Abstract
In 2019, the world experienced the rapid outbreak of the Covid-19 pandemic creating an alarming situation worldwide. The virus targets the respiratory system causing pneumonia with other symptoms such as fatigue, dry cough, and fever which can be mistakenly diagnosed as pneumonia, lung cancer, or TB. Thus, the early diagnosis of COVID-19 is critical since the disease can provoke patients’ mortality. Chest X-ray (CXR) is commonly employed in healthcare sector where both quick and precise diagnosis can be supplied. Deep learning algorithms have proved extraordinary capabilities in terms of lung diseases detection and classification. They facilitate and expedite the diagnosis process and save time for the medical practitioners. In this paper, a deep learning (DL) architecture for multi-class classification of Pneumonia, Lung Cancer, tuberculosis (TB), Lung Opacity, and most recently COVID-19 is proposed. Tremendous CXR images of 3615 COVID-19, 6012 Lung opacity, 5870 Pneumonia, 20,000 lung cancer, 1400 tuberculosis, and 10,192 normal images were resized, normalized, and randomly split to fit the DL requirements. In terms of classification, we utilized a pre-trained model, VGG19 followed by three blocks of convolutional neural network (CNN) as a feature extraction and fully connected network at the classification stage. The experimental results revealed that our proposed VGG19 + CNN outperformed other existing work with 96.48 % accuracy, 93.75 % recall, 97.56 % precision, 95.62 % F1 score, and 99.82 % area under the curve (AUC). The proposed model delivered superior performance allowing healthcare practitioners to diagnose and treat patients more quickly and efficiently.
Keywords: Pneumonia; Lung cancer; COVID-19, TB, Lung opacity; X-ray images; Deep learning, VGG19 +CNN; Multiclass diseases classification
1. Introduction
Numerous studies have discussed the efficacy of computer-aided diagnoses in the medical context, based on collaboration between medical researchers and computer scientists. Certain computer-aided diagnosis systems in medicine may be classified as expert systems since they seek to replicate the decisions of medical professionals. In addition, computer-aided detection systems in medicine can process complicated and large clinical data [1]. Computer-aided detection systems can also assist clinicians to gain new insights into data and apply the knowledge to improve diagnostic accuracy. As a result, the systems are considered intelligent systems since they employ a process of feedback to continuously enhance their performances. Large clinical data is complicated to analyze. Intelligent Computer-aided diagnosis systems using data mining, artificial intelligence (AI), and deep learning methodologies are beneficial in diagnosing an array of illnesses and medical disorders.
In the last century, researchers have accumulated substantial knowledge regarding human anatomy and physiology.
In recent years, chest X-rays (CXR), ultrasounds, and magnetic resonance imaging (MRI) have played vital roles in enhancing the accurate diagnosis of human diseases. Significant improvements in healthcare and medical research have helped people to improve their quality of life as new technologies have facilitated the accurate diagnoses of patients’ ailments and diseases. In the last few decades, medical experts faced challenges in conducting an accurate diagnosis of diseases, which compounded unnecessary healthcare and malpractice claims for both the doctor and patients. Machine learning, deep learning, and statistical analysis are effective tools for computer-aided diagnosis. These tools are used in solving difficult computer vision tasks in medical imaging, such as segmenting lungs, classifying lung diseases, and so on. With recent developments in deep learning, machines can perform equally or better than humans on a wide range of activities. For example, deep learning can be used to calculate the treatment outcomes, such as cancer therapy. With huge-labelled datasets and deep learning-based approaches, promising findings are developed in the categorization of thoracic disorders using a CXR modality. In addition, machine learning is the model that can learn and make decisions based on a vast number of input data sets. Artificial intelligence performs activities that require human intellect, such as voice recognition, translation, and the ability to analyze colors and shapes by evaluating the incoming data and making predictions. A combination of machine learning algorithms, known as deep learning, has demonstrated remarkable success in various sectors, particularly in the healthcare sector [2], [3]. Deep learning models can accurately predict and categorize numerous diseases, such as tuberculosis (TB), lung cancer, pneumonia, and currently COVID-19 using images, without human intervention. As the network becomes larger, data representation becomes deeper, making deep learning to be more effective, contrary to classical machine learning. Consequently, the model automatically collects characteristics and generates more accurate outcomes. Since the models use a combination of non-linear functions rather than linear functions, deep learning algorithms are more accurate than typical machine learning methods.
In late 2019, the coronavirus (COVID-19) pandemic invaded the planet, leading to an alarming scenario. The virus was first formally discovered in Wuhan, China in December 2019, and the World Health Organization (WHO) designated it as an emergency health problem at the beginning of 2020. By March 2020, WHO classified it as a pandemic [4]. The Coronavirus causes pneumonia, persistent cough, high fever, and fatigue, among other symptoms. Reverse transcription-polymerase chain reaction (RT-PCR) is employed to identify positive cases of the virus. However, it can take several hours, even days, to generate results using this form of diagnosis. RT-PCRs are both time-consuming and expensive. Subsequently, experts are facing significant challenges in developing alternatives via detection technologies. AI is being used to automate the diagnosis of many diseases today, and AI has been proven to achieve superior performance during automatic image categorization using various machine learning algorithms. The detection based on the image processing is based on the classification of the features extracted from the CXR or CT, as shown in Fig. 1 . Furthermore, machine learning specifies models that have the capability of learning and making decisions based on a massive input of data samples.
Fig. 1.
Image processing-based classification model.
In the context of deep learning, the extraction and classification of features from images is the primary goal. Deep learning has been a huge success in a wide range of industries, including healthcare [3]. In addition, deep learning can develop models that can accurately predict and diagnose illnesses using images. It has been effective in diagnosing TB [5], [6], [7], [8], [9], [10], pneumonia [11], [12], [13], [14], [15], [16], [17], [18], lung cancer [19], [20], [21], [22], [23], and COVID-19 diagnosis, without need of human expertise. Unlike traditional machine learning, the fundamental reason behind using deep learning techniques is its ability to build the model of input as the size of network deeply grows. Because of this, the model automatically gathers data and generates findings that are more accurate. Deep learning models, in contrast to typical machine learning algorithms, describe features using a sequence of non-linear functions that are incorporated to optimize the accuracy of the utilized model.
The remaining part of this paper is organized as follows: Section 2 presents the research-related works and background on chest diseases detection and classification based deep learning. The details of the materials and methods, the dataset for the study, data preprocessing, and the proposed deep learning framework are described in Section 3. Section 4 incorporates the results, including the experimental parameters and the performance metrics for our proposed system, with comparisons to current state-of-the-art systems of chest diseases detection and classification. Finally, conclusions and possible ongoing future work are given in Section 5.
2. Literature review
In most countries, chest computed tomography (CT) and X-ray pictures are widely utilized as a feasible option for identification of COVID-19. However, COVID-19 identification is a complex process, which requires clinical imaging of patients [24], [25], [26], [27], [28], [29], [30]. Lung cancer represents a major source of mortality in humans. The immediate diagnosis could improve human survival [19], [20], [21], [22]. Applying machine learning and image processing have presented considerable promise for lung cancer diagnosis. In this section, an exhaustive evaluation of deep learning models for TB, COVID-19, lung cancer, and pneumonia are discussed.
Transferring learning tools, such as VGG-16, ResNet-50, and InceptionV3, to clinical pictures of lung illnesses and COVID-19 has offered promising results [24]. It is discovered that pneumonia is among major symptoms of COVID-19. Transfer learning helps discover that the same virus causes pneumonia and COVID-19. A study demonstrates that the information obtained by a model trained to detect viral pneumonia may be applied to identify COVID-19 [24]. As a result, Haralick features can be used to facilitate feature extraction. This approach involves statistical analyses that focus on a specific area of COVID-19 diagnosis. In comparison to the traditional classifications, transfer learning has consistently proven to offer statistically significant outcomes [24]. Some studies developed and analyzed a fully automated COVID-19 detection framework utilizing CTX. To diagnose COVID-19, the visual features were extracted from volumetric chest CT images using COVID-19 neural network approach. The outcomes show that the approach has outperformed the existing work. Pre-trained models-based convolutional neural network (CNN) architecture such as Inception-ResNetV2, ResNet152, ResNet50, InceptionV3, and ResNet101 was used in related work to identify COVID-19 pneumonia based on the CXR images. Among the existing models, the ResNet50 exhibited the most accurate classification outcomes [28]. The comparison and modelling were based on CT images of 101 pneumonia, 88 COVID-19 patients, and 86 healthy cases from two areas in China. A details relation extraction neural network (DRENet) learning-based CT diagnostic algorithm identified COVID-19 patients. The model correctly distinguished between COVID-19 patients with a recall of 0.93, AUC of 0.99, and accuracy of 0.96. The research showed that deep learning based on CT scans may help to detect COVID-19 patients and automatically identify possible abnormal changes. Another study categorized COVID-19 CXR images by applying modified MobileNet and a ResNet architecture. With this approach, characteristics from multiple CNN layers are dynamically combined to overcome the gradient vanishing problem. The proposed approaches outperform over the current methods by 99.3 % on the CT image dataset and by 99.6 % on the CXR. [27].
Some studies developed a model to distinguish between critical and severe COVID-19 instances using deep learning characteristics and radiomics based on d-Resnet [29], [31]. These authors studied 217 individuals in three Chinese hospitals, 82 with extreme severity and 135 with serious disease. The patients were grouped into two (174 patients) training and (43 patients) testing. The authors created a 3-dimensional deep learning network using the clipped segments and multivariable logistic regression to integrate relevant radiomics characteristics and deep learning scores. To test the robustness of their methods, they used stratified analysis, cross-validation, decision curve analysis, and survival analysis. An AUC of 0.909 distinguishes between critical patients in the test and training groups [31]. Another study applied InceptionV3, NASNet, Xception, DenseNet, MobileNet, VGGNet, InceptionResNetV2, and ResNet for classifying the COVID-19, which was tested on the mixed dataset of CXR and CT images. DenseNet121 offered the best performance with an accuracy of 99 % [29].
Image segmentation is used to categorize chest CTX into pneumonia, COVID-19, and normal illnesses using four CNN base learners, a modified stack ensemble model, and Naive Bayes as the meta-learner in one research. For COVID-19, pneumonia, and normal classes, the suggested technique beats current techniques by 0.9867 on standard datasets and 0.98 Kappa on the same datasets [32] based on CT scans. By reducing manually labelled CT images, the suggested technique may accurately detect COVID-19 infections and rule out the case of COVID-19. Based on the positive qualitative and quantitative results, we envision our created approach is widely used in large-scale clinical trials [33]. The convolution neural networks are effective in converting 360 X-ray and CT scan pictures into a categorization on a binary class pneumonia-based translation of decision tree, Inception V2, and VGG-19 models. Compared to decision tree (60 %) models and Inception V2 (78 %), the fine-tuned version VGG-19 (91 %) exhibits the greatest increase in training and validation precision [33].
The GSA-DenseNet121-COVID-19 is a unique mixed CNN architecture that utilizes DenseNet121 and the optimization technique as gravitational search (GSA). The DenseNet121-COVID-19 could identify COVID-19 better than other DenseNet121, which could only diagnose 94 % of the test set. The suggested method was contrasted to an Inception-v3 CNN architecture and manual analysis when computing hyperparameter estimates. The GSA-DenseNet121-COVID-19 outclassed the comparison technique, which could only categorize 95 % of the test set samples [34].
EfficientNet-based pre-trained models were lowered using kernel principal component analysis. Then, multiple retrieved features were merged using a feature fusion technique. Finally, stacked ensemble meta-classifiers were used to classify the model into two stages.
Predictions were made in the first step using a support vector machine (SVM) and a random forest, which were then pooled and fed into the second stage. Next, a logistic regression classifier divides the X-ray and CT data into 2 classes (COVID, NON-COVID). The model’s performance was compared to other CNN-based pre-trained models. The new model outperforms previous approaches and may be used by clinicians for point-of-care diagnosis [35]. In a comparable work, researchers used ResNet32 and the deep transfer learning technique to categorize COVID-19-infected patients, and the results were published. Comparing the COVID-19 classifier to earlier supervised learning models, experimental data demonstrated that it delivered superior outcomes when compared to previous learning models [36].
A cutting-edge attention-based deep learning model with VGG-16 and a fine-tuned classification process was designed using a unique deep learning model that uses a convolution layer of the VGG-16 models for COVID-19. The experimental analysis shows steady and promising performance after comparing the suggested approach to the existing models [38]. The integrated stacking deep convolutional network using pre-trained models like ResNet101 and XceptionV3 was applied for InstaCovNet-19. The accuracy of 0.99 for 3 classes (Normal, Pneumonia, COVID-19) and 0.9953 for 2 classes (COVID, non-COVID) is achieved. In ternary classification, the suggested model obtained 98 % accuracy, whereas binary classification achieved 100 % precision and 98 % recall [38].
The CNN is used to implement binary and multiclass classification. The model was trained on 3877 CT and X-ray pictures, of which 1917 were of Covid-19 affected people. The binary classifier achieved a 99.64 % accuracy and exhibited a 99.58 % recall, a 99.56 % precision, a 99.59 % F1 score, and a 100 % ROC. The model was trained with 6077 photographs. A total of 1917 of which were of Covid-19 infected patients, 1960 healthy people, and 2200 pneumonia patients. The suggested technique obtained 98.28 % accuracy, 98.25 % recall (or sensitivity), 98.22 % precision, 98.23 % F1-score, and 99.87 % ROC for multiclass classification [40].
The early detection of lung cancer increases survival chances from 14 % to 49 %. Although CT approaches are found to deliver more accuracy than X-rays, a conclusive diagnosis that relies on many imaging modalities. An artificial DNN can spot lung cancer in CT images. Therefore, studies have proposed an adaptive boosting technique and a DenseNet to classify the lung image as normal or malignant. A total of 201 lung pictures have been included in the training dataset, with 85 percent of them being utilized for training and 15 percent being used for testing and classification. The proposed approach was shown to achieve a 90 % accuracy in testing [22]. The MLP classifier offered a higher accuracy of 88.55 % than the alternative classifiers, according to the outcome of the analysis of a study [21]. The CNN, DNN, and sparse auto-encoder deep neural networks were employed to identify lung cancer calcification. CT scans of benign and malignant lung nodules were classified using these networks. The Lung Image Database Consortium image collection (LIDC) database examined the networks where accuracy was 84.15 %, sensitivity 83.96 %, and a specificity 84.32 % [20]. The CNN was the most accurate of the three networks. Another work called applied Optimal Deep Neural Network (ODNN) and Linear Discriminate Analysis (LDA) to evaluate CT lung images that reduces the dimensionality of deep features. The ODNN is used with CT scans and optimized using the Gravitational Search Algorithm to classify lung cancer, thereby offering a 96.2 % as sensitivity, 94.2 % as specificity, and 94.56 % as accuracy [19].
Since medical specialists’ face challenges in distinguishing between Covid-19 and pneumonia, one study utilized an artificial neural network, ensemble classifier, SVM, and KNN for categorization. However, a RNN with a LSTM has been proposed as a deep learning architecture to identify lung conditions. Outcomes of the experiments demonstrated the resilience and effectiveness of the suggested model [18]. Another work uses ensemble of InceptionResNet_V2, ResNet50, and MobileNet_V2 for classifications. The outcomes revealed that the ResNet50, MobileNet_V2, and InceptionResNet_V2 models provide an F1 score of 94.84 %, which is higher than other models [17]. In addition, the CNN with pre-trained weights is utilized to categorize Covid-19, pneumonia, and healthy individuals using transfer learning techniques. Those who have active SARS-CoV-2 and pneumonia were accurately categorized in the dataset, which is one of the most important discoveries of that work [16]. Another study examined the potential of using machine learning to delineate and pinpoint pneumonia in CXR using RetinaNet and Mask R-CNN as an ensemble for the identification and localization of pneumonia, thereby achieving a recall of 0.793 for a large dataset [15]. For a variety of lung diseases, the transfer learning approach was used to capture images on CXR and CT. As COVID-19 resembles pneumonic viral lung illness, COVID-19 detection is challenging and relies on a thorough examination of a patient’s clinical pictures. The goal is attained using a novel architecture trained to identify virus-related pneumonia for COVID-19 detection. When compared to traditional categories, the findings of transfer learning are strikingly different [14].
One study develops the CNN model from scratch to extract characteristics from an image of pneumonia infected person’s chest X-ray and categorize it. This concept might alleviate some of the issues associated with dealing with medical images. It is difficult to obtain a significant number of pneumonia datasets for this classification assignment due to the limited availability of such data. The multiple data augmentation strategies were used to increase the training and validation classification accuracy of the proposed model. This has achieved a significant precision of 0.94814 the validation phase [13]. The transfer learning system automatically differentiates between 3883 CXR pictures classified as exhibiting pneumonia and 1349 that are designated normal. As an initialization, the suggested technique makes use of weights pre-trained on ImageNet using the Xception Network. When compared to current approaches, the model is competitive to obtain 0.84, 0.91, 0.99, 0.97 as precision, recall, F1, and ROC respectively [12]. In a separate study, researchers studied 180 X-ray images of persons who had been infected with COVID-19. The research attempted to employ the most successful systems, such as ResNet50V2 and Xception networks to detect the virus. Overall, the suggested model achieved a 91.4 % accuracy for all classes and a 99.50 % accuracy for instances of COVID-19 [11]. Using a CXR dataset from the National Library of Medicine Shenzhen No.3 Hospital, researchers developed a DCNN model to detect tuberculosis. This dataset was compared with a non-TB-specific chest X-ray dataset of a different population. The DCNN offered an AUC as 0.9845 and 0.8502. The AUC of the supervised DCNN model in the CXR dataset, on the other hand, was much lower, at 0.7054, than in the other datasets. A total of 36.51 % of aberrant radiographs in the CXR dataset associated to tuberculosis were predicted by the final DCNN model [5].
Another study combined ResNet and depth-ResNet in predicting severity scores and an analysis of TB’s likelihood. A depth-ResNet of 92.70 % and ResNet-50 of 67.15 % were produced for the TB detection. The study used the overall severity probability, different likelihoods for high severity (1 to 3 scores), and low severity (4 and 5 scores), where scores of 1 to 5 were converted into the probabilities of 0.9, 0.7, 0.50, 0.30, and 0.2. A 75.88 % and 85.29 %, respectively are the averaged accuracies for both approaches [6]. Other studies proposed three standard designs in the ensemble technique, namely AlexNet, GoogleNet, and ResNet. As a result, a new classifier for TB categorization has been developed from scratch. A combined dataset of publicly accessible standard datasets is used to train and test the suggested approach. Accuracy of 88 % and the AUC of 0.93 %, which is better than most existing approaches, are achieved [7].
The hierarchical feature extraction for abnormality detection method uses two levels of hierarchy to classify characteristics into healthy and unhealthy categories. Two levels of feature extraction are identified: level one is handmade geometrical feature extraction, and level two is typical statistical feature extraction and textural feature extraction from segmented lung fields. They were tested on 800 CXR images derived from two public datasets to verify their performance. AUC = 0.99 0.01 for Shenzhen and 0.95 0.06 for Montgomery, which illustrated that the two TB detection approaches offered a promising performance as compared to the existing techniques, as demonstrated by the obtained findings. Furthermore, Friedman's posthoc multiple comparison methods are demonstrated to statistically validate the suggested method [8]. In [43], [44] COVID-19 classification-based detection based on machine learning was introduced. The common reviewed studies about the chest diseases detection and classification are summarized in Table 1 .
Table 1.
Comparative analysis of existing work.
| Disease | Study | Method | Medical Image | Performance |
||
|---|---|---|---|---|---|---|
| Acc. | Prec. | Sens. | ||||
| COVID-19 | [24] | VGG-16, ResNet-50, InceptionV3 | CXR + CT | 93 | 91 | 90 |
| [25] | VGG-19 + ResNet-50 | CT | 94 | 95 | 90 | |
| [26] | DRE-Net | CT | 86 | 96 | 93 | |
| [27] | Modified ResNet | CXR + CT | 99.3 | 99.7 | 99.1 | |
| [28] | ResNet50 | CXR | 96.1 | 76.5 | 91.8 | |
| [29] | DenseNet121 | CXR + CT | 98 | 96 | 96 | |
| [30] | VGG-16 | CXR | 98.67 | 100 | 98 | |
| [31] | d-Resnet-10 network | CT | 81.4 | 79.8 | 87.5 | |
| [32] | VGG + CNN | CT | 96.2 | 97.3 | 94.5 | |
| [33] | VGG-16, InceptionV2, DT | CXR + CT | 91 | 94 | 97 | |
| [34] | GSA-DenseNet121 | CXR | 98.38 | 98.5 | 98.5 | |
| [35] | Deep learning Meta classifier | CXR + CT | 99 | 99 | 99 | |
| [36] | ResNet32 + DTL | CT | 93 | 95 | 91 | |
| [37] | VGG-16 | CXR | 79.58 | 92 | 95 | |
| [38] | InstaCovNet-19 | CXR | 99.08 | 99 | 99 | |
| [39] | CNN | CXR + CT | 98.28 | 98.22 | 98.25 | |
| Lung Cancer | [22] | FPSO-CNN | CT | 95.62 | 96.32 | 97.93 |
| [21] | Multi-layer Perceptron (MLP) | CT | 88.55 | 86.59 | 89.84 | |
| [20] | CNN | CT | 84.15 | 84.32 | 83.96 | |
| [19] | MGSA | CT | 94.56 | 94.2 | 96.2 | |
| Pneumonia | [18] | RNN-LSTM | CXR | 95.04 | 88.89 | 95.41 |
| [17] | ResNet50 + MobileNetV2 + InceptionResNetV2 | CXR | 95.09 | 95.53 | 94.43 | |
| [16] | CNN with pre-trained weights on ImageNet | CXR | 91 | 92 | 87 | |
| [15] | RetinaNet and Mask R-CNN | CXR | 83.80 | 75.8 | 79.3 | |
| [14] | Transfer learning | CXR + CT | 94.9 | 93 | 93 | |
| [13] | CNN | CXR | 93.73 | – | – | |
| [12] | Xception Network pre-trained weights on ImageNet | CXR | 97.3 | 84.3 | 99 | |
| [11] | Xception + ResNet50V2 | CXR | 99.50 | 92.69 | 80.53 | |
| Tuberculosis | [5] | DCNN | CXR | 98.45 | 82 | 72 |
| [6] | Depth-ResNet | CT | 85.29 | – | 84.16 | |
| [7] | Ensemble (AlexNet, GoogleNet and ResNet) | CXR | 88.24 | 88.0 | 88.42 | |
| [8] | (SVM + FOSF + GLCM) | CXR | 99.40 | 99.42 | 99.40 | |
| Lung Opacity | [41] | Xception | CXR | 95.71 | – | 97.19 |
| [42] | ResNet2 | CXR | 88.68 | – | – | |
3. Proposed methdology
The human respiratory system is attacked by a variety of lung illnesses. These diseases include pneumonia, tuberculosis, lung cancer, and lung opacity, among others. These diseases can cause similar effects on human lungs therefore X-ray images are commonly employed for diagnosing these diseases. AI in the form of deep learning algorithms has increasingly played a key role in diseases identification and classification. Deep learning facilitates the diagnosis process and saves time for healthcare providers.
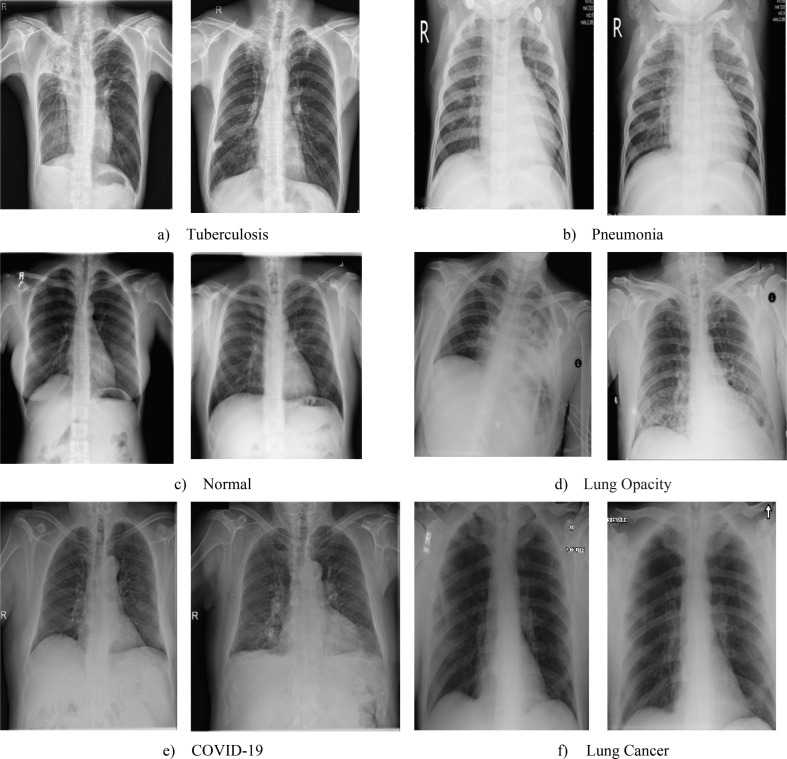
The study presents a multiclass deep learning classification model to identify the most common chest diseases. The aim of the research work is to design a deep learning framework and classify multi-class of Pneumonia, Lung Cancer, TB, Lung Opacity, and most recently COVID-19. A thorough search of the literature shows that our study is the first attempt to use the single deep learning framework, incorporating and classifying all these six classes at a time. Fig. 2 represented the proposed framework in a block diagram. The framework is divided into three phases as pre-processing, feature extraction, and classification. As observed in Fig. 3 , X-ray pictures were used as inputs, and the categorization of the input X-ray image on a disease level was the final output of the model.
Fig. 2.
Diagram of the proposed framework.
Fig. 3.
Chest X-ray images: (a) Tuberculosis images, (b) Pneumonia images, (c) Normal images, (d) Lung Opacity images, (e) COVID-19 images, (f) Lung cancer images.
During the first phase, the input images undergo pre-processing function such as normalization, resizing, and data image splitting into 80 % training and 20 % validation at random. Then, deep learning algorithms are used during the second and third stages. The second phase involves feature extraction, which is performed using VGG19 and CNN techniques. The fully connected network technique is employed during the image categorization step.
3.1. Dataset
For the experimental purpose, in addition to healthy cases, tremendous X-ray images of pneumonia, TB, lung cancer, lung opacity, and most recently Covid-19 were accessed and collected from reliable sources. To begin with, for COVID-19, 3615 CXR images from various sources of public datasets and published studies [45], [46], [47], [48], [49], [50], [51] were included in this study.
Secondly, 5856 CXR images of pneumonia were extracted from the Radiological Society of North America (RSNA), the Italian Society of Medical and Interventional Radiology (SIRM), and Radiopaedia which are publicly available for research purpose [52], [53]. To distinguish COVID-19 from pneumonia as a part of our experiments, these datasets have been utilized for training our proposed deep model. Furthermore, the Radiological Society of North America [52] represents 6012 CXR images of Lung opacity whilst [54], [55] indicate the datasets resource for a total of 20,000 X-ray images of lung cancer. The fourth dataset describes 5870 CXR images of pneumonia cases obtained from a variety of research articles [53], [54]. Ultimately, the total of 1400 X-ray images for tuberculosis [56], [57] were collected and employed in our research.
80,000 specifies the total number of CXR images used in our experiments. Samples of chest X-ray images for COVID-19, normal, pneumonia, TB, lung opacity and lung cancer are shown in Fig. 3. The number of patients for each disease dataset with respect to ages: ages were frequently between 38 and 65 for the COVID-19 dataset, 26 and 62 for the pneumonia dataset, 28 and 58 for the lung cancer dataset, and for normal patients the ages were between 33 and 58 years.
3.2. Dataset Pre-processing
Some pre-processing processes were employed to adjust the input data to meet the requirements of the deep learning model: 1) The images were resized; 2) the images were normalized; 3) the images were converted to an array to be employed an input in the model’s next phase. To ensure that the variation of the images meets the requirements to train the proposed model, the data were randomly divided into validation and training subsets at 20 % and 80 %, respectively. To meet the criteria of the framework, all images were scaled to 224 * 224 * 3.
After normalizing each pixel in the image to the interval [0,1], all images were transformed to array data representation.
3.3. Proposed deep learning VGG19 + CNN model
Supervised deep learning for multiclass classification of the most common chest diseases is presented in this research. For classification, we used a pre-trained model, VGG19, and the CNN as a feature extraction model which has fully connected.
The use of X-ray pictures to identify specific forms of chest ailments is demonstrated using a VGG19 followed by a CNN model. The model is depicted in detail in Fig. 4 . X-ray chest pictures with a dimension of 224 * 224 * 3 were used as input data for our model. The VGG19 pre-trained model is followed by three CNN blocks during the feature extraction stage. VGG19 is designed to provide great accuracy for large-scale image applications. [58] feature architecture was used that comprised 19 CNN with 3 convolution filters and 1 stride. Multiple deep learning models were merged with the VGG19 to improve picture categorization accuracy. A convolution layer with a ReLU as an activation function is included in each CNN block. Following these three CNN blocks, a batch normalization and a max-pooling layer were applied, which were then followed by a dropout layer, as indicated in Fig. 4.
Fig. 4.
The model architecture.
In the feature extraction step, the output was turned into a one-dimensional data vector, which was then used as an input in the classification stage after being modified through the flattening layer. The remaining components of the categorization step are comprised of three thick layers, each having 512, 256, and 128 neurons. It is a thick layer with six neurons and the SoftMax activation function that generates the final classification output. This layer is responsible for classifying the output image into one of the six chest diseases classes: pneumonia, tuberculosis, lung cancer, and lung opacity. A total of 24,622,470 model parameters are span into two categories. First were the trainable parameters (24,622,342), which were revised throughout the training process. The best value for these parameters was required to ensure the training accuracy. The second category was the untrainable parameters (1 2 8), which were those that did not change at the time of training. Fig. 5 illustrate the pseudo-code for the proposed framework.
Fig. 5.
Pseudo-Code for the proposed framework.
4. Results
With the help of Python 3 and the Keras framework, we were able to create a classification model for chest disease. The model was simulated on a Google Colab Pro edition [59] that has storage of 2 TB, RAM 25 GB, and CPU-P100. The ImageDataGenerator class in Keras [60] was used during the pre-processing stage, which included picture scaling, normalization, and conversion to an array of data.
The suggested multi-chest illnesses classification deep learning model input was created using the outcome of the pre-processing step. An optimizer and appropriate fit algorithms were used with 5000 epochs to train and validate the model. Eight iterations and 32 batch sizes were employed in each epoch. With the greatest precision, the performance metrics formulae were entered into the validation data outputs. The Adam [58] optimizer was employed, with a learning rate of 0.000009 (LR). Our suggested deep learning model’s code was published on the GitHub website [61].
Precision, loss, F1-Score, accuracy, precision, the AUC, and recall were applied to measure the performance of the model. The accuracy was determined in terms of the number of instances with correct predictions out of the total number of instances. The positive predictive value represented the precision. The accuracy was determined in line with the number of samples that was positive versus those forecasted as positive. Th F1-Score was calculated based on the harmonic mean of precision. According to this, the percentage of samples predicted to be positive from the total number of samples which considered to be the true positive rate. These matrices are defined as in equations (1), (2), (3), (4) [62]:
| (1) |
| (2) |
| (3) |
| (4) |
Where the actual positive and negative parameters are denoted by and respectively. The False positive and false negative values are denoted by and , respectively. In addition, a confusion matrix is computed for the proposed model.
Fig. 6 shows how the performance of the varying epochs during the training and validation phases.
Fig. 6.
Performance metrics changes with epochs in training and validation phases.
4.1. Experimental results
The suggested VGG19 + CNN model validation metric values are shown in Table 2 . These values represent those acquired during the highest accuracy validation iteration. The accuracy , precision (), F1-score (), and recall (Sensitivity) () were calculated using equations 1–4.
Table 2.
The performance validation of the VGG19 + CNN model.
| Methods | Loss | Acc | Pre | AUC | F1 | Recall |
|---|---|---|---|---|---|---|
| VGG19 + CNN model | 0.1792 | 96.48 | 97.56 | 99.82 | 95.62 | 93.75 |
4.2. Comparative anaysis
To the best of our knowledge, there is no recent existing research has employed a single deep learning model for evaluating and classifying the following chest diseases together: Tuberculosis, Pneumonia, Lung Opacity, Lung cancer, and COVID-19 images. To show the effectiveness of the proposed model, Table 3 introduces a comparative analysis of fifteen existing works. The comparison process was evaluated based on the architecture performance, the number of classes, and their deep learning approaches.
Table 3.
The performance comparison between the proposed VGG19 + CNN and existing related work.
| Ref | Number of Classes | Method | Medical Image |
Performance |
||
|---|---|---|---|---|---|---|
| Acc. | Prec. | Sens. | ||||
| [24] | 3 | VGG-16, ResNet-50, InceptionV3 | CXR + CT | 93 | 91 | 90 |
| [25] | 3 | VGG-19 + ResNet-50 | CT | 94 | 95 | 90 |
| [26] | 3 | DRE-Net | CT | 86 | 96 | 93 |
| [28] | 2 | ResNet50 | CXR | 96.1 | 76.5 | 91.8 |
| [36] | 2 | ResNet32 + DTL | CT | 93 | 95 | 91 |
| [31] | 2 | d-Resnet-10 network | CT | 81.4 | 79.8 | 87.5 |
| [21] | 2 | Multi-layer Perceptron (MLP) | CT | 88.55 | 86.59 | 89.84 |
| [20] | 2 | CNN | CT | 84.15 | 84.32 | 83.96 |
| [16] | 3 | CNN with pretrained weights on ImageNet | CXR | 91 | 92 | 87 |
| [15] | 2 | RetinaNet and Mask R-CNN | CXR | 83.80 | 75.8 | 79.3 |
| [14] | 3 | Transfer learning | CXR + CT | 94.9 | 93 | 93 |
| [13] | 2 | CNN | CXR | 93.73 | – | – |
| [6] | 1 Class with 5 Levels of Severity |
Depth-ResNet | CT | 85.29 | – | 84.16 |
| [7] | 2 | Ensemble (AlexNet, GoogleNet and ResNet) | CXR | 88.24 | 88.0 | 88.42 |
| [42] | 3 | ResNet2 | CXR | 88.68 | – | – |
| Proposed | 6 | VGG19 + CNN | CXR | 96.48 | 97.56 | 93.75 |
4.3. Architecture performance
Accuracy, precision, and recall (sensitivity) are the major parameters used to measure the performance of the model. The accuracy of the proposed framework produced the highest results with 96.48 overcoming the rest models in Fig. 7 .
Fig. 7.
Competitive analysis based on Accuracy.
As revealed in Fig. 8 , the best precision value was 97.56 with our proposed model. However, Fig. 9 confirms that our proposed model achieves the highest sensitivity of 93.75 compared to others.
Fig. 8.
Competitive analysis based on Precision.
Fig. 9.
Competitive analysis based on Sensitivity.
As presented in Fig. 10 , various architectures of individual pre-trained models, transfer learning, and ensemble techniques based on deep learning have been investigated and compared with our multi-class proposed framework. The results show that the proposed VGG19-CNN achieved the best performance. ResNet50 [28] was better than Transfer Learning [14]. However, the Ensemble model [7] records the lowest.
Fig. 10.
Competitive analysis based on a variety of utilized deep learning approaches.
Moreover, Table 3 illustrates the proposed multi-class framework used to classify-six classes of the most popular chest diseases: tuberculosis, pneumonia, lung opacity, lung cancer, and COVID-19, in addition to normal cases. The model significantly outperformed binary classes presented by [28], [21], [15], and [7]. Likewise, the model got over multi-class as observed by [25], [16], [14], and [42].
The confusion matrix for the VGG19 + CNN proposed model is shown in Fig. 11 , revealing that the VGG19 + CNN model can successfully classify the six chest diseases with the highest ratio to the COVID-19, starting from lung opacity, normal chest, lung cancer, pneumonia, and lastly the tuberculosis disease.
Fig. 11.
The confusion matrix.
A multiclass deep learning classification model has been used in this work to incorporate and classify-six classes of COVID-19, lung opacity, TB, lung cancer, and pneumonia using the VGG19 + CNN approach. The architecture of the model was based on VGG19 + CNN for feature extraction and a fully linked network for classification.
The recall, accuracy, AUC, F1 score, and precision of the suggested model were all tested. The findings showed that the VGG19 + CNN provided satisfactory classification performance with 96.48 % accuracy, as shown in Table 3.
Based on X-ray images, the VGG19 + CNN can identify various chest disorders with 96.48 % accuracy, 93.75 % recall, 97.56 % precision, 95.62 % F1 score, and 99.82 % AUC. We expect that the deep learning model will contribute to the development of a model for diagnosing chest disorders from CXR chest pictures, improving patient outcomes, and saving lives.
5. Conclusion and future work
In this study, a multi class chest diseases classification based a deep learning architecture was developed and evaluated for classifying TB, lung opacity, lung cancer, pneumonia, normal, and COVID-19 using CXR images. In terms of classification, a pre-trained model, VGG19 followed by three blocks of convolutional neural network (CNN) as a feature extraction and fully connected network at the classification stage were introduced. The experimental results revealed that our proposed VGG19 + CNN outperformed other existing work with 96.48 % accuracy, 93.75 % recall, 97.56 % precision, 95.62 % F1 score, and 99.82 % area under the curve (AUC).
The CT scan can accurately detect the aberrant pattern even before symptoms appear. Therefore, employing a combination of CXR and CT images is a potential enhancement parameter for the future work. Moreover, the identification of the region of interest (ROI) in conjunction with severity levels classification based on a powerful segmentation model is another direction for the exploration of the future work.
Funding statement
This research was funded by the Lancaster University, U.K.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported in part by The Engineering and Physical Sciences Research Council (EPSRC) under Grant EP/P009727/1, and in part by the Leverhulme Trust under Grant RF-2019-492.
Footnotes
Peer review under responsibility of Faculty of Engineering, Alexandria University.
References
- 1.Yanase J., Triantaphyllou E. A systematic survey of computer-aided diagnosis in medicine: Past and present developments. Expert Syst. Appl. 2019;38:112821. doi: 10.1016/j.eswa.2019.112821. [DOI] [Google Scholar]
- 2.Khan M.A. An IoT Framework for Heart Disease Prediction Based on MDCNN Classifier. IEEE Access. 2020;8:34717–34727. doi: 10.1109/ACCESS.2020.2974687. [DOI] [Google Scholar]
- 3.Khan M.A., Quasim M.T., Alghamdi N.S., Khan M.Y. A Secure Framework for Authentication and Encryption Using Improved ECC for IoT-Based Medical Sensor Data. IEEE Access. 2020;8:52018–52027. doi: 10.1109/ACCESS.2020.2980739. [DOI] [Google Scholar]
- 4.Albahli S. Efficient gan-based chest radiographs (CXR) augmentation to diagnose coronavirus disease pneumonia. Int. J. Med. Sci. 2020;17(10):1439–1448. doi: 10.7150/IJMS.46684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sathitratanacheewin S., Sunanta P., Pongpirul K. Deep learning for automated classification of tuberculosis-related chest X-Ray: dataset distribution shift limits diagnostic performance generalizability. Heliyon. 2020;6(8):e04614. doi: 10.1016/j.heliyon.2020.e04614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao X.W., James-Reynolds C., Currie E. Analysis of tuberculosis severity levels from CT pulmonary images based on enhanced residual deep learning architecture. Neurocomputing. 2020;392:233–244. doi: 10.1016/j.neucom.2018.12.086. [DOI] [Google Scholar]
- 7.Hooda R., Mittal A., Sofat S. Automated TB classification using ensemble of deep architectures”. Multimed. Tools Appl. 2019;78(22):31515–31532. doi: 10.1007/s11042-019-07984-5. [DOI] [Google Scholar]
- 8.Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Automatic detection of tuberculosis related abnormalities in Chest X-ray images using hierarchical feature extraction scheme. Expert Syst. Appl. 2020;158 doi: 10.1016/j.eswa.2020.113514. [DOI] [Google Scholar]
- 9.Lopes U.K., Valiati J.F. Pre-trained convolutional neural networks as feature extractors for tuberculosis detection. Comput. Biol. Med. 2017;89:135–143. doi: 10.1016/j.compbiomed.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Kumar J.S., Alias Balamurugan S.A., Sasikala S. Analysis of Deep Learning Techniques for Tuberculosis Disease. SN Comput. Sci. 2021;2(4):1–10. doi: 10.1007/s42979-021-00680-y. [DOI] [Google Scholar]
- 11.Rahimzadeh M., Attar A. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of Xception and ResNet50V2. Inform. Med. Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luján-García J.E., Yáñez-Márquez C., Villuendas-Rey Y., Camacho-Nieto O. A transfer learning method for pneumonia classification and visualization. Appl. Sci. 2020;10(8) doi: 10.3390/APP10082908. [DOI] [Google Scholar]
- 13.Stephen O., Sain M., Maduh U.J., Jeong D.U. An Efficient Deep Learning Approach to Pneumonia Classification in Healthcare. J. Healthc. Eng. 2019 doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lascu M.R. Deep Learning in Classification of Covid-19 Coronavirus, Pneumonia and Healthy Lungs on CXR and CT Images. J. Med. Biol. Eng. 2021;41(4):514–522. doi: 10.1007/s40846-021-00630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirazitdinov I., Kholiavchenko M., Mustafaev T., Yixuan Y., Kuleev R., Ibragimov B. Deep neural network ensemble for pneumonia localization from a large-scale chest x-ray database. Comput. Electr. Eng. 2019;78:388–399. doi: 10.1016/j.compeleceng.2019.08.004. [DOI] [Google Scholar]
- 16.Luján-García J.E., Moreno-Ibarra M.A., Villuendas-Rey Y., Yáñez-Márquez C. Fast COVID-19 and pneumonia classification using chest X-ray images. Mathematics. 2020;8:9. doi: 10.3390/MATH8091423. [DOI] [Google Scholar]
- 17.El Asnaoui K. Design ensemble deep learning model for pneumonia disease classification. Int. J. Multimed. Inf. Retr. 2021;10(1):55–68. doi: 10.1007/s13735-021-00204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goyal S., Singh R. Detection and classification of lung diseases for pneumonia and Covid-19 using machine and deep learning techniques. J. Ambient Intell. Humaniz. Comput. 2021;(0123456789) doi: 10.1007/s12652-021-03464-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakshmanaprabu S.K., Mohanty S.N., Shankar K., Arunkumar N., Ramirez G. Optimal deep learning model for classification of lung cancer on CT images. Futur. Gener. Comput. Syst. 2019;v92:374–382. doi: 10.1016/j.future.2018.10.009. [DOI] [Google Scholar]
- 20.Song Q.Z., Zhao L., Luo X.K., Dou X.C. Using Deep Learning for Classification of Lung Nodules on Computed Tomography Images. J. Healthc. Eng. 2017 doi: 10.1155/2017/8314740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh G.A.P., Gupta P.K. Performance analysis of various machine learning-based approaches for detection and classification of lung cancer in humans. Neural Comput. Appl. 2019;31(10):6863–6877. doi: 10.1007/s00521-018-3518-x. [DOI] [Google Scholar]
- 22.Kalaivani N., Manimaran N., Sophia S., Devi D.D. Deep Learning Based Lung Cancer Detection and Classification. IOP Conf. Ser. Mater. Sci. Eng. 2020;994(1):7731–7776. doi: 10.1088/1757-899X/994/1/012026. [DOI] [Google Scholar]
- 23.ALzubi J.A., Bharathikannan B., Tanwar S., Manikandan R., Khanna A., Thaventhiran C. Boosted neural network ensemble classification for lung cancer disease diagnosis. Appl. Soft Comput. J. 2019;80:579–591. doi: 10.1016/j.asoc.2019.04.031. [DOI] [Google Scholar]
- 24.Perumal V., Narayanan V., Rajasekar S.J.S. Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl. Intell. 2021;51(1):341–358. doi: 10.1007/S10489-020-01831-Z/FIGURES/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li L., Xu Z., et al. Artificial Intelligence Distinguishes COVID-19 from Community Acquired Pneumonia on Chest CT. Radiology. 2020;296(2):E65–E71. doi: 10.1148/RADIOL.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song Y., Zheng S., Li L., Zhang X., Zhang X., Huang Z., Chen J., Wang R., Zhao H., Chong Y., Shen J., Zha Y., Yang T. Deep learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT images. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2021 doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia G., Lam H.K., Xu Y. Classification of COVID-19 chest X-Ray and CT images using a type of dynamic CNN modification method. Comput. Biol. Med. 2020;134 doi: 10.1016/j.compbiomed.2021.104425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narin A., Kaya C., Pamuk Z. Automatic detection of coronavirus disease (COVID-19) using X-ray images and deep convolutional neural networks. Pattern Anal. Appl. 2021;24(3):1207–1220. doi: 10.1007/s10044-021-00984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassania S.H., Kassanib P.H., Wesolowskic M.J., Schneidera K.A., Detersa R. Automatic Detection of Coronavirus Disease (COVID-19) in X-ray and CT Images: A Machine Learning Based Approach. Biocybern. Biomed. Eng. 2021;41(3):867–879. doi: 10.1016/j.bbe.2021.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singh R.K., Pandey R., Babu R.N. COVIDScreen: explainable deep learning framework for differential diagnosis of COVID-19 using chest X-rays. Neural Comput. Appl. 2021;33(14):8871–8892. doi: 10.1007/s00521-020-05636-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C., Dong D., Li L., Gong W., Li X., Bai Y., Wang M., Hu Z., Zha T., Tian J. Classification of Severe and Critical Covid-19 Using Deep Learning and Radiomics. IEEE J. Biomed. Heal. Informatics. 2020;24(12):3585–3594. doi: 10.1109/JBHI.2020.3036722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.J. Shi, X. Yuan, M. Elhoseny, X. Yuan, Weakly Supervised Deep Learning for Objects Detection from Images 8 (2020) 231–242, doi: 10.1007/978-3-030-45099-1_18.
- 33.Dansanan D., Kumar R., Bhattacharjee A., Hemanth D.J., Gupta D., Khanna A., Castillo O. Early diagnosis of COVID-19-affected patients based on X-ray and computed tomography images using deep learning algorithm. Soft Comput. 2020;(0123456789) doi: 10.1007/s00500-020-05275-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Ezzat D., Hassanien A.E., Ella H.A. An optimized deep learning architecture for the diagnosis of COVID-19 disease based on gravitational search optimization. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravi V., Narasimhan H., Chakraborty C., Pham T.D. Deep learning-based meta-classifier approach for COVID-19 classification using CT scan and chest X-ray images. Multimed. Syst. 2021;(0123456789) doi: 10.1007/s00530-021-00826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathak Y., Shukla P.K., Tiwari A., Stalin S., Singh S. Deep Transfer Learning Based Classification Model for COVID-19 Disease. Irbm. 2020;1:1–6. doi: 10.1016/j.irbm.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sitaula C., Hossain M.B. Attention-based VGG-16 model for COVID-19 chest X-ray image classification. Appl. Intell. 2021;51(5):2850–2863. doi: 10.1007/s10489-020-02055-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gupta A., Anjum, Gupta S., Katarya R. InstaCovNet-19: A deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021;99:106859. doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thakur S., Kumar A. X-ray and CT-scan-based automated detection and classification of covid-19 using convolutional neural networks (CNN) Biomed. Signal Process. Control. 2021;69 doi: 10.1016/j.bspc.2021.102920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.K.F. Monowar, M.A.M. Hasan, J. Shin, Lung Opacity Classification With Convolutional Neural Networks Using Chest X-rays, 2020 11th International Conference on Electrical and Computer Engineering (ICECE), (2020), 169-172, doi: 10.1109/ICECE51571.2020.9393135.
- 41.Latif G. FTA. Lung Opacity Pneumonia Detection with Improved Residual Networks. J. Med. Biol. Eng. 2021;41(5):581–591. [Google Scholar]
- 42.BIMCV-COVID19, 2022, https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590858128006-9e640421-6711 (accessed 5 February 2022).
- 43.Shahin, Osama R., Hamoud H. Alshammari, Ahmed I. Taloba, Rasha M. Abd El-Aziz, Machine Learning Approach for Autonomous Detection and Classification of COVID-19 Virus, Comput. Electr. Eng. 101 (2022) 108055. [DOI] [PMC free article] [PubMed]
- 44.Shahin, Osama R., Rasha M. Abd El-Aziz, Ahmed I. Taloba, Detection and classification of Covid-19 in CT-lungs screening using machine learning techniques, J. Interdisciplinary Math. 25(3) (2022) 791-813.
- 45.ml-workgroup covid-19-image-repository, 2022, Public https://github.com/ml-workgroup/covid-19-image-repository/tree/master/png, (accessed 5 February 2022).
- 46.COVID-19 DATABASE, 2022, https://sirm.org/category/senza-categoria/covid-19/, (accessed 5 February 2022).
- 47.https://eurorad.org, (accessed 5 February 2022).
- 48.ieee8023 covid-chestxray-dataset, 2002 https://github.com/ieee8023/covid-chestxray-dataset, (accessed 5 February 2022).
- 49.COVID-19 Chest X-Ray Image Repository, 2002, https://figshare.com/articles/COVID-19_Chest_X-Ray_Image_Repository/12580328, (accessed 5 February 2022).
- 50.Armiro COVID-CXNet, 2022, https://github.com/armiro/COVID-CXNet, (accessed 5 February 2022).
- 51.RSNA Pneumonia Detection Challenge, Can you build an algorithm that automatically detects potential pneumonia cases?, 2022, https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data, (accessed 5 February 2022).
- 52.Chest X-Ray Images (Pneumonia) 5,863 images, 2 categories, 2022, https://www.kaggle.comC/paultimothymooney/chest-xray-pneumonia, (accessed 5 February 2022).
- 53.Elshennawy N.M., Ibrahim D.M. Deep-Pneumonia Framework Using Deep Learning Models Based on Chest X-Ray Images. Diagnostics. 2020;10:649. doi: 10.3390/diagnostics10090649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.J. Shiraishi, S. Ka tsuragawa, J. Ikezoe, T. Ma tsumoto, T. Kobayashi, K.-I. Komatsu, M. Ma tsui, H. Fujita, Y. Kodera, K. Doi, Development of a digital image database for chest ra diographs with and without a lung nodule: receiver operating char acteristic analysis of radiologists’ detection of pulmonar y nodules, Am. J. Roentgenol. 174 (2000) 71–74. [DOI] [PubMed]
- 55.Jaeger S., Karargyris A., Candemir S., Folio L., Siegelman J., Callaghan F., Xue Z., Palaniappan K., Singh R.K., Antani S., Thoma G., Wang Y.X., Lu P.X., McDonald C.J. Automatic tuberculosis screening using chest radiographs. IEEE Trans. Med. Imaging. 2014;33(2):233–245. doi: 10.1109/TMI.2013.2284099. PMID: 24108713. [DOI] [PubMed] [Google Scholar]
- 56.Candemir S., Jaeger S., Palaniappan K., Musco J.P., Singh R.K., Xue Z., Karargyris A., Antani S., Thoma G., McDonald C.J. Lung segmentation in chest radiographs using anatomical atlases with nonrigid registration. IEEE Trans. Med. Imaging. 2014 Feb;33(2):577–590. doi: 10.1109/TMI.2013.2290491. PMID: 24239990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.K. Simonyan, A. Zisserman, Very deep convolutional networks for large-scale image recognition, arXiv (2014), arXiv:1409.1556.
- 58.Bisong E. Springer; 2019. Building Ma chine Learning and Deep Learning Models on GoogleCloud Platform. [Google Scholar]
- 59.Gulli A., Pal S. Packt Publishing Ltd; 2017. Deep Lear ning with Kera s. [Google Scholar]
- 60.D.P. Kingma, J. Ba, Adam: A Method for Stochastic Optimization, 2014arXiv preprint ar Xiv:1412.6980.
- 61.GitHub: A Deep Learning Architecture for Multi-Class Lung Diseases Classification using Chest X-ray Images: https://github.com/abunaif544/vgg.git, (accessed August 20, 2022).
- 62.Khan M.A., Algarni F. A Healthcare Monitoring System for the Diagnosis of Heart Disease in the IoMT Cloud Environment Using MSSO-ANFIS. IEEE Access. 2020;8:122259–122269. doi: 10.1109/ACCESS.2020.3006424. [DOI] [Google Scholar]