Abstract
Nineteen isolates belonging to a cryptic genospecies of Haemophilus (referred to here as genital strains) isolated from genital tract infections (6 strains) and from neonatal infections (13 strains) were studied for fimbrial genes. Sixteen strains exhibit peritrichous fimbriae observed by electron microscopy. By PCR with primers corresponding to the extreme ends of the Haemophilus influenzae type b (Hib) hifA and hifD genes and Southern blotting, a hifA-like gene (named ghfA) and a hifD-like gene (named ghfD) were identified in 6 of the 19 strains. Five of these six strains were from the genital tracts of adults, and one was from a neonate. For each gene, the nucleotide sequence was identical for the six strains. A hifE-like gene (named ghfE) was amplified from only one of the 19 genital strains of Haemophilus, but the ghfE probe gave a signal in Southern hybridization with the five other strains positive for ghfA and ghfD. Therefore, these strains may carry a ghfE-like gene. The Hib fimbrial gene cluster is located between the purE and pepN genes as previously described. For the 13 genital Haemophilus strains that lack fimbrial genes, this region corresponds to a noncoding sequence. Another major fimbrial gene designated the fimbrin gene was previously identified in a nontypeable H. influenzae strain. A fimbrin-like gene was identified for all of our 19 genital strains. This gene is similar to the ompP5 gene of many Haemophilus strains. Therefore, other, unidentified genes may explain the piliation observed in electron microscopy on genital Haemophilus strains which do not possess LKP-like fimbrial genes. Fimbrial genes were significantly associated with strains isolated from the genital tract. They may confer on the strain the ability to survive in the genital tract.
Haemophilus influenzae strains are gram-negative rods which colonize human respiratory and genital mucosa. Although they are commensal bacteria, they can cause serious respiratory tract infections, meningitis, and genital and neonatal infections. Adhesion to epithelial cells is the first step in host colonization by many bacteria (5, 9). Adhesion can be mediated by nonfimbrial and fimbrial structures. Most nontypeable strains of Haemophilus (95%) adhere to human epithelial cells either by high-molecular-weight surface proteins (84%) or by Hia protein (16%) (4, 34, 35). Short, thin surface fibrils involved in H. influenzae type b (Hib) adherence to human epithelial cells were recently identified (31, 32). Both Hib and nontypeable H. influenzae (NTHi) strains commonly express fimbriae, which are polymeric structures composed of a major structural protein associated with several other minor proteins. The fimbrial proteins of a large number of gram-negative bacteria have been purified, and their sequences have been determined. The major fimbrial proteins from different species are often very similar (14, 15, 36, 38). Fimbria-mediated adhesion to cells has also been reported for H. influenzae strains that colonize the nasopharynx and that are responsible for various infections, including meningitis, chronic bronchitis, otitis media, and Brazilian purpuric fever (3, 12, 21, 29, 37).
The genes coding for the major fimbrial subunit (hifA genes) in several Hib strains and in some NTHi strains responsible for various diseases have been characterized. The deduced amino acid sequences of these proteins are about 80% similar, and the amino acid sequences are particularly well conserved at the N- and C-terminal ends (2, 7, 10, 15, 33, 38, 42). The genes from some Hib strains coding for minor proteins that constitute fimbriae (hifD and hifE genes) have been characterized (18, 39). These two genes map downstream from the hifA gene. The complete fimbrial gene cluster in Hib has been identified and lies between the purE and pepN genes (11, 39). The adhesive component in H. influenzae fimbriae has not been clearly defined; it could be the major subunit (HifA) or HifE, a minor subunit located at the tips of fimbriae (17, 40).
Another fimbrial gene in a strain of NTHi has been characterized. It encodes a fimbrin protein, an adhesin homologous to OmpA proteins of many gram-negative bacteria (30).
In the last 20 years, urogenital, mother-infant, and neonatal infections caused by Haemophilus strains have been reported with increasing frequency (1, 16, 23, 41). The members of a group of Haemophilus strains isolated from neonatal and genital tract infections (referred to here as genital strains) and commonly identified as H. influenzae biotype IV have several unusual features (19, 24, 26). Genetic analysis of these strains demonstrates that they constitute a cryptic genospecies which forms a monophyletic unit with H. influenzae and Haemophilus haemolyticus, most closely related to H. haemolyticus (25, 27). These isolates may have a specific tropism for the genital tract. They were not detected among a large number of H. influenzae isolates recovered from invasive, respiratory, and other infections in children and adults (1, 20). Most express peritrichous fimbriae, adhere better to HeLa cells than to HEp-2 cells and do not cause agglutination of human erythrocytes expressing the AnWj antigen (28), contrasting with numerous fimbriated H. influenzae strains responsible for respiratory infections and meningitis (3, 12, 21, 37). Therefore, if these fimbriae are involved in the colonization of the genital tract, they may differ from those produced by strains isolated from the respiratory tract.
Our objective was to explore a group of 19 strains genetically assigned to the cryptic genital Haemophilus genospecies (27) for the presence of fimbrial genes. In addition, this population was investigated for the presence of the gene coding for fimbrin. When present, these genes were sequenced, and the sequences were compared to those of previously described genes coding for proteins that are components of the fimbriae of H. influenzae. If no such gene was detected, the absence of these genes was confirmed by Southern blotting experiments and the nucleotide sequence between the purE and pepN genes was analyzed. For each isolate, the expression and the morphology of fimbriae were studied by electron microscopy.
MATERIALS AND METHODS
Bacterial strains.
Thirty-one Haemophilus strains were studied, including 1 H. influenzae biotype I strain, 1 H. influenzae biotype II strain, 1 H. influenzae biotype III strain, 6 H. influenzae biotype IV sensu stricto strains, 2 H. haemolyticus strains, 1 H. influenzae biogroup Aegyptius strain, and 19 strains previously assigned to the cryptic genital Haemophilus genospecies on the basis of DNA-DNA hybridization and small-subunit ribosomal DNA sequencing (25, 27). Genital strains of Haemophilus from the United States were kindly provided by J. M. Musser (Department of Pathology, Baylor College of Medicine, Houston, Tex.). The anatomic and geographic origins of these strains are reported in Table 1. Strains were stored at −80°C in Schaedler-vitamin K3 broth (bioMérieux, Marcy l’Etoile, France) with 10% glycerol. A heavily fimbriated acapsular variant of H. influenzae (strain 770235), kindly provided by L. van Alphen (Department of Medical Microbiology, University of Amsterdam, Amsterdam, The Netherlands), was used as a control. This strain harbors long, thick, hemagglutination-positive (LKP) pili. The nucleotide sequences of the genes coding for proteins constituting the fimbriae of this strain have been reported by van Ham et al. (39).
TABLE 1.
Identification, origin, piliation, and presence of fimbrial genes for Haemophilus strains
| Strain | Identificationa | Biotype (capsular type) | Origin | Anatomic site | Presence ofc:
|
|||
|---|---|---|---|---|---|---|---|---|
| Fimbriae | ghfA | ghfD | ghfE | |||||
| Control strain 770235 | H. influenzae | I (b) | The Netherlands | Cerebrospinal fluid | + | + | + | + |
| Collection strains | ||||||||
| CIP 52152 | H. influenzae | I (b) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 102514 | H. influenzae | II (NTb) | Unknown | Unknown | + | 0 | 0 | 0 |
| CIP 102284 | H. influenzae | III (NT) | France | Sputum | ND | 0 | 0 | 0 |
| CIP 5424 | H. influenzae | IV (NT) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 5483 | H. influenzae | IV (d) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 5494 | H. influenzae | IV (b) | United States | Cerebrospinal fluid | + | + | + | + |
| CIP 5484 | H. influenzae | IV (e) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 52154 | H. influenzae | IV (d) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 52155 | H. influenzae | IV (e) | Unknown | Unknown | 0 | 0 | 0 | 0 |
| CIP 103290 | H. haemolyticus | Unknown | Unknown | + | 0 | 0 | 0 | |
| CIP 102348 | H. haemolyticus | France | Sputum | 0 | 0 | 0 | 0 | |
| CIP 52129 | H. influenzae biogroup Aegyptius | United States | Conjunctiva | 0 | + | 0 | + | |
| Clinical isolates | ||||||||
| 189 | Genital Haemophilus | United States | Amniotic fluid | +d | 0 | 0 | 0 | |
| 421 | Genital Haemophilus | United States | Blood of neonate | 0 | 0 | 0 | 0 | |
| 422 | Genital Haemophilus | United States | Blood of neonate | 0 | 0 | 0 | 0 | |
| 427 | Genital Haemophilus | United States | Amniotic fluid | 0 | 0 | 0 | 0 | |
| 799 | Genital Haemophilus | United States | Blood of neonate | + | 0 | 0 | 0 | |
| 847 | Genital Haemophilus | United States | Scrotal abcess | + | 0 | 0 | 0 | |
| 911 | Genital Haemophilus | United States | Cerebrospinal fluid of neonate | + | 0 | 0 | 0 | |
| 1595 | Genital Haemophilus | United States | Blood of neonate | + | 0 | 0 | 0 | |
| 1610 | Genital Haemophilus | United States | Blood of neonate | + | 0 | 0 | 0 | |
| 3N | Genital Haemophilus | France | Gastric fluid of neonate | +d | 0 | 0 | 0 | |
| 10N | Genital Haemophilus | France | Gastric fluid of neonate | + | 0 | 0 | 0 | |
| 12N | Genital Haemophilus | France | Gastric fluid of neonate | + | 0 | 0 | 0 | |
| 15N | Genital Haemophilus | France | Gastric fluid of neonate | + | + | + | 0 | |
| 16N | Genital Haemophilus | France | Gastric fluid of neonate | + | 0 | 0 | 0 | |
| 10U | Genital Haemophilus | France | Urethra (male) | + | + | + | 0 | |
| 11PS | Genital Haemophilus | France | Urethra (male) | + | + | + | 0 | |
| 26E | Genital Haemophilus | France | Uterus | + | + | + | + | |
| PIZ | Genital Haemophilus | France | Endocervix | + | + | + | 0 | |
| 2406 | Genital Haemophilus | France | Vagina | + | + | + | 0 | |
Genital Haemophilus strains were phenotypically identified as H. influenzae biotype IV, except strain 2406, which was phenotypically identified as H. parainfluenzae.
NT, nontypeable.
+, present; 0, absent; ND, not determined. The presence of fimbriae was determined by electron microscopy; the presence of fimbrial genes (ghfA, ghfD, and ghfE) was assessed by PCR and confirmed by Southern blotting.
This strain expressed fimbriae only after enrichment.
Electron microscopy.
The presence and appearance of fimbriae were examined by electron microscopy before and after selection of piliated Haemophilus strains as previously described (28). Bacteria grown to stationary phase were washed three times in saline buffer, settled onto 400-mesh copper grids coated with carbon film, negatively stained with a 1.5% uranyl acetate solution in distilled water as previously described (28), and then examined with a JEOL 1010 electron microscope at 80 kV.
Genomic DNA extraction.
Each strain was subcultured on four chocolate agar plates (15 by 15 cm) for 18 to 24 h at 37°C in 8% CO2. The cultures were checked visually for purity, harvested in 15 ml of buffer (40 mM Tris, 2 mM EDTA, pH 8), and lysed by adding 150 μl of a 25% (wt/vol) aqueous solution of sodium dodecyl sulfate and 22.5 μl of 2% self-digested pronase (Sigma, St. Louis, Mo.). The mixture was incubated overnight at 37°C, and DNA was extracted and purified as previously described (6), dialyzed on 0.025-μm-pore-size filters (Millipore, St Quentin en Yvelines, France), and diluted in water to a concentration of 10 μg/ml.
PCR assay.
One hundred nanograms of DNA was used for PCR. Eleven primers were used to amplify genes coding for a major fimbrial subunit (Eurogentec, Seraing, Belgium) (Tables 2 and 3). Primers N1, N34, N51, N69, C189, C198, and C207 were designed to correspond to conserved sequences in the hifA genes of Hib 770235, AO2, and Eagan, NTHi M37 and 1128, and H. influenzae biogroup Aegyptius, described previously (Fig. 1 and Table 2) (2, 8, 10, 39, 42). Primers were named as follows: the letters N and C indicate the localization of the primer sequence in the 5′ and 3′ ends of the gene, respectively, and the number following the letter corresponds to the position of the first or the last translated codon, respectively (Fig. 1). The Crypt N and Crypt C primers were designed to correspond to two regions that are different in the major fimbrial protein genes of genital Haemophilus strains and of H. influenzae (Fig. 1 and Table 2). Primers fim5 and fim3 were deduced from a gene coding for the fimbrial subunit (fimbrin) of the NTHi 1128 strain sequenced by Sirakova et al. (30) (Table 3).
TABLE 2.
Nucleotide sequences of PCR primers used to amplify a hifA-like gene from genital Haemophilus strains
| Primera | Nucleotide sequence |
|---|---|
| N1b | 5′ CTCTTTATGGAGCAATTTATTATG 3′ |
| N34b | 5′ GGTAAGGTTGTTGAGAATACTTGT 3′ |
| N51 | 5′ AGTGTAGTATTAAATGATGTGGTTAAA 3′ |
| N69 | 5′ GCAATGCCAACGCCATTTACGATT 3′ |
| C186 | 5′ GTAATATTGGGCGATAAAGTGGAG 3′ |
| C198 | 5′ GAGAACATGAAATGGTCGTCAACG 3′ |
| C207b | 5′ TTATTCGTAAGCAATTTGGAAATTTACTGA 3′ |
| Crypt Nb | 5′ ACTGATTGTGCTGTAGCAAATGGA 3′ |
| Crypt Cb | 5′ AGTTTTACCATTTAATTCAGTTGC 3′ |
Primers N1, N34, N51, and N69 were identical to the leading strand; primers C186, C198, C207, and Crypt C were identical to the lagging strand.
This primer gave an amplification product.
TABLE 3.
Nucleotide sequences of PCR primers used to amplify a fimbrin-like gene and genes coding for minor fimbrial proteins from genital Haemophilus strains
| Primera | Nucleotide sequence | Positionb |
|---|---|---|
| fim5 | 5′ GGACATCAAAATGAAAAAAACTGC 3′ | 396 |
| fim3 | 5′ TTATTTAGTACCGTTTACTGCGAT 3′ | 1485 |
| hifD5 | 5′ CAAAAAACACCCAAAAAATTAACC 3′ | 5467 |
| hifD3 | 5′ AGTTATAACTGCACTTGAAAAGTT 3′ | 6112 |
| hifE5 | 5′ ATGAAAACTTTAACAACATACGCA 3′ | 6138 |
| hifE3 | 5′ ATTGATATGACATTGTGAAAGTGG 3′ | 7443 |
| purE | 5′ GACCCCATCACAACGGCAATTTGT 3′ | 447 |
| pepN | 5′ CTGTGACCGTAAAATCTGGTTGTT 3′ | 7017 |
Primers fim5, hifD5, hifE5, and purE were identical to the leading strand; primers fim3, hifD3, hifE3, and pepN were identical to the lagging strand.
FIG. 1.

Amino acid sequence deduced from major fimbrial protein gene sequence of genital Haemophilus strain. The sequence is compared with previously reported HifA sequences (2, 8, 13, 38, 42). Amino acid residues chosen as the basis of primer sequences are double underlined, and the name of each primer is shown above the line. Dots represent identity. Dashes indicate gaps introduced to optimize alignment. H. Aegyptius; H. influenzae biogroup Aegyptius; genital H., genital cryptic strains of Haemophilus.
Two sets of primers were used to amplify the genes coding for minor proteins. Each set of primers was defined at the extreme ends of the hifD and hifE genes of Hib strain 770235 (39) (Table 3).
In addition, two primers (purE and pepN) (Table 3) correspond to the ends of the purE and pepN genes, respectively. These two genes were previously demonstrated to flank the Hib fimbrial gene cluster (11, 39). We used these primers to verify that a fimbrial cluster was not present in Haemophilus genital strains which did not possess previously described major or minor fimbrial protein genes.
The PCR mixture (20 μl) contained primers (0.5 μM each), genomic DNA (100 ng), deoxynucleoside triphosphates (100 μM each) (Boehringer, Mannheim, Germany), Taq polymerase (1.5 U) (Appligène, Illkirch, France), MgCl2 (1.5 mM), 10 mM Tris HCl (pH 8.3), and 50 mM KCl. The PCR consisted of a first long denaturation step (5 min at 95°C); 25 cycles each of 1 min of denaturation at 95°C, 2 min of annealing at 50°C, and 2 min of elongation at 72°C; and then a 10-min final elongation step (Cetus 480; Perkin-Elmer Cetus, Norwalk, Conn.).
PCR products were analyzed by agarose gel electrophoresis for 1 h at a constant voltage (110 V). Gels contained 1% agarose (Eurogentec) in TBE buffer (pH 8.0) (89 mM Tris, 89 mM borate, 2.5 mM EDTA). A 100-bp ladder (Pharmacia Biotech, Saclay, France) was used as the molecular size standard. Gels were stained with ethidium bromide (1 μg/ml) (Bioprobe System, Montreuil, France) for 30 min.
DNA sequencing.
PCR products were purified by a 15-min spin at 500 × g on a Microcon 100 (Amicon, Beverly, Mass.) to remove unincorporated deoxynucleoside triphosphates, primers, and salts. Nucleotide sequences were determined by the dideoxy chain termination method of Sanger et al. (29a) with Thermo Sequenase dye terminator cycle sequencing premixed version 2 (Amersham, Les Ulis, France) and PCR primers with an Abi Prism 377 sequencer (Perkin-Elmer) according to the manufacturer’s instructions.
Southern blotting.
To confirm the absence of major or minor fimbrial protein genes for strains which did not give a PCR product with the fimbrial primers, probes for major and minor fimbrial protein genes labeled with digoxigenin (DIG) were synthesized by PCR with alkali-labile DIG-11–2′-deoxyuridine-5′-triphosphate (DIG-11–dUTP) (Boehringer). For this PCR assay 30 μM dTTP was replaced by 30 μM DIG-11–dUTP. One probe was prepared with the N1 and C207 primers and DNA from strain 11PS, which carries the hifA-like gene observed in genital strains. Another probe was constructed with the N1 and C207 primers and the Hib reference strain 770235. Two other probes were prepared with a genital strain of Haemophilus: the first was prepared from strain 11PS by using the hifD5 and hifD3 primers, and the second was prepared from strain 26E by using the hifE5 and hifE3 primers.
Two micrograms of DNA from each of the 31 strains of Haemophilus was digested with 100 U of EcoRI overnight. The restriction fragments were resolved on a gel containing 0.8% agarose for 20 h at a constant voltage (30 V). The DNA was transferred by capillarity with 20× SSC (trisodium citrate, 0.3 M; NaCl, 3 M; pH 7) onto a positively charged nylon membrane (Boehringer Mannheim). The DNA on the membrane was then tested for hybridization at 65°C with the various probes in a hybridization buffer (sodium dodecyl sulfate, 1%; NaCl, 1 M; Tris, 50 mM [pH 7.5], blocking reagent, 1% [wt/vol]). The hybridized probe was immunodetected with anti-DIG Fab fragments conjugated to alkaline phosphatase and visualized with the chemiluminescent substrate disodium 3-(4-methoscyspiro-{1,2-dioscetane-3,2′-(5′-chloro)tricyclo-[3 · 3 · 7 · 73,7]decan}-4-yl)phenylphos-phate (Boehringer). Light emission was recorded on Hyperfilm MP (Amersham).
Nucleotide sequence accession numbers.
The sequence of the genital Haemophilus major fimbrial protein gene (ghfA) has been deposited in the EMBL sequence database under accession no. AJ000653. The accession numbers of the hifA-like gene sequences of Hib strain 5494 and H. influenzae biogroup Aegyptius strain 52129 are AJ000636 and AJ000637, respectively. The accession numbers corresponding to the genes coding for minor fimbrial proteins are AJ006783 for the ghfD gene of the genital strains 15N, 10U, 11PS, 26E, PIZ, and 2406 and AJ006784 for the ghfE gene of the genital strain 26E. The accession number of the omp gene of the genital strain 16N is AJ007317.
RESULTS
Characterization of a genital Haemophilus major fimbrial gene with primers corresponding to conserved sequences in hifA genes of Hib.
PCR assay with the primers N34 and C207 (Table 2; Fig. 1) amplified a fragment of about 550 bp from DNA of the control strain (Hib 770235). A comparable fragment was obtained from 6 of the 19 genital Haemophilus strains. These six strains were all from French patients; five were from the genital tracts of adult patients (strains 10U, 11PS, 26E, PIZ, and 2406), and only one was from a neonate (strain 15N) (Table 1). A fragment was also amplified from two H. influenzae reference strains: the Hib CIP 5494 strain, which is of biotype IV sensu stricto, and the H. influenzae biogroup Aegyptius CIP 52129 strain (Table 1). Except for CIP 52129, all strains for which a fragment was amplified exhibited peritrichous piliation under our culture conditions (Table 1).
To determine the 5′ end of the hifA-like gene, a PCR was performed with primers N1 and C207, which bracket the entire gene (Table 2; Fig. 1). An amplified fragment of about 650 bp was obtained for the same nine strains that gave an amplified fragment with primers N34 and C207: six genital Haemophilus strains and the control strain (Fig. 2A) and H. influenzae strains CIP 5494 and CIP 52129. No amplified product was obtained for the other strains. The PCR fragments obtained with primers C207 and N1 or N34 from the control strain Hib 770235 had slightly lower electrophoretic mobilities than the fragments amplified from the genital Haemophilus strains (Fig. 2A). No DNA amplification was obtained from any strain except the control strain Hib 770235 by using primer N51 or N69 with primer C186 or C198 (Fig. 1; Table 2).
FIG. 2.
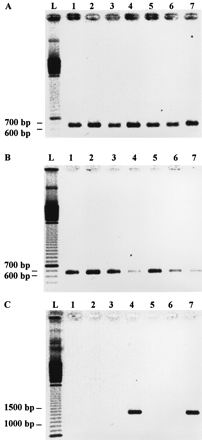
PCR products obtained with fimbrial primers corresponding to the extreme ends of the three genes coding for the three proteins constituting the fimbria. (A) Primers N1 and C207 were used to amplify a hifA-like gene fragment. The fragment amplified from the control strain Hib 770235 (lane 7) had a slightly lower electrophoretic mobility than the fragments obtained from genital Haemophilus strains (lanes 1 to 6). (B) PCR products obtained with hifD5 and hifD3. (C) PCR products obtained with hifE5 and hifE3. Lanes 1 to 6, genital cryptic Haemophilus strains 15N, 10U, 11PS, 26E, PIZ, and 2406, respectively; lane 7, Hib control strain 770235; lane L, 100-bp ladder.
The fragments amplified from all strains with primers N1 and C207 were sequenced. For the six cryptic strains, the nucleotide sequences were strictly identical.
The genital Haemophilus PCR fragment included a 621-bp open reading frame which we designate ghfA (for genital Haemophilus fimbria gene A). ghfA has 72% identity with the hifA gene of the control strain Hib 770235. It is 18 bp shorter than the Hib 770235 hifA gene, consistent with the small difference observed in electrophoretic mobility in agarose gels (Fig. 2A). It has a G+C content of 34%. It encodes a 207-amino-acid protein, named GhfA, which includes a predicted 20-amino-acid leader sequence. The leader sequence is typical of prokaryotic secreted proteins (43) and is identical to HifA leader sequences previously described for NTHi strain 1128, H. influenzae biogroup Aegyptius strain F3031 and Hib strains 770235, M43, and Eagan (2, 10, 13, 15, 38, 42). The remaining 561 bp of ghfA encodes a mature protein of 187 amino acids with a calculated molecular mass of 19,652 Da. GhfA is 63% identical and 71% similar to Hib strain 770235 HifA described by van Ham et al. (38).
The nucleotide sequence of the PCR fragment from cryptic Haemophilus shows 69 and 74% identity with those of two other strains we sequenced: Hib CIP 5494 and H. influenzae biogroup Aegyptius CIP 52129, respectively. There are 63 and 64% identity and 65 and 72% similarity between the translated sequences, respectively.
To try to identify a hifA-like gene in the 13 genital Haemophilus strains that did not give an amplified fragment, additional primers, Crypt N and Crypt C (Fig. 1; Table 2), were synthesized to correspond to regions of low sequence similarity between H. influenzae and genital Haemophilus strains. An amplification product was obtained only with the same six cryptic strains which gave amplified fragments with N34 (or N1) and C207 and with the H. influenzae biogroup Aegyptius strain. No amplification product was obtained for the Hib control strain (770235) or for the Hib biotype IV sensu stricto strain (CIP 5494).
To determine more accurately how many of the genital strains of Haemophilus contain a hifA-like gene, Southern hybridizations were performed with two probes. The first was the ghfA gene. Hybridization was obtained only for the nine strains giving a PCR fragment with the N1 and C207 primers (Fig. 3). The hybridization band intensities for Hib strain 770235, Hib strain CIP 5494, and H. influenzae biogroup Aegyptius strain CIP 52129 were lower than those for the six genital Haemophilus strains. This is consistent with the percentages of identity between the ghfA probe and the hifA or hifA-like genes: 100% for genital Haemophilus strains, 63% for Hib 770235, 69% for Hib CIP 5494, and 74% for H. influenzae biogroup Aegyptius CIP 52129. The second probe prepared from the Hib control strain 770235 was a hifA probe. A similar result was obtained, but the hybridization signal was more intense for Hib 770235 than for the other strains, also in accordance with the percentages of identity between the hifA probe and the ghfA or hifA-like genes. The size of the restriction fragment of the genital strain 2406 hybridizing with the probes was different from those for the other genital strains (Fig. 3). This strain exhibits the phenotypic characteristics of Haemophilus parainfluenzae, whereas the other genital Haemophilus strains are phenotypically identified as H. influenzae biotype IV. Nevertheless, previous DNA-DNA hybridization experiments indicated that this strain belongs to the genital Haemophilus genospecies (100.0% relative binding with the genital Haemophilus reference strain) and not to H. parainfluenzae (14.3% relative binding with the type strain of H. parainfluenzae) (22).
FIG. 3.
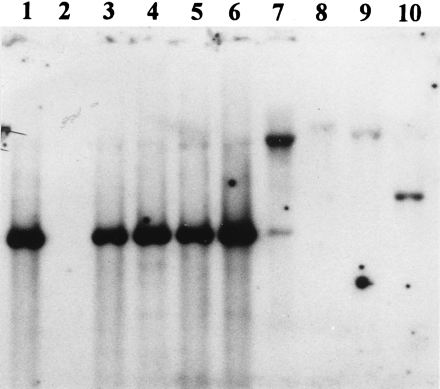
Southern blots obtained with the major fimbrial protein gene of genital Haemophilus (ghfA) labeled with DIG as a probe. Hybridization was observed only with DNA from strains giving a PCR product with primers N1 and C207 (Hib strains 770235 and 5494, H. influenzae biogroup Aegyptius strain 52129, and genital Haemophilus strains 15N, 10U, 11PS, 26E, PIZ, and 2406). Lanes 1 to 7, genital cryptic Haemophilus strains 15N, 16N, 10U, 11PS, 26E, PIZ, and 2406, respectively; lane 8, Hib control strain 770235; lanes 9 and 10, Hib strain CIP 5494 and H. influenzae biogroup Aegyptius strain CIP 52129, respectively.
Identification of genes coding for minor fimbrial proteins for genital Haemophilus.
Two sets of primers were used to amplify the genes coding for the two minor fimbrial proteins. The hifD5 and hifD3 primers (Table 3), corresponding to the extreme ends of the hifD gene of the Hib strain 770235, were used to amplify the hifD-like gene, and hifE5 and hifE3 (Table 3), corresponding to the extreme ends of the hifE gene of the control strain, were used to amplify the hifE-like gene.
With the hifD5 and hifD3 primers, a PCR fragment of about 650 bp was obtained for eight strains: the Hib control strain 770235, the six genital strains having the ghfA gene (Fig. 2B), and the H. influenzae biotype IV sensu stricto strain CIP 5494 (Table 1). Only the H. influenzae biogroup Aegyptius strain was positive for a hifA-like gene and negative for a hifD-like gene. The PCR fragments of the six genital strains of Haemophilus were sequenced. The sequences were strictly identical for the six strains. The sequence included a 648-bp open reading frame, which we designate ghfD (for genital Haemophilus fimbria gene D), that encodes a 218-amino acid-protein (GhfD). The amino acid sequence of GhfD was compared with the HifD sequence of Hib strain 770235; it was 79% identical and 82% similar.
With the hifE5 and hifE3 primers, a PCR fragment of 1,308 bp was obtained only for the genital strain 26E; it was named ghfE (Fig. 2C). The derived amino acid sequence was 53% identical and 59% similar to the Hib 770235 HifE sequence. No amplification was obtained for the genital strains 15N, 10U, 11PS, PIZ, and 2406, which possess ghfA and ghfD genes, but hybridization with the ghfE probe was observed for these strains.
For the other strains of genital Haemophilus, no amplification was obtained with the hifD5-hifD3 and hifE5-hifE3 primers. The absence of these genes was confirmed by Southern blotting.
Analysis of the region between the purE and pepN genes for ghfA-negative strains.
By using primers derived from the conserved flanking regions (purE and pepN genes), a PCR fragment of about 450 bp was obtained for all ghfA-negative genital strains of Haemophilus, except for two (genital strains 1595 and 3N) for which the PCR fragment was of about 250 bp. A 250-bp fragment was also amplified for the two strains of H. haemolyticus (CIP 102348 and CIP 103290).
The 450-bp fragment from the genital strain 16N was sequenced. It had 95% identity with a noncoding nucleotide sequence between the purE and pepN genes previously described for the fimbria-negative strain H. influenzae Rd. This fragment included a 126-bp sequence which shows some identities with a part of the purK genes of several H. influenzae strains (determined with FASTA software). The 250-bp PCR fragments from strain 1595 and one of the two strains of H. haemolyticus were sequenced. The sequences were identical and correspond to the noncoding sequence of H. influenzae Rd.
Characterization of a gene in our genital Haemophilus collection possibly involved in adherence and previously described as a fimbrial gene in an NTHi strain.
The primers fim5 and fim3 (Table 3) were deduced from the sequence of the gene coding for the fimbrin previously studied by Sirakova et al. (30). A fragment of about 1.1 kb was amplified for all genital strains of Haemophilus, for the 10 strains of H. influenzae sensu stricto, for the strain of H. influenzae biogroup Aegyptius, and for a strain of H. haemolyticus. Only one strain of H. haemolyticus (strain 102348) was negative. This PCR fragment was sequenced for the genital strain 16N. The sequence of the fimbrial gene in an NTHi strain (strain 1128) described by Sirakova et al. (30) is 84% identical to the sequence of the PCR fragment from 16N. A search for homology by using the FASTA program indicated that the fimbrin-like proteins encoded by these genes were similar to several H. influenzae outer membrane P5 proteins.
Electron microscopic observation of the fimbriae.
Fourteen of the 19 genital Haemophilus strains showed abundant peritrichous piliation (Table 1). There was no notable difference between the appearances of the fimbriae on strains having LKP-like fimbrial genes and those on the genital strains lacking these genes. The appearances of fimbriae before or after several contacts with human erythrocytes were the same. Nevertheless, two of the five nonpiliated strains (189 and 3N) exhibited fimbriae after this enrichment. No evident difference was observed between the electron microscopic appearances of these fimbriae and those of the 14 other strains.
DISCUSSION
The aim of this study was to identify and sequence genes coding for proteins participating in fimbria biogenesis in a particular group of Haemophilus strains that are specifically associated with genital, maternofetal, and neonatal infections.
As previous studies demonstrate about 80% similarity between the primary sequences of the major fimbrial subunits in all Hib and NTHi strains, including H. influenzae biogroup Aegyptius (2, 7, 10, 42), we used primers corresponding to conserved sequences at the 3′ and 5′ ends of previously described hifA genes. A hifA-like gene (named ghfA) was found in only 6 of the 19 genital Haemophilus strains studied. These PCR results were confirmed by Southern blotting with the hifA and ghfA genes as probes. This gene had 71 to 80% identity with the known hifA genes of various Haemophilus strains, percentages of identity which are consistent with those observed between H. influenzae genes encoding the major fimbrial subunit (2, 7, 10, 42). No amplification was obtained with the two H. haemolyticus strains, although this genospecies is the most closely related to genital Haemophilus genospecies strains (25, 27). ghfA codes for a protein (GhfA) whose sequence has 63% identity and 71% similarity to the translated amino acid sequence of the Hib 770235 hifA gene (38). It is 3 to 9 amino acids shorter than known HifA proteins in Haemophilus strains and includes two cysteines, 40 residues apart, and a penultimate tyrosine, a characteristic of H. influenzae HifA proteins (2, 8, 10, 13, 38, 42). Differences between the amino acid sequences were not localized to any one region but were distributed throughout the protein. In addition, the FASTA software (GenBank) found an identity of 62 to 68% between GhfA and major fimbrial subunit precursors of several H. influenzae strains. Therefore, GhfA appears to be a member of a family of major fimbrial proteins which includes almost all HifA proteins characterized to date from Hib, NTHi, and H. influenzae biogroup Aegyptius strains. There are also identities of 35% with the Escherichia coli F17 fimbrial protein precursor and 36% with the Klebsiella pneumoniae fimbrial subunit type 3 precursor.
Geluk et al. suggest that the complete fimbria gene cluster is either always present or entirely absent (11). Southern blotting suggest the presence of ghfE (a hifE-like gene) in the six genital Haemophilus strains which possess ghfA and ghfD. Nevertheless, at least one of the extreme ends (hifE5 or hifE3 sequences) of the gene in five of these six strains differs, because no amplification was obtained with these primers.
For 13 of the 19 genital strains, no PCR product was obtained with any of the primers used. This result was confirmed by Southern blotting with the three fimbrial gene probes and by sequencing of the products amplified with primers defined on the two genes surrounding the fimbrial cluster (purE and pepN). These data confirm the absence of LKP-like fimbrial genes from most (68%) of the genital Haemophilus strains. However, 10 of these 13 genital Haemophilus strains were piliated (Table 1). Therefore, other genes coding for proteins that constitute these fimbriae must be located elsewhere on the chromosome.
Because genital Haemophilus strains are commonly phenotypically identified as H. influenzae biotype IV, we also sequenced the PCR fragment of an H. influenzae biotype IV sensu stricto strain (Hib CIP 5494) for comparison. The nucleotide sequence is 69% identical to that of ghfA. The translated sequences are 63% identical and 65% similar. These percentages correspond to the identities and similarities observed between GhfA and H. influenzae HifA proteins as assessed by using FASTA software.
van Ham et al. demonstrated that HifA mediated H. influenzae-specific adherence (40). They found that the hydrophilicity pattern of HifA was different from that of HifD, which is a minor subunit of fimbriae and is closely related to HifA but which is not required for adherence. They suggested, therefore, that hydrophilic domains of HifA are responsible for binding to a specific eukaryotic receptor. Recently, McCrea et al. (17) suggested that the adhesion is mediated by the HifE protein localized at the tips of fimbriae rather than by the HifA major subunit, because hemagglutination by their Hib Eagan strain was inhibited by the presence of antibodies directed against HifE. Cryptic genital Haemophilus strains do not agglutinate erythrocytes (28). Nevertheless, the three hydrophilic domains previously observed in HifA were also identified in GhfA from the cryptic Haemophilus strains. This suggests that the three hydrophilic domains of HifA may be involved in an interaction other than binding to a specific eukaryotic receptor and that the adhesive site may not be localized on the HifA protein. These observations are in part in agreement with a previous study (2) in which an LKP hifA-like gene was amplified by PCR from a strain responsible for otitis that did not agglutinate erythrocytes. In this strain, pili were morphologically different from LKP pili, and an additional fimbrial subunit gene (fimbrin gene) having a low sequence similarity with the LKP gene has since been identified by Sirakova et al. (30). By using primers derived from this fimbrin gene, an omp gene which has 84% identity with the fimbrin gene described by Sirakova et al. and about 85% identity with the ompP5 genes of many H. influenzae strains was amplified for all our genital strains. Thus, the product of this gene could be an adhesin commonly observed for Hib, NTHi, and genital strains of Haemophilus, but it seems doubtful that it is the major protein constituting fimbriae. Therefore, other genes, as yet unidentified, may explain the piliation observed by electron microscopy on genital Haemophilus strains which do not carry LKP-like fimbrial genes.
Genital Haemophilus strains that possess the ghfA and ghfD genes were isolated more often from mucosal cells of adults (5 of the 6 strains) than from neonates (only 1 of the 13 strains). In addition, ghfA and ghfD were present only in French strains. Previous genetic studies give no evidence for differences between French and U.S. strains (25, 27). Therefore, the apparent relationship between the geographic origin of strains and the presence of ghf genes may reflect the fact that most strains isolated from urogenital epithelial cells were from French adults, whereas most of the strains isolated in the United States were from neonates. Thus, these fimbrial genes may confer on the strain the ability to adhere to urogenital epithelial cells and to persist in the ecological and physiological conditions of the adult genital tract. In contrast, ghfA and ghfD do not appear to be essential for colonizing neonates at birth. The study of a larger number of strains may confirm this observation.
ACKNOWLEDGMENTS
We thank L. van Alphen for providing Hib strain 770235, J. M. Musser for providing U.S. strains of genital Haemophilus, and H. Dabernat for providing some French strains of genital Haemophilus.
This research was supported by the Université François Rabelais.
REFERENCES
- 1.Albritton W L, Brunton J L, Meier M, Bowman M N, Slaney L A. Haemophilus influenzae: comparison of respiratory tract isolates with genitourinary tract isolates. J Clin Microbiol. 1982;16:826–831. doi: 10.1128/jcm.16.5.826-831.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bakaletz L O, Ahmed M A, Kolattukudy P E, Lim D J, Forney L J. Cloning and sequence analysis of a pilin-like gene from an otitis media isolate of nontypeable Haemophilus influenzae. J Infect Dis. 1992;165:S201–S203. doi: 10.1093/infdis/165-supplement_1-s201. [DOI] [PubMed] [Google Scholar]
- 3.Bakaletz L O, Tallan B M, Hoepf T, De Maria T F, Birck H G, Lim D J. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect Immun. 1988;56:331–335. doi: 10.1128/iai.56.2.331-335.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barenkamp S J, Leininger E. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect Immun. 1992;60:1302–1313. doi: 10.1128/iai.60.4.1302-1313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beachey E H. Bacterial adherence: adhesin-receptor interactions mediating the attachment of bacteria to mucosal surfaces. J Infect Dis. 1981;143:325–345. doi: 10.1093/infdis/143.3.325. [DOI] [PubMed] [Google Scholar]
- 6.Brenner D J, McWhorter A C, Leete Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clemans D L, Marrs C F, Patel M, Duncan M, Gilsdorf J R. Comparative analysis of Haemophilus influenzae hifA (pilin) genes. Infect Immun. 1998;66:656–663. doi: 10.1128/iai.66.2.656-663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman T, Grass S, Munson R., Jr Molecular cloning, expression, and sequence of the pilin gene from nontypeable Haemophilus influenzae M37. Infect Immun. 1991;59:1716–1722. doi: 10.1128/iai.59.5.1716-1722.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forney L J, Marrs C F, Bektesh S L, Gilsdorf J R. Comparison and analysis of the nucleotide sequences of pilin genes from Haemophilus influenzae type b strains Eagan and M43. Infect Immun. 1991;59:1991–1996. doi: 10.1128/iai.59.6.1991-1996.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geluk F, Eijk P P, van Ham S M, Jansen H M, van Alphen L. The fimbria gene cluster of nonencapsulated Haemophilus influenzae. Infect Immun. 1998;66:406–417. doi: 10.1128/iai.66.2.406-417.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilsdorf J R, Chang H Y, McCrea K W, Bakaletz L O. Comparison of hemagglutinating pili of Haemophilus influenzae type b with similar structures of nontypeable Haemophilus influenzae. Infect Immun. 1992;60:374–379. doi: 10.1128/iai.60.2.374-379.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilsdorf J R, Marrs C F, McCrea K W, Forney L J. Cloning, expression, and sequence analysis of Haemophilus influenzae type b strain M43p+ pilin gene. Infect Immun. 1990;58:1065–1072. doi: 10.1128/iai.58.4.1065-1072.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krogfelt K A. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesins of Escherichia coli. Rev Infect Dis. 1991;13:721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 15.Langermann S, Wright A. Molecular analysis of the Haemophilus influenzae type b pilin gene. Mol Microbiol. 1990;4:221–230. doi: 10.1111/j.1365-2958.1990.tb00589.x. [DOI] [PubMed] [Google Scholar]
- 16.Martel A Y, St Laurent G, Dansereau L A, Bergeron M G. Isolation and biochemical characterization of Haemophilus species isolated simultaneously from oropharyngeal and anogenital areas. J Clin Microbiol. 1989;27:1486–1489. doi: 10.1128/jcm.27.7.1486-1489.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCrea K W, Watson W J, Gilsdorf J R, Marrs C F. Identification of two minor subunits in the pilus of Haemophilus influenzae. J Bacteriol. 1997;179:4227–4231. doi: 10.1128/jb.179.13.4227-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCrea K W, Watson W J, Gilsdorf J R, Marrs C F. Identification of hifD and hifE in the pilus gene cluster of Haemophilus influenzae type b strain Eagan. Infect Immun. 1994;62:4922–4928. doi: 10.1128/iai.62.11.4922-4928.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy T F, Kirkham C, Sikkema D J. Neonatal, urogenital isolates of biotype 4 nontypeable Haemophilus influenzae express a variant P6 outer membrane protein molecule. Infect Immun. 1992;60:2016–2022. doi: 10.1128/iai.60.5.2016-2022.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musser J M, Barenkamp S J, Granoff D M, Selander R K. Genetic relationships of serologically nontypable and serotype b strains of Haemophilus influenzae. Infect Immun. 1986;52:183–191. doi: 10.1128/iai.52.1.183-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pichichero M E. Adherence of Haemophilus influenzae to human buccal and pharyngeal epithelial cells: relationship to piliation. J Med Microbiol. 1984;18:107–116. doi: 10.1099/00222615-18-1-107. [DOI] [PubMed] [Google Scholar]
- 22.Quentin R, Dubarry I, Martin C, Cattier B, Goudeau A. Evaluation of four commercial methods for identification and biotyping of genital and neonatal strains of Haemophilus species. Eur J Clin Microbiol Infect Dis. 1992;11:546–549. doi: 10.1007/BF01960812. [DOI] [PubMed] [Google Scholar]
- 23.Quentin R, Goudeau A, Burfin E, Pinon G, Berger C, Laugier J, Soutoul J H. Infections materno-foetales àHaemophilus influenzae. Press Med. 1987;16:1181–1184. [PubMed] [Google Scholar]
- 24.Quentin R, Goudeau A, Wallace R J, Jr, Smith A L, Selander R K, Musser J M. Urogenital, maternal and neonatal isolates of Haemophilus influenzae: identification of unusually virulent serologically nontypable clone families and evidence for a new Haemophilus species. J Gen Microbiol. 1990;136:1203–1209. doi: 10.1099/00221287-136-7-1203. [DOI] [PubMed] [Google Scholar]
- 25.Quentin R, Martin C, Musser J M, Pasquier-Picard N, Goudeau A. Genetic characterization of a cryptic genospecies of Haemophilus causing urogenital and neonatal infections. J Clin Microbiol. 1993;31:1111–1116. doi: 10.1128/jcm.31.5.1111-1116.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quentin R, Musser J M, Mellouet M, Sizaret P Y, Selander R K, Goudeau A. Typing of urogenital, maternal, and neonatal isolates of Haemophilus influenzae and Haemophilus parainfluenzae in correlation with clinical source of isolation and evidence for a genital specificity of H. influenzae biotype IV. J Clin Microbiol. 1989;27:2286–2294. doi: 10.1128/jcm.27.10.2286-2294.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quentin R, Ruimy R, Rosenau A, Musser J M, Christen R. Genetic identification of cryptic genospecies of Haemophilus causing urogenital and neonatal infections by PCR using specific primers targeting genes coding for 16S rRNA. J Clin Microbiol. 1996;34:1380–1385. doi: 10.1128/jcm.34.6.1380-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenau A, Sizaret P Y, Musser J M, Goudeau A, Quentin R. Adherence to human cells of a cryptic genospecies of Haemophilus responsible for genital and neonatal infections. Infect Immun. 1993;61:4112–4118. doi: 10.1128/iai.61.10.4112-4118.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sable N S, Connor E M, Hall C B, Loeb M R. Variable adherence of fimbriated Haemophilus influenzae type b to human cells. Infect Immun. 1985;48:119–123. doi: 10.1128/iai.48.1.119-123.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sirakova T, Kolattukudy P E, Murwin D, Billy J, Leake E, Lim D, De Maria T, Bakaletz L. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect Immun. 1994;62:2002–2020. doi: 10.1128/iai.62.5.2002-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. Geme J W, III, Cutter D. Influence of pili, fibrils, and capsule on in vitro adherence by Haemophilus influenzae type b. Mol Microbiol. 1996;21:21–31. doi: 10.1046/j.1365-2958.1996.6241331.x. [DOI] [PubMed] [Google Scholar]
- 32.St. Geme J W, III, Cutter D, Barenkamp S J. Characterization of the genetic locus encoding Haemophilus influenzae type b fibrils. J Bacteriol. 1996;178:6281–6287. doi: 10.1128/jb.178.21.6281-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.St. Geme J W, III, Falkow S. Isolation, expression, and nucleotide sequencing of the pilin structural gene of the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect Immun. 1993;61:2233–2237. doi: 10.1128/iai.61.5.2233-2237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.St. Geme J W, III, Falkow S, Barenkamp S J. High-molecular-weight proteins of nontypeable Haemophilus influenzae mediate attachment to human epithelial cells. Proc Natl Acad Sci USA. 1993;90:2875–2879. doi: 10.1073/pnas.90.7.2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St. Geme J W, III, Kumar V V, Cutter D, Barenkamp S J. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect Immun. 1998;66:364–368. doi: 10.1128/iai.66.1.364-368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Strom M S, Lory S. Structure-function and biogenesis of the type IV pili. Annu Rev Microbiol. 1993;47:565–596. doi: 10.1146/annurev.mi.47.100193.003025. [DOI] [PubMed] [Google Scholar]
- 37.van Alphen L, van den Berghe N, Geelen-van den Broek L. Interaction of Haemophilus influenzae with human erythrocytes and oropharyngeal epithelial cells is mediated by a common fimbrial epitope. Infect Immun. 1988;56:1800–1806. doi: 10.1128/iai.56.7.1800-1806.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Ham S M, Mooi F R, Sindhunata M G, Maris W R, van Alphen L. Cloning and expression in Escherichia coli of Haemophilus influenzae fimbrial genes establishes adherence to oropharyngeal epithelial cells. EMBO J. 1989;11:3535–3540. doi: 10.1002/j.1460-2075.1989.tb08519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Ham S M, van Alphen L, Mooi F R, van Putten J P M. The fimbrial gene cluster of Haemophilus influenzae type b. Mol Microbiol. 1994;13:673–684. doi: 10.1111/j.1365-2958.1994.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 40.van Ham S M, van Alphen L, Mooi F R, van Putten J P M. Contribution of the major and minor subunits to fimbria-mediated adherence of Haemophilus influenzae to human epithelial cells and erythrocytes. Infect Immun. 1995;63:4883–4889. doi: 10.1128/iai.63.12.4883-4889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallace R J, Baker C J, Quinones F, Hollis D G, Weaver R E, Wiss K. Nontypeable Haemophilus influenzae (biotype 4) as a neonatal, maternal, and genital pathogen. Rev Infect Dis. 1983;5:123–136. doi: 10.1093/clinids/5.1.123. [DOI] [PubMed] [Google Scholar]
- 42.Whitney A M, Farley M M. Cloning and sequence analysis of the structural pilin gene of Brazilian purpuric fever-associated Haemophilus influenzae biogroup aegyptius. Infect Immun. 1993;61:1559–1562. doi: 10.1128/iai.61.4.1559-1562.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H C. Proteolytic processing of signal peptides. In: Strauss A W, Boime I, Kreil G, editors. Protein compartmentalization. New York, N.Y: Springer-Verlag; 1986. pp. 33–59. [Google Scholar]


