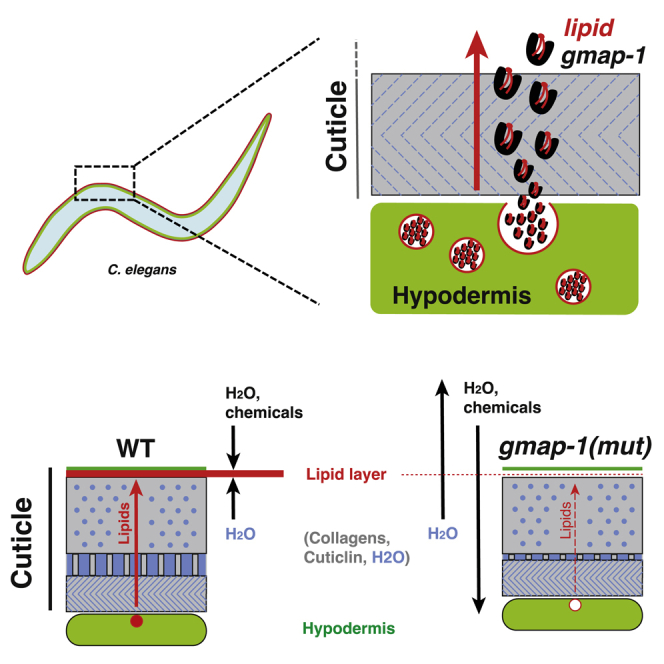
Summary
The cuticle of C. elegans is impermeable to chemicals, toxins, and pathogens. However, increased permeability is a desirable phenotype because it facilitates chemical uptake. Surface lipids contribute to the permeability barrier. Here, we identify the lipid transfer protein GMAP-1 as a critical element setting the permeability of the C. elegans cuticle. A gmap-1 deletion mutant increases cuticular permeability to sodium azide, levamisole, Hoechst, and DiI. Expressing GMAP-1 in the hypodermis or transiently in the adults is sufficient to rescue this gmap-1 permeability phenotype. GMAP-1 protein is secreted from the hypodermis to the aqueous fluid filling the space between collagen fibers of the cuticle. In vitro, GMAP-1 protein binds phosphatidylserine and phosphatidylcholine while in vivo, GMAP-1 sets the surface lipid composition and organization. Altogether, our results suggest GMAP-1 secreted by hypodermis shuttles lipids to the surface to form the permeability barrier of C. elegans.
Subject areas: Biological sciences, molecular biology, physiology
Graphical abstract
Highlights
-
•
GMAP-1 is secreted by the hypodermis toward the cuticle of Caenorhabditis elegans
-
•
GMAP-1 binds and shuttle phosphoglycerides
-
•
GMAP-1 sets the lipid composition of the cuticle
-
•
While healthy, gmap-1 mutant displays high cuticular permeability
Biological sciences, molecular biology, physiology
Introduction
The nematode cuticle is a thick and resistant hydroskeleton that acts as a barrier controlling exchanges between the external environment and the internal medium (Chisholm and Hsiao, 2012; Chisholm and Xu, 2012; Page and Johnstone, 2007). Its composition, organization, and properties are tailored to the lifestyle of each species or developmental stage. The cuticle of C. elegans - a free-living nematode - is impermeable to chemicals, toxins, and pathogens. However, increased permeability is a desirable phenotype because it facilitates chemical uptake, leading to increased sensitivity to agents and reduced risks of false negatives during small molecule screening (Burns et al., 2010). Permeability defects can arise from deficiencies in a subset of collagens (Sandhu et al., 2021), surface coat glycosylation (Partridge et al., 2008), or lipid biogenesis (Bada Juarez et al., 2019; Kage-Nakadai et al., 2010; Loer et al., 2015). These genes involved in permeability defects are putative targets to fight pathogenic nematodes.
In C. elegans, the cuticle is secreted by hypodermis cells and fully replaced at each moult of its 4 larval stages. In addition, cuticular components are continuously produced and exported during intermoult and during adulthood (Chisholm and Xu, 2012; Page and Johnstone, 2007). Although the cuticle varies across stages, its layering from top to bottom always includes the outermost hydrophilic surface coat which is covered by glycoproteins; the epicuticle that contains lipids; and the cuticle proper which is made of several layers predominantly made of collagen (Blaxter, 1993; Cox et al., 1981b; Johnstone, 2000; Peixoto and De Souza, 1995). The exact location of the permeability barrier is yet to be defined but is expected to include the lipids present in the epicuticle (Bada Juarez et al., 2019; Kage-Nakadai et al., 2010; Loer et al., 2015).
Which lipids constitute the barrier, and how these lipids are secreted by the hypodermis and shuttled to the epicuticle is unknown. Here, we identify the lipid transfer protein GMAP-1 as a critical element setting the permeability of C. elegans cuticle by its effect on the lipid composition and the lipid organization of the epicuticle.
Results
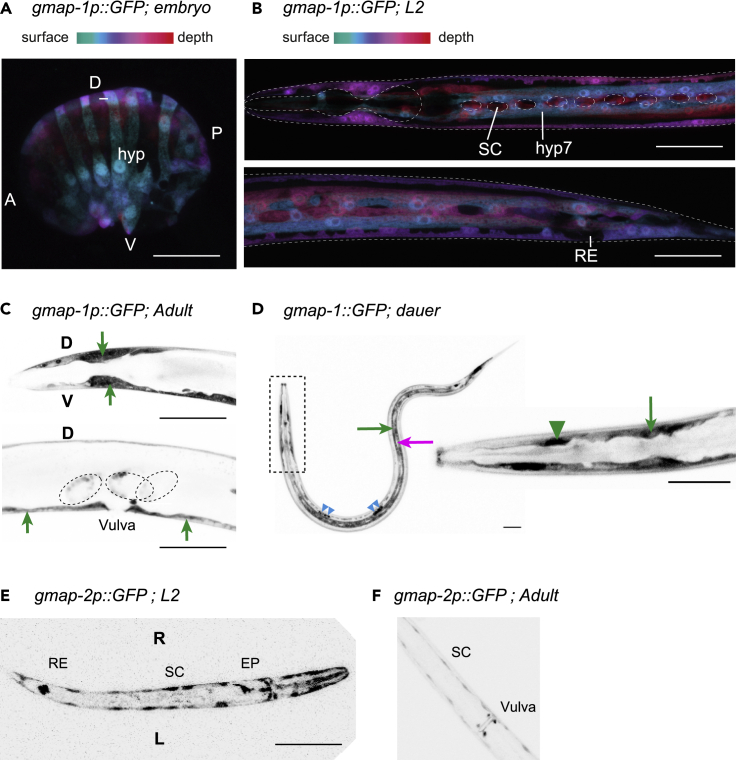
The C. elegans cuticle is fully replaced at each moult of its 4 larval stages. Numerous genes show oscillating expression during C. elegans moulting cycle (Turek and Bringmann, 2014). Among those genes, 3 proteins loosely related to human GM2 Activator Protein (GM2AP) are induced during moult, when the new cuticle is synthesized: C34 × 107.4/gmap-1, C05 × 107.1/gmap-2 and C05 × 107.2/gmap-3 (Figure S1). Human GM2AP is a lysosomal lipid transfer protein that extracts GM2 ganglioside from membranes and presents it to b-hexosaminidase A for degradation (Sandhoff and Harzer, 2013). According to time-resolved transcriptome and single-cell transcriptomes (Boeck et al., 2016; Cao et al., 2017; Packer et al., 2019), C34 × 107.4/gmap-1 is expressed in embryonic hypodermal cells during ventral enclosure, when hypodermal sheet spreads to wrap around the embryo. In larvae and adults, it is expressed in hypodermis cells including hyp2 and hyp7 cells; in sheath glia including ILsh, AMsh, and PHsh cells; and in Socket cells. To characterize gmap-1 expression pattern, we generated a transcriptional reporter strain, where 3772 bp of gmap-1 promoter drives GFP expression. In this strain, GFP was expressed in the hypodermal syncytium (hyp7) and other hypodermal cells of the head and tail. We also observed GFP expression in hypodermis precursors at all developmental stages from late embryo to adulthood, including at the dauer stage, an alternative 3rd larval stage of C. elegans promoted by harsh conditions. However, GFP was never observed in specialized epithelial cells such as the seam cells, vulval epithelium, excretory pore, excretory duct, and rectal epithelium (Figures 1A–1D). On a separate locus, an operon co-transcribes C05 × 107.1/gmap-2 and C05 × 107.2/gmap-3 from the gmap-2p promoter. A 2052 bp fragment of this gmap-2 promoter drives GFP expression only in specialized epithelial cells including seam cells, rectal epithelium, vulval epithelium, and excretory pore (Figures 1E and 1F). In conclusion, gmap1 and gmap-2/3 genes are both expressed in epithelial cells although with non-overlapping expression patterns.
Figure 1.
Expression profile of gmap-1, gmap-2 and gmap-3 in C. elegans
The expression pattern of gmap-1 was determined using a GFP transcriptional reporter strain bearing a [gmap-1p::SL2 GFP] transgene.
(A) In comma stage embryo, GFP expression was observed in the hypodermal precursor cells (hyp) before ventral closure. GFP fluorescence is reported in 3D using depth color-coded projection. Scale bar = 50 μm. A: anterior, P: posterior, V: ventral, D: dorsal.
(B) During the second larval stage, GFP expression was observed in the hypodermis syncytium (hyp7), in hypodermal cells of the head and tail, but not in the rectal epithelium (RE) or seam cells (SC). GFP fluorescence is reported in 3D using depth color-coded projection. Scale bar = 100 μm.
(C) In adults, GFP expression was observed in the hypodermis of the head (green arrows) and in the hyp7 hypodermal syncytium surrounding the vulva (green arrow) but not in the vulval epithelium itself. It was also observed in embryos, prior to egg laying (scattered circle). Scale bar = 100 μm.
(D) We observed GFP was strongly expressed in dauer hypodermis (green arrows) but not in seam cells (magenta arrow). It was observed in unidentified head’s glia (green arrowhead), as well as in coelomocytes (blue arrowheads). Scale bar = 100 μm.
(E and F) The expression pattern of the gmap-2, gmap-3 operon was determined using a transcriptional reporter strain bearing a [gmap-2p::GFP] transgene: (E) GFP expression was observed in the seam cells (SC), the excretory pore (EP) and the rectal epithelium (RE) of the L1 stage (F) GFP expression was observed in the seam cells and vulval cells of adults. Scale bar = 100 μm. Images are representative of over 30 animals observed over 2 or 3 independent sampling.
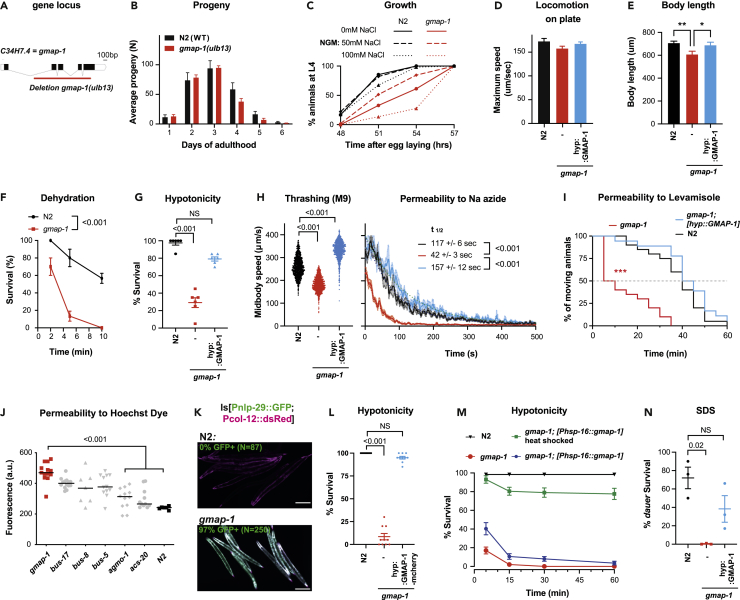
To determine the function of these lipid transfer proteins in the hypodermis of nematodes, we generated a 1515 bp deletion allele of gmap-1 using CRISPR-Cas9 (Figure 2A). The number of eggs produced by gmap-1(ulb13), and their hatching was similar to N2 (the C. elegans control strain) (Figure 2B). Postembryonic development from egg laying to L4 larval stage took ∼54 h for N2 at 20°C on regular cultivation plates in presence of Escherichia coli. This growth was delayed by 3hrs in gmap-1 (Figure 2C). The maximum locomotion speed of gmap-1 on agar plate was similar to N2 (Figure 2D). Because gmap-1 is mostly expressed in the hypodermis, and as the cuticle is produced by the hypodermis, we reasoned gmap-1 might contribute to cuticle formation. Several mutants of the cuticular collagen genes affect the morphology of C. elegans (Page and Johnstone, 2007). Grown in regular conditions, gmap-1 mutant animals showed a slightly shorter body length than N2 (Figure 2E) but no other morphological phenotype. Short body length can result from defects in collagen production and release by the hypodermis, defects in TGFβ signaling in hypodermis that regulate collagen production, impaired feeding, or impaired neuromuscular function (Johnstone, 2000; Nystrom et al., 2002; Page and Johnstone, 2007). To ascertain that the short body length phenotype resulted from the lack of GMAP-1 function in the hypodermis, we rescued gmap-1 specifically in the hypodermis using the agmo-1 promoter, previously shown to be specifically expressed in hypodermal cells (Loer et al., 2015). Correct expression of the gmap-1 cDNA transgene was verified using a bicistronic expression cassette where cytoplasmic EGFP is placed in an operon with gmap-1. Expression of GMAP-1 in the hypodermis was sufficient to rescue the short body length phenotype of gmap-1 (Figure 2E, blue).
Figure 2.
gmap-1 deletion increases cuticular permeability
(A) Schematic representation of the gmap-1 gene and the localization of the 1515 bp deleted by CRISPR in gmap-1(ulb-13).
(B) Analysis of the progeny produced by N2 and gmap-1 over the first 6 days of adulthood.
(C) The percentage of individuals reaching L4 stage determined 48 h after an egg laying window for ∼70 N2 and gmap-1. The animals were grown at 20°C in presence of fresh bacteria either on regular NGM plates (50nM NaCl) or at indicated NaCl concentrations.
(D) The maximum forward speed of N2 and gmap-1 determined on NGM plates in presence of OP50 bacteria (N = 60 across 3 replicates; One-way ANOVA followed by Dunnet’s comparison).
(E) Body length measured in young adults 3 h post L4 stage for N2, gmap-1, and gmap-1(ulb13) carrying Ex[agmo-1p::gmap-1SL2GFP] (N = 60 across 3 replicates; One-way ANOVA followed by Dunnet’s comparison, ∗:p value <0.05; ∗∗:p value <0.01).
(F) Survival to dehydration measured for adult worms after 5 min of contact to dried 2% agar pad covered with mineral oil (N = 30 across 3 replicates; 2-way RM ANOVA with Geisser correction).
(G) Survival to hypotonicity measured for adult worms after 15 min exposure to deionized water (N = 200 across 10 replicates). Survival was measured for N2, gmap-1(ulb13) mutants, and gmap-1(ulb13) rescued in the hypodermis by the expression of Ex[agmo-1p::gmap-1SL2GFP]. One-way ANOVA followed by Kruskal-Wallis test and Dunn’ correction for multiple comparisons.
(H) Thrashing speed in M9 buffer was measured for N2, gmap-1(ulb13) mutants, and gmap-1(ulb13) carrying Ex[agmo-1p::gmap-1SL2GFP]. The induction of paralysis by 5 mM sodium azide was measured over 10 min. The half-life is calculated by one-phase decay non-linear fit. (N = ≥80 across 5-6 replicates per group; One-way ANOVA followed by Dunnet’s comparison).
(I) The percentage of individuals paralyzed in presence of 0.4 mM Levamisole was measured over 1 h for N2, gmap-1(ulb13) mutants, and gmap-1(ulb13) carrying Ex[agmo-1p::gmap-1SL2GFP]. (N = 80 across 4 replicates, Log rank p value compared to N2, ∗∗∗:p value <0.001).
(J) The median cuticular permeability to Hoechst 33342 was measured for 20 individuals of the indicated genotypes. Fluorescence was quantified in the head after 30 min exposure to Hoechst 33342. One-way ANOVA followed by Kruskal-Wallis test and Dunn’ correction for multiple comparisons.
(K) A transcriptional reporter strain carrying Is[nlp-29p::GFP; col-12p::DsRed] was crossed with gmap-1 mutants and N2 controls and grown on regular NGM plates + OP50 E. coli. Images are representative of the induction of nlp-29p::GFP, observed in 94.4% (N = 250) of gmap-1(ulb13) but in none of the N2 controls (N = 87). Scale bar = 200 μm.
(L) Hypodermal expression of GMAP-1-mcherry was assessed for its ability to rescue hypersensitivity to deionized water of gmap-1 mutants. Survival in deionized water was quantified for N2, gmap-1 and in a strain co-expressing cytoplasmic mEGFP and GMAP-1-mCherry under an hypodermis promoter [agmo-1p::eGFP; agmo-1p::gmap-1-mCherry]. One-way ANOVA followed by Kruskal-Wallis test and Dunn’ correction for multiple comparisons.
(M) Acute expression of GMAP-1mCherry in adults was assessed for its ability to rescue hypersensitivity to deionized water of gmap-1 mutants. Survival in deionized water was quantified for N2, gmap-1 and in a strain expressing GMAP-1-mCherry under a heat-shock promoter [phsp-16::gmap-1-mCherry] maintained at 15°C and exposed to 33°C for 5 min at late L4 stage. As negative control, the [phsp-16::gmap-1-mCherry] strain was maintained at 15°C, instead. One-way ANOVA followed by Kruskal-Wallis test and Dunn’ correction for multiple comparisons.
(N) Survival after 30 min of exposure to 1% SDS was measured for 60 dauers in 3 replicates. dauer survival was measured for N2, gmap-1(ulb13) mutants, and gmap-1(ulb13) carrying Ex[agmo-1p::gmap-1SL2GFP]. One-way ANOVA followed by Kruskal-Wallis test and Dunn’ correction for multiple comparisons. All data are represented as Mean +/− SEM.
Lipids located at the surface of the cuticle in a layer called the epicuticle play a major role in the permeability barrier formed by the cuticle (Blaxter, 1993; Riddle et al., 1997). As GM2AP are lipid transfer proteins, we evaluated whether gmap-1 invalidation could reduce the impermeability of its cuticle. The cuticle prevents the desiccation of the animal in dried environment. Placed on a dried agar pad covered with mineral oil, >80% of gmap-1 animals died of dehydration within 5 min, while ∼80% of N2 remained alive (Figure 2F). The cuticle prevents internal solutes to leak out from C. elegans. Around 75% of gmap-1 animals placed in deionized water died within 15 min, while N2 remained alive in deionized water for hours (Figure 2G). Specific expression of GMAP-1 in hypodermis was sufficient to rescue hypersensitivity of gmap-1 to deionized water (Figure 2G, blue). In line with a defect of gmap-1 to control its internal milieu, we observed that gmap-1 growth was slowed in hypotonic or hypertonic conditions compared to the optimal conditions (NGM in Figure 2C). We next tested the cuticular permeability of gmap-1 to 3 molecules of different molecular weights: Sodium Azide (NaN3; 65 g/mol), Levamisole (204.3 g/mol), and Hoechst 33258 (533.9 g/mol). Depletion of intracellular ATP by NaN3 -an oxidative phosphorylation uncoupler-leads to worm paralysis. Soaked in the M9 buffer, the thrashing speed of gmap-1 was below that of N2 (Figure 2H, left). In presence of 5 mM NaN3, this thrashing speed quickly decreased: paralysis happened faster in gmap-1 than in N2 or in gmap-1 rescued in the hypodermis (Figure 2H, right). The cholinergic agonist levamisole causes muscular hypercontraction and paralysis. Soaked in M9 buffer supplemented with 0.4 mM levamisole, 50% of gmap-1 individuals were paralyzed within 5 min while it took 40 min for N2 or gmap-1 rescued in the hypodermis (Figure 2I). Finally, the cuticle normally prevents the cell-permeable nucleic acid-staining dye Hoechst 33258 to stain the nuclei of hypodermis cells located below the cuticle. When stained with 1 μg/mL Hoechst 33258 for 15 min, we observed increased cuticular permeability to Hoechst 33258 in gmap-1 mutants (Figure 2J). Other mutants with cuticular permeability defects were previously reported (Choi et al., 2016; Kage-Nakadai et al., 2010; Sandhu et al., 2021; Xiong et al., 2017). The Hoechst dye intake in gmap-1 was equal to bus mutants but higher than acs-20 and agmo-1 (Figure 2J).
In response to cuticle injury, osmotic shock or pathogenic bacteria, expression of the antimicrobial peptide NLP-29 is induced in the hypodermis. In worms grown on a non-pathogenic strain of E. coli (OP50), the transcriptional reporter strain IG274 carrying a nlp-29 promoter::GFP transgene does not express GFP (Pujol et al., 2008). On OP50, we observed a significant induction of Pnlp-29::GFP in gmap-1 mutants: GFP expression was detected in 94.4% (N = 250) of gmap-1 mutants as opposed to none of the controls worms (N = 87) (Figure 2K). This increased Pnlp-29::GFP expression was observed at all postembryonic development stages of gmap-1 mutants, suggesting that gmap-1 cuticular barrier is compromised from the first larval stage to adulthood. To examine if gmap-1 is required developmentally, we expressed it acutely using a heat-shock-inducible promoter. First, to visualize GMAP-1 expression, we fused it C-terminally to mCherry. This GMAP-1-mCherry fusion protein remained functional: its expression in the hypodermis under the agmo-1 promoter rescued the sensitivity of gmap-1 mutant to hypotonic stress (Figure 2L). Next, we generated a transgenic strain expressing GMAP-1-mCherry under the hsp-16.41 promoter in a gmap-1(ulb13) mutant background. Maintained at 15°C, the gmap-1(ulb-13);[phsp-16::GMAP-1-mcherry] strain did not show any expression of GMAP-1-mcherry and did not rescue gmap-1 hypersensitivity to hypotonic stress. The same strain was exposed 30 min to 33°C at the end of the 4 th larval stage. In these conditions, GMAP-1-mCherry was visible in Day 1 adults 16hrs later. This acute expression of GMAP-1-mCherry during adulthood was sufficient to fully restore an impermeable cuticle in gmap-1 mutants (Figure 2M), suggesting that GMAP-1 expression alters the cuticular properties of adults. Finally, we determined whether gmap-1 also affects cuticle permeability at dauer stage. Dauer larvae present a thicker and more resistant cuticle and can resist exposure to 1% SDS (Cassada and Russell, 1975). In contrast to N2, gmap-1-deficient dauer was hypersensitive to 1% SDS (Figure 2N). Expression of GMAP-1 in the hypodermis rescued the SDS hypersensitivity of gmap-1 dauer. In conclusion gmap-1 expression in the hypodermis is required to maintain an impermeable cuticle at all developmental stages.
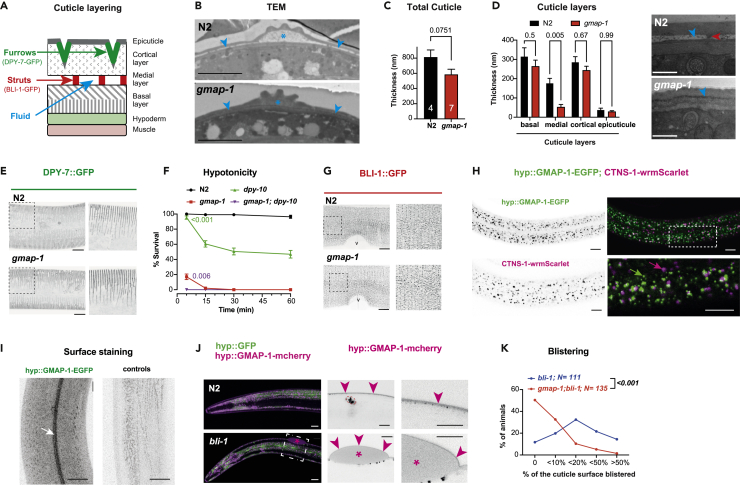
Given all these observations suggesting cuticular defects in gmap-1, we next explored the cuticle ultrastructure. The composition, structure, and thickness of C. elegans cuticle vary across larval stages (Cox et al., 1981a, 1981b, 1981c, 1981b, 1981c). Nevertheless, the cuticle is always made of several layers distinguishable by electron microscopy. The outermost coat layer is covered by glycoproteins. Below the coat is the epicuticle that contains lipids. In the adults, a >500nm thick extracellular matrix below the epicuticle is made of three layers (from top to bottom: cortical, medial, and basal) varying in their organization and composition. Insoluble proteins called cuticulins are found in the cortical layer, while variable cross-linked collagen proteins are found in all three layers (Figure 3A). To preserve ultrastructure independently of the speed of penetration of fixative and osmium, we used high-pressure freezing and freeze substitution. By transmission electron microscopy (TEM), we observed the cuticle at several locations along the body of animals cut transversally at the midbody. Triad-shaped alae are lateral cuticular ridges that appear in adults after the last moult. We observed similar alae in N2 and gmap-1, suggesting the animals were correctly staged. However, the medial layer fluid below the alae was severely reduced in gmap-1 (Figure 3B, blue star). The total thickness of the cuticle of gmap-1 mutant was reduced by ∼45% compared to N2 (Figure 3C). Although all cuticular layers were thinner in gmap-1 mutants, thinning was most obvious for the medial layer (Figure 3D, left). In many locations, the typically unstained medial layer was not observed in gmap-1 cuticle (Figures 3B and 3D, right; blue arrowheads). Of note, due to higher permeability of the cuticle, we observed stronger osmium staining in gmap-1 in TEM done after chemical fixation (Figure S2).
Figure 3.
GMAP-1 is secreted from hypodermis and contributes to the cuticular ultrastructure
(A) The adult cuticle is organized in several layers, including coat, epicuticle, cortical, medial, and basal layers. The furrows are circumferential indentations within the cortical layer labeled by DPY-7-GFP. The struts are collagen pillars within the medial layer labeled by BLI-1-GFP (Chisholm and Xu, 2012; Lints and Hall, 2009; Thein et al., 2003; Tong et al., 2009).
(B) Representative electron micrographs of 4 N2 and 7 gmap-1 individuals are presented for adult worms cut at midbody, including the alae. The blue star indicates the fluid accumulation in the medial layer below the alae in N2 and its reduction in gmap-1. The blue arrowheads indicate the fluid accumulation in the medial layer of N2 and its absence in gmap-1. Scale bars = 3 μm.
(C) The mean thickness of the whole cuticle and of each of its layer was quantified at the midbody of 4 N2 and 7 gmap-1 individuals. Unpaired t-test.
(D) Left, the mean thickness of each of the cuticle layers was quantified at the midbody of 4 N2 and 7 gmap-1 individuals observed at high magnification. It shows a reduced medial layer in gmap-1 mutants compared to N2. two-Way ANOVA with Sidak’s correction for multiple comparisons test. Right, representative electron microgram of the same cuticle area in gmap-1 mutants compared to N2. The blue arrowhead indicates the fluid accumulation in the medial layer, red arrowhead indicates the strut in N2. Scale bars = 500 nm.
(E) Marked by DPY-7-GFP, the annular furrows are observed as ventral and dorsal annular stripes along the body of the animal and were similarly organized in ∼20 gmap-1 and N2 observed at high magnification. Bellow, zoom ups show higher magnification of the cuticle. Scale bar = 20 μm.
(F) The potentially additive effects of dpy-10 and gmap-1 were assessed in dpy-10;gmap-1 double mutants. Survival in deionized water was quantified for N2, gmap-1, dpy-10 and gmap-1;dpy-10 double mutants over 1 h. At 5 min, we observe additive effects of gmap-1 and dpy-10 on survival to deionized water. The p values are indicated for single mutants compared to the double mutant (RM two-way ANOVA with Geisser’s correction, and Tukey’s for multiple comparisons).
(G) Marked by BLI-1-GFP, the struts are observed as regularly spaced puncta, forming two rows on each side of each furrow and were similarly organized in ∼20 gmap-1 and N2 observed at high magnification. Bellow, zoom ups show higher magnification of the cuticle. The vulva location is indicated by v. Scale bar = 20 μm.
(H) Confocal imaging shows the subcellular localization of the GMAP-1-EGFP (green) and CTNS-1-wrmScarlet (Magenta) proteins in the hypodermis. In the lateral hyp7 syncytium, GMAP-1-EGFP accumulates in mixed-size cytoplasmic vesicles (green arrow). In the same confocal plane, CTNS-1-wrmScarlet marker for lysosomes was observed in separate cytoplasmic vesicles (magenta arrows) not marked with EGFP. Scale bar = 20 μm.
(I) GMAP-1-EGFP also marks the surface of the animal, likely the cuticle, including the longitudinal alae (arrow). As controls N2 animals not expressing GMAP-1-EGFP are used in the same imaging conditions. Scale bar = 20 μm.
(J) In the bli-1 strain, GMAP-1-mCherry appears outside the hypodermis, within the blister (magenta asterisk). The right insets show single medial confocal images, stressing GMAP-1-mCherry sublocalization within the cuticle. In N2 it marks the cuticle, in bli-1 it marks the surface of the blisters (magenta arrowheads) as well as the fluid inside the blisters (magenta asterisk). GMAP-1-mCherry also weakly marked the coelomocytes (surrounded in dotted red): scavenger cells known to endocytose proteins secreted in pseudocoelom, suggesting a small fraction is secreted toward the pseudocoelomic cavity. Scale bar = 20 μm.
(K) The size of day 1 adults blisters was evaluated in bli-1 and in gmap-1;bli-1 double mutants. Individuals are categorized according to the percentage of their body covered by blister(s) or absence of blisters. Wilcoxon rank-sum test with continuity correction. All data are represented as Mean +/− SEM.
A reduced amount or an absence of specific cuticular collagens can result in reduced cuticle thickness, reduced body length, and increased cuticular permeability (Johnstone, 2000; Nystrom et al., 2002; Schultz et al., 2014). Given the alterations of the gmap-1 cuticle, we sought to determine the amount and organization of the cortical and medial layer collagen fibers. The adult cortical layer is patterned with circumferential-oriented ridges and furrows (Figure 3A). Furrows do not form in dpy-7 and dpy-10 mutants and dpy-7 and dpy-10 are associated with increased cuticular permeability (McMahon et al., 2003; Sandhu et al., 2021). A DPY-7-GFP fusion protein localizes to the furrows that appear as regular transversally striped patterns. These furrow patterns were similar in gmap-1 and N2 controls (Figure 3E). Together with the shortening of the gmap-1 body length, we observed that the distance between furrows was reduced in gmap-1 (1.76μm ± 0.07, N = 7) compared to N2 controls (1.99μm ± 0.14, N = 5), suggesting a reduced elasticity of the cuticle in gmap-1. If gmap-1 and dpy-10 independently set the barrier function, we should observe additive disruption of the barrier in gmap-1;dpy-10 double mutants. In the hypotonicity assay, the cuticle of dpy-10 was weakly permeable compared to gmap-1: >98% of gmap-1 but only 40% of dpy-10 died after 15 min of exposure to distilled water. After 5 min exposure to distilled water, we observed additive effects in gmap-1;dpy-10 double mutants, suggesting GMAP-1 and DPY-10 acts independently to establish the barrier (Figure 3F). As the gmap-1 medial layer was severely affected, we explored its organization. Within the medial layer of the adult C. elegans, “struts” containing BLI-1 collagen connect the cortical and basal layers, and do not form in bli-1 mutants (J. M. Kramer, personal communication and (De Melo et al., 2002)). Observed with a BLI-1-GFP fusion protein, struts appeared as regularly spaced puncta on each side of the furrows in gmap-1 as in N2 controls (Figure 3G). Therefore, despite the thinning of the medial layer in gmap-1, the organization and amount of BLI-1 collagen fibers did not appear modified. Altogether, these results suggest GMAP-1 act in parallel to collagen secretion to set the permeability of the cuticle.
In mammals, GM2AP is sorted into lysosomes by Sortilin receptor (Lefrancois et al., 2003). To visualize the subcellular distribution of GMAP-1 protein, we fused GMAP-1 C-terminally to EGFP. To determine whether GMAP-1-EGFP fusion protein was sorted to the lysosomes, we expressed it under the control of the agmo-1 hypodermal promoter in a strain expressing a lysosomal marker. As lysosome marker strain, we used a strain where wrmScarlet is knocked in frame with ctns-1. Prior studies confirmed that CTNS-1 cysteine transporter specifically localizes to lysosomes in C. elegans (Kalatzis et al., 2001; Mangahas et al., 2008). GMAP-1-EGFP was observed in cytoplasmic vesicles of mixed size (Figure 3H), these vesicles poorly colocalized with CTNS-1-wrmScarlet, suggesting GMAP-1-EGFP mostly marked a different organelle. However, GMAP-1 fused to mCherry was observed in vesicles of mixed size and shape, including tubules, not observed with GMAP-1-EGFP. Therefore, some proportion of GMAP-1 -or cleaved mCherry-might travel to an acidic compartment, where EGFP is extinguished but mCherry can accumulate. We observed GMAP-1-GFP and GMAP-1-mCherry signals within the cuticle, including the lateral alae (Figure 3I). Therefore, previous results suggest GMAP-1 is released to the cuticle of C. elegans. Similarly, keratinocytes secrete ∼30% of their GM2AP to the extracellular space (Glombitza et al., 1997). Onchocerca volvulus and Trichinella spiralis nematodes secrete GM2APs to the extracellular space (Bruce et al., 2006; Njume et al., 2019). As GMAP-1 sublocalization within the <1μm thick cuticle could not be determined in N2 by light microscopy, we took advantage of the blistered phenotype of bli-1 mutants. Mutants for the collagen gene bli-1 lack the struts connecting basal and cortical layers of the cuticle and present blisters. Blisters are regions where cortical and basal layers of the cuticle detach from one another; fluid accumulates inside this expanded medial layer forming cuticle bumps onto the surface of the animals (De Melo et al., 2002; Page and Johnstone, 2007). In blistered animals, we observed GMAP-1-mCherry marked the fluid inside the blister (Figure 3J, asterisks). GMAP-1-mCherry was also observed above blisters, in cortical and/or epicuticle layers (Figure 3J, magenta arrowheads). As the fluid within blister and medial layer is not made of collagen, this accumulation of GMAP-1-mCherry inside blisters suggests that GMAP-1-mCherry moves freely within an aqueous fluid filling the cuticle. In conclusion, GMAP-1 is secreted from the hypodermis toward the fluid of the cuticle and contributes to sculpt the cuticular ultrastructure.
The mesh of filamentous and globular collagen fibers observed in the cuticle leaves an empty space filled by this aqueous fluid (Peixoto and De Souza, 1995). Given the GMAP-1-mCherry accumulating in the blister fluid and the reduced thickness of the medial layer in gmap-1, we hypothesized GMAP-1 might contribute to the formation and/or expansion of the medial layer by an effect on fluid production or fluid retention within the cuticle. If gmap-1 was involved in fluid control, it should reduce fluid accumulation in blisters as it does in medial layers of gmap-1. To test this hypothesis, we crossed bli-1 mutants with gmap-1 mutants and quantified the formation of blisters in young adults. We observed gmap-1;bli-1 double mutant developed smaller blisters or no blisters in day 1 adults compared to the same age bli-1 single mutants, blistering all along their body (Figure 3K). As strut density and distribution are not affected in gmap-1 mutants, this reduced blistering suggests GMAP-1 contributes to fluid accumulation inside blisters.
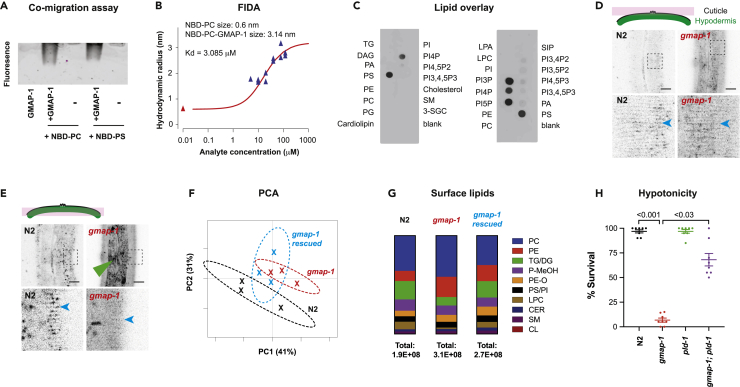
The human GM2AP presents GM2 ganglioside to β-hexosaminidase A for degradation. Accumulation of GM2 ganglioside in neuronal lysosomes causes severe neurological conditions (Furst and Sandhoff, 1992; Liu et al., 1997; Sandhoff and Harzer, 2013). However, GMAP-1 lacks the domain-promoting interaction with β-hexosaminidase A. In other nematodes, GMAP-1 orthologous proteins do not promote GM2 degradation (Bruce et al., 2006; Njume et al., 2019). In vitro, GM2AP is a rather unspecific lipid-binding protein that binds several sphingolipids and phosphoglycerides with low affinity (Ran and Fanucci, 2009; Schwarzmann et al., 2015). To assess its lipid-binding activity in vitro, a histidine-tagged version of C. elegans GMAP-1 was expressed in insect cells and purified using a combination of affinity and size exclusion chromatography (Figure S3). We determined whether the purified protein was able to bind the neutral lipids phosphatidylcholine (PC) and phospatidylserine (PS). To avoid steric issues frequently observed in lipid overlay assay, the lipid-binding assays were performed in solution using nitrobenzoxadiazole (NBD) fluorescently labeled PC and PS (NBD-PC and NBD-PS, respectively). Recombinant C. elegans GMAP-1 bound to NBD-PC and NBD-PS (Figure 4A). Since PC and PS are neutral lipids, their binding to nematode’ GMAP-1 is likely to rely on hydrophobic interactions, as previously observed for human GM2AP binding to a range of lipids (Wright et al., 2005). The binding of NBD-PC to GMAP-1 in solution was confirmed by Flow Induced Dispersion Analysis (FIDA) (Otzen et al., 2021). Assuming 1:1 binding stoichiometry, FIDA determined a dissociation constant (Kd) for NBD-PC and GMAP-1 around 3 μM, (Figure 4B). On lipid overlay assays, we observed that C. elegans GMAP-1 could also bind several phosphoglycerides, including PS and phosphatidylinositol phosphates (PIP), but not PC (Figure 4C).
Figure 4.
GMAP-1 carries phosphoglycerides and its deletion modifies the lipid composition of the epicuticle
(A) Purified GMAP-1-8xHis was incubated in solution with NBD-PC and NBD-PS. GMAP-1 complex with NBD-PG was visualized in a native gel following migration at 100V for 1h. Fluorescence imaging reveals that GMAP-1-8xHis binds and carries NBD-PC and NBD-PS.
(B) NBD-PC binding to purified GMAP-1 was confirmed by Flow Induced Dispersion Analysis (FIDA) which allows size-based characterization of biomolecules and their complexes in solution. The hydrodynamic radius of free NBD-PC was determined to 0.6 nm. The apparent size of NBD-PC increased steadily with GMAP-1 concentrations and was used for generating a binding curve. The complex size was determined to 3.14 nm, in line with the hydrodynamic radius of a 30 kDa protein with a flexible His tag. The dissociation constant (Kd) of the interaction was determined around 3 μM, assuming 1:1 binding stoichiometry.
(C) Purified GMAP-1 binds phosphoglycerides lipids on membrane strip, including phosphatidylserine (PS), phosphatidylinositols PI4P, PI3P, and PI5P.
(D and E). Surface lipids of the epicuticle were visualized by DiI staining of the cuticle for 12 N2 and 15 gmap-1 mutants. (D) A confocal plane at the surface of the cuticle highlights the furrows (blue arrowheads) that are less contrasted in gmap-1 than in N2. The bottom panel shows a zoom up of the area indicated by a square in the top panels. (E) A confocal plane at the level of hyp7 underneath the cuticle highlights the staining of hyp7 (green arrowhead) in gmap-1 but not in N2 and the reduced staining of the furrows in gmap-1. Scale bar = 20 μm.
(F) Analysis of the lipid composition of the epicuticle: PCA plot of the external cuticle layer extracted by lipodisq for N2 (black), gmap-1 mutants (red), and gmap-1 rescued in hypodermis (blue).
(G) Contribution of each lipid family (indicated by their color code on the right) on the total lipid extracted from the surface of the C. elegans cuticle from the N2, gmap-1, and gmap-1 rescued strains.
(H) Survival to hypotonicity was measured for adult worms after 15 min exposure to deionized water (N = 160 across 8 experimental replicates). Survival was measured for N2 controls, gmap-1(ulb13) and pld-1(ok2222) single mutants and gmap-1;pld-1 double mutants. One-way ANOVA followed by Kruskal-Wallis test. Data are represented as Mean +/− SEM.
As GMAP-1 bound lipids in vitro and localized to fluids within the cuticle in vivo, we hypothesized that it might serve as lipid transfer protein to shuttle lipids toward the epicuticle where lipids are known to contribute to the permeability barrier. To test this hypothesis, we stained the epicuticle with DiI, a lipophilic dye whose fluorescence is substantially enhanced in lipid environments (Lubart et al., 2020; Schultz and Gumienny, 2012). The cuticle of N2 adults consistently retained DiI fluorescence on its surface, therefore staining annuli and alae (Figure 4D). In contrast, the DiI staining of gmap-1 cuticle appeared disorganized: the annuli labeling was less compact, and the surface staining was diffuse (Figure 4D, blue arrowhead). In gmap-1, DiI also stained the plasma membrane and intracellular vesicles of the underlying hypodermis, something never observed in N2 (Figure 4E, green arrowhead). A weak staining of the gut was observed in both N2 and gmap-1. As DiI fluorescence is strongly enhanced in lipid environments, these results suggested that lipids of the cuticle are disorganized in gmap-1 and that the permeability of gmap-1 cuticle to a lipophilic dye (DiI: 933.9 g/mol) is strongly increased, allowing DiI to access and stain the hypodermis.
To determine if the lipid composition of the cuticle was modified in gmap-1, we extracted lipids from the cuticle surface using lipodisq nanoparticles, previously reported to extract surface lipids of C. elegans without affecting worm survival (Bada Juarez et al., 2019). Next, lipids were extracted from lipodisq and characterized using LC-MS and LC-MS/MS. Multivariate analysis of the LC-MS chromatograms by Principal Component Analysis (PCA) determines 2 multivariate axes that represent together 72% of the variance observed in the full dataset made of 3 technical replicates for each strain: N2, gmap-1, and gmap-1 rescued in the hypodermis. A 2-dimensions PCA representation showed a clear segregation of N2 from gmap-1 mutants, while gmap-1 rescued in the hypodermis overlaid with N2 and gmap-1 (Figure 4F). m/z masses corresponding to PE C36:2 and PE C35:3 were identified among the most discriminant features and appeared to increase in gmap-1. MetGem generates molecular networks based on the hypothesis that chemically similar compounds would share similar fragmentation patterns (Olivon et al., 2018). Using MetGem 1.3.6, we identified lipids from the same families forming clusters of related spectra (Figure S4A). We identified phosphatidylcholine (PC), Di- and Tri-Glycerides (DG/TG), phosphatidylethanolamines (PE), Phosphatidic acid Methyl ester (PMeOH), LysoPhosphatidylCholines (LPC), phosphatidylethanolamines Ether (PE-O), phosphatidylserine (PS), phosphatidylinositol (PI), Ceramides (Cer), sphingomyelin (SM) and cardiolipine (CL). These lipid families correspond to previously reported lipids of C. elegans cuticle (Bada Juarez et al., 2019; Blaxter, 1993; Gounaris et al., 1996; Proudfoot et al., 1990; Riddle et al., 1997). Based on the detection of identified lipids in LC-MS/MS operated in positive mode, the relative proportion of DG/TG and LPC decreased in gmap-1, while PC and PE increased in gmap-1 compared to N2 and to gmap-1 rescued in the hypodermis (Figure 4G and Table S1). In each lipid family, the individual lipids appeared modulated in both directions (Figure S4A). However, one-third of the identified PE lipids were upregulated in gmap-1 compared to N2 and to rescued strain and none in the opposite direction, suggesting gmap-1 had a robust effect, increasing the amount of several PE (Figure S4B). If phosphoglycerides (PG) amount or composition are involved in the defective permeability barrier observed in gmap-1, increasing PG or reducing PE presence in the cuticle could restore impermeability in gmap-1. To test this hypothesis, we took advantage of pld-1 mutants where PG transphosphatidylation or degradation by Phospholipase D/PLD-1 does not occur, increasing PG and modifying PG composition. We crossed gmap-1 with pld-1 and tested the single and double mutants for their survival to hypotonic stress. Although not fully rescued, the gmap-1; pld-1 were more resistant than the gmap-1 single mutants to hypotonic stress (Figure 4H). Altogether, these results suggest that reduced TG amount or modified PG composition of the epicuticle of gmap-1 contributes to the increased permeability of gmap-1.
Discussion
Because of its rapid life cycle, small size, and hermaphroditism, C. elegans can be used to screen libraries of small molecules in a high-throughput fashion for bioactivity in the context of a whole animal and over its entire life (Burns et al., 2006). However, a screen for >1000 drugs revealed that fewer than 10% of them accumulate in worms after 6h incubation (Burns et al., 2010). In that context, mutant strains bearing compromised cuticles were shown to facilitate chemical uptake, leading to increased sensitivity to agents and reduced risk of false negatives (Xiong et al., 2017). However, the use of these mutants depends on a trade-off between enhanced cuticle permeability and reduced animal fitness. We determined that gmap-1 mutants have increased permeability to molecules with variable size and water solubility: Sodium Azide (65 g/mol), Levamisole (204.3 g/mol), Hoechst 33,258 (533.9 g/mol), and the poorly soluble DiI (933.9 g/mol). Its permeability places gmap-1 among the most permeable mutants together with bus-5, bus-8, and bus-17. However, the bus mutants affect numerous other phenotypes including locomotion (Gravato-Nobre et al., 2005; Partridge et al., 2008). In comparison, gmap-1 fitness is largely unaffected in laboratory conditions including its reproduction, growth, and locomotion. Therefore, we suggest gmap-1 is an advantageous alternative to more deleterious mutants for pharmacological and toxicological assays.
Previously described permeability mutants suggest multiple mechanisms can lead to higher permeability of the cuticle. Among the increased permeability mutants, bus-5, bus-8, and bus-17 alter the glycosylation of coat surface proteins (Partridge et al., 2008); dpy-7 and dpy-10 alter a subset of collagens of the cortical layer (Sandhu et al., 2021); pod-1, fasn-1, acs-20, mboa-1, and agmo-1 alter the biogenesis of long-chain fatty acids, cholesterol esters or ester-linked lipids (Bada Juarez et al., 2019; Kage-Nakadai et al., 2010; Li and Paik, 2011; Loer et al., 2015; Watschinger and Werner, 2013). Although we cannot fully exclude that gmap-1 could affect collagens or glycosylation, the localization of GMAP-1, its lipid-binding properties and its effect on lipid content and organization at the surface of the cuticle as well as genetic interactions with DPY-10-collagen pathway or PLD-1 lipid pathway suggest that GMAP-1 acts independently of collagens or glycosylation pathways but dependent of the PLD-1 pathway. In addition, we did not observe changes in the amount or the organization of 2 major collagen components of the cortical layer (DPY-7-GFP) and of struts (BLI-1-GFP).
Lipids are essential elements of permeability barriers of multicellular organisms exemplified by cutin, suberin and waxes of the leaf surfaces of plants or by the stratum corneum of human skin (Lillywhite, 2006; Philippe et al., 2020). In nematodes, lipids produced and secreted by the hypodermis get enriched in the epicuticle layer where they contribute to the permeability barrier by forming a lipid bilayer (Blaxter, 1993; Mullaney and Ashrafi, 2009; Riddle et al., 1997). In vitro, GMAP-1 binds some phosphoglycerides (PG) including PS and PC in the μM range; in vivo, PG composition is altered in the cuticle of gmap-1. Higher PE/PC ratio, as we observe in gmap-1, correlates with higher permeability of plasma membranes in other biological models (Berglund et al., 2004; Li et al., 2006). In line with a role for PG amount or composition in cuticular impermeability of C. elegans, we show that increasing/modulating the cuticular PG composition rescues gmap-1 cuticular impermeability. We also observe that the strongly hydrophobic TG is reduced in gmap-1. Surface TG contributes to the resistance of parasitic nematodes to desiccation (Wharton et al., 2008). Instead of being localized in the epicuticle, these TG were observed in exudates coating the surface of desiccated nematodes (Lee, 2002). TG might therefore be involved in C. elegans impermeability by forming a lipid coat above the epicuticle and the glycoprotein coat.
In mammals, GM2AP presents GM2 gangliosides to β-hexosaminidase A for their degradation within the lysosomes (Furst and Sandhoff, 1992; Sandhoff and Harzer, 2013). Nematode’ GMAP-1 function appears to diverge from mammalian GM2APs. In nematodes, GMAP-1 orthologs do not interact with β-hexosaminidase A and do not participate in GM2 degradation (Bruce et al., 2006; Njume et al., 2019). C. elegans GMAP-1 did not bind glycolipids in the overlay assay (SM). Of note, we did not detect glycolipids in C. elegans cuticle, in agreement with previous data (Blaxter, 1993). Instead of a role in lysosomes, we observed GMAP-1 was secreted apically by the hypodermis from mixed-size intracellular organelles that are not lysosomes. Therefore, our results suggest GMAP-1 is secreted and may act extracellularly: GMAP-1-EGFP and GMAP-1-mCherry were observed in the cuticle and in the fluid filling the space between collagen fibers of the cuticle. In this fluid, GMAP-1 could serve as lipid transfer proteins to shuttle lipids between hypodermis and epicuticle located >500nm apart. It may be especially important in adults to maintain the surface lipids composition in absence of moult. Accordingly, we observed that acute expression of GMAP-1 in adult fully rescued the gmap-1 permeability phenotype. In humans, GM2AP and the sequence-related Saposins (SAPs) and Surfactant Protein B (SP-B) are found in the extracellular space. Within the stratum corneum of the skin, SAPs present GlcCer to β-glucocerebrosidase for degradation into ceramides and contribute to the rearrangement of lipids into smooth, linear lipid arrays (Doering et al., 1999; Fujita et al., 1996; Holleran et al., 1994).
Currently, our results converge to suggest GMAP-1 acts as a lipid transfer protein within the hypodermis and/or within the cuticle. Future work will be necessary to establish the GMAP-1 site of action. Within hypodermis organelles GMAP-1 may present lipids to an unknown enzymatic activity or reorganize lipids to facilitate their release. Also, GMAP-1 might shuttle lipids extracellularly to the epicuticle and/or rearrange lipids of the epicuticle. Although apparently distinct, these functions are not necessarily exclusive: SP-B acts both within the alveolar epithelium to generate the concentric lipid bilayer stacks characteristic of lamellar bodies and within alveoli to form and stabilize an extracellular phospholipid film at the air-liquid interface to drastically reduce surface tension in the lung (Clark et al., 1995; Nogee et al., 1993; Sever et al., 2021; Stahlman et al., 2000).
Limitations of study
In the current study, we fell short to identify which lipids are shuttled by GMAP-1 in C. elegans cuticle.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal Anti-polyHistidine antibody | Sigma | Cat#H1029 |
| Rabbit anti-mouse peroxidase conjugate | Sigma | Cat#A9044 |
| Bacterial and virus strains | ||
| E. Coli DH5 alpha | Thermofischer | Cat#18265017 |
| AcMNPV (Bac10:KO1629,Δv-cath/chiA-LoxP:DsRed) | Kolesnikova et al. (2022) | N/A |
| E. coli OP50 | CGC | https://cgc.umn.edu/strain/OP50 |
| Chemicals, peptides, and recombinant proteins | ||
| GMAP-1-8X Histidine | This study | N/A |
| cOmplete™ His tag purification resin | Merck | Cat# 5893682001 |
| Superdex® 200 10/300 GL | Cytiva | Cat# GE17-5175-01 |
| PIP Micro-Strips | Echelon Biosciences | Cat#P-6001 |
| PIP Micro-Strips | Echelon Biosciences | Cat#P-6002 |
| NBD-PC | Merck | Cat# 810131P |
| NBD-PS | Merck | Cat# 810198C |
| Cas9-NLS Purified Protein | IDT-DNA | Cat# 1081058 |
| Alt-R® CRISPR-Cas9 tracrRNA, 5 nmol | IDT | Cat#1072532 |
| Critical commercial assays | ||
| Vivaspin 2 centrifugal concentrators | Merck | Cat# Z614211 |
| Experimental models: Cell lines | ||
| Adapted Sf9 cells | Thermofisher | Cat# B82501 |
| Grace Medium supplemented with 10% FCS | Thermofisher | Cat# 11595030 |
| Adapted High Five cells | Thermofisher | Cat# B85502 |
| Express Five medium | Thermofisher | Cat#10486025 |
| Experimental models: Organisms/strains | ||
| bus-5 (br19) X | CGC | DC19 |
| bus-8 (e2883) X. | CGC | CB6177 |
| bus-17(br2) X. | CGC | CB7431 |
| acs-20 (e3031) IV. | CGC | CB7198 |
| agmo-1(e3016) III. | CGC | LC144 |
| bli-1(e769) II. | CGC | CB769 |
| pld-1(ok2222) II. | CGC | RD1737 |
| gmap-1(ulb13) | This study | OQ192 |
| gmap-1(ulb13); bli-1(e769) | This study | OQ396 |
| gmap-1(ulb13); pld-1(ok2222) | This study | OQ396 |
| gmap-1(ulb13); Ex[pgmap-1::egfp + ccrfp] | This study | OQ194 |
| gmap-1(ulb13); Ex[pgmap-1::gmap-1(cDNA)::sl2 gfp + ccrfp] | This study | OQ195 |
| gmap-1(ulb13); Ex[pgmap-1::gmap-1(cDNA)::egfp + ccrfp] | This study | OQ196 |
| gmap-1(ulb13); Ex[pagmo-1::gmap-1(cDNA)::sl2 gfp + ccrfp] | This study | OQ197 |
| gmap-1(ulb13); Ex[pC05 × 107.1::egfp + ccrfp] | This study | OQ198 |
| gmap-1(ulb13) X.; Ex[pAgmo-1::mEGFP + pAgmo-1::gmap-1::mCherry] | This study | OQ286 |
| Ex[pAgmo-1::mEGFP + pgmap-1::gmap-1::mCherry] | This study | OQ288 |
| bli-1(e769) II.; Ex[pAgmo-1:gmap-1::mCherry + pAgmo-1::mEGFP] | This study | OQ397 |
| frIs7 [nlp-29p::GFP + col-12p::dsRed] IV | CGC | IG274 |
| gmap-1(ulb13) X.; frIs7 [nlp-29p::GFP + col-12p::dsRed] IV. | This study | OQ283 |
| gmap-1(ulb13) X; frIs7 [nlp-29p::GFP + col-12p::dsRed] IV. | This study | OQ284 |
| qxIs722 [dpy-7p::dpy-7::SfGFP (single copy)] | CGC | XW18042 |
| gmap-1(ulb13) X; qxIs722 [dpy-7p::dpy-7::SfGFP (single copy)] | This study | OQ347 |
| cgEx198 [(pJC14) bli-1::GFP + unc-119(+)]. | CGC | CH1445 |
| gmap-1(ulb13); cgEx198 | This study | OQ398 |
| gmap-1(ulb13); Ex[pgmap-1::gmap-1::sl2 gfp + ccRFP] | This study | OQ399 |
| knockin[ctns-1::wrmScarlet]+ Ex[pgmap-1::gmap-1-GFP-10ng/ul] | This study | OQ449 |
| cas9 knockin [ctns-1::wrmScarlet] | Stigloher lab | PHX5270 |
| gmap-1(ulb13); dpy-10(e928) | This study | OQ428 |
| gmap-1(ulb13); Ex[phsp-16.41::gmap-1::mCherry + ccRFP] | This study | OQ449 |
| Oligonucleotides | ||
| pGMAP-1Fow AttB4 GGGGACAACTTTGTATAGAAA AGTTGGTGGCCCATGAAACAGTTGAC |
This study | N/A |
| pGMAP-1Rev AttB1 GGGGACTGCTTTTTTGTACAA ACTTGTCATTAACGCAAGTCGTGTTTC |
This study | N/A |
| pagmo1 Fwd AttB4 GGGGACAACTTTGTATA GAAAAGTTGTTAACTTGGCAAACA GTTGGAAGC |
This study | N/A |
| pagmo1 Rev AttB1 GGGACTGCTTTTTTGTAC AAACTTGGCCTCTTTTCATTTGG TTAAAATTT |
This study | N/A |
| pC05 × 107.1 Fwd AttB4 GGGGACAACTTTGT ATAGAAAAGTTGATACAATTTTTCTTT CCTCC |
This study | N/A |
| pC05 × 107.1 Rev AttB1 GGGACTGCTTTTTTGTACA AACTTGGAAGTTACAAGAACAG TATAGAAAAAG |
This study | N/A |
| GMAP-1 cDNA (stop) Fwd AttB1 GGGGACAAGTTTGTA CAAAAAAGCAGGCTATGACGACCCG AAGAATCGTAC |
This study | N/A |
| GMAP-1 cDNA (stop) Rev AttB2 GGGGACCACTTTGT ACAAGAAAGCTGGGTTTAGTTTGGCTG GGACACCG |
This study | N/A |
| GMAP-1 Fwd + BamH1 ATCTAATTTAGGATCCATGAC GACCCGAAGAATCGTAC |
This study | N/A |
| GMAP-1 Rev + EcoR1 GTTATACCTCTAGAATTCGTT TGGCTGGGACACCGTC |
This study | N/A |
| Translational reporter GMAP-1 Fwd GACTGTAAATCATACGATTTGGTGG | This study | N/A |
| Translational reporter GMAP-1 Rev TGCCCAGTTCACATCTACCAATATTG | This study | N/A |
| gmap-1 Ext FWD TGCGTTAATGACGACCCGAAG |
This study | N/A |
| gmap-1 Int FWD CAGAGTGAAGATTGCGCTTCAAAGTTTTAAAAC |
This study | N/A |
| gmap-1 Ext REV AGTTTGGCTGGGACACCGTC |
This study | N/A |
| acs-20(tm3232) Ext FWD ACTACGGAACCAAAGACCCT |
This study | N/A |
| acs-20(tm3232) Int FWD CCTTAGATTTTTGCCCGCTTCTGC | This study | N/A |
| acs-20(tm3232) Ext REV ACTCGAACAACTGACGCCAT |
This study | N/A |
| pld-1(ok2222) Ext FWD GCCAAGTATTTCCGATTCCACC |
This study | N/A |
| pld-1(ok2222) Int REV CAACAGAGAAAGAAGGATCGGC | This study | N/A |
| pld-1(ok2222) Ext REV GTAATTCGGTTCCGAGAACACG |
This study | N/A |
| Recombinant DNA | ||
| pAC8_MF | Kolesnikova et al. (2022) | Addgene Plasmid #139768 |
| pAC8_MF modified linker for expression of a protein fused to a C-terminal 8 His tag (BamHI-HindIII fragment) GGATCCCGGTCCGAAGCGCGCGGAATTCTCTAGAGAAA ACCTGTACTTCCAAGCGCACCATCACCATCACCATCATC ACTGAACTAGTGCCTGCAGTCTCGACAAGCTT |
This study | N/A |
| Software and algorithms | ||
| ImageJ/fiji | Schindelin et al. (2012) | N/A |
| Tierpsy | Javer et al. (2018) | N/A |
| Fidabio software suite | Fida Biosystems ApS | N/A |
| Prism 9.2.0 | GraphPad | https://www.graphpad.com/scientific-software/ |
| Illustrator 26.3.1 | Adobe | https://www.adobe.com/products/illustrator.html |
| ApE | Wayne Davis | https://jorgensen.biology.utah.edu/wayned/ape/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Patrick Laurent (patrick.laurent@ulb.be).
Materials availability
The C. elegans strains generated in this study will be deposited in the Caenorhaditis Genetics Center (CGC). All plasmids generated from this study are available upon request to the lead author.
Experimental model and subject details
C. elegans was used in the current research and was grown according to standard procedures as described in the methods. The C. elegans strains used are detailed in key resources table. No human subjects or mice were involved in the study.
Method details
Molecular biology
Constructs were assembled using the Gateway recombination system (Invitrogen). All PCR reactions were performed either with Phusion DNA polymerase (Invitrogen) or Q5 high fidelity polymerase (New England Biolabs). Constructs were assembled using the Gateway recombination system (Invitrogen). Amplification of the promoter of gmap-1 was done using the primer set pGMAP-1 Fwd AttB4 as attb4-containing forward primer and pGMAP-1Rev AttB1 as attb1-containing reverse primer for position 1 gateway cloning. For amplification of the gmap-1 cDNA from C. elegans total cDNA preparation, the following primers were used GMAP-1 cDNA (stop) Fwd AttB1 as attb1-containing GMAP-1 cDNA (stop) Rev AttB2 as attb2-containing reverse primer. The amplification of gmap-2/3 promoter was carried out using the following primers: pC05 × 107.1 Fwd AttB4 as attb-4-containing forward primer and pC05 × 107.1 Rev AttB1 as attb1-containing reverse primers. To identify hypodermal specific expression, the amgo-1 promoter was amplified from C. elegans genomic DNA using pagmo1 Fwd AttB4 as attb-4-containing forward primer and pagmo1 Rev AttB1 as attb1-containing reverse primers. The translational reporter, pgmap-1:gmap-1 gDNA:gfp was amplified directly from the H12 fosmid using the following primers: translational reporter GMAP-1 Fwd as forward primer and Translational reporter GMAP-1 Rev as reverse primer. All PCR reactions were performed either with Phusion DNA polymerase (Invitrogen) or Q5 high fidelity polymerase (New England Biolabs) and cloned into pDNR vectors by BP clonase (Invitrogen). Primer sequences, templates are listed in S1 List.
The full-length cDNA coding for GMAP-1 (Wormbase ID: C34 × 107.4) was amplified using the GMAP-1 Fwd + BamH1 as forward and GMAP-1 Rev + EcoR1 as reverse primers and was inserted between the BamHI and EcoRI sites of a modified pAC8_MF transfer plasmid (Addgene Plasmid #139768) harboring a C-terminal 8x His tag, yielding the pAC8_MF_GMAP-1 plasmid.
C. elegans strains
Worm strains were propagated and maintained on Nematode Growth Medium (NGM) agar plates seeded with either OP50 or HB101 E. coli as food source. Worms were propagated at 20°C unless otherwise stated. The worm strains, mutants, and transgenes are indicated in key resources table. The deletion for gmap-1(ulb-13) corresponds to a 1515 bp deletion flanked by ACCTATCCAAAGCTT and TGCCAAGACATTGAA. Some strains were obtained from the C. elegans Genetics Center (CGC). Transgenic animals were obtained by microinjection, the injection mix contained 10 or 30 ng/μL of the construct for transgenesis +30 ng/μL of unc-122P:RFP as marker of transgenesis and 50 or 30 ng/μL DNA mass ladder. For all strains used, stable transgenic lines were selected based on the unc-122P:RFP marker, fluorescent in coelomocytes as visualized under the fluorescent stereoscope.
Behavioral and morphological analysis
Behavior was recorded on day 1 adults, on NGM plates in presence of OP50 using video recording unit. Videos extraction of behavioral features including was done using Tierpsy (Javer et al., 2018). Tierpsy is freely available at https://github.com/Tierpsy/tierpsy-tracker. Worms were placed in the new plates 30 min prior to video recording. Maximal speed of locomotion was recorded in response to exposure to 1% O2.
Dehydration assay
Dried agar pads were prepared as follow: 100 μL of hot 2% agarose was placed on a 50X22-mm glass coverslip. A second coverslip is quickly placed on the drop and lightly pressed. After the agarose solidified, the coverslips are slide apart, and the agar pad is left to dry overnight at room temperature. 700 Halocarbon oil was placed on the dry agar pad and day 1 adult worms were gently pushed down to adhere to the dry agar. Animals were left 2, 5 or 10 min, then recovered in M9 buffer. Worms were transferred on an NGM plate with OP50 for recovery and checked for survival 1 h later.
Hypotonicity assay
For hypotonicity assays, L4s were picked onto a fresh NGM plate seeded with OP50 E. Coli and allowed to develop overnight to young gravid adults. 10 worms were picked and placed in 100 μL of deionized H20. The total number of ruptured worms was then counted after 15 min. N represents the total number of worms counted. Worms were picked and counted using a Nikon SMZ 745T stereoscope.
Acute expression of GMAP-1 in adults
All strains, including N2, gmap-1 and gmap-1(ulb13); Ex[hsp-16.41::GMAP-1-mCherry] were maintained at 15C from eggs to late adulthood. The acute expression of [hsp-16.41::GMAP-1-mCherry] was induced by soaking wrapped plates containing the L4 stage for 30 min in a water bath set at 33C, then returned to 15C. 16 h later, the induction of mCherry was verified under stereomicroscope in Day 1 adults. Another plate containing gmap-1(ulb13); Ex[hsp-16.41::GMAP-1-mCherry] animals but maintained at 15C and not exposed to 33C was used as controls and did not show visible expression of mCherry under stereomicroscope.
Sodium azide assay
Animals were synchronized by selecting L4 larva the day prior to the assay. The following day, 15–20 animals were transferred to region of the plate off-food to remove excess of remaining OP50 E. coli present in their body surface. Animals were then transferred into a 100 μL drop of freshly prepared 5 mM Sodium Azide (NaN3) (#S2002,Sigma-Aldrich, MO, USA) diluted with M9 buffer, or M9 buffer only for control groups. The 100 μL M9 or Sodium Azide drops were centered into 6 well plates (#734–2777,VWR, Leuven, BE) that were used to perform recordings with a 3 array camera connected to a computer equipped with Motif Software (Loopbio, Wien, Austria). Recordings were done for a 900 s period at 15 fps (4024 × 3036 pixels) and the experiments were replicated 6 times for Sodium azide and 2 times for M9 control groups, the position of each group was randomized to avoid location-dependent effects. Recordings were cropped to only detect the drop area and later processed with a custom python script to generate a masked video based on a set threshold to detect the animal body surface. Videos were analyzed with Tierpsy detection software as previously described (Martineau et al., 2020). We used the speed_midbody feature as a proxy to calculate the thrashing speed of a worm population over time. We then pooled the data for the different replicates and represented it as a time-course graph. Data analysis was done using GraphPad Prism, we plotted the curves and applied a no—linear one-phase exponential decay and from that we obtained the average half-lives representing the hypersensitivity to Sodium Azide for each genotype.
Levamisole paralysis assay
Animals were synchronized by selecting L4 larva the day prior to the assay. The following day, 20 animals were transferred to region of the plate off-food to remove excess of remaining OP50 E. coli present in their body surface. Animals were then transferred into a 10 μL drop of M9 buffer centered in each well of a 24-well plate (#734–2779, VWR, Leuven, BE) then we added 1 mL of Levamisole 0.4 mM diluted with M9 buffer to each of the wells. Recordings were done for 1 h at 15 fps (4024 × 3036 pixels), videos were then manually analyzed by counting the number of animals moving within the well every 5 min. Animals were counted as paralyzed when they were not able to perform at least one body bend (thrashing) in less than 30 s. The experiments were replicated 4 times for each genotype.
SDS assay
Dauer survival in SDS was assayed by washing of worms (N2, gmap-1 and from a starved NGM plate using 1 mL of 1% SDS into a 1.5 mL microfuge tube. The worms were allowed to incubate for 30 min in 1% SDS after which they were spun at 2500 x g for 5 min. The supernatant was discarded, and worms were resuspended in 100 μL of M9 or SDS and transferred to the side of a fresh NGM plate. The number of worms emerging into OP50 were counted using the Nikon SMZ 745T stereoscope.
NLP-29 assay
The IG274 strain containing Is[nlp-29:GFP; col-12p:dsRed] (Pujol et al., 2008) was crossed with gmap-1 mutants, and 2 independent lines were selected from these crossings. Worms were reared in standard laboratory conditions in NGM plates seeded with OP50 E. coli and were synchronised by selecting L4 animals the day prior to the assay. The day of the assay worms were anesthetized and mounted into agarose pads for imaging. To acquire multiple animals in the same field of view we used the Zeiss V16 mesoscope and performed a tile acquisition of the region of interest where animals lying. Imaging conditions were set for the IG274; gmap-1 strain and kept equal for all the other samples. The total number of animals were counted based on the col-12p:dsRed fluorescence and then the fraction of GFP + animals was counted based on the nlp-29p:GFP reporter expression for each experimental group.
Hoechst 33342 staining assay
Hoechst 33342 (membrane permeable but cuticle impermeable) was used to assess the integrity of the cuticular barrier as previously described. Briefly, worms were incubated in 1 μg/mL Hoechst 33342 in M9 buffer at room temperature for 15 min, followed by several washes with M9 buffer. Identical parameters (lens and magnifiers, filters, exposure time, resolution) were used. Fluorescence micrographs were processed using the ImageJ software for quantification of fluorescence intensity.
DiI staining assay
Lipophilic dye DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine) staining was performed as in (Schultz and Gumienny, 2012). (Briefly, 20–30 mid-to late-L4-stage larvae were washed once in 1 mL M9 buffer containing 0.5% Triton X-100 (100ML; Sigma, St. Louis, MO) and twice in 1 mL M9 buffer. Once all excess buffer had been removed, 400 μL of 30 μg/mL DiI (60,010; Biotium) in M9 buffer was added, and larvae were incubated on a rocker for 3 h at room temperature. After the incubation, worms were pipetted directly from the DiI solution onto NGM-agar plates heavily seeded with OP50 E. coli. The plates were kept in the dark for 30–45 min while worms swam from DiI solution onto OP50 E. coli. Worms that had successfully moved onto OP50 E. coli were chosen for imaging. Epifluorescence microscopy images were taken using 60-msec exposures, to maximize fluorescence intensity dynamic range. This resulted in nearly undetectable levels of fluorescence in N2 animals. Images were scored for intensity by a researcher blinded to genotypes.
Imaging
C. elegans were synchronized for imaging by doing egg-laying windows. Worms were reared at 20–23°C. Synchronized animals were mounted on 2–4% agarose pads and anesthetized with 25 mM NaN3 dissolved in M9 solution, images were acquired right after the animals were anesthetized and body movements ceased. Images were acquired at the Light Microscopy Facility LiMiF (http://limif.ulb.ac.be).
For cuticle and hypodermis images, we used a LSM780NLO confocal system fitted on an Observer Z1 inverted microscope. (Carl Zeiss, Oberkochen, Germany). The following fluorophores excitation (Ex) and detection wavelengths (DW) were used: for GFP (Ex: 488 nm – DW: 493–569 nm), for wrmScarlet/RFP-derivates and DiI (Ex: 543 nm – DW: 570–695 nm). Laser power and detector gain settings were adjusted to maximize signal-to-noise ratio and minimize saturation when possible. Images were saved in.lsm Zeiss file format.
For IG274 assay imaging we used the Zeiss AxioZoom V16 mesoscope equipped with a Hamamatsu Camera and a PanNeoFluar Z 1.0X objective. 2 channel tiles were acquired resulting in a 16 tiles array with a final resolution of 7827 × 7765 px (6.51 × 6.46 mm). RFP channel settings [Ex. 587/Em. 610 at 100 ms Exposure]. GFP channel settings [Ex. 488/Em. 509 at 350 ms Exposure]. Images were saved in.czi Zeiss file format.
Electron microscopy analysis
For the EM analysis of both wild type and gmap-1 mutant worms, two different methods were employed for the fixing of the worm; the high pressure freezing/freeze substitution (HPF/FS) and chemical fixation method. HPF/FS was done as previously described (Markert et al., 2020; Stigloher et al., 2011). Electron micrographs were taken on a JEOL JEM-1400 Flash at 120kV with a Matataki digital camera. For the chemical fixation, worms were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde in 0.1 M cacodylate buffer and also analyzed by transmission EM.
Recombinant protein production
The pAC8_MF_GMAP-1 transfer vector was co-transfected in Sf9 cells with linearized AcMNPV viral DNA (Bac10:KO1629,Δv-cath/chiA-LoxP:dsRed) to generate the recombinant baculovirus (Kolesnikova et al., 2022) and the corresponding virus was amplified. For protein production, Hi Five cells cultivated in Express-five serum free medium were infected at a density of 1.0 × 106 cells/mL and MOI of 2 (Detailed protocols for baculovirus expression can be found in (Osz-Papai et al., 2015). After 3 days of infection, cells were pelleted by centrifugation (1000 x g for 10 min) and the culture medium was incubated with Ni affinity resin (cOmplete His tag purification resin) for 12 h under gentle agitation using 0.5 mL of resin for 0.25 L of culture supernatant. The resin was first extensively washed with Tris 20 mM pH7,5, 150 mM KCl and 5 mM imidazole and then with 4 column volume (CV) of the same buffer containing 35 and 100 mM imidazole (Figure S4A). The bound protein was eluted with 500 mM imidazole, concentrated to 0.5 mL using a 10 kDa cut-off ultrafiltration device and further purified using an s200pg size exclusion column (Figure S4B). The purification yielded 0.2 mg of protein at a concentration of 0.1 mg/mL. For storage, 10% glycerol were added, and the protein was flash-frozen in liquid nitrogen.
Lipid binding assays
Lipid binding of GMAP-1 was assessed in lipid overlay assays as well as fluorescent lipid binding in solution. For protein-lipid overlay assays, PIP Micro-Strips (Echelon Biosciences Inc., Salt Lake City, UT) containing 100 pmol of various phospholipids, spotted and immobilized on a nitrocellulose membrane were used. The nitrocellulose strips were blocked in TBS-T 3% fatty acid free BSA (Sigma, Germany) for 1 h at room temperature (RT). Then, the strips were incubated with approximately 50 ng of GMAP-1 in TBS-T 3% fatty acid free BSA at RT for 1h. The strips were then washed (3x, 10 min) with TBS-T/BSA and gentle agitation at room temperature. Following the washes, the strips were incubated in a 1: 5,000 dilution of monoclonal mouse anti-His antibody (Sigma, Germany) in blocking buffer for 1 h. After three washes of 10 min each, rabbit anti-mouse peroxidase labeled secondary antibody (Sigma, Germany) was added (1: 5,000 in blocking buffer) and incubated for an additional hour at room temperature. After washing (3x, 10 min), the membranes were incubated for 10 min with the chemiluminescent substrate and revealed by chemiluminescence using a chemiluminescence scanner (LiCOR biosciences, USA). We noticed the absence of binding for PC, although GMAP-1 bind NBD-PC in solution. This might be explained by limitations of this assay, as the lipids are fixed on solid supports preventing a 3D interaction with the protein. For the fluorescence lipid binding assay, about 400 ng of GMAP-1 was incubated with NBD-PC or NBD-PS for 1 h in the dark at room temperature as previously described (Kumar et al., 2018). The mixture ran on a 6% native gel at 100 V for 1 h after which the fluorescence was measured using a chemiluminescence scanner (Amersham, Sweden). The binding of NBD-PC to GMAP-1 was confirmed by Flow Induced Dispersion Analysis (FIDA) conducted at 25°C on a FIDA 1 instrument using 488 nm laser-induced fluorescence detection, with FIDA standard capillaries as described in (Pedersen et al., 2019). The affinity measurement was performed at a fixed concentration of NBD-PC (1250 nM) titrated against varying concentrations of C34 × 107 protein (0, 5, 10, 20, 40, 80 and 120 μM) in duplicates. Sample analysis was performed by filling the capillary with 4 μL of buffer solution (20 mM Tris pH 7.5, 150 mM KCl, 5% glycerol), followed by injection of 40 nL NBD-PC pre-incubated with C34 × 107.4 protein. Data analysis with a 1:1 model yielded a Kd of 3.085 μM, sizes of the indicator and of the complex of 0.6 and 3.14 nm respectively with an R2 of 0.975.
Lipidomics
Synchronised populations of young adult N2, gmap-1 and gmap-1 rescued in hypodermis strains grew on NGM plates in the presence of E. coli. A culture plate with only E. coli served as a control. Worms were harvested by washing the plates with 10 mM Tris 5 mM NaCl pH 8 followed by three rounds of washing and centrifugation (2500 g, 2 min, 4°C). Epicuticle lipids were extracted using Lipodisq nanoparticles as described in (Bada Juarez et al., 2019) with an SMA polymer containing a molar ratio of styrene:maleic acid of 3:1. The SMA solution (25% w/v in 50 mM Tris buffer pH 8) was added to the washed worms at a final concentration of 12.5% v/v in 50 mM Tris buffer pH 8 and incubated for 30 min at 30–40°C. The sample was centrifuged, and the supernatant was kept for further analysis. Lipids were extracted from the Lipodisq samples according to the method of Bligh and Dyer (1959). Briefly, 200 μL Lipodisq-containing sample was mixed vigorously for 1 h, with 2 mL chloroform/methanol (2:1 v/v). Phase separation was achieved by adding 1 mL of miiliQ water. The extract was left for 10 min at room temperature and then centrifuged (1,000g, 10 min) and the lower phase was retrieved, washed twice with a methanol/water mix (1:1 v/v) and dried under a stream of N2 gas. Finally, the sample was reconstituted with 100 μL of a mixture of chloroform/methanol (2:1 v/v) before injection by LC-MS/MS. A reverse phase chromatography coupled with the new high-resolution mass spectrometer was used. A Column Zorbax XDB C18 (50 × 4.6 mm; 1.8 μm) at 40°C was used with a gradient phase of acetate ammonium pH 5, 5 mM (solvent A) and isopropanol (solvent B). Briefly, 2 μL of sample were injected in an LC1200 (Agilent Technologies) with a flow of 0.5 mL/min and the following gradient: 0 min 90/10%; 10 min 20/80%; 25 min 20/80%; 27 min 90/10%; 30 min 90/10%. The source was at 350°C, drying gas 7 L/min, Nebulizer 50 L/min, Vcap 4500 V, Fragmentor 210 V. The fragmentation was operated with a fixed energy of 25 eV in autoMSMS mode on a QTOF 6520 (Agilent Technologies); MS range – 200–20000 m/Z; MS/MS range 50–2000 m/Z. ProteoWizard MSConvert tools (Version 3.03.9393, 64-bit) was used to convert MS and MS/MS raw data to.mzXML file. Data pre-processing were performed with Mzmine 2.53. The following parameter were applied. Range time of interest was set at 3–30 min being this part of the chromatograms the richest in the focused compounds. Mass detection was performed by fixing the noise level at 1000 for MS1 and 50 for MS2. ADAP Chromatogram Builder was employed to build chromatograms by setting the parameters as follows: minimum group size of three scans, group intensity = 50 and minimum intensity = 50. The m/z tolerance was fixed at 24 ppm. Baseline cut-off algorithm was used for deconvolution by fixing a minimum feature height at 200 and a peak duration range at 0–5 min. The deconvolution was performed by fixing a value of 0.1 Da as m/z range for MS2 scan pairing and 0.2 min as RT range for MS2 scan pairing. Isotopic peak grouper was then performed with an m/z tolerance of 24 ppm, an RT tolerance of 0.2 min and a maximum charge of 2. Join aligner for data alignment was used to generate a peak list. The parameters were set as follows: m/z tolerance = 24 ppm, weight for m/z = 75, RT tolerance = 0.5 min, weight for RT = 25. Feature list filter was built by fixing m/z scan at 350–2000, RT range at 1–30, peak duration range at 0–5 min. Gap filled step was performed with 10% of intensity tolerance, m/z tolerance = 24 ppm and RT tolerance = 0.25. A filter to keep only peaks with MS2 scan (GNPS) was performed and data were then exported as.mgf files for spectra and.csv files as sample metadata. Molecular networks were built with MetGem 1.3.6 software (https://metgem.github.io/). Standard parameters were used to import Data “.mgf” and “.csv”: m/z tolerance was set at 0.02 and minimum matched peaks were set at 4. A cosine score (CS) above 0.4 has been applied to filter edges; maximum neighbor number (top K) of 10 and a max. connected component size of 1000 was set to build the networks. Annotation of each nodes/each metabolite was performed according to a lipidomic database for identification (MS-DIAL) (Olivon et al., 2018). For each metabolite, the intensity of each replicate was summed. The surface of the node is colored accordingly to the relative intensity of the metabolite in each genotype (Figure S4A.).
Quantification and statistical analysis
The unpaired T-test was used for analysis to calculate means between strains. One-way ANOVA test followed by Kruskal-Wallis test and Dunn’s correction was used for analysis of multiple comparisons. In some instances, a two-way ANOVA with Geisser’s, Sidak’s or Tukey’s correction was used for multiple comparisons. The Wilcoxon’s rank test with continuity correction was used for analysis of blisters. Where applicable, experiments were performed in triplicates and the SEM was determined.
Acknowledgments
LV and PL are respectively directors of research and research associates of the Belgian National Fund for Scientific Research (FRS-FNRS). FNN received a stipend from “Les Amis de l’Institut Pasteur à Bruxelles.” In vitro studies were funded by Instruct-ERIC through PID7256 and 20749 and to an internship to FNN at IGBMC, Strasbourg together with support from the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05. V.P and C.S. are supported by the DFG-grant GRK2581-P06, G.F. with grant FA1046/3-1 and CS with grant STI700/1-1. The Transmission Electron Microscope JEOL JEM-1400Flash was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) – 426173797 (INST 93/1003-1 FUGG). We thank the Analytical Platform of the Faculty of Pharmacy (APFP) and the Imaging Facility of the Faculty of Medecine (LiMiF) which are ULB Platforms supported by the FRS-FNRS. We thank Jean-Marie Vanderwinden (LiMiF) for imaging advice; Nadia Messadeq (IGBMC Imaging Plat-form) for preliminary EM analysis and Fabien Alpy (IGBMC Functional Genomics and Cancer Dept) for fruitful discussions on protein/lipid interactions. Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions
Conceptualization, L.V. and P.L; Methodology, F.N.N., A.P., P.V.A., P.L.; Validation, C.B., C.S., P.V.A., A.P., L.V., P.L.; Formal analysis, F.N.N., P.L.; Investigation, F.N.N, A.R., M.S., V.P., G.F., A.B., C.D., P.P., S.P., C.B.; Resources, A.P., P.V.A., C.S., L.V., P.L.; Writing – Original Draft, F.N.N., L.V., P.L.; Writing – Review and Editing, F.N.N., A.R., A.P., P.V.A., C.S., L.V., P.L.; Visualization, F.N.N., A.R., P.L.; Supervision, S.M.G, J.S., A.P., L.V., P.L.; Funding Acquisition, F.N.N., L.V., P.L.
Declaration of interests
The authors declare no competing interests.
Published: November 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105357.
Supporting citations
The following references appear in the supplemental information: Schindelin et al. (2012); Kolesnikova et al. (2022).
Supplemental information
Data and code availability
All necessary data has been included alongside the manuscript. No codes were used in this study.
References
- Bada Juarez J.F., O'Rourke D., Judge P.J., Liu L.C., Hodgkin J., Watts A. Lipodisqs for eukaryote lipidomics with retention of viability: sensitivity and resistance to Leucobacter infection linked to C.elegans cuticle composition. Chem. Phys. Lipids. 2019;222:51–58. doi: 10.1016/j.chemphyslip.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Berglund A.H., Larsson K.E., Liljenberg C.S. Permeability behaviour of lipid vesicles prepared from plant plasma membranes--impact of compositional changes. Biochim. Biophys. Acta. 2004;1682:11–17. doi: 10.1016/j.bbalip.2004.01.001. [DOI] [PubMed] [Google Scholar]
- Blaxter M.L. Cuticle surface proteins of wild type and mutant Caenorhabditis elegans. J. Biol. Chem. 1993;268:6600–6609. [PubMed] [Google Scholar]
- Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Boeck M.E., Huynh C., Gevirtzman L., Thompson O.A., Wang G., Kasper D.M., Reinke V., Hillier L.W., Waterston R.H. The time-resolved transcriptome of C. elegans. Genome Res. 2016;26:1441–1450. doi: 10.1101/gr.202663.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A.F., Gares M.P., Selkirk M.E., Gounaris K. Functional characterisation of a nematode secreted GM2-activator protein. Mol. Biochem. Parasitol. 2006;147:224–229. doi: 10.1016/j.molbiopara.2006.02.014. [DOI] [PubMed] [Google Scholar]
- Burns A.R., Kwok T.C.Y., Howard A., Houston E., Johanson K., Chan A., Cutler S.R., McCourt P., Roy P.J. High-throughput screening of small molecules for bioactivity and target identification in Caenorhabditis elegans. Nat. Protoc. 2006;1:1906–1914. doi: 10.1038/nprot.2006.283. [DOI] [PubMed] [Google Scholar]
- Burns A.R., Wallace I.M., Wildenhain J., Tyers M., Giaever G., Bader G.D., Nislow C., Cutler S.R., Roy P.J. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat. Chem. Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- Cao J., Packer J.S., Ramani V., Cusanovich D.A., Huynh C., Daza R., Qiu X., Lee C., Furlan S.N., Steemers F.J., et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science. 2017;357:661–667. doi: 10.1126/science.aam8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassada R.C., Russell R.L. The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev. Biol. 1975;46:326–342. doi: 10.1016/0012-1606(75)90109-8. [DOI] [PubMed] [Google Scholar]
- Chisholm A.D., Hsiao T.I. The Caenorhabditis elegans epidermis as a model skin. I: development, patterning, and growth. Wiley Interdiscip Rev Dev Biol. 2012;1:861–878. doi: 10.1002/wdev.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm A.D., Xu S. The Caenorhabditis elegans epidermis as a model skin. II: differentiation and physiological roles. Wiley Interdiscip Rev Dev Biol. 2012;1:879–902. doi: 10.1002/wdev.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M.K., Son S., Hong M., Choi M.S., Kwon J.Y., Lee J. Maintenance of membrane integrity and permeability depends on a patched-related protein in Caenorhabditis elegans. Genetics. 2016;202:1411–1420. doi: 10.1534/genetics.115.179705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark J.C., Wert S.E., Bachurski C.J., Stahlman M.T., Stripp B.R., Weaver T.E., Whitsett J.A. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc. Natl. Acad. Sci. USA. 1995;92:7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G.N., Kusch M., DeNevi K., Edgar R.S. Temporal regulation of cuticle synthesis during development of Caenorhabditis elegans. Dev. Biol. 1981;84:277–285. doi: 10.1016/0012-1606(81)90395-x. [DOI] [PubMed] [Google Scholar]
- Cox G.N., Kusch M., Edgar R.S. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J. Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G.N., Staprans S., Edgar R.S. The cuticle of Caenorhabditis elegans. II. Stage-specific changes in ultrastructure and protein composition during postembryonic development. Dev. Biol. 1981;86:456–470. doi: 10.1016/0012-1606(81)90204-9. [DOI] [PubMed] [Google Scholar]
- De Melo J.V., De Souza W., Peixoto C.A. Ultrastructural analyses of the Caenorhabditis elegans DR 847 bli-1(n361) mutant which produces abnormal cuticle blisters. J. Submicrosc. Cytol. Pathol. 2002;34:291–297. [PubMed] [Google Scholar]
- Doering T., Holleran W.M., Potratz A., Vielhaber G., Elias P.M., Suzuki K., Sandhoff K. Sphingolipid activator proteins are required for epidermal permeability barrier formation. J. Biol. Chem. 1999;274:11038–11045. doi: 10.1074/jbc.274.16.11038. [DOI] [PubMed] [Google Scholar]
- Fujita N., Suzuki K., Vanier M.T., Popko B., Maeda N., Klein A., Henseler M., Sandhoff K., Nakayasu H., Suzuki K. Targeted disruption of the mouse sphingolipid activator protein gene: a complex phenotype, including severe leukodystrophy and wide-spread storage of multiple sphingolipids. Hum. Mol. Genet. 1996;5:711–725. doi: 10.1093/hmg/5.6.711. [DOI] [PubMed] [Google Scholar]
- Fürst W., Sandhoff K. Activator proteins and topology of lysosomal sphingolipid catabolism. Biochim. Biophys. Acta. 1992;1126:1–16. doi: 10.1016/0005-2760(92)90210-m. [DOI] [PubMed] [Google Scholar]
- Glombitza G.J., Becker E., Kaiser H.W., Sandhoff K. Biosynthesis, processing, and intracellular transport of GM2 activator protein in human epidermal keratinocytes. The lysosomal targeting of the GM2 activator is independent of a mannose-6-phosphate signal. J. Biol. Chem. 1997;272:5199–5207. doi: 10.1074/jbc.272.8.5199. [DOI] [PubMed] [Google Scholar]
- Gounaris K., Smith V.P., Selkirk M.E. Structural organisation and lipid composition of the epicuticular accessory layer of infective larvae of Trichinella spiralis. Biochim. Biophys. Acta. 1996;1281:91–100. doi: 10.1016/0005-2736(96)00022-3. [DOI] [PubMed] [Google Scholar]
- Gravato-Nobre M.J., Nicholas H.R., Nijland R., O'Rourke D., Whittington D.E., Yook K.J., Hodgkin J. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 2005;171:1033–1045. doi: 10.1534/genetics.105.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran W.M., Ginns E.I., Menon G.K., Grundmann J.U., Fartasch M., McKinney C.E., Elias P.M., Sidransky E. Consequences of beta-glucocerebrosidase deficiency in epidermis. Ultrastructure and permeability barrier alterations in Gaucher disease. J. Clin. Invest. 1994;93:1756–1764. doi: 10.1172/JCI117160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javer A., Ripoll-Sánchez L., Brown A.E.X. Powerful and interpretable behavioural features for quantitative phenotyping of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018;373:20170375. doi: 10.1098/rstb.2017.0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone I.L. Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends Genet. 2000;16:21–27. doi: 10.1016/s0168-9525(99)01857-0. [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E., Kobuna H., Kimura M., Gengyo-Ando K., Inoue T., Arai H., Mitani S. Two very long chain fatty acid acyl-CoA synthetase genes, acs-20 and acs-22, have roles in the cuticle surface barrier in Caenorhabditis elegans. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalatzis V., Cherqui S., Antignac C., Gasnier B. Cystinosin, the protein defective in cystinosis, is a H(+)-driven lysosomal cystine transporter. EMBO J. 2001;20:5940–5949. doi: 10.1093/emboj/20.21.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnikova O., Zachayus A., Pichard S., Osz J., Rochel N., Rossolillo P., Kolb-Cheynel I., Troffer-Charlier N., Compe E., Bensaude O., et al. HR-Bac, a toolbox based on homologous recombination for expression, screening and production of multiprotein complexes using the baculovirus expression system. Sci. Rep. 2022;12:2030. doi: 10.1038/s41598-021-04715-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N., Leonzino M., Hancock-Cerutti W., Horenkamp F.A., Li P., Lees J.A., Wheeler H., Reinisch K.M., De Camilli P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018;217:3625–3639. doi: 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.L. Taylor & Francis; 2002. The Biology of Nematodes. [Google Scholar]
- Lefrancois S., Zeng J., Hassan A.J., Canuel M., Morales C.R. The lysosomal trafficking of sphingolipid activator proteins (SAPs) is mediated by sortilin. EMBO J. 2003;22:6430–6437. doi: 10.1093/emboj/cdg629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Paik Y.K. A potential role for fatty acid biosynthesis genes during molting and cuticle formation in Caenorhabditis elegans. BMB Rep. 2011;44:285–290. doi: 10.5483/BMBRep.2011.44.4.285. [DOI] [PubMed] [Google Scholar]
- Li Z., Agellon L.B., Allen T.M., Umeda M., Jewell L., Mason A., Vance D.E. The ratio of phosphatidylcholine to phosphatidylethanolamine influences membrane integrity and steatohepatitis. Cell Metab. 2006;3:321–331. doi: 10.1016/j.cmet.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lillywhite H.B. Water relations of tetrapod integument. J. Exp. Biol. 2006;209:202–226. doi: 10.1242/jeb.02007. [DOI] [PubMed] [Google Scholar]
- Lints R.a.H., Hall D. WormAtlas. 2009. The cuticle. [DOI] [Google Scholar]
- Liu Y., Hoffmann A., Grinberg A., Westphal H., McDonald M.P., Miller K.M., Crawley J.N., Sandhoff K., Suzuki K., Proia R.L. Mouse model of GM2 activator deficiency manifests cerebellar pathology and motor impairment. Proc. Natl. Acad. Sci. USA. 1997;94:8138–8143. doi: 10.1073/pnas.94.15.8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loer C.M., Calvo A.C., Watschinger K., Werner-Felmayer G., O'Rourke D., Stroud D., Tong A., Gotenstein J.R., Chisholm A.D., Hodgkin J., et al. Cuticle integrity and biogenic amine synthesis in Caenorhabditis elegans require the cofactor tetrahydrobiopterin (BH4) Genetics. 2015;200:237–253. doi: 10.1534/genetics.114.174110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubart Q., Hannestad J.K., Pace H., Fjällborg D., Westerlund F., Esbjörner E.K., Bally M. Lipid vesicle composition influences the incorporation and fluorescence properties of the lipophilic sulphonated carbocyanine dye SP-DiO. Phys. Chem. Chem. Phys. 2020;22:8781–8790. doi: 10.1039/c9cp04158c. [DOI] [PubMed] [Google Scholar]
- Mangahas P.M., Yu X., Miller K.G., Zhou Z. The small GTPase Rab2 functions in the removal of apoptotic cells in Caenorhabditis elegans. J. Cell Biol. 2008;180:357–373. doi: 10.1083/jcb.200708130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markert S.M., Skoruppa M., Yu B., Mulcahy B., Zhen M., Gao S., Sendtner M., Stigloher C. Overexpression of an ALS-associated FUS mutation in C. elegans disrupts NMJ morphology and leads to defective neuromuscular transmission. Biol. Open. 2020;9:bio055129. doi: 10.1242/bio.055129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martineau C.N., Brown A.E.X., Laurent P. Multidimensional phenotyping predicts lifespan and quantifies health in Caenorhabditis elegans. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon L., Muriel J.M., Roberts B., Quinn M., Johnstone I.L. Two sets of interacting collagens form functionally distinct substructures within a Caenorhabditis elegans extracellular matrix. Mol. Biol. Cell. 2003;14:1366–1378. doi: 10.1091/mbc.e02-08-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney B.C., Ashrafi K. C. elegans fat storage and metabolic regulation. Biochim. Biophys. Acta. 2009;1791:474–478. doi: 10.1016/j.bbalip.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njume F.N., Ghogomu S.M., Shey R.A., Gainkam L.O.T., Poelvoorde P., Humblet P., Kamgno J., Robert A., Mutesa L., Lelubre C., et al. Identification and characterization of the Onchocerca volvulus excretory secretory product Ov28CRP, a putative GM2 activator protein. PLoS Negl. Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogee L.M., de Mello D.E., Dehner L.P., Colten H.R. Brief report: deficiency of pulmonary surfactant protein B in congenital alveolar proteinosis. N. Engl. J. Med. 1993;328:406–410. doi: 10.1056/NEJM199302113280606. [DOI] [PubMed] [Google Scholar]
- Nyström J., Shen Z.Z., Aili M., Flemming A.J., Leroi A., Tuck S. Increased or decreased levels of Caenorhabditis elegans lon-3, a gene encoding a collagen, cause reciprocal changes in body length. Genetics. 2002;161:83–97. doi: 10.1093/genetics/161.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivon F., Elie N., Grelier G., Roussi F., Litaudon M., Touboul D. MetGem software for the generation of molecular networks based on the t-SNE algorithm. Anal. Chem. 2018;90:13900–13908. doi: 10.1021/acs.analchem.8b03099. [DOI] [PubMed] [Google Scholar]
- Osz-Papai J., Radu L., Abdulrahman W., Kolb-Cheynel I., Troffer-Charlier N., Birck C., Poterszman A. Insect cells-baculovirus system for the production of difficult to express proteins. Methods Mol. Biol. 2015;1258:181–205. doi: 10.1007/978-1-4939-2205-5_10. [DOI] [PubMed] [Google Scholar]
- Otzen D.E., Buell A.K., Jensen H. Microfluidics and the quantification of biomolecular interactions. Curr. Opin. Struct. Biol. 2021;70:8–15. doi: 10.1016/j.sbi.2021.02.006. [DOI] [PubMed] [Google Scholar]
- Packer J.S., Zhu Q., Huynh C., Sivaramakrishnan P., Preston E., Dueck H., Stefanik D., Tan K., Trapnell C., Kim J., et al. A lineage-resolved molecular atlas of C. elegans embryogenesis at single-cell resolution. Science. 2019;365:eaax1971. doi: 10.1126/science.aax1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page A., Johnstone I.L. WormBook. 2007. The cuticle; pp. 1–15. [DOI] [Google Scholar]
- Partridge F.A., Tearle A.W., Gravato-Nobre M.J., Schafer W.R., Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Pedersen M.E., Østergaard J., Jensen H. Flow-induced dispersion analysis (FIDA) for protein quantification and characterization. Methods Mol. Biol. 2019;1972:109–123. doi: 10.1007/978-1-4939-9213-3_8. [DOI] [PubMed] [Google Scholar]
- Peixoto C.A., De Souza W. Freeze-fracture and deep-etched view of the cuticle of Caenorhabditis elegans. Tissue Cell. 1995;27:561–568. doi: 10.1016/s0040-8166(05)80065-5. [DOI] [PubMed] [Google Scholar]
- Philippe G., Sørensen I., Jiao C., Sun X., Fei Z., Domozych D.S., Rose J.K. Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020;55:11–20. doi: 10.1016/j.pbi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Proudfoot L., Kusel J.R., Smith H.V., Harnett W., Worms M.J., Kennedy M.W. The surface lipid of parasitic nematodes: organization, and modifications during transition to the mammalian host environment. Acta Trop. 1990;47:323–330. doi: 10.1016/0001-706x(90)90033-v. [DOI] [PubMed] [Google Scholar]
- Pujol N., Cypowyj S., Ziegler K., Millet A., Astrain A., Goncharov A., Jin Y., Chisholm A.D., Ewbank J.J. Distinct innate immune responses to infection and wounding in the C. elegans epidermis. Curr. Biol. 2008;18:481–489. doi: 10.1016/j.cub.2008.02.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran Y., Fanucci G.E. Ligand extraction properties of the GM2 activator protein and its interactions with lipid vesicles. Biophys. J. 2009;97:257–266. doi: 10.1016/j.bpj.2009.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R. In: C. elegans II, 2nd edition. Riddle D.L., Blumenthal T., Meyer B.J., Priess J.R., editors. 1997. Introduction to C. elegans. [Google Scholar]
- Sandhoff K., Harzer K. Gangliosides and gangliosidoses: principles of molecular and metabolic pathogenesis. J. Neurosci. 2013;33:10195–10208. doi: 10.1523/JNEUROSCI.0822-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu A., Badal D., Sheokand R., Tyagi S., Singh V. Specific collagens maintain the cuticle permeability barrier in Caenorhabditis elegans. Genetics. 2021;217:iyaa047. doi: 10.1093/genetics/iyaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R.D., Bennett E.E., Ellis E.A., Gumienny T.L. Regulation of extracellular matrix organization by BMP signaling in Caenorhabditis elegans. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz R.D., Gumienny T.L. Visualization of Caenorhabditis elegans cuticular structures using the lipophilic vital dye DiI. J. Vis. Exp. 2012:e3362. doi: 10.3791/3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzmann G., Breiden B., Sandhoff K. Membrane-spanning lipids for an uncompromised monitoring of membrane fusion and intermembrane lipid transfer. J. Lipid Res. 2015;56:1861–1879. doi: 10.1194/jlr.M056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sever N., Miličić G., Bodnar N.O., Wu X., Rapoport T.A. Mechanism of lamellar body formation by lung surfactant protein B. Mol. Cell. 2021;81:49–66.e8. doi: 10.1016/j.molcel.2020.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlman M.T., Gray M.P., Falconieri M.W., Whitsett J.A., Weaver T.E. Lamellar body formation in normal and surfactant protein B-deficient fetal mice. Lab. Invest. 2000;80:395–403. doi: 10.1038/labinvest.3780044. [DOI] [PubMed] [Google Scholar]
- Stigloher C., Zhan H., Zhen M., Richmond J., Bessereau J.L. The presynaptic dense projection of the Caenorhabditis elegans cholinergic neuromuscular junction localizes synaptic vesicles at the active zone through SYD-2/liprin and UNC-10/RIM-dependent interactions. J. Neurosci. 2011;31:4388–4396. doi: 10.1523/JNEUROSCI.6164-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thein M.C., McCormack G., Winter A.D., Johnstone I.L., Shoemaker C.B., Page A.P. Caenorhabditis elegans exoskeleton collagen COL-19: an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 2003;226:523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- Tong A., Lynn G., Ngo V., Wong D., Moseley S.L., Ewbank J.J., Goncharov A., Wu Y.C., Pujol N., Chisholm A.D. Negative regulation of Caenorhabditis elegans epidermal damage responses by death-associated protein kinase. Proc. Natl. Acad. Sci. USA. 2009;106:1457–1461. doi: 10.1073/pnas.0809339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek M., Bringmann H. Gene expression changes of Caenorhabditis elegans larvae during molting and sleep-like lethargus. PLoS One. 2014;9 doi: 10.1371/journal.pone.0113269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watschinger K., Werner E.R. Alkylglycerol monooxygenase. IUBMB Life. 2013;65:366–372. doi: 10.1002/iub.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wharton D.A., Petrone L., Duncan A., McQuillan A.J. A surface lipid may control the permeability slump associated with entry into anhydrobiosis in the plant parasitic nematode Ditylenchus dipsaci. J. Exp. Biol. 2008;211:2901–2908. doi: 10.1242/jeb.020743. [DOI] [PubMed] [Google Scholar]
- Wright C.S., Mi L.Z., Lee S., Rastinejad F. Crystal structure analysis of phosphatidylcholine-GM2-activator product complexes: evidence for hydrolase activity. Biochemistry. 2005;44:13510–13521. doi: 10.1021/bi050668w. [DOI] [PubMed] [Google Scholar]
- Xiong H., Pears C., Woollard A. An enhanced C. elegans based platform for toxicity assessment. Sci. Rep. 2017;7:9839. doi: 10.1038/s41598-017-10454-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All necessary data has been included alongside the manuscript. No codes were used in this study.