, male; NA, not applicable; PFS, progression‐free survival; TMB, tumor mutation burden; TPS, tumor proportion score.
Secondary finding associated with hereditary tumor
One of the 60 patients had BRCA2 Q1361* mutation as a secondary finding associated with hereditary breast and ovarian cancer, after checking the family history of neoplastic diseases. The patient was a 70‐year‐old woman with squamous cell carcinoma of the lung who had undergone surgical treatment for left breast cancer at age 48. Her sister was diagnosed with breast cancer at age 55 and her paternal aunt was diagnosed with uterine cancer at age 75. The prevalence of tumor diseases in her family tree is shown in (Figure 4). The BRAC2 Q1361* mutation was considered to be associated with hereditary breast and ovarian cancer in the analysis of tumor tissue alone, but the patient did not wish to receive genetic counseling and her germline mutation of BRAC2 Q1361* was not examined using normal tissue.
FIGURE 4.

Pedigree of a family with strong hereditary breast and ovarian cancer history. Red arrows point to the lung cancer case that underwent CGP panel analysis. The case received the operation for the left breast cancer in a 48‐year‐old. The younger sister had breast cancer at the age of 55. Her father had lung cancer at age 70 and colorectal cancer at age 72, and her paternal aunt had a history of uterine cancer
No oncogenic mutation in thymic cancer exhibiting exceptional response to TS‐1
Seven of the 60 patients had thymic tumors. Among them, in one case no genetic alterations were detected in the CGP panel. The patient received CBDCA plus paclitaxel as the first‐line treatment, but the disease progressed after 7 months, therefore he received tegafur + gimeracil + oteracil as the second‐line treatment and currently his disease has been in remission for 9 years.
DISCUSSION
Among patients with malignant thoracic diseases, mainly lung cancer, six (10%) patients with an insurance‐approved indication at evidence level A, based on CGP panel results, received molecularly targeted drugs. In addition, high TMB was detected in 4/6 (66.7%) small‐cell carcinomas, making pembrolizumab a new treatment option for TMB‐high small‐cell carcinomas where treatment is limited. In five cases, HER2 mutations eligible for trastuzumab deruxtecan treatment at evidence level B were detected. MET ex.14 skipping mutation was detected as a new driver mutation in specimens with EGFR‐TKI resistance, leading to the introduction of a new therapeutic agent. Compared to other cancer types, CGP panel testing has a higher probability of leading to promising treatments in lung cancer and CGP panels may be more useful for lung cancer. In this study, we clarified the significance of multiple NGS panels because a CGP panel in clinical practice has a high probability of detecting TMB‐high in SCLC, and also because a druggable driver mutation can be detected in cases with EGFR mutations that were previously screened by assays other than NGS.
Of the six cases that led to molecularly targeted agents at evidence level A, two cases of MET ex.14 skipping mutations were detected by the CGP panel because the same mutation had not been searched for using RT‐PCR. As pre‐treatment NGS‐based gene panels become more prevalent in the future, such cases are expected to become less frequent. All four cases in which EGFR mutations were detected had undergone RT‐PCR‐based EGFR testing at least once. Cases 1 and 2 demonstrated Ex.19 deletion and L858R mutation, respectively, which were major activating EGFR mutations and therefore variants covered by RT‐PCR. 20 Case 1 results may have been false negative because the specimen used was the biopsy‐forceps washing fluid, which probably contained a low percentage of cancer cells. In case 2, the mutation was caused by a two‐base substitution, therefore it is possible that the primers specific for the L858R mutation could not bind and RT‐PCR was not successful. 21 Cases 3 and 4 demonstrated variants that were not covered by the RT‐PCR‐based EGFR assay and by the Oncomine Dx target test, respectively. Therefore, the mutations could only be detected by the CGP panel. 22 , 23 , 24 All four patients survived for >2 years and nearly 8 years without EGFR‐TKIs, suggesting that EGFR mutation is a favorable prognostic factor regardless of EGFR‐TKIs. 25 ALK fusion has also been reported to be a favorable prognostic factor. 26 Long‐term survivors of advanced lung cancer are not likely to have undergone an NGS panel at presentation and should be actively considered for a CGP panel.
The number of approved chemotherapeutic regimens for SCLC is smaller than for NSCLC. 27 Recently, the use of pembrolizumab was approved for TMB‐high solid tumors. 28 With regard to the efficacy of pembrolizumab in SCLC treated with >2 lines of chemotherapy, a previous study reported an objective response rate of 19.3% (95% confidence interval 11.4–29.4); two of 83 patients showed complete response, and 14 patients showed partial response. The median duration of response was not reported (range 4.1–35.8 months, plus sign indicates ongoing response). 29 This trial excluded cases with SCLC in which the anti‐PD‐L1 antibody drugs atezolizumab 30 and durvalmab, 31 which are currently approved for first‐line induction, were used. However, pembrolizumab, an anti‐PD‐1 antibody drug, may be effective in cases in which anti‐PD‐L1 antibody drugs are ineffective. 32 , 33 The CGP panel is useful because it adds a new treatment option, pembrolizumab. Interestingly, in this study, the CGP panel detected high TMB in 66.7% of cases with SCLC.
ICIs are approved for all malignant thoracic diseases except thymic tumors and are recommended unless there is a specific reason for avoiding their use. 34 As genome profiling progresses, mutations that negatively correlate with the effects of ICIs have been reported, STK11 and KEAP1 being representative of such mutations. 16 , 17 NEF2L2 has been reported to form a complex with KEAP1 and exhibit intracellular bioactivity. 35 NSCLCs with these oncogenic mutations have been reported to be resistant to ICIs. 16 , 17 , 36 Similarly, in the present study, we observed a trend toward reduced efficacy of ICIs in patients with these mutations. If unnecessary ICI administration can be avoided by genome profiling, it may be possible to avoid a reduction in the quality of life due to immune‐related adverse events. Therefore, we believe it is worthwhile to conduct the CGP panel prior to the start of treatment, rather than after.
The availability of specimen volume is an issue in CGP panel testing in patients with advanced‐stage lung cancer who have had an NGS panel performed at the time of initial diagnosis. At diagnosis, physicians rely on bronchoscopic biopsy specimens in nearly 60% of patients with advanced‐stage lung cancer. 37 , 38 F1 CDx requires at least 1 mm3 of tissue and the NCC OncoPanel requires 10 unstained slides with a minimum size of 4 mm2 (16 mm2 is recommended). 14 These tumor volumes are often difficult to obtain from bronchoscopic biopsy specimens, and the possibility of obtaining specimens that can withstand two NGS panels is much lesser. Surgical biopsy specimens accounted for 35% of specimens in this study, while bronchoscopic biopsy specimens accounted for only 25% (Table 1). In eight cases, a re‐biopsy for gene panel evaluation was required, suggesting that specimen collection is an important issue in thoracic malignancies. In addition, a surgical biopsy specimen may be able to withstand multiple gene panels, 23 and it is important to consider a genomic biopsy policy that aims not only at diagnosis but also at genomic analysis. 38
This study had several limitations. First, this was a single‐center, retrospective, controlled study with a limited number of cases, therefore the statistical significance of mutations as a factor for poor treatment response to ICIs could not be fully investigated. Second, the sample predominantly included cases with lung cancer, and the significance of CGP panels in other malignant thoracic diseases could not be adequately studied. Third, since this was a retrospective study, there was a selection bias for cases in which a CGP panel was performed. To examine the usefulness of the CGP panel, it would be helpful to examine the impact of the CGP panel on clinical practice by prospectively examining all cases with thoracic malignant diseases over a period of time. Fourth, the reach rate for clinical trials based on CGP panel results is likely to be influenced by region. Since clinical trials for cancer drugs are more common in Tokyo than in other parts of Japan, it is conceivable that the reach rate for clinical trials may also be higher in Tokyo.
CONCLUSION
In the present study, the CGP panel detected favorable genetic alterations, including druggable mutations, in 12 (20%) of 60 patients. TMB‐high SCLC responded to pembrolizumab, whereas MET ex.14 skipping mutation was resistant to EGFR‐TKI. Compared with other cancer types, lung cancer is rich in molecular‐targeted agents, therefore the usefulness of a CGP panel may be greater. Mutations in STK‐11, KEAP1, and NEF2L2 may be useful for predicting the effect of ICIs, and the importance of conducting a CGP panel before the start of treatment in clinical practice was suggested.
AUTHORS' CONTRIBUTIONS
K.K., N.S., T.K., T.Y., K.H., S.N., Y.K., F.F., T.I., Y.Y., M.K., T.W., T.Y., S.Y., T.H., T.I., M.T., F.I., and K. Nishimura made substantial contributions to the study conception, study design, study protocol, data interpretation, and writing or critically reviewing the manuscript for important intellectual content. Material preparation, data collection, and analysis were performed by K.K. The first draft of the manuscript was written by K.K., N.S., T.K., T.Y., K.H., S.N., Y.K., F.F., T.I., Y.Y., T.W., T.Y., S.Y., T.H., T.I., M.T., F.I., K. Nishimura, and K. Nishino commented on the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
Dr. Kunimasa reports honoraria for lecture from AstraZeneca, Chugai Pharma, Novartis, Ono Pharmaceutical, Eli Lilly, and Pfizer Merk. Dr. Sugimoto reports honoraria for lecture from MSD, Eli Lilly, Chugai Pharma, Taiho Pharmaceutical, Dai‐ichi Sankyo, and Ono Pharmaceutical. Dr. Tamiya reports grants from Ono Pharmaceutical, Bristol‐Myers Squibb, and Boehringer Ingelheim, and honoraria for lectures from Taiho Pharmaceutical, Eli Lilly, Asahi Kasei Pharmaceutical, MSD, Boehringer Ingelheim, AstraZeneca, Chugai Pharmaceutical, Ono Pharmaceutical, and Bristol‐Myers Squibb. Dr. Nishimura reports honoraria for a lecture from AstraZeneca. Dr. Nishino reports a grant from Nippon Boehringer Ingelheim and honoraria for lectures from Chugai Pharma, AstraZeneca, Nippon Boehringer Ingelheim, Eli Lilly Japan, Roche Diagnostics, Novartis, and Pfizer Merk. The other authors have no conflict of interest.
ACKNOWLEDGMENTS
We would like to acknowledge to all patients with thoracic malignancies who received CGP panels in our hospital. The study was approved by the ethics committee of the Osaka International Cancer Center (#22046).
Kunimasa K, Sugimoto N, Kawamura T, Yamasaki T, Honma K, Nagata S, et al. Clinical application of comprehensive genomic profiling panel to thoracic malignancies: A single‐center retrospective study. Thorac Cancer. 2022;13(21):2970–2977. 10.1111/1759-7714.14643
REFERENCES
- 1. Kohno T. Implementation of "clinical sequencing" in cancer genome medicine in Japan. Cancer Sci. 2018;109(3):507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kou T, Kanai M, Yamamoto Y, Kamada M, Nakatsui M, Sakuma T, et al. Clinical sequencing using a next‐generation sequencing‐based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017;108(7):1440–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sunami K, Ichikawa H, Kubo T, Kato M, Fujiwara Y, Shimomura A, et al. Feasibility and utility of a panel testing for 114 cancer‐associated genes in a clinical setting: a hospital‐based study. Cancer Sci. 2019;110(4):1480–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takeda M, Takahama T, Sakai K, Shimizu S, Watanabe S, Kawakami H, et al. Clinical application of the FoundationOne CDx assay to therapeutic decision‐making for patients with advanced solid tumors. Oncologist. 2021;26(4):e588–e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne liquid CDx, a novel 324‐gene cfDNA‐based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One. 2020;15(9):e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sunami K, Bando H, Yatabe Y, Naito Y, Takahashi H, Tsuchihara K, et al. Appropriate use of cancer comprehensive genome profiling assay using circulating tumor DNA. Cancer Sci. 2021;112(9):3911–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li MM, Datto M, Duncavage EJ, Kulkarni S, Lindeman NI, Roy S, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Leichsenring J, Horak P, Kreutzfeldt S, Heining C, Christopoulos P, Volckmar AL, et al. Variant classification in precision oncology. Int J Cancer. 2019;145(11):2996–3010. [DOI] [PubMed] [Google Scholar]
- 10. Luchini C, Lawlor RT, Milella M, Scarpa A. Molecular tumor boards in clinical practice. Trends Cancer. 2020;6(9):738–44. [DOI] [PubMed] [Google Scholar]
- 11. Yamaguchi T, Nishiura H. Predicting the epidemiological dynamics of lung cancer in Japan. J Clin Med. 2019;8(3):326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Middleton G, Fletcher P, Popat S, Savage J, Summers Y, Greystoke A, et al. The National Lung Matrix Trial of personalized therapy in lung cancer. Nature. 2020;583(7818):807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akamatsu H, Ninomiya K, Kenmotsu H, Morise M, Daga H, Goto Y, et al. The Japanese lung cancer society guideline for non‐small cell lung cancer, stage IV. Int J Clin Oncol. 2019;24(7):731–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yatabe Y, Sunami K, Goto K, Nishio K, Aragane N, Ikeda S, et al. Multiplex gene‐panel testing for lung cancer patients. Pathol Int. 2020;70(12):921–31. [DOI] [PubMed] [Google Scholar]
- 15. Johnson DB, Nebhan CA, Moslehi JJ, Balko JM. Immune‐checkpoint inhibitors: long‐term implications of toxicity. Nat Rev Clin Oncol. 2022;19(4):254–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marinelli D, Mazzotta M, Scalera S, Terrenato I, Sperati F, D'Ambrosio L, et al. KEAP1‐driven co‐mutations in lung adenocarcinoma unresponsive to immunotherapy despite high tumor mutational burden. Ann Oncol. 2020;31(12):1746–54. [DOI] [PubMed] [Google Scholar]
- 17. Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan‐tumor genomic biomarkers for PD‐1 checkpoint blockade‐based immunotherapy. Science. 2018;362(6411):eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mukai Y, Ueno H. Establishment and implementation of cancer genomic medicine in Japan. Cancer Sci. 2021;112(3):970–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kunimasa K, Sugimoto N, Tamiya M, Inoue T, Kawamura T, Kanzaki R, et al. Dacomitinib overcomes afatinib‐refractory carcinomatous meningitis in a lung cancer patient harbouring EGFR ex.19 deletion and G724S mutation; a case report. Invest New Drugs. 2022;40(5):1137–40. [DOI] [PubMed] [Google Scholar]
- 20. Kimura H, Kasahara K, Kawaishi M, Kunitoh H, Tamura T, Holloway B, et al. Detection of epidermal growth factor receptor mutations in serum as a predictor of the response to gefitinib in patients with non‐small‐cell lung cancer. Clin Cancer Res. 2006;12(13):3915–21. [DOI] [PubMed] [Google Scholar]
- 21. Roma C, Esposito C, Rachiglio AM, Pasquale R, Iannaccone A, Chicchinelli N, et al. Detection of EGFR mutations by TaqMan mutation detection assays powered by competitive allele‐specific TaqMan PCR technology. Biomed Res Int. 2013;2013:385087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coleman N, Woolf D, Welsh L, McDonald F, MacMahon S, Yousaf N, et al. EGFR exon 20 insertion (A763_Y764insFQEA) mutant NSCLC is not identified by Roche Cobas version 2 tissue testing but has durable intracranial and extracranial response to Osimertinib. J Thorac Oncol. 2020;15(10):e162–e5. [DOI] [PubMed] [Google Scholar]
- 23. Kunimasa K, Nishino K, Kukita Y, Matsumoto S, Kawachi H, Kawamura T, et al. Late recurrence of lung adenocarcinoma harboring EGFR exon 20 insertion (A763_Y764insFQEA) mutation successfully treated with osimertinib. Cancer Genet. 2021;256–257:57–61. [DOI] [PubMed] [Google Scholar]
- 24. Zhang H, Shao YW, Xia Y. Responsiveness to full‐dose Afatinib in a patient with lung adenocarcinoma harboring EGFR S768I and V769L mutations. J Thorac Oncol. 2019;14(2):e25–e7. [DOI] [PubMed] [Google Scholar]
- 25. Li WY, Zhao TT, Xu HM, Wang ZN, Xu YY, Han Y, et al. The role of EGFR mutation as a prognostic factor in survival after diagnosis of brain metastasis in non‐small cell lung cancer: a systematic review and meta‐analysis. BMC Cancer. 2019;19(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Blackhall FH, Peters S, Bubendorf L, Dafni U, Kerr KM, Hager H, et al. Prevalence and clinical outcomes for patients with ALK‐positive resected stage I to III adenocarcinoma: results from the European thoracic oncology platform Lungscape project. J Clin Oncol. 2014;32(25):2780–7. [DOI] [PubMed] [Google Scholar]
- 27. Ganti AKP, Loo BW, Bassetti M, Blakely C, Chiang A, D'Amico TA, et al. Small cell lung cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2021;19(12):1441–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marcus L, Fashoyin‐Aje LA, Donoghue M, Yuan M, Rodriguez L, Gallagher PS, et al. FDA approval summary: Pembrolizumab for the treatment of tumor mutational burden‐high solid tumors. Clin Cancer Res. 2021;27(17):4685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chung HC, Piha‐Paul SA, Lopez‐Martin J, Schellens JHM, Kao S, Miller WH Jr, et al. Pembrolizumab after two or more lines of previous therapy in patients with recurrent or metastatic SCLC: results from the KEYNOTE‐028 and KEYNOTE‐158 studies. J Thorac Oncol. 2020;15(4):618–27. [DOI] [PubMed] [Google Scholar]
- 30. Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, et al. First‐line atezolizumab plus chemotherapy in extensive‐stage small‐cell lung cancer. N Engl J Med. 2018;379(23):2220–9. [DOI] [PubMed] [Google Scholar]
- 31. Paz‐Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, et al. Durvalumab plus platinum‐etoposide versus platinum‐etoposide in first‐line treatment of extensive‐stage small‐cell lung cancer (CASPIAN): a randomised, controlled, open‐label, phase 3 trial. Lancet. 2019;394(10212):1929–39. [DOI] [PubMed] [Google Scholar]
- 32. Xu Z, Hao X, Yang K, Wang Q, Wang J, Lin L, et al. Immune checkpoint inhibitor rechallenge in advanced or metastatic non‐small cell lung cancer: a retrospective cohort study. J Cancer Res Clin Oncol. 2022. [online ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu S, Shukuya T, Tamura J, Shimamura S, Kurokawa K, Miura K, et al. Heterogeneous outcomes of immune checkpoint inhibitor Rechallenge in patients with NSCLC: a systematic review and meta‐analysis. JTO Clin Res Rep. 2022;3(4):100309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ettinger DS, Wood DE, Aisner DL, Akerley W, Bauman JR, Bharat A, et al. Non‐small cell lung cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(5):497–530. [DOI] [PubMed] [Google Scholar]
- 35. Baird L, Llères D, Swift S, Dinkova‐Kostova AT. Regulatory flexibility in the Nrf2‐mediated stress response is conferred by conformational cycling of the Keap1‐Nrf2 protein complex. Proc Natl Acad Sci USA. 2013;110(38):15259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scalera S, Mazzotta M, Corleone G, Sperati F, Terrenato I, Krasniqi E, et al. KEAP1 and TP53 frame genomic, evolutionary, and immunologic subtypes of lung adenocarcinoma with different sensitivity to immunotherapy. J Thorac Oncol. 2021;16(12):2065–77. [DOI] [PubMed] [Google Scholar]
- 37. Kunimasa K, Matsumoto S, Nishino K, Nakamura H, Kuhara H, Tamiya M, et al. Improvement strategies for successful next‐generation sequencing analysis of lung cancer. Future Oncol. 2020;16(22):1597–606. [DOI] [PubMed] [Google Scholar]
- 38. Kunimasa K, Matsumoto S, Nishino K, Honma K, Maeda N, Kuhara H, et al. Comparison of sampling methods for next generation sequencing for patients with lung cancer. Cancer Med. 2022;11(14): 2744–54. [DOI] [PMC free article] [PubMed] [Google Scholar]


