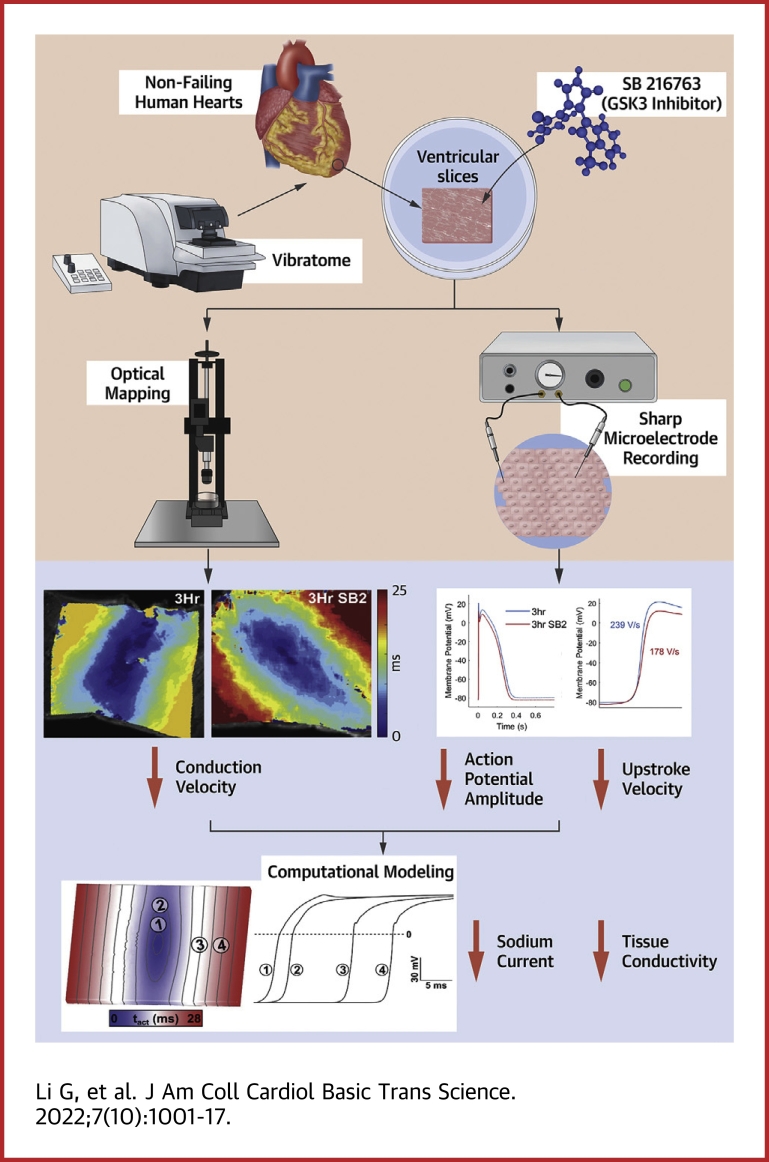
Visual Abstract
Key Words: electrophysiology, GSK-3 inhibitor, human cardiac slices, SB216763
Abbreviations and Acronyms: ABC, active β-catenin; APD, action potential duration; BDM, 2,3-butanedione monoxime; cDNA, complementary DNA; CV, conduction velocity; Cx43, connexin 43; dVm/dtmax, maximum upstroke velocity; GNa, sodium-channel conductance; GOF, gain of function; GSK-3, glycogen synthase kinase 3; INa, sodium current; LV, left ventricle; NaV1.5, pore-forming α-subunit protein of the voltage-gated cardiac sodium channel; PCR, polymerase chain reaction; RMP, resting membrane potential; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SB2, SB216763
Highlights
-
•
Human cardiac slices cultured with SB216763 showed reduced conduction velocity and decreased excitability, as measured by dVm/dtmax, at 3 and 24 hours compared to slices cultured in control media.
-
•
Both overall and nuclear β-catenin levels increased, whereas NaV1.5 decreased, in human cardiac slices cultured with SB216763.
-
•
Induced stabilization of β-catenin in adult murine hearts decreases conduction velocity and increases susceptibility to arrhythmias.
Summary
Glycogen synthase kinase 3 (GSK-3) inhibition has emerged as a potential therapeutic target for several diseases, including cancer. However, the role for GSK-3 regulation of human cardiac electrophysiology remains ill-defined. We demonstrate that SB216763, a GSK-3 inhibitor, can acutely reduce conduction velocity in human cardiac slices. Combined computational modeling and experimental approaches provided mechanistic insight into GSK-3 inhibition-mediated changes, revealing that decreased sodium-channel conductance and tissue conductivity may underlie the observed phenotypes. Our study demonstrates that GSK-3 inhibition in human myocardium alters electrophysiology and may predispose to an arrhythmogenic substrate; therefore, monitoring for adverse arrhythmogenic events could be considered.
Glycogen synthase kinase 3 (GSK-3) is a ubiquitous, constitutively active kinase downstream of multiple regulatory pathways. Active GSK-3 generally represses many intracellular processes and is itself inhibited by a variety of cellular stimuli to achieve downstream signaling effects.1 In the past 30 years, GSK-3 has emerged as a therapeutic target for neurodegenerative diseases and mental disorders, such as Alzheimer disease, Rett syndrome, and bipolar disorder, with multiple GSK-3 inhibitors currently in use or in clinical trials.2, 3, 4 In the heart, GSK-3 has been implicated in both congenital and acquired cardiovascular diseases, including cardiomyocyte hypertrophy, diabetic cardiomyopathy, acute cardiac ischemia, and heart failure.5, 6, 7, 8 Many of these conditions show altered GSK-3 activity in addition to being associated with arrhythmias.9, 10, 11 The clinical use of lithium, a classic mood stabilizer and known GSK-3 inhibitor, has been reported to cause cardiac conduction abnormalities ranging from benign electrocardiographic changes to life-threatening arrhythmias.12 Additionally, SB216763 (SB2), a small-molecule GSK-3 inhibitor, has been shown to rescue both structural and electrophysiologic defects in murine and zebrafish models of arrhythmogenic cardiomyopathy.9,13 However, despite growing evidence for GSK-3 as a regulator of cardiac electrophysiology, the effects and mechanisms remain poorly defined.
One major signaling pathway that is regulated by GSK-3 is the canonical Wnt signaling pathway. In the absence of Wnt ligands, a destruction complex including GSK-3β phosphorylates β-catenin, the main transcriptional effector that initiates transcription of Wnt-responsive genes. Phosphorylation of β-catenin targets the protein for ubiquitination and degradation, resulting in relatively low cytoplasmic levels of β-catenin during homeostasis.14 Upon canonical Wnt ligand binding to cell surface receptors, parts of the GSK-3β–associated destruction complex are recruited to the cell membrane, preventing the destruction complex from phosphorylating β-catenin. As a result, β-catenin accumulates in the cytoplasm, translocates into the nucleus, and activates downstream target genes.15 Therefore, pharmacologic inhibition of GSK-3 or disease-associated decrease of GSK-3 activity can result in activation of the Wnt signaling pathway. Previous studies have shown that both developmental and adult perturbation of β-catenin can affect cardiac electrophysiology and increase arrhythmia susceptibility.16, 17, 18 Specifically, both Scn5a, the gene encoding the major cardiac sodium-channel subunit NaV1.5 (the pore-forming α-subunit protein of the voltage-gated cardiac sodium channel), and Gja1, the gene encoding for cardiac gap junction connexin 43 (Cx43), are transcriptionally regulated by Wnt/β-catenin.18, 19, 20, 21, 22 β-catenin is also involved in cell-cell adhesion at the adherens junctions, where it binds to N-cadherin.23,24 It has been shown that β-catenin accumulates at the intercalated discs of hearts with hypertrophic cardiomyopathy and that GSK-3β is simultaneously decreased.25, 26, 27 Additionally, β-catenin directly associates with Cx43 at the intercalated discs.19,28 Therefore, because of its role in the regulation of β-catenin, it can be hypothesized that decreased GSK-3 activity may regulate Wnt/β-catenin–mediated electrophysiologic reprogramming.
In this study, we used our previously published methodology for culturing human left ventricular (LV) organotypic slices from nonfailing donor hearts to demonstrate that acute treatment with SB2 decreases the conduction velocity (CV) after only 3 hours in culture. Reduced CV was associated with decreased maximum upstroke velocity (dVm/dtmax) of cardiac action potentials. The ability to couple experimental models of human myocardium with computational models has been a powerful tool for elucidating drug effects.29,30 Simulations conducted in a computational model of human myocardium showed that the observed reduction in CV and dVm/dtmax can be explained by decreased sodium-channel conductance (GNa) and tissue conductivity. Consistent with the modeling results, we found that NaV1.5 protein was decreased in slices treated with SB2 at 3 hours. β-catenin protein levels increased significantly in the chromatin-bound nuclear fraction, whereas surprisingly, levels were unchanged in other subcellular compartments. To determine the effects of chronically increased β-catenin protein levels on electrophysiology, we used a genetic mouse model with stabilized β-catenin and showed conduction slowing along with arrhythmia susceptibility. We demonstrate here that GSK-3 inhibition can alter cardiac conduction, resulting in a proarrhythmic substrate in both mouse and human hearts, and that therapeutic targeting of GSK-3 should consider these possible adverse cardiovascular effects.
Methods
Human tissue acquisition
Nonfailing donor hearts rejected from transplantation were obtained from Mid-America Transplant Services in St. Louis, Missouri. Informed consent was obtained by Mid-America Transplant Services for all tissue used in this study. Experimental protocols were approved by the Washington University in St. Louis Institutional Review Board and are in accordance with all research guidelines on human tissue. Living donor hearts were procured by an experienced cardiothoracic surgeon, perfused, and transported to the research laboratory while in Belzer University of Wisconsin Cold Storage Solution (Bridge to Life).
Human cardiac slice preparation
Heart samples were processed within 30 minutes of procurement. A 1-cm by 1-cm cube of LV tissue was cut in Belzer University of Wisconsin Cold Storage Solution (Bridge to Life) from a region near the left anterior descending artery and circumflex artery. Premade, cooled 4% agarose gel was glued to the tissue holder of a vibrating microtome (7000 smz-2, Campden Instruments), and the tissue cube was mounted, endocardium up, using Histoacryl glue (B. Braun). Slices were cut tangential to the endocardium with a thickness of 400 μm, at a rate of 0.04 cm/s, in a 4 °C bath of modified Tyrode’s solution with excitation contraction uncoupler 2,3-butanedione monoxime (BDM) (NaCl, 140 mmol/L; KCl, 6 mmol/L; glucose, 10 mmol/L; 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mmol/L; MgCl2, 1 mmol/L; CaCl2, 1.8 mmol/L; BDM, 10 mmol/L; pH 7.4) supplied with 100% oxygen.
Culture of human cardiac slice
Slices were cultured on porous Transwell inserts (PICM0RG50, Millipore) in 6-well culture plates with 1.1 mL of culturing medium (medium 199 [M4530, Sigma]) supplemented with 1% penicillin-streptomycin (15140163, Thermo Fisher Scientific), 1× insulin-transferrin-selenium supplement (41400045, Thermo Fisher Scientific), and 10 mmol/L BDM each. Culture plates were placed in a 37 °C incubator with humidified air and 5% CO2 for 3 or 24 hours before slices were used for electrophysiology or saved for histology, gene expression, and protein. SB2, an adenosine triphosphate–competitive inhibitor of GSK-3, was added to culture media to reach a concentration of 343 nmol/L, 10 times the half maximal inhibitory concentration reported in cell-free assays. Dimethyl sulfoxide was used as vehicle control. Adjacent slices from the same heart were used as controls to minimize endocardium-to-epicardium and heart-to-heart variability.
Optical mapping
Changes in the membrane potential of human ventricular slices and murine Langendorff perfused hearts were recorded using optical mapping described previously.31,32 Slices were superfused with modified Tyrode’s solution (NaCl, 140 mmol/L; KCl, 4.5 mmol/L; glucose, 10 mmol/L; 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mmol/L; MgCl2, 1 mmol/L; CaCl2, 1.8 mmol/L; pH 7.4) at 37 °C, and murine hearts were perfused with a different modified Tyrode’s solution (NaCl, 128.2 mmol/L; KCl, 4.7 mmol/L; glucose, 11.1 mmol/L; NaH2PO4, 1.19 mmol/L; NaHCO3, 20 mmol/L; MgCl2, 1.05 mmol/L; CaCl, 1.3 mmol/L; pH 7.4) at 37 °C. Blebbistatin (Cayman Chemical), an excitation-contraction uncoupler, was used to eliminate motion artifacts in the recorded optical signals. Changes in the membrane potential were detected by Di-4-ANEPPS (Life Technologies), a voltage-sensitive dye. A green LED light source with a wavelength of 520 ± 5 nm was used to excite the voltage-sensitive dye. The emitted fluorescence was filtered by a long-pass filter at 650 nm and collected by a complementary metal-oxide-semiconductor camera (SciMedia). The recorded data were analyzed using an open-source custom MATLAB program (MathWorks). Human ventricular slices and murine hearts were electrically stimulated at different cycle lengths to obtain adaptation and restitution curves, with the stimulus amplitude being twice the capture threshold with a pulse duration of 2 ms. Using surface coordinates and time of the maximum derivatives (dVm/dtmax) of the optical signals, activation maps were generated and used to calculate the CV. Longitudinal CV was calculated by the distance traveled over the number of time isochrones along multiple lines originating from the site of stimulation. Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome.
Sharp microelectrode recording
Microelectrode recordings were obtained from slices of human hearts at baseline, 3 hours in culture with dimethyl sulfoxide (vehicle control), and 3 hours in culture with SB2. Thin glass pipettes with high resistance (10-20 MΩ) filled with 3 mol/L KCl were used to record action potentials from a minimum of 8 single cells from different regions of each slice (technical replicates). To decrease noise from motion artifacts, blebbistatin was used to arrest motion and allow for stable microelectrode recording without requiring the use of floating electrodes. Electrical signals were digitized by a Power 1401 analog-to-digital converter. Action potentials of sampled cells with a resting membrane potential (RMP) of <−75 mV were averaged. The RMP, action potential amplitude, dVm/dtmax, and action potential duration (APD) at 90% repolarization are reported. Analysis of microelectrode recordings was performed using a custom MATLAB script. Cells with at least 20 action potentials and stable RMP were included in the data analysis. For each action potential in each cell, the peak potential value and time were calculated. The RMP was then calculated for each action potential using a histogram, where the maximum frequency value was defined as the RMP. The magnitude (amplitude) of each action potential was then defined as the difference between the peak potential value and the RMP. The maximum change in voltage with respect to time (dVm/dtmax) was calculated using a first derivative. The point in time that corresponds to this value was then used as the start time for the APD calculation. The end time was when the membrane potential decreased to 10% of the amplitude value. Investigators were blinded to the sample group allocation during the experiment and analysis of the experimental outcome.
Histology and immunohistochemistry
Immunohistochemistry was performed on optimal cutting temperature compound–embedded frozen sections (Sakura 4583) with a combination of sarcomeric α-actinin antibody (1:100, Abcam ab9465), NaV1.5 antibody (1:100, Cell Signaling Technology 14421), PCM1 antibody (1:1,000, Sigma HPA023370), active β-catenin (ABC) antibody (1:50, Developmental Studies Hybridoma Bank, University of Iowa PY489-B-catenin), pan β-catenin antibody (1:100, BD 610153), and Cx43 antibody (1:50, Thermo Fisher Scientific 71-0700). Secondary antibodies included Alexa Fluor 488 (1:200, Abcam ab150077, ab150113, and ab150121) and Alexa Fluor 568 (1:200, Abcam ab175471). Slides were treated with TrueBlack Lipofuscin Autofluorescence Quencher (Biotium) and then with DAPI (Sigma 28718-90-3).
Western blotting
Protein lysates were prepared from human LV slices and used for immunoblotting as previously described.33 Briefly, ventricular slices were homogenized in ice-cold 1× radioimmunoprecipitation assay buffer lysis buffer (50 mmol/L Tris-HCl, 150 mmol/L NaCl, 5 mmol/L EDTA, 1% Triton-X, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate) with protease inhibitor cocktail tablet (1 tablet per 50 mL lysis buffer, Roche). Samples were centrifuged at 13,200g for 10 minutes at 4 °C. The supernatants were used for electrophoresis after protein analysis via bicinchoninic acid assay (Thermo Fisher Scientific 23225). Protein fractionation was performed using a subcellular protein fractionation kit for tissues (Thermo Fisher Scientific 87790). Immunoblotting was performed using anti–β-catenin (1:1,000, BD Biosciences 610153), anti-Cx43 (1:250, Invitrogen 71-0700), anti-NaV1.5 (1:1,000, Abcam ab56240), anti–phospho-Cx43 (1:1,000, Cell Signaling Technology 3511), anti–nonphosphorylated β-catenin (1:1,000, Cell Signaling Technology 8814), anti–Na,K-ATPase (1:1,000, Cell Signaling Technology 23565), anti-lamin A/C (1:1,000, Cell Signaling Technology 2032), anti-histone H3 (1:1,000, Cell Signaling Technology 9715), and anti-GAPDH (1:2,000, Cell Signaling Technology 14C10) antibodies. Secondary detection was performed using horseradish peroxidase antibody (1:5,000, Abcam ab6721, ab205719) and Clarity Western enhanced chemiluminescence Substrate (Bio-Rad).
Reverse transcription quantitative polymerase chain reaction
Total RNA was isolated from murine hearts using Trizol (Invitrogen) and DNase treated using Turbo DNA-Free Kit (Invitrogen). RNA concentrations were determined using NanoDrop One (Thermo Fisher Scientific). First-strand complementary DNA (cDNA) was synthesized using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Primers were designed on Integrated DNA Technologies website and validated using bulk human or mouse cDNA. Gene expression was assayed using the PowerSYBR Green PCR MasterMix (Applied Biosystems) and quantified using the QuantStudio Flex 6 Real-Time polymerase chain reaction (PCR) system (Applied Biosystems). Relative fold changes were then calculated using the comparative CT method, with the most stable reference genes chosen for analysis using NormFinder and geNorm.34, 35, 36
Computational modeling
Electrophysiology simulations were conducted in a 3-dimensional virtual organotypic slice model of human ventricular tissue (dimensions: 0.8 × 0.6 × 0.04 cm; average tetrahedral edge length: 123.3 μm). At the cellular scale, the O’Hara Rudy dynamic human ventricular action potential model (midmyocardial variant) provided a realistic representation of membrane kinetics.37 As in previous work,38 to facilitate realistic electrophysiologic behavior at the tissue scale, the O’Hara Rudy dynamic version of the sodium current (INa) model was replaced with the INa formulation of ten Tusscher et al.39 To mimic the experimental preparation, a uniform vector field parallel to the y-axis of the slice (ie, 0.6-cm edge) was used to represent myocardial fiber orientations. Conductivity tensor values were calibrated to match simulated CV with experimental observations (see the Results section for specific parameter values). All stimuli were delivered via transmembrane current injection to a central region on the top face of the block (strength: 120 μA/cm2; duration: 10 ms; diameter: 250 μm). Wavefront propagation was simulated on a standard Linux workstation (Intel i7-8086K 12-core central processing unit at 4.00 GHz, 64 GB random-access memory) by solving the monodomain equation using the finite element method in the CARP software package.40,41 A version of this software that is free for noncommercial academic use is available42; all model and parameter files necessary to reproduce all simulation data in this study will be made available upon request.
Mice information
The inducible cardiomyocyte-specific β-catenin gain-of-function (GOF) mouse model was made by combining mice expressing MerCreMer under control of the α-myosin heavy chain promoter (αMHC-MCM43) with Ctnnb1fl(ex3).44 Cardiomyocyte-specific expression of Cre recombinase was induced by administration of tamoxifen chow (Teklad) for 10 days in 8-week-old mice. Excision of β-catenin exon 3 results in stabilized, nondegradable β-catenin. A washout period of 4 weeks after induction was used to simulate a chronic stabilized β-catenin state while also allowing recovery from the possibility of confounding tamoxifen-related cardiomyopathy.45 Littermate control mice treated with tamoxifen or αMHC-MCM+/o/Ctnnb1fl(ex3)/+ without induction were used as controls. Mice of both sexes were used in all studies, and although experiments were not powered to specifically detect sex differences, there were no obvious differences in any parameters between sexes. Animal protocols were approved by the Animal Studies Committee at Washington University, and animals were handled in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Statistical analysis
All data are expressed as mean ± SEM. For the comparison of more than 2 experimental groups, statistical significance of observed mean differences was evaluated using a 1- or 2-way analysis of variance, followed by Tukey’s post hoc test for multiple pairwise comparisons. For within-group comparisons (correlated samples), repeated-measures analysis of variance was used. For comparison of 2 experimental groups, an equal variance Student’s t-test was used, and a 2-tailed P value of <0.05 was considered statistically significant. All data were analyzed with parametric tests, given the small sample sizes for each experiment with insufficient power to reject the null hypothesis of distribution normality.
Results
GSK-3 inhibition by SB2 reduces CV in human myocardium
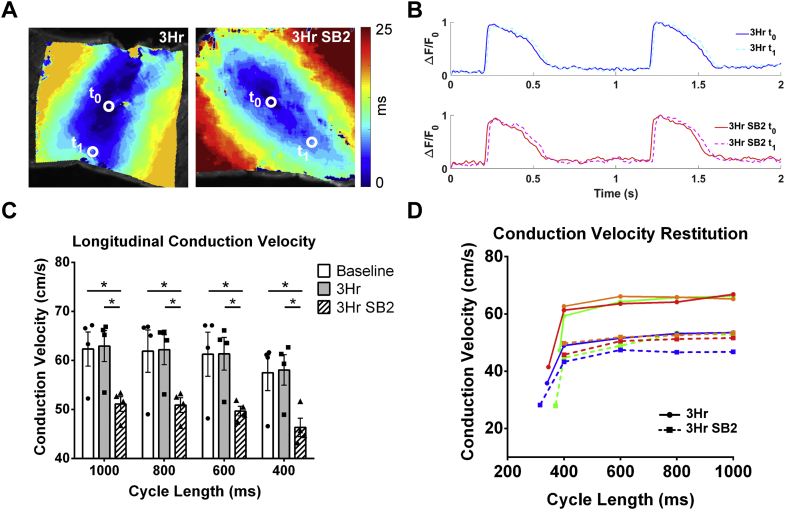
GSK-3 dysregulation can occur in the setting of acquired cardiovascular disease in association with arrhythmias.10,46 In addition, there have been case reports of associated cardiac arrhythmias in patients with neurologic disorders that were prescribed the GSK-3 inhibitor lithium,12 and there is growing evidence for a role of GSK-3 in the regulation of cardiac electrophysiology in animal models.9,16 To determine whether acute GSK-3 inhibition has an effect on cardiac conduction in the human heart, we performed optical mapping on human organotypic slices at baseline (0 hours), and after culture with either control (vehicle) or SB2 (343 nmol/L in culture media). To minimize potential variability among hearts and among different regions within the same heart, adjacent slices were used for control and drug treatment. Given the different time scales of various biological processes that could potentially underlie changes in physiology, we performed experiments over a range of timepoints, from 15 minutes to 24 hours. Compared to baseline, CV did not change 15 minutes after the addition of SB2 into the physiologic bath, suggesting that SB2 does not directly inhibit the sodium channel (data not shown). However, we determined that CV was decreased by 3 hours of culture with SB2 when compared to baselines and to adjacent slices from the same heart cultured with vehicle for 3 hours. A representative activation map shows slower electrical activation and action potential propagation in SB2-cultured slices compared with 3-hour control (Figures 1A and 1B). CV of SB2-cultured slices was slower by 18% compared to the 0-hour baseline and 3-hour control (baseline: 62 cm/s; 3 hours: 63 cm/s; 3-hour SB2: 51 cm/s at 1,000-ms cycle length) across all stimulation cycle lengths (Figure 1C). CV restitution of control and SB2-treated slices for each individual heart showed a similar magnitude of decrease in CV and no change in the restitution slope (Figure 1D). We identified a similar-magnitude CV decrease after 24 hours in culture with SB2 when compared to baseline and to adjacent slices from the same heart cultured with vehicle for 24 hours (baseline: 61 cm/s; 24 hours: 60 cm/s; 24-hour SB2: 47 cm/s), and this decrease persisted at all stimulation cycle lengths (Supplemental Figures 1A and 1B). Importantly, CV remained unchanged between baseline and 24-hour control slices (Supplemental Figure 1B), suggesting that our culture technique preserves human cardiac electrophysiology. CV restitution plots from control and SB2-treated slices from each individual heart show similar decreases in CV with no change in the restitution slope caused by SB2 (Supplemental Figure 1C). Because CV was reduced at 3 and 24 hours in culture with SB2 but not at 15 minutes, SB2 is unlikely to have an immediate effect on ionic conductance leading to reduced CV and likely acts through cell signaling processes.
Figure 1.
Conduction Velocity of Human LV Slices Is Reduced by Short-Term Culture With SB2
(A) Representative activation maps of a control slice (left) compared to an SB2-treated slice (right) from the same heart. Electrical activation from time t0 to t1 is slower in the SB2-treated slice as seen by the greater number of isochrones to reach the same distance. (B) Control optical action potentials (top) at t0(blue) and t1(dashed cyan) shows faster propagation compared to SB2 optical action potentials (bottom) from t0(red) to t1(dashed magenta). (C) Conduction velocities quantified from activation maps show no difference between baseline and 3-hour control but reduced conduction velocity in 3-hour SB2-treated slices (baseline: 62 cm/s; 3-hour control: 63 cm/s; 3-hour SB2: 51 cm/s) at pacing cycle lengths of 1,000 ms, 800 ms, 600 ms, and 400 ms. (D) Conduction velocity restitution of each individual heart shows reduced conduction velocity at all stimulation cycle lengths but no change in restitution slope in SB2-treated slices. Each color represents an individual heart for 3-hour vehicle (solid line) or 3-hour SB2 (dashed line). Statistics were performed using repeated-measures analysis of variance followed by Tukey’s post hoc test for multiple pairwise comparisons. ∗P < 0.05. 3HR = 3-hour; SB2 = SB216763.
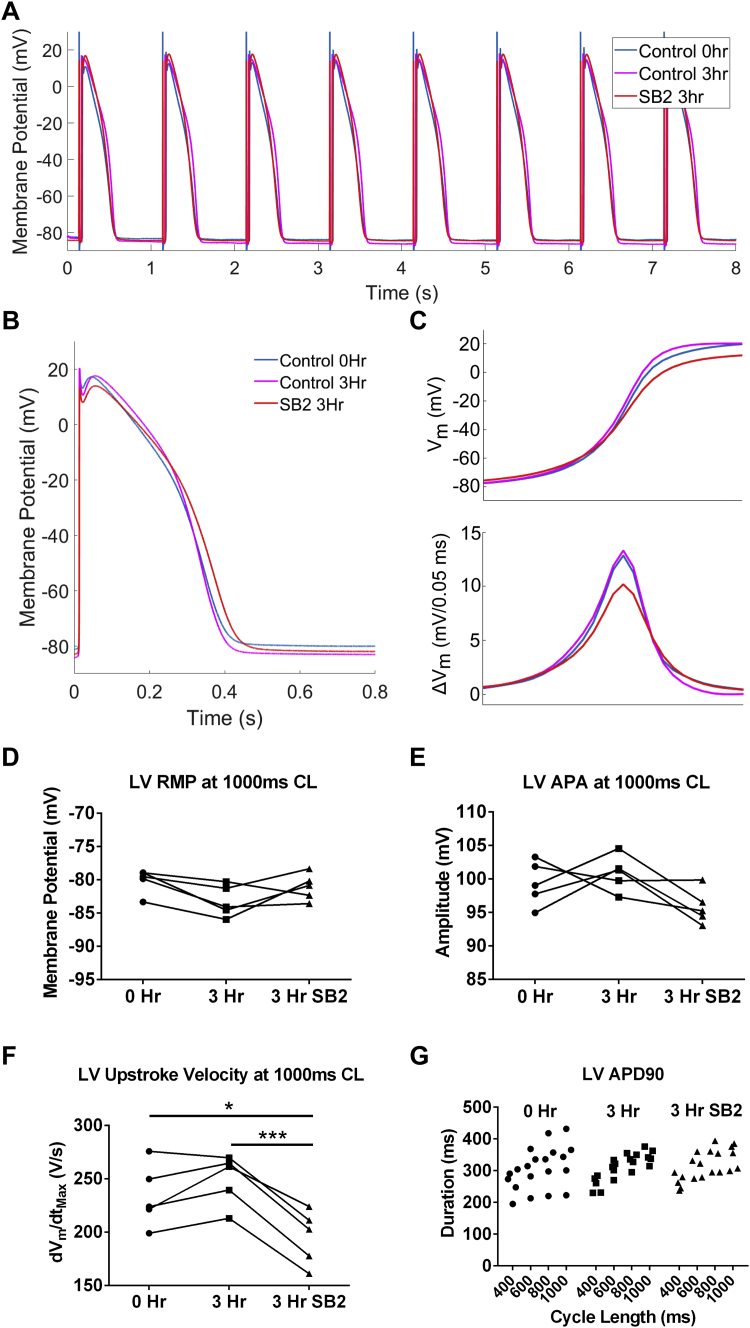
Action potential excitability is decreased by SB2 in human myocardium
To determine more precisely how acute application of SB2 affects cardiac action potentials in a manner that is expected to lead to CV slowing, we performed sharp microelectrode recordings on human organotypic slices. Representative averaged action potentials of slices paced at 1,000 ms from 1 human heart are shown for 0 hours, 3 hours in culture, and 3 hours in culture with SB2 (Figures 2A and 2B). A magnified view to allow for closer inspection of the averaged action potential upstroke reveals that the dVm/dtmax is decreased in SB2-cultured slices (Figure 2C). This reduction was consistently observed in SB2-treated slices from each individual heart (Supplemental Figure 2). Although the RMP, action potential amplitude, and APD at 90% repolarization (APD90) were unchanged between all conditions, dVm/dtmax was decreased by 22% (250 V/s in vehicle control vs 195 V/s in SB2-treated slices) (Figures 2D to 2G). One major determinant regulating cardiac CV is sodium current density and kinetics. The dVm/dtmax of action potentials has been shown to be a direct surrogate of maximum sodium current density and/or channel activation/inactivation.47 Therefore, the observed decrease in dVm/dtmax caused by SB2 suggests either a decrease in sodium current density and/or a leftward shift in channel inactivation contributing to the observed reduction in CV.
Figure 2.
Action Potential Upstroke Velocity Is Decreased in SB2-Treated Slices
(A) Representative action potentials from control without culture (blue), 3 hours in culture (magenta), and 3-hour SB2-cultured (red) slices paced at a cycle length of 1,000 ms. (B) Averaged action potential of all recorded myocytes from slices for each condition. (C) (Top) Zoom of action potential upstroke from B shows decreased dVm/dtmax in SB2-treated slices compared to control and 3-hour cultured control (baseline: 234.1 V/s; 3-hour control: 249.7 V/s; 3-hour SB2: 195.4 V/s). Action potential upstroke change in membrane potential (ΔVm) (bottom) over the same time period shows a significant decrease in dVm/dtmax for SB2 treatment and not a decrease for the entire duration. (D to G) RMP, action potential amplitude, APD90, and APD restitution are not changed between all conditions for each individual heart, whereas dVm/dtmax is significantly decreased in action potentials from slices cultured with SB2 compared with control and cultured control. Statistics were performed using repeated-measures analysis of variance followed by Tukey’s post hoc test for multiple pairwise comparisons. ∗P < 0.05; ∗∗∗P < 0.001. 3Hr = 3 hours; APA = action potential amplitude; APD90 = 90% action potential duration; CL = cycle length; dVm/dtmax = maximum upstroke velocity; LV = left ventricle; RMP = resting membrane potential; SB2 = SB216763.
Computational modeling reveals that reduced GNa and tissue conductivity could account for observed decreases in CV and action potential excitability
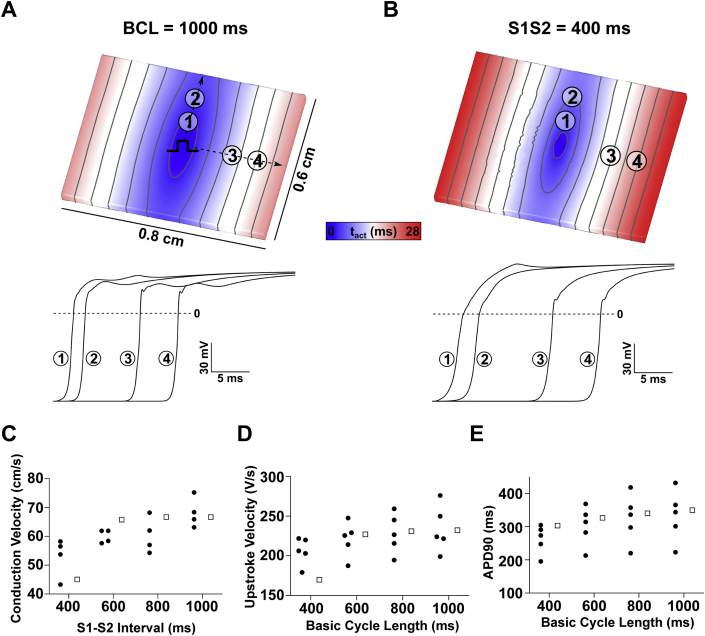
To determine whether reduced GNa can account for the magnitude of conduction slowing, we carried out simulations in a virtual organotypic slice. Conductivity tensor values were calibrated to match experimental CV observations (longitudinal: σL = 0.234 S/m; transverse: σT = 0.0176 S/m); all other input parameters were unchanged from published versions, as described in the Methods section. At baseline, all output parameters predicted by the model at various stimulation cycle lengths matched well with experimentally measured values (Figures 3A to 3E) (eg, at the 1,000-ms basic cycle length, CV = 67 cm/s; APD90 = 350 ms; dVm/dtmax = 232 V/s). Therefore, we are confident that the computational model can be used to predict drug effects on cardiac slice electrophysiology.
Figure 3.
Computational Simulations Conducted in Virtual Cardiac Organotypic Slices Accurately Reproduce Experimental Electrophysiologic Data
(A) Activation sequence in response pacing at a basic cycle length (BCL) of 1,000 ms (top) and membrane voltage over time at points indicated in top graphic (bottom). (B) The same as A except that the excitation in response to a stimulus (S2) was delivered 400 ms later. (C) Longitudinal conduction velocity restitution relationship shows reasonable correspondence between simulations (white squares) and experiments (black circles) at all S1-S2 intervals. (D) Adaptation relationships for upstroke velocity (dVm/dtmax) and (E) 90% action potential duration (APD90) show that the model reproduces values seen in experiments for all BCL values tested.
Both CV and dVm/dtmax are influenced by GNa and sodium-channel kinetics (eg, shifts in voltage-dependent activation/inactivation).48 Therefore, we first asked if changes in 1 or both of these parameters might explain our experimental observations. Using a bisection approach, we found that an 18% CV reduction (as observed experimentally) could be elicited in the model by either a 53.6% GNa decrease or a -8.14-mV shift in INa inactivation kinetics; however, both of these changes resulted in a much more dramatic dVm/dtmax reduction than we observed ex vivo (rows 1 and 2 of Table 1). Likewise, adjusting GNa and INa inactivation shift to different values (19.2% and -3.14 mV, respectively) produced better agreement with upstroke velocity behavior seen ex vivo, but this resulted in divergent CV behavior (rows 3 and 4 of Table 1). Therefore, neither decreased GNa nor shifted INa inactivation kinetics can fully account for the observed drug effects in the experimental data. In contrast, when we examined the effects of either input in conjunction with reduced tissue conductivity (σi), we found combinations that gave rise to simulations with reductions in CV and dVm/dtmax that closely matched those seen ex vivo (rows 5 and 6 of Table 1).
Table 1.
Summary of Simulation Results in Models With Selected Combinations of Reduced GNa, Shifted INa Inactivation Curve, and Reduced σi
| Row | Parameters Modified to Match Ex Vivo CV and dVm/dtmax |
In Silico Percent Error From Target Values |
|||
|---|---|---|---|---|---|
| GNa Reduction, % | INa Inactivation Shift | σi Reduction, % | CV | dVm/dtmax | |
| 1 | 53.60 | n/a | n/a | -0.63 | -40.64 |
| 2 | n/a | -8.14 | n/a | -0.63 | -40.19 |
| 3 | 19.20 | n/a | n/a | 28.42 | -0.05 |
| 4 | n/a | -3.14 | n/a | 28.42 | -0.50 |
| 5 | 25.00 | n/a | 40.00 | -0.63 | 1.13 |
| 6 | n/a | -4.00 | 40.00 | -0.63 | 0.59 |
Rows 1 to 4 show that neither GNa reduction nor shifted INa inactivation kinetics can fully explain the experimentally observed changes in CV and dVm/dtmax. In contrast, when either of the 2 parameters is reduced in combination with σi reduction (rows 5 and 6), model behavior closely matching the experiments is observed. Bold indicate close match (<2% error from target values); italic indicate ≥2% error.
σi = tissue conductivity; CV = conduction velocity; dVm/dtmax = maximum upstroke velocity; GNa = sodium-channel conductance; INa = sodium current; n/a = not applicable.
Acute GSK-3 inhibition by SB2 decreases NaV1.5 protein
To gain further insight into the underlying molecular mechanism(s) for the observed decrease in CV and dVm/dtmax revealed through combining experimental and computational approaches, we performed Western blotting and reverse transcription-quantitative PCR (RT-qPCR) to determine the expression levels of relevant proteins and transcripts. First, we analyzed the protein abundance of Cx43, the major ventricular connexin responsible for cell-to-cell electrical coupling and a major contributor to tissue conductivity. In addition to total amounts, phosphorylated isoforms of Cx43 have been shown to be involved in altered intercellular communication and to influence gap junction assembly.49 Specifically, phosphorylation of Cx43 at serine 368 results in Cx43 mislocalization and reduced channel conductance.50,51 In our experimental system, Cx43 total amounts were not significantly changed between conditions (Supplemental Figure 3A), and additionally, we found no difference in Cx43 phosphorylated at serine 368 between baseline, 3-hour control, and 3-hour SB2 by Western blot (data not shown). Consistent with the Western blot results, immunostaining demonstrated that Cx43 is primarily localized at intercalated discs, with no observable differences in Cx43 localization between baseline, 3-hour control, and 3-hour SB2 (Supplemental Figures 3B to 3D). Therefore, the observed reduction in CV caused by tissue conductivity is not likely attributable to changes in Cx43 protein amount, serine 368 phosphorylation, or localization. Because adherens junctions and intercalated discs can also be regulated by β-catenin,23,24,28 we also performed immunostaining for β-catenin and determined that disc localization of β-catenin was unchanged by SB2 (Supplemental Figures 3B to 3D). Together, these data suggest that CV changes are not caused by altered disc localization of either Cx43 or β-catenin.
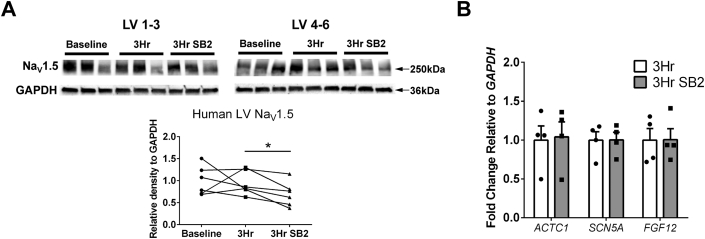
Given the reduced dVm/dtmax in SB2-cultured cardiac slices, we examined sodium-channel protein abundance and found that NaV1.5 was decreased by 30% after SB2 treatment when compared to control slices (Figure 4A). To determine whether this decrease was caused by transcriptional down-regulation, we performed RT-qPCR on 3-hour–treated human slices for SCN5A, which encodes NaV1.5. Interestingly, we determined that SCN5A was not decreased, indicating mechanism(s) independent of transcription underlying the protein level change with acute SB2 treatment (Figure 4B). FGF12, the human homolog of mouse Fgf13 that encodes for fibroblast growth factor 13, has been shown to be down-regulated by Wnt activation52 and has been previously shown to directly modulate sodium current density.53 Therefore, we asked whether this gene is altered by acute GSK-3 inhibition. Similar to SCN5A, FGF12 was not changed by acute 3-hour SB2 treatment (Figure 4C).
Figure 4.
Short-Term SB2 Culture of LV Slices Results in Changes in Nav1.5 Protein But Not Gene Transcription
(A) Western blot for NaV1.5 and GAPDH with quantification based on band density relative to GAPDH shows a decrease in NaV1.5 protein (30% average decrease) in the SB2 condition compared to 3-hour control. (B) Reverse transcription quantitative polymerase chain reaction shows no changes between 3 hours in culture and 3 hours in culture with SB2 for ACTC1 (negative control) or electrophysiology-related transcripts (SCN5A) and FGF12, a known regulator of cardiac sodium current. Statistics in A were performed using repeated-measures analysis of variance followed by Tukey’s post hoc test for multiple pairwise comparisons. Statistics in B were performed using Student’s t-test. ∗P < 0.05. NaV1.5 = pore-forming α-subunit protein of the voltage-gated cardiac sodium channel; other abbreviations as in Figure 2.
In addition to reduced total protein amounts, mislocalization of NaV1.5 could also contribute to CV decreases. Immunostaining revealed that NaV1.5 is localized to the intercalated discs and sarcolemma, with no major differences between localization between baseline, 3-hour control, and 3-hour SB2 (Supplemental Figures 3E to 3G). These results suggest that CV slowing and reduced action potential upstrokes are, at least partially, caused by decreased Nav1.5 protein and reduced sodium conductance.
Acute GSK-3 inhibition increases chromatin-bound β-catenin in human myocardium
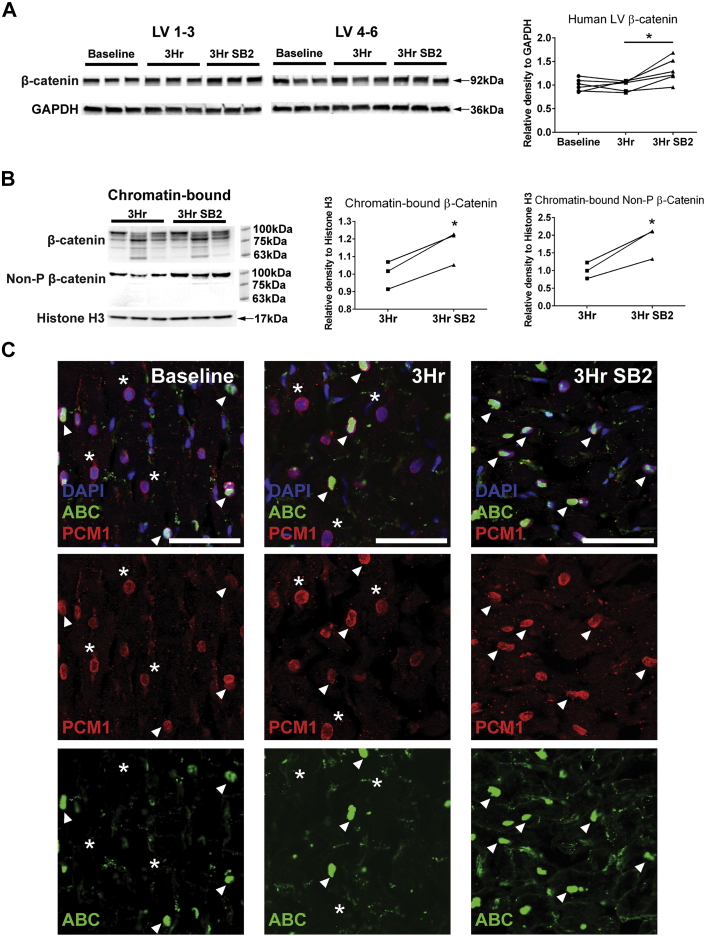
Inhibition of GSK-3 results in β-catenin accumulation and eventual trafficking to the nucleus, where canonical Wnt/β-catenin signaling targets can be activated.54,55 Therefore, we hypothesized that pharmacologic inhibition of GSK-3 through SB2 in the human cardiac slices would result in β-catenin stabilization and eventual downstream activation of the Wnt signaling pathway. We first examined whether inhibition of GSK-3 via SB2 can acutely lead to increased β-catenin levels in the adult human heart. Compared to time zero (baseline) and 3-hour control (vehicle only), slices cultured in SB2 for 3 hours showed a 31% increase in β-catenin protein (Figure 5A). β-catenin is known to localize within the cytoplasm, in the nucleus, and at intercalated discs. Therefore, to further investigate how different pools of β-catenin are affected by treatment with SB2, we fractionated proteins from human slices into cytoplasmic, membrane-bound, soluble nuclear, and chromatin-bound nuclear pools (Supplemental Figure 4A). Within the cytoplasm, membrane fraction, and soluble nuclear fraction, β-catenin levels were not significantly changed after 3 hours in culture between control and SB2 slices (Supplemental Figures 4B to 4D). In contrast, overall β-catenin as well as the nonphosphorylated active form of β-catenin were both significantly increased in the chromatin-bound fraction, suggesting that acute GSK-3 inhibition by SB2 results in β-catenin translocation into the nucleus (Figure 5B). Surprisingly, in addition to a band at 92 kDa, the β-catenin in the chromatin-bound fraction had multiple bands of lower molecular weights compared with the other subcellular fractions. This result is consistent with studies in other cell types that have shown that truncated forms of β-catenin exist exclusively in the nucleus and are involved in gene transactivation.56, 57, 58
Figure 5.
Acute SB2 Treatment Increases β-Catenin Protein in Human LV Slices
(A) Western blot of β-catenin and GAPDH levels with quantification based on band density relative to GAPDH shows an increase in β-catenin (31% average increase) in SB2-treated slices. (B) Both chromatin-bound β-catenin and nonphosphorylated β-catenin levels increased with respect to histone H3 in slices cultured with SB2. In contrast to other subcellular fractions, chromatin-bound β-catenin shows multiple bands of lower molecular weights in addition to the primary band at 92 kDa. (C) ABC (green) is present in human myocyte nuclei, marked by PCM1 (red) and DAPI (blue), in baseline, 3-hour control, and 3-hour SB2 conditions. White arrowheads indicate β-catenin–positive cardiomyocyte nuclei, whereas white asterisks mark β-catenin–negative cardiomyocyte nuclei. Scale = 50 μm. Statistics were performed using repeated-measures analysis of variance followed by Tukey’s post hoc test for multiple pairwise comparisons. ∗P < 0.05. ABC = active β-catenin; DAPI = 4′,6-diamidino-2-phenylindole; non-P = nonphosphorylated; other abbreviations as in Figure 2.
To further confirm the cell fractionation findings, we used immunofluorescence to determine whether ABC (phospho-Y489-β-catenin described as nuclear bound59) localizes within cardiomyocyte nuclei by assessing colocalization with the myocyte nuclei marker pericentriolar material 1 (PCM1). In all conditions, many myocyte nuclei are positive for ABC staining, whereas there are few to no PCM1-negative, noncardiomyocyte nuclei positive for ABC in either the presence or absence of SB2 (Figure 5C). Taken together with the increased chromatin-bound β-catenin seen after SB2 treatment (Figure 5B), these results suggest that β-catenin is increased specifically within the cardiomyocyte nuclei of SB2-cultured slices. β-catenin presence in the nucleus is tightly regulated and, before this study, thought to be scarcely detected in healthy hearts, where Wnt/β-catenin signaling is quiescent. However, we show here that significant amounts of chromatin-bound β-catenin exist in adult human hearts at baseline, with increased nuclear β-catenin levels seen in response to acute GSK-3 inhibition. Despite seeing an increase in chromatin-bound β-catenin and nuclear ABC at the acute 3-hour timepoint, key Wnt target genes, including TCF7L2, LEF1, and AXIN2, were unchanged (Supplemental Figure 5). This indicates that up-regulation of nuclear β-catenin did not acutely result in transcriptional changes associated with active Wnt signaling within the same timeframe of the conduction phenotype, suggesting that conduction effects of GSK-3 inhibition occur, at least partially, through additional post-transcriptional mechanisms.
Induced genetic stabilization of β-catenin in adult cardiomyocytes reduces CV and predisposes to arrhythmias
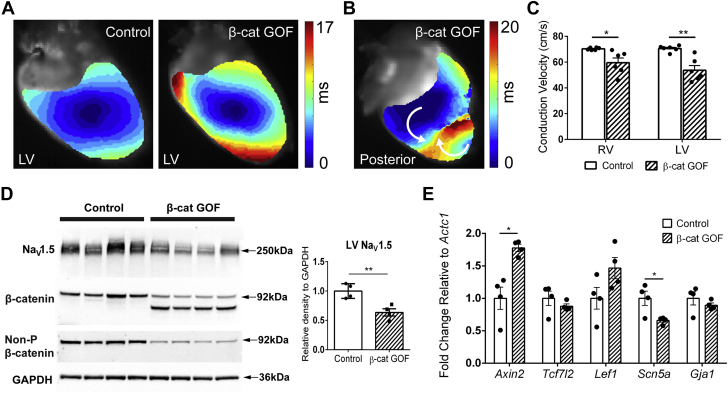
Our laboratory and others have previously shown that developmental chronic β-catenin stabilization in mouse cardiomyocytes results in up-regulation of the Wnt signaling pathway and down-regulation of Scn5a, together with reduced NaV1.5 protein and sodium current density.17,18,22 Therefore, we asked whether adult-induced, chronically increased β-catenin in cardiomyocytes, which partially recapitulates GSK-3–mediated pharmacologic effects, is proarrhythmic. To determine whether cardiac conduction is regulated by β-catenin levels, we used a tamoxifen-inducible mouse model to conditionally delete β-catenin exon 3 in cardiomyocytes of adult mice (αMHC-MCM+/o/Ctnnb1fl(ex3)/+, β-catenin GOF), which prevents β-catenin degradation through loss of phosphorylation sites targeting the protein for ubiquitination. Mice were harvested 4 weeks after an 8-day induction period to investigate the effects of chronic β-catenin stabilization while also allowing recovery from the possibility of confounding tamoxifen-related cardiomyopathy.45 Using optical mapping techniques, we found that electrical activation within tamoxifen-induced β-catenin GOF ventricles is significantly slower when compared to control (combination of the same genotype noninduced control mice and control genotype induced mice) (54 vs 71 cm/s, respectively) (Figure 6A). β-catenin GOF hearts were susceptible to both spontaneous and stimulation-induced ventricular tachycardia (5/6 hearts), whereas littermate controls were completely free from arrhythmias (0/6 hearts). A representative electrical activation map during an episode of ventricular tachycardia shows multiple wave fronts, abnormal propagation patterns, and a line of functional block (Figure 6B). CV was significantly reduced by 15% and 24% in both the right ventricle and LV of β-catenin GOF hearts, respectively (Figure 6C).
Figure 6.
Myocyte-Specific β-Catenin GOF and Acute GSK-3 Inhibition by SB2 Reduces Conduction Velocity in Murine Hearts
(A) Representative activation maps showing slowed electrical activation in β-catenin GOF hearts (right) compared to noninduced controls (left). (B) Activation map showing ventricular tachycardia induced from electrical stimulation of β-catenin GOF ventricles. The white arrows show irregular propagation of 2 different wave fronts, which results in a functional block. Five of 6 β-catenin GOF and 0 of 6 control hearts had spontaneous or stimulation-induced ventricular tachycardia. (C) Conduction velocities of RVs and LVs are significantly reduced in β-catenin GOF hearts compared to controls. (D) Western blot shows expression of stabilized β-catenin (lower-molecule-weight band associated with loss of exon 3) in β-catenin GOF mice and decreased NaV1.5 protein in β-catenin GOF compared to control hearts. (E) Transcription of Wnt/β-catenin target gene Axin2 is increased, and Scn5a is decreased, in β-catenin GOF compared to control hearts. Statistics were performed using Student’s t-test: ∗P < 0.05; ∗∗P < 0.01. β-cat = β-catenin; GOF = gain of function; RV = right ventricle; other abbreviations as in Figure 2.
Next, we sought to determine the mechanism(s) underlying the chronic β-catenin–mediated conduction phenotype and compare to the acute SB2-treated human results. We first verified that β-catenin GOF hearts express both the wild-type band, presumably in noncardiomyocytes, and a truncated β-catenin lacking exon 3 (stabilized β-catenin), resulting in the additional lower-molecular-weight band (Figure 6D). Nonphosphorylated β-catenin antibody, which specifically recognizes residues encoded by exon 3, only detects the wild-type β-catenin from noncardiomyocytes (Figure 6D). Consistent with the 3-hour SB2-treated human LV slices, NaV1.5 protein is significantly decreased in β-catenin GOF compared to control hearts, relative to GAPDH (Figure 6D). In contrast to 3-hour SB2-treated human LV slices, 1 downstream effector of the canonical Wnt signaling pathway, Axin2, was significantly up-regulated, and Scn5a was down-regulated, in β-catenin GOF hearts (Figure 6E). These results show that increased β-catenin in adult cardiomyocytes leads to conduction slowing and susceptibility to ventricular arrhythmias that are likely mediated, as least partially, through decreased NaV1.5, which likely results from both transcriptional and nontranscriptional mechanisms.
Discussion
GSK-3 inhibition has emerged as a potential therapy for a broad spectrum of diseases, ranging from cancer to diabetes to Alzheimer disease.4,6,60 In the heart, studies involving conditional deletion of GSK-3 in mouse models have provided additional evidence that inhibition of GSK-3 can be cardioprotective in settings of myocardial infarction and ischemia.8,61 Recent evidence implicating GSK-3 in the regulation of cellular electrophysiology led us to examine the effects of GSK-3 inhibition on cardiac electrophysiology in the human heart. The ability to measure electrophysiology in 3D cardiac tissue engineered platforms has been important in studying drug effects and heritable arrhythmias.62,63 Our study used organotypic slice culture as a platform for accessing the physiologic response of the human heart to acute GSK-3 inhibition. The unique advantage of the slice culture model for investigating cardiac electrophysiology and molecular mechanisms over isolated cardiomyocytes and induced pluripotent stem cell–derived tissues is a mature, 3-dimensional multicellular model with intercellular connections. We show, for the first time to our knowledge, the effects of GSK-3 inhibition on human cardiac tissue electrophysiology, and an acute timepoint of 3 hours is sufficient to reduce CV and action potential excitability. Finally, via analysis of simulations conducted in virtual organotypic slices, we formulate and subsequently test the hypothesis that reduced sodium-channel expression underlies this effect.
GSK-3 is well known to regulate many different ion channels, and specifically, GSK-3β regulates various calcium, chloride, sodium, and potassium ion channels in the nervous system.64 A study in adrenal chromaffin cells showed that, in addition to regulating GSK-3, lithium may also directly inhibit sodium-channel function.65 Lithium-induced proarrhythmic effects have been described to occur through dosage-dependent block of peak sodium current.12 Conflicting evidence exists on the effects of GSK-3 inhibition on cardiac electrophysiology. In this study, we used a specific inhibitor of GSK-3, namely, SB2, and demonstrated that acute GSK-3 inhibition slows CV in tissue slices from nonfailing human hearts while not affecting APD, suggesting changes to tissue excitability while sparing calcium handling. Additionally, we demonstrated that SB2 reduced the maximum action potential upstroke velocity and NaV1.5 protein in human cardiac tissue. In contrast to these findings, other studies have found SB2 can be beneficial in treating cardiomyopathy progression in cell lines, as well as in murine and zebrafish models of arrhythmogenic cardiomyopathy, through trafficking-mediated reversal of mislocalization of membrane proteins including Cx43 and NaV1.5.9,13 Multiple factors could possibly account for the differences we observed, including differences in model platform used (normal vs disease tissue) and, importantly, the drug dosages and timepoints of the analysis. In our studies, we used adult human organotypic slices from nonfailing donor hearts without gross structural phenotypes, whereas other studies used cell lines, zebrafish, and mouse hearts with desmosomal protein sequence variants that lead to severe structural remodeling. The dosage and chronicity of SB2 used in our human slices also differed from previous animal studies, with an acute application (3 hours in culture vs weeks of daily intraperitoneal injection) and a much lower dosage (10 times the half maximal inhibitory concentration vs 2.5 mg/kg). Additionally, GSK-3β was shown to be elevated in both mouse and human patient samples of arrhythmogenic cardiomyopathy but not in other cardiovascular pathologies.9 Therefore, it is possible that the effects of SB2 may depend on initial levels of GSK-3 activity, and inhibition of hyperactivity vs homeostatic levels may yield different outcomes.
Chromatin-bound β-catenin levels within the cell increased after acute GSK-3 inhibition (Figure 5B). As a tightly regulated transcriptional cofactor, β-catenin localization to the nucleus is typically thought to be associated with transcriptional changes. However, in our acute SB2-treated human slices, Wnt target genes were unchanged, potentially indicating an indirect, nontranscriptional effect of β-catenin at this acute timepoint (Supplemental Figure 5, Figure 4C). However, in our chronic β-catenin GOF mice, where a similar CV reduction is seen after 1 month of induction, Axin2, a Wnt target gene, is up-regulated, and decreased NaV1.5 protein is associated with down-regulation of Scn5a transcript (Figures 6D and 6E). Therefore, it is likely that transcriptional processes may be triggered through more prolonged stabilization of β-catenin, which may be seen with chronic exposure to SB2. Other more rapid processes, such as phosphorylation, SUMOylation, and ubiquitination of NaV1.5,66,67 may mediate the more acute conduction slowing before the transcriptional effects of increased β-catenin. However, in more chronic exposure, these processes may work in parallel with β-catenin–mediated transcriptional regulation to inhibit GNa. Indeed, it is often the case that cell signaling pathways amplify an initial signal, acting at multiple points downstream to inhibit cellular processes leading to pleiotropic effects. Interestingly, we found that β-catenin exists in human cardiomyocyte nuclei at baseline, and multiple β-catenin isoforms of varying molecular weights are bound to chromatin and are not present in other cellular fractions. Consistent with our data presented here, a recent study showed that chromatin-bound β-catenin exists in a repressive complex with GATA4 in cardiomyocytes at baseline, and only in the presence of pathologic stimuli does β-catenin become transcriptionally active.68 Therefore, whether the increase in chromatin-bound β-catenin and/or different β-catenin isoforms observed in the nucleus have significance in gene regulation in the adult human heart remains an area of future investigation.
This paper highlights the possibilities of coupling experimental and computational models of human cardiac electrophysiology to delineate the mechanistic effects of drugs. Using this approach, we observed an average 30% decrease in NaV1.5 protein in human tissue slices cultured with SB2 ex vivo, which could account for at least the magnitude of decrease in dVm/dtmax but would fall short of the observed reduction in CV. Simulation showed that σi must be decreased by 40% in conjunction with a 25% GNa decrease to fully recapitulate the experimentally observed behavior. Simulations showed that σi reduction combined with a modest negative shift in INa channel inactivation kinetics could also explain the particular combination of dVm/dtmax and CV decrease we saw ex vivo, but overall, the reduced NaV1.5 protein is most consistent with GNa decrease. Finally, it is noteworthy that, aside from a single calibration step (ie, the adjustment of baseline σi values to match CV), our model consists entirely of previously published formulations that were derived independent of the present experimental data. As such, we find it quite reassuring that our virtual organotypic slices accurately recapitulated complex electrophysiologic relationships (APD90 and upstroke velocity adaptation). We are thus optimistic that this computational platform could be used in future studies to test (and generate) mechanistic hypotheses for future work in this field.
Study Limitations
Although the methodology used to culture human cardiac organotypic slices enabled the detection of acute electrophysiological changes in response to SB2, assessing chronic changes would likely require additional modification to the human slice culturing methodology, such as pacing and/or mechanical stretch, to preserve slice viability during longer-term culture. Additional limitations of an ex vivo model system include the inability to integrate direct tissue effects with indirect effects due to the sympathetic and parasympathetic nervous system and circulating factors.
Conclusions
Overall, this study uses human organotypic ventricular slices as a robust preclinical platform for studying the effects of drugs on human electrophysiology. We demonstrate that GSK-3 inhibition acutely affects human cardiac electrophysiology in a proarrhythmic manner, partially through NaV1.5 down-regulation. Our study expands the collective understanding of how GSK-3 inhibitors may increase arrhythmogenic potential in patients and, taken together with clinical case reports,12 may warrant frequent monitoring for possible proarrhythmic effects in clinical trials targeting therapeutic GSK-3 inhibition in the treatment of noncardiac diseases.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: Understanding the effects of GSK-3 inhibition on the human heart holds implications for developing noncardiac therapeutics targeting the GSK-3 pathway. A better understanding of how human cardiac electrophysiology is affected by drug treatment will provide safer recommendations for future clinical trials and the everyday use of this class of drugs.
TRANSLATIONAL OUTLOOK: Organotypic cardiac slices provide a robust experimental model of the human heart, with mature cells in their native cellular organization, allowing for the mechanistic study of the effects of drugs on cardiac physiology. In addition to electrophysiology, this platform can be applied to examine mechanobiology, metabolism, transcriptomics, and proteomics under various experimental conditions or disease states.
Funding Support and Author Disclosures
This study has received grants from the National Science Foundation (NSF) and the National Institutes of Health (NIH): NSF GRFP DGE-1745038 and NIH T32 HL125241-03 to Ms Brumback, NIH T32HL007081 to Dr Jimenez, NIH T32HL134635 to Mr Zhang, and NIH R01 HL 130212 and UH3 HL 141800 to Dr Rentschler. Dr Rentschler holds a Career Award for Medical Scientists from the Burroughs Wellcome Fund. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank Dr Michael K. Pasque and Mid-America Transplant Services for procurement of heart tissue and the families of the organ donors for their generous gifts that make this research possible. They also thank the National Science Foundation (NSF) and the National Institutes of Health (NIH) for their ongoing support
Footnotes
Junichi Sadoshima, MD, served as Guest Associate Editor for this paper. Michael Bristow, MD, PhD, served as Guest Editor-in-Chief for this paper.
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures, please see the online version of this paper.
Appendix
References
- 1.Beurel E., Grieco S.F., Jope R.S. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–131. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jope R.S. Glycogen synthase kinase-3 in the etiology and treatment of mood disorders. Front Mol Neurosci. 2011;4:1–11. doi: 10.3389/fnmol.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorge-Torres O.C., Szczesna K., Roa L., et al. Inhibition of Gsk3b reduces Nfkb1 signaling and rescues synaptic activity to improve the Rett syndrome phenotype in Mecp2-knockout mice. Cell Rep. 2018;23:1665–1677. doi: 10.1016/j.celrep.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Mines M.A., Beurel E., Jope R.S. Regulation of cell survival mechanisms in Alzheimer’s disease by glycogen synthase kinase-3. Int J Alzheimers Dis. 2011;2011:861072. doi: 10.4061/2011/861072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haq S., Choukroun G., Kang Z.B., et al. Glycogen synthase kinase-3β is a negative regulator of cardiomyocyte hypertrophy. J Cell Biol. 2000;151:117–130. doi: 10.1083/jcb.151.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lal H., Ahmad F., Woodgett J., Force T. The GSK-3 family as therapeutic target for myocardial diseases. Circ Res. 2015;116:138–149. doi: 10.1161/CIRCRESAHA.116.303613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y., Feng W., Xue W., et al. Inactivation of GSK-3β by metallothionein prevents diabetes-related changes in cardiac energy metabolism, inflammation, nitrosative damage, and remodeling. Diabetes. 2009;58:1391–1402. doi: 10.2337/db08-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhai P., Sciarretta S., Galeotti J., Volpe M., Sadoshima J. Differential roles of GSK-3β during myocardial ischemia and ischemia/reperfusion. Circ Res. 2011;109:502–511. doi: 10.1161/CIRCRESAHA.111.249532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chelko S.P., Asimaki A., Andersen P., et al. Central role for GSK3β in the pathogenesis of arrhythmogenic cardiomyopathy. JCI Insight. 2016;1:85923. doi: 10.1172/jci.insight.85923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haq S., Choukroun G., Lim H., et al. Differential activation of signal transduction pathways in human hearts with hypertrophy versus advanced heart failure. Circulation. 2001;103:670–677. doi: 10.1161/01.cir.103.5.670. [DOI] [PubMed] [Google Scholar]
- 11.Zhou J., Ahmad F., Parikh S., et al. Loss of adult cardiac myocyte GSK-3 leads to mitotic catastrophe resulting in fatal dilated cardiomyopathy. Circ Res. 2016;118:1208–1222. doi: 10.1161/CIRCRESAHA.116.308544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darbar D., Yang T., Churchwell K., Wilde A.A.M., Roden D.M. Unmasking of Brugada syndrome by lithium. Circulation. 2005;112:1527–1531. doi: 10.1161/CIRCULATIONAHA.105.548487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asimaki A., Kapoor S., Plovie E., et al. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med. 2014;6:240ra74. doi: 10.1126/scitranslmed.3008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nusse R., Clevers H. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Li V.S.W., Ng S.S., Boersema P.J., et al. Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell. 2012;149:1245–1256. doi: 10.1016/j.cell.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Gras E., Lombardi R., Giocondo M.J., et al. Suppression of canonical Wnt/beta-catenin signaling by nuclear plakoglobin recapitulates phenotype of arrhythmogenic right ventricular cardiomyopathy. J Clin Invest. 2006;116:2012–2021. doi: 10.1172/JCI27751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillers B.S., Chiplunkar A., Aly H., et al. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ Res. 2015;116:398–406. doi: 10.1161/CIRCRESAHA.116.304731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li G., Khandekar A., Yin T., et al. Differential Wnt-mediated programming and arrhythmogenesis in right versus left ventricles. J Mol Cell Cardiol. 2018;123:92–107. doi: 10.1016/j.yjmcc.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai Z., Fischer A., Spray D.C., Brown A.M., Fishman G.I. Wnt-1 regulation of connexin43 in cardiac myocytes. J Clin Invest. 2000;105:161–171. doi: 10.1172/JCI7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang W., Cho H.C., Marbán E. Wnt signalling suppresses voltage-dependent Na+ channel expression in postnatal rat cardiomyocytes. J Physiol. 2015;593:1147–1157. doi: 10.1113/jphysiol.2014.285551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang N., Huo R., Cai B., et al. Activation of Wnt/β-catenin signaling by hydrogen peroxide transcriptionally inhibits NaV1.5 expression. Free Radic Biol Med. 2016;96:34–44. doi: 10.1016/j.freeradbiomed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huo R., Hu C., Zhao L., et al. Enhancement of β-catenin/T-cell factor 4 signaling causes susceptibility to cardiac arrhythmia by suppressing NaV1.5 expression in mice. Heart Rhythm. 2019;16:1720–1728. doi: 10.1016/j.hrthm.2019.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haÿ E., Laplantine E., Geoffroy V., et al. N-cadherin interacts with axin and LRP5 to negatively regulate Wnt/beta-catenin signaling, osteoblast function, and bone formation. Mol Cell Biol. 2009;29:953–964. doi: 10.1128/MCB.00349-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson W.J., Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luckey S.W., Walker L.A., Smyth T., et al. The role of Akt/GSK-3beta signaling in familial hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2009;46:739–747. doi: 10.1016/j.yjmcc.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masuelli L., Bei R., Sacchetti P., et al. Beta-catenin accumulates in intercalated disks of hypertrophic cardiomyopathic hearts. Cardiovasc Res. 2003;60:376–387. doi: 10.1016/j.cardiores.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Stauffer B.L., Konhilas J.P., Luczak E.D., Leinwand L.A. Soy diet worsens heart disease in mice. J Clin Invest. 2006;116:209–216. doi: 10.1172/JCI24676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spagnol G., Trease A.J., Zheng L., et al. Connexin43 carboxyl-terminal domain directly interacts with β-catenin. Int J Mol Sci. 2018;19:1–12. doi: 10.3390/ijms19061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kernik D.C., Morotti S., Wu H., et al. A computational model of induced pluripotent stem-cell derived cardiomyocytes incorporating experimental variability from multiple data sources. J Physiol. 2019;597:4533–4564. doi: 10.1113/JP277724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang P.-C., DeMarco K.R., Aghasafari P., et al. A computational pipeline to predict cardiotoxicity: from the atom to the rhythm. Circ Res. 2020;126:947–964. doi: 10.1161/CIRCRESAHA.119.316404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang C., Qiao Y., Li G., et al. Human organotypic cultured cardiac slices: new platform for high throughput preclinical human trials. Sci Rep. 2016;6:1–13. doi: 10.1038/srep28798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lang D., Sulkin M., Lou Q., Efimov I.R. Optical mapping of action potentials and calcium transients in the mouse heart. J Vis Exp. 2011;55:1–6. doi: 10.3791/3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khandekar A., Springer S., Wang W., et al. Notch-mediated epigenetic regulation of voltage-gated potassium currents. Circ Res. 2016;119:1324–1338. doi: 10.1161/CIRCRESAHA.116.309877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen C.L., Jensen J.L., Ørntoft T.F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 35.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 36.Vandesompele J., De Preter K., Pattyn F., et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):1–12. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O’Hara T., Virág L., Varró A., Rudy Y. Simulation of the undiseased human cardiac ventricular action potential: model formulation and experimental validation. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Priest J.R., Gawad C., Kahlig K.M., et al. Early somatic mosaicism is a rare cause of long-QT syndrome. Proc Natl Acad Sci U S A. 2016;113:11555–11560. doi: 10.1073/pnas.1607187113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ten Tusscher K.H.W.J., Noble D., Noble P.J., Panfilov A.V. A model for human ventricular tissue. Am J Physiol Heart Circ Physiol. 2004;286:H1573–H1589. doi: 10.1152/ajpheart.00794.2003. [DOI] [PubMed] [Google Scholar]
- 40.Vigmond E.J., Hughes M., Plank G., Leon L.J. Computational tools for modeling electrical activity in cardiac tissue. J Electrocardiol. 2003;36(suppl):69–74. doi: 10.1016/j.jelectrocard.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Vigmond E.J., Weber dos Santos R., Prassl A.J., Deo M., Plank G. Solvers for the cardiac bidomain equations. Prog Biophys Mol Biol. 2008;96:3–18. doi: 10.1016/j.pbiomolbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.openCARP. Accessed May 19, 2022. Available at: https://opencarp.org/
- 43.Sohal D.S., Nghiem M., Crackower M.A., et al. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 44.Harada N., Tamai Y., Ishikawa T., et al. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. EMBO J. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koitabashi N., Bedja D., Zaiman A.L., et al. Avoidance of transient cardiomyopathy in cardiomyocyte-targeted tamoxifen-induced MerCreMer gene deletion models. Circ Res. 2009;105:12–15. doi: 10.1161/CIRCRESAHA.109.198416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masarone D., Limongelli G., Rubino M., et al. Management of arrhythmias in heart failure. J Cardiovasc Dev Dis. 2017;4:1–20. doi: 10.3390/jcdd4010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kléber A.G. The shape of the electrical action-potential upstroke: a new aspect from optical measurements on the surface of the heart. Circ Res. 2005;97:204–206. doi: 10.1161/01.RES.0000177922.62341.f2. [DOI] [PubMed] [Google Scholar]
- 48.Boyle P.M., Franceschi W.H., Constantin M., et al. New insights on the cardiac safety factor: unraveling the relationship between conduction velocity and robustness of propagation. J Mol Cell Cardiol. 2019;128:117–128. doi: 10.1016/j.yjmcc.2019.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Márquez-Rosado L., Solan J.L., Dunn C.A., Norris R.P., Lampe P.D. Connexin43 phosphorylation in brain, cardiac, endothelial and epithelial tissues. Biochim Biophys Acta. 2012;1818:1985–1992. doi: 10.1016/j.bbamem.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ek-Vitorin J.F., King T.J., Heyman N.S., Lampe P.D., Burt J.M. Selectivity of connexin 43 channels is regulated through protein kinase C-dependent phosphorylation. Circ Res. 2006;98:1498–1505. doi: 10.1161/01.RES.0000227572.45891.2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hund T.J., Lerner D.L., Yamada K.A., Schuessler R.B., Saffitz J.E. Protein kinase Cepsilon mediates salutary effects on electrical coupling induced by ischemic preconditioning. Heart Rhythm. 2007;4:1183–1193. doi: 10.1016/j.hrthm.2007.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kinoshita-Ise M., Tsukashima A., Kinoshita T., Yamazaki Y., Ohyama M. Altered FGF expression profile in human scalp-derived fibroblasts upon WNT activation: implication of their role to provide folliculogenetic microenvironment. Inflamm Regen. 2020;40:1–10. doi: 10.1186/s41232-020-00141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C., Hennessey J.A., Kirkton R.D., et al. Fibroblast growth factor homologous factor 13 regulates Na+ channels and conduction velocity in murine heart. Circ Res. 2011;109:775–782. doi: 10.1161/CIRCRESAHA.111.247957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 55.Clevers H., Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 56.Morgan R.G., Ridsdale J., Payne M., et al. LEF-1 drives aberrant β-catenin nuclear localization in myeloid leukemia cells. Haematologica. 2019;104:1365–1377. doi: 10.3324/haematol.2018.202846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Goretsky T., Bradford E.M., Ye Q., et al. Beta-catenin cleavage enhances transcriptional activation. Sci Rep. 2018;8:1–15. doi: 10.1038/s41598-017-18421-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Su H., Sureda-Gomez M., Rabaneda-Lombarte N., et al. A C-terminally truncated form of β-catenin acts as a novel regulator of Wnt/β-catenin signaling in planarians. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1007030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhee J., Buchan T., Zukerberg L., Lilien J., Balsamo J. Cables links Robo-bound Abl kinase to N-cadherin-bound beta-catenin to mediate Slit-induced modulation of adhesion and transcription. Nat Cell Biol. 2007;9:883–892. doi: 10.1038/ncb1614. [DOI] [PubMed] [Google Scholar]
- 60.McCubrey J.A., Steelman L.S., Bertrand F.E., et al. GSK-3 as potential target for therapeutic intervention in cancer. Oncotarget. 2014;5:2881–2911. doi: 10.18632/oncotarget.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murphy E., Steenbergen C. Inhibition of GSK-3beta as a target for cardioprotection: the importance of timing, location, duration and degree of inhibition. Expert Opin Ther Targets. 2005;9:447–456. doi: 10.1517/14728222.9.3.447. [DOI] [PubMed] [Google Scholar]
- 62.Blazeski A., Lowenthal J., Wang Y., et al. Engineered heart slice model of arrhythmogenic cardiomyopathy using plakophilin-2 mutant myocytes. Tissue Eng Part A. 2019;25:725–735. doi: 10.1089/ten.tea.2018.0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blazeski A., Lowenthal J., Zhu R., Ewoldt J., Boheler K.R., Tung L. Functional properties of engineered heart slices incorporating human induced pluripotent stem cell-derived cardiomyocytes. Stem Cell Rep. 2019;12:982–995. doi: 10.1016/j.stemcr.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sopjani M., Millaku L., Nebija D., Emini M., Rifati-Nixha A., Dërmaku-Sopjani M. The glycogen synthase kinase-3 in the regulation of ion channels and cellular carriers. Curr Med Chem. 2019;26:6817–6829. doi: 10.2174/0929867325666181009122452. [DOI] [PubMed] [Google Scholar]
- 65.Yanagita T., Maruta T., Uezono Y., et al. Lithium inhibits function of voltage-dependent sodium channels and catecholamine secretion independent of glycogen synthase kinase-3 in adrenal chromaffin cells. Neuropharmacology. 2007;53:881–889. doi: 10.1016/j.neuropharm.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 66.Tang B., Hu Y., Wang Z., et al. UBC9 regulates cardiac sodium channel Nav1.5 ubiquitination, degradation and sodium current density. J Mol Cell Cardiol. 2019;129:79–91. doi: 10.1016/j.yjmcc.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marionneau C., Abriel H. Regulation of the cardiac Na+ channel NaV1.5 by post-translational modifications. J Mol Cell Cardiol. 2015;82:36–47. doi: 10.1016/j.yjmcc.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 68.Iyer L.M., Nagarajan S., Woelfer M., et al. A context-specific cardiac β-catenin and GATA4 interaction influences TCF7L2 occupancy and remodels chromatin driving disease progression in the adult heart. Nucleic Acids Res. 2018;46:2850–2867. doi: 10.1093/nar/gky049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.